Abstract
RNA modifications are crucial for gene expression in eukaryotes; however, the regulatory role of 5’ 7-methylguanosine (m7G) cap, the first modification of mRNA, remains unknown in the protozoan parasite Toxoplasma gondii. Here, we show that the mRNA capping machinery of Toxoplasma consists of three distinct enzymes: RNA triphosphatase, guanylyltransferase, and guanine-N7-methyltransferase, which together add m7G cap to RNA, recognized by cap-binding protein, TgeIF4E. Biochemical and genetic studies show that among three capping enzymes, RNA triphosphatase (TgCet) is unique and a member of the tunnel family of metal-dependent phosphohydrolases, which is structurally and mechanistically distinct from the human RNA triphosphatase. Using conditional knockdown, we show that TgCet is essential for mRNA capping, and its depletion generates widespread changes in m7G-capped transcripts, resulting in the complete arrest of parasite replication both in culture and in mouse host, thereby protecting them from lethal infection. Finally, the therapeutic potential of TgCet was evaluated using two compounds, Myricetin and 3,4-dicaffeoylquinic acid, reported to inhibit Trypanosoma Cet enzyme. However, only Myricetin demonstrated selective inhibition of TgCet activity and effectively blocked parasite growth in culture. Overall, this study highlights the essential role of TgCet-mediated mRNA capping, establishing RNA triphosphatase as a promising drug target for Toxoplasma infection.
Similar content being viewed by others
Introduction
Toxoplasma gondii is an extremely successful obligate intracellular apicomplexan parasite that causes lifelong chronic infections in almost all warm-blooded animals, including humans, and severe disease in fetuses and immunocompromised individuals1. An estimated one-third of the global human population is chronically infected with T. gondii2. Following strong host immune response during the acute infection, the rapidly multiplying tachyzoites differentiate into a slow-growing encysted bradyzoite and develop chronic infection3. A fast conversion of bradyzoites into tachyzoites leads to severe mortality in immunosuppressed individuals if not treated. The standard treatment drugs for toxoplasmosis are few and have severe side effects on long-term use, target the tachyzoite stage of the parasite, and are ineffective against encysted bradyzoite in the tissues, necessitating the identification of new drug targets and therapeutics4.
The complex multi-host life cycle of Toxoplasma involves changes between life stages with distinct morphologies, metabolisms, and reproductive niches. The parasite utilizes an intricate transcriptional, post-transcriptional, and epigenetic network to regulate gene expression required to thrive in these rapidly changing environments and hosts5,6. Growing evidence suggests that RNA processing, especially co-transcriptional and post-transcriptional mechanisms, are crucial for regulating gene expression during life cycle stage transitions and environmental adoption; however, the underlying mechanism is yet to be discovered in apicomplexan parasites.
In eukaryotes, 5′ mRNA capping is the first and one of the essential co-transcriptional modifications to produce mature mRNA7. The 5′ cap is essential throughout the life cycle of the mRNA in coordinating various functional processes accomplished by interacting with the cap-binding complex (CBC). The nuclear CBC facilitates splicing, polyadenylation, and export into the cytoplasm, whereas the cytoplasmic CBC, containing eukaryotic initiation factor 4F (eIF4F), binds to the cap and recruits the 40S ribosomal subunit to initiate translation8.
All eukaryotic mRNA contains a 5′ cap structure of 7-methylguanosine (m7G) linked via 5′ to 5′ triphosphate bridge to the first transcribed nucleotide (m7GpppN). The m7GpppN cap structure is formed by a series of three enzymatic steps: (i) hydrolysis of the 5′ triphosphate (pppN) end of the nascent RNA to a diphosphate (ppN) by RNA triphosphatase; (ii) transfer of a guanine monophosphate (GMP) nucleotide to the 5′-diphosphate RNA (GpppN-guanosine cap) by RNA guanylyltransferase; and (iii) addition of a methyl group to the N7 amine of the guanine cap (m7GpppN) by guanine-N7 methyltransferase. These three catalytic activities, collectively called “capping enzymes,” are encoded by separate genes in yeast, whereas in metazoans, the first two capping steps are catalyzed by a single enzyme consisting of two functional domains, the N-terminal triphosphatase and the C-terminal guanylyltransferase8,9,10,11. The structures and mechanisms of the mammalian and fungal capping enzymes have been well elucidated; however, studies on the capping enzymes of protozoa12,13,14,15, except Trypanosoma16,17, are mostly limited to their biochemical characterization.
While guanylyltransferase (GTase) and methyltransferase (guanine-N7 MTase) are conserved, RNA triphosphatase (TPase) has a distinct structure and mechanism of action among eukaryotes. Consequently, RNA triphosphatases (TPase) are classified into two families: (i) the divalent cation-dependent TPase of triphosphate tunnel metalloenzyme (TTM) family found in fungi18,19,20 and protozoa12,13,14,15,16 and (ii) the divalent cation-independent TPase of metazoans and plants of cysteine-phosphatase superfamily found in metazoan and plants21,22,23,24,25. The TTM-type TPases are essential for growth in fungi23,26,27 and Trypanosoma brucei protozoan parasite17, and as a result, RNA triphosphatases are considered potential drug targets for these infections. The TPase protein essentiality studies have been carried out only in tissue culture; however, its essentiality has never been tested for pathogen survival in the host. Additionally, the impact of lack of capping in the TPase-deficient pathogens on genome-wide m7G-capped RNA abundance, mRNA expression, turnover, and stability has never been studied.
Here, we report a detailed characterization of Toxoplasma capping enzymes and investigate the consequences of RNA triphosphatase depletion on transcript homeostasis and parasite survival. Toxoplasma has a functional three-component capping system consisting of separate TTM-type RNA triphosphatase (TgCet), guanylyltransferase (TgCeg), and guanine-N7 methyltransferase (TgCmt). The m7G RNA generated using capping enzymes binds to the translation initiation factor, TgeIF4E, and is successfully translated into protein in the parasite. We used an auxin-inducible degron method to generate TgCet conditional knockdown parasites and show that rapid depletion of TgCet leads to downregulation of global m7G-capped transcripts of essential genes, which resulted in the death of the parasite in the culture and in the mouse host. Lastly, we predicted the structure of TgCet and evaluated the inhibitory effect of two compounds (previously shown to target RNA triphosphatases)17 on TgCet protein, parasites, and mouse infection model. Overall, the study shows the therapeutic potential of TgCet with possibly minimal or no impact on structurally similar proteins in the host.
Results
T. gondii encodes three separate mRNA capping enzymes
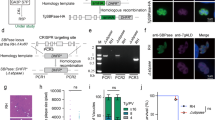
To examine the presence of 7-methylguanosine (m7G) cap in Toxoplasma, we performed immunofluorescence analysis using an anti-m7G antibody in the asexual stages (tachyzoite and bradyzoite) of the parasite. Analysis of intracellular parasites revealed predominant punctate staining for m7G at the nuclear periphery and cytoplasm of tachyzoite (Tz) and bradyzoite (Bz) (Fig. 1A). To detect whether the m7G mark is present on RNA, we performed an immuno-dot blot using an anti-m7G antibody on total RNA and genomic DNA (gDNA) extracted from filter-purified tachyzoites. The results demonstrated that the m7G mark was exclusively present in RNA (Fig. 1B). Collectively, these results show that m7G cap RNA is a feature of Toxoplasma.
A Immunofluorescence analysis of m7G mark in tachyzoites and bradyzoites (n = 3). Tz tachyzoite and Bz bradyzoite. Scale bar 5 µm. B Immuno-dot blot analysis of m7G levels in the indicated amount of RNA and Toxoplasma genomic DNA (gDNA) (n = 3). EtBr staining serves as a loading control. C Predicted three capping genes of Toxoplasma (TgCet, TgCeg, and TgCmt) with ToxoDB number and CRISPR score. D Schematic depicting TgCet: the triphosphatase tunnel (364–728 aa) with conserved amino acids, TgCeg: the catalytic domain (111–255 aa) with conserved amino acids and C-terminal domain 330–480 aa), and TgCmt: the conserved SAM binding motif. E–H T. gondii capping enzymes functionally complement yeast (S. cerevisiae) counterparts (n = 3). S. cerevisiae Δcet1, or Δceg1, or Δabd1 strain was transformed with pYES3 (TRP1) plasmid containing TgCet, TgCeg, or TgCmt gene. Triple deletion yeast strain was transformed with TgCet, TgCeg, and TgCmt genes containing plasmids. A control transformation was performed with the pYES3 plasmid lacking an insert. Single Trp+ transformants were patched to agar plates lacking tryptophan (−Trp) and then patched on agar medium containing FOA (−Trp+FOA). FOA-resistant colonies were picked and streaked on −Trp+FOA agar medium.
To identify capping enzymes, which could add m7G structure to RNA, we performed BLASTP homology searches of the Toxoplasma genome28 (http://toxodb.org/toxo/) using amino acid sequences of yeast Cet1, Ceg1, and Abd1 proteins as queries. The search analysis revealed three separate RNA capping enzymes in Toxoplasma that are similar to yeast. The identified candidate Toxoplasma RNA triphosphatase, guanylyltransferase, and guanine-N7 methyltransferase were named TgCet (TGME49_224650), TgCeg (TGME49_305320), and TgCmt (TGME49_272720), respectively (Fig. 1C). The MEGA analysis was performed to determine how closely the Toxoplasma capping enzymes (TgCEs) are related to the alveolates’ and other eukaryotes’ capping proteins. Phylogenetic analysis revealed a distinction between two apicomplexan classes, Conoidasida and Aconoidasida, for all three capping proteins. Members of the Sarcocystidae family were found to encode the largest triphosphatase and methyltransferase proteins (Supplementary Fig. 1).
The putative TgCet gene encodes a 920-aa polypeptide and has characteristic features of fungal triphosphatases9, including two glutamate-containing metal-binding motifs, homologs of β strands that comprise the active site tunnel, and conserved hydrophilic amino acids required for catalysis. (Fig. 1D and Supplementary Fig. 2). The putative TgCeg is a 509-aa protein that contains two conserved domains, a nucleotidyl transferase (NTase) domain and a C-terminal oligonucleotide-binding domain (OB)29,30,31. The NTase domain has six conserved motifs (I, III, IIIa, IV, V, and VI)32 (Supplementary Fig. 3) with a lysine-containing KxDG motif I (Fig. 1D), which comprises the active site of GTP-binding and nucleotidyl transfer33,34. The putative TgCmt encodes 1283-aa polypeptide with large N-terminal extension and conserved glycine-rich sequence in the SAM-binding motif (Fig. 1D). Among all three capping enzymes, TgCeg and TgCmt are significantly larger than yeast’s TPase and guanine-N7 MTase; however, each of the three Toxoplasma capping enzyme orthologue is essential for parasites according to a genome-wide CRISPR screen (Fig. 1C), suggesting that m7G capping of RNA is important and required for parasite fitness.
Owing to the sequence similarity between Toxoplasma capping enzymes (TgCEs) and yeast enzymes, we performed yeast complementation to test whether TgCEs function in the cap-synthetic pathway and sustain the growth of yeast cells that lack one or more capping enzymes. We separately cloned the TgCet, TgCeg, and TgCmt genes into a yeast 2μ TRP1 pYES3 plasmid, and the function of each of these genes was tested by plasmid shuffle in S. cerevisiae Δcet1 or Δceg1 or Δabd1 cells that contain respective gene on a CEN URA3 plasmid (Supplementary Tables 1 and 2). The mutant strain cannot survive on a medium containing 5-FOA, a drug that selects against the URA3 plasmid unless it is transformed with a second plasmid containing a functional homolog from another source. We found that 2μ TgCet, TgCeg, and TgCmt supported the growth of Δcet1, Δceg1, and Δabd1 cells, respectively (Fig. 1E–G). Similarly, 2μ TgCet + TgCeg + TgCmt supported the growth of triple mutant yeast cells (Fig. 1H), which was further validated by expression of all three genes (Supplementary Fig. 4). These results demonstrate that Toxoplasma encodes biologically active capping enzymes.
Toxoplasma RNA triphosphatase TgCet shows metal-dependent triphosphatase activity
To gain insight into the expression, localization, and biochemical function of TgCet, full-length His6-TgCet protein of ~100 kDa was purified (Fig. 2A, Supplementary Fig. 5, and Supplementary Table 1) and used to generate specific anti-TgCet antibodies. The expression and localization studies revealed that TgCet is robustly expressed (Fig. 2B) in both the asexual stages and localized in the nucleus of the tachyzoite and bradyzoite stages (Fig. 2C), as demonstrated using anti-TgCet antibodies. Further, we tested the triphosphatase activity of TgCet using a detailed biochemical characterization. The recombinant wild type (WT) TgCet catalyzed the release of 32Pi from [γ-32P]ATP in the presence of manganese (Fig. 2D); however, the ATP hydrolysis was ineffective in the presence of magnesium (Fig. 2D). The extent of ATP hydrolysis was proportional to TgCet concentration (Fig. 2E). No ATP hydrolysis was observed without divalent cation (Fig. 2D). Next, we performed the assay to determine the optimal condition using a linear range of TgCet (10 ng to 500 ng). The optimal activity was detected in 15 min with 100 ng TgCet input protein at 30 °C (Supplementary Fig. 6). To test whether a higher concentration of divalent cations stimulates the activity of TgCet, we performed an ATPase assay using 10 mM of ions. 10 mM Mg2+ could enhance the TgCet activity (as opposed to 5 mM presented in Fig. 2D). However, Ca2+ and Zn2+ failed to support TgCet activity even at higher concentrations (Fig. 2F). With the optimal reaction condition (100 ng TgCet, Supplementary Fig. 6), the extent of Pi release was determined as a function of time. The optimal activity of TgCet protein was observed ~15 min of incubation (Fig. 2G). This optimized condition was used for the subsequent experiments. No significant change in the TgCet activity was observed from 10 °C to 70 °C (Fig. 2H). TgCet was found to be catalytically active in a wide pH range (5.5–10) with an optimal activity from pH 7 to 9 (Fig. 2I). We also tested the specificity for NTP hydrolysis using two different triphosphorylated nucleoside substrates. The rate of release of 32Pi from [γ-32P]ATP was similar to the rate of conversion of [α-32P]ATP to [α-32P]ADP and [α-32P]UTP to [α-32P]UDP in a parallel reaction mixture containing the same concentration of TgCet (Fig. 2J). Two conserved glutamate residues corresponding to the metal binding sites of the tunnel (Fig. 1 and Supplementary Fig. 2) were replaced by alanine (E366A and E726A) (Figs. 1D, 2K), and enzyme activities were compared as a linear range of protein input. ATPase activity of the E366A mutant was found to be 70% less than wild-type TgCet (Fig. 2L), whereas in comparison, E726A showed <5% activity (Fig. 2L). These results suggest that both residues are important for TgCet activity; however, in addition to E366, other amino acid residues may be important for coordinating the metal-ion binding, as complete loss of the activity was not observed. The loss or decrease in activity in the mutant proteins, at the higher concentration (100 ng), was not due to the change in the secondary structure but mutations in the catalytically important amino acid residues (Supplementary Fig. 7). Finally, we performed an RNA triphosphatase assay using [γ-32P]RNA as a substrate to confirm that TgCet indeed has RNA triphosphatase activity (Fig. 2M). Together, these results demonstrated that TgCet belongs to the family of triphosphate tunnel metalloenzymes (TTMs).
The expression and localization of TgCet. Coomassie blue-stained SDS-PAGE gel of recombinant full-length His-TgCet protein (A), Western blot analysis of TgCet expression levels in tachyzoites and bradyzoites (B). Immunofluorescence analysis in RH/ME49 parasites with α-TgCet antibody (C). Loading controls: TgAldolase (TgALD) - parasite proteins, TgSAG1 and TgBAG1, tachyzoite- and bradyzoite stage-specific markers, respectively. Scale bar 5 µm. D Manganese- and Magnesium-dependent activity of TgCet. TPase reaction mixtures contained 1.5 mM [γ−32P]ATP, 0.5 μg of TgCet, and either MgCl2 or MnCl2 as specified or no added divalent cation. The positions of [γ−32P]ATP and 32Pi are indicated. E TgCet titration. TPase reaction mixtures contained 5 mM MnCl2 and TgCet as specified. F TgCet enzyme activity with different divalent cations. The reaction mixtures contained 1.5 mM of [γ−32P]ATP, 0.5 μg of TgCet, and 10 mM divalent cation as specified. The Pi release was calculated as a function of a divalent cation. G TgCet enzyme activity at various incubation durations. TPase reactions were incubated for indicated time points, and Pi release was quantified. H TgCet activity at different temperatures. TPase reactions were incubated at specified temperatures, and Pi release was estimated. I TgCet activity at variable pH. TPase reactions were performed in the Tris buffer with pH as indicated, and Pi release was estimated. J ATPase activity of TgCet with different NTP substrates. The reaction mixtures contained 100 ng of TgCet, 5 mM MnCl2, and NTP substrate as specified. K Coomassie blue-stained SDS-PAGE gel of recombinant His-tag WT and mutant TgCet proteins (E366A and E726A) as indicated. L TPase activity of WT and mutant TgCet. The reaction mixtures contained 1.5 mM of [γ−32P]ATP, 5 mm MnCl2, and either WT or mutant proteins as indicated. The Pi release was plotted as a function of the input enzyme. M. RNA triphosphatase activity of TgCet. Figure 2A-M: n = 3 replicates. Figures 2F-J and 2L: mean ± SD, one way ANOVA; Dunnett’s comparison test; ns not significant; p values: *<0.05; *** <0.001; ****<0.0001. Source data are provided as a source data file.
Characterization of Toxoplasma guanylyltransferase TgCeg
The full-length His6-TgCeg protein of ~60 kDa was purified (Fig. 3A, Supplementary Fig. 5, and Supplementary Table 1) and used to generate anti-TgCeg antibodies. TgCeg is robustly expressed in the asexual stages (Fig. 3B) and localized in the nucleus of the tachyzoite and bradyzoite stages (Fig. 3C). All known guanylyltransferases accomplish nucleotidyl transfer through a covalent enzyme-(lysyl-N)-GMP intermediate that can be detected by label transfer from [α-32P]GTP to the enzyme. To determine the guanylyltransferase activity of TgCeg, protein was incubated with [α-32P]GTP and a divalent cation, which resulted in the formation of an SDS-stable ~60 kDa enzyme-GMP intermediate (Fig. 3D). TgCeg activity requires a divalent cation cofactor, either manganese or magnesium; however, enzyme activity was more effective in the presence of manganese than magnesium (Fig. 3D). Enzyme-guanylate formation was linear with respect to TgCeg concentration (Fig. 3E). We mutated conserved Lys (essential for GMP interaction during the guanylyltransferase reaction for GTases)35, Tyr, Glu, and Gly residues to alanine (K133A, T134A, D135A, and G136A) of TgCeg and compared enzyme activities (Fig. 3F). None of the mutant proteins showed GTase activity (Fig. 3G); however, after long autoradiographic exposure, a trace of residual activity was observed for T134A (Fig. 3G). Loss of activity in the TgCeg mutant proteins was not due to a change in the secondary structure, as demonstrated by circular dichroism (Supplementary Fig. 8). We conclude that the observed guanylyltransferase activity is intrinsic of TgCeg.
A–C. The expression and localization of TgCeg. Coomassie blue-stained SDS-PAGE gel of recombinant full-length His-TgCeg protein (A), Western blot analysis of TgCeg expression levels in tachyzoites and bradyzoites (B). Immunofluorescence analysis in RH/ME49 parasites with α-TgCeg antibody (C). Loading controls: TgAldolase (TgALD) - parasite proteins, TgSAG1 and TgBAG1, tachyzoite- and bradyzoite stage-specific markers, respectively. Tz: tachyzoite and Bz: bradyzoite. Scale bar 5 µm. DManganese- and Magnesium-dependent GTase activity of TgCeg. The reaction mixtures contained 1 mM [α−32P]GTP, 0.5 μg of TgCeg, and either MgCl2 or MnCl2 as specified or no added divalent cation. The reactions were resolved on SDS PAGE gel, visualized by autoradiography, and the TgCeg-GMP adduct was assessed. E TgCeg titration. The reaction mixtures contained 2 mM MnCl2 and TgCeg as specified. F Coomassie blue-stained SDS-PAGE gel of recombinant His-tag WT and mutant TgCeg proteins (K133A, T134A, D135A, and G136A) as indicated. G GTase activity of WT and mutant TgCeg. The reaction mixtures contained 1 mM of [α−32P]GTP, 2 mm MnCl2, and either WT or mutant proteins as indicated. TgCeg-GMP intermediate was visualized by autoradiography (left panel). TgCeg-GMP intermediate quantification (right panel). Figure 3A-G: n = 3 replicates. Figure 3G: mean ± SD, one way ANOVA; Dunnett’s comparison test; ns not significant; p values: *<0.05; *** <0.001; ****<0.0001. Source data are provided as a source data file.
Characterization of Toxoplasma guanine-N7 methyltransferase TgCmt
His6-TgCmt 716-1283 (subscript denotes amino acid coordinates) protein of ~60 kDa was purified (Fig. 4A, Supplementary Fig. 5) and used to generate anti-TgCmt antibodies. The expression and localization studies showed TgCmt expressed in the asexual stages (Fig. 4B) and primarily localized in the nucleus of the tachyzoite and bradyzoite stages (Fig. 4C). To test the guanine-N7 methyltransferase activity of TgCmt, we first generated 32 mer RNA (pppN32RNA) using in vitro transcription and sequentially treated with TgCet to generate ppN32, TgCeg to generate GpppN32, and TgCmt in the presence of methyl donor S-adenosyl methionine (SAM) to generate m7GpppN32, as shown in Fig. 4D. The individual reaction was spotted on the membrane and capping of RNA substrate was determined using anti-m7G antibody. The immune-blot analysis revealed that TgCmt could successfully add m7G to the guanylated RNA (Fig. 4E). Vaccinia Capping Enzyme (VCE) was used as a positive control to generate m7GpppN32 (Fig. 4E). The specificity of TgCmt to use SAM was tested using sinefungin, a structural analog of SAM and inhibitor of methyltransferases36,37. Immunoblot analysis using anti-m7G antibody revealed that sinefungin inhibits m7GpppN32 synthesis in a concentration-dependent manner (Fig. 4F). Together, these results demonstrate that TgCEs are biochemically active and function in a cap-synthetic pathway in Toxoplasma.
A–C The expression and localization of TgCmt. Coomassie blue-stained SDS-PAGE gel of recombinant His6- TgCmt716-1283 protein (A), Western blot analysis of TgCmt expression levels in tachyzoites and bradyzoites (B). Immunofluorescence analysis in RH/ME49 parasites with α-TgCmt antibody (C). Loading controls: TgAldolase (TgALD) - parasite proteins, TgSAG1 and TgBAG1, tachyzoite- and bradyzoite stage-specific markers, respectively. Tz tachyzoite, and Bz bradyzoite. Scale bar 5 µm. D Experimental workflow showing steps involved to generate 32-mer (N32) RNA variants, such as triphosphate RNA (pppN32RNA), diphosphate RNA (ppN32RNA), Guanylated RNA (GpppN32RNA), 7-methylguanosine RNA (m7GpppN32RNA). E Immuno-dot blot analysis of RNA with α-m7G antibody. RNA variants were generated by incubating IVT RNA with or without capping enzymes as specified. Vaccinia capping enzyme (VCE) was used as a positive control. Methylene blue stain serves as an RNA loading control. Graphical representation is Mean ± S.D. (n = 3 replicates) analyzed by one-way ANOVA. F SAM-dependent RNA methylation. A capping reaction was performed with or without Sinefungin at the specified concentration. A–F: n = 3 replicates. E: mean ± SD, one way ANOVA; Dunnett’s comparison test; ns not significant; p values: *<0.05; *** <0.001; ****<0.0001. Source data are provided as a source data file.
Toxoplasma CEs generate productive capped-transcripts
The eukaryotic translation initiation factor, eIF4E, binds to the m7G cap of mRNA and initiates translation38,39. Similarly, we wanted to test whether in vitro-generated capped RNA could be recognized by eIF4E and productively translated into protein in Toxoplasma. BLASTP search using Plasmodium40 and human41 eIF4E revealed three eIF4E-like homologs (TGME49_223410, TGME49_315150, and TGME49_312560) in T. gondii genome35. Of these three homologs, TGME49_223410 showed highest amino acid similarity, including conserved residues required for m7G interaction with Plasmodium (e score: 1e-69) and human protein (e score: 1e-14) (Supplementary Fig. 9). While the eIF4E protein is highly conserved in eukaryotes, TgeIF4E clusters with fungi and other protozoan parasites, and the metazoans form a separate clade (Supplementary Fig. 10). The full-length TgeIF4E-His6 protein of ~26 kDa was purified (Fig. 5A and Supplementary Fig. 5) and used to generate specific antibodies. TgeIF4E is robustly expressed in the asexual stages (Fig. 5B) and, as expected, localized in the cytoplasm of the tachyzoite and bradyzoite stages (Fig. 5C).
A–C The expression and localization of TgeIF4E (n = 3). Coomassie blue-stained SDS-PAGE gel of recombinant full-length TgeIF4E-His protein (A), Western blot analysis of TgeIF4E expression levels in tachyzoites and bradyzoites (B). Immunofluorescence analysis in RH/ME49 parasites with α-TgeIF4E antibody (C). Loading controls: TgAldolase (TgALD) - parasite proteins, TgSAG1 and TgBAG1, tachyzoite- and bradyzoite stage-specific markers, respectively. Tz: tachyzoite and Bz: bradyzoite. Scale bar 5 µm. Microscale thermophoresis analysis of labeled TgeIF4E with pppN32RNA (D), GpppN32RNA (E), and m7GpppN32RNA (F) (n = 2). G Experimental workflow showing generation luciferase (FLuc) RNA variants with or without 5′-cap and 3′-poly(A) tail, such as pppFLuc, pppFLuc-[A]n, and m7GpppFLuc-[A]n. VCE generated m7GpppFLuc-[A]n was used as a positive control. H T. gondii tachyzoites were transfected with luciferase RNAs synthesized with and without 5′-cap and 3′-poly(A) tails as specified. Parasites were transfected with m7GpppFLuc-[A]n generated by VCE as a positive control or mock-transfected with no RNA as a negative control. Luciferase activity was determined 20 h post-transfection. The readings were normalized with VCE RNA (m7GpppFLuc-[A]n). H: n = 3 replicates, mean ± SD, one way ANOVA; Dunnett’s comparison test; ns not significant; p values: *<0.05; *** <0.001; ****<0.0001. Source data are provided as a source data file.
To determine the interaction between TgeIF4E with capped or non-capped RNA variants generated using TgCEs, a microscale thermophoresis (MST) assay was performed using fluorescently labeled TgeIF4E. As measured, TgeIF4E showed strong binding affinity (KD = 8.01 ± 1 nM) towards m7GpppN32RNA (Fig. 5D–F), and no binding affinity was measured for pppN32RNA (Fig. 5D–F) and GpppN32RNA (Fig. 5D–F). These results confirm the m7G cap specificity of TgeIF4E. Further, to evaluate m7G RNA was utilized by Toxoplasma to promote translation, we generated luciferase transcript variants in vitro with and without 5′-m7G cap (using TgCEs) and 3′-poly(A)tails (Fig. 5G). We found that luciferase activity was detected after transfection of capped and polyadenylated RNA (m7G-FLuc-[A]n), but not after transfected with either only IVT RNA (pppFLuc) or capped RNA (m7G-FLuc) or polyadenylated RNA (pppFLuc-[A]n)14 (Fig. 5H). As a positive control, 5’capped and 3′polyadenyaled FLuc RNA was generated using VCE and PolyA polymerase and transfected. However, after comparing the luciferase activity, we found that TgCEs are less effective than VCE (Fig. 5H). Together, these results demonstrate that in vitro-generated capped RNA using TgCEs can be utilized by Toxoplasma to promote translation in vivo.
Depletion of TgCet impairs overall m7G levels and arrest parasite replication
Toxoplasma RNA triphosphatase is a metalloenzyme unique to the parasite. To better understand the role of this protein in the transcription-associated processes, we endogenously tagged TgCet at the C-terminus with mini auxin-inducible degron (mAID) fused with three copies of HA (3HA) epitope (TgCet-mAID-3HA) in RH strain parasites expressing TIR1, which allows rapid degradation of the TgCet-mAID-HA protein (Fig. 6A) upon the addition of indole 3-acetic acid (IAA). The resulting TgCet-mAID-HA strain was confirmed by diagnostic PCR (Fig. 6B). Western blot analysis using anti-HA antibody revealed complete loss of the TgCet -mAID-HA protein in as little as 1 h following addition of 500 μM IAA in the culture medium (Fig. 6C). Using IF analysis, we also observed no staining for TgCet-mAID-HA in the 1 h IAA treated parasites (Fig. 6D). Consistent with the role in the mRNA capping, depletion of TgCet for 8 h diminished m7G capped RNA in the parasite (Fig. 6E, F). We assessed the m7G levels of total parasite RNA (100 ng as determined by the calibration curve Supplementary Fig. 11A) of control vs treated parasites at 2 h, 4 h, and 8 h; however, we could only observe a significant decline (~30%) in m7G levels after 8 h of TgCet depletion. Hence, we selected an 8 h IAA treatment period for all the further experiments.
A Strategy for tagging of TgCet protein in RH-TIR1-3FLAG parental line. B Diagnostic PCR for confirmation of 3′ integration of mAID-3HA in the TgCet gene locus (n = 3). C Western blot analysis of TgCet-mAID-HA expression levels in tachyzoites with or without IAA at indicated time-points (n = 3). D Immunofluorescence analysis of TgCet-mAID-HA grown with IAA or vehicle for 1 h (n = 3). Scale bar 5 µm. E, F Immuno-dot blot and quantification analyzes of m7G levels of RNA extracted from TgCet-mAID-HA parasite (−IAA/+IAA). G TgCet-mAID-3HA parasites were transfected with TgTub-FLuc plasmid DNA. Luciferase activity was measured at indicated time-points. H RT-qPCR analysis of FLuc mRNA in TgCet-mAID-HA −IAA and +IAA parasites. I Percent parasite viability assessed with trypan blue in TgCet-mAID-HA −IAA and +IAA parasites. J Representative IFA images of TgCet-mAID-HA tachyzoites treated with either vehicle, IAA, or CHX and incubated with puromycin prior to parasite fixation (n = 3). Scale bar, 5 µm. K Parasite replication. TgCet-mAID-3HA parasites were grown in HFF monolayers for 18 h with IAA or vehicle. L TgCet-mAID-3HA parasites were grown with IAA or vehicle for 8 h. Number of parasites with defective morphology per vacuole was counted and plotted as a percentage of normal and abnormal parasites. M Crystal violet-stained images of plaques formed by RH-TIR1 and TgCet-mAID-3HA parasites on HFF monolayer treated with IAA or vehicle. Quantification of plaque numbers (N) and plaque areas (O). P Quantification of plaques after IAA withdrawal at indicated time-points in TgCet-mAID-3HA parasites. Q Parasite invasion. The graph shows the percentage invasion of RH-TIR1 and TgCet-mAID-3HA parasites on HFF monolayer. R Parasite egress. The graph shows the percentage of egress of RH-TIR1 and TgCet-mAID-3HA parasites cultured with IAA or vehicle, followed by calcium ionophore treatment. E–I: n = 3 replicates; mean ± S.D; two-way ANOVA with Tukey’s post hoc test; ns not significant; p values: *<0.05; *** <0.001; ****<0.0001. Figure 6K-R; n = 3 replicates; mean ± S.D; one-way ANOVA; Dunnett’s comparison test; ns not significant; p values: *<0.05; *** <0.001; ****<0.0001. Source data are provided as a source data file.
To investigate the impact of TgCet loss on mRNA capping, we designed a reporter plasmid incorporating the firefly luciferase gene (FLuc) under the TgTub promoter. Upon transfection of this FLuc plasmid, in control parasites, the FLuc RNA will be capped and translated into a functional FLuc protein, whereas TgCet-depleted parasites would show impaired capping of FLuc transcript, leading to reduced FLuc protein. As measured, the luciferase activity was decreased after 4 h, and a drastic reduction in the activity was observed after 8 h of Cet depletion (Fig. 6G). To test whether the decrease in FLuc levels is proportional mRNA levels, RT-qPCR analysis was performed, and the relative mRNA levels of FLuc were found to be significantly reduced at 4 h and 8 h of IAA treatment (Fig. 6H, Supplementary Fig. 11B, C, Supplementary Table 1, and Supplementary data 1). To test whether the observed reduction in FLuc levels was not due to parasite death, parasite viability was tested using trypan blue staining. No significant change in the mean viability of the parasite was observed within the 8 h of IAA treatment (Fig. 6I), suggesting a specific effect of TgCet depletion. Furthermore, the effect of TgCet depletion on global translation was examined using the puromycin incorporation into nascent translated proteins. No reduction in the global translation (Fig. 6J) was observed even after 8 h of TgCet depletion, indicating sustained translation of mRNAs. Collectively, these results show that depletion of TgCet leads to reduced RNA capping in the parasite.
We next examined the effect of TgCet depletion on parasite-specific processes. The complete arrest of parasite replication was observed in the TgCet-depleted parasites tested using a standard parasite counting assay (Fig. 6K). A significant number of parasites displayed morphological defects upon TgCet depletion (Fig. 6L). The impact of TgCet depletion on parasite growth was tested using plaque assays. Unlike the parental strain, TgCet-depleted parasites produced no visible plaques (Fig. 6M–O). To determine whether parasites can recover from a transient loss of TgCet expression, we conducted a plaque assay that used six different IAA treatment regimens (Fig. 6P). After an initial 24 h of growth, parasites were treated with IAA or vehicle for 1/2/4/8/12/24 h; at this point, the media was replaced with fresh IAA (or vehicle) and incubated for 5 days. The number of plaques formed was similar for 1 h IAA or vehicle-treated parasites (Fig. 6P). Two hours of IAA-treated TgCet-mAID-HA parasites showed a 50% reduction in the number of plaques formed, whereas no plaques were observed when treated for 4 h or 8 h or 12 h, or 24 h IAA (Fig. 6P). These results indicate that parasites could not recover after depletion of capping for >4 h. The depletion of TgCet significantly decreased the invasion efficiency of the parasite (Fig. 6Q); however, a marginal difference was observed in the ability of parasites to egress upon inducing egress using calcium ionophore (Fig. 6R). Together, these data provide compelling evidence that TgCet is essential for Toxoplasma viability and proliferation.
Depletion of TgCet perturbs gene expression
Given that m7G RNA capping governs overall mRNA metabolism, we reasoned that the loss of parasite viability upon TgCet depletion could be due to a defect in the capped mRNA abundance and gene expression. We investigated the consequences of TgCet depletion (the absence of capping) on gene expression using cap sequencing, which was aimed at capturing all the m7G-capped mRNA population to provide a transcriptome-wide profile that will reveal the identity and relative levels of capped RNA. The m7G-capped mRNAs were enriched using sequential enzyme treatments (Fig. 7A) on 8 h IAA- or vehicle-treated TgCet-mAID-HA parasites (in biological duplicates). In the first step for the enrichment capped mRNA, total RNAs were treated with RppH (in the presence of NEBuffer2) to remove triphosphate-ended RNAs without affecting m7G-RNAs (Fig. 7B). In the second step, recovered RNAs were treated with Xrn1 to remove rRNA and 5’-monophosphate-ended RNA (Fig. 7C). In the third step, RNAs were treated with RppH in the presence of Thermopol buffer to convert m7G-capped mRNA (decapping) to monophosphate-ended mRNA (Fig. 7D). These decapped mRNAs were subjected to library preparation followed by sequencing. RNA-seq library generated from all the RNA samples showed no obvious 5′–3′ bias, as demonstrated by 5′–3′ coverage plots using Qualimap42 (Supplementary Fig. 12). The transcriptomic data were utilized for differential gene expression analysis (DESeq2) (Supplementary data 2).
A Experimental workflow showing steps involved to enrich capped mRNA. Total RNA was isolated from TgCet-mAID-3HA parasites grown with IAA or vehicle. RNA was treated with the indicated enzymes and subsequently used for library construction and RNA sequencing. B Agarose gel image showing rRNA levels in ±RppH+Xrn1 treatment in TgCet-mAID-3HA parasites grown with IAA or vehicle (n = 3). C Immuno-dot blot showing m7G RNA levels in ±RppH treatment (Thermopol buffer 2) in TgCet-mAID-3HA parasites grown with IAA or vehicle (n = 3). Methylene blue stain serves as a loading control. D Immuno-dot blot showing m7G RNA levels in ±RppH treatment (NEBuffer 2) in TgCet-mAID-3HA parasites grown with IAA or vehicle (n = 3). Methylene blue stain serves as a loading control. E Volcano plot showing differentially expressed genes in TgCet-mAID-3HA parasites with IAA or vehicle. Genes with a log2 fold change ≥1 and p ≤ 0.05 are presented (n = 2, Benjamini–Hochberg test, p value in DESeq2). F Validation of altered gene expression in TgCet parasites (−IAA/ + IAA) in a subset of genes, including H3, H2A1, H2Bb, H2Ba, PYK, PGK, ENR, PGD, RPS12, and RPS27 RT-qPCR. Gene expression was normalized to 18 s rRNA levels (n = 2 replicates). G RT-qPCR of H3, H2A1, H2Bb, H2Ba, PYK, PGK, ENR, PGD, RPS12, and RPS27 genes in TgCet parasites (−Actinomycin D/+ Actinomycin D). 18 s rRNA levels were used for normalization (n = 2 replicates). Gene ontology analysis of downregulated (H), and upregulated (I) expressed genes with respect to cellular components and biological processes. Genes with a log2 fold change ≥1 and p ≤ 0.05 are presented (n = 2, Benjamini–Hochberg test, p value in DESeq2). Source data are provided as a source data file.
The significantly altered capped transcripts were identified by DESeq2 (FDR ≤ 0.05; log2 fold change ±1), resulting in 185 and 186 genes whose abundance was decreased and increased, respectively, upon depletion of TgCet (Fig. 7E). The transcript length analysis revealed that transcripts of >1000 bp were over-represented (Supplementary Fig. 13) and <500 bp were found to be less abundant (downregulated). The genes with significantly different gene expressions relevant to the study are shown in the volcano plot (Fig. 7E). Validation of RNA-seq data was confirmed using RT-qPCR analysis (Fig. 7F, Supplementary Fig. 11B, C, Supplementary Table 1, and Supplementary data 1) of four each of downregulated (H3, H2A1, H2Bb, and H2Ba) and over-represented (PYK, PGK, ENR, and PGD) candidate genes, and two unchanged genes (RPS12 and RPS27). Cap-mediated dysregulation of gene expression was further compared with Actinomycin D (to inhibit all transcription). Indeed, we observed a good correlation regarding mRNA expression of candidate genes between IAA (Fig. 7F) and Actinomycin D-treated parasites (Fig. 7G, Supplementary Fig. 11B, C, Supplementary Table 1, and Supplementary data 1), suggesting a common gene expression pattern upon transcription inhibition. To understand what processes the differentially expressed genes might be involved in, we performed gene ontology (GO) enrichment analyzes (Supplementary data 2). We could identify clear terms for cellular components (CC) and biological processes (BP) for the significantly dysregulated genes. The multiple GO terms were identified for the downregulated genes, but were mostly related to DNA packaging, cell membrane, and parasite invasion. The most significant downregulated genes (Fig. 7H) were histones (H1 like, H2A1, H2Ba, H2Bb, H2Ax, H3, and H4) and inner membrane complex proteins (IMC16, IMC17, IMC20, IMC22, IMC26, IMC34, IMC35). Transcripts related to protein phosphorylation (Fig. 7F, G) were found to be significantly downregulated (Rhoptry kinases-ROPs, aurora kinase, calcium-dependent protein kinase, cGMP, cAMP, and AMP-dependent kinases). Several host cell invasion genes, ROP-1,-15,-29,-34,-37, GRA-5,-12,-16,-62,-63, MIC-3,-4,-10, SRS-20,-29, and-35 were found to be downregulated (Fig. 7E), which was consistent with significantly reduced invasion for TgCet-depleted parasites. However, transcripts related to transporter activities (phosphate transporters -PT2, MC family transporter-SFT2, RIC1, GOT1, Aquaporin2, and ABC) and transferase activity (glucosamine transferases, SET methytransferases, dehydrogenases, esterases) were found to be significantly over-represented (Fig. 7I). Further analysis revealed glucose (PYK1, ENO1, GAPDH1, ENR, and ACC1) and glutamate metabolism (glutathione synthase, glutamate cysteine ligase) pathways over-represented upon loss of capping (Fig. 7G). Overall, lack of mRNA capping mediated through TgCet depletion results in dysregulation of gene expression of many parasite processes, with a major impact on shorter transcripts with higher turnover.
Depletion of TgCet protects mice from lethal toxoplasmosis
TgCet protein is essential for parasite fitness in tissue culture; however, to test its essentiality in establishing an infection in the host, we performed mouse infection studies. First, we tested whether TgCet-mAID-HA could be depleted in vivo (Fig. 8A). Mice were infected with 50 tachyzoites of RH TgCet-mAID-HA parasites intraperitoneally and then treated orally with 200 mg/kg/day IAA43 or vehicle from day 2 to 4 post-infection (Fig. 8A). On days 5 and 6 pi, the mice were sacrificed, and peritoneal exudate cells (PECs) were collected for IF microscopy. Parasites from IAA-treated mice showed depleted TgCet-mAID-HA levels but normal levels of a control protein, TgIMC1, as compared to parasites from the vehicle-treated mice (Fig. 8B–D). This experiment confirmed the successful in vivo depletion of TgCet-mAID-HA protein as early as 3 days after IAA treatment. Next, to determine the effect of prolonged depletion of TgCet on parasite survival in vivo, mice were infected with RH TgCet-mAID-HA parasites and then treated with IAA or vehicle orally for 15 days to deplete TgCet (Fig. 8E). The depletion TgCet was confirmed by IF microscopy in one of the sacrificed mice at 5 days of IAA treatment, as demonstrated above. No mortality was observed in uninfected mice supplemented with IAA (Fig. 8F). All mice receiving the vehicle control treatment succumbed to lethal toxoplasmosis by day 11 post-infection (Fig. 8F). Conversely, IAA treatment (i.e., TgCet depletion) rescued all mice from lethal toxoplasmosis, indicating that TgCet is necessary for acute infection in T. gondii (Fig. 8F). Severe morbidity and complete mortality were seen in the control treatment group compared to the IAA treatment group (Fig. 8G, F). By day 2 pi, the control group showed weight loss, a sign of illness, and continued to lose up to a quarter of their initial weight by the time of death. The IAA treatment group showed no weight loss. Suppressing TgCet expression during the acute phase of infection effectively blocked replication of T. gondii since discontinuation of IAA treatment following day 15 did not result in morbidity (Fig. 8G), mortality (Fig. 8F), or parasite presence (tested by collecting PECs -data not shown) as monitored for an additional 15 days. Overall, these results show the essential role of TgCet for parasite survival and replication in the mouse host.
A, B In vivo assessment of TgCet-mAID-3HA knockdown. BALB/c mice were challenged with 50 TgCet-mAID-3HA parasites intraperitoneally and treated with IAA or vehicle from day 2 to 6. On days 5 and 6, mice were sacrificed, and peritoneal exudate cells were collected for IF microscopy. Fixed cells were probed for parasites (rabbit α-TgIMC1 and α-rabbit IgG Alexa Fluor 594) and Cet-mAID-3HA (mouse α-TgHA and α-mouse IgG Alexa Fluor 488). Scale bar, 5 μm. C, D Quantification of TgCet-mAID-3HA (D) and TgIMC1 expression (E) in (B). The abundance of each protein (mean fluorescence intensity ± SD) was quantified and averaged from n = 50 parasites for control and +IAA groups (n = 2). Statistical significance was assessed using one-way ANOVA; Dunnett’s comparison test; ns not significant; p values: *<0.05; *** <0.001; ****<0.0001. E Experimental design of in vivo test of TgCet essentiality. Related to (B) (n = 10 mice per group and n = 2 trials). F Survival curve of BALB/c mice infected with 50 TgCet-mAID-3HA parasites intraperitoneally and treated with IAA or vehicle for 15 days. The Gehan-Breslow-Wilcoxon test was used to compare differences between the survival curves, p < 0.0001 (IAA vs control). No mortality was observed in uninfected mice supplemented with IAA, as indicated by pink line. G Mean body weight ± SD of BALB/c mice infected with 50 TgCet-mAID-3HA parasites intraperitoneally and treated with IAA or vehicle for 15 days. Source data are provided as a source data file.
Structural analysis of T. gondii RNA triphosphatase
The tunnel family RNA triphosphatases are potential antiinfective targets owing to the complete divergence in structures and mechanisms of the RNA triphosphatases of the unicellular pathogen and the mammalian host. We showed that the mechanism of action of TgCet is different from its host counterpart; however, we are yet to know the structural details of this protein, which can help develop a parasite-specific inhibitor. To gain insight into the structure of TgCet, we first attempted to perform comparative modeling of TgCet with known PDB structures of other RNA triphosphatases; however, we failed to obtain the structure owing to poor (<40%) sequence homology. Next, we retrieved Alphafold predicted structure of TgCet using the AlphaFold database44 (https://alphafold.ebi.ac.uk/entry/S8F0G9). The obtained structure showed a moderate to high confidence score (the per-residue model confidence score: 40–95) for the region corresponding to the triphosphate tunnel (361–729aa) and a high predicted error (PAE) for the other regions of TgCet (Supplementary Fig. 14A, B). Finally, using a ColabFold45, the structure of the triphosphate tunnel region (361–729aa) was predicted (Supplementary Fig. 14C). The predicted structure comprised 10 anti-parallel β barrels (green) with 3 α helices (blue) surrounding the active tunnel site (Fig. 9A, B).
A Predicted structure of TgCet (361–729aa) using Collabfold. B The structure contains a ten-stranded anti-parallel β-barrel with three α-helices surrounding the active-site tunnel. C The predicted structure of TgCet in green was superimposed on S. cerevisiae Cet1 colored in yellow (1D8H). D Conservation of active site residues of ScCet1, TgCet, TcCet1, and hTTP. E The predicted structure of TgCet in green was superimposed on T. cruzi Cet1 colored in blue (6L7W). F The predicted structure of TgCet in green was superimposed on human thiamine triphosphates, colored in red (3TVL).
Structural comparison with S. cerevisiae RNA triphosphatase (ScCet1) revealed a nearly identical structure (for triphosphatase tunnel) of the two enzymes (Fig. 9C) with 9 of 15 side chain positions important for Cet1 activity conserved in TgCet (Fig. 9D). The triphosphatase tunnel structure of TgCet also showed high structural similarity, including active site residues with the recently deciphered crystal structure of Trypanosoma cruzi RNA triphosphatase (TcCet1). (Fig. 9D, E). Importantly, both glutamate residues (E366 and E726) essential for the catalytic activity of TgCet were found to align with the residues predicted to form the metal-binding site within the tunnel of ScCet1 and TcCet1 (Supplementary Fig. 15A–E).
Next, we checked whether any host protein has similar structure as of TgCet. Using psi BLAST43, we found human thiamine triphosphatase (hTTP) is the only TTM with the available structure that resembles TgCet predicted structure46 (Fig. 9F). Structural comparison with hTTP47 revealed that the metal binding sites are identical between the two enzymes (Fig. 9D); however, TgCet lacks the C-terminal plug-in helix48,49, which ensures a topologically closed structure and provides substrate specificity only for thiamine triphosphatase (Fig. 9F). Collectively, these results demonstrate the overall similarity in the structure of TgCet with other TTMs. Although the central tunnel structure of TTM is conserved amongst TgCet and hTTP, the substrate recognition and specificity vary between them, and this raises the possibility of development of an inhibitor that specifically targets the entry of RNA substrate into TgCet with minimal to no effect on the hTTP.
Determination of therapeutic potential of TgCet
To evaluate the potential of TgCet as a therapeutic target, we selected two compounds: Myricetin (Flavonoid) and 3,4-dicaffeoylquinic acid (3,4-diCQA) (Quinic acid ester) (Supplementary Fig. 16A). These compounds actively inhibited recombinant Trypanosoma RNA triphosphatase (TbCet) at submicromolar concentrations17. To determine if TgCet is the molecular target for Myricetin and 3,4-diCQA, we performed an ATPase assay using recombinant TgCet (Supplementary Fig. 16A). We first calculated the half-maximal inhibitory concentration (IC50) of both Myricetin and 3,4-diCQA against TgCet (Fig. 10A, B). Both compounds exhibited concentration-dependent inhibition, with IC50 values of 0.4 µM for Myricetin and 5.07 µM for 3,4-diCQA. As both compounds inhibited TgCet activity, we further tested their inhibitory effects on the asexual stage of the parasites.
Determination of the median inhibitory concentration (IC50) of Myricetin (A) and 3,4-diCQA (B) for TgCet protein using a 10-dose response curve (n = 3). C Evaluation of the half-maximal effective concentration (EC50) of Myricetin against RH tachyzoites (n = 3). D IFA showing the effect of Myricetin and 3,4-diCQA on the development of in vitro-induced bradyzoite cysts (n = 3). Scale bar 5 μm. E Cyst size was measured in individual cysts after compound treatment or DMSO. F Bradyzoite viability upon compound treatment as assessed by plaque assay. G Effect of Myricetin on lethal toxoplasmosis was tested using BALB/c mice infected with the RH strain. Survival curve of mice from four groups. First- DMSO with infection (blue line), second—Myricetin with infection (red line), third- pyrimethamine with infection (black line), and Myricetin without infection (green line). The Gehan-Breslow-Wilcoxon test was used to compare differences between the survival curves. Ns not significant. Mortality due to acute toxoplasmosis in DMSO (first group, H) and Myricetin (second group, I) treated mice confirmed by IFA (n = 3). Figures 10E and 10F: n = 3 replicates; mean ± S.D; one-way ANOVA; Dunnett’s comparison test; ns not significant; p values: *<0.05; *** <0.001; ****<0.0001. Source data are provided as a source data file. J Effect of lack of capping mediated by TgCet depletion on the parasite. The intricate mRNA capping process orchestrates a precise equilibrium in mRNA levels, thereby ensuring the regular growth and replication of the parasite, evident in both in vitro (A) and in vivo (B). The absence of TgCet results in impaired mRNA capping and leads to parasite replication arrest (C) and rendering the parasites non-viable, ultimately preventing lethal toxoplasmosis (D). Leveraging insights from both in vitro and in vivo investigations, the strategic use of a small molecule inhibitor, specifically designed to target the TgCet RNA entry tunnel (E), emerges as a promising approach to block TgCet effectively. This inhibitor approach could mimic the observed effects in (D) and offer a potential avenue for controlling lethal T. gondii infection. The illustration was created in BioRender. Mitra, P. (2025) https://BioRender.com/opn4jhz.
The impact of these compounds on tachyzoite growth was evaluated using a plaque assay. Both compounds were tested over a concentration range from 0.001 µM to 50 µM. Myricetin demonstrated the ability to inhibit the growth of RH parasites, with an effective concentration (EC50) of 0.58 µM (Fig. 10C). In contrast, 3,4-diCQA showed no effect on tachyzoite growth, even at the highest concentration tested (50 µM). Further, we evaluated the impact of these compounds on bradyzoite growth by measuring the cyst size of bradyzoites differentiated in vitro for three days and treated with the compounds for two days (Supplementary Fig. 16B). The cyst size, as an indicator of bradyzoite growth, decreased significantly with Myricetin at EC50 and 3xEC50 concentrations (Fig. 10D, E). To examine whether the bradyzoites in these cysts were viable, we disrupted cysts, trypsinized to release bradyzoites, and then cultured them on HFF monolayers for plaque formation assessment (Fig. 10F). Myricetin-treated bradyzoites demonstrated reduced viability, as evidenced by significantly fewer plaques. In contrast, 3,4-diCQA showed no effect on bradyzoite growth as plaque numbers were comparable to control (Fig. 10F). Since Myricetin was effective against TgCet protein and both asexual stages of T. gondii, we further examined its potential protective effect against lethal toxoplasmosis using a mouse infection model.
To test Myricetin’s efficacy in vivo, BALB/c mice were infected with 100 RH tachyzoites and treated with Myricetin (100 mg/kg)50,51 or pyrimethamine (50 mg/kg)52 for 10 days post-infection (Fig. 10G). Despite its potent in vitro activity, Myricetin did not enhance survival in infected mice (mean survival = 12 days), as demonstrated by the presence of an acute infection (Fig. 10H) like control (Fig. 10I), suggesting limited efficacy in acute toxoplasmosis. No Myricetin-induced toxicity was observed in uninfected mice orally supplemented with 100 mg/kg Myricetin (Fig. 10G). These results demonstrate that Myricetin could serve as a promising candidate that yet requires optimization for in vivo applications.
Discussion
A defining feature of eukaryotic gene expression is the addition of an m7G cap to nascent pre-mRNAs shortly after the initiation of synthesis53. We aimed to understand the capping process in Toxoplasma to examine parasite-specific variations and to find the contribution of this process to a broader understanding of mRNA metabolism and cellular responses. T. gondii encodes three separate mRNA capping enzymes: triphosphatase, guanylyltransferase, and cap methyltransferase, similar to that of fungi and distinct from the two enzyme capping systems of metazoans and plants. Biochemical characterization of the triphosphatase TgCet firmly categorized it in the family of metal-dependent phosphohydrolases identified in fungi, DNA viruses, and some protozoa. Despite low similarity with other homologs, guanylyltransferase TgCeg and the methyltransferase TgCmt are similar in structure and mechanism to the typical guanylyltransferases and cap methyltransferase enzymes found in all eukaryotes. The capping functions of TgCEs were verified biochemically by testing the productive translation of the in vitro generated capped reporter RNA and genetically through complementation of S. cerevisiae strains lacking endogenous enzymes. The depletion of TgCet resulted in global defects in gene expression and complete arrest of parasite replication (tissue culture and mouse host), highlighting the essential role of triphosphatase in the parasite. The therapeutic potential of TgCet was established using of two compounds previously shown to target RNA triphosphatases.
In eukaryotes, capping is facilitated by direct recruitment of the capping enzymes to the transcription machinery via interactions with the phosphorylated C-terminal domain (CTD) of Rpb1, the largest subunit of RNAP II54. In many organisms, guanylyltransferase and methyltransferase interact directly with the phosphorylated RNAP II CTD; however, in S. cerevisiae, triphosphatase and in S. pombe, triphosphatase and guanylyltransferase bind the phosphorylated RNAP II CTD independently29,55. These enzymes preferred to bind a CTD substrate that was phosphorylated at Ser5 or doubly phosphorylated at Ser2 and Ser5 in the highly conserved heptapeptide repeats Y1S2P3T4S5P6S7. T. gondii Rbp1-CTD contains a mixture of 10 heptapeptide YSPxSPx sequences instead of conserved YSPTSPS; however, Ser2 and Ser5 residues are conserved in those 10 heptapeptide repeats. The role of Ser5-56 and Ser2-57phosphorylation has been shown in transcription initiation and elongation, respectively, in T. gondii. Hence, it will be interesting to test which T. gondii capping enzyme interacts with Ser5 and/or Ser2-phosphorylated CTD of Rpb158. Besides, T. gondii triphosphatase and methyltransferase are exceptionally large proteins with N-terminal extensions, suggesting additional functions or more regulatory roles in coordinating cap formation with RNA synthesis59,60. Also, it is intriguing to test the distribution of TgRNAP II on chromatin without capping to test whether RNAP II transits to the elongation phase, pauses, or transcribes at a low speed or pausing followed by premature transcription termination61.
Auxin-induced effective depletion demonstrated that TgCet is an essential protein. Despite the effective depletion of TgCet in 1 h, its impact on capped mRNA was found to be marginal even after 8 h of depletion, suggesting factors beyond TgCet activity might contribute to the persistence of capped mRNA. The stability of pre-existing capped mRNA, a compensatory mechanism that preserved the Cap structure, and the slower turnover rate of certain capped mRNAs could result in a cumulative delay in observing the full effects of TgCet depletion on cap removal. Depletion of TgCet had no impact on nascent protein synthesis; however, transiently transfected luciferase showed a drastic decrease in both mRNA and protein levels. The profound effect on luciferase could be due to strong tubulin promoter driving the expression of luciferase or higher turnover of such reporter genes that require continuous transcription and thereby capping. Depletion of TgCet results in a significant reduction in invasion efficiency and a marginal difference in the egress of the parasite. While the observed defect in invasion correlates well with the transcriptomic data showing the downregulation of several invasion-related genes, the minimal effect on egress could be due to less turnover of egress transcripts and yet-to-be-identified post-transcriptional regulation of gene expression, which might confer higher stability to such transcripts. The reduced transcription and increased mRNA storage in schizont-stage parasites were also observed in Plasmodium spp62. Additionally, it is possible that such transcripts in Toxoplasma could be regulated through an alternative capping mechanism, supporting the observed marginal difference in mRNA capping despite TgCet knockdown being effective. For instance, cytoplasmic mRNA capping of the decapped mRNA in Trypanosoma by TbCe1 (Cytoplasmic capping enzyme)63. While TbCe1 homolog could not be identified in T. gondii, the presence of a highly divergent protein with a similar function can not be ruled out.
Cap sequencing upon TgCet depletion revealed global gene expression defects with a relatively similar number of genes that were downregulated and over-represented. The transcript length analysis revealed that over-represented genes encode mostly longer transcripts (>1000 bp), and downregulated genes encode mostly shorter transcripts (>500 bp). For example, we observed significant downregulation of expression of all histone variants (short mRNAs). Generally, longer mRNAs are occupied by multiple ribosomes (polysome occupancy), which often correlates with higher translation efficiency and stability of the mRNA. This increased translation activity may contribute to greater mRNA stability, as translated transcripts are typically protected from degradation mechanisms64,65. On the contrary, shorter mRNAs may bind fewer ribosomes and show rapid turnover. Histone mRNAs are also known to lack a poly(A) tail66,67, further contributing to their shorter half-lives. Hence, the interplay of mRNA length, ribosome engagement, and decay pathway dynamics could also explain why longer transcripts remain more stable and abundant while shorter mRNAs undergo more rapid turnover, resulting in lower abundance. Altogether, the lack of mRNA capping highlights the intricate relationship between transcript characteristics and the surrounding cellular environment, which together influence the stability of transcripts in Toxoplasma.
The most significantly downregulated transcripts were related to chromatin maintenance, structural integrity, key kinases, and host cell invasion, which explains why a halt of parasite replication occurs even with a transient depletion of TgCet. However, the transcripts related to transporter activities, transferase activity, glucose, and glutamate metabolism were over-represented upon loss of capping. The global transcriptome data (ToxoDB) in T. gondii suggest that in TgCet-depleted parasites, most downregulated genes were highly expressed, and the over-represented genes expressed at low levels. These findings are consistent with the recent study in S. cerevisiae, where the impact of defective capping (depletion of guanylyltransferase) on genome-wide mRNA abundance revealed the reduction of highly expressed mRNAs and the accumulation of lowly expressed and more stable mRNAs68. In yeast, such cap-defective transcripts are detected by an Npl3-mediated surveillance mechanism that triggers decapping and subsequent mRNA degradation. In Toxoplasma, the mRNA surveillance mechanism and mRNA degradation pathways are yet to be studied.
Generally, glucose and glutamate are the sole physiological nutrient sources in carbon metabolism, obligatory for parasite growth and survival69,70. Hence, unsurprisingly, we observed high transcript levels of carbon metabolism genes in cap-defective parasites. Such transcripts are stabilized probably through a posttranscriptional mechanism that maintains the steady-state mRNA levels of these transcripts for such obligatory parasite function. In such a case, the transcript stabilization could outweigh the lowered level of de novo transcription. Stabilizing such transcripts would eventually limit the decline in the transcription rate and ensure some level of advantage to the parasite to survive under deprivation of either carbon source. The reasons for this stability might be attributed to RNA binding proteins, specificity in the decay pathways, other mRNA modifications, or RNA stability elements such that even in the absence of productive transcription71, these mechanisms could contribute to maintaining existing mRNA pools of certain genes. This interplay between stabilization and degradation processes is essential for the dynamic regulation of gene expression and might explain the adaptation and survival mechanisms in the parasite. Hence, in the choice to live or die, the parasite prefers to use its energy on basic metabolic processes for its survival than on the process required for its replication, and this explains why DNA replication and packaging genes were downregulated in TgCet-depleted parasites.
The complete differences between the tertiary structures, active sites, and chemical mechanisms of the RNA triphosphatase component of the mRNA capping system in pathogenic fungi, viruses, and protozoa and those of their metazoan hosts highlight TPase as a target for anti-infective drug discovery23,24,25,72. The triphosphate tunnel metalloenzyme (TTM)-type TPases are essential for the growth of S. cerevisiae (ScCet1)23, S. pombe (SpPct1)26, the human-pathogenic fungus Aspergillus fumigatuts (AfTriA)27, and the human-pathogenic protozoa T. brucei (TbCet1)16. To our knowledge, it has not been determined whether TTM-type TPase is essential for pathogen growth in the host. Here, we employed an auxin-inducible degron approach to show that Toxoplasma TgCet (TTM-type) is essential for the growth of parasites in both the culture and the mouse host. TgCet showed robust manganese-dependent NTPase activity in an exceptionally wide range of temperatures and pH. The crystal structures of ScCet123 and TbCet146 revealed that the TTM active site comprises essential amino acids that either coordinate a metal ion or the γ-phosphate or stabilize the tunnel architecture. Most of these active site residues in the triphosphate tunnel are conserved in TgCet, and the alanine mutations of a few of these residues showed a dramatic reduction in the activity, suggesting a similar function as observed for ScCet173 and TbCet146 in interacting with the 5′-end of the triphosphate RNA substrate.
The recent studies on biochemical screening for small-molecule inhibitors showed that Trypanosoma TbCet1 could be inhibited using a nanomolar concentration of various classes of phenolic compounds17, whereas kribellosides, metabolites from actinomycetes, selectively inhibit ScCet174. Based on this study, we selected Myricetin and 3,4-dicaffeoylquinic acid (3,4-diCQA) compounds, which inhibited TbCet1 activity at submicromolar concentrations. Only Myricetin exhibited potent inhibition against TgCet protein and asexual parasite stages (tachyzoites and bradyzoites). Despite the target-specific inhibition of Myricetin in vitro, its lack of efficacy in treating lethal toxoplasmosis in mice could be due to factors such as poor bioavailability, rapid efflux, or metabolic breakdown75,76. According to inhibitor studies in Trypanosoma, these inhibitors displayed great potency and selectivity for the target enzyme, indicating separate screening against pathogen-specific TTM-type TPases. Additionally, the high-resolution crystal structure of TgCet, along with screening for small-molecule inhibitors, will provide critical insights into the basis for TgCet inhibition. The identified compounds can then be tested for parasite growth inhibition in the culture and mouse (natural Toxoplasma host). It is tempting to speculate that these parasite-specific inhibitors may not inhibit human (host) thiamine triphosphatase (hTTP), the only known TTM with similar metal binding sites48, owing to the presence of alpha helix at the c-terminal end of hTTP, which positions the thiamine in a way that prevents the entry of non-specific substrates like pppRNA, thereby ensuring specificity49 (Fig. 10J).
Overall, our work presents a detailed characterization of three-component mRNA-capping machinery in Toxoplasma, where RNA triphosphatase is an essential component with complete divergence in the structure and catalytic mechanism from human ortholog. Lack of capping mediated through RNA triphosphatase depletion results in perturbation of gene expression, which is detrimental to the parasite, both in vitro and in vivo, and renders RNA triphosphatase an attractive therapeutic target for Toxoplasma infection.
Methods
Ethics statement
The institutional ethics committee has approved using laboratory research protocols (IBSC/April2024/NIAB/ASD001) and animals (IAEC/NIAB/2024/11/ASD).
Parasite culture
T. gondii tachyzoites of RH and ME49 strains (Supplementary Table 3) were maintained in human foreskin fibroblast cells (HFFs, ATCC) in DMEM containing 10% fetal bovine serum, 10 µg/ml gentamicin, 1% penicillin-streptomycin, and 2 mM L-glutamine at 37 °C and 5% CO2. Tachyzoite to bradyzoite stage differentiation was carried out by incubating the ME49 tachyzoites in bradyzoite induction medium (RPMI pH 8.2) at 37 °C for 5 days without CO277.
Cloning, expression, and purification of TgCet, TgCeg, TgCmt, and TgeIF4E
The E. coli codon-optimized ORF of T. gondii Cet, Ceg, Cmt2146-3849 (subscript denotes nucleotide coordinates), and eIF4E were synthesized by Life Technologies (Supplementary Fig. 5). All three genes were initially cloned into the pMK-RQ vector (Life Technologies) between NdeI-EcoRI sites, which were further subcloned into the pET-28a or pET-21a (Novagen, USA), and recombinant proteins were expressed in E. coli BL21 Rosetta as N-terminal (TgCet, TgCeg, and TgCmt) and C-terminal (TgeIF4E) 6-xHis-tag and purified on a nickel-nitrilotriacetic acid-agarose resin column as described previously78. Briefly, E. coli transformed with pET28a-TgCet/TgCeg/TgCmt/TgeIF4E was grown in 10 ml of Luria-Bertani (LB) medium supplemented with 50 µg/ml kanamycin and 34 µg/ml chloramphenicol overnight at 37 °C. Subsequently, 10 ml of the overnight culture was added to 1000 ml of LB containing the same antibiotics and incubated at 37 °C with vigorous shaking. When the OD600 reached 0.6, IPTG was added to the culture to a final concentration of 1 mM, and the cells were further incubated at 25 °C for 16 h. The cells were then harvested by centrifugation, and the pellets were resuspended in 50 ml of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 100 μg/ml lysozyme, 10 mM imidazole, 0.1% Triton X-100, and 0.3 mM PMSF, pH 8.0). After centrifugation, the protein was purified from the supernatant with the use of a Ni2+-NTA agarose (Qiagen) and step-eluted with 1-ml aliquots of 20–300 mm imidazole in lysis buffer. The quality of the eluted protein was analyzed using SDS-PAGE. The appropriate elutes containing the protein were then dialyzed in 1X PBS and stored at −80 °C. TgCet mutants (E366A and E726A) and TgCeg mutants (K133A, T134A, D135A, and G136A) were generated using a specific set of primers (Supplementary Table 1) following the Stratagene (#210515) site-directed mutagenesis protocol. Recombinant mutant protein purification was performed as for wild-type proteins.
Polyclonal antibody raising
Mouse polyclonal antibodies to recombinant TgCet, TgCeg, TgCmt, and TgeIF4E were generated by primary injection with 30 μg of purified recombinant protein in Freund’s complete adjuvant (#F5881, Sigma) followed by four boosts of 20 μg each in Freund’s incomplete adjuvant (#F5506, Sigma) at 2-week intervals. Serum was collected after day 60 post immunization. Polyclonal antibodies for Tg- IMC178, SAG179, BAG180, CST180, and Aldolase (ALD) antibodies were used from previous studies.
Immunoblotting
Filter-purified 2 × 105 parasites were suspended in SDS-PAGE sample buffer and boiled for 10 min before being run on a single lane of a 10% polyacrylamide gel. The gel was then transferred to a 0.2 μm PVDF membrane (BioRad) using a Trans-Blot System (BioRad) for 12 h at 30 V. The PVDF membrane was blocked in 5% (w/v) non-fat milk in PBS for 60 min before being probed with a primary antibody (α- TgCet/TgCeg/TgCmt/TgeIF4E-1:500; αHA (Sigma #H3663)-1:5,000; α-TgSAG1/TgBAG1/TgALD-1:2,000) in non-fat milk overnight at 4 °C. The blot was washed 3x with PBS plus Tween-20 detergent (PBST; 0.1% Tween-20) before probing with either HRP-conjugated α-rabbit (Santa Cruz #Sc-2537) or α-mouse-IgG (Santa Cruz #Sc-2005). The blot was washed and developed using the Clarity Western ECL kit (BioRad) and visualized on a ChemiDoc Imager (Biorad).
Immunofluorescence (IF) staining
HFFs were grown on glass coverslips until confluent and subsequently infected with T. gondii RH/ME49/TgCet-mAID-HA tachyzoites. The infected HFFs were fixed with methanol-free 4% paraformaldehyde in PBS, permeabilized in 0.1% Triton X-100 in PBS for 15 min at room temperature, and blocked with 5% (w/v) bovine serum albumin (BSA, Sigma) in PBS for 60 min at RT. Primary and secondary antibodies were diluted in 1% (w/v) BSA in PBS. Samples were first incubated with the primary antibody (α- m7G/TgCet/TgCeg/TgCmt/TgeIF4E-1:100; αHA-1:1,000; αTgIMC1/TgSAG1/TgCST1/TgBAG1 −1:2,000) at 25 °C for 60 min, washed 5x with PBS, and then incubated with fluorescent secondary antibodies (1:1000) and 4’,6-diamidino-2-phenylindole (DAPI; 300 nM) at 25 °C for 60 min. Secondary antibodies (Invitrogen #A11001, #A11008, #A11005, #A11012) were conjugated to either Alexa Fluor (AF) green or red fluorophores and specific to the species of primary antibody used. Samples were then washed 5x with PBS before mounting the coverslip on a glass slide using Vectashield medium (Vector Laboratories). IF staining was visualized using a Leica confocal microscope with a 100X oil immersion objective. Images were processed using LAS X software (Leica Microsystems). A similar protocol was employed for bradyzoite IF staining.
RNA dot-blot
RNA and genomic DNA were isolated from filter-purified parasites using RNeasy kit (Qiagen) and DNeasy Blood and Tissue kit (Qiagen), respectively. For m7G RNA estimation, indicated amounts of nucleic acids were spotted on the Hybond-N + membrane (Sigma) and fixed to the membrane with a UV crosslinker (Bioanalytik Jena). The dried membrane was stained with ethidium bromide or methylene blue stain for 15 min at room temperature to stain the spotted nucleic acids. The membrane was blocked for 30 min in 5% (w/v) skim milk in PBS and washed 3x with PBS. The membrane was first incubated with α-m7G antibody (Sigma #MABE419 Clone H20 1:1000) at 4 °C for 12 hrs, washed 5x with PBS, and then incubated with HRP-conjugated anti-mouse secondary antibody (Invitrogen) at 25 °C for 60 min. The membrane was washed and developed using the Clarity Western ECL kit and visualized on a ChemiDoc Imager.
For estimation of m7G levels in the TgCet depleted parasites, capped luciferase RNA ranging from 12.5 ng to 400 ng (prepared as described in Luciferase expression assay “method” section) was spotted on the membrane, and dot blots were developed using α-m7G antibody as mentioned above. A (Supplementary Fig. 11A) calibration graph was generated in GraphPad Prism by plotting the concentration of m7G-capped RNA on the x-axis and pixel intensity on the y-axis (Supplementary Fig. 11A). The m7G levels in the TgCet-mAID-HA parasites (−IAA/+IAA) were determined using 100 ng RNA at indicated time points following the abovementioned procedure. For dot blot quantification, the m7G spot intensity was normalized to the respective methylene blue control, and the ratio of the intensity of IAA-treated RNA to control RNA was plotted. The Cap sequencing experiment followed a similar procedure to determine the m7G level of RNA samples treated with RppH and XrnI from control and IAA-treated parasites.
Triphosphatase assay
Triphosphatase activity was assayed12,13 by quantifying the release of 32Pi from γ32P-labeled ATP. Standard reaction mixtures (10 µl) containing 50 mM Tris HCl (pH 7.5), 5 mM DTT, 1–10 mM MnCl2 or MgCl2/10 mM ZnCl2 or CaCl2, 1.5 mM ATP, and TgCet protein as specified (20–500 ng) were incubated for 30 min at 30 °C. The reaction mixtures were applied to a polyethyleneimine-cellulose thin-layer chromatography (TLC) plate, which was developed with 0.75 M potassium phosphate (pH 4.3). The release of 32Pi from [γ32P]ATP was quantitated by scanning the TLC plate with a Phosphor Imager (Typhoon, Cytiva). 5 mM MnCl2 and 100 ng TgCet protein were used to test the time-dependent, temperature-dependent, and pH-dependent RNA triphosphatase activity. Triphosphatase activity in the presence of different NTPs was assayed using 1.5 mM [γ32P]ATP or 1.5 mM [α32P]ATP, or 1.5 mM [α32P]UTP, and 100 ng TgCet protein. RNA triphosphatase reaction mixture containing 2 µM γ-32P labeled triphosphate-terminated N32 RNA (pppN32 RNA), 50 mM Tris HCl (pH 7.5), 5 mM DTT, 5 mM MnCl2, and 100 ng TgCet protein was incubated for 30 min at 30 °C. RNA 5’ triphosphatase activity was measured similarly to Pi release in ATPase assay. The pppN32 RNA was prepared using dsDNA (N32 mer), 1 mM each of CTP, UTP, GTP, 220 µm [γ32P]ATP, and T3 RNA polymerase (Ambion).
Guanylyltransferase assay
Guanylyltransferase activity was measured12,13 in reaction mixtures (20 µl) containing 50 mM Tris HCl (pH 8.0), 5 mM DTT, 5 mM MnCl2, 1 mM [α32P]GTP, and TgCeg protein (0.5–4 µg) that were incubated for 30 min at 37 °C. The reactions were stopped by adding SDS-PAGE sample buffer and boiling for 10 min. The samples were electrophoresed through a 10% polyacrylamide gel, and the GT-GMP intermediate complex was determined by autoradiography using a phosphor imager.
Preparation of m7GpppN32 RNA
Template dsDNA (N32 mer) and RNA variants were prepared as described previously in ref. 81. The dsDNA (Supplementary Table 1) was used to generate pppN32 RNA using MEGAScript T3 Transcription Kit (Ambion). The pppN32 RNA was treated with TgCet (as described in the triphosphatase assay) to obtain ppN32 RNA, and the ppN32 RNA was treated with TgCeg (as described in the Guanylyltransferase assay) to obtain GpppN32 RNA. To obtain m7GpppN32 RNA from GpppN32 RNA, GpppN32 RNA was incubated with TgCmt protein in a reaction mixture containing capping buffer for 30 °C for 3 h. The (VCE, NEB) was used in the reaction to convert pppN32 RNA to m7GpppN32 RNA. The obtained RNA samples were heated at 95 °C for 2 min and purified using an RNA cleanup and concentrator kit (Zymo).
Luciferase expression assay
For reporter RNA transfection, the firefly luciferase gene (FLuc) was amplified (from the pmirGLO plasmid, Promega) by PCR using the forward primer (Supplementary Table 1) containing the T3 promoter sequence and the starting sequence from the luciferase gene. The obtained DNA was gel-purified and subjected to IVT using the MEGAScript T3 Transcription Kit (Ambion). The reaction was incubated at 37 °C for 3 h, treated with DNaseI at 37 °C for 1 h, and IVT luciferase RNA was purified using the RNA cleanup kit. The luciferase RNA (ppp-FLuc RNA) was treated sequentially with TgCet, TgCeg, and TgCmt proteins or VCE to obtain m7G-FLuc RNA. The Poly(A) tailing of ppp-Fluc RNA-[A]n or m7G-FLuc RNA-[A]n was performed using E. coli Poly(A) Polymerase (NEB# M0276). Around 20 µg of each RNA variant was used to transfect 107 RH parasites using the Gene Pulser Xcell Total System (Biorad, #1652660). Transfected parasites were immediately transferred to a new flask containing a confluent HFF monolayer. Parasites were harvested 18 h post-infection, and luciferase activity was determined using the Luciferase Reporter Assay System (Promega, # E1910).
For reporter DNA transfection, FLuc gene was amplified (Supplementary Table 1) from the pmirGLO plasmid and cloned between BglII and Nde1 restriction sites in plasmid pTUB1:YFP-mAID-3HA33, replacing YFP coding sequence. Around 10 µg of reporter plasmid construct was used to transfect 107 TgCet-mAID-HA parasites. Transfected parasites were immediately transferred to a new flask containing a confluent HFF monolayer and incubated for 24 h before supplementing with vehicle or IAA and then harvested at different time points as indicated. The luciferase activity was determined as mentioned above. Plasmid, pTUB1:YFP-mAID-3HA, DHFR-TS:HXGPRT was a gift from David Sibley (Addgene plasmid # 87259; http://n2t.net/addgene:87259; RRID: Addgene_87259).
Microscale thermophoresis (MST) assay
MST assays were performed with a Monolith NT.115 (Nanotemper). His-tagged TgeIF4E protein was labeled with RED-MALEIMIDE 2nd generation dye (#MO L014) according to the protocol provided by Nanotemper. Dilutions of the RNA variants (pppN32, GpppN32, and m7GpppN32) in a range of 0.18 nM to 600 nM were prepared in a buffer containing 50 mM HEPES, pH 7.2, 1 mM EDTA, 1 mM DTT, and 100 mM KCl. The reactions were incubated at RT for 15 min, followed by the loading of reaction mixtures into glass capillaries (Nanotemper). The fluorescence intensity for each reaction was measured by keeping 40% infrared laser power and 60% light-emitting diode. The fluorescence values were analyzed using the Affinity Analysis software version 2.3 (Nano Temper) to determine the binding affinity (dissociation constant: KD) between TgeIF4E and the RNA variants used.
Generation of auxin-inducible TgCet-mAID-HA transgenic parasites
TgCet-mAID-HA transgenic parasites were generated by CRISPR/Cas9-mediated site-specific gene editing using the T. gondii line RH TIR1-3FLAG82 (Supplementary Table 3). A CRIPSR/Cas9 plasmid with a specific guide RNA (gRNA) targeting the 3′ end of Cet was generated from the pSAG1::Cas9-U6::sgUPRT plasmid83 by a Q5 Hot Start site-directed mutagenesis kit (NEB) and primers (Supplementary Table 1) to induce double-strand DNA breaks and direct the insertion of the PCR fragment. The PCR fragment containing the mAID-3HA tag and the HXGPRT selection was amplified from the vector pTUB1:YFP-mAID-3HA using the primers (Supplementary Table 1) with 40 bp of homology with the 3′ end of Cet and Q5 polymerase (NEB) to facilitate direct insertion of the PCR fragment and double homologous recombination. Ten μg each of TgCet-mAID-HA amplicon and pSAG1::Cas9-U6::sgRT were transfected into 107 T. gondii RH TIR1-3FLAG parasites by electroporation using the Gene Pulser Xcell Total System (Biorad, #1652660). Transfected parasites were drug-selected using mycophenolic acid (25 μg/ml) and xanthene (50 μg/ml) for two growth cycles before cloning out by serial dilution. Endogenous tagging of TgCet-mAID-HA was verified using sequencing, diagnostic PCR, immunoblotting, and IF staining. The auxin-inducible degradation of TgCet-mAID-HA was tested by culturing these parasites in a medium containing 500 μM indole-3-acetic acid (IAA) (Sigma, I2886), followed by immunoblotting and IF staining using α-HA antibody. Plasmids pSAG1::CAS9-U6::sgUPRT was a gift from David Sibley (Addgene plasmid # 54467; http://n2t.net/addgene:54467; RRID: Addgene_54467).
Puromycin incorporation assay
Nascent protein synthesis levels in TgCet parasites were assessed using puromycin labeling84. TgCet-mAID-HA parasites were grown on HFF monolayers in the presence of a vehicle or IAA for the specified time points. Cells were treated with 10 µg/ml puromycin for 30 min prior to paraformaldehyde fixation, followed by IFA. Complete inhibition of protein synthesis in the control was achieved by treating the cells with 100 µg/ml cycloheximide (CHX) for 8 h before adding puromycin and followed by IFA. An anti-puromycin antibody (Sigma #ZMS1016) was used to stain the newly synthesized proteins, and an anti-HA antibody was used to visualize the TgCet-mAID-HA protein.
Trypan blue staining
Parasite viability was assessed using the standard trypan blue exclusion method. Specifically, 20 μL of 0.4% trypan blue solution was mixed with an equal volume of parasite suspension and incubated for 3 min. Approximately 150 parasites were counted using a Neubauer chamber under a 40× objective lens of a light microscope. Parasite viability was calculated as the ratio of live tachyzoites to the total number of tachyzoites and was expressed as the mean viability at indicated time points.
Plaque assay
HFFs were grown in 6-well plates until confluent and subsequently infected with 100 parasites per well. After 24 h, the medium was removed and the wells were treated with 500 μM IAA or an equivalent volume of MeOH vehicle. Plates remained undisturbed for 5 days before the infected HFFs were fixed with 100% ice-cold MeOH for 20 min and stained with a 1% crystal violet solution for 20 min. Plaques (zone of lysis: white) could be visualized against intact cells (purple). The plaque areas were quantified using ImageJ. Three independent experiments were performed with similar results. For the rescue experiment, a similar plaque assay procedure was followed; however, the IAA-containing medium was replaced with normal parasite medium at the indicated time points.
Replication assay
HFFs were grown on glass coverslips in a 6-well plate until confluent and subsequently infected with 104 parasites per well. After 12 h, the medium was removed, and the wells were treated with 500 μM IAA or an equivalent volume of MeOH vehicle. 18 h later, infected HFFs were fixed, permeabilized, stained with α-TgIMC1 (1:2000) and DAPI, and subjected to IF staining. The number of parasites per vacuole was determined by counting the parasites from 50 random vacuoles. A total of three independent replicates were performed. A similar procedure was followed to count the morphologically defective parasites.
Invasion assay
Filter-purified 103 TgCet-mAID-HA parasites were incubated with a medium containing IAA or MeOH at 37 °C for 30 min. Subsequently, parasites were allowed to infect HFFs grown on glass coverslips for 30 min at 37 °C before fixation with 4% paraformaldehyde. Standard non-permeabilizing IFA was performed by staining extracellular parasites with rabbit α-TgIMC1 (1:2000) antibody, followed by permeabilization with PBS/Triton and staining with mouse α-TgIMC1 (1:2000). HFFs were stained with AF-conjugated secondary antibodies (anti-mouse IgG AF 488 and anti-rabbit IgG AF 568) and DAPI. Parasites (green/red) were counted from fifty random fields. The relative efficiency of attachment and invasion of IAA-treated parasites was expressed as a mean percentage of the control treatment from three independent experiments.
Egress assay
Filter-purified TgCet-mAID-HA parasites were inoculated on HFF monolayers grown on glass coverslips. After 30 h, the medium was replaced with DMEM containing IAA or MeOH, and cells were further incubated for 8 h. Parasite egress was initiated by adding 3 μM calcium ionophore A23187 to infected HFFs for 2 min. The infected HFFs were fixed, permeabilized, and stained with α-TgGRA2 (1:1000) and α-TgIMC1 (1:2000). A total of 150 vacuoles from each experiment (n = 3) were examined to count the intact or collapsed vacuoles. Vacuoles containing > 2 parasites were considered intact.
CAP sequencing
TgCet-mAID-HA parasites (5 × 107) were harvested from infected HFFs cultured in DMEM supplemented with either IAA or MeOH vehicle for 8 h. Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen) and subjected to a series of enzymatic treatments followed by further purification. Specifically, 5 μg of RNA was first incubated with RppH (NEB) in NEB Buffer 2 at 37 °C for 1 h, followed by treatment with Xrn1 (NEB) in NEB Buffer 3 at 37 °C for 1 h. These treatments were designed to enrich 5′m7G and 5′OH RNA species while depleting ribosomal RNA (rRNA). Subsequently, RNA was further processed by incubating with RppH (NEB) in Thermopol Buffer at 37 °C for 1 h to convert m7G-RNA to 5′ phosphate (p-RNA). Conversion to p-RNA was confirmed by dot blotting with an α-m7G antibody. The quality of the recovered RNA was assessed using the TapeStation 4150, with RNA Integrity Number (RIN) values below 2 indicating effective rRNA depletion. RNA quantification (Supplementary data 2) was performed using a Qubit fluorometer (Thermo Fisher, #Q33238). The cDNA libraries were constructed using the TruSeq Kit (Illumina, Catalog #20020597) and sequenced as paired-end reads on the Illumina NovaSeq 6000 platform. The quality of raw FASTQ reads for the sample was initially assessed using FastQC version 0.11.9 with default parameters. Preprocessing of the raw reads was performed with Fastp version 0.20.1. Following preprocessing, the reads were reassessed for quality using FastQC85,86, and results were summarized using MultiQC87. The processed reads were aligned against the T. gondii reference genome (GCF_000006565.2) using STAR v2.7.988. The rRNA and tRNA features were removed from the GTF file. For transcript quantification, the T. gondii genome was reindexed using RSEM version 1.3.3.489. The STAR-aligned BAM files were then utilized for quantification with RSEM’s rsem-calculate-expression command, specifying the parameters --paired-end and --no-bam-output. The resulting transcript abundance estimates were imported into R using the Bioconductor package “tximport”. QualiMap42 was used to determine the annotated transcript’s depth of coverage based on the alignment files (BAM) and GTF annotation files. “regularized log” transformation in DESeq290 (Supplementary data 2) was used for principal component and clustering analysis. Normalization and differential expression analysis were performed using DESeq2. An adjusted p value (FDR) threshold ≤ 0.05 and a log2 fold change cutoff of ±1.0 were used for statistical estimation of gene expression. Differential expression results were visualized with Volcano plots generated using EnhancedVolcano91 and MA plots created with the ggmaplot function from the ggpubr R package92.
For Gene Ontology (GO) annotation, the protein sequences were obtained based on the DESeq2 result and were subjected to the ToxoDB database28 to generate GO terms. A threshold of p value 0.05 was set to define statistically significant GO terms, which were used to generate GO plots.
RNA Isolation and quantitative RT-PCR (RT-qPCR)
Total RNA was extracted from the samples using the RNeasy Mini Kit (Qiagen), which includes gDNA Eliminator spin columns. RNA concentration and quality were assessed using a Nanodrop and agarose gel electrophoresis (for 28S and 18S RNA). The cDNA synthesis was performed using SuperScript III First-strand Synthesis System (Invitrogen). Briefly, 1 µg RNA was added to a reaction mix containing random hexamers, dNTP mix, SuperScript III reverse transcriptase (Invitrogen) in the RT buffer and subjected to thermocycling program: 10 min at 25 °C, 50 min at 50 °C, 5 min at 85 °C. Reverse transcriptase minus (RT-) negative control was performed to rule out genomic DNA contamination of the RNA sample. No RNA template negative control (NTC) was also used to check for reagent contamination. The RT-qPCR was performed on the Bio-Rad CFX96 Real-Time PCR System using cDNA, gene-specific primers (Supplementary Table 1) with SYBR green PCR Master Mix (Applied Biosystems). All RT-qPCR reactions for candidate genes yielded a single product of 140 bp, whereas 18 s rRNA gave an 88 bp DNA product (Supplementary Fig. 11B). PCR efficiency was checked by diluting the original cDNA in a ratio of 1 to 10 to generate a 6 series of dilutions ranging from 10 ng to 100 fg (Supplementary Fig. 11C). The Log (cDNA in ng/reaction) was plotted against averaged Ct values from two technical replicates. The standard curve equation y = Ax + B and R2 was obtained in a GraphPad Prism. The PCR efficiency (E, %) was calculated as E = (10−1/A − 1) × 100. qPCR reactions were performed using 10 ng cDNA with the following conditions: 2 min at 50 °C, 10 min at 95 °C, and 40 cycles of 10 s at 95 °C and 30 s at 60 °C. A melting curve analysis was performed starting at 65 °C–95 °C. For each run, 2 NTC reactions were used to rule out any reagent contamination (Supplementary data 1). The expression data were normalized to the non-capped 18S rRNA gene, and relative transcript levels were calculated using the 2−ΔΔCT method93.
Mouse infection
The auxin-inducible degradation of protein in mice was performed as described previously in ref. 94 with some modifications. To test TgCet depletion in vivo, 6-week BALB/c male mice (n = 4) were injected intraperitoneally (i.p.) with 50 tachyzoites of RH TgCet-mAID-HA (2 groups). From the second day of post-infection (pi), of the two groups, one group was given IAA, and the other group was administered an equivalent volume of MeOH vehicle in the drinking water containing 5% sucrose. The IAA was administered in two ways: i) in drinking water (0.5 mg/ml) and ii) by oral gavage (12.5 mg/mL). The total treatment period was 4 days (2 days pi to 5 days pi). On days 5 and 6 pi, 2 mice each were sacrificed by CO2 asphyxiation, and the peritoneal exudate cells (PECs) were collected and examined by IF staining using α-HA and α-TgIMC1 antibodies to test the TgCet protein depletion. After successful depletion of TgCet-mAID-HA in IAA-administered mice, a long-term (30-day) survival experiment was performed by following a similar experimental procedure. In the 30-day survival experiment, each group had 10 mice (2 groups RH TgCet-mAID-HA). Mice from two groups were injected with 50 tachyzoites of RH TgCet-mAID-HA. On the second day of pi, one of two groups was given IAA, and the other group was administered an equivalent volume of MeOH vehicle in the drinking water containing 5% sucrose. The third group had no infection control with IAA. Mice were weighed and monitored daily; PECs were collected on days 6 and 12; IAA treatment was given for 15 days in the surviving mice; and the surviving mice were sacrificed on day 30. Relative weight loss was calculated based on the initial body weight on the day of infection. The overall experimental setup and procedures used are indicated within the relevant figures.
Structure predictions
The alphafold structure46 of TgCet (S8F0G9) was obtained from ToxoDB. The disordered region was manually removed, and amino acid sequence corresponding to 361–729aa, which consists of 10β barrel tunnels, was used as input sequence for structure prediction using GoogleCollab notebook45 https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFol
d.ipynb#scrollTo=rowN0bVYLe9n. The obtained structure of Cet was superimposed with PDB structures of RNA triphosphatase of Saccharomyces cerevisiae (1D8H)23, Trypanosoma cruzi (6L7W)28 and thiamine triphosphatase93 of Homo sapiens (3VTL) using PyMol (v2.x).
Yeast complementation
For complementation assays, S. cerevisiae Δcet1, Δceg1, Δabd1, and three genes chromosomal copy deletion mutant strains (Supplementary Table 2) carrying the wild-type copy in a plasmid with URA marker were utilized50,51. Full-length T. gondii cet, ceg, and C-terminus of cmt genes were cloned (Supplementary Table 1) into pYES3/CT vector between KpnI-EcoRI, BamHI-XhoI, and KpnI-EcoRI, respectively. The cet, ceg, and abd yeast mutant strains were transformed with respective plasmid carrying respective T. gondii gene or empty plasmid. Triple deletant mutant strain was transformed with plasmids carrying all three T. gondii genes or empty vector. Transformants were selected on SD–Trp plates with or without 5-Fluoroorotic Acid (5-FOA). Plasmid DNA was isolated from FOA-resistant triple-deletant complemented strain using an Easy Yeast Plasmid Isolation Kit (Clontech), and gene-specific PCR was performed to validate the expression of TgCEs (Supplementary Fig. 4).
TgCet inhibition assay
The enzyme inhibition assay for TgCet was performed using an ATPase assay activity kit (Sigma #MAK113). For determination of IC50 values, 100 ng TgCet protein was incubated in a 10-fold dilution (50 µM to 0.02 µM) of Myricetin (Sigma # 476275) or 3,4-Dicaffeoylquinic acid (3,4-diCQA) (Sigma #SMB00131) in RNA triphosphatase buffer containing 5 mM ATP at 37 °C for 15 min. The reaction was stopped using a malachite green stop solution, and incubated at RT for 5 min, and the absorbance was measured at 620 nm. Enzyme activity was calculated as per the kit manual. All experiments were repeated three times with internal duplicates.
Tachyzoite lytic assay
Parasiticidal effect of Myricetin and 3,4-diCQA was assessed using a standard plaque assay in 24-well plate. HFFs were infected with freshly harvested RH parasites (105/well), and after 12 h of incubation, the medium was removed, and the wells were treated with 10-fold dilutions (50 µM to 0.02 µM) of Myricetin and 3,4-diCQA (prepared in 300 μl DMEM). Plates were incubated for 72 h at 37 °C, followed by crystal violet staining to determine the plaques. EC50 values were calculated based on a number of plaques, considering the mean of three biological replicates, with each replicate containing two technical replicates. Both the host cell (HFFs) toxicity of Myricetin and EC50 value estimation were performed using CellTiter 96 AQueous One Solution Reagent (Promega #G3582) as per manufacturer’s instructions.
In vitro bradyzoite growth assays
To monitor the effects of Myricetin and 3,4-diCQA on bradyzoite growth, 3-day-old intracellular ME49 bradyzoites were treated with 10-fold dilutions (50 µM to 0.02 µM) of either compound and incubated for 2 days in the bradyzoite growth medium without CO2 at 37 °C. Subsequently, the infected monolayers were subjected to IFA using bradyzoite stage-specific antibodies TgCST1 (1:200) and TgBAG1 (1:200). Cyst size was determined by measuring the area in microns. To test the viability of Myricetin-treated bradyzoites, infected HFFs were scraped and passed through 23 g needle to liberate the cyst, followed by trypsin treatment (0.25 mg/ml) for 10 min to release bradyzoites. Harvested bradyzoites were inoculated onto confluent HFF monolayers and cultured for 10 days, followed by crystal violet staining to determine the plaques.
In vivo efficacy of Myricetin
To test the effect of Myricetin on lethal toxoplasmosis, six-week-old BALB/c male mice (n = 10) were taken in four different groups. Mice from three groups were intraperitoneally injected with 100 tachyzoites of RH. From 2nd until 12th day of pi, one group was orally administered Myricetin at 100 mg/Kg body weight52,53, whereas the second and third groups were orally given pyrimethamine (50 mg/Kg body weight)54 and 1% DMSO, respectively. The fourth group served as no-infection Myricetin control. All mice were weighed and monitored daily for 40 days. PECs were collected from dead mice and examined by IF staining using α-TgIMC1 antibodies. All survived mice were sacrificed on day 40, and PECs were collected and examined for the presence of tachyzoites by IFA.
Data analyses
All data analyses, including graph preparation and statistics, were performed using GraphPad Prism 9. Immunoblot quantification was performed using ImageJ (Version 6).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The Cap sequencing data for the TgCet-mAID-3HA transgenic parasite line have been deposited in NCBI https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1064470 (control - SRR27549198 and treatment -SRR27560546). Source data are provided with this paper.
References
Tenter, A. M., Heckeroth, A. R. & Weiss, L. M. Toxoplasma gondii: from animals to humans. Int J. Parasitol. 30, 1217–1258 (2000).
Montoya, J. G. & Liesenfeld, O. Toxoplasmosis. Lancet 363, 1965–1976 (2004).
Cerutti, A., Blanchard, N., Besteiro, S. The bradyzoite: a key developmental stage for the persistence and pathogenesis of toxoplasmosis. Pathogens 9, 234 (2020).
Montazeri, M. et al. Drug Resistance in Toxoplasma gondii. Front. Microbiol. 9, 2587 (2018).
Radke, J. R. et al. The transcriptome of Toxoplasma gondii. BMC Biol. 3, 26 (2005).
Holmes, M. J., Augusto, L. D. S., Zhang, M., Wek, R. C. & Sullivan, W. J. Jr. Translational control in the latency of apicomplexan parasites. Trends Parasitol. 33, 947–960 (2017).
Furuichi, Y. Discovery of m(7)G-cap in eukaryotic mRNAs. Proc. Jpn Acad. Ser. B Phys. Biol. Sci. 91, 394–409 (2015).
Ramanathan, A., Robb, G. B. & Chan, S. H. mRNA capping: biological functions and applications. Nucleic Acids Res. 44, 7511–7526 (2016).
Shuman, S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 66, 1–40 (2001).
Shuman, S. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 3, 619–625 (2002).
Shuman, S. RNA capping: progress and prospects. RNA 21, 735–737 (2015).
Ho, C. K. & Shuman, S. A yeast-like mRNA capping apparatus in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 98, 3050–3055 (2001).
Hausmann, S., Vivares, C. P. & Shuman, S. Characterization of the mRNA capping apparatus of the microsporidian parasite Encephalitozoon cuniculi. J. Biol. Chem. 277, 96–103 (2002).
Hausmann, S. et al. Yeast-like mRNA capping apparatus in Giardia lamblia. J. Biol. Chem. 280, 12077–12086 (2005).
Simoes-Barbosa, A., Hirt, R. P. & Johnson, P. J. A metazoan/plant-like capping enzyme and cap modified nucleotides in the unicellular eukaryote Trichomonas vaginalis. PLoS Pathog. 6, e1000999 (2010).
Ho, C. K. & Shuman, S. Trypanosoma brucei RNA triphosphatase. Antiprotozoal drug target and guide to eukaryotic phylogeny. J. Biol. Chem. 276, 46182–46186 (2001).
Smith, P., Ho, C. K., Takagi, Y., Djaballah, H. & Shuman, S. Nanomolar Inhibitors of Trypanosoma brucei RNA Triphosphatase. mBio 7, e00058–00016 (2016).
Takagi, T., Moore, C. R., Diehn, F. & Buratowski, S. An RNA 5’-triphosphatase related to the protein tyrosine phosphatases. Cell 89, 867–873 (1997).
Ho, C. K., Schwer, B. & Shuman, S. Genetic, physical, and functional interactions between the triphosphatase and guanylyltransferase components of the yeast mRNA capping apparatus. Mol. Cell Biol. 18, 5189–5198 (1998).
Yamada-Okabe, T. et al. Isolation and characterization of the Candida albicans gene for mRNA 5’-triphosphatase: association of mRNA 5’-triphosphatase and mRNA 5’-guanylyltransferase activities is essential for the function of mRNA 5’-capping enzyme in vivo. FEBS Lett. 435, 49–54 (1998).
Wen, Y., Yue, Z. & Shatkin, A. J. Mammalian capping enzyme binds RNA and uses protein tyrosine phosphatase mechanism. Proc. Natl. Acad. Sci. USA 95, 12226–12231 (1998).
Changela, A., Ho, C. K., Martins, A., Shuman, S. & Mondragon, A. Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme. EMBO J. 20, 2575–2586 (2001).
Lima, C. D., Wang, L. K. & Shuman, S. Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell 99, 533–543 (1999).
Schwer, B., Lehman, K., Saha, N. & Shuman, S. Characterization of the mRNA capping apparatus of Candida albicans. J. Biol. Chem. 276, 1857–1864 (2001).
Shuman, S. The mRNA capping apparatus as drug target and guide to eukaryotic phylogeny. Cold Spring Harb. Symp. Quant. Biol. 66, 301–312 (2001).
Pei, Y., Schwer, B., Saiz, J., Fisher, R. P. & Shuman, S. RNA triphosphatase is essential in Schizosaccharomyces pombe and Candida albicans. BMC Microbiol. 1, 29 (2001).
Monteiro, M. C. & De Lucas, J. R. Study of the essentiality of the Aspergillus fumigatus triA gene, encoding RNA triphosphatase, using the heterokaryon rescue technique and the conditional gene expression driven by the alcA and niiA promoters. Fungal Genet. Biol. 47, 66–79 (2010).
Kissinger, J. C., Gajria, B., Li, L., Paulsen, I. T. & Roos, D. S. ToxoDB: accessing the Toxoplasma gondii genome. Nucleic Acids Res. 31, 234–236 (2003).
Takase, Y., Takagi, T., Komarnitsky, P. B. & Buratowski, S. The essential interaction between yeast mRNA capping enzyme subunits is not required for triphosphatase function in vivo. Mol. Cell Biol. 20, 9307–9316 (2000).
Gu, M., Rajashankar, K. R. & Lima, C. D. Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA capping apparatus. Structure 18, 216–227 (2010).
Chu, C. et al. Structure of the guanylyltransferase domain of human mRNA capping enzyme. Proc. Natl. Acad. Sci. USA 108, 10104–10108 (2011).
Wang, S. P., Deng, L., Ho, C. K. & Shuman, S. Phylogeny of mRNA capping enzymes. Proc. Natl. Acad. Sci. USA 94, 9573–9578 (1997).
Cong, P. & Shuman, S. Covalent catalysis in nucleotidyl transfer. A KTDG motif essential for enzyme-GMP complex formation by mRNA capping enzyme is conserved at the active sites of RNA and DNA ligases. J. Biol. Chem. 268, 7256–7260 (1993).
Shuman, S. & Schwer, B. RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol. Microbiol. 17, 405–410 (1995).
Saha, N., Schwer, B. & Shuman, S. Characterization of human, Schizosaccharomyces pombe, and Candida albicans mRNA cap methyltransferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J. Biol. Chem. 274, 16553–16562 (1999).
Zheng, S. et al. Mutational analysis of Encephalitozoon cuniculi mRNA cap (guanine-N7) methyltransferase, structure of the enzyme bound to sinefungin, and evidence that cap methyltransferase is the target of sinefungin’s antifungal activity. J. Biol. Chem. 281, 35904–35913 (2006).
Bhattacharya, A. et al Genomewide Analysis of Mode of Action of the S-Adenosylmethionine Analogue Sinefungin in Leishmania infantum. mSystems 4, e00416–19 (2019).
Yanagiya, A. et al. Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol. Cell. 46, 847–858 (2012).
Haimov, O. et al. Dynamic interaction of eukaryotic initiation factor 4G1 (eIF4G1) with eIF4E and eIF1 underlies scanning-dependent and -independent translation. Mol. Cell Biol. 38, e00139–18 (2018).
Tuteja, R. Identification and bioinformatics characterization of translation initiation complex eIF4F components and poly(A)-binding protein from Plasmodium falciparum. Commun. Integr. Biol. 2, 245–260 (2009).
Tomoo, K. et al. Structural basis for mRNA Cap-Binding regulation of eukaryotic initiation factor 4E by 4E-binding protein, studied by spectroscopic, X-ray crystal structural, and molecular dynamics simulation methods. Biochim. Biophys. Acta 1753, 191–208 (2005).
Okonechnikov, K., Conesa, A. & García-Alcalde, F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32, 292–294 (2016).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Varadi, M. et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Takagi, Y., Kuwabara, N., Dang, T. T., Furukawa, K. & Ho, C. K. Crystal structures of the RNA triphosphatase from Trypanosoma cruzi provide insights into how it recognizes the 5’-end of the RNA substrate. J. Biol. Chem. 295, 9076–9086 (2020).
Song, J., Bettendorff, L., Tonelli, M. & Markley, J. L. Structural basis for the catalytic mechanism of mammalian 25-kDa thiamine triphosphatase. J. Biol. Chem. 283, 10939–10948 (2008).
Bettendorff, L. & Wins, P. Thiamine triphosphatase and the CYTH superfamily of proteins. FEBS J. 280, 6443–6455 (2013).
Delvaux, D. et al. Structural determinants of specificity and catalytic mechanism in mammalian 25-kDa thiamine triphosphatase. Biochim. Biophys. Acta 1830, 4513–4523 (2013).
Zhao, Z. et al. Myricetin relieves the symptoms of type 2 diabetes mice and regulates intestinal microflora. Biomed. Pharmacother. 153, 113530 (2022).
Yao, Q., Li, S., Li, X., Wang, F. & Tu, C. Myricetin modulates macrophage polarization and mitigates liver inflammation and fibrosis in a murine model of nonalcoholic steatohepatitis. Front. Med. 7, 71 (2020).
Aguirre-Cruz, L., Velasco, O. & Sotelo, J. Nifurtimox plus pyrimethamine for treatment of murine toxoplasmosis. J. Parasitol. 84, 1032–1033 (1998).
Galloway, A. & Cowling, V. H. mRNA cap regulation in mammalian cell function and fate. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 270–279 (2019).
Hsin, J. P. & Manley, J. L. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 26, 2119–2137 (2012).
Schneider, S., Pei, Y., Shuman, S. & Schwer, B. Separable functions of the fission yeast Spt5 carboxyl-terminal domain (CTD) in capping enzyme binding and transcription elongation overlap with those of the RNA polymerase II CTD. Mol. Cell Biol. 30, 2353–2364 (2010).
Deshmukh, A. S., Mitra, P. & Maruthi, M. Cdk7 mediates RPB1-driven mRNA synthesis in Toxoplasma gondii. Sci. Rep. 6, 35288 (2016).
Deshmukh, A. S., Mitra, P., Kolagani, A. & Gurupwar, R. Cdk-related kinase 9 regulates RNA polymerase II-mediated transcription in Toxoplasma gondii. Biochim. Biophys. Acta Gene Regul. Mech. 1861, 572–585 (2018).
Varshney, D. et al. mRNA Cap methyltransferase, RNMT-RAM, promotes RNA Pol II-dependent transcription. Cell Rep. 23, 1530–1542 (2018).
Wang, S. P. & Shuman, S. Structure-function analysis of the mRNA cap methyltransferase of Saccharomyces cerevisiae. J. Biol. Chem. 272, 14683–14689 (1997).
Bueren-Calabuig, J. A., GB, M., Cowling, V. H. & Pisliakov, A. V. Mechanism of allosteric activation of human mRNA cap methyltransferase (RNMT) by RAM: insights from accelerated molecular dynamics simulations. Nucleic Acids Res. 47, 8675–8692 (2019).
Muniz, L., Nicolas, E. & Trouche, D. RNA polymerase II speed: a key player in controlling and adapting transcriptome composition. EMBO J. 40, e105740 (2021).
Painter, H. J. et al. Genome-wide real-time in vivo transcriptional dynamics during Plasmodium falciparum blood-stage development. Nat. Commun. 9, 2656 (2018).
Ignatochkina, A. V., Iguchi, J. A., Kore, A. R. & Ho, C. K. Trypanosome mRNA recapping is triggered by hypermethylation originating from cap 4. Nucleic Acids Res. 52, 10645–10653 (2024).
Chu, C. Y. & Rana, T. M. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 4, e210 (2006).
Decker, C. J., Teixeira, D. & Parker, R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179, 437–449 (2007).
Davila Lopez, M. & Samuelsson, T. Early evolution of histone mRNA 3’ end processing. RNA 14, 1–10 (2008).
Chen, D. et al. Polyadenylation of histone H3.1 mRNA promotes cell transformation by displacing H3.3 from gene regulatory elements. iScience 23, 101518 (2020).
Zanin, O. et al. Ceg1 depletion reveals mechanisms governing degradation of non-capped RNAs in Saccharomyces cerevisiae. Commun. Biol. 6, 1112 (2023).
Blume, M. et al. A Toxoplasma gondii gluconeogenic enzyme contributes to robust central carbon metabolism and is essential for replication and virulence. Cell Host Microbe 18, 210–220 (2015).
Nitzsche, R., Zagoriy, V., Lucius, R. & Gupta, N. Metabolic cooperation of glucose and glutamine is essential for the lytic cycle of obligate intracellular parasite Toxoplasma gondii. J. Biol. Chem. 291, 126–141 (2016).
Arraiano, C. M. et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 34, 883–923 (2010).
Gong, C., Smith, P. & Shuman, S. Structure-function analysis of Plasmodium RNA triphosphatase and description of a triphosphate tunnel metalloenzyme superfamily that includes Cet1-like RNA triphosphatases and CYTH proteins. RNA 12, 1468–1474 (2006).
Bisaillon, M. & Shuman, S. Structure-function analysis of the active site tunnel of yeast RNA triphosphatase. J. Biol. Chem. 276, 17261–17266 (2001).
Igarashi, M. et al. Kribellosides, novel RNA 5’-triphosphatase inhibitors from the rare actinomycete Kribbella sp. MI481-42F6. J. Antibiot. 70, 582–589 (2017).
Bak, M. J., Das Gupta, S., Wahler, J. & Suh, N. Role of dietary bioactive natural products in estrogen receptor-positive breast cancer. Semin. Cancer Biol. 40-41, 170–191 (2016).
Li, J., Xiang, H., Huang, C. & Lu, J. Pharmacological actions of myricetin in the nervous system: a comprehensive review of preclinical studies in animals and cell models. Front. Pharmacol. 12, 797298 (2021).
Sugi, T. et al. Toxoplasma gondii cyclic AMP-dependent protein kinase subunit 3 is involved in the switch from tachyzoite to bradyzoite development. mBio 7, e00755–16 (2016).
Mitra, P., Deshmukh, A. S., Gurupwar, R. & Kashyap, P. Characterization of Toxoplasma gondii Spt5 like transcription elongation factor. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 184–197 (2019).
Deshmukh, A. S. et al. Toxoplasma gondii induces robust humoral immune response against cyst wall antigens in chronically infected animals and humans. Micro. Pathog. 152, 104643 (2021).
Mitra, P., Deshmukh, A. S., Banerjee, S., Khandavalli, C. & Choudhury, C. A functionally divergent transcription elongation factor 1-like protein in Toxoplasma gondii. FEBS Lett. 596, 112–127 (2022).
Trotman, J. B., Schoenberg, D. R. RNA cap methyltransferase activity assay. Bio. Protoc. 8, e2767 (2018).
Brown, K. M., Long, S., Sibley, L. D. Plasma membrane association by N-acylation governs PKG function in Toxoplasma gondii. mBio 8, e00375–17 (2017).
Shen, B., Brown, K. M., Lee, T. D. & Sibley, L. D. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. mBio 5, e01114 (2014).
Holmes, M. J. et al. mRNA cap-binding protein eIF4E1 is a novel regulator of Toxoplasma gondii latency. mBio 15, e0295423 (2024).
Zhang, F., & Kang, H. M. FASTQuick: rapid and comprehensive quality assessment of raw sequence reads. Gigascience 10, giab004 (2021).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Ewels, P., Magnusson, M., Lundin, S. & Kaller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 12, 323 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Ge, S. X., Son, E. W. & Yao, R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinforma. 19, 534 (2018).
Huerta-Cepas, J. et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122 (2017).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2^(-ΔΔCt) method. Methods 25, 402–408 (2001).
Brown, K. M. & Sibley, L. D. Essential cGMP signaling in Toxoplasma is initiated by a hybrid P-Type ATPase-guanylate cyclase. Cell Host Microbe 24, 804–816.e806 (2018).
Acknowledgements
This work is funded by the NIAB core grant (C0022) and DST-SERB (CRG/2023/004341/BHS) to ASD. KA and PK acknowledge DBT and UGC, respectively, for graduate studies fellowships provided by the Government of India. We thank Prof. Stewart Shuman, Sloan Kettering Institute, USA, for yeast mutant strains. We thank NIH Biodefense and Emerging Infections Research Sources Repository, NIAID, NIH for the following parasite strains: Toxoplasma gondii, RH-88, NR-223; Toxoplasma gondii, ME49 (B7 Clone), NR-20729; Toxoplasma gondii, RH-TIR1-3FLAG, NR-51145. We thank Dr. David Sibley for the plasmids pSAG1::CAS9-U6::sgUPRT (Addgene, #54467) and pTUB1:YFP-mAID-3HA, DHFR-TS:HXGPRT (Addgene # 87259).
Author information
Authors and Affiliations
Contributions
Kalyani R. Aswale: Data curation, investigation, analysis, and writing manuscript. Poonam Kashyap: Data curation and analysis. Abhijit S. Deshmukh: Conceptualization, funding acquisition, investigation, and writing manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks C. Kiong Ho and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Aswale, K.R., Kashyap, P. & Deshmukh, A.S. RNA triphosphatase-mediated mRNA capping is essential for maintaining transcript homeostasis and the survival of Toxoplasma gondii. Nat Commun 16, 5452 (2025). https://doi.org/10.1038/s41467-025-59867-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59867-z