Abstract
Huntington’s disease and other disorders of the basal ganglia create challenges for biomolecule-based medicines given the poor accessibility of these deep brain structures following intracerebral or intravascular delivery. Here, we found that low dose, low volume delivery of unbiased AAV libraries into the globus pallidus allowed recovery of novel capsids capable of broad access to key deep brain and cortical structures relevant for human therapies. One such capsid, AAV-DB-3, provided transduction of up to 45% of medium spiny neurons in the adult NHP striatum, along with substantial transduction of relevant deep layer neurons in the cortex. Notably, AAV-DB-3 behaved similarly in mice as in NHPs and potently transduced human neurons derived from induced pluripotent stem cells. Thus, AAV-DB-3 provides a unique AAV for network level brain gene therapies that translates up and down the evolutionary scale for preclinical studies and eventual clinical use.
Similar content being viewed by others
Introduction
Recombinant adeno-associated viruses (rAAVs) are a promising therapeutic modality to treat neurodegenerative diseases for their high safety profile, ability to infect non-dividing cells, and to facilitate long-term transgene expression. Naturally occurring serotypes are effective at targeting different cells of the nervous system with variable tropism determined by capsid identity and route of administration1,2. Broad transduction of the central nervous system (CNS) can be achieved through non-invasive intravenous (IV) delivery of AAV9 and AAVrh10 and numerous modified capsid variants designed to cross the blood-brain barrier (BBB)3,4,5,6,7,8,9. However, this approach lacks specificity and has low targeting efficiency with substantial peripheral virus burden that can contribute to immune responses and liver toxicity10,11,12,13,14. Direct, intraparenchymal delivery of AAV confers higher specificity and targeting efficiency, but tissue distribution is largely limited to the site of injection, with large brain structures requiring multiple injections15,16,17 and methods to enhance delivery2,18.
Huntington’s disease (HD)19 is an autosomal dominant disorder characterized by progressive degeneration and transcriptional dysregulation in the basal ganglia with subsequent changes in cortical regions of the brain20,21,22. The basal ganglia comprise a collection of ganglia and nuclei located deep within the brain that form connections important for movement, decision making, and reward behaviors. Major functional components include the caudate nucleus, globus pallidus, putamen, substantia nigra, subthalamic nucleus and ventral pallidum. The globus pallidus (“pale globe”) represents a central figure in the basal ganglia circuitry and is divided into two functionally distinct segments, internal (GPi) and external (GPe). The GPi and GPe receive synaptic input from the striatum (caudate and putamen) and project to the thalamus and subthalamic nucleus, respectively. Together, affected regions in HD pose unique challenges for therapeutic delivery: relevant structures lie deep within the brain and span large areas that cannot be fully targeted by a single, direct delivery.
Despite these challenges, much effort has been put toward development of therapies for HD, including strategies to knockdown the mutant huntingtin (Htt) gene product or indirectly impact disease pathogenesis23,24,25,26,27. Preclinical studies in mice have shown therapeutic benefit following a single, direct injection of AAV into the striatum, however this paradigm is not suited for the larger structures of the non-human primate (NHP) or human brain with currently available capsids. A clinical trial is underway to treat HD that targets huntingtin transcripts (HTT) with an AAV5-delivered artificial miRNA at three sites in the caudate nucleus and putamen per hemisphere using convection-enhanced delivery28 (NCT04120493).
To overcome the challenges for AAV gene therapies for disorders that impact cortical and subcortical structures, such as HD21,29, we devised an unbiased approach to identify highly potent capsids capable of transducing our target brain regions following delivery to the globus pallidus. Our libraries were based on three parental AAV serotypes, processed in parallel in the same animals, and the resulting capsids did not require further optimization to improve their trafficking and targeting capabilities. Notably, our most potent capsid, AAV-DB-3, was greater than two orders of magnitude more potent in NHP brain than the AAV5 capsid being assessed clinically for HD. AAV-DB-3 also efficiently transduces human iPSC-derived neurons and retains the properties of broad distribution in mice, supporting its application to pre-clinical testing and relevant dose range finding studies important for advancing to HD patients.
Results
In vivo screen of a peptide modified AAV library
A peptide-modified AAV capsid library approach was leveraged to identify highly performant capsids variants targeting deep brain structures after low-dose infusion into the globus pallidus (GP). Three parental serotypes (AAV1, AAV2, and AAV9) were modified to create the capsid variant libraries used in this work. Each parental serotype was modified by insertion of a 7-mer amino acid sequence into the variable region of loop 8 at positions 590, 587, and 588 in AAV1, AAV2, and AAV9, respectively. The insertions are a sequence of semi-random DNA bases “NNKNNKNNKNNKNNKNNKNNK” where “N” represents DNA bases A, C, T or G and “K” represents G or T. Our AAV selection methodology consisted of several rounds of in vivo selection in adult NHPs, NHPs 1–5 (refer to Supplementary Table S1 for all NHP information), accompanied by Illumina amplicon sequencing to resolve and quantify enrichment of capsid variants (Fig. 1A). The estimated diversity of the initial screening library used in this work was ~ 6.8 million. After two rounds of in vivo target tissue enrichment screening, the total number of remaining capsid variants across brain regions of interest was 42,487, a reduction of more than two orders of magnitude from the initial screening library. To assess individual capsid variant performance, amplicon sequencing was performed. Analysis of the NGS results identified 161 top-performing capsid variants based on broad expression across various brain regions and/or selective enrichment in target regions (caudate nucleus and putamen). A final round of screening using this small library of top performing capsids was performed in both an adult rhesus macaque and African green monkey to assess cross-species performance (Fig. 1B). In the analysis of amplicon sequencing data of this final screen, capsid variants were prioritized for validation based on detection across brain regions relevant to HD, focusing on caudate and putamen (Fig. 1C). AAV RNA detection was also considered in selecting top candidates. Based on these considerations, two AAV2 variants and one AAV1 variant were selected for validation and named AAV-DB-1, AAV-DB-2, and AAV-DB-3 (Fig. 1C).
A Schematic depicting the AAV capsid selection pipeline used in this work. AAV peptide display libraries were designed to contain a semi-random heptapeptide insert in multiple parental AAV serotypes (AAV1, AAV2, and AAV9), yielding an initial ~ 6.8 million capsid variants. Enrichment and selection of capsids is achieved by sequential passages through primate brain. The first round of enrichment (black arrows) involved a single infusion of ‘Round 1’ input library composed of peptide-modified AAV capsids from three parental serotypes. The input library was injected by bilateral intraparenchymal infusion into the globus pallidus of a rhesus macaque. After two days in life, CNS regions of interest were collected. DNA was extracted from collected tissues and recovered AAV variant sequences were used to generate a new (Round 2) AAV input library. A second round of screening (gray arrows) was initiated by bilateral GP infusion of the Round 2 input library into rhesus macaque. For round 2 screening, both viral DNA and RNA were collected from tissues. Peptide modifications of successfully enriched capsids were identified using Illumina sequencing of viral amplicons. Capsid performance was assessed using a combination of total detection in target tissues and enrichment, which measures the ratio of a capsid’s relative abundance in a target region vs the input library. B 161 top-ranking capsids across all parental serotypes were identified to generate a third input vector library, hereafter referred to as the optimized pool. A final round of screening was initiated by injection of the optimized pool input vector into the GP of an adult rhesus macaque and African green monkey. Tissues collected from these primates became the basis for DNA- and RNA-based amplicon sequencing. C Three highly enriched capsid variants were identified based on a combination of overall detection across brain regions (AVG enrichment), and/or targeted enrichment in caudate and putamen (AVG CP enrichment), and were selected for fluorescence validation. Rd, round; NGS, next-generation sequencing; DB1, AAV-DB-1; DB2, AAV-DB-2; DB3, AAV-DB-3; Cd, caudate; Pu, putamen; STh, subthalamic nucleus; SN, substantia nigra; Th, thalamus; Cx, cortex. Panels (A and B) created in BioRender. Lewandowski, B. (2025) https://BioRender.com/y3ikqcc.
AAV-DB candidate validation in adult NHPs
The deep-brain targeting capsid variants (AAV-DB) selected for further validation were packaged with distinct reporter transgenes driven by the ubiquitous CAG promoter. AAV-DB-1, 2, and 3 vectors expressing different fluorescent payloads were mixed and injected at a dose of 7.5E+10 vg each into the GP of an adult rhesus macaque, NHP 6 (Fig. 2A). After approximately 3 weeks, the animal was euthanized, and tissue processed for histology. Expression of fluorescent reporters in the caudate, putamen and cortex was assessed using direct fluorescence or by in situ hybridization (FISH) for their respective transcripts (Fig. 2B). AAV -DB-1, 2 and 3 transduced numerous cells in the GP, putamen, and caudate, with particularly robust transgene expression as assessed by RNA FISH (Fig. 2C, panels i, iii, v) or direct fluorescence (Fig. 2C, panels ii, iv, vi) for AAV-DB-2 and DB-3. Transduction was remarkably widespread in the caudate nucleus and putamen, reaching distances greater than a centimeter from the infusion site. AAV-DB-1, DB-2, and DB-3 also transduced cells in the substantia nigra reticulata (Fig. 2C panels vii, viii).
A A mix of AAV-DB-1.mRuby3, AAV-DB-2.mNeonGreen, and AAV-DB-3.mTFP1 was infused into the GP of a rhesus macaque at 7.5E+10 vg/AAV. B Renderings of the NHP putamen and caudate52 are intersected with planes indicating the anatomical positions for direct fluorescence or FISH analysis. C AAV-DB-1, AAV-DB-2, and AAV-DB-3 transduction in the globus pallidus (i, ii), putamen (iii, iv), caudate (v, vi), and substantia nigra (vii, viii). Cell transduction was analyzed by FISH (i, iii, v, vii), and direct fluorescence of expressed fluorophores (ii, iv, vi, viii). D mTFP1 fluorescence in the fibers of the internal capsule. E Direct fluorescence of mTFP1 positive neurons in motor and premotor cortex. Scale bars: (A–D) 50 µm, (E) 200 µm. C–E are representative images from a single rhesus macaque (Table S1). Panel (A) Created in BioRender. Lewandowski, B. (2025) https://BioRender.com/y3ikqcc Panel (B) used 3D reconstructions adapted from ref. 52.
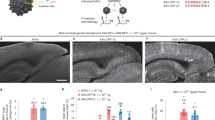
In tissues outside the basal ganglia, AAV-DB-3.mTFP1 robustly transduced cortical layer 5/6 projection neurons, as evidenced by axonal labeling in the internal capsule and cell body labeling in the cerebral cortex (Fig. 2D, E and Supplementary Fig. S1). In contrast, AAV-DB-1 and AAV-DB-2 did not transduce neurons in the cortex (Supplementary Fig. S1). Cortical layer 5/6 projection neurons are particularly vulnerable in HD, and transduction of these cells therefore makes AAV-DB-3 a particularly attractive capsid variant for HD gene therapies21. There were scattered transduced cells in other brain areas, including thalamus and the cingulate cortex (Supplementary Fig. S1), but transgene expression was largely confined to the basal ganglia and connected brain areas and limited predominately to the ipsilateral side of the infusion. AAV-DB candidates were also mixed and injected at a lower dose of 3.0E+10 vg each into another adult rhesus macaque, NHP 7. AAV-DB-3 again showed widespread transduction in the striatum and motor cortex, similar to the 7.5E+10 vg dose (Fig. 3A, B). There was little to no transduction observed in dorsal root ganglia or liver, which are common off-target tissues for CSF-or IV-delivered AAVs10,30,31 (Supplementary Fig. S2). Based on AAV-DB-3.mTFP1’s widespread transgene expression across basal ganglia regions and superior cerebral cortex transduction relative to AAV-DB-1 and AAV-DB-2, we selected AAV-DB-3 as the top AAV capsid variant for deep brain applications and further testing in mouse strains and neurons derived from human iPSCs.
A, B Fluorescence tilescans showing the extent of AAV-DB-3.mTFP1 transgene expression in motor cortex and putamen following injection of 7.5E+10 (A) or 3.0E+10 (B) vg into the GP. Scale bars 500 µm, insets 50 µm. A, B are each representative images from a single rhesus macaque (Supplementary Table S1).
To further examine cell types transduced by AAV-DB-3.mTFP1, we performed co-labeling experiments by RNA-FISH focusing on neurons in the caudate nucleus and putamen, as medium spiny neurons (MSNs) are among the more vulnerable neurons in HD and a critical cell type for other basal ganglia diseases. RNA FISH for mTFP1 and co-labeling with the neuronal marker NeuN revealed that the majority of the transduced cells in the caudate and putamen were neurons (Fig. 4A). Two major classes of MSNs, D1-MSNs and D2-MSNs, express D1 or D2 dopamine receptors (encoded by DRD1 and DRD2, respectively). RNA FISH for mTFP1, DRD1, and DRD2 showed that dopamine receptor expressing MSNs in the caudate and putamen were the most highly transduced cell type (Fig. 4B). In the caudate and putamen, up to 45% of all DRD1- or DRD2-expressing MSNs were positive for mTFP1 transcripts (Fig. 4C). Although transgene RNA expression was highest in MSNs, we cannot rule out the possibility that AAV-DB-3 also transduced non-neuronal cells in the caudate and putamen with low levels of RNA expression undetectable by fluorescence methods or RNA FISH. Note that the doses used in these animals were relatively low (3.0E+10 or 7.5E+10 total vg) compared to conventional viruses like AAV2 or AAV5 applied using convection-enhanced delivery18,32, which was not required here, given the low volumes of infusate. Higher doses may provide even greater coverage of MSNs. Taken together, transgene expression data in adult, old-world NHPs showed potent transduction of a significant percentage of neurons of the basal ganglia and cerebral cortex after a single, low-dose infusion of AAV-DB-3.
A Representative images from a single rhesus macaque (Supplementary Table S1) of RNA FISH detection of the neuronal specific marker NeuN and mNeonGreen or mTFP1 transcripts in AAV-DB-2.mNeonGreen or AAV-DB-3.mTFP1 transduced cells, respectively, in caudate and putamen. B Representative images from a single rhesus macaque of RNA FISH labeling of DRD1 or DRD2 medium spiny neuron (MSN) subpopulations transduced by AAV-DB-3.mTFP1. C Quantification of the percentage of AAV-DB3 transduced cells among the indicated MSN subtypes counted from 284 total images taken across the ventral-dorsal axis of the putamen and caudate of three NHPs (NHPs 6, 9, and 10; Supplementary Table S1). A total of 22,510 cells and 8579 MSNs were profiled in this analysis. Scale bars, 50 µm. Data shown as mean with points for each animal. MSNs, medium spiny neurons. Source data are provided in the Source Data file.
Comparison of AAV-DB-3 and AAV5 biodistribution following GP delivery in rhesus macaques
AAV5 is a well-characterized capsid for delivery to the caudate nucleus and putamen in NHPs32,33 and, as noted above, is currently in clinical trials for the treatment of Huntington’s disease (NCT04120493). To directly compare AAV-DB-3 to AAV5, we delivered identical doses and volumes of AAV-DB-3.mTFP1 to the left GP and AAV5.mNeonGreen to the right GP in two rhesus macaques, NHPs 9 and 10. After four weeks in life, tissue was processed for histology (Fig. 5 and Supplementary Fig. S3). Fluorescence images were collected from the caudate nucleus, putamen, and other brain areas from 7 coronal planes numbered i-vii (Fig. 5A and Supplementary Fig. S3A). At the injection sites in the left and right GP, both AAV-DB-3 and AAV5 transduced cells (Fig. 5B, panel iv; Supplementary Fig S3B, panel iv). However, AAV-DB-3 transduced cells much further from the injection site than AAV5. Of note, AAV-DB-3 broadly transduced cells in the caudate nucleus and putamen, particularly through planes ii–iv, whereas AAV5 transduced just a few small patches of cells at this dose (Fig. 5B panels ii–iv, Supplementary Fig. S3B panels ii–iv). AAV-DB-3 also transduced scattered cells in the thalamus and pons, highlighting AAV-DB-3’s ability to spread over long distances through deep brain structures (Fig. 5B, panel v, Fig. S3B, panel v). AAV-DB-3 transduction in the caudate nucleus reached as far as plane i, over a cm away from the GP injection site. In the substantia nigra, AAV-DB-3 transduced cells in the pars compacta and robustly labeled axon terminals in the pars reticulata originating from MSNs in the caudate nucleus and putamen (Fig. 5B panel vii, Supplementary Fig. S3B, panel vii). Higher magnification images of the caudate nucleus and putamen revealed cells transduced by AAV-DB-3 with MSN morphology along the entire rostro-caudal axis (Supplementary Fig. S4A). Cortical layer 5/6 projection neurons were also observed in multiple regions of motor, premotor, prefrontal, and cingulate cortex (Supplementary Fig. S4B). Finally, we compared AAV-DB-3.mTFP1 and AAV5.mNeonGreen transgene mRNA expression by reverse transcription digital droplet PCR (RT-ddPCR) (Fig. 5C). AAV-DB-3 transgene mRNA expression was on average 92-fold higher than AAV5 in GP, 531-fold higher in caudate, 184-fold higher in putamen, and 271-fold higher in motor cortex (Fig. 5C). The RT-ddPCR results underscore AAV-DB-3’s ability to deliver high levels of transgene mRNA expression in key brain areas for HD gene therapy following GP delivery.
7.5E+10 vg of AAV-DB-3.mTFP1 or AAV5.mNeonGreen vectors were delivered to the left or right globus pallidus, respectively. A 3D model with the location of each brain slice viewed in subsequent panels (i, ii, iii, iv, v, vi, vii); Slices are 4 mm apart. Bi–vii Tile scan images showing direct fluorescence across structures. Scale bars 500 μm, inset scale bars 50 μm. C mTFP1 and mNeonGreen transcript quantification by RT-ddPCR in globus pallidus, caudate, putamen, and motor cortex (NHPs 9 and 10; Supplementary Table S1). Data shown as mean with points for each animal. Acb, accumbens; Cd, caudate; GPe, external globus pallidus; GPi, internal globus pallidus; HC, hippocampus; LGN, lateral geniculate nucleus; Pn, pons; Pu, putamen; SNc, substantia nigra pars compacta; SNr, substantia nigra pars reticulata; Th, thalamus; ac anterior commissure; ic, internal capsule. Source data are provided in the Source Data file. A used 3D reconstructions adapted from ref. 52.
Caudate nucleus and thalamus delivery of AAV-DB-3 in rhesus macaque
Given AAV-DB-3’s neuronal tropism and ability to spread through the brain, we investigated other deep brain delivery sites. AAV-DB-3.mTFP1 (7.5E+10 vg) was injected into the left caudate nucleus and right VA/VL thalamus in a single rhesus macaque, NHP 11 (Supplementary Fig. S5). The caudate nucleus injection robustly transduced cells around the injection site (Supplementary Fig. S5A). However, the spread in the caudate nucleus was limited compared to GP injections, with few if any cells transduced in the putamen (Supplementary Fig. S5A). VA/VL thalamus delivery robustly transduced cells not only around the injection site, but also cortical layer 5/6 projection neurons in motor and cingulate cortex (Supplementary Fig. S5B). Neither direct caudate nucleus injection nor VA/VL thalamus injection of AAV-DB-3 resulted in the broad transduction of the caudate nucleus and putamen observed following GP injection in previous experiments. Finally, we assessed neuroinflammation by GFAP and IBA1 immunofluorescence in the caudate nucleus and putamen in NHPs 9 and 10 vs the right hemisphere of NHP 11, which had very little AAV transduction (Supplementary Fig. S6). NHPs 9 and 10 had low, baseline expression of GFAP and IBA1, similar to NHP 11 (Supplementary Fig. S6), suggesting limited toxicity following AAV-DB-3.mTFP delivery.
AAV-DB-3 transduction properties in mice
To evaluate the evolutionary conservation of AAV-DB-3’s characteristic broad biodistribution, we injected C57/BL6 mice in the mouse equivalent of the external globus pallidus (GPe) with AAV-DB-3.mTFP1. Three weeks after injection, brain tissue was harvested for histological analysis. In addition to abundant GPe neurons transduced around the injection site (Fig. 6A, B), we observed mTFP1 expression in numerous MSNs in the striatum (Fig. 6C). We also observed abundant neuronal transduction in the motor, cingulate, and medial prefrontal cortices (Fig. 6A, D) and the thalamus (Fig. 6E). These results largely mirror the NHP transgene expression results and support an evolutionarily conserved mechanism for AAV-DB-3 biodistribution. In parallel, two groups of C57/BL6 mice were injected in the striatum or thalamus (Supplementary Fig. S7). The brains were collected, and tissue sections were processed in the same manner as above. Our results after striatal injections show a vast number of labeled striatal MSNs (Supplementary Fig. S7B), pallidostriatal neurons in the GPe (Supplementary Fig. S7F), and numerous neurons located in somatosensory, frontal, and motor cortices (Supplementary Fig. S7E), as well as in thalamo-striatal neurons (Supplementary Fig. S7C, D). In mice injected in the thalamus (Supplementary Fig. S7G), we observed numerous neurons located in several thalamic nuclei (i.e, reticular nucleus, lateral geniculate nucleus, and ventral thalamus) (Supplementary Fig. S7H, J), as well as in the somatosensory, motor and frontal association cortices (Supplementary Fig. S7I) and the substantia nigra (Supplementary Fig. S7K).
A Low magnification image showing the distribution of mTFP1 direct fluorescence. B mTFP1-positive cells in the GP. C mTFP1-positive cells in the striatum. D mTFP1-positive cells in the cortex. E mTFP1-positive cells in the thalamus. Scale bars: (A) 1.5 mm, (B–E) 200 µm. B–E are representative images from 3 mice.
AAV-DB-3 efficiently transduces human neurons
We further evaluated the transduction efficiency of AAV-DB-3 in human neurons derived from induced pluripotent stem cells (iPSCs). iPSC-derived cortical neurons were generated using dual SMAD inhibition, which results in neurons resembling layer 5/6 pyramidal neurons34. The cells were transduced at day 40 of differentiation with AAV-DB-3.mTFP1 or its parental serotype AAV1.eGFP at different multiplicities of infection (MOIs; 1E+3, 1E+4, and 1E+5). Ten days post transduction, qualitative analysis by microscopy revealed more abundant transduction of AAV-DB-3 compared to AAV1 at equivalent doses (Fig. 7A). At MOI of 1E+3 quantification from micrographs demonstrated a 10-fold increase in the percent of transduced MAP2-positive cells for AAV-DB-3 over AAV1, resulting in a 60-fold increase in transgene expression as quantified by RT-qPCR (Fig. 7B, C). To assess AAV-DB-3’s performance in a second type of human-iPSC-derived neurons, NGN2 cultures were generated that resemble cortical layer 2/3 excitatory neurons35,36. As expected, NGN2-derived neurons expressed NGN2 and MAP2 as observed by immunofluorescence (Supplementary Fig. S8A). AAV-DB-3.mTFP1 was applied at an MOI of 1E+4, and 5–7 days later, cells were sorted to quantify percent of neurons transduced (Supplementary Fig. S8B). mTFP1 expression was detected in nearly 90% of sorted cells. Thus, AAV-DB-3 potently transduced human iPSC-derived neurons generated with two distinct differentiation protocols.
A Direct fluorescence of AAV1.eGFP and AAV-DB-3.mTFP1 in iPSC-derived excitatory neurons 10 days post-transduction with indicated MOIs. Nuclei were stained with Hoechst 33258. Scale bars, 50 µm. B Quantitation of percent transduction of AAV-DB3.mTFP relative to AAV1.eGFP. Five or more images were quantified per dose. 3857 and 3333 cells were counted in the AAV1 and AAV-DB-3 treatment groups respectively. Data are represented as mean +/− SEM. Results were significant by 2-way ANOVA for AAV treatment (54.43% of total variation, F (1, 26) = 68.03, p < 0.0001) and dose (24.77% of total variation, F (2, 26) = 15.48, p < 0.0001). C Quantitative PCR for transgene expression. All samples were normalized to GAPDH and are presented relative to AAV1.eGFP at the 1 E3 MOI. Data are represented as mean +/− SEM with points representing 3 biological replicates performed in triplicate. Results were significant by 2-way ANOVA for AAV treatment (40.66% of total variation, F (1, 14) = 22.77, p = 0.0003) and dose (34.35% of total variation, F (2, 14) = 9.617, p = 0.0024). Source data are provided in the Source Data file.
Discussion
AAV gene therapies for disorders with dysfunction in deep brain areas have shown early promise despite the challenges with delivery to these regions. Of note, in children with aromatic L-amino acid decarboxylase (AADC) deficiency, delivery of an AAV2 vector expressing AADC to the substantia nigra and ventral tegmental area was well tolerated and led to clinical improvements37. In Parkinson's Disease (PD) patients, AAV2 gene therapies with AADC or glial cell-derived neurotrophic factor (GDNF) payloads delivered to the putamen were well tolerated and, for AADC, showed possible stabilization of motor function17,38. As noted previously, a gene therapy for HD with an AAV5 vector expressing a huntingtin-lowering microRNA delivered to the caudate and putamen is also in clinical trials (NCT04120493). However, these approaches rely on large infusion volumes and convection-enhanced delivery via multiple cannula trajectories per hemisphere to achieve adequate deep brain coverage. For example, in the AAV2 AADC trial, patients in the highest dose group received 900 µL of test article per hemisphere via 2-3 cannula trajectories, and coverage of the putamen was around 42% based on intraoperative MRI scans17. These injection volumes necessitate long surgical times, typically under MRI guidance, increasing patient burden and creating logistical challenges when scaling a therapy to a larger number of patients.
An alternative to direct intraparenchymal delivery of gene therapies to deep brain areas is IV-delivery of AAVs with engineered capsids that cross the blood-brain barrier (BBB). Naturally occurring AAV variants poorly penetrate the BBB, with most viral particles accumulating in the liver and other peripheral organs14,30,39. In mice, peptide-modified AAV9 capsid variants were identified with a dramatically enhanced ability to cross the BBB5,6. However, this ability was not retained in all mouse strains or in nonhuman primates40,41,42. Building on this work, new AAV variants with greater potential for clinical application have been identified through a variety of approaches, including in vivo screens in NHPs and human transferrin receptor binding8,9,43,44. Other emerging approaches may involve identifying receptors that facilitate BBB penetrance45,46 or combining machine learning with in vivo or in vitro screens47,48. These efforts show promise, and progress continues towards identifying more potent BBB-penetrant AAV capsids. If sufficiently potent capsids are developed, challenges remain for IV delivery, including safety concerns around peripheral exposure to high doses of AAV12,13 as well as manufacturing the high doses required.
Here, we identified capsid variants capable of broad transduction of neurons in deep brain areas in NHPs following low-volume intraparenchymal infusion in the GP. The most potent variant, AAV-DB-3, transduced cells across multiple basal ganglia and cortical regions in NHPs, including MSNs in the caudate nucleus and putamen. AAV-DB-3 also potently transduced neurons in the mouse brain as well as human IPSC-derived neurons differentiated into regionally distinct cortical neurons. The cross-species validation of AAV-DB-3 in two NHP species, mice, and human neurons contrasts with results for other engineered capsids that show highly species-specific performance9,40. The pattern of AAV-DB-3’s spread in the NHP and mouse brain points to retrograde transport along axons, but additional studies will be required to characterize the mechanism of AAV-DB-3’s remarkable spread in the brain. AAV-DB-3 primarily expressed transgene in neurons rather than other cell types outside of the GP; this tropism may be intrinsic to the AAV-DB-3 capsid but may also point to a mechanism of uptake involving axonal transport. Of note, AAV-DB-3’s parental serotype is AAV1 and not AAV9, commonly used as the sole serotype for similar capsid screens, and AAV-DB-3 would not have emerged if our initial capsid screen did not contain multiple serotypes in addition to the commonly used AAV9. Taken together, our results suggest that delivery of AAV-DB-3 to the GP may provide an alternative to high-volume convection-enhanced delivery or high-dose IV delivery for AAV gene therapies targeting deep brain areas. Infusions in the GP are expected to be clinically feasible, as functional neurosurgeons have been placing electrodes in the GPi for deep brain stimulation for 30 years49. One potential application for AAV-DB-3 is HD, for which various cargos may be packaged targeting HTT or other implicated genes, including MSH3 or PIAS126,27,50.
Methods
Peptide modified AAV library description and construction
Plasmids used for the AAV library construction were generated by cloning AAV2 Rep and AAV Cap of different serotypes into an ITR-containing plasmid using Gibson assembly. Two unique restriction enzyme (RE) sites were built flanking the peptide insertion site in the AAV Cap gene. Oligonucleotides encoding the random 7-mer (NNK)7 were synthesized by Integrated DNA Technologies (Coralville, IA). The oligonucleotide library was cloned into the RE-digested recipient plasmids using Gibson assembly. Pooled plasmids were transformed into Endura electrocompetent cells (LGC, Middlesex, UK) and plasmid isolation performed using Qiagen Giga Prep Endofree kits (QIAGEN, Germantown, MD). For the second round library generation, viral genomic DNA from collected tissues (caudate, putamen, subthalamic nuclei, substantia nigra, thalamus and cortex) was isolated by QIAamp DNA mini kits (QIAGEN, Germantown, MD) and the variable regions of the AAV library amplified using serotype-specific primers and the NEB Q5 2x Master Mix (New England Biolabs, Ipswich, MA). Amplicons were incorporated into recipient plasmids by Gibson assembly and transformed into Endura electrocompetent cells (LGC, Middlesex, UK).
AAV library virus production
AAVs were generated in HEK 293 cells transfected with pAd helper and pAAV.Lib plasmids, the latter at an average of 1000 plasmid copies per cell. Recombinant virus was harvested and purified by two rounds of cesium chloride gradient centrifugation, and AAVs titered post-purification by ddPCR (Supplementary Table S2). Plasmids encoding the final capsids for evaluation were used in the final studies, each encoding a different fluorophore as noted in the figure legends. In all cases, the sequences were cloned into loop 8 of the VP3, which was at amino acid positions 590, 587, and 588 for AAV1, AAV2, and AAV9, respectively.
NHP animals and procedures
This study was conducted in accordance with the Guide for the Care and Use of Laboratory Animals (National Research Council), and all procedures were approved by the CHOP Research Institute Animal Care and Use Committee. Animals were housed at the CHOP Research Institute under a 12 h light:dark cycle under normal temperature and humidity conditions, with ad libitum access to purified drinking water and twice daily feedings with Purina LabDiet Certified Primate Diet (5048) enriched with fruits and vegetables. For anesthesia, NHPs were fasted overnight, sedated, intubated, and maintained on isoflurane. The head and chest were shaved, and electrocardiogram (ECG) electrodes were attached to the ventral thorax. A dose of buprenorphine sustained release (SR; 0.2 mg/kg subcutaneously (SC)) and meloxicam (0.2 mg/kg SC) was administered prior to surgery. During surgery, ECG, oxygen saturation (SpO2), end tidal carbon dioxide (ETCO2), non-invasive blood pressure (NIBP), rectal temperature, heart rate, and respiratory rate were all monitored continuously. Fluid support was administered throughout the procedure through an intravenous catheter placed in the saphenous vein. Thermal support was provided via a warmwater recirculating blanket and several layers of insulating material wrapped around the animal. The GP was targeted by ClearPoint MRI-guided infusion using a 3 T research MRI unit (Siemens Trio; SYNGO Vb17). A ClearPoint SmartFlow cannula was attached to a BD syringe controlled by a Harvard Apparatus infusion pump. The cannula was primed with test article and inserted into the brain to the desired depth before infusing 30 µL of test article at a rate of 1.0 µL/min. Following the infusion and a 10 min dwell period, the cannula was removed, the skin sutured, and the animal recovered.
AAV9.eGFP (3.0E+13 vg total, a mid-range dose for CSF-delivered AAV931) was infused as a positive control for DRG transduction10. Infusion was to the left lateral ventricle using a stereotactic device for NHPs using coordinates calculated from baseline MRI scans and confirmed by administration of a small volume of Iso-Vue M contrast under fluoroscopic guidance. After a three-minute dwell period, the needle was removed, the skin sutured, and the animal recovered. Buprenorphine SR was given as analgesia, and the animals were monitored postoperatively for pain and welfare.
For euthanasia, NHPs were fasted overnight, sedated, intubated, and maintained on isoflurane. Upon confirmation of deep anesthesia (stage III, plane 3), a thoracotomy was done prior to cardiac perfusion of ice-cold PBS. Brains were removed and processed into 4 mm coronal slabs in a rhesus macaque brain matrix. For AAV library experiments, tissue samples from basal ganglia, thalamus, cerebral cortex, and substantia nigra were micro-dissected and flash frozen for analysis. For fluorophore experiments, brain slabs, DRGs, and other tissues were postfixed in 4% paraformaldehyde. Additional samples for molecular analysis were banked, including liver samples used for ddPCR. For NHP information in tabular form, see Supplementary Table S1. For primer information, see Supplementary Table S2.
Amplicon sequencing of AAV libraries
To quantify input library composition and viral transduction in target tissues (caudate, putamen, subthalamic nuclei, substantia nigra, thalamus and cortex), amplicons were created from the peptide modified region of the viral Cap gene and sequenced. Genomic DNA (gDNA) or RNA from the tissues was isolated using a QIAamp DNA mini kit or RNeasy Plus kit (QIAGEN, Germantown, MA). Sequence encoding the modified capsid insertion was amplified using serotype-specific primers with PCR cycles less than 30 by using Q5 2x Master Mix (NEB, Ipswich, MA). Two microliters of 1st round of PCR products were input to total 50 µl reaction of the second round PCR to introduce Illumina i5 and i7 sequencing indices. PCR products were run on an agarose gel and purified by the MinElute Gel Extraction Kit (QIAGEN, Germantown, MA) to recover NGS amplicon libraries. Libraries were sequenced on an Illumina Novaseq 6000 using a 350-cycle reagent kit and paired-end read chemistry, yielding 175 bp paired-end read outputs. Target read depth was variable and allocated according to library identity and diversity. Read depth targets ranged from 2 to 10 million reads per sample.
Amplicon sequencing data analysis
Illumina amplicon sequencing data were processed using a custom Python / R pipeline. Illumina FASTQs were processed to identify and discard low-quality reads by testing for perfect string matches against constant regions of the amplicons. Passing reads were parsed to tabulate the occurrence of unique sequences encoding the variants’ heptamer inserts. Variant abundance was quantified as the UMI collapsed read count for each unique sequence, and a minimal performance threshold was established by removing any variants in a dataset with < 100 UMI counts. Additionally, potential sequencing errors were filtered out by removing any variants that had < 1% the UMI counts and were within 1 edit distance of another variant. Variant performance was assessed using a combination of two metrics: abundance (UMI counts) and an enrichment score. The enrichment score reports the percent change in a capsid variant’s relative abundance in a tissue vs the input virus, with values > 0 indicating increased representation in the tissue sample:
Capsid variant performance was assessed broadly across DNA and RNA data, UMI counts and enrichment metrics, and targeted single-region and broad cross-region analyses. Selection of capsids for fluorescent validation involved a collaborative assessment of all available data.
ddPCR for AAV genomes
DNA was isolated from NHP liver samples using the QIAamp DNA kit (Qiagen). ddPCR was performed on the isolated DNA using the ddPCR Supermix for Probes (BioRad) on the QX200 ddPCR instrument (BioRad) with primer-probe sets for the CAG promoter in each fluorophore transgene and NHP gDNA. Values were averaged from two technical repeats.
Mouse biodistribution studies
All procedures were approved by the CHOP Institutional Animal Care and Use Committee. Mice were maintained on a 12 h light/dark cycle at normal temperature and humidity conditions, and ad libitum access to water and food. Adult C57BL/6 mice (Jackson Laboratories, Bar Harbor ME, JAX strain #000664: (n = 3) were anesthetized with inhaled isoflurane and then injected with AAV-DB-3.mTFP1 at 1.0E+10 vg. Injections targeting the GPe (from bregma in mm: AP, − 0.46; ML, ± 1.85; DV, 3.7; n = 3, 2 F/1 M) were done at a rate of 25 nl/min. Injections targeting the thalamus (from bregma in mm: AP, − 1.70; ML ± 1.25; DV, 3.3; n = 2, 2 M) or striatum (from bregma in mm: AP, + 0.5; ML ± 1.75; DV, 3.2, 2.6; n = 3, 1 F/2 M) were performed using the same dose and injection rate. Three weeks after injection, mice were anesthetized with isoflurane and transcardially perfused with 0.05 M PBS, pH 7.4, followed by 50 ml of 4% w/v paraformaldehyde in phosphate buffer (0.1 M, pH 7.4). Brains were cryoprotected and sagittally sectioned. For each brain, the sites of injection were verified, and sections analyzed using a Leica DM6000B epifluorescence microscope for low-power and tiled images. Z-stacks were acquired using a Leica SP8 confocal microscope.
Direct fluorescence imaging
4 mm thick coronal slabs of the entire NHP cerebrum were fix in 4% PFA and 30% sucrose cryoprotected. 40 μm thick slices were collected with a Leica SM2010R microtome with a Physitemp BFS-30MP cold stage. Slices were washed with 0.1 M PBS and nucleus counterstain with 1:5000 Hoechst 33258 solution. Then, slices were mount in Superfrost plus slides and lipofuscin autofluorescence quenched with TrueBlack (Biotium, #23007). Images were collected using a Leica DM6000B equipped with a Hamamatsu ORCAflash2.0 monochrome camera. 33258 Hoechst, mTFP1, mNeonGreen, and mRuby3 fluorophores were analyzed using Leica LED 405, Chroma 39001 AT-ECFP/C, Chroma 49003 ET-EYFP, and Leica Y3 ET filter cubes respectively.
RNA-FISH
4 mm coronal NHP brain slabs were post-fixed in 4% PFA, sucrose cryoprotected, and OCT-embedded before 16 µm thick slices were collected on Superfrost Plus slides using a Leica CM1950 or CM3050S cryostats. Fluorescent in situ hybridization was performed using RNAscope Multiplex Fluorescent Reagent Kit v2 Assay (Advanced Cell Diagnostics, Cat. 323100-USM) following manufacturer guidelines. For quantitation, three RNAscope probes recognizing DRD1 (Advanced Cell Diagnostics Cat. 549041), DRD2 (Cat. 549031-C2), and mTFP1 (Cat. 500271-C3) were used to label regions of the caudate and putamen from two NHPs. Probe validation beyond commercial validation was not done. Specific Opal fluorophores were associated to each RNAscope probe (DRD1-C1-Opal520, DRD2-C2-Opal570, and mTFP1-C3-Opal620). One slice per slab was analyzed. Random non-overlapping 40X images were collected using a Leica SP8 confocal microscope. Confocal images comprised of 12 z-planes (11 µm) were flattened by maximum projection prior to quantification. Forty-two total images were collected from these regions and quantified using an unbiased automated cell counting approach. Puncta in all channels greater than .002 µm2 were detected using QuPath with the following relative fluorescence units (RFU) minimum pixel intensity thresholds DRD1 (Opal520) ≥ 25, DRD2 (Opal570) ≥ 25, mTFP1 (Opal620) ≥ 50. Detected puncta were then counted, and counts tables were processed using a custom R computational pipeline. Nuclei were defined as being DRD1 or DRD2 positive neurons if they contained ≥ 50 puncta in the respective channel. If nuclei contained > 60% puncta of either Opal520 or Opal570-positive they were assigned that DRD1 or DRD2 identity accordingly. If a nucleus containing ≥ 50 puncta did not meet this threshold it was classified as DRD1 and DRD2 double positive. Nuclei of all types were defined as positive for mTFP1 transduction if they were found to contain ≥ 2 mTFP1 puncta. The above settings were selected to achieve consistency with human counting results and for their ability to clearly differentiate positive nuclei from non-cellular background signal.
RT-ddPCR quantification of mTFP1 and mNeonGreen transcripts
OCT-embedded brain tissue was cryosectioned, mounted on glass slides, and brain regions of interest collected using disposable scalpels. RNA was isolated using the RNease FFPE kit (Qiagen), DNAse-treated using the TURBO DNA-free kit (Invitrogen), and reverse transcribed using Maxima H Minus Reverse Transcriptase (Thermo Scientific) according to manufacturers’ instructions. ddPCR was performed on the isolated DNA using the ddPCR Supermix for Probes (BioRad) on the QX200 ddPCR instrument (BioRad) with primer-probe sets for each fluorophore transgene (Supplementary Table S2).
Human iPSC-derived neuron studies
iPSC-derived cortical excitatory neurons were generated from iPSCs as previously described34. Virus transduction was performed at Day 40 of differentiation during a scheduled half-media change. Ten days later, transgene expression was determined via RT-qPCR using standard methods. The data represent 3 biological replicates done in triplicate. For microscopy, iPSC-derived cortical neurons were seeded on coverslips, and transduced as above, and 10 days later, cells were fixed with 4% PFA and immunofluorescence performed using a MAP2 primary antibody (Abcam ab5392, 1:5000). Antibody validation beyond commercial validation was not done. Fluorescence was assessed using a Leica SP8 confocal microscope. Quantification of transduction was performed using QuPath. Quantification steps included nuclear segmentation (QuPath: watershed cell detection plugin) to define the bounds of each nucleus, followed by nuclear-centroid-based fluorescence intensity measurement (QuPath: IntensityFeaturesPlugin). Measurements, including fluorescence intensities and nuclei size, were exported and indexed by image ID and nucleus ID. A custom R script was created to process and visualize results. Neuronal cells were defined by nuclear size, neuronal marker (MAP2) intensity, and nuclear stain intensity. Transduced neurons were defined as neurons with AAV1 or DB3 marker fluorescence intensity above a threshold of 10. Uniform thresholding was applied to all images.
For the iNeuron studies, stable dox-inducible NGN2 human iPSCs (sourced from the CHOP Human Pluripotent Stem Cell Core) were seeded onto Matrigel (Corning 354277) coated 6-well plates at a density of 1.0 × 105 cells per well in mTESR1 (Stem Cell Technologies 85850) supplemented with 10 μM Y-27632 (Tocris 1254). The following three days, basal media was fully replenished, supplemented with 2 μg/mL dox (Sigma D9891), 100 nM LDN-193189 (Tocris 6053), 10 μM SB431542 (Tocris 1614), 2 μM XAV939 (Tocris 3748). Basal media was replenished on day 0 with KSR [knockout serum: KO DMEM (ThermoFisher Scientific 10829-018), 15% KSR (ThermoFisher Scientific 10828028), NEAA (ThermoFisher Scientific 10370088), Glutamax (ThermoFisher Scientific 35050061), ß-mercaptoethanol (ThermoFisher Scientific 21985-023)], a 50/50 mix of KSR and NIM [neural induction media: DMEM/F12 (ThermoFisher Scientific 10829-018), Glutamax, NEAA, 0.16% Dextrose (Sigma D9434), 1x N2 (ThermoFisherScientific 17502048)] on day 1, and NIM on day 2. Puromycin was included on days 1 and 2 at 0.5 μg/mL (ThermoFisher Scientific A11138-03). On day 3, cells were dissociated with Accutase (ThermoFisher Scientific A11105-01) and plated at 4 × 105 cells per well onto PLO (Sigma P3655) and laminin (Gibco 23017015) coated 12-well plates or glass coverslips in NBM [neurobasal media: neurobasal (ThermoFisher Scientific 21103049), Glutamax, NEAA, B27 (ThermoFisher Scientific 17504044), N2] supplemented with 20 ng/mL BDNF (R&D Systems 248-BDB-050/CF), 20 ng/mL GDNF (R&D Systems 212-GD-050-CF), 2 ug/mL dox, and 10uM Y-27632. Neurons were maintained long term in NBM supplemented with 20 ng/mL BDNF, 20 ng/mL GDNF, and 2 μg/mL dox with 50% media changes every other day. Cells were treated once with 2 µM AraC (Sigma C6645) at day 8. Dox was withdrawn at day 14. Immunofluorescence to characterize the cells was performed using a MAP2 primary antibody (BioLegend PCK-554P, 1:5000) and an NGN2 primary antibody (Cell Signaling 13144S, 1:300). Antibody validation beyond commercial validation was not done. Viral transduction (MOI of 1E + 4) was performed at DIV15, coinciding with a 50% media change. After 5–7 days, AAV-DB-3 transduced neurons were dissociated into single cells using 20 U/mL papain (Worthington Biochemical, LK003178), 5 mM MgCl, and 5 μg/mL DNase (Worthington Biochemical, LS006333) and resuspended in NBM supplemented with 20 ng/mL GDNF, 20 ng/mL BDNF, and 10 μM Y-27632. Neurons were passed through a 35 μm filter and analyzed using the BD FACSAria Fusion (BD Biosciences) with an 80 μm nozzle. mTFP1 fluorescence was detected with a 405 laser and the 450/50 filter. The dissociated cell population was gated using FSC/SSC and SSC-W/SSC-A to remove debris and select for single cells, respectively. Untransduced cells were used to set all gates and to set the boundary for mTFP-positive populations. Data was analyzed using FlowJo version 10.6.2. For cell population abundance, post-sort fractions recovered 6.7–16.7% of the initial neuron population. The purity of the sorted populations was determined by quantifying the proportion of mTFP-positive cells in the neuron population, which represents DB3 transduced neurons.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Additional data, beyond what is reported here, are subject to legal and commercial restrictions and are available from the authors upon request in accordance with The Children’s Hospital of Philadelphia’s institutional policies. Source data are provided in this paper.
Code availability
The computational tools for capsid performance evaluation and automated fluorescence image quantification are publicly accessible via GitHub (https://github.com/DavidsonLabCHOP/DB3_Publication_2025.git) and Zenodo (https://doi.org/10.5281/zenodo.14851572)51.
References
Zhou, K., Han, J., Wang, Y., Zhang, Y. & Zhu, C. Routes of administration for adeno-associated viruses carrying gene therapies for brain diseases. Front. Mol. Neurosci. 15, 988914 (2022).
Lonser, R. R., Akhter, A. S., Zabek, M., Elder, J. B. & Bankiewicz, K. S. Direct convective delivery of adeno-associated virus gene therapy for treatment of neurological disorders. J. Neurosurg. 134, 1751–1763 (2020).
Foust, K. D. et al. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat. Biotechnol. 27, 59–65 (2009).
Zhang, H. et al. Several rAAV vectors efficiently cross the blood-brain barrier and transduce neurons and astrocytes in the neonatal mouse central nervous system. Mol. Ther. 19, 1440–1448 (2011).
Deverman, B. E. et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209 (2016).
Chan, K. Y. et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017).
Goertsen, D. et al. AAV capsid variants with brain-wide transgene expression and decreased liver targeting after intravenous delivery in mouse and marmoset. Nat. Neurosci. 25, 106–115 (2022).
Huang, Q. et al. An AAV capsid reprogrammed to bind human Transferrin Receptor mediates brain-wide gene delivery. Science. 384, 1220–1227 (2024).
Stanton, A. C. et al. Systemic administration of novel engineered AAV capsids facilitates enhanced transgene expression in the macaque CNS. Med 4, 31–50 e38 (2023).
Hinderer, C. et al. Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an adeno-associated virus vector expressing human SMN. Hum. Gene Ther. 29, 285–298 (2018).
Hudry, E. et al. Liver injury in cynomolgus monkeys following intravenous and intrathecal scAAV9 gene therapy delivery. Mol. Ther. 31, 2999–3014 (2023).
Ertl, H. C. J. Immunogenicity and toxicity of AAV gene therapy. Front. Immunol. 13, 975803 (2022).
Duan, D. Lethal immunotoxicity in high-dose systemic AAV therapy. Mol. Ther. 31, 3123–3126 (2023).
Thomsen, G. et al. Biodistribution of onasemnogene abeparvovec DNA, mRNA and SMN protein in human tissue. Nat. Med. 27, 1701–1711 (2021).
McBride, J. L. et al. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington’s disease. Mol. Ther. 19, 2152–2162 (2011).
Worgall, S. et al. Treatment of late infantile neuronal ceroid lipofuscinosis by CNS administration of a serotype 2 adeno-associated virus expressing CLN2 cDNA. Hum. Gene Ther. 19, 463–474 (2008).
Christine, C. W. et al. Safety of AADC gene therapy for moderately advanced Parkinson disease: Three-year outcomes from the PD-1101 trial. Neurology 98, e40–e50 (2022).
Hadaczek, P. et al. Convection-enhanced delivery of adeno-associated virus type 2 (AAV2) into the striatum and transport of AAV2 within monkey brain. Hum. Gene Ther. 17, 291–302 (2006).
A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell 72, 971–983 (1993).
Rikani, A. A. et al. The mechanism of degeneration of striatal neuronal subtypes in Huntington disease. Ann. Neurosci. 21, 112–114 (2014).
Pressl, C. et al. Selective vulnerability of layer 5a corticostriatal neurons in Huntington’s disease. Neuron 112, 924–941 (2024).
Matsushima, A. et al. Transcriptional vulnerabilities of striatal neurons in human and rodent models of Huntington’s disease. Nat. Commun. 14, 282 (2023).
Harper, S. Q. et al. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc. Natl. Acad. Sci. USA 102, 5820–5825 (2005).
Kordasiewicz, H. B. et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron 74, 1031–1044 (2012).
Monteys, A. M., Wilson, M. J., Boudreau, R. L., Spengler, R. M. & Davidson, B. L. Artificial miRNAs targeting mutant huntingtin show preferential silencing in vitro and in vivo. Mol. Ther. Nucleic Acids 4, e234 (2015).
Ochaba, J. et al. PIAS1 Regulates Mutant Huntingtin Accumulation and Huntington’s Disease-Associated Phenotypes In Vivo. Neuron 90, 507–520 (2016).
Morozko, E. L. et al. PIAS1 modulates striatal transcription, DNA damage repair, and SUMOylation with relevance to Huntington’s disease. Proc. Nat.l Acad. Sci. USA 118, e2021836118 (2021).
Bankiewicz, K. S. et al. AAV viral vector delivery to the brain by shape-conforming MR-guided infusions. J. Control Release 240, 434–442 (2016).
Lee, H. et al. Cell type-specific transcriptomics reveals that mutant huntingtin leads to mitochondrial RNA release and neuronal innate immune activation. Neuron 107, 891–908 (2020).
Ballon, D. J. et al. Quantitative whole-body imaging of I-124-labeled adeno-associated viral vector biodistribution in nonhuman primates. Hum. Gene Ther. 31, 1237–1259 (2020).
Hordeaux, J. et al. Adeno-associated virus-induced dorsal root ganglion pathology. Hum. Gene Ther. 31, 808–818 (2020).
Spronck, E. A. et al. Intrastriatal administration of AAV5-miHTT in non-human primates and rats is well tolerated and results in miHTT transgene expression in key areas of huntington disease pathology. Brain Sci. 11, 129 (2021).
Samaranch, L. et al. MR-guided parenchymal delivery of adeno-associated viral vector serotype 5 in non-human primate brain. Gene Ther. 24, 253–261 (2017).
Waxman, E. A. et al. Reproducible differentiation of human pluripotent stem cells into two-dimensional cortical neuron cultures with checkpoints for success. Curr. Protoc. 3, e948 (2023).
Zhang, Y. et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798 (2013).
Fernandopulle, M. S. et al. Transcription factor-mediated differentiation of human iPSCs into neurons. Curr. Protoc. Cell Biol. 79, e51 (2018).
Pearson, T. S. et al. Gene therapy for aromatic L-amino acid decarboxylase deficiency by MR-guided direct delivery of AAV2-AADC to midbrain dopaminergic neurons. Nat. Commun. 12, 4251 (2021).
Rocco, M. T. et al. Long-term safety of MRI-guided administration of AAV2-GDNF and gadoteridol in the putamen of individuals with Parkinson’s disease. Mol. Ther. 30, 3632–3638 (2022).
Zincarelli, C., Soltys, S., Rengo, G. & Rabinowitz, J. E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 16, 1073–1080 (2008).
Hordeaux, J. et al. The neurotropic properties of AAV-PHP.B are limited to C57BL/6 J mice. Mol. Ther. 26, 664–668 (2018).
Hordeaux, J. et al. The GPI-Linked protein LY6A drives AAV-PHP.B transport across the blood-brain barrier. Mol. Ther. 27, 912–921 (2019).
Huang, Q. et al. Delivering genes across the blood-brain barrier: LY6A, a novel cellular receptor for AAV-PHP.B capsids. PLoS ONE 14, e0225206 (2019).
Chuapoco, M. R. et al. Adeno-associated viral vectors for functional intravenous gene transfer throughout the non-human primate brain. Nat. Nanotechnol. 18, 1241–1251 (2023).
Gonzalez, T. J. et al. Cross-species evolution of a highly potent AAV variant for therapeutic gene transfer and genome editing. Nat. Commun. 13, 5947 (2022).
Shay, T. F. et al. Human cell surface-AAV interactomes identify LRP6 as blood-brain barrier transcytosis receptor and immune cytokine IL3 as AAV9 binder. Nat. Commun. 15, 7853 (2024).
Huang, Q. et al. An AAV capsid reprogrammed to bind human transferrin receptor mediates brain-wide gene delivery. Science 384, 1220–1227 (2024).
Eid, F. E. et al. Systematic multi-trait AAV capsid engineering for efficient gene delivery. Nat. Commun. 15, 6602 (2024).
Zhu, D. et al. Optimal trade-off control in machine learning-based library design, with application to adeno-associated virus (AAV) for gene therapy. Sci. Adv. 10, eadj3786 (2024).
Siegfried, J. & Lippitz, B. Bilateral chronic electrostimulation of ventroposterolateral pallidum: a new therapeutic approach for alleviating all parkinsonian symptoms. Neurosurgery 35, 1126–1129 (1994).
Moss, D. J. H. et al. Identification of genetic variants associated with Huntington’s disease progression: a genome-wide association study. Lancet Neurol. 16, 701–711 (2017).
Davidson Lab CHOP and bclewandowski. DavidsonLabCHOP/DB3_Publication_2025: DB3_Publication_2025 v1.0.0. Zenodo. https://doi.org/10.5281/zenodo.14851572 (2025).
Saleem, K. S. et al. High-resolution mapping and digital atlas of subcortical regions in the macaque monkey based on matched MAP-MRI and histology. Neuroimage 245, 118759 (2021).
Acknowledgements
We thank Laurence Busque, Ashley Robbins, Melanie McFadden and the members of the CHOP Comparative Medicine Services Core and ClearPoint team members for their contributions to this work.
Author information
Authors and Affiliations
Contributions
Conception, D.E.L., Y.H.C., L.T., P.T.R., and B.L.D.; Study design and execution, D.E.L., Y.H.C., L.T., P.T.R., M.S.K., B.C.L., E.M.C., S.A., I.H.O., D.L., C.M.F., C.C., L.X., and B.L.D.; writing and editing; D.E.L., Y.H.C., L.T., P.T.R., B.C.L., E.M.C., and B.L.D.
Corresponding author
Ethics declarations
Competing interests
L.T., Y.H.C., D.E.L., P.T.R., M.S.K., and B.L.D. are founders of Latus Bio and have stock options. D.E.L. and P.T.R. are employees of Latus Bio. B.L.D. has sponsored research or serves an advisory role for Carbon Bio, Resilience, Roche and Latus Bio. The other authors declare that they have no competing interests. A patent has been submitted; PCT/US2023/021905, ‘Adeno-associated viral vectors for targeting deep brain structures’, B.L.D., P.T.R., M.S.K., Y.H.C., L.T., and D.E.L.
Peer review
Peer review information
Nature Communications thanks Aravind Asokan, Jeffrey Kordower, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Leib, D.E., Chen, Y.H., Tecedor, L. et al. Optimized AAV capsids for basal ganglia diseases show robust potency and distribution. Nat Commun 16, 4653 (2025). https://doi.org/10.1038/s41467-025-60000-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-60000-3