Abstract
KPC and NDM co-producing carbapenem-resistant Klebsiella pneumoniae (KN-CRKP) showed an upward trend; nevertheless, global systematic and comprehensive analyses profiling remain lacking. 968 local CRKP were collected from 6 provinces in China, and 64,354 genomes were retrieved from GenBank. All 413 genomes of KN-CRKP were obtained from 32 countries, including 16 subtypes of KN-CRKP. The top three CRKP subtypes, K2N1-CRKP, K2N5-CRKP and K3N1-CRKP, exhibited distinct geographic distributions, with K2N1-CRKP and K2N5-CRKP primarily circulating in China while K3N1-CRKP showed predominant prevalence in USA. Meanwhile, ST11-KL64, ST11-KL47, and ST258-KL107, were the three most prevalent ST and KL, and 64.3% of ST11-KL64 KN-CRKP belonged to hypervirulent strains. Genomes revealed Clone Group 1, accounting for 55.0% of KN-CRKP, shifting from KL47 to KL64 and carrying more hypervirulence genes, has a significant advantage in adhesion, invasion, and proliferation, and its dispersal was the primary driver contributing to the worldwide spread of KN-CRKP. Furthermore, ST11 KN-CRKP was generally formed by KPC-producing CRKP acquiring blaNDM-carrying plasmid and novel hybrid plasmids co-encoding KPC and NDM have occurred. Aztreonam/avibactam and cefiderocol were promising antimicrobial agents against KN-CRKP. The global KN-CRKP research, spanning from 2005 to 2024, provides valuable insights into the global transmission, dynamics, and treatment of KN-CRKP.
Similar content being viewed by others
Introduction
Carbapenem-resistant Klebsiella pneumoniae (CRKP), which the World Health Organization has listed as a high-priority pathogen (https://www.who.int/publications/i/item/9789240093461) in 2024, belonging to carbapenem-resistant Enterobacterales (CRE), has emerged and circulated worldwide, causing infections with high mortality rates that present a crucial public health threat1,2,3. The pivotal mechanism involving carbapenem resistance is the production of carbapenemases, of which K. pneumoniae carbapenemase (KPC) and New Delhi metallo-β-lactamase (NDM), encoded by blaKPC and blaNDM, respectively, are the two most prevalent types globally, apart from OXA-48-like4,5. The blaKPC-carrying CRKP (KPC-CRKP) was the most common and focused on multi-locus sequence type (MLST) ST258, ST512, and ST11 in USA2, Europe5, and China6, respectively, whereas the blaNDM-carrying CRKP (NDM-CRKP) was the second common CRKP but with more dispersed STs6,7. Significantly, KPC and NDM-co-producing CRKP (KN-CRKP) have been increasingly documented in recent years, mostly in individual strains, especially in China, which exhibits pan-β-lactam resistance and results in few therapeutic alternatives8,9,10.
Studies show that KN-CRKP involves several subclasses, including blaKPC-2- and blaNDM-1-co-carrying CRKP (K2N1-CRKP), blaKPC-2- and blaNDM-5-co-carrying CRKP (K2N5-CRKP) and blaKPC-3- and blaNDM-1-co-carrying CRKP (K3N1-CRKP), as having caused sporadic outbreaks in hospitals11,12. Importantly, compared to patients infected with blood-borne KPC-CRKP, patients infected with blood-borne KN-CRKP showed a markedly higher 30-day mortality rate (56.0% vs 32.5%)10.
According to the analysis of publicly available sequences of KPC-producing CRE and NDM-producing CRE, KPC- and NDM-co-producing CRE accounted for 0.7% of CRE, and 69.1% of these belonged to KN-CRKP13. Moreover, the proportion of KN-CRKP in CRKP has risen from 0 in 2010 to 4.4% in 2021 in another global study14. Likely, a multi-center, longitudinal study conducted in China on CRKP revealed the proportion of KN-CRKP was 1.2% in 1017 CRKP between 2016 and 202015.
Previous investigation revealed blaKPC and blaNDM were mainly located on separate plasmids, and KN-CRKP was generally formed by that a KPC-CRKP strain acquired blaNDM-harboring plasmid, based on plasmid transferability and homology8. Furthermore, blaKPC- and blaNDM-co-existing plasmids of CRKP have been detected to be capable of being stably transmitted, alerting great compatibility for blaKPC and blaNDM16. With the spread and evolution of KN-CRKP, it will pose a great challenge to public health and clinical treatment. More in-depth forming mechanisms remain to be thoroughly investigated.
The prevalence of KN-CRKP from various public genomes recently has been described simply in several studies13,14,15, however, global systematic and comprehensive analyses profiling of KN-CRKP, remain undetermined. In the current study, we conducted an extensive analysis for global KN-CRKP (413 genomes) to understand their epidemiology, resistance and virulence characterizations, dissemination and cloning, evolutionary mechanisms, and novel β-lactam-β-lactamase inhibitor (BL/BLI) combinations providing a new perspective on the development of effective approaches to control infections with KN-CRKP.
Results
Distribution and subtypes diversity of global KN-CRKP strains
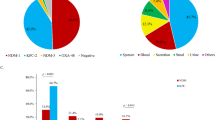
To investigate the prevalence and distribution of KN-CRKP, global genome data were retrieved from the NCBI GenBank database during 1980- 2024 (Fig. 1A). A total of 388 non-duplicate KN-CRKP genomes, spanning 32 countries worldwide and 22 provinces of China, were obtained from 2005 to 2024. Among the 388 KN-CRKP, China accounted for the highest number (43.0%, 167/388), followed by the United States (25.0%, 97/388), Brazil (6.7%, 26/388), Greece (3.6%, 14/388), and Vietnam (3.4%, 13/388). In China, KN-CRKP strains were found mostly in Zhejiang (19.2%, 32/167), Beijing (13.8%, 23/167), Shandong (13.2%, 22/167), Jiangsu (12.6%, 21/167) and Hubei (7.8%, 13/167). According to the current study, the first KN-CRKP strain XL-20059286, belonging to ST11-KL64 and co-producing blaKPC-2 and blaNDM-1, was discovered in 2005 in Zhejiang, China. The ratio of KN-CRKP globally in K. pneumoniae significantly increased from 0.03% to 3.10% during 2014–2023, especially from 2017 to 2023 (Fig. 1D). However, the number of KN-CRKP presented a declining trend in China during 2021–2022 (Fig. 1D), that this is probably due to the COVID-19 pandemic and subsequent control policies. 13 different subtypes were identified in KN-CRKP, according to various combinations of the blaKPC and blaNDM types that already existed in this study. The most prevalent subtypes were K2N1-CRKP (53.9%, 209/388), followed by K2N5-CRKP (18.8%, 73/388) and K3N1-CRKP (14.9%, 58/388) (Fig. 1C). K2N1-CRKP and K3N1-CRKP were the most prevalent subtypes in China and USA, respectively. Meanwhile, K2N5-CRKP showed a rising tendency (Fig. 1B).
A Distribution of 388 KN-CRKP strains globally between 2005 and 2024. The pie charts show the top five countries (left) and provinces (right, in China) with the most separation B Percentage variations of carbapenemase genes from 2005 to 2024 in 167 strains KN-CRKP from China on Genbank. Source data are provided as a Source Data file. C Carbapenemase genes and their proportion of 388 strains KN-CRKP worldwide. D Number of global and China’s KN-CRKP strains between 2005 and 2024. Source data are provided as a Source Data file. E Carbapenemase genes and number of local KN-CRKP. Abbreviations: KN-CRKP, KPC and NDM co-producing carbapenem-resistant Klebsiella pneumoniae.
In the process of CRE surveillance for 6 provinces in China between 2017 and 2024, 25 KN-CRKP isolates were retrieved and accounted for 2.6% of the 968 non-duplicate CRE isolates (Fig. 1E). blaKPC-2- and blaNDM-13-co-carrying CRKP (K2N13-CRKP, 10 strains) comprised 40.0% of the 25 KN-CRKP, followed by K2N1-CRKP (36.0%, 9/25), K2N5-CRKP (12.0%, 3/25), blaKPC-33- and blaNDM-1-co-carrying CRKP (K33N1-CRKP) (8.0%, 2/25) and blaKPC-14- and blaNDM-1-co-carrying CRKP (K14N1-CRKP) (4.0%, 1/25). To our knowledge, we first reported K2N13-CRKP, K14N1-CRKP, and K33N1-CRKP in K. pneumoniae. Furthermore, the in-hospital mortality rate among patients with KN-CRKP infections was 40.0% (10/25).
In total, 413 KN-CRKP strains were identified, 388 from global data and 25 from our surveillance, and related comprehensive analysis was conducted below (Supplementary Figs. 1, 11, Supplementary Data Dataset 1). Additionally, respiratory tract samples (27.1%, 83/306), urine (20.3%, 62/306), and blood (20.3%, 62/306) were the top three sample types from patients among the 306 known clinical isolates of KN-CRKP, and unexpectedly, 8.8% (27/306) of them was from nonclinical samples.
Resistance- and virulence-associated gene carriage in KN-CRKP
Apart from carbapenemase genes blaKPC and blaNDM, blaOXA-48, blaOXA-181, blaVIM-1, blaVIM-24, and blaIMP-4 also were detected in 8, 5, 2, 2, and 1 K2N1-CRKP strains, respectively, which indicated K2N1-CRKP has a great compatibility, compared to other KN-CRKP subtypes. Moreover, blaCTX-M-65 (30.0%, 124/413), blaCTX-M-15 (26.6%, 110/413) and blaSHV-12 (16.9%, 70/413) belonged to extended-spectrum beta-lactamases (ESBLs) genes, of which carrying rate was 60.8% (251/413) in KN-CRKP strains. aadA2 (51.1%, 211/413), rmtB (37.5%, 155/413), aac (6’)-Ib-cr (29.3%, 121/413), aph3-Ia (21.1%, 87/413), aac(3)-IId (17.2%, 71/413) and armA (12.8%, 53/413) mediated aminoglycosides resistance, and the carriage rate of aminoglycosides-resistant genes in KN-CRKP was 98.3% (406/413). Sulfonamides-resistant gene sul, quinolones-resistant gene qnrS1, tetracyclines-resistant gene tet(A) and fosfomycin-resistant gene fosA3 occurred in 84.0% (347/413), 37.3% (154/413), 36.1% (149/413), and 14.0% (58/413) KN-CRKP strains, respectively. The mutations of membrane protein OmpK35 (OmpK35-17%, 38.7%, 160/413) and OmpK36 (OmpK36GD, 53.0%, 219/413) also hinder the entrance of carbapenems, raising the degree of resistance to carbapenems17. Of the 413 KN-CRKP strains, 5 of them carried the mobile colistin resistance genes mcr-1.1 and mcr-9.1, respectively. Concurrently, 58 truncated mgrB and 4 truncated pmrB also were identified in the 413 KN-CRKP strains. Given the above genes, 15.7% (65/413) KN-CRKP carried colistin resistance genes. However, the percentage of KN-CRKP that carried tigecycline resistance gene tmexCD-toprJ was only 4.4% (18/413).
Hypervirulence-associated genes, rmpA (7.0%, 29/413), rmpA2 (23.0%, 95/413), iroBCDN (1.5%, 6/413), iutAiucABCD (27.8%, 115/413) and peg344 (17.9%, 74/413), were identified in KN-CRKP, and 7.0% (29/413) KN-CRKP strains simultaneously contained the above 4 different hypervirulence genes except for iroBCDN. 23.0% KN-CRKP strains (95/413) carrying both iucA and rmpA/rmpA2 were classified as hypervirulent KN-CRKP (hv-KN-CRKP). Additionally, the type VI secretion system (T6SS), usually containing 13 core components and mediating hostile or collaborative communications either intra- and inter-bacteria or between bacteria and eukaryotes, was closely relevant to the hypervirulence of CRKP. All KN-CRKP strains included 9 core components, TssA, TssE, TssF, TssG, TssI, TssJ, TssK, TssL and TssM, and 90.6% KN-CRKP (374/413) possessed 13 complete core components (Supplementary Fig. 2).
Enrichment of genomic characterizations by MLST and KL
A total of individual 61 STs were distinguished by blasting 7 housekeeping genes in 413 KN-CRKP strains (Supplementary Fig. 3A). 73.4% of strains were represented by 8 main STs, namely, ST11 (42.4%, 175/413), ST258 (7.0%, 29/413), ST147 (5.3%, 22/413), ST16 (5.1%, 21/413), ST22 (3.6%, 15/413), ST512 (3.6%, 15/413), ST39 (3.1%, 13/413) and ST307 (3.1%, 13/413), whereas the remaining 53 STs scattered in 110 strains (Supplementary Fig. 4). MST showed ST of KN-CRKP strains exhibited differences in geographical and temporal distribution (Supplementary Fig. 5A, B). ST11, ST258, ST147, ST15 and ST22 were observed to occur in distinct epidemiological years, implying the possibility of dissemination. In China, ST11 was distinctly prominent, accounting for 79.2% (152/192), followed by ST22 (7.3%, 14/192) and eleven more. In USA, 35 STs were distinguished in 97 stains, in which ST258 (26.8%, 26/97) and ST147 (13.4%, 13/97) were the two most common types.
Capsular and lipopolysaccharide O antigen genotyping showed 38 different K loci (KLs) and 9 different O loci (OLs) were distinguished (Supplementary Fig. 3B, C). The three most prevalent KLs were KL64 (28.3%, 117/413), KL47 (11.3%, 47/413) and KL107 (9.9%, 41/413), and the three most prevalent OLs were O1/O2v1 (45.3%, 187/413), O1/O2v2 (27.8%, 115/413) and OL101 (10.2%, 42/413), respectively. All OLs of KL64 strains were O1/O2v1. Likely, the most common OL of KL107 and KL47 strains was O1/O2v2 (90.2%, 37/41) and OL101 (89.4%, 42/47) respectively. KL47 and KL64 strains were mainly distributed in China, and KL64 KN-CRKP strains have a significant increase among 2021–2023, compared with KL47 strains (Supplementary Fig. 4B). However, KL107 strains mainly were isolated from USA (63.4%, 26/41).
To better know the enrichment of KN-CRKP in ST and KL, carbapenemase type, countries, and hypervirulent genes of KN-CRKP were presented by scatter plots with box (Fig. 2A–C). ST11-KL64 (23.7%, 98/413) was the most prominent ST-KL type, followed by ST11-KL47 (11.4%, 47/413) and ST258-KL107 (6.3%, 26/413). All 98 ST11-KL64 KN-CRKP strains, isolated from China (96.9%, 95/98) and Uruguay (2.0%, 2/98), mainly co-produced KPC-2 and NDM-1 (61.2%, 60/98), and KPC-2 and NDM-5 (24.5%, 24/98), and 64.3% of those (63/98) was defined as hypervirulent strains, accounting for 66.3% of all hv-KN-CRKP. Among the ST11-KL47 CRKP strains isolated in China, 65.9% (29/44) co-produced KPC-2 and NDM-5 carbapenemases, with only five strains exhibiting hypervirulent phenotypes. By contrast, 84.6% of ST258-KL107 strains (22/26) co-produced KPC-3 and various NDM. 96.2% (25/26) of ST258-KL107 strains were from USA, and only one strain was hypervirulent. Therefore, ST11-KL64 KN-CRKP strains with hypervirulent genes were the most common KN-CRKP and mainly from China, posing a great threat to human health.
Data shown presented the distribution of carbapenemase genes (A), country (B), hypervirulent genes (C) and Clone group (D) by ST and KL. Each circle represented a genome colored based on the different types of the four classes. Barplots summarize the number of genomes from each KL (top) and ST (right) and are colored based on the different types of the four classes. Gray means absence. Source data are provided as a Source Data file. Abbreviations: CG Clone Group, MLST Multi-locus sequence type, KL K loci.
Transmission and cloning of KN-CRKP
RhierBAPS revealed that 413 KN-CRKP strains were divided into 11 distinct clades (Clade 1–11, Fig. 3). Clade 1 (55.2%, 228/413) was the most prevalent, followed by Clade 2 (10.9%, 45/413) and Clade 5 (9.9%, 41/413). Whole genome SNPs analysis exhibited the KN-CRKP sequences were classified as 22 clone groups (CG 1-22, Supplementary Figs. 3D, 6 and 7). CG 1, 3, 4, 5, 7, 11, and 20 occurred in China while CG1 and the other 15 CGs in USA. In the local KN-CRKP, K32335, K78844 and KGX21 were assigned to CG4, CG10 and CG5, respectively, while the remaining 22 strains were assigned to CG1. Furthermore, ST and KL of CG1 strains mostly concentrate on ST11-KL64 (43.2%, 98/227), ST11-KL47 (20.7%, 47/227), ST258-KL107(11.5%, 26/227) and ST512 -KL107 (6.6%, 15/227) (Fig. 2D). Interestingly, strains from CG1 (55.0%, 227/413) were found in 16 countries, including China (68.3%, 155/227) and USA (13.3% 28/227), where strains were highly consistent with those of Clade 1 (both have 227 identical strains). CG1 has an overwhelming proportion relative to other groups (Supplementary Fig. 3D), suggesting that the expansion of CG1 strains was the main driver of the rise in KN-CRKP strains.
The phylogenetic tree was annotated by clade, Clone group, carbapenemase types, ST, serotype, virulence genes, sample, year, and country from inner to outer, separately. Different colors of branches denote different Clone groups. Abbreviations: CG Clone Group, MLST Multi-locus sequence type, KN-CRKP KPC and NDM co-producing carbapenem-resistant Klebsiella pneumoniae.
To further trace the origin and dissemination of CG1 strains, dated phylogeny and phylogeography were drawn (Fig. 4A). CG1 strains likely first emerged in Brazil in 1954 (95% highest posterior density [HDP] interval 1940–1968), and then spread to other continents, including Europe, Asia, and North America (Fig. 4D). The correlation coefficient and R2 for the root-to-tip genetic divergence against time were both 1, suggesting a strong linear relationship between accumulated mutations and sampling time (Fig. 4B). Therefore, under a strict clock model, bayesian phylogenetic analysis to estimate the substitution rate was performed. The median molecular clock rate was estimated to be 3.8001 × 10−4 (95% HPD interval 3.1436–4.4749 × 10−4) substitutions/site/year. According to a Bayesian skyline plot (BSP), strains of CG1 rose before 2010, and declined twice as quickly by the end of 2020 and in 2015—two periods of dramatic declines that may have been caused by COVID-19 and the Zika virus, respectively, and then start gradually to rise again in 2022 (Fig. 4C). Moreover, the prominent serotype KL64 strains of CG1 had occurred in China since 1993 and gradually became the prominent serotype, whereas the prominent serotype of CG1 remained KL107 in USA (Fig. 4A).
A Dated phylogeny of CG1 strains was illustrated and blue bars along branches denoted 95% highest posterior probabilities. The color of the tipnode and tiplab represented different countries and serotypes, respectively. B Regression of root-to-tip distance against sampling time showing temporal signal in CG1. Source data are provided as a Source Data file. C. Effective population size of CG1 strains based on the population structure. Shaded areas showed 95% highest posterior probabilities. D Phylogeography analyses of CG1 strains. Arrows showed the direction and dates of transmission. KN-CRKP isolate originated in Brazil, and then spread to the rest of the world. The radius of the blue circles, which indicate the main outbreak zones, corresponds to the total number of strains examined in each country. Abbreviations: CG Clone Group, KN-CRKP KPC and NDM co-producing carbapenem-resistant Klebsiella pneumoniae.
To evaluate the differences in core genes and associated function between CG1 and non-CG1, pan-genome analysis was performed for 413 KN-CRKP strains (Fig. 5A). The strains belonging to CG1 and the other strains were easily separated in the evolutionary tree. Pan-genome analysis showed the core genes and accessory genes were 3224 and 31860, respectively, and CG1 and non-CG1 have a distinct difference in core genes. Moreover, the pan-genome is closed and representative, as shown by the exponential value of the fitted curve being less than 0.5. The number of shared core genes between CG1 and non-CG1 was 2378, and the remaining core genes of CG1 was 1433. The 1433 genes were subjected to KEGG pathway enrichment to find out why CG1 was prominent (Fig. 5B). KEGG analysis suggested the genetic differences of CG1 were mainly enriched in carbohydrate metabolism, amino acid metabolism, cellular community and membrane transport (p < 0.05), involving in the survival of bacteria, growth and adaptation, compared with non-CG1. Moreover, genes related to signal transduction, signaling and cellular processes in CG1 outnumbered that of non-CG1, which contributed to adhesion, invasion and proliferation of bacteria, such as fimbria adhesin gene ecpD and iron transport-related gene cirA.
A Pan-genome analysis of 413 KN-CRKP by roary. The cells with blue color represent the presence of genes. The red box and Venn diagram indicated the difference in core genes between CG1 (n = 227) and non-CG1 (n = 186). Source data are provided as a Source Data file. As the number of genomes increased, the number of gene clusters was displayed at the top right. Means and standard errors (vertical line) were obtained by repeating 100 times random input orders of the genomes. The continuous curves were generated by the least-squares fit. B Differential gene function annotation of the top 30 listed in Clone group 1. Level 2 pathways were annotated with different colors, and the number of level 3 pathways was shown by step height. Abbreviations: CG Clone Group, KN-CRKP KPC and NDM co-producing carbapenem-resistant Klebsiella pneumoniae.
Dissemination and evolution of local KN-CRKP
To determine whether propagation and evolution happened in local KN-CRKP strains, the network of pairwise alignments was performed by using ANI (Fig. 6A). Three propagation cases of identical strains were also discovered in the same hospital. The first case of dissemination occurred in 2018, involving three strains, K11253, K11312 and K67367, isolated from three patients from three distinct wards on different days. The second propagations were detected in ten patients from various wards in the same hospital spanning a ten-month period. The ten strains involved in this dissemination, carrying blaKPC-2 and blaNDM-13, were initially reported by us in another article18. The concerned strains of the most recent propagations were K64102 and K67034, simultaneously carrying blaKPC-33 and blaNDM-1, which appeared in two lung transplantation recipients from the same wards within approximately three months. Additionally, all strains from the second and last propagations, belonging to CG 1, contained hypervirulent plasmids and were designated as hv-KN-CRKP.
A Networks of local KN-CRKP strains and their homologous strains B Networks of blaKPC- and blaNDM-carrying plasmids. C Phylogenetic analysis of blaKPC- or blaNDM-carrying plasmids harboured by KN-CRKP. Plasmids of the phylogenetic trees from left to right were blaKPC-carrying plasmids and blaNDM-carrying plasmids, respectively. The blue lines on the tree indicated tree scale, and the shades of tiplab were the plasmids co-carrying blaKPC and blaNDM. Connections of the two trees denoted the two separated plasmids located on the same genomes of KN-CRKP. D Formation mode of ST11 KN-CRKP. Created in BioRender. Mato, F. (2025) https://BioRender.com/dc61lmf. Abbreviations: CG Clone Group, MLST Multi-locus sequence type, KN-CRKP KPC and NDM co-producing carbapenem-resistant Klebsiella pneumoniae; KPC-CRKP, blaKPC-carrying CRKP.
To investigate the origin of local KN-CRKP strains, strains of the same genus from the same patients were analyzed, and the potential strains were sequenced by NGS. Four ST11 KN-CRKP strains, K55475, K58559, K64102, and K68336, could be traced back to the blaKPC-carrying originating orthologs from the same patients with the administration of ceftazidime/avibactam. Moreover, the difference in genome and replicons revealed the four KN-CRKP strains were generated by acquiring blaNDM-carrying plasmid based on KPC-CRKP (Supplementary Fig. 8, Supplementary Data Dataset 2). Growth curve revealed there was no significant difference in fitness cost between KN-CRKP and the relative KPC-CRKP, except for blaKPC-33- and blaNDM-1-co-carrying K64102 (Supplementary Fig. 9).
Characterization of bla KPC- or bla NDM-carrying plasmids in KN-CRKP
79 KN-CRKP strains, including 71 strains from China and 47 ST11 strains, have complete genomes (59 global and 20 local KN-CRKP), which of plasmids carrying blaKPC or blaNDM were analyzed. IncFII(pHN7A8)/IncR (43.0%, 34/79) was the most prevalent replicon in 14 kinds of plasmids carrying blaKPC, and meanwhile, IncX3 (21.5%, 17/79) was the most prevalent replicon in 19 kinds of plasmids carrying blaNDM (Supplementary Fig. 10). Combining analysis of MLST and KL, ST11-KL64-IncFII(pHN7A8)/IncR-IncX3 genome was dominant for 79 complete genomes, and these KN-CRKP were typical in China.
Importantly, the co-existence of blaKPC and blaNDM in one hybrid plasmid was observed in ST11, ST409 and ST1049 strains, implying the convergent tendency of blaKPC and blaNDM genes. Replicons of these hybrid plasmids included IncM1 (5), IncFII(pHN7A8)/IncN/IncR (4), IncC/IncFII(pHN7A8)/IncR (1), IncFII(pHN7A8)/IncN2 (1) and IncC (1). Moreover, the chromosome of strain CHS5 (CP110688.1) also concurrently carried blaKPC and blaNDM, where plasmid replicon IncFIB(K)/IncFII(K)/IncFII(pKP91)/repB(R1701) was also detected.
To further characterize the plasmid, phylogenetic trees were drawn (Fig. 6C). Highly homologous plasmids, including local and public plasmids, were assembled into one cluster and generally contained the same replicon, revealing plasmid horizontal transfer. Moreover, blaKPC- or blaNDM-carrying plasmids were also clustered together with blaKPC- and blaNDM-co-carrying plasmids. For example, blaKPC-carrying plasmid ON111449.1 was clustered with blaKPC- and blaNDM-co-carrying ON111447.1, and blaNDM-carrying plasmid ON111450.1 was clustered with blaKPC- and blaNDM-co-carrying ON111448.1. That is, the convergent plasmids can be generated by acquiring mobile genetic elements (MGE) carrying blaKPC or blaNDM on blaNDM- or blaKPC-carrying plasmids. Moreover, some blaNDM-carrying plasmids (3.8%, 3/79) also harbored hypervirulent genes. Plasmid self-transferability revealed 51.9% blaKPC-carrying plasmids (41/79), 65.8% blaNDM-carrying plasmids (52/79), and all blaNDM- and blaKPC-co-carrying plasmids have self-transferable potential, owing to possessing four complete modules for transfer, except for CP163100.1. Conjugation experiments showed only blaNDM-carrying plasmids in 24 local KN-CRKP could be transferred to E. coli J53, and others were not detected. Homologous plasmid (identity and coverage ≥ 90%) tracing showed homologous plasmids of blaKPC-carrying plasmids almost exclusively distributed within Klebsiella genus, whereas the majority of homologous plasmids of blaNDM-carrying plasmids distributed outside of Klebsiella. Plasmid-plasmid networks (pairwise ANI ≥ 95%) showed that plasmids carrying blaKPC had a higher degree of connectivity compared to plasmids carrying blaNDM, and the latter had a larger range of homologous plasmids (Fig. 6B). Therefore, the formation mechanism of ST11 KN-CRKP was presented, according to the development of local KN-CRKP and plasmids analyses (Fig. 6D).
Novel BL/BLI combinations offering alternatives for KN-CRKP
AST showed all 25 non-duplicate KN-CRKP strains were resistant to imipenem, meropenem, piperacillin/tazobactam, cefepime, and ceftazidime/avibactam, exhibiting pan-β-lactam resistance. To deal with the intractable KN-CRKP strains, cefiderocol and BL/BLI combinations were chosen to perform AST for the 25 KN-CRKP strains (Table 1). Rates of resistance to imipenem/relebactam, meropenem/taniborbactam, cefepime/taniborbactam, meropenem/vaborbactam, cefepime/zidebactam and meropenem/nacubactam were 100.00%, 96.0%, 92.0%, 100.00%, 96.0% and 100.00%, respectively, exhibiting poor antibacterial activity. Fortunately, the incidences of susceptibility to both aztreonam/avibactam and cefiderocol were 96.0%. Consequently, aztreonam/avibactam and cefiderocol could be considered the last alternatives for treating KN-CRKP.
Except for co-carrying blaKPC-33 and blaNDM-1 K64102, the cefiderocol MIC of KN-CRKP strains increased to 2–8 folds, compared to their orthologs carrying blaKPC-2. Likely, except for K64102, the imipenem and meropenem MIC of KN-CRKP strains also increased to 2–4 folds, compared to their orthologs carrying blaKPC-2. Therefore, the existence of blaNDM-1 contributed to the increase of cefiderocol, imipenem, and meropenem MIC.
Discussion
With the growth of global expansion of KPC-CRKP and NDM-CRKP2,19,20, KN-CRKP has rapidly increased in recent years, particularly in China and USA. Several studies have reported8,13,14,15 that the percentage of KN-CRKP in CRKP was 0.34% before 2017; afterwards, the proportion of KN-CRKP in CRKP was between 1.2% and 4.4% with a significant increase, similar to that in the current study. However, resistance, virulence, dissemination, and evolution characterizations of KN-CRKP lacked thorough analyses, which were necessary for the prevention and treatment of KN-CRKP, owing to its pan-β-lactam resistance and rapid growth. Considering local and worldwide KN-CRKP, 413 genomes of KN-CRKP, isolated from 32 countries of Asia, USA, Europe, and Africa between 2005 and 2024, were involved in rounded analyses.
KN-CRKP has emerged as a global dissemination trend, presenting substantial therapeutic challenges in clinical practice due to its broad multidrug-resistant profile. Since the first KN-CRKP strain XL-20059286 was isolated from China: Zhejiang in 2005, the number of KN-CRKP exhibited global proliferation trends from 0.03% to 3.10%, especially in 2017–2023. K2N1-CRKP, K2N5-CRKP, and K3N1-CRKP, mostly distributed in China for the first two types, and USA for the last type, were the three most prominent subtypes in 16 kinds of KN-CRKP, and the K2N1-CRKP showed great compatibility for more carbapenemase genes. Particularly, two K33N1-CRKP strains, isolated from two different patients and belonging to the same clone, were mistaken for NDM-CRKP by APB/EDTA method, resulting in improper treatment. Additionally, more than 60% of KN-CRKP strains carried a set of genes causing resistance to aminoglycosides, ESBL genes, and sul while less than one-seventh of KN-CRKP strains carried the fosfomycin-resistant gene fosA3. Although only less than one-fifth of KN-CRKP carried colistin- and tigecycline-resistant genes, CRKP was capable of rapidly evolving resistance and promoting exacerbation or chronicity of CRKP infections after using the two agents21,22,23. Out of the cefiderocol and seven kinds of BL/BLIs, only aztreonam/avibactam and cefiderocol had great activity against KN-CRKP in local CRKP strains. Therefore, aztreonam/avibactam and cefiderocol can act as the main agents against KN-CRKP.
KN-CRKP showed significant differences in ST, KL, virulence and countries, and CG1 emerged as the principal dissemination vector for KN-CRKP strains globally, especially hypervirulent ST11-KL64 KN-CRKP. Genome analyses revealed ST11-KL64, ST11-KL47 and ST258-KL107, corresponding to O1/O2v1, OL101 and O1/O2v2, respectively, were the three most prevalent STs. Among them, ST11-KL64 and ST11-KL47 are mainly distributed in China, and ST258-KL107 is mainly distributed in USA. Remarkably, about two-thirds of ST11-KL64 KN-CRKP belonged to hypervirulent strains, increasing the therapeutic difficulty for infections with ST11-KL64 hv-KN-CRKP. CG1 strains based on whole genome SNPs and Clade 1 strains based on RhierBAPS classification are extremely consistent, according to transmission and evolution analyses of KN-CRKP. CG1 possessed the most strains in 22 Groups, and these strains were isolated from different countries and years, implying the clonal spread of KN-CRKP. Origin analyses revealed CG1 strains derived from Brazil in 1954 and then spread worldwide. CG1 population also increased after the epidemic of COVID-19. More significantly, in line with the KL of CRKP2, the dominant KL of ST11 CG1 strains in China has shifted from KL47 to KL64, with hypervirulence. Furthermore, the 413 genomes of KN-CRKP were representative owing to a closed pan-genome. KEGG pathway enrichment showed CG1 strains have a great advantage in adhesion, invasion, and proliferation, compared with non-CG1 strains. Consequently, CG1 strains have a great advantage over antibiotic resistance, pathogenicity, environmental adaptability, and dissemination, and hindering the dissemination of CG1 strains is extremely necessary to control the epidemic of KN-CRKP.
ST11 KN-CRKP was probably generated by KPC-CRKP acquiring blaNDM-carrying plasmid. The genomic analysis of blaKPC- or blaNDM-carrying plasmids of KN-CRKP revealed the two genes mostly located on two different plasmids, and the most prevalent replicon type for blaKPC-carrying plasmids and blaNDM-carrying plasmids was IncFII(pHN7A8)/IncR and IncX3, respectively, especially in ST11 KN-CRKP. This was likely explained by that blaKPC-carrying IncFII(pHN7A8)/IncR plasmids of K. pneumoniae carrying blaKPC-2 were the most prevailing plasmid type, and most of these were confined to Asia, especially in China, and blaNDM-carrying IncX3 plasmids were the most common plasmid in other genera of CRE carrying blaNDM24,25,26,27. Notably, blaKPC and blaNDM are also concurrently located in one plasmid by transposition recombination via plasmid fusion or the transfer of MGE. Plasmid self-transferability and homology revealed blaNDM-carrying plasmids were easy to transfer among different genera and had a broader host range, therefore, KN-CRKP was generally formed by KPC-CRKP acquiring blaNDM-carrying plasmid. This finding was in line with the formation of KN-CRKP in vivo in this study and previously reported by Wang et al.8. Another study also showed 28.0% NDM-CRKP carried IV-A CRISPR targeting the specific sequences of blaKPC-carrying plasmid, which also supported the above view28. In other words, blocking the blaNDM-carrying plasmid from entrancing KPC-CRKP was vital to the reduction of KN-CRKP.
One fifth of KN-CRKP isolates were derived from bloodstream infections in this study. According to the Blood Bacterial Resistant Investigation Collaborative System (BRICS), the proportion of KN-CRKP was only 0.5% in 794 CRKP bloodstream isolates across 19 provinces of China between 2014 and 201929. However, 10.3% KN-CRKP was identified out of 242 CRKP strains from bloodstream infection between 2020 and 2022 in a teaching hospital in Nanjing, China10. The latter most likely had a nosocomial transmission that led to the KN-CRKP rapidly spreading and 56.0% mortality. Likely, in the second propagation of our local KN-CRKP strains, belonging to CG 1, 8 of 10 patients suffered from bloodstream infections and had a 50.0% mortality. Therefore, bloodstream infections with KN-CRKP require further consideration.
To address the dissemination of KN-CRKP, a series of measures should be urgently implemented, especially for CG 1 strains. First, effective and urgent prevention and surveillance in hospital settings are necessary for clonal transmission, particularly in immunocompromised patients. Second, patients with KPC-CRKP during the use of ceftazidime/avibactam and carbapenems should be performed to identify carbapenemases genes, owing to promoting the blaNDM-carrying plasmid to enter KPC-CRKP, especially in mixed infection. Third, more valid antimicrobial agents should be continuously developed, with the emergence of drug resistance, though aztreonam/avibactam and cefiderocol were effective barriers to most KN-CRKP. In a phase 3 multinational, open-label, randomized controlled trial30, aztreonam-avibactam demonstrated favorable tolerability profiles and significantly reduced 28-day all-cause mortality in patients with severe Gram-negative bacterial infections. Furthermore, meta-analyses31 and multinational cohort studies32 revealed that cefiderocol exhibited superior 30-day survival outcomes compared to conventional antimicrobial therapies for multidrug-resistant Gram-negative pathogens. However, aztreonam/avibactam and cefiderocol are not yet commercially available in most countries, such as China. Moreover, high costs and slow progress in clinical approval also limit the application of the two drugs. Therefore, the combination of aztreonam and ceftazidime/avibactam is currently applied as an alternative to aztreonam/avibactam in clinical practice.
The principal strength of this study resides in its being the rare large size of genomic surveillance study on KN-CRKP conducted to date, spanning from 2005 to 2024 while systematically elucidating genomic profiles, transmission dynamics, and evolutionary trajectories. Previous studies only partially conducted the genomic analyses of limited strain collections or single outbreak events. In contrast, our study implements a comprehensive global analytical framework for KN-CRKP characterization. Several findings were identified from the above analysis. First, the study revealed a global increase in KN-CRKP prevalence between 2005 and 2024. Three predominant subtypes - K2N1-CRKP, K2N5-CRKP, and K3N1-CRKP-were mostly distributed in China and the United States. Accordingly, the top three STs were ST11-KL64, ST11-KL47, and ST258-KL107. Particularly, about two-thirds of ST11-KL64 KN-CRKP were hypervirulent strains. Notably, phylogenetic reconstruction identified a high-transmission cluster designated Clone Group 1 (CG1) and possessed a significant advantage in adhesion, invasion, and proliferation. We firstly identify the critical clone group “Clone Group 1”, as essential for further interventions. Additionally, KPC-CRKP typically acquired blaNDM-bearing plasmid to form ST11 KN-CRKP, and new hybrid plasmids co-encoding KPC and NDM have been discovered. Finally, aztreonam/avibactam and cefiderocol were the two most promising antimicrobial agents against KN-CRKP.
The present study is limited by several factors. First, although the current study presented the greatest number of KN-CRKP globally until now, the biases of strains in isolation time and nations might exist owing to detectability, differences in surveillance, and accessibility of data among different countries and regions. AST on novel β-lactam-β-lactamase inhibitor combinations were only performed on 25 available KN-CRKP strains, even though the small sample size is not uncommon in this type of study due to the rarity of CRKP strains co-carrying blaKPC and blaNDM genes, therefore reducing the robustness of our study. The conclusions should be interpreted with circumspection, thus, further studies are still needed to provide insights into our knowledge. Second, CG 1 showed a significant predominance, and its interaction with the host and treatment strategies needs further study. Third, other combinations of carbapenemases were not analyzed in this study. We found blaNDM and blaOXA-48-like-co-carrying CRKP strains are increasing with a different mechanism from KN-CRKP, and will be studied further.
In summary, we performed research on global KN-CRKP over the longest period, involving 32 nations. Our research showed that CG1 has a significant edge in adhesion, invasion, and proliferation, and its dispersal was the primary factor contributing to the worldwide spread of KN-CRKP. Moreover, ST11 KN-CRKP was generally formed by KPC-CRKP acquiring blaNDM-carrying plasmid and novel hybrid plasmids encoding KPC and NDM have occurred. Cefiderocol and aztreonam/avibactam have strong antibacterial potential against KN-CRKP. Overall, this study provides valuable insights into the global transmission and dynamics of KN-CRKP.
Methods
Local surveillance and global collection of KN-CRKP
To investigate the epidemiology of KN-CRKP, all 968 non-duplicate CRKP strains were isolated from 6 provinces, including Beijing, Hebei, Guangxi, Hunan, Shanxi, and Henan Province of China, from Jan. 1, 2017 to Dec. 31, 2024. The 968 non-duplicate CRKP strains were isolated from K. pneumoniae infected patients, and the first isolated CRKP was collected for each patient. Species identification was performed using matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) at China-Japan Friendship Hospital. The blaKPC and blaNDM carbapenemases genes were detected by real-time fluorescent PCR33, and KN-CRKP was subjected to whole-genome sequencing. Moreover, 64,354 K. pneumoniae strains were retrieved from the GenBank of NCBI covering the period from 1980 to Dec. 31, 2024. Quality assessment of all non-redundant genomes of KN-CRKP was conducted by CheckM v1.1.10 34 (genomes completeness ≥95% and contamination ≤5%) and Kleborate v2.4.135 (N50 length ≥50,000 bp, and coverage depth ≥50x).
Antibiotic sensitivity testing (AST)
The minimal inhibitory concentrations (MICs) of imipenem, meropenem, tigecycline, colistin, and ceftazidime/avibactam were assessed using the microdilution broth method (bio-KONT, Ltd., China), with E. coli ATCC 25922 as the quality control strain. Owing to the limited therapeutic alternatives, cefiderocol and novel BL/BLIs, including aztreonam/avibactam, imipenem/relebactam, meropenem/taniborbactam, cefepime/taniborbactam, meropenem/vaborbactam, cefepime/zidebactam, meropenem/nacubactam,were also chosen to perform AST. Meanwhile, the MICs of piperacillin/tazobactam, cefepime, amikacin, and aztreonam were determined using N335 susceptibility cards and the Vitek-2 system (bioMerieux, France). The breakpoints of tigecycline and aztreonam/avibactam were interpreted by the U.S. FDA36 and EUCAST37 on AST, respectively. The breakpoints of meropenem/taniborbactam, meropenem/nacubactam, cefepime/taniborbactam, and cefepime/zidebactam were defined by referring to the corresponding β-lactam breakpoints. The breakpoints of other antimicrobial agents were defined following the standards of the Clinical Laboratory Standards Institute (CLSI, 2024)38. As recommended by CLSI 202438, Carbapenemase inhibitor APB (3-aminophenylboronic acid) and EDTA enhancement method (APB/EDTA method) were performed to assess the production of carbapenemases.
Whole-genome sequencing (WGS) and annotation
The genomic DNA of 25 local KN-CRKP and four homologous KPC-CRKP was extracted by Bacterial genome DNA Kit from Tiangen, China (product standard: DP302), then sequenced by Illumina HiSeq 2500 platform. Quality control, adapter trimming, and low-quality filtering were performed for raw data by using fastp39 to obtain clean data. De novo assembly was conducted for clean data by using SPAdes Genome Assembler v3.13.140. Meanwhile, 20 of these KN-CRKP were conducted nanopore sequencing on Nanopore Flow Cell (R10.4.1). Quality control, adapter trimming, and low-quality filtering were performed for raw data by using PoreChop (https://github.com/rrwick/Porechop) to obtain clean data. Long and short reads were used as input for Unicycler41 to perform hybrid genome assembly for obtaining the complete genomes. These genomes were annotated using Prokka v1.13.742. MLST, serotype, resistance, and virulence genes of all KN-CRKP were determined by Kleborate v2.4.135 and Kaptive v1.2.043, and plasmid identification was identified by the CGE server (https://www.genomicepidemiology.org/services/). peg344 and type VI secretion system (T6SS) were confirmed using BLAST + 2.14.0 (identity ≥ 90 and evalue ≤ 0.00001). CRKP carrying both iucA and rmpA/rmpA2 were classified as hypervirulent CRKP, according to previous studies44. The Minimum Spanning Tree (MST) of MLST was built by PHYLOViZ45 and annotated by country, year and Clonal Group. Scatter plots of ST and KL were generated by ggplot2 v3.5.0. The transferability of blaKPC- or blaNDM-carrying plasmids was performed by oriTfinder46.
Cluster and phylogenetic analysis of KN-CRKP
Separate clades of heterogeneous populations were hierarchically clustered using rhierbaps in R-4.1.2. To trace the transmission of KN-CRKP2, Clone Group clustering was simultaneously performed by GraphSNP47 utilizing the different single nucleotide polymorphism (SNP) identified by Snippy 4.6.0 (https://github.com/tseemann/snippy) and removed recombination by Gubbins v2.4.148, with cutoff ≤ 21 SNPs. The phylogenetic analysis included 25 K. pneumoniae genomes from this study and 388 K. pneumoniae genomes obtained from GenBank globally, with HS11286 (GCA_000240185.2) as the reference strain.
The maximum-likelihood phylogenetic tree of 413 genomes of KN-CRKP was built to evaluate the dissemination and evolution by RaxML v8.2.1249 from the alignment generated by Snippy 4.6.0 and filtered by removing recombination using Gubbins v2.4.148. For the phylogenetic trees of homologous plasmids (both identity and coverage ≥ 90%) carrying blaKPC or blaNDM, the alignments were generated by Mafft v750 and then used for building a tree by IQ-Tree v2.3.251. The iTOL (https://itol.embl.de) was used for visualization.
Evolution, dissemination, and function analyses of Clone Group 1
Bayesian Markov Chain Monte Carlo was performed using BEAST252 to analyze the alignment of putative substitution mutations identified by Snippy and Gubbins in 227 CG1 strains, running 500 million iterations and sampled from every 5000 steps, which make the ESS > 200 (Effective Sample Size) and parameters convergence. Based on the above results of BEAST2, the time-scaled phylogenetic tree was built, and time of the most recent common ancestor (tMRCA) and mutation rates also were estimated. Root-to-tip regression analysis was conducted to identify that the maximum-likelihood tree had sufficient temporal signal, using TempEst version 1.5.3 (https://github.com/beast-dev/Tempest). Tracer version 1.7.1 (https://github.com/beast-dev/tracer/releases) was used to evaluate the operation convergence and effective population size under Bayesian Skyline reconstruction. Phylogenies and phylogeography were then reconstructed using Figtree v1.4.4 (https://github.com/rambaut/figtree) and spreaD353. Furthermore, Roary54 and KofamScan55 were performed to function analyses and alignment between CG1 and non-CG1. Moreover, the acquirement of blaNDM-carrying plasmids in 4 local KN-CRKP and their homologous strains carrying blaKPC were identified by genome alignment.
Plasmid conjugation and fitness cost
Plasmid conjugation experiments were conducted for locally collected KN-CRKP strains. Azide-resistant E. coli J53 was employed as the recipient strain. Briefly, both donor and recipient were adjusted to a McFarland standard of 0.5 and then mixed at a ratio of 1:3, and the mixture had an 18 h incubation at 37 °C and was selected on China blue agar (CBA) plates containing both meropenem (1 mg/L) and azide (120 mg/L). The above transconjugants were confirmed by PCR and AST. To evaluate the fitness cost of the evolutionary strains, the growth curve assay was performed (n = 3), as previously described8. All tests were repeated in triplicate.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Complete sequences of 25 local KN-CRKP strains from this study can be accessed at the NCBI database under the BioProject number PRJNA1152235. Source data are provided with this paper.
References
Wang, M. et al. Clinical outcomes and bacterial characteristics of carbapenem-resistant Klebsiella pneumoniae complex among patients from different global regions (CRACKLE-2): a prospective, multicentre, cohort study. Lancet. Infect. Dis. 22, 401–412 (2022).
Wang, Q. et al. Expansion and transmission dynamics of high risk carbapenem-resistant Klebsiella pneumoniae subclones in China: an epidemiological, spatial, genomic analysis. Drug Resist. Updat. 74, 101083 (2024).
Group, I. P. C. Global burden associated with 85 pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Infect. Dis. 24, 868–895 (2024).
Argimon, S. et al. Rapid genomic characterization and global surveillance of Klebsiella using pathogenwatch. Clin. Infect. Dis. 73, S325–S335 (2021).
Feilong, Z., Wenting, Y., Binghuai, L. & Bin, C. Dynamic global variation in resistance and hypervirulence of carbapenem-resistant Klebsiella pneumoniae between 2010 and 2023. J. Infect. 90, 106493 (2025).
Kazmierczak, K. M., Karlowsky, J. A., de Jonge, B. L. M., Stone, G. G. & Sahm, D. F. Epidemiology of Carbapenem resistance determinants identified in meropenem-nonsusceptible enterobacterales collected as part of a Global Surveillance Program, 2012 to 2017. Antimicrob. Agents Chemother. 65, e0200020 (2021).
Zhang, Y. et al. Large-scale comparative analysis reveals phylogenomic preference of bla(NDM-1) and bla(KPC-2) transmission among Klebsiella pneumoniae. Int. J. Antimicrob. Agents 64, 107225 (2024).
Gao, H. et al. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine 51, 102599 (2020).
Liu, H. et al. Prevalence of ST1049-KL5 carbapenem-resistant Klebsiella pneumoniae with a bla(KPC-2) and bla(NDM-1) co-carrying hypertransmissible IncM1 plasmid. Commun. Biol. 7, 695 (2024).
Li, J. et al. Clinical and molecular characteristics of patients with bloodstream infections caused by KPC and NDM co-producing Carbapenem-resistant Klebsiella pneumoniae. Infect. Drug Resist. 17, 1685–1697 (2024).
Posteraro, B. et al. In-depth characterization of multidrug-resistant NDM-1 and KPC-3 co-producing Klebsiella pneumoniae bloodstream isolates from Italian hospital patients. Microbiol. Spectr. 12, e0330523 (2024).
Huang, Y. et al. Detection of carbapenem-resistant hypervirulent Klebsiella pneumoniae ST11-K64 co-producing NDM-1 and KPC-2 in a tertiary hospital in Wuhan. J. Hosp. Infect. 131, 70–80 (2023).
Sellera, F. P., Lincopan, N., Fuentes-Castillo, D., Stehling, E. G. & Furlan, J. P. R. Rapid evolution of pan-beta-lactam resistance in Enterobacterales co-producing KPC and NDM: insights from global genomic analysis after the COVID-19 pandemic. Lancet Microbe 5, e412–e413 (2024).
Guo, H. et al. Global emergence of carbapenem-resistant Klebsiella pneumoniae co-carrying multiple carbapenemases. Comput. Struct. Biotechnol. J. 21, 3557–3563 (2023).
Hu, F. et al. Carbapenem-resistant Klebsiella pneumoniae capsular types, antibiotic resistance and virulence factors in China: a longitudinal, multi-centre study. Nat. Microbiol. 9, 814–829 (2024).
Sun, S. et al. Emergency of the plasmid co-carrying bla(KPC-2) and bla(NDM-1) genes in carbapenem-resistant hypervirulent Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 36, 26–32 (2024).
Wong, J. L. C. et al. OmpK36-mediated Carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo. Nat. Commun. 10, 3957 (2019).
Zhang, F. et al. Two outbreak cases involving ST65-KL2 and ST11-KL64 hypervirulent carbapenem-resistant Klebsiella pneumoniae: similarity and diversity analysis. Commun. Biol. 7, 1602 (2024).
Kazmierczak, K. M., de Jonge, B. L. M., Stone, G. G. & Sahm, D. F. Longitudinal analysis of ESBL and carbapenemase carriage among Enterobacterales and Pseudomonas aeruginosa isolates collected in Europe as part of the International Network for Optimal Resistance Monitoring (INFORM) global surveillance programme, 2013-17. J. Antimicrob. Chemother. 75, 1165–1173 (2020).
van Duin, D. et al. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect. Dis. 20, 731–741 (2020).
Jin, X. et al. Resistance evolution of hypervirulent carbapenem-resistant Klebsiella pneumoniae ST11 during treatment with tigecycline and polymyxin. Emerg. Microbes Infect. 10, 1129–1136 (2021).
van Duin, D. et al. Tigecycline therapy for carbapenem-resistant Klebsiella pneumoniae (CRKP) bacteriuria leads to tigecycline resistance. Clin. Microbiol. Infect. 20, O1117–O1120 (2014).
Xie, M. et al. Clinical use of tigecycline may contribute to the widespread dissemination of carbapenem-resistant hypervirulent Klebsiella pneumoniae strains. Emerg. Microbes Infect. 13, 2306957 (2024).
Cai, M. et al. Tracking intra-species and inter-genus transmission of KPC through global plasmids mining. Cell Rep. 43, 114351 (2024).
Bloomfield S., et al. Mobility of antimicrobial resistance across serovars and disease presentations in non-typhoidal Salmonella from animals and humans in Vietnam. Microbial. Genomics 8, mgen000798 (2022).
Acman, M. et al. Role of mobile genetic elements in the global dissemination of the carbapenem resistance gene bla(NDM). Nat. Commun. 13, 1131 (2022).
Wu W., et al. NDM metallo-beta-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32, e00115-18 (2019).
Long, J. et al. Genomic insights into CRISPR-harboring plasmids in the Klebsiella genus: distribution, backbone structures, antibiotic resistance, and virulence determinant profiles. Antimicrob. Agents Chemother. 67, e0118922 (2023).
Zhou, K. et al. A point mutation in recC associated with subclonal replacement of carbapenem-resistant Klebsiella pneumoniae ST11 in China. Nat. Commun. 14, 2464 (2023).
Carmeli, Y. et al. Aztreonam-avibactam versus meropenem for the treatment of serious infections caused by Gram-negative bacteria (REVISIT): a descriptive, multinational, open-label, phase 3, randomised trial. Lancet Infect. Dis. 25, 218–230 (2025).
Risco-Risco C., Henriquez-Camacho C., Herrera-Rueda M., Barberan J., Andaluz-Ojeda D. Cefiderocol versus best available therapy in the treatment of critically Ill patients with severe infections due to resistant Gram-negative bacteria: a systematic review and meta-analysis. Antibiotics 13, 1048 (2024).
Giacobbe, D. R. et al. Use of cefiderocol in adult patients: descriptive analysis from a prospective, multicenter, cohort study. Infect. Dis. Ther. 13, 1929–1948 (2024).
Zhang, F. et al. In-host intra- and inter-species transfer of bla(KPC-2) and bla(NDM-1) in Serratia marcescens and its local and global epidemiology. Int. J. Antimicrob. Agents 64, 107327 (2024).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Lam, M. M. C. et al. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12, 4188 (2021).
Marchaim, D. et al. Major variation in MICs of tigecycline in Gram-negative bacilli as a function of testing method. J. Clin. Microbiol 52, 1617–1621 (2014).
EUCAST. Breakpoints for aztreonam-avibactam May 2024. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Addenda/Aztreonam-avibactam_addendum_22_May_2024.pdf (2024).
CLSI.Performance standards for antimicrobial susceptibility testing. 34sted. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute. (2024).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A. & Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinforma. 70, e102 (2020).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595 (2017).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Wyres, K. L. et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Micro Genom. 2, e000102 (2016).
Yang, X. et al. Molecular epidemiology of carbapenem-resistant hypervirulent Klebsiella pneumoniae in China. Emerg. Microbes Infect. 11, 841–849 (2022).
Ribeiro-Goncalves, B., Francisco, A. P., Vaz, C., Ramirez, M. & Carrico, J. A. PHYLOViZ Online: web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res. 44, W246–W251 (2016).
Li, X. et al. oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 46, W229–W234 (2018).
Permana, B., Beatson, S. A. & Forde, B. M. GraphSNP: an interactive distance viewer for investigating outbreaks and transmission networks using a graph approach. BMC Bioinforma. 24, 209 (2023).
Croucher, N. J. et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15 (2015).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Katoh, K., Rozewicki, J. & Yamada, K. D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166 (2019).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Bouckaert, R. et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 (2014).
Bielejec, F. et al. SpreaD3: interactive visualization of spatiotemporal history and trait evolutionary processes. Mol. Biol. Evol. 33, 2167–2169 (2016).
Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015).
Aramaki, T. et al. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252 (2020).
Acknowledgements
This work was supported by the Beijing Natural Science Foundation (7242124) and the CAMS Innovation Fund for Medical Sciences (CIFMS; No.2021-I2M-1-030) granted to LBH.
Author information
Authors and Affiliations
Contributions
Z.F.L., L.X.M., L.Z.H., L.Z.Y., and L.H.B. designed the research project. Z.F.L. and L.Z.H. performed the experiments. Z.F.L., L.X.M., L.Z.H., L.Z.C., Y.X.R., L.Q., M.Y.Q., and L.H.B. analyzed and discussed the data. Z.F.L., L.Z.H. wrote original draft. Z.F.L., L.X.M., L.Z.H., L.Z.Y., F.Y.Y., and L.H.B. reviewed and discussed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study was approved by the Ethics Committee of the China-Japan Friendship Hospital (CJFH, 2022-KY-054).
Peer review
Peer review information
Nature Communications thanks Yi-Rong Li, Maurizio Sanguinetti and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, F., Liu, X., Li, Z. et al. Tracking international and regional dissemination of the KPC/NDM co-producing Klebsiella pneumoniae. Nat Commun 16, 5574 (2025). https://doi.org/10.1038/s41467-025-60765-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-60765-7