Abstract
Sexual dysmorphism in the number and distribution of meiotic crossovers is seen across species but is poorly understood. Here, we disrupt multiple anti-crossover pathways in hermaphrodite Arabidopsis and analyze thousands of female and male progeny genomes. The greatest crossover increase is seen in zyp1 recq4 mutants, with a 12-fold rise in females and 4.5-fold in males. Additional manipulation of crossover regulators does not further increase crossovers but shifts the balance between crossover pathways, suggesting competition for a shared, limited precursor pool. While wild-type crossover patterns differ between sexes, mutant crossover landscapes converge on a unique distinct profile, which we term Crossover Potential (COP). COP can be accurately predicted using only sequence and chromatin features. We propose that COP reflects the density of eligible recombination precursors, which is determined by genomic features and is thus identical across sexes, with sexual dimorphism resulting solely from differential regulation of their maturation into crossovers.
Similar content being viewed by others
Introduction
In sexual reproduction, meiotic recombination generates reciprocal exchanges between homologous chromosomes called crossover (CO), enhancing genetic diversity1,2. Notably, in the context of plant breeding, COs introduce favorable gene combinations and break up unfavorable linkages, allowing breeders to improve crop varieties. However, COs are limited in number, with typically 1-3 per chromosome per meiosis3,4, despite a large excess of molecular precursors. CO formation is initiated by programmed DNA double-strand breaks (DSBs) which are repaired by using the homologous chromosome as a template. The number of early recombination intermediates is estimated to be 10-25 fold greater than the eventual number of COs in mammals and plants5,6,7,8,9. CO frequencies are not homogenous along chromosomes, creating a landscape of recombination with high peaks and large valleys. COs are also subjected to interference, in which one CO inhibits the formation of another nearby, effectively limiting the frequency of close double COs10,11,12,13. Intriguingly, CO frequency and distribution can differ markedly between females and males of the same species, a phenomenon called heterochiasmy14. The factors and mechanisms that shape CO distribution along the genome and differently in females and males are not well understood. In particular, it is unclear what is the relative contribution of precursor distribution versus a differential likelihood of precursor maturation to CO is in the final landscape.
In most eukaryotes, two molecular pathways contribute to the maturation of recombination intermediates into CO, defining two classes of COs. Class I COs are promoted by the ZMMs protein, are sensitive to interference, and account for the majority of COs; class II COs are ZMM-independent and promoted by nucleases, including MUS81. Class II is a minor pathway in most eukaryotes, including plants, and is not or much less sensitive to interference11,15.
A series of factors that limit either class I or class II COs are known in Arabidopsis. One is the dosage of the ZMM protein HEI10, whose overexpression (HEI10oe) almost doubles the number of class I COs16,17. Another key factor regulating class I CO is the central part of the synaptonemal complex (SC), including the transverse filament ZYP118,19,20. The SC forms a structure that connects the homologous chromosomes all their length during the meiotic prophase. Mutation of ZYP1 eliminates CO interference and heterochiasmy and increases class I CO numbers ~twice18,19, and has a cumulative effect with HEI10oe on the number of class I CO21. On the other hand, the RECQ4A and RECQ4B helicases (called here RECQ4 for simplicity) redundantly prevent the formation of class II CO3,22. The combination of recq4 mutation and HEI10oe showed an additive effect with an increase in both class I and class II COs16, suggesting that class I and class II CO can be manipulated independently. The AAA-ATPase protein FIGL1 also limits class II COs by regulating strand invasion23,24 with has a synergetic effect with recq4, reaching the largest increase of (class II) CO described to date3,23,25,26. Additional proteins, such as FLIP, SNI, TOP3/RMI1, HSBP, HCR1 and HCR3 have anti-CO roles in Arabidopsis, being partners or regulators of the functions described above25,27,28,29,30,31,32. Several combinations of anti-CO factors including higher-order ones, remained to be tested to explore the upper limit of crossover frequencies.
Here, we manipulated one, two, three, or four of these CO regulators in Arabidopsis and examined the CO frequency in thousands of progenies derived from female and male crosses. Notably, COs in zyp1 recq4 showed a 12-fold increase in females and a 4.5-fold increase in males, accompanied by only a slight reduction in fertility. Manipulating additional CO regulators in zyp1 recq4 did not further increase CO frequency but altered the ratio of class I and class II COs, suggesting an upper limit had been reached. The relative distributions of COs along the genome in various mutant combinations converge to a common profile in both females and males, which diverges markedly from the wild-type distribution. We termed this profile Crossover Potential (COP) and showed that it can be accurately predicted using a few sequence and chromatin features. We propose that COP reflects the density of recombination precursors, which is determined by genomic features and is identical in both sexes, with sexual dimorphism in crossover landscapes arising solely from differences in precursor maturation into COs.
Results
HEI10oe , zyp1 and recq4 act differently in boosting CO numbers
We analyzed the number and distribution of COs in both female and male meiosis in wild-type, single and multi-mutants, by generating biallelic- Col/Ler F1 hybrids and reciprocally crossing them with wild-type Col (Fig. 1a, and Figure S1–S5). The obtained female and male-derived progenies (Back-cross BC1 populations) were analysed by whole genome Illumina sequencing to detect CO transmitted by female and male gametes, respectively (Fig. 1b, c). This method allows CO measurement with high accuracy, with some limitations. First, the CO position cannot be determined in regions lacking reliable markers (e.g., centromeric regions); but note that the presence/absence of CO in such areas can be reliably assessed by analyzing flanking markers. Second, with our stringent parameters, double-COs less than 90 kb apart, if they exist, and terminal CO less than 45 kb away from the telomere would be missed (see “methods”). In parallel, we counted the number of MLH1/HEI10 co-foci, which marks class I COs sites at diplotene/diakinesis33,34, in the Col/Ler F1 hybrids of each genotype. The MLH1/HEI10 focus counts were done in meiocytes both from ovules (female meiocytes) and anthers (male meiocytes) at the diplotene/diakinesis stage (Fig. 2a–m). At these stages, MLH1 and HEI10 foci almost perfectly co-localize (Table S1, supplementary data 2). Comparison of the number of MLH1/HEI10 foci (class I CO, counted the Col/Ler F1 hybrids) and genetic crossovers detected by sequencing (class I + class II COs, produced in the F1 hybrids and transmitted to the BC1 populations), allows the estimation of the contribution of each pathway (Fig. 2n). Note that the mean number of CO per gamete is expected to be half the number of cytological CO (or chiasma). This is because a chiasma affects only two of the four chromatids of a chromosome pair, and a gamete inherits a single chromatid35 (see Figure S11 in21). Thus, under the assumption that each MLH1 focus is converted into a CO and that all chiasma are marked by MLH1/HEI10, the frequency of genetic CO per gamete/BC1 is expected to be half of the frequency of MLH1/HEI10 foci per cell (1 focus=0.5 CO, indicated by a dashed line in Fig. 2n). If a proportion of chiasmata/CO is nor marked by MLH1/HEI10 (i.e., class II COs), the frequency of genetic CO per gamete is expected to be higher than half of the frequency of MLH1/HEI10 foci per cell (an upward deviation from the dashed line in Fig. 2n).
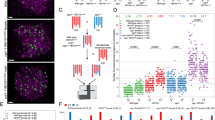
a Schematic representation of the experimental design. Col/Ler hybrid plants, either wild-type or carrying mutations, were crossed with wild-type Col in both directions. The resulting plant populations (BC1) were sequenced to score the COs transmitted by the female and male gametes of the hybrids. Created in BioRender. Lian, Q. (https://BioRender.com/m7vjhfg). b The number of COs per female and male gametes for each genotype. Each point represents the number of COs in one BC1/gamete. Circles and triangles are females and males, respectively. A black line indicates the mean and its value is specified at the top of the graph. The BC1/gamete population size (n) is indicated for each genotype. Two-sided Mann-Whitney test was used to evaluate the differences in CO numbers between female and male meiosis (***: < 10−3). See also Figure S5 for chromosome per chromosome analysis (c) Representative transmitted chromatid sets are shown for selected samples. The genotype and the name of the sample are specified. Red and Blue indicate Col and Ler genotypes, a transition marking a crossover. White points indicate centromeres. d The average number of COs per F2 sample (for recq4 figl1, yellow) or pseudo-F2 (sum of female and male averages, for all other genotypes).
a Counts of MLH1/HEI10 foci in female and male meiocytes of wild type and mutants Col/Ler F1 hybrids. Each dot is an individual cell. Red bars and numbers indicate the mean number of MLH1/HEI10 co-foci. Circles and triangles are females and males, respectively. One or two plants were used for each genotype (see source data file for details). b–m Representative immunostaining of REC8 (purple), MLH1 (green) and HEI10 (red) in wild type (b–e), zyp1 HEI10oe (f–i) and zyp1 recq4 (j–m) male diplotene meiocytes. MLH1-HEI10 co-foci indicate class I CO sites. The maximum intensity projection is shown and the complete Z-stack of the cells is available as supplementary data 2. The number of analyzed cells correspond to the number of points in Fig. 2a. Scale bar=2 µm. n Comparison of the numbers of genetic COs (Fig 1a) and MLH1/HEI10 foci per cell (Fig. 2a) in females and males of various genotypes. The number of samples are identical to Figs. 1a and 2a. Females are shown by circle, and males are shown by triangle. Error bars are the 90% confidence intervals of the mean. The dashed line indicates the number of genetic COs expected to be observed under the hypotheses that each MLH1/HEI10 foci is converted into a CO and all COs are marked by MLH1/HEI10 (no class II COs).
In the wild type, we observed an average of 2.8 COs per female gamete, and 5.2 per male (p < 10−6 Mann-Whitney test), confirming the previously observed heterochiasmy36,37. The number of MLH1 foci matches 2-fold the number of genetic COs, suggesting that in both female and male wild-type, the vast majority of crossovers are class I COs (Fig. 2n). Class II COs were suggested to represent ~15% of all meiotic COs in Arabidopsis, based on the observation of residual chiasmata in the zmm mutants9. The almost perfect match between the number of foci and the number of genetic COs that we observed suggests a more minor contribution of the class II pathway in wild-type meiosis.
In single mutants, HEI10oe and zyp1, the average CO number per transmitted gamete was substantially increased compared with the wild type, consistent with previous results17,18. In HEI10oe, the CO number in females (6.2, 2.2-fold/wild type, p < 10−6) remains lower than in males (9.9, 1.9-fold/wild type, p < 10−6. Difference in fold increases female vs male, Mann-Whitney p = 0.001). In zyp1, the CO number is equal in female (7.1, 2.5-fold/wild type, p < 10−6) and male meiosis (7.0, 1.3-fold/wild type, p < 10−6. zyp1 female vs zyp1 male, p = 0.41. Differences in fold increase female vs male, p < 10−6). In both cases, MLH1/HEI10 foci were also increased in female and male meiocytes of the F1 (p < 10−4), matching the number of COs in their progenies (Fig. 2a, n). Thus, overexpressing HEI10 or mutating ZYP1 exclusively increases class I COs. However, while heterochiasmy and CO interference are still present in HEI10oe, both are abolished in zyp1 (Fig. 1b, Figure S6).
Single recq4 mutation increased COs to higher levels and, remarkably, inverted heterochiasmy, reaching 17.5 in females (6.3-fold/wild type, p < 10−6) and 14 in male gametes (4.5-fold/wild type, p < 10−6) (Fig. 1b)3. In contrast to HEI10oe and zyp1 in which MLH1/HEI10 foci are increased, the number of MLH1/HEI110 foci was unchanged in both female or male recq4 meiocytes compared to wild-type (p = 0.89 and p = 0.09) (Fig. 2), consistent with the role of RECQ4 in limiting specifically class II COs22. This is reflected in Fig. 2n, by a vertical deviation from the dashed line, with the number of genetic COs largely exceeding the expected number according to MLH1/HEI10 focus counts. Accordingly, as class II COs are not subjected to interference, CO interference is not detectable in recq4 (Figure S6). The absence of interference is also reflected by a higher variation in the number of COs per gamete in recq4 (and combinations, see below) than in the wild type (Fig. 2b). Thus, recq4 boosts class II CO, with a more prominent effect in female than male meiosis.
Maximizing CO numbers
HEI10oe increases class I COs while recq4 increases class II COs. Combining recq4 HEI10oe, we observed an average of 23 COs in female and 17.2 in male gametes, higher than both single mutants (p < 10−6) (Fig. 1b). This is consistent with previous studies performed in F2 population16. Under the hypothesis of additive effects, simply summing the wild-type counts with the gains observed in each single mutant, we would have expected 20.9 COs in female gametes [2.8 + (6.2 − 2.8) + (17.5 − 2.8)] and 18.7 in male gametes [5.2 + (9.9 − 5.2) + (14.0 − 5.2)], which is similar to the observed values. In both female and male meiosis of F1s, the numbers of MLH1/HEI10 foci in recq4 HEI10oe are not modified compared to HEI10oe (p = 0.47 and p = 0.55) (Fig. 2a, n), suggesting that class I COs are unaffected by the recq4 mutation. Altogether, this shows that combining recq4 and HEI10oe leads to parallel increases of class I COs and class II COs provoked by respectively by HEI10oe and recq4 and thus to an additive effect on the total number of COs.
Next, we combined the two factors that individually increase class I COs, HEI10oe and zyp121. Following the same logic as above, the hypothesis of an additive effect predicts 11.7 COs in female (2.8 + 3.4 + 4.3) and 10.5 COs in male gametes (5.2 + 4.7 + 1.8). In zyp1 HEI10oe, we observed an average of 14.7 COs in male gametes, which is higher than single mutants (p < 10−6) and than the additive prediction, and 19.6 in females, which is almost twice the prediction. MLH1/HEI10 foci are largely increased in F1 female and male meiosis, reaching ~29.5 in both sexes, more than each single mutant and 3-4-fold the wild-type (Fig. 2a). This indicates that HEI10oe and zyp1 synergistically augment class I COs21. In male zyp1 HEI10oe meiocytes, the number of MLH1/HEI10 foci (29.3) matches the frequency of genetic COs (Figs. 1b, 2a, n), suggesting that all COs in male zyp1 HEI10oe are of class I COs. In contrast, in female zyp1 HEI10oe, the MLH1/HEI10 foci counts were 29.8, which corresponds to 14.9 COs per gamete, while 19.6 COs were observed (Figs. 1b, 2a, n). This suggests that in addition to the large boost in class I COs, class II COs are also increased in zyp1 HEI10oe female meiosis, leading to an inversion of heterochiasmy.
In the last combination of two factors, zyp1 recq4, the COs number per gamete was massively increased to 34.7 in females and 23.5 in males, almost twice the number measured in recq4, the highest single mutant (p < 10−6) (Fig. 1b). This is significantly higher than the predictions under an additive hypothesis, 21.8 in females [2.8 + (7.1 − 2.8) + (17.5-2.8)] and 15.8 in males [5.2 + (7.0 − 5.2) + (14 − 5.2)], indicating a synergetic effect. The high CO levels in zyp1 recq4 are unprecedented (compared to wild-type, 12.4-fold in females and 4.5-fold in males), being above all previously described mutants, including the champion to date recq4 figl1 (Fig. 1b−d)16,17,18,21,22,26. The variance of CO per gamete is also large, with the extreme case of a female gamete with 76 COs distributed among the five chromosomes, while the highest number observed in a wild-type female gamete is 6 COs (Fig. 1b, c). In zyp1 recq4, the number of MLH1/HEI10 foci (12.5 in females, 12.7 in males) are stable compared with zyp1 (12.3 in females, 12.7 in males) (Fig. 2a, n) and do not match the number of COs, suggesting that the vast majority of CO are class II COs (Fig. 2a, n). Mutating RECQ4 in the zyp1 mutant thus increases class II COs, like it does in wild-type, but with a much stronger effect (Fig. 2n). Altogether, we conclude that in zyp1 recq4 class I COs are increased similarly to the single zyp1 while class II COs are boosted by a synergistic effect of the two mutations, which is particularly marked in female meiosis. This also points to a role of ZYP1 in preventing class II COs, in addition to its described role in regulating class I COs.
The results described above show that all the combinations of two mutations among HEI10oe, zyp1, and recq4 have more COs than the single mutants, predicting that the triple mutant should have even more COs. However, the CO numbers in zyp1 recq4 HEI10oe did not increase significantly compared to zyp1 recq4, reaching 34.0 COs in females (p = 0.68) and 25.6 in males (p = 0.06), respectively (Fig. 1b), indicating that a certain upper limit might have been reached. Interestingly, the number of MLH1/HEI10 foci is increased when adding HEI10oe to zyp1 recq4 in both female and male meiocytes (p < 10−6), but not the total number of COs, suggesting that an increase in class I COs is compensated by a decrease in class II COs. This further supports the conclusion that an upper limit has been reached and that the two pathways compete for a large but limited number of CO precursors.
FIGL1 is another anti-class II CO factor that acts independently from RECQ43,26. Adding figl1 mutation to zyp1 recq4 HEI10oe is thus expected to increase COs further. However, the number of COs in zyp1 recq4 HEI10oe figl1 mutant was not increased and instead was decreased compared to zyp1 recq4 HEI10oe (p < 10−6) (Fig. 1b, d). This further supports the idea of an upper limit in CO formation and shows that the figl1 mutation can increase or decrease COs depending on the context.
Inverted heterochiasmy in hyper-recombinant mutants
Heterochiasmy is defined as a different rate of meiotic recombination between the two sexes of the same species38. In wild-type Arabidopsis, the frequency of COs is higher in males (ratio female/male=0.54, p < 10−6), with a similar ratio in HEI10oe (0.63, p < 10−6). In zyp1, heterochiasmy is abolished with similar CO frequency in female and male gametes (p = 0.47) (Fig. 1b). Intriguingly, the heterochiasmy is inverted, with more COs in females than males in single recq4 (1.25, p = 0.00026) and all double and triple mutants studied here: zyp1 HEI10oe (1.33, p < 10−6), recq4 HEI10oe (1.34, p < 10−6), zyp1 recq4 (1.48, p < 10−6), zyp1 recq4 HEI10oe (1.33, p < 10−6), and zyp1 recq4 HEI10oe figl1 (1.32, p = 1.02 10−6) (Fig. 1b). This shows that while CO levels are higher in males than in females in wild-type, the potential for CO formation is higher in females.
Fertility and meiosis are only marginally affected in zyp1 recq4
In most eukaryotes, CO frequencies are limited to 1-3 per chromosome, leading to the idea that a higher frequency of COs could be detrimental, possibly disturbing chromosome segregation during meiotic divisions and fertility. We estimated the fertility by counting the number of seeds per fruit (silique). Different mutants showed varying degrees of reduction in fertility, but the fertility is poorly correlated with CO frequency (Fig. 1b, Fig. 3a). The fertility of recq4 was not significantly reduced compared to the wild type, despite a massive increase in COs (6.3-fold in females, 2.7-fold in males), consistent with previous results3,16 (Fisher’s LSD test, p > 0.9999, Fig. 1b, Fig. 3a). In zyp1, fertility is modestly but significantly affected (p < 0.0001, Fig. 3a), probably because of the failure to ensure the obligate CO18,19. Strikingly, zyp1 recq4 showed a fertility similar to zyp1 (p = 0.9913, Fig. 3a, b), showing that the dramatic increase in COs is not associated with a further reduction in fertility. Despite a ~ 12-fold elevated CO frequency in female meiosis, the fertility of zyp1 recq4 is still ~75% of wild-type (Fig. 3a, b). Further, analyses based on genome coverage by sequencing did not detect any aneuploids in the progeny of zyp1 recq4 (n = 2 × 186) suggesting that chromosome missegregation at meiosis is absent or rare (Fig. 3c). However, chromosome spreads of male meiocytes revealed abnormal chromosome structures in ~16% of metaphase I cells, suggestive of abnormal recombination intermediates, which might cause reduced fertility in zyp1 recq4 (Fig. 3e, f, k, l, p).
a Quantification of fertility. Each dot represents the fertility of an individual plant, measured as the number of seeds per fruit averaged on ten fruits. The red bar shows the mean. All plants were grown in parallel, and the wild-type controls were siblings of the mutants. The number n of analyzed plants is indicated. Letters (a–c) above each data point group indicate statistically significant differences among genotypes, as determined by one-way ANOVA followed by Tukey’s multiple comparisons test (p < 0.05). Genotypes sharing the same letter are not significantly different from one another. b Representative cleared fruits of wild type, zyp1 recq4, zyp1 recq4 HEI10oe and zyp1 recq4 HEI10oe figl1 mutants in Col/Ler background. c The percentage of aneuploid samples detected in each population. The proportion of aneuploid samples in each population is shown on top of the bars. d–o DAPI-stained meiotic chromosome spreads from Col/Ler male meiocytes in wild type (d, j), zyp1 req4 (e, f, k, i) HEI10oe (g, m), zyp1 recq4 HEI10oe (h, n) and zyp1 recq4 HEI10oe figl1 (i, o). d–i Metaphase I. j–o Anaphase I. Scale bar = 10 µm. Red arrows pointed out abnormal chromosome connections, fragments and chromosome threads. p Quantification of different chromosome behaviors at metaphase I in wild type, zyp1 recq4, HEI10oe, zyp1 recq4 HEI10oe and zyp1 recq4 HEI10oe figl1. Cells were categorized according to normal (5 bivalents) and abnormal chromosome behavior (fragmentation, univalent and chromosome connection). The number of analyzed cells is indicated above the bar.
HEI10oe showed a significant fertility reduction (p < 0.0001, Fig. 3a), consistent with previous studies16, possibly due to a chromosomal rearrangement associated with the transgene in the HEI10oe C2 line17, which is heterozygous in the hybrid context. Consistently chromosome connections were observed in 34% of HEI10oe meiotic cells (Fig. 3g, m, p). Aneuploids were also detected in the progeny of all genotypes with HEI10oe (Fig. 3c) whereas no aneuploids were detected in recq4 and zyp1 recq4 progenies (Fig. 3c). Fertility of zyp1 recq4 HEI10oe figl1 was not reduced compared with zyp1 recq4 HEI10oe (p = 0.9961, Fig. 3a, b), suggesting that plants can still maintain a certain level of fertility even with disturbed strand-invasion and high recombination.
Meiotic chromosome fragmentation and reduced fertility in zyp1 mus81
As shown above, mutating ZYP1 in certain contexts (recq4, female HEI10oe) provokes an increase in COs but not MLH1/HEI10 foci, suggesting an anti-class II CO function for ZYP1, in addition to its established role in regulating class I COs. Mutating the nuclease MUS81 in anti-class II factors in Arabidopsis provoked meiotic catastrophe22,23,39,40, presumably because DNA joint molecules remain unrepaired. Similarly, we observed some chromosome fragments in 40% of anaphase I meiotic cells in zyp1 mus81 (Figure S8). In addition, fertility in zyp1 mus81 was reduced by 25% compared to wild type or mus81 (Figure S8j). This suggests that ZYP1 prevents the formation of joint molecules that need MUS81 to be repaired. However, this role is likely minor or redundant as no increase of class II COs is observed in the single zyp1 mutant (Fig. 2n)18.
Revealing the distribution of Crossover Potential
Next, we examine the distribution of COs along the chromosomes to test whether or not the increase observed in the different mutants is homogeneous (Fig. 4). In all the mutant combinations with large CO increases, COs tend to accumulate in distal regions in both females and males. It is especially striking when looking at the highest recombining genotypes zyp1 recq4 and zyp1 recq4 HEI10oe (Fig. 4a, b). The same is observed when merging the female and male data (pseudo-F2), and in the previously analyzed recq4 figl1 F2 populations (Fig. 4c)26. Intriguingly, the crossover landscapes in the different hyper-recombining mutants were highly consistent with each other, with common peaks and valleys (see also below). Looking at the local fold increase compared to the wild type (Figure S9), we detected common hotspots of CO increase in mutants, some above 200-fold in zyp1 recq4 and zyp1 recq4 HEI10oe, notably in distal regions in females where CO frequency is low in wild-type and high in the mutants. In contrast, regions in the periphery of the centromeres where CO frequencies are relatively high in wild-type meiosis, correspond to recalcitrant zones with no or limited increase in the mutants.
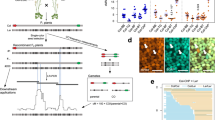
a, b The distribution (sliding window-based, window size 1 Mb, step size 50 kb) of COs along chromosomes in female (a) and male (b) of wild type, HEI10oe, zyp1, recq4, zyp1 HEI10oe, recq4 HEI10oe, zyp1 recq4, zyp1 recq4 HEI10oe and zyp1 recq4 HEI10oe figl1. c The distribution (sliding window-based, window size 1 Mb, step size 50 kb) of COs along chromosomes in F2 or pseudo F2 of wild type, HEI10oe, zyp1, recq4, zyp1 HEI10oe, recq4 HEI10oe, recq4 figl1, zyp1 recq4, zyp1 recq4 HEI10oe and zyp1 recq4 HEI10oe figl1. d The zoom of the CO position in the centromere proximal regions (Non-Recombining Zones, NRZs). Each point is a CO, circles, triangles and squares are females, males and F2s, respectively. The vertical dashed lines in red indicates the position of marker COs of NRZs, lines in black shows the middle position of centromeres.
Zooming on closer centromere-proximal region, we examined the Non-Recombining Zones (NRZs) previously defined as the interval spanning the centromere with a complete absence of CO in the wild type, as scored in 3,613 gametes containing a total of 14,397 COs41 (Fig. 4d). Strikingly, with such an unprecedented increase in CO numbers in the mutants described here, the NRZs persisted in resisting recombination and barely had any CO, despite our dataset containing a total of 70,022 COs. No COs were detected in NRZs in zyp1 recq4 (10,825 COs), and only one CO was found in each population of recq4 (3,675 COs), recq4 figl1 (15,543 COs), and zyp1 recq4 HEI10oe (10,991 COs) (Fig. 4d). This strongly suggests that other mechanisms, that do not depend on HEI10, ZYP1, RECQ4 or FIGL1, prevent CO formation in the NRZs.
Crossover landscapes in various mutants appear to have similar landscapes (Figs. 4, 5). To explore this formally, and compare the shape of the distributions independently of the absolute frequencies, we normalized the data by calculating the distribution of relative frequency per chromosome (i.e., what proportion of COs that occurred in this chromosome occurred in a given interval) (Fig. 5 and Figure S10–12). In wild-type females, the density of COs was the highest in the periphery of centromeres, and low at chromosome ends (Fig. 5a). In the wild-type males, COs are also frequent in the periphery of centromeres, but chromosome ends are also CO-rich (Fig. 5b). Consequently, female and male distributions are poorly correlated in the wild type (Spearman’s correlation r = 0.45, Fig. 5h). Both female and male zyp1 recq4 distributions differ markedly from the wild-type distributions (R = −0.14 and R = 0.36, Fig. 5a–b, d, f), but are strikingly similar to each other (r = 0.93, Fig. 5a, b, i). Further, the CO distributions in various mutants were similar to each other, but distinct from the wild-type, in both females (e.g., r = 0.86, zyp1 HEI10oe vs zyp1 recq4; Fig. 5a, d–e, Figure S10) and males (e.g., r = 0.88, recq4 HEI10oe vs zyp1 recq4; Fig. 5b, f, g, Figure S11). The CO landscape in the F2 populations of recq4 figl1 and zyp1 recq4 are also strikingly consistent (r = 0.97, recq4 figl1 vs zyp1 recq4; r = 0.97, zyp1 recq4 HEI10oe vs zyp1 recq4; Fig. 5c, j, k, and Figure S12)26. CO landscapes in recq4, zyp1 HEI10oe, recq4 HE10oe, recq4 figl1, zyp1 recq4, zyp1 recq4 HEI10oe are all very similar, with correlations in the range 0.75–0.92 (Figure S12).
a, b The distribution (scaled per chromosome, sliding window-based, window size 1 Mb, step size 50 kb) of relative CO frequency along chromosomes in female (a) and male (b) of wild type, recq4, zyp1 HEI10oe, recq4 HEI10oe, zyp1 recq4 and zyp1 recq4 HEI10oe. c The distribution (scaled per chromosome, sliding window-based, window size 1 Mb, step size 50 kb) of COs along chromosomes in F2 or pseudo F2 of wild type, recq4, zyp1 HEI10oe, recq4 HEI10oe, recq4 figl1, zyp1 recq4 and zyp1 recq4 HEI10oe. d–k Spearman’s correlation tests of relative CO frequencies (each point in an interval, as defined as Fig. 5a–c) comparing between genotypes and sexes. The blue line was added by using “lm” method, with 0.95 confidence intervals marked by gray shading.
The convergence towards a similar pattern in females and males in different mutants suggests the existence of a common underlying feature. We propose that this distribution of Crossover Potential (COP) represents the distribution of eligible recombination intermediates (i.e., DSBs that are being repaired with the homologous chromosomes, potentially leading to a CO). We then combined the 21,104 female and 15,188 male COs from the various mutant combinations (recq4, zyp1 HEI10oe, recq4 HEI10oe, zyp1 recq4, zyp1 recq4 HEI10oe, and zyp1 recq4 HEI10oe figl1) to create high-resolution maps of COP in females and males, separately (Fig. 6a). The female and male COP maps are strikingly similar, the female curve being slightly above the male one. When normalized, the two COP curves almost perfectly overlap (Fig. 6b, Spearman’s correlation r = 0.91), showing that the distribution of COP is shared between the two sexes. These features of COP strikingly deviate from those of wild-type COs, which are more frequent in males than females and very differently distributed. We then combined the female and male COs of the mutants listed above, together with the recq4 figl1 F2 population, totaling 49,482 COs, and generated a high-resolution universal COP map (Fig. 6c). The COP is not homogenous along chromosomes with null values in the centromeric regions and globally higher frequencies toward chromosome ends, and sharp peaks and valleys.
a The distribution (sliding window-based, window size 300 kb, step size 50 kb) of CO frequency along chromosomes in females and males, respectively, by merging recq4, zyp1 HEI10oe, recq4 HEI10oe, zyp1 recq4, zyp1 recq4 HEI10oe and zyp1 recq4 HEI10oe figl1. b The distribution (scaled per genome, sliding window-based, window size 300 kb, step size 50 kb) of relative precursor frequency along chromosomes in females and males, respectively, by merging recq4, zyp1 HEI10oe, recq4 HEI10oe, zyp1 recq4, zyp1 recq4 HEI10oe and zyp1 recq4 HEI10oe figl1. c The distribution (scaled per genome, sliding window-based, window size 100 kb and 300 kb, step size 50 kb) of relative precursor frequency along chromosomes in F2s, by merging females and males of recq4, zyp1 HEI10oe, recq4 HEI10oe, zyp1 recq4, zyp1 recq4 HEI10oe and zyp1 recq4 HEI10oe figl1, and F2s of recq4 figl1. The vertical dashed lines in black shows the middle position of centromeres. d Spearman’s correlation test shows the comparison with features along chromosomes, with differences in color and length according to the correlation scale. SNPs (SNPs density between Col and Ler), INV_TRANS (inversions and translocations between Col and Ler), Synteny (collinearity between Col and Ler), Genes, TEs and GC (expressed gene in meiocytes, TE and GC density), ATAC and DNase (chromatin accessibility, ATAC-seq and DNase-seq, log2(Tn5/gDNA) and log2(DNase/gDNA), in floral tissues), H3K4me1/2/3, H3K9me2, H3K27me1 (euchromatin, heterochromatin, and Polycomb histone marks, ChIP-seq, log2(ChIP/input), in flower buds), mCG, mCHG and mCHH (DNA methylation in CG, CHG, and CHH contexts, proportion methylated cytosine, in male meiocytes), MNase (nucleosome occupancy, MNase-seq, log2(MNase/gDNA), in buds). e The cumulated proportion of variation that can be explained with the features at the genome scale. The top seven most important features are colored, for which the cumulative proportion of variation that can be explained reaches the plateau. f The chromosomal distribution of observed and predicted precursor maps. The precursor profiles of individual chromosomes were predicted using profiles of the top seven most important features from the other three chromosomes. g The Spearman’s correlation test between the predicted and observed precursor distributions. The training-testing dataset is differentiated in shapes.
Genomic features can predict the CO potential
We next wondered what could shape the COP landscape. We performed correlation analysis with 17 different genomic features (Fig. 6d), including GC content, meiotically expressed genes42, transposable elements, sequence divergence between the two parental lines, chromatin accessibility, euchromatic and heterochromatic histone modification marks, DNA methylation, and nucleosome occupancy36. The distribution of COP is strongly positively correlated with euchromatic histone modification markers (H3K4me3, r = 0.85), density of expressed genes in meiocytes (r = 0.84), open chromatin (DNase, r = 0.75), and synteny (r = 0.78). Oppositely, significant negative correlations were found with DNA methylation (mCHG, r = −0.88), heterochromatic histone modification marks (H3K9me2, r = −0.86), density of TEs (r = −0.85) and inversions and translocations (r = −0.61). This result suggests that COP is favored by certain open chromatin states and disfavored by sequence divergence.
We then employed a machine-learning algorithm (random forest) to measure the capacity of the 17 features to predict the COP landscape. Overall, 94% of the genome-wide variation can be explained by a model developed with all 17 features (Fig. 6e). Among the 17 features, SNP density was assessed as the most predictive, which alone explained 6.4% of the genome-wide variation (Fig. 6e) and 28.4% of the variation along chromosome arms (Figure S13b). By adding features step by step in the order of importance, we found that the top seven and eight features can explain 93% of the genome-wide variation and 90.6% of the variation along chromosome arms, respectively (Fig. 6d, Figure S13b). To explore the predictive performance in the COP landscape of the model, we next performed 4 times cross-validation by using three chromosomes as the training dataset and one chromosome as the testing dataset (Fig. 6f, g, Figure S13c, d). Considering the top seven and eight features for genome-wide and chromosome arms, the model trained from the training dataset worked well with the testing dataset, resulting in a strong correlation (r = 0.9, Fig. 6f, g, and r = 0.86, Figure S11c, d) between the prediction and observation. The capacity of these features to accurately predict the landscape suggests that the COP might be entirely determined by (epi)genomic characteristics, including chromatin state and sequence divergence.
Discussion
Meiotic crossover frequencies are naturally limited to a few per chromosome. Here, we have shown an unprecedented elevation of meiotic crossovers in Arabidopsis by simultaneously mutating the synaptonemal complex ZYP1 and the anti-recombination helicase RECQ4. We observed an average of 34.7 and 23.5 COs per female and male zyp1 recq4 gametes, respectively. This corresponds to 58 COs per generation in zyp1 recq4, to be compared with 8.1 in wild-type and 50 in the previous champion recq4 figl13,26. The fertility is only moderately affected in zyp1 recq4 and no genomic instability was observed, opening a possibility to manipulate recombination for the benefit of plant breeding. Higher CO levels could be leveraged to enhance genetic mixing of genetic information in the early steps of breeding, reduce the size of introgression to elite lines, and facilitate the identification of genetic determinants of traits26. The mutation of RECQ4 increases CO numbers in rice, tomato, and pea43,44, suggesting that manipulation of this factor can be useful in a wide variety of crops. However, it should be noted that the mutation of some anti-crossover factors, such as FIGL1, can have little effect on fertility in Arabidopsis and a much more dramatic effect in crop species44. Similarly, the recq4 mutation does not reduce fertility in Arabidopsis, rice, and tomato but does affect fertility in peas and wheat, which may limit its usefulness44,45. Mutation of ZYP1 in rice also increased class I CO number, but a ZYP1 RNAi reduced them in Barley46,47, calling for testing the effects of zyp1, and zyp1 recq4 in more species.
Mutating both ZYP1 and RECQ4 provoked a remarkable elevation in CO numbers, beyond what was expected under the hypothesis of simple additive effects of both mutations on class I and class II COs, respectively. These massive increases correspond to class II COs, as indicated by MLH1/HEI10 not marking extra COs. This points to a function of ZYP1 in preventing class II COs, which is revealed only when RECQ4 is absent, in addition to its documented function in regulating class I COs18,19. It suggests that recombination precursors (DNA double-strand breaks) could be increased in the absence of ZYP1, as shown when the homologous protein Zip1 is defective in yeast48,49. If the same scenario happens in zyp1 recq4, the additional DSBs would produce additional recombination intermediates, that would be repaired as non-crossover in the presence of RECQ4, but converted into class II COs in its absence. Another non-exclusive possibility is a role of the synaptonemal complex in preventing the formation of aberrant recombination intermediates (e.g., Multiple invasions from a single DSB). This is supported by the slight chromosome fragmentation observed in zyp1 mus81, pointing to the role of the nuclease MUS81 in repairing recombination intermediates in the absence of ZYP1. The more extensive fragmentation observed in rice zyp1 mus8150 suggests that this function could be more prominent in this species. Finally, one may speculate a role of ZYP1 in channeling the repair of a proportion of DSBs to the sister chromatid, leading to an increase of inter-homolog intermediate in zyp1 that would synergize with the anti-CO effect of RECQ4.
Combining HEI10oe, zyp1 or recq4 two by two systematically increased CO beyond the level of the single mutants. This predicted that combining the three should increase even further CO frequencies. However, adding HEI10oe to zyp1 recq4, the double mutant with the strongest effect, did not further increase COs. Interestingly, HEI10oe did increase the number of MLH1/HEI10 foci in both sexes in zyp1 recq4, without changing the total number of COs. This suggests that overexpressing HEI10 does, as expected, increase the number of class I COs, but that this is at the expense of class II COs. This suggests that a maximum number of COs has been reached and that at these high levels, the class I and class II pathways compete for a limited number of precursors. Intriguingly, while figl1 can enhance CO frequencies in recq43,26, adding the figl1 mutation to zyp1 recq4 HEI10oe does not increase COs, but decreases the CO frequency. We suspect that in this context, aberrant recombination molecules are formed, limiting their possibility to be matured into COs. In any case, it appears that the CO frequency observed in zyp1 recq4 corresponds to some kind of maximum. This maximum corresponds to ~70 chiasmata/COs per female meiocyte (average of 35 COs per gamete) and ~48 chiasmata/COs per male meiocyte (average of 24 COs per gamete). It is possible that this does not correspond to an absolute maximum, and playing with additional factors, for example, related to chromatin state or mismatch detection, may further enhance CO frequencies51,52. Based on DMC1/RAD51 foci, it is estimated that ~200 DSBs are produced per male meiocyte53. If we consider that 1/3 are repaired on the sister (absence of bias, one sister chromatid and two homologous chromatids)54, and that the class II pathway is not biased in the repair of CO precursors (1:1 CO/NCO), we would expect ~66 COs, to compare with the estimated ~48 chiasmata in zyp1 recq4 males. If all the considerations are correct, this suggests that additional mechanisms prevent DSB maturation into COs. Intriguingly, we observed an inversion of heterochiasmy in all mutants in which class II COs are deregulated (all the genotypes with the recq4 mutation, and female zyp1 HE10oe), with more COs in females than in males. One parsimonious hypothesis is that in all contexts, including wild-type, more DSBs are formed in females than males making female meiosis in mutants more prone to class II COs, but that the CO designation process in wild-type which is linked to the length of the SC and impassive to DSB counts would lead to fewer COs in females than males.
Strikingly, in female and male meiosis of various mutants, the CO distribution converged to a common profile, leading us to propose the concept of Crossover Potential (COP), whose density map is revealed when the CO designation process is deregulated. In this view, the COP determines where COs might occur, combining the local capacity to experience DNA double-strand breaks and the availability of a viable repair template on the homologous chromosome. Accordingly, open chromatin markers (positively correlated) and sequence divergences (negatively correlated) can together efficiently predict COP. It would be interesting to establish the COP in other species with different (epi)genomic characteristics compared to Arabidopsis (e.g., higher transposon or polymorphism density) and to test if similar features can predict it. COP would be responsible for the local placement of COs in genes observed in many species55,56,57,58,59. Note that PRDM9, which drives DSB positions in many mammals, dramatically affects COP distribution60. Importantly, COP very partially dictates the final CO distribution because of the CO designation process61,62 that determines the fate of eligible recombination intermediates. A striking example is the sexual dimorphism in the hermaphrodite Arabidopsis, where female and male CO landscapes markedly differ despite an identical genome and COP map. The limited influence of COP on the global CO distribution explains why the megabase-scale CO landscape is largely independent of sequence divergence36. While COP appears to be determined at the (epi)genomic level (chromatin state and sequence divergence), the CO designation process is a chromosomal event influenced by the higher-order spatial organization in the synaptonemal complex21,63,64,65. The CO distribution is thus determined by the combination of the COP, which is particularly important at the local scale and can dismiss some regions, and the designation process that shapes the global chromosomal landscape and dictates CO counts and heterochiasmy.
Methods
Plant materials and growth conditions
Arabidopsis thaliana plants were cultivated in Polyklima growth chambers (16-h day, 21.5 °C, 280 µM; 8-h night, 18 °C: 60% humidity). The following Arabidopsis lines were used in this study: Wild type Col-0(186AV1B4) and Ler − 1 (213AV1B1) from the Versailles stock center (http://publiclines.versailles.inra.fr/). This study used zyp1-1 in Col and zyp1-6 in Ler18, recq4a in Col (recq4a-4, N419423)66, recq4a-W387X in Ler22, recq4b in Col (recq4b-2, N511330)66, figl1-19 in Col (SALK_089653), figl1-12 in Ler23 and mus81-2 in Col67. The HEI10 over-expression line is Col HEI10 line C217. Genotyping of the mutants was carried out by PCR amplification (Supplementary Table 2).
Generation of multi-mutants
The combinations of different mutants (Col/Ler hybrids) were obtained by a series of crossing schemes (Table S2 and Figs. S1–4). zyp1 HEI10oe was produced previously21. These eight different mutants and their sister wild-type control plants were backcrossed with Col plants as males and females to get the BC1 populations for sequencing and subsequent CO analysis. BC1 plant populations were grown for three weeks (16-h day/ 8-h night) and four days in the dark. 100–150 mg leaf samples from individual plantlets were collected from the BC1 populations68. Each plantlet was used as an individual sample for DNA extraction, library preparation and sequencing. The complete list of samples is provided in the Source data file of Fig. 1.
CO identification and analysis
In this study, the female and male population of wild type (95 and 89 plants), HEI10oe (48 and 46 plants), zyp1 (42 males), recq4 HEI10oe (142 and 139 plants), zyp1 recq4 (186 and 186 plants), zyp1 recq4 HEI10oe (190 and 177 plants), zyp1 recq4 HEI10oe figl1 (138 and 134 plants), were sequenced by Illumina HiSeq3000 (2 × 150 bp) conducted by the Max Planck-Genome-center (https://mpgc.mpipz.mpg.de/home/). The raw sequencing data of the female and male population of wild type (428 and 294 plants, ArrayExpress number: E-MTAB-1125436), HEI10oe (144 and 141 plants, ArrayExpress number: E-MTAB-1169621 and zyp1 (272 and 225 plants, ArrayExpress number: E-MTAB-959318, E-MTAB-1169621) from previous studies were also included in this study. In total, we analyzed female and male populations of 523 and 383 wild type, 192 and 187 HEI10oe, 272 and 267 zyp1, 142 and 138 recq4, 117 and 116 zyp1 HEI10oe, 142 and 139 recq4 HEI10oe, 186 and 186 zyp1 recq4, 190 and 177 zyp1 recq4 HEI10oe, 138 and 134 zyp1 recq4 HEI10oe figl1 plants. The number of COs, sequencing depth and source of each individual sample is provided in the Source data file of Fig. 1.
The raw sequencing data were quality-controlled using FastQC v0.11.9 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The sequencing reads were aligned to the Arabidopsis thaliana Col-0 TAIR10 reference genome69,70, using BWA v0.7.15-r114071, with default parameters. A set of Sambamba v0.6.872 commands was used for sorting and removing duplicated mapped reads. The creation of the high-confidence SNP marker list between Col and Ler, meiotic CO detection (a sliding window-based method, with a window size of 30 kb and a step size of 15 kb), A double-CO must be supported by at least five windows (total of 90 kb), and a terminal CO by at least two windows (45 kb). Check and filtering of low covered and potential contaminated samples were performed according to previous protocols18,21,36,73,74. Samples of each population were randomly selected to check predicted COs manually by inGAP-family v1.074.
The Coefficient of Coincidence (CoC) was calculated using MADpattern v1.164,75, with 10 intervals. The chromosome 4 was excluded from interference analyses because of a translocation associated with the HEI10 transgene16 and potential inversion in our Ler line18.
For CO distribution analysis, we refined the position of the marker COs of each NRZ boundary against the Col-TAIR10 reference genome used in this study. In addition, it should be noted that a translocation associated with the HEI10 C2 transgene at the short arm and a megabase-scale inversion at the long arm of chromosome 4, which may introduce bias of the CO distribution and thus chromosome 4 was excluded16.
To define hot and cold zones of CO burst from hyper recombination mutants, we examined COs in 1-Mb windows with 50-kb sliding and defined (i) the hot zones as common regions in zyp1 HEI10oe and at least 3 other mutants, with at least a two-fold increase than the median compared with wild-type, and (ii) the cold zones as common regions in zyp1 HEI10oe and at least 3 other mutants, with at most a half-fold increase than the median compared with wild-type.
For the genome-wide CO potential profile analysis by machine-learning algorithm, the chromosomal profiles of CO potential, genomic and epigenomic features were estimated in 300-kb windows with 50-kb steps along chromosomes. Then, all the random forest models were trained using randomForest v4.6-14 package in R, with the setting of “mtry=3, importance=TRUE, na.action=na.omit, ntree=2000”.
Aneuploidy screening by whole–genome sequencing
The genome was first cut into non-overlapping 100 kb windows, whose sequencing depth was estimated by Mosdepth v0.2.776 with parameters of “-n –fast-mode –by 100000”. Then, pairwise testing of sequencing depths between chromosomes of individuals was performed using the Mann–Whitney test, and the p values were adjusted using the fdr method. A pair of tested chromosomes with fold change > 1.2 and p value < 1e − 20 was considered aneuploid.
Meiocyte RNA-seq analysis
The RNA-seq dataset of Arabidopsis thaliana meiocytes from a previous study42 was downloaded from the NCBI SRA database (SRR5209212 and SRR5209213). First, quality checking of the raw sequencing reads was performed by using FastQC. Then, HISAT2 v2.1.077 was used to align the reads against the Col-0 TAIR10 reference genome. Gene expression was normalized as TPM, which was calculated by StringTie v2.0.678 with default parameters.
Cytology and image processing
Meiotic chromosome spreads were performed as previously described79. Chromosomes were stained with DAPI (2 μg/ml). Slides were observed using a Zeiss Axio Imager Z2 microscope. Images were acquired under a 100 × oil immersion objective, and processed with ZEN software. Immunolocalization performed on meiocytes with preserved three-dimensional structures was performed from 0.8–1.2 mm pistils (female meiocytes embedded in ovules) and anthers from 0.35–0.45 mm flower buds (male meiocytes), respectively18,21. Three primary antibodies were used for MLH1 foci number counting: anti-REC8 raised in rat80 (laboratory code PAK036, dilution 1:250), anti-MLH1 raised in rabbit81 (PAK017, 1:200), and anti-HEI10 raised in chicken82 (PAK046, 1:5000). Secondary antibodies (dilution 1:250) were Abberior STAR ORANGE Goat anti-rat IgG (STORANGE-1007), STAR RED Goat anti-chicken IgY (STRED-1005) and STAR GREEN Goat anti-rabbit IgG (STGREEN-1002).Images were acquired under Laica THUNDER Imager system and Abberior instrument facility line (https://abberior-instruments.com/). For Leica THUNDER Imager system, 555-nm, 635-nm and 475-nm excitation lasers were used for STAR Orange, STAR Red and STAR GREEN, respectively. Images were deconvoluted using Large Volume Computational Clearing (LVCC) mode in a Laica LAS X 5.1.0 software. For Abberior instrument facility line, images were acquired with 561- and 640-nm excitation lasers (for STAR Orange and STAR Red, respectively) and a 775-nm STED depletion laser. Confocal images were taken with the same instrument with a 485-nm excitation laser (for STAR GREEN/ Alexa488). Images were deconvoluted by Huygens Essential (version 21.10, Scientifific Volume Imaging, https://svi.nl/). Deconvoluted pictures were imported to Imaris x64 9.6.0 (https://imaris.oxinst.com/, Oxford Instruments, UK) for foci counting. The Spots module was used for MLH1 foci counting in diplotene and diakinesis cells. The vast majority of MLH foci colocalize with HEI10 foci (Table S1). Only double MLH1/HEI10 foci present on chromosomes were taken into account.
Statistics & reproducibility
No statistical method was used to predetermine sample size. No data were excluded from the analyses. The experiments were randomized by genetic segregation of mutant and wild-type alleles in plant populations. The Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The list of identified COs in the female and male populations of wild type, HEI10oe, zyp1, recq4, zyp1 HEI10oe, recq4 HEI10oe, zyp1 recq4, zyp1 recq4 HEI10oe and zyp1 recq4 HEI10oe figl1 can be accessed in Supplementary Data 1. The number of COs, sequencing depth and source of each individual sample is provided in the Source data file of Fig. 1. The raw sequencing data generated in this study have been deposited in the ArrayExpress EMBL-EBI database under accession codes E-MTAB-14424, E-MTAB-14425, E-MTAB-14426, E-MTAB-14427, E-MTAB-14428 and E-MTAB-14430. Previously published datasets, deposited in the ArrayExpress EMBL-EBI database under accession codes E-MTAB-11254, E-MTAB-11696 and E-MTAB-9593, were also used in this study. Source data are provided with this paper.
References
Zickler, D. & Kleckner, N. Meiosis: dances between homologs. Annu. Rev. Genet. 57, 1–63 (2023).
Veller, C., Kleckner, N. & Nowak, M. A. A rigorous measure of genome-wide genetic shuffling that takes into account crossover positions and Mendel’s second law. Proc. Natl Acad. Sci. 116, 1659–1668 (2019).
Fernandes, J. B., Séguéla-Arnaud, M., Larchevêque, C., Lloyd, A. H. & Mercier, R. Unleashing meiotic crossovers in hybrid plants. Proc. Natl Acad. Sci. 115, 2431–2436 (2018).
Lian Q., Maestroni L., Gaudin M., Llorente B., Mercier R. Meiotic recombination is confirmed to be unusually high in the fission yeast Schizosaccharomyces pombe. iScience 26, 107614 (2023).
Lange, J. et al. The landscape of mouse meiotic double-strand break formation, processing, and repair. Cell 167, 695–708.e616 (2016).
Moens, P. B., Marcon, E., Shore, J. S., Kochakpour, N. & Spyropoulos, B. Initiation and resolution of interhomolog connections: crossover and non-crossover sites along mouse synaptonemal complexes. J. Cell Sci. 120, 1017–1027 (2007).
Baudat, F. & de Massy, B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 15, 565–577 (2007).
Cole, F., Keeney, S. & Jasin, M. Comprehensive, fine-scale dissection of homologous recombination outcomes at a hot spot in mouse meiosis. Mol. cell 39, 700–710 (2010).
Mercier, R., Mezard, C., Jenczewski, E., Macaisne, N. & Grelon, M. The molecular biology of meiosis in plants. Annu Rev. Plant Biol. 66, 297–327 (2015).
Muller, H. J. The mechanism of crossing-over. Am. Naturalist 50, 193–221 (1916).
Otto, S. P. & Payseur, B. A. Crossover interference: shedding light on the evolution of recombination. Annu. Rev. Genet. 53, 19–44 (2019).
Zickler, D. & Kleckner, N. A few of our favorite things: pairing, the bouquet, crossover interference and evolution of meiosis. Semin Cell Dev. Biol. 54, 135–148 (2016).
Girard C., Zwicker D., Mercier R. The regulation of meiotic crossover distribution: a coarse solution to a century-old mystery? Biochem Soc Trans, 51, BST20221329 (2023).
Lenormand, T. The Evolution of Sex Dimorphism in Recombination. Genetics 163, 811–822 (2003).
Hunter N. et al. Meiotic recombination: the essence of heredity. Cold Spring Harbor perspectives in biology 7, a016618 (2015).
Serra, H. et al. Massive crossover elevation via combination of HEI10 and recq4a recq4b during Arabidopsis meiosis. Proc. Natl Acad. Sci. 115, 2437–2442 (2018).
Ziolkowski, P. A. et al. Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination. Genes Dev. 31, 306–317 (2017).
Capilla-Pérez, L. et al. The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis. Proc. Natl Acad. Sci. 118, e2023613118 (2021).
France, M. G. et al. ZYP1 is required for obligate cross-over formation and cross-over interference in Arabidopsis. Proc. Natl Acad. Sci. 118, e2021671118 (2021).
Vrielynck, N. et al. SCEP1 and SCEP2 are two new components of the synaptonemal complex central element. Nat. Plants 9, 2016–2030 (2023).
Durand, S. et al. Joint control of meiotic crossover patterning by the synaptonemal complex and HEI10 dosage. Nat. Commun. 13, 5999 (2022).
Séguéla-Arnaud, M. et al. Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc. Natl Acad. Sci. 112, 4713–4718 (2015).
Girard, C. et al. AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLOS Genet. 11, e1005369 (2015).
Kumar, R., Duhamel, M., Coutant, E., Ben-Nahia, E. & Mercier, R. Antagonism between BRCA2 and FIGL1 regulates homologous recombination. Nucleic Acids Res 47, 5170–5180 (2019).
Fernandes J. B. et al. FIGL1 and its novel partner FLIP form a conserved complex that regulates homologous recombination. 14, e1007317 (2018).
Capilla-Pérez, L. et al. Enhanced recombination empowers the detection and mapping of Quantitative Trait Loci. Commun. Biol. 7, 829 (2024).
Zhu, L. et al. Natural variation identifies SNI1, the SMC5/6 component, as a modifier of meiotic crossover in Arabidopsis. Proc. Natl Acad. Sci. 118, e2021970118 (2021).
Kim J. & Park J. Arabidopsis heat shock factor binding protein is required to limit meiotic crossovers and HEI10 transcription. 41, e109958 (2022).
Seguela-Arnaud, M. et al. RMI1 and TOP3α limit meiotic CO formation through their C-terminal domains. Nucleic acids Res. 45, 1860–1871 (2017).
Nageswaran D. C. et al. High Crossover Rate1 encodes protein phosphatase X1 and restricts meiotic crossovers in Arabidopsis. 7, 452–467 (2021).
Kim, H. et al. Control of meiotic crossover interference by a proteolytic chaperone network. Nat. Plants 10, 453–468 (2024).
Li, X. et al. Regulation of interference-sensitive crossover distribution ensures crossover assurance in Arabidopsis. Proc. Natl Acad. Sci. USA 118, e2107543118 (2021).
Chelysheva, L. et al. An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenet Genome Res 129, 143–153 (2010).
Duroc, Y. et al. The kinesin AtPSS1 promotes synapsis and is required for proper crossover distribution in meiosis. PLoS Genet 10, e1004674 (2014).
Tease, C. Cytological detection of crossing-over in BUdR substituted meiotic chromosomes using the fluorescent plus Giemsa technique. Nature 272, 823–824 (1978).
Lian, Q. et al. The megabase-scale crossover landscape is largely independent of sequence divergence. Nat. Commun. 13, 3828 (2022).
Giraut, L. et al. Genome-Wide Crossover Distribution in Arabidopsis thaliana Meiosis Reveals Sex-Specific Patterns along Chromosomes. PLOS Genet. 7, e1002354 (2011).
Lenormand, T. & Dutheil, J. Recombination difference between sexes: a role for haploid selection. PLOS Biol. 3, e63 (2005).
Crismani, W. et al. FANCM limits meiotic crossovers. Science 336, 1588–1590 (2012).
Singh, D. K. et al. The FANCC-FANCE-FANCF complex is evolutionarily conserved and regulates meiotic recombination. Nucleic Acids Res 51, 2516–2528 (2023).
Fernandes, J. B. et al. Structural variation and DNA methylation shape the centromere-proximal meiotic crossover landscape in Arabidopsis. Genome Biol. 25, 30 (2024).
Huang, J. et al. Meiocyte-Specific and AtSPO11-1-Dependent Small RNAs and Their Association with Meiotic Gene Expression and Recombination. Plant Cell 31, 444–464 (2019).
de Maagd, R. A. et al. CRISPR/Cas inactivation of RECQ4 increases homeologous crossovers in an interspecific tomato hybrid. Plant Biotechnol. J. 18, 805–813 (2020).
Mieulet, D. et al. Unleashing meiotic crossovers in crops. Nat. Plants 4, 1010–1016 (2018).
Bazile, J. et al. TaRECQ4 contributes to maintain both homologous and homoeologous recombination during wheat meiosis. Front Plant Sci. 14, 1342976 (2023).
Wang, M. et al. The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22, 417–430 (2010).
Barakate, A. et al. The synaptonemal complex protein ZYP1 is required for imposition of meiotic crossovers in barley. Plant Cell 26, 729–740 (2014).
Lee, M.-S. et al. The synaptonemal complex central region modulates crossover pathways and feedback control of meiotic double-strand break formation. Nucleic acids Res. 49, 7537–7553 (2021).
Mu, X., Murakami, H., Mohibullah, N. & Keeney, S. Chromosome-autonomous feedback down-regulates meiotic DNA break competence upon synaptonemal complex formation. Genes Dev. 34, 1605–1618 (2020).
Mu, N. et al. MUS81 is required for atypical recombination intermediate resolution but not crossover designation in rice. N. Phytologist 237, 2422–2434 (2023).
Underwood, C. J. et al. Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation. Genome Res. 28, 519–531 (2018).
Blackwell A. R., et al. MSH2 shapes the meiotic crossover landscape in relation to interhomolog polymorphism in Arabidopsis. EMBO J. 21, e104858 (2020).
Kurzbauer, M.-T. et al. ATM controls meiotic DNA double-strand break formation and recombination and affects synaptonemal complex organization in plants. Plant Cell 33, 1633–1656 (2021).
Goldfarb, T. & Lichten, M. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLOS Biol. 8, e1000520 (2010).
Choi, K. et al. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 45, 1327–1336 (2013).
Mancera, E., Bourgon, R., Brozzi, A., Huber, W. & Steinmetz, L. M. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454, 479–485 (2008).
Zhang H., Lundberg M., Ponnikas S., Hasselquist D. & Hansson B. Male-biased recombination at chromosome ends in a songbird revealed by precisely mapping crossover positions. G3 Genes|Genomes|Genetics, 14, jkae150 (2024).
Singhal, S. et al. Stable recombination hotspots in birds. Science 350, 928–932 (2015).
Johnston, S. E. Understanding the genetic basis of variation in meiotic recombination: past, present, and future. Mol. Biol. Evolution 41, msae112 (2024).
Paigen, K. & Petkov, P. M. PRDM9 and its role in genetic recombination. Trends Genet.: TIG 34, 291–300 (2018).
Bishop, D. K. & Zickler, D. Early decision: meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117, 9–15 (2004).
Gray, S. & Cohen, P. E. Control of meiotic crossovers: from double-strand break formation to designation. Annu Rev. Genet 50, 175–210 (2016).
Fozard, J. A., Morgan, C. & Howard, M. Coarsening dynamics can explain meiotic crossover patterning in both the presence and absence of the synaptonemal complex. Elife 12, e79408 (2023).
Zhang, L., Liang, Z., Hutchinson, J. & Kleckner, N. Crossover patterning by the beam-film model: analysis and implications. PLOS Genet. 10, e1004042 (2014).
Wang, T. et al. HEI10 is subject to phase separation and mediates RPA1a degradation during meiotic interference-sensitive crossover formation. Proc. Natl Acad. Sci. USA 120, e2310542120 (2023).
Hartung, F., Suer, S. & Puchta, H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 104, 18836–18841 (2007).
Berchowitz, L. E., Francis, K. E., Bey, A. L. & Copenhaver, G. P. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 3, e132 (2007).
Rowan, B. A., Patel, V., Weigel, D. & Schneeberger, K. Rapid and Inexpensive Whole-Genome Genotyping-by-Sequencing for Crossover Localization and Fine-Scale Genetic Mapping. G3 Genes|Genomes|Genet. 5, 385–398 (2015).
Lamesch, P. et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic acids Res. 40, D1202–D1210 (2012).
The Arabidopsis Genome I. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Tarasov, A., Vilella, A. J., Cuppen, E., Nijman, I. J. & Prins, P. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034 (2015).
Qi, J., Chen, Y., Copenhaver, G. P. & Ma, H. Detection of genomic variations and DNA polymorphisms and impact on analysis of meiotic recombination and genetic mapping. Proc. Natl Acad. Sci. 111, 10007–10012 (2014).
Lian, Q., Chen, Y., Chang, F., Fu, Y. & Qi, J. inGAP-family: accurate detection of meiotic recombination Loci and causal mutations by filtering out artificial variants due to genome complexities. Genomics, Proteom. Bioinforma. 20, 524–535 (2022).
White M. A., Wang S., Zhang L. & Kleckner N. Quantitative Modeling And Automated Analysis Of Meiotic Recombination. In: Meiosis (ed Stuart D. T.). Springer New York (2017).
Pedersen, B. S. & Quinlan, A. R. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 34, 867–868 (2018).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Kovaka, S. et al. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20, 278 (2019).
Cromer L., et al. Patronus is the elusive plant securin, preventing chromosome separation by antagonizing separase. 116, 16018–16027 (2019).
Cromer, L. et al. Centromeric cohesion is protected twice at meiosis, by SHUGOSHINs at anaphase I and by PATRONUS at interkinesis. Curr. Biol. 23, 2090–2099 (2013).
Chelysheva, L. et al. An easy protocol for studying chromatin and recombination protein dynamics during arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenetic genome Res. 129, 143–153 (2010).
Chelysheva, L. et al. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet 8, e1002799 (2012).
Acknowledgements
This work was supported by core funding from the Max Planck Society, and Alexander von Humboldt Fellowships to Q.L. and J.J. We thank Ian Henderson for kindly providing the HEI10 C2 line.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
R.M. lead the project. J.J. produced the genetic material, analyzed the cytology experiments and fertility (wild type, zyp1, recq4, zyp1 recq4, zyp1 HEI10oe, recq4 HEI10oe, zyp1 recq4 HEI10oe and zyp1 mus81). Q.L. analyzed the sequencing data and performed recombination, interference, and aneuploidy analyses. S.D. developed the protocol for female immunolocalization, produced the genetic material and analyzed the cytology experiments and fertility (zyp1 recq4 HEI10oe figl1). J.J., Q.L., and R.M. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing interests: A Patent was granted to Institut National de la Recherge Agronomique (INRA) on the use of RECQ4 to manipulate meiotic recombination in crops, with RM listed among the inventors (US10,920,237/EP3149027). A patent was filed by the Max Planck Society on the combined use of RECQ4 and ZYP1 to manipulate recombination in crops, with R.M. J.J., Q.L., and S.D. listed as inventors (EP23179262. 14.06.2023).
Peer review
Peer review information
Nature Communications thanks Yan He, Yingxiang Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jing, J., Lian, Q., Durand, S. et al. Maximizing meiotic crossover rates reveals the map of Crossover Potential. Nat Commun 16, 5306 (2025). https://doi.org/10.1038/s41467-025-60663-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-60663-y