Abstract
Approaches for the discovery of molecular glues remain limited. Here we report a phenotypic screening approach in which cytotoxins whose mechanisms require ubiquitination show a gain of viability following pharmacological inhibition of the Ubiquitin-like modifier activating enzyme (UBA1/UAE). This approach reveals PRLX-93936 and BMS-214662 as molecular glues that directly target the E3 ligase TRIM21 to induce degradation of nucleoporin proteins and inhibit nuclear trafficking. The cytotoxicity of these agents correlates strongly with TRIM21 expression, suggesting re-evaluation of these clinically tested agents in patients with TRIM21-high cancers. Relative to recently disclosed TRIM21-targeting glues, PRLX-93936 and newly-synthesized analogs represent a distinct structural series, lack known cellular off-targets, and offer greatly enhanced potency. Additionally, we elaborate PRLX-93936 to a heterobifunctional degrader that uses wild-type TRIM21 to degrade a multimeric protein. Together, our work creates opportunities for targeted protein degradation and enables the design of additional TRIM21-targeting glues and Proteolysis-Targeting Chimeras (PROTACs).
Similar content being viewed by others
Introduction
The mechanistic elucidation of small molecules like lenalidomide, indisulam, and CR8 as molecular glues that induce an E3 ligase to ubiquitinate and degrade a target protein has led to a surge of interest in targeted protein degradation (TPD)1,2,3,4,5,6,7. This concept has led to the identification of additional molecular glues and the development of proteolysis targeting chimeras (PROTACs), which comprise an E3-targeting small molecule, a linker, and a target protein ligand to achieve TPD8,9. While molecular glues remain challenging to discover, recent work has begun to define design strategies and screening approaches capable of identifying molecular glues in a more systematic way10,11,12,13,14. Recently, we reported a variant of an established approach15 in which pharmacological inhibition of Cullin RING ligases (CRLs) revealed small molecules whose cytotoxic mechanisms involved CRL-mediated protein degradation16. While CRL family substrate receptors like Cereblon, VHL, and DCAF15 mediate most molecular glues and PROTACs identified to date, E3 ligases represent a large and biologically diverse protein family, and expanding the repertoire of E3 ligases that can be leveraged for TPD remains of broad interest17,18.
TRIM21 is an E3 ubiquitin ligase that has been well studied in immunology, where it is known to oligomerize and ubiquitinate immunoglobulin G (IgG)-coated pathogens to mediate their rapid degradation19,20,21,22. The ability of TRIM21 to degrade antibody-bound cargoes has also been harnessed for the TRIM-AWAY platform, in which cellular delivery of an antibody against a target protein can induce its TRIM21-mediated degradation23,24. Recent reports have also explored the potential of this approach as a gene therapy, for example by delivering viruses encoding TRIM21-nanobody fusion proteins that target aggregation-prone proteins like Tau25,26. During the course of our work, the first TRIM21-targeting small molecules were reported. Hydroxy-acepromazine27, HGC65228, and PRLX-93936/BMS-21466229 were shown to function as molecular glues that induce degradation of nuclear pore proteins and impair nuclear transport. TRIM21 ligands lacking glue activity, including HGC1g, were also recently disclosed28,30. Excitingly, hydroxy-acepromazine also was elaborated to PROTACs (or, TRIMTACs) that selectively induced degradation of target proteins when they were forced into protein aggregates or condensates but not when the target protein was monomeric27. This finding likely reflects TRIM21’s biological preference for ubiquitinating large multimeric complexes and suggests TRIM21 could be an optimal E3 ligase in contexts where clearance of protein aggregates could be therapeutic25,26,27.
In this work, we implement a screening strategy in which pharmacological inhibition of all E3 ligases independently leads to the identification of PRLX-93936, a cytotoxin of unknown mechanism of action31,32, and BMS-21466233, a farnesyltransferase inhibitor, as molecular glue degraders targeting TRIM21. These molecules are structurally unrelated to hydroxy-acepromazine or HGC652/HGC1g, and newly-synthesized analogs of PRLX-93936 show substantially superior cellular potency. Like other known TRIM21 glues, we show that PRLX-93936 and BMS-214662 require TRIM21 activity for cytotoxicity and induce rapid degradation of a wide swath of nuclear pore proteins, leading to loss of nuclear trafficking and cell death. We also confirm that genetic disruption of NUP98’s autoproteolysis domain effectively prevents PRLX-93936-mediated nucleoporin degradation and cell death, further supporting a direct interaction between TRIM21 and NUP98. Evaluation of 30 PRLX-93936 analogs establishes the SAR of this series and enables the identification of a TRIM21-targeting PROTAC, or TRIMTAC, (34) that selectively degrades an engineered protein aggregate. Notably, in contrast to hydroxy-acepromazine-derived TRIMTACs27, 34 does not require expression of a TRIM21 mutant for efficacy. Together, these studies define the anticancer mechanism of PRLX-93936 and BMS-214662 and provide high-quality glues and TRIMTACs to propel the development of degraders of multimeric protein aggregates.
Results
Recently, we reported a screening approach that used pharmacological inhibition of neddylation to identify small molecules whose mechanism of cytotoxicity requires Cullin RING Ligase activity16. A limitation of this approach is that only the Cullin RING subfamily of E3 ligases are inactivated. We imagined that inactivating a wider range of E3 ligases could potentially spotlight additional degraders whose mechanisms target non-CRL E3 ligases. Small-molecule inhibition of the ubiquitin-like modifier activating enzyme (UBA1/UAE) prevents activation of the large majority of E3 ligases and is currently being pursued as an anticancer strategy34,35. While sustained treatment with the UBA1 inhibitor TAK-243 was potently cytotoxic to the acute myeloid leukemia cell line OCI-AML-3 (EC50 15 nM), treatment for 10 h was better-tolerated (Supplementary Fig. 1a). Short-term treatment with TAK-243 thus provided a meaningful window during which cells could be maintained in a viable state without E3 ligase activity.
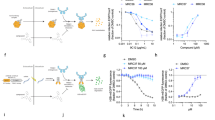
We next established a comparative cell viability assay in 384-well plates in which the impact of small molecules was compared with and without the addition of TAK-243; we then evaluated 1754 known bioactive small molecules using this assay (Fig. 1a, b; Supplementary Table 1). Only 5 molecules were identified that showed <50% viability and gained at least 30% viability with TAK-243 co-treatment (Fig. 1b). Hit validation across a wide concentration-response range confirmed only one of the five, PRLX-93936, as having diminished cytotoxicity when co-treated with TAK-243 (Fig. 1c, d; Supplementary Fig. 1b). This low confirmed hit rate likely reflects both the uncommon nature of ubiquitination-dependent cytotoxic mechanisms and the requirement for induction of cell death within 10 h. PRLX-93936 cytotoxicity could also be rescued by the proteasome inhibitor bortezomib, supporting a mechanism of cytotoxicity dependent on the proteasome (Fig. 1d).
a Schematic of the synthetic rescue screening strategy. Desired molecules show reduction of cytotoxicity when E1 Ubiquitin Activating Enzyme activity is impaired by the UBA1 inhibitor TAK-243, indicative of a mechanism of cell death reliant on ubiquitination. Created in BioRender. Adams, D. (2025) https://BioRender.com/66b3l1h. b Dot–plot representing the performance of 1754 bioactive small molecules in the synthetic rescue screen performed using OCI-AML-3 cells treated for 10 h. x-axis, viability measured for each library molecule with only DMSO vehicle treatment; y-axis, viability measured for each library molecule co-treated with TAK-243 (200 nM). Hits (red oval) were significantly cytotoxic as single agents (viability <50%, x-axis) and showed at least 30% greater viability in combination with TAK-243. Red circle, PRLX-93936. Each point represents data from a single well. c Structures of PRLX-93936 and erastin. d Cell viability (CellTiter-Glo) following treatment with PRLX-93936 alone or in combination with TAK-243 (200 nM) or Bortezomib (500 nM) for 10 h in OCI-AML-3 cells (n = 2 independent experiments, each with at least 2 wells per concentration). e, f Pearson correlation plots (www.depmap.org) establishing that TRIM21 expression (log2(Transcripts per million+1) is correlated with PRLX-93936 cytotoxicity (log2(Fold Change) (e) and that the cytotoxicity of PRLX-93936 and BMS-214662 is uniquely well correlated with TRIM21 among the small molecules within the database (f). Pearson correlation p-values were derived from two-sided tests with no correction for multiple comparisons. Top 1000 correlates are shown. Each correlation represents data from at least 453 cell lines. g Structures of BMS-214662 and BMS-225975. h Cell viability (CellTiter-Glo) following treatment with PRLX-93936 alone or in combination with TAK-243 (200 nM) or Bortezomib (500 nM) for 10 h in OCI-AML-3 cells (n = 2 independent experiments, each with at least 2 wells per concentration). Source data are provided as Source Data files.
PRLX-93936 is a cytotoxin of unknown mechanism of action that has been linked to diminished HIF pathway signaling31,32. Publicly-available cancer cell line profiling data (depmap.org) indicated that PRLX-93936 induced wide-ranging cytotoxic responses in cancer cell lines, with many potently killed and others fully resistant (Supplementary Fig. 1c). Correlation of cell killing potency with expression of single transcripts has previously provided mechanistic understanding for molecules of unknown mechanism-of-action, including the molecular glue CR84,36. Analysis of publicly available transcriptomic profiles revealed that sensitivity to PRLX-93936 was uniquely correlated with the expression of TRIM21, a member of the TRIM family of E3 ligases (Fig. 1e, Supplementary Fig. 1c). Conversely, among the thousands of small molecules in the DepMap database, PRLX-93936 was best-correlated with TRIM21 transcript levels, highlighting the strong association between PRLX-93936 and TRIM21 (Fig. 1f).
A second molecule, the farnesyl transferase inhibitor (FTI) BMS-214662, also appeared well-correlated with TRIM21 expression level (Fig. 1f, g). Interestingly, work from Bristol Myers Squibb and others has established that this molecule has an additional uncharacterized apoptotic mechanism not observed for closely related FTI inhibitors, including its N-methyl analog BMS-225975 (Fig. 1g)33,37,38. We next confirmed that BMS-214662, like PRLX-93936, induced rapid cell death in OCI-AML-3 cells that could be suppressed by co-treatment with either TAK-243 or bortezomib (Fig. 1h). Together these data suggested that PRLX-93936 and BMS-214662 share a cytotoxic mechanism of action that involves TRIM21-mediated ubiquitination.
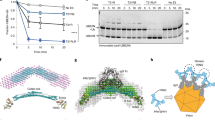
We next confirmed strong expression of TRIM21 in both JURKAT and OCI-AML-3 cells and used CRISPR/Cas9 targeting to knockout TRIM21 in both cell lines (Fig. 2a, b). Strikingly, while WT cells were highly sensitive to PRLX-93936 (EC50 ca. 100 nM), TRIM21 KO in both Jurkat and OCI-AML-3 cells led to full resistance to PRLX-93936 at concentrations as high as 50 μM (Fig. 2c, d; Supplementary Fig. 2a). TRIM21 KO had no discernible impact on proliferation in these cell lines, consistent with TRIM21 not being a known dependency across hundreds of cancer cell lines (Supplementary Fig. 2b, depmap.org). Likewise, both TRIM21 KO cell lines showed >100-fold increase in EC50 for BMS-214662 (Fig. 2e, f); farnesyl transferase inhibition may explain cell killing observed at high concentrations. Notably, while PRLX-93936 shares some structural features with erastin, an inducer of ferroptosis, erastin sensitivity was unaffected by TRIM21 KO (Fig. 2g). The cytotoxicity of farnesyl transferase inhibitor BMS-225975, which differs from BMS-214662 by only addition of a methyl group, was also unaffected by TRIM21 KO (Fig. 2h).
a, b Western blot following CRISPR/Cas9 targeting of TRIM21 in OCI-AML-3 cells (a) and JURKAT cells (b). Representative of n = 1 independent experiments. c, d Cell viability following treatment with PRLX-93936 for 24 h in OCI-AML-3 cells (c) or JURKAT cells (d). e, f Cell viability measurements as described for c, d but using BMS-214662. g Cell viability following treatment with erastin for 72 h in OCI-AML-3 cells. h Cell viability following treatment with BMS-225975 for 72 h in JURKAT cells. All cell viability measurements used CellTiter-Glo. All CTG data represents n = 2 independent experiments, each with at least 2 wells per condition, except h which is n = 1 with 2 wells per condition. Source data are provided as Source Data Files.
We also evaluated the impact of overexpression of TRIM21 on PRLX-93936 and BMS-214662 sensitivity. OCI-AML-3 cells expressing a TRIM21-FLAG allele became ca. 10-fold more sensitive to both PRLX-93936 and BMS-214662 (Fig. 3a–c). TRIM21 expression had no discernible impact on proliferation (Supplementary Fig. 2c), also consistent with prior reports that stable TRIM21 overexpression is nontoxic23. In contrast, expression of an established catalytically inactive triple TRIM21 mutant (C16A/C31A/H33W; TRIM21CA) partially suppressed the cell killing induced by these probes (Fig. 3a–c)39,40. Since TRIM21 enzymatic activity is known to be enhanced by its oligomerization, this catalytically inactive allele may induce a dominant-negative effect and partially mimic TRIM21 loss-of-function. We also overexpressed TRIM21 in C33A cells, a cervical cancer line that both publicly available transcriptomics data (www.depmap.org) and immunoblotting support as not expressing TRIM21 (Fig. 3d). While parental C33A cells were resistant to PRLX-93936 and BMS-214662 at concentrations up to 10 μM, overexpression of TRIM21 greatly sensitized C33A cells to these probes, with EC50 values comparable to those seen in WT OCI-AML-3 or Jurkat cells (ca. 100 nM; Fig. 3e, f). As expected, expression of the catalytically inactive TRIM21 CA mutant allele had no impact on sensitivity (Fig. 3e, f). As a final example, overexpression of TRIM21 in HEK293T cells also induced extraordinary sensitivity to PRLX-93936 and BMS-214662 (Fig. 3g–i). Together these gain- and loss-of-function genetic manipulations indicate that TRIM21 catalytic activity is both necessary and sufficient for PRLX-93936 and BMS-214662 to induce cell death.
a, d, g Western blot following lentiviral overexpression of TRIM21-FLAG or inactive mutant TRIM21CA-FLAG in OCI-AML-3 cells (a), C33A cells (d), or HEK293T cells (g). Representative of n = 1 independent experiment. b, e, h Cell viability following treatment with PRLX-93936 for 24 h in TRIM21-expressing OCI-AML-3 cells (b) or 72 h in TRIM21-expressing C33A (e) or HEK293T cells (h) (n = 2 independent experiments, each with at least 2 wells per concentration, except (h), which shows two replicate wells from n = 1 independent experiment). c, f, i Cell viability following treatment with BMS-214662 for 24 h in TRIM21-FLAG-expressing OCI-AML-3 (c), C33A cells (f), or HEK293T cells (i). N = 2 independent experiments, each with at least 3 wells per concentration, except (i) which shows two independent replicate wells from one experiment. j, k CETSA experiment monitoring the impact of PRLX-93936 (j; n = 1) or BMS-214662 (k; n = 2 independent experiments) on the thermal denaturation of TRIM21-FLAG in OCI-AML-3 cells. l Immunoprecipitation assay in which TRIM21 is captured by an IgG-coated resin in the presence of PRLX-93936 analog 1 (see Supplementary Table 2 and Fig. 6a for characterization of this analog) at indicated concentrations. IP = Immunoprecipitation, FT = Flow-through. IP and FT/Input samples from the same experiment were run on separate gels in parallel. Representative of n = 1 independent experiments. Source data are provided as Source Data Files.
To evaluate whether these small molecules directly bind TRIM21, we performed cellular thermal shift assays (CETSA)41. PRLX-93936 induced clear stabilization of TRIM21-FLAG by 1-2 °C, consistent with a direct binding interaction (Fig. 3j; Supplementary Fig. 2d). BMS-214662 stabilized TRIM21-FLAG by a larger margin, providing additional evidence that these molecules induce cell death by the direct binding to TRIM21 and subsequent ubiquitination and proteasomal degradation of an essential protein (Fig. 3k, Supplementary Fig. 2e). As recently-reported TRIM21-targeting glues have been demonstrated to bind to TRIM21’s PRYSPRY domain, we hypothesized that PRLX-93936 and BMS-214662 also likely bound within this established pocket27,30. To support this idea, we immobilized endogenous TRIM21 from cell lysates onto IgG-coated resin; IgG is a high-affinity endogenous ligand of the TRIM21 PRYSPRY domain42. Treatment with 1, a close analog of PRLX-93936 (Supplementary Table 2), or BMS-214662 dose-responsively displaced TRIM21 from the resin, supporting a competitive interaction with the TRIM21 PRYSPRY domain (Fig. 3l; Supplementary Fig. 2f, g). Additionally, we observed that HGC1g, a recently reported high-affinity ligand for the TRIM21 PRYSPRY domain that lacks molecular glue properties30, fully and dose-responsively suppressed the cytotoxicity of PRLX-93936 and 1 (Supplementary Fig. 2h). Together these studies establish PRLX-93936 as directly engaging TRIM21 at its PRYSPRY domain.
To identify the protein or proteins degraded by PRLX-93936 and BMS-214662, we next performed an unbiased proteomic analysis. We initially evaluated PRLX-93936 treatment in both JURKAT and OCI-AML-3 cells to identify proteins comparably depleted across these equally sensitive cell lines. Strikingly, a cluster of nucleoporin proteins were strongly downregulated in both JURKAT and OCI-AML-3 cells (Fig. 4a, b, Supplementary Fig. 3a, b). Strongest effects were seen for Nucleoporin 214 (NUP214) and Nucleoporin 88 (NUP88), which are essential proteins known to directly interact at the cytoplasmic face of the nuclear pore (Fig. 4b, d)43,44. However, numerous additional essential nucleoporin proteins–NUP98, NUP188, NUP155, and NUP35, among others–were also reduced (Fig. 4a, Supplementary Fig. 3c). Importantly, changes in NUP214 and NUP88 levels were not observed when the proteomics experiment was repeated using TRIM21 KO OCI-AML-3 cells, indicating that the reduction of these nucleoporins is TRIM21-dependent (Fig. 4c, e). Additionally, proteomic analysis revealed that nucleoporin degradation was rapid, with levels of some nucleoporins reduced by half within an hour (Fig. 4f, Supplementary Fig. 3e).
a Label-free LC/MSMS quantitation (LFQ) of protein levels following PRLX-93936 treatment (500 nM, 6 h) in Jurkat cells (x-axis) and OCI-AML-3 sgScramble (sgSCR) cells (y-axis), with nucleoporin proteins highlighted in orange. b, c Volcano plot highlighting alterations in protein levels by LFQ following treatment with PRLX-93936 in OCI-AML-3 sgSCR cells (b) and OCI-AML-3 sgTRIM21 cells (c) with nucleoporin proteins labeled in blue. FC = fold change. d, e LFQ intensity values for specific nucleoporins noted panels a-c in both sgSCR and sgTRIM21 OCI-AML-3 cells. f Heatmap showing fold change in nucleoporin abundance by LFQ after treatment of OCI-AML-3 cells with PRLX-93936 (500 nM) for 1, 2, or 4 h. Represents mean of n = 3 independent replicates. g Volcano plot as in b except OCI-AML-3 sgSCR cells are treated with BMS-214662 (1 μM, 4 h). h, i LFQ intensity values for specific nucleoporins in both sgSCR and sgTRIM21 OCI-AML-3 cells following treatment with BMS-214662 (1 μM, 4 h). Scatter and volcano plot points represent mean of n = 3 independent replicates. Volcano plot P-values derived from two-sided t tests, no correction for multiple comparisons. Bar graph points represent one of n = 3 independent replicates. j Western blot demonstrating reduction of NUP214 and NUP88 protein levels with increasing concentrations of PRLX-93936 treatment (4 h, OCI-AML-3 cells). k Western blot evaluating the impact of pre-treatment (2 h) with the proteasome inhibitor bortezomib (5 μM) or UBA1 inhibitor TAK-243 (0.5 μM) prior to addition of PRLX-93936 (1 μM, 4 h). Western blot results in (j, k) representative of n = 1 independent experiments. l Quantitation of immunofluorescence imaging of RANBP1 reported as the ratio of nuclear to cytoplasmic signal intensity in the indicated C33A overexpression cell lines. LMB = Leptomycin B; PRLX = PRLX-93936; BMS = BMS-214662. All treatments 6 h. Points represent average of 2 wells/condition with >900 cells quantified/condition from n = 2 independent experiments. m Representative images of RANBP1 subcellular localization following the treatments in l (n = 2 independent experiments). Panels (a, b) and (g, h) include data from the same experiment. Source data are provided as Source Data Files.
We then repeated these proteomics experiments with BMS-214662. As with PRLX-93936, multiple nucleoporins were degraded by BMS-214662 after 4 h in OCI-AML-3 cells, with NUP88 and NUP98 most strongly affected (Fig. 4g, h). Critically, nucleoporin levels were unaffected by BMS-214662 treatment in OCI-AML-3 cells lacking TRIM21, further supporting TRIM21’s essential role in mediating nucleoporin degradation (Fig. 4i, Supplementary Fig. 3f). Moreover, BMS-225975, which differs from BMS-214662 only by addition of a methyl group but lacks TRIM21-dependent cell killing (Fig. 2h), had no impact on nucleoporin levels (Supplementary Fig. 3g).
We also used western blotting as an orthogonal approach to validate our proteomics results. Levels of both NUP214 and NUP88 were substantially depleted following treatment with PRLX-93936 for 4 h in OCI-AML-3 cells (Fig. 4j). In contrast, reductions in NUP214 and NUP88 were not observed when PRLX-93936 was co-treated with the E1 inhibitor TAK-243 or the proteasome inhibitor bortezomib, indicating the nucleoporin loss was ubiquitin- and proteasome-dependent (Fig. 4k).
Loss of numerous essential nucleoporin proteins would be expected to impair nuclear trafficking. We next evaluated nuclear export in C33A cells using immunofluorescence detection of RANBP1 as a common marker of nuclear-to-cytoplasmic trafficking. C33A cells express TRIM21 at undetectable levels and are resistant to PRLX-93936/BMS-214662 unless TRIM21 is overexpressed (Fig. 3d–f). In C33A cells, PRLX-93936 and BMS-214662 had no impact on the subcellular localization of RANBP1; however, a canonical nuclear export inhibitor, the XPO1-targeting natural product Leptomycin B, led to strong nuclear accumulation of RANBP1 (Fig. 4l, m, Supplementary Fig. 3h). In contrast, treatment of TRIM21-overexpressing C33A cells with multiple concentrations of either PRLX-93936 or BMS-214662 induced nuclear accumulation of RANBP1 comparable to Leptomycin B treatment (Fig. 4l, m, Supplementary Fig. 3h). Additionally, nuclear accumulation of RANBP1 was not observed in cells overexpressing catalytically inactive TRIM21 (Fig. 4l). Importantly, this TRIM21-dependent nucleoporin degradation mechanism helps to rationalize past unexplained observations of extensively altered subcellular protein localization following BMS-214662 treatment38. Together, these data highlight that the observed loss of multiple essential nucleoporins leads to lethal nuclear trafficking deficits that are dependent on catalytically active TRIM21.
Recent studies, including two published during review, have also noted TRIM21-mediated degradation of nucleoporins by small molecules including hydroxy-acepromazine, HGC652, PRLX-93936, and BMS-21466227,28,29. These studies also provided genetic evidence that hydroxy-acepromazine induced degradation of multiple nucleoporins by recruiting TRIM21 specifically to NUP98. We confirmed that hydroxy-acepromazine indeed caused TRIM21-dependent cell death in OCI-AML-3 cells, albeit with 100-fold diminished potency relative to PRLX-93936 and BMS-214662 (Fig. 5b, c). Additionally, we established that targeted disruption of the autoproteolysis domain of NUP98 was sufficient to prevent cell killing by PRLX-93936 and BMS-214662. Delivery of recombinant Cas9 and five independent single-guide RNAs (sgRNAs) targeting this region of NUP98 led to isolation of A549 cells that were strongly resistant to both PRXL-93936 and BMS-214662 (Fig. 5d–e). These resistant cells did not show resistance to the unrelated cytotoxic agent TAK-243, supporting a selective effect (Fig. 5f). Each of these five distinct sgRNAs led to detection of a 200 kDa NUP98 proteoform in these PRLX-93936-resistant cells, indicating that the autoproteolysis of NUP98 had been impaired (Fig. 5g). Impaired autoproteolysis of NUP98 was recently demonstrated as a resistance mechanism to hydroxy-acepromazine27. Finally, these PRLX-93936-resistant cells did not show reduction in NUP214 or NUP88 levels following treatment with PRLX-93936, providing additional support for nucleoporin degradation as essential for this cytotoxic mechanism (Fig. 5h).
a Structure of hydroxy-acepromazine. b, c Viability (CellTiter-Glo) of OCI-AML-3 cells (WT or TRIM21 KO) following 24 h treatment with the indicated concentrations of hydroxy-acepromazine (b) or PRLX-93936 (c). d–f A549 cells were subjected to CRISPR/Cas9 targeting using 5 sgRNAs targeting the autoproteolysis domain of NUP98 and then exposed to PRLX-93936 (1 μM) for 7 days. Surviving cells were then exposed to the indicated concentrations of PRLX-93936 (d), BMS-214662 (e) or TAK-243 (f) and cell viability was measured after 72 h with CellTiter-Glo. All CellTiter-Glo experiments represent n = 2 independent experiments with at least 2 wells per condition. g Western blot of NUP98 in WT A549 as well as A549 cells targeted with the five independent NUP98 sgRNAs. A NUP98 proteoform (~200 kDa) indicative of impaired autoproteolysis is highlighted. h Western blot of NUP214 and NUP88 following PRLX-93936 treatment (1 μM /12 h) in A549 cells resistant to PRLX-93936 following treatment with four of the NUP98 sgRNAs reported in d-g. Western blotting results in (g, h) representative of n = 1 independent experiments. Source data are provided as Source Data Files.
We then performed medicinal chemistry optimization on the PRLX-93936 template both to identify analogs with improved potency and to elaborate PRLX-93936 to TRIMTACs that degrade a cellular protein aggregate. We synthesized a collection of 30 PRLX-93936 derivatives and evaluated their ability to induce TRIM21-mediated cell death in both OCI-AML-3 and JURKAT cells (see Supplementary Table 2 for data for all analogs). Initial analogs made modifications to the piperazine ring of PRLX-93936. Paralleling results observed during evaluation of PRLX-93936 analogs as HIF1 pathway antagonists31, we found that modifications that altered the terminal piperazine amine functionality, either by alkylation, substitution with oxygen, or conversion to an amide, strongly abrogated potency (Fig. 6a, Supplementary Table 2). However, analogs that added a methyl substituent to the piperazine carbon backbone were tolerated, with some leading to clear improvement in cell killing potency (Fig. 6a, Supplementary Table 2). Separation of methyl diastereomers 1 and 8 revealed that the (S)-methyl-piperazine 1 showed clearly improved potency relative to the (R)-configured 8 and represents the most potent TRIM21-targeting glue in this series (EC50 3 nM for cytotoxicity to OCI-AML-3)(Fig. 6a). Additionally, each cytotoxic analog was evaluated in TRIM21 KO cells, which were uniformly resistant up to 2 μM, supporting TRIM21’s essential role in the observed cell killing (Supplementary Table 2).
a–c Viability (CellTiter-Glo) of OCI-AML-3 cells following 24 h treatment with the indicated concentrations of the drawn PRLX-93936 analogs. n = 3 independent biological replicates are shown, each representing the mean of two independent wells. d Representative images of an A549 PML-eGFP-BRD4(BD2) clone expressing TRIM21-FLAG treated with indicated compounds for 8 h. 31–35 each 10 μM; see Supplementary Fig 3d for structures of 31–33. Representative of n = 1 independent experiments. e Quantification of EGFP foci in A549 PML-eGFP-BRD4(BD2) clone expressing TRIM21-FLAG treated with indicated compounds in dose for 8 h. Points indicate two independent wells derived from n = 1 independent experiment with >50 quantified cells per dose. f Representative images as in (d). 34, 10 μM; HGC1g, 10 μM; BTZ, Bortezomib, 2 μM. Representative of n = 2 independent experiments. g Image quantification as in (e) for the experiment described in (f). N = 2 independent experiments are shown, each point represented as the mean of two independent wells and >100 quantified cells per dose. h Structures of 34 and 35. Absolute configuration of the chiral axis of 34/35 is unknown, and the assignment is arbitrary. Source data are provided as Source Data Files.
Additionally, in confirming the enantiopurity of these analogs by analytical chiral SFC, we noted that these molecules in fact were a racemic mixture of stereoisomers. Past reports for related quinazolinone systems and calculations performed on PRLX-93936 have established that these molecules exist as atropisomers with rotational barriers of >35 kcal/mol, making them indefinitely stable near room temperature45,46. We used preparatory chiral SFC to separate the two PRLX-93936 atropisomers (9 and 10) and confirmed that no interconversion occurred over 96 h (Supplementary Fig. 4a). Strikingly, all cellular activity resided in one atropisomer, which was roughly twice as potent as the racemic mixture of PRLX-93936 (Fig. 6b). Similarly, only one atropisomer showed cell killing among the enantiomeric pair of 11 and 12 (Supplementary Fig. 4c). Remaining analogs were evaluated as racemates unless noted.
Further analysis of modified piperazine-containing analogs revealed that substitution of the piperazine methyl group of 1 with ethyl or isopropyl was tolerated but somewhat less potent than 1 (13, 14, Supplementary Table 2). In contrast, numerous analogs that added a second methyl group around the piperazine were inactive (15–17, Supplementary Table 2). Additionally, while either (R)- or (S)-methyl substitution of the piperazine was tolerated, gem-dialkyl substitution at this position was deleterious, as cyclopropyl analog 18 showed greatly diminished potency relative to 1 (Supplementary Table 2).
We also evaluated modifications to the quinazolinone and aryl rings of PRLX-93936. Substitution of the quinazolinone with methyl at each of the available four positions led to analogs with diminished potency, with substitution at the 5- and 6-positions somewhat less disruptive than substitution at the 7- and 8-positions (19–22, Supplementary Fig. 4b). In contrast, when methyl was added to each of the four positions of the aryl ring, a wide spectrum of outcomes was observed. Methyl substitution at the 3-position was not tolerated; addition at the 5- or 6-positions led to modest losses in potency; and addition of methyl at the 4-position led to a 3-fold gain in potency relative to PRLX-93936 (23–26, Fig. 6c, Supplementary Table 2). Subsequent combination of the (S)-methyl piperazine (as in 1) with the 3-methyl moiety of 24 gave 27, which did not show further potency increase relative to 1 alone (Supplementary Table 2). Notably, bulkier isopropyl substituents could be accommodated at C4, C5 and C6 of the aryl ring, highlighting that this region is broadly tolerant of expansion (28–30, Supplementary Table 2).
We next sought to evaluate whether the PRLX-93936 scaffold could be leveraged for the development of PROTACs that degrade protein aggregates. While our SAR data highlighted multiple positions where substituents could be added while improving potency, it remained unclear which positions could successfully accommodate the necessary linker moiety and facilitate ternary complex formation. We generated an initial set of five heterobifunctionals that connected PRLX-93936 via a flexible linker to JQ-1, a ligand used frequently in PROTACs to promote degradation of BET bromodomain proteins. Of these five, three added the linker moiety to piperazine functionality (31–33), while one added the linker at C5 of the aryl ring, which uniquely led to separable disatereomeric atropisomers 34 and 35 that were evaluated separately (Supplementary Fig 4d). To test these candidate PROTACs, we used a recently-reported assay in which bromodomain 2 of the BRD4 protein is fused both to PML, a protein known to form nuclear condensates, and enhanced green fluorescent protein (EGFP)27. Expression of this fusion protein in A549 cells also expressing wild-type TRIM21 gave rise to cells with prominent green nuclear puncta, indicative of the expected aggregation of the fusion protein (Fig. 6d). Treatment of these cells for 8 h with 31, 32, or 33 had little to no impact of EGFP+ nuclear puncta; in contrast, 34 gave near-complete loss of EGFP+ nuclear puncta with an EC50 of 1.4 μM (Fig. 6d, e, Supplementary Fig. 4e,f). Notably, no such degradation was observed in cells treated with diastereomeric atropisomer 35, highlighting the stereospecificity of this effect (Fig. 6d, e, Supplementary Fig. 4e, f). Additionally, the TRIM21 ligand HGC1g or the proteasome inhibitor bortezomib were sufficient to prevent degradation of the EGFP fusion protein, confirming TRIM21-mediated proteasomal degradation of this aggregation-prone fusion protein by the PRLX-93936-derived TRIMTAC 34 (Fig. 6f, g, Supplementary Fig. 4g).
Discussion
We have established that PRLX-93936 and BMS-214662 induce cell death via TRIM21-mediated degradation of nucleoporins. Genetic manipulations support TRIM21 as necessary and sufficient for the observed cell death across multiple cell lines. Extensive proteomic analyses identified a wide range of nucleoporins as degraded following treatment with PRLX-93936 and BMS-214662. Additionally, CRISPR targeting of NUP98’s autoproteolysis domain provided strong resistance to these small molecules. This finding supports that NUP98 plays an essential role in the observed nucleoporin degradation and may directly contact TRIM21 in the presence of PRLX-93936 and BMS-214662. Notably, NUP98 levels were unchanged in TRIM21 KO cells (Supplementary Fig. 3d), supporting that TRIM21 does not regulate NUP98 levels in the absence of small molecule glues.
Very recent work has established other small molecules that induce cell death via TRIM21-mediated nucleoporin degradation, including hydroxy-acepromazine27 and HGC65230. These molecules, like PRLX-93936 and BMS-214662, share the ability to degrade multiple nucleoporins and other proteins, demonstrating how loss of nuclear pore function can induce broad and rapidly lethal disruption of the proteome. Interestingly, these four series of TRIM21-targeting glues share almost no structural similarity, highlighting how distinct scaffolds can equally function to induce nucleoporin degradation. Notably, analog 1 offers substantially enhanced potency for cell killing relative to hydroxy-acepromazine or HGC652.
A unique aspect of PRLX-93936 and BMS-214662 is that these molecules have previously been investigated clinically in cancer. While clinical data for PRLX-93936 have only been presented at conferences47,48, published reports indicate that BMS-214662 was well tolerated and showed robust objective responses in a subset of acute myeloid leukemia and myelodysplastic syndrome patients49. Apoptotic responses were observed in tumors following treatment with BMS-214662 but not with other farnesyl transferase inhibitors, suggesting that the observed clinical responses were unlikely to result from farnesyl transferase inhibition. Together with our observation that high expression of TRIM21 strongly predicts PRLX-93936 and BMS-214662 sensitivity (Fig. 1), these findings suggest the potential for clinical re-evaluation of these agents in subsets of patients whose tumors have elevated TRIM21 expression.
Recent work with hydroxy-acepromazine has demonstrated that TRIM21-targeting glues can be elaborated to TRIMTACs that degrade proteins engineered to reside within nuclear condensates27. However, the low affinity of hydroxy-acepromazine for wild-type TRIM21 necessitated the use of a mutant TRIM21 allele to observe protein degradation. Our optimization of highly potent analogs of PRLX-93936 has now enabled us to identify a TRIMTAC capable of leveraging wild-type TRIM21 to degrade an aggregated protein. The design strategies and scope of aggregation-prone proteins amenable to TRIM21-mediated degradation remain to be established; however, TRIM21’s unique biological preference for ubiquitination of antibody-coated pathogens and other large multimeric assemblies suggests that TRIM21 may be ideally suited for degrading aggregated proteins25,26,27. Establishing PRLX-93936 and BMS-214662 as highly potent TRIM21-targeting molecular glues and evolving PRLX-93936 into a TRIMTAC (34, Fig. 6h) creates opportunities for modulating TRIM21 for cancer therapy and in a wide range of targeted protein degradation applications.
Methods
Statistical analysis
All statistical analyses were performed in GraphPad Prism unless otherwise stated. Replicate description and statistical tests are described in the associated figure legends. Unless otherwise stated, normality was assumed for statistical tests that require this assumption (Pearson correlation, t-test). All statistical tests were two-tailed, and all P values are reported as exact values unless otherwise noted. No corrections for multiple comparisons were made.
Cell lines
Cell lines were incubated at 37 °C with 5% CO 2 under humidified conditions. Lenti-X 293 T cells were purchased from Takara Bio. JURKAT E6-1 (TIB-152) and A549 were a gift of Dr. Mohamed Abazeed (Northwestern). OCI-AML-3 cells were a gift from Dr. David Wald (CWRU). C33a cells were a gift from Dr. John Pink (CWRU). HEK 293 T cells were a gift from Dr. Derek Taylor (CWRU). OCI-AML-3, JURKAT cell lines were maintained in glutamine-containing RPMI-1640 supplemented with 10% fetal bovine serum (FBS), and 1% penicillin/streptomycin. Lenti-X 293 T, HEK 293 T, and A549 cells were maintained in DMEM medium supplemented with 10% FBS, 2 mM L-glutamine, and 1% penicillin/streptomycin. C33a cells were maintained in glutamine-containing EMEM medium supplemented with 10% FBS and 1% penicillin/streptomycin.
High-throughput chemical screen
OCI-AML-3 cells were seeded in a sterile tissue culture 384-well plate (Corning, 3765) at 1000 cells and 50 μl of media per well using an EL406 Microplate Washer Dispenser (Biotek). Screening library molecules (a collection of 1754 molecules derived from the Sigma LOPAC1280 and Selleck Bioactive Compound Library L1700 collections) were maintained as a 3 mM DMSO stock in Abgene storage 384-well plates (ThermoFisher Scientific; AB1055). Transfer of 100 nL of each library molecule to assay plates was achieved using a solid pin tool, resulting in a final screening concentration of 6 μM. DMSO was used as a negative control within each plate. From each library plate, two replicate assay plates were generated that were identical except one plate was co-treated with 200 nM TAK-243 (added directly to cells and media 1 h before dispensing of library compounds). Plates were then incubated at 37 °C for 10 h, at which time 5 μL of CellTiter-Glo reagent (Promega, G7572) was added to each well using the EL406 Microplate Washer Dispenser (Biotek). Plates were allowed to equilibrate for 10 min with shaking at room temperature before luminescence was immediately read using a Synergy Neo2 Multimode microplate reader (Biotek). Viability was calculated relative to DMSO wells within each plate, and hits were called on the basis of at least 50% reduction in viability in the DMSO conditions as well as a % viability gain of at least 30% in the TAK-243 condition.
Cell viability assay
OCI-AML-3 and JURKAT cells were plated at a density of 1000 cells in 50 μl of culture medium in 384-well plates and treated with inhibitors same day before incubation at 37 °C. For C33a, A549, and HEK 293 T cells, 500 cells in 50 μl in 384-well plates were used. After 24 h (OCI-AML3, JURKAT) or 72 h (C33a, A549, HEK 293 T), 5 μl of CellTiter-Glo (Promega) was added to each well, and the contents were mixed on an orbital shaker at room temperature for 10 min. Luminescence was recorded using a Biotek Synergy Neo2 microplate reader with Gen5 3.03.14 software. Dose curves were fitted using a four-parameter variable slope model in GraphPad Prism.
Correlation analysis of cancer cell line profiling data
PRISM (23Q2) drug sensitivity and Expression Public (23Q2) gene expression data were retrieved from the Cancer Dependency Map (DepMap). PRISM (23Q2) data are expressed as a log2 fold change for each cell line and Expression Public (23Q2) data are expressed as log2(Transcripts per Million+1). Pearson correlation coefficients and P values were calculated within the DepMap Custom Analysis tool by comparing drug sensitivity to gene expression profiles across cell lines with a minimum of 453 cell lines used for each correlation. Statistical tests associated with the Pearson correlation were two-sided and no correction for multiple comparisons was implemented.
Lentivirus production and stable cell line generation
Lentiviral ORF-containing plasmids were synthesized and sequence validated by Vectorbuilder. Lenti-X 293T cells were plated in 6 well plates and allowed to attach overnight at 37 °C. Cells were then co-transfected with lentiviral plasmids, psPAX2 (a gift from Didier Trono, Addgene #12260), and pMD2.G (a gift from Didier Trono, Addgene #12259) using Lipofectamine 2000 (Invitrogen, 11668027) per manufacturer’s instructions. After 6 h, media was replaced, and cells were incubated for 48 h. Lentivirus containing media was collected and filtered through a 0.45 μm filter before being directly added to cells with 8 μg/mL polybrene (Sigma Aldrich TR-1003-G). After 24 h at 37 °C, media were replaced, and cells were incubated for another 24 h before selection with puromycin or blasticidin.
A549 PML-eGFP-BRD4(BD2) clones were generated by limiting dilution in 96-well plates and cultured over two weeks with a media change on day 7. Clones were then screened by fluorescence imaging on an Operetta High Content Imaging and Analysis System (Perkin Elmer) for intermediate PML-eGFP-BRD4(BD2) expression and consistent focus morphology. Selected clones were then transduced with lentivirus containing a TRIM21-FLAG allele.
Western blotting
Cells were washed twice with phosphate buffered saline (PBS) and lysed in Pierce RIPA Buffer (Thermo Fisher Scientific, 89900) supplemented with 1X cOmplete™ Mini EDTA-free Protease Inhibitor Cocktail (Roche, 11836170001). OCI-AML-3 cells were washed twice with PBS and resuspended in PBS supplemented with 1X cOmplete™ Mini EDTA-free Protease Inhibitor Cocktail before being lysed with two freeze-thaw cycles in liquid nitrogen. Lysates were clarified by centrifugation at 20,000g for 20 min at 4 °C. Soluble protein concentrations were quantified with a Pierce BCA Assay (Thermo Fisher Scientific, 23225). Lysates were normalized by protein concentration and separated on 4–12% or 3-8% gradient gels (Invitrogen, NW04125BOX or TA03815BOX) before being transferred to PVDF membranes using the XCell II Blot Module (Thermo Fisher Scientific, EI9051). Membranes were blocked in 5% milk in TBST and incubated in primary antibody overnight at 4 °C. Primary antibodies used: anti-β-actin (Sigma-Aldrich, A3854, 1:10,000), anti-TRIM21 (Proteintech, 12108-1-AP, 1:1000), anti-FLAG M2 (Sigma-Aldrich, F3165), anti-NUP214 (Abcam, ab70497, 1:500), anti-NUP88 (Santa Cruz Biotechnology, sc-365868), anti-NUP98 (Abclonal, A0530). Membranes were then incubated with secondary antibody conjugated to horseradish peroxidase (Cell Signaling Technology, 7076 or 7074, 1:1000). Membranes were developed using either SuperSignal West Pico Plus Chemiluminescent Substrate (Thermo Fisher Scientific, 34580) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, 34095) before imaging using a LI-COR Odyssey Fc Imaging System.
Liquid chromatography–tandem mass spectrometry
After incubation, cells were washed twice with PBS and resuspended in PBS supplemented with 1X cOmplete™ Mini EDTA-free Protease Inhibitor Cocktail (Roche, 11836170001). Cells were lysed with a Fisher Scientific Sonic Dismembrator Model 60 using 15 × 1-s pulses at a power level of 3 at 4 °C. Protein concentrations were quantified using a Pierce BCA Protein Assay kit (Thermo Fisher Scientific, 23225), and colorimetric development was measured using a Biotek Synergy Neo2 microplate reader. 50 μg of protein was denatured in 8 M Urea, reduced with 10 mM dithiothreitol, and alkylated with 25 mM iodoacetamide. Samples were then diluted with 100 mM Ammonium Bicarbonate and digested with 1 μg of Lys-C (Promega, V1671) overnight at room temperature. Peptides were desalted on C18 reverse-phase spin columns (BioPureSPN Mini FastEq, TARGA C18, 120 Å, The Nest Group, HUM S18R) before elution with 0.1% formic acid in 80% acetonitrile and 20% water. Peptides were dried and reconstituted in 0.1% formic acid in water for LC-MS/MS analysis.
Samples were analyzed by LC-MS using a timsTof Pro2 instrument (Bruker) equipped with a NanoElute UHPLC system. A 5 µl aliquot of each digest was injected onto a ThermoScientific (0.5 x 5 mm) Acclaim Pepmap C18, 5-μm, trapping column. Liquid chromatographic elution was performed using a flow rate of 0.3 μl/min on a reverse phase column (ReproSil AQ C18, 75 μm x 150 mm, 1.9-μm 120-Å). Peptide elution was performed using a binary gradient of mobile phase A, 0.1% formic acid and mobile phase B, 0.1% formic acid in acetonitrile. Each sample was analyzed using a linear gradient starting at 2% at 0 minutes and increasing to 35% B in 50 min, followed by an increase to 90% B in 2 min, then holding at 90% for 5 min before re-equilibration at 2% B. The electrospray voltage was 1500 V. A PASEF-DDA method was utilized for peptide identification. MS1 scans were carried out with a resolution of 30,000 measuring masses between 100-1700 Da with 1/k0 values between 0.6 and 1.6vs/cm2. MS2 scans between 100 and 1700 Da at a resolution of 30,000 was performed on precursor between charge states of 2-5 with targeted intensities of 20000, an intensity threshold of 2500 au and 10 PASEF MS/MS scans were performed in each cycle with cycle times of 1.2 sec. The peptides were fragmented using CID with an isolation window of 2 Da and collision energies ranging from 20 eV (1/k0 value of 0.6 Vs/cm2) to 59 eV (1/k0 value of 1.6 Vs/cm2). Dynamic exclusion was enabled for 30 s.
The LC-MS/MS data were searched against the human SwissProtKB database downloaded on 3-23-2022 (26,576 entries) using the program PEAKSOnline v11. These searches were performed considering full LysC peptides with no more than 2 missed cleavage sites. The MS1 and MS2 mass accuracies were set to 20ppm and 0.05 Da, respectively, carbamidomethylating was considered as a fixed modification, and oxidation of methionine and protein acetylation were considered as variable modifications. Deep learning boost was enabled. The results were filtered using a reverse decoy database strategy using percolator and the PSM, Peptide, and protein FDR rates were set to 1%. Positively identified proteins were required to be identified by a minimum of 2 peptides with at least one of these being a unique peptide identification.
Label-free quantitation was performed with PeaksOnline using ID-directed LFQ. Match between runs was enabled with a mass tolerance of 20 ppm, retention time shift set to auto, and a 1/k0 tolerance of 0.05. Quantitation was performed on unique peptides with a feature intensity of at least 150. The LFQ output produced after this step was used for bar graphs in Fig. 4, S2, and the heatmap in Fig. 4F. Data used for Volcano plots in Fig. 4 and S2 were further processed as described below. The LFQ output was uploaded into Perseus V2.0.11 and the intensity values were log2 transformed and the data was filtered to remove proteins with <2 quantitative values in at least one of the groups. After matrix reduction, missing values were imputed using a normal distribution with parameters of width = 0.3, downshift = 1.8, and mode = separately for each column. The LFQ intensities were used to determine the abundance ratios of the protein across groups and significance was determined using a two-sided t-test derived p-value (no correction for multiple comparisons was performed). Volcano plots in Fig. 4, S2 show proteins that were detected in all three DMSO technical replicates pre-imputation step. Each proteomics experiment consisted of n = 3 independent biological replicates per drug condition.
Recombinant protein production
A cDNA sequence corresponding to human TRIM21 PRY-SPRY amino acids 287-465 was cloned into a pET-6X-His vector to generate human 6X-His TRIM21 PRY-SPRY. E. coli BL21 DE3 cells were transformed and induced with 0.4 mM IPTG in TB media supplemented with Ampicillin at OD600 = 0.8 overnight at 18 °C. Cleared cell lysates were prepared by sonication of cell pellets in Lysis Buffer (50 mM Tris, 1 M NaCl, 2 mM TCEP, pH=8.0) supplemented with cOmplete protease inhibitors (Roche) followed by centrifugation at 15,000g at 4 °C for 1 h. The supernatant was loaded onto a Ni-NTA column and washed with Wash Buffer (50 mM Tris-HCl, 300 mM NaCl, 1 mM TCEP, 20 mM imidazole, pH 8.0). Proteins were eluted with Elution Buffer (50 mM Tris-HCl, 300 mM NaCl, 1 mM TCEP, 400 mM imidazole, pH 8.0). The eluted 6X-His TRIM21 PRYSPRY was further purified by size-exclusion chromatography and fractionated in PBS, pH 7.4. Fractions containing purified protein were pooled and concentrated to ~166 μg/mL before being supplemented with 0.5 mM TCEP.
IgG pulldown of TRIM21
For pulldown of endogenous TRIM21 from cell lysates, wild-type (WT) OCI-AML-3 cells were washed 2X with PBS before being resuspended in Immunoprecipitation (IP) Buffer composed of 50 mM Tris pH 8.0, 200 mM NaCl, and 0.1% TERGITOL solution (NP40S, Sigma) supplemented with 1X cOmplete™ Mini EDTA-free Protease Inhibitor Cocktail (Roche, 11836170001) at a concentration of 3 grams cell pellet weight/mL. Cell suspensions were lysed by 2X freeze thaw cycles in liquid nitrogen. Lysates were clarified by centrifugation at 20,000g for 20 min at 4 °C and soluble protein concentrations were quantified by Pierce BCA Assay (Thermo Fisher Scientific, 23225). Lysates were diluted to 1 mg/mL and 0.5 mg of total soluble protein was loaded onto 20 μl of Anti-FLAG M2 magnetic bead slurry (M8823, Millipore Sigma) with DMSO or indicated compounds. For pulldown of recombinant TRIM21, 20 μg of purified 6X-His TRIM21 PRY-SPRY was diluted in PBS supplemented with 0.5mM TCEP to a concentration of 20 μg/mL and loaded onto 40μl of Anti-FLAG M2 magnetic bead slurry (M8823, Millipore Sigma) with DMSO or indicated compounds. Bead-lysate mixtures were incubated with end-over-end rotation at 4 °C for 3 h before washing 5X with IP buffer containing DMSO or indicated compounds. Bead elution was performed by incubation at 95 °C in 15 μl of 2X LDS sample buffer (NP0007, Invitrogen) for 3 min. Eluted proteins were then separated on a 4–12% gradient gel (Invitrogen, NW04125BOX) before being transferred to PVDF membranes using the iBlot 2 Gel Transfer Device (Invitrogen, IB21001) or stained with InstantBlue Coomassie Protein Stain (Abcam, ISB1L). Membranes were processed as described in the Western Blotting section.
Immunofluorescence
C33a eGFP and TRIM21-FLAG cells were plated at a density of 6000 cells per well in a black, clear-bottom 96-well plate (PerkinElmer, 50-209-9831) and allowed to attach overnight at 37 °C. Cells were then treated with respective inhibitors for 6 h. Cells were fixed in 4% PFA for 20 min, washed with PBS, and stained with anti-RANBP1 (Abcam, ab97659) at a 1:500 dilution and DAPI (Sigma-Aldrich, D8417) at a 1:20,000 dilution. Cells were imaged using an Operetta High Content Imaging and Analysis System (Perkin Elmer), with eight fields captured per well and two wells per condition at ×20 magnification. Images were analyzed using Harmony software on the Columbus data server (PerkinElmer). In brief, live cells were identified and their nuclear regions established using DAPI staining. Around these nuclear regions, the cytoplasmic region was defined according to the region of RANBP1 staining not overlapping with the nucleus. The nuclear-to-cytoplasmic ratio of total signal intensity per well for RANBP1 was calculated based on these criteria.
Live fluorescence microscopy (high-content imaging)
A549 PML-eGFP-BRD4(BD2) TRIM21-FLAG cells were plated at a density of 2000 cells in phenol red-free complete medium in a black, clear-bottom 384-well plate (Falcon, 353962) and allowed to attach overnight at 37 °C. Cells were pre-treated with indicated rescue compounds for 2 h and then treated with indicated experimental compounds for 8 h. Cells were stained with Hoechst 33342 (Thermo Fisher Scientific, H3570) 15 min before imaging. Cells were then imaged using an Operetta High Content Imaging and Analysis System (Perkin Elmer), with five fields captured per well and two wells per condition at ×20 magnification. Images were analyzed using Harmony software on the Columbus data server (PerkinElmer). In brief, live cells were identified and their nuclear regions established using Hoechst staining. Cell boundaries were established using the eGFP channel and eGFP+ foci were called and quantified using the Harmony Software. Data for each molecule was normalized to DMSO wells.
Generating PRLX-93936-resistant cells with CRISPR–Cas9 targeting of NUP98-96
Short guide RNA oligonucleotides targeting exon 17 of NUP98-96 were synthesized by Synthego with default modifications containing the following sequences (all 5’-3’): GAUUUCACUAUUGGUCGGAA, UCUCUGAUUUCACUAUUGGU, UUAGCAAGGUCAUCCAUAGA, UUACUAUACUAUUCCAUCUA, and CACAGGUAUUAUUCUCACUA. Recombinant SpCas9-2NLS purchased from Synthego was mixed with each guide for 15 min at room temperature to allow for ribonucleoprotein complex formation (20 pmol Cas9:60 pmol single guide RNA per 1 million cells). Each mixture was added to 1 million A549 cells and delivered via electroporation using the Lonza 4D-Nucleofector system and an SF Cell Line 4D-Nucleofector kit according to the manufacturer’s instructions. After electroporation, the cells were allowed to proliferate for 72 h before selection for resistant clones using 1 μM PRLX-93936. After 7 days of selection, the surviving cells were maintained in 1 μM PRLX-93936 and harvested for analysis by western blotting and cell viability assays.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The proteomics data generated in this study have been deposited in the PRIDE database under accession code PXD062839 (Whole cell proteomics experiments in OCI-AML-3 and JURKAT cells). All data that support the findings in this paper are included in the Main Text, Figures, Supplementary Information, or available from the authors. The complete sets of microscopy image data that support Figs. 4l, 6E, and 6G are not included due to large file size but are available from the authors. Source data are provided with this paper.
References
Han, T. et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 356, https://doi.org/10.1126/science.aal3755 (2017).
Kronke, J. et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 (2014).
Lu, G. et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309 (2014).
Slabicki, M. et al. The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K. Nature 585, 293–297 (2020).
Nomura, D. K. & Dey, M. Advances and opportunities in targeted protein degradation. Cell Chem. Biol. 28, 887–888 (2021).
Sasso, J. M. et al. Molecular glues: the adhesive connecting targeted protein degradation to the clinic. Biochemistry 62, 601–623 (2023).
Tsai, J. M., Nowak, R. P., Ebert, B. L. & Fischer, E. S. Targeted protein degradation: from mechanisms to clinic. Nat. Rev. Mol. Cell Biol. https://doi.org/10.1038/s41580-024-00729-9 (2024).
Winter, G. E. et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381 (2015).
Bekes, M., Langley, D. R. & Crews, C. M. PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov. 21, 181–200 (2022).
Dewey, J. A. et al. Molecular glue discovery: current and future approaches. J. Med Chem. 66, 9278–9296 (2023).
Domostegui, A., Nieto-Barrado, L., Perez-Lopez, C. & Mayor-Ruiz, C. Chasing molecular glue degraders: screening approaches. Chem. Soc. Rev. 51, 5498–5517 (2022).
Dong, G., Ding, Y., He, S. & Sheng, C. Molecular glues for targeted protein degradation: from serendipity to rational discovery. J. Med. Chem. 64, 10606–10620 (2021).
Oleinikovas, V., Gainza, P., Ryckmans, T., Fasching, B. & Thoma, N. H. From thalidomide to rational molecular glue design for targeted protein degradation. Annu. Rev. Pharm. Toxicol. 64, 291–312 (2024).
Toriki, E. S. et al. Rational chemical design of molecular glue degraders. ACS Cent. Sci. 9, 915–926 (2023).
Mayor-Ruiz, C. et al. Rational discovery of molecular glue degraders via scalable chemical profiling. Nat. Chem. Biol. 16, 1199–1207 (2020).
Chen, Y. F. et al. C646 degrades Exportin-1 to modulate p300 chromatin occupancy and function. Cell Chem. Biol. 31, 1363–1372 e1368 (2024).
Belcher, B. P., Ward, C. C. & Nomura, D. K. Ligandability of E3 ligases for targeted protein degradation applications. Biochemistry 62, 588–600 (2023).
Schapira, M., Calabrese, M. F., Bullock, A. N. & Crews, C. M. Targeted protein degradation: expanding the toolbox. Nat. Rev. Drug Discov. 18, 949–963 (2019).
Holwek, E., Opinc-Rosiak, A., Sarnik, J. & Makowska, J. Ro52/TRIM21—From host defense to autoimmunity. Cell Immunol. 393-394, 104776 (2023).
James, L. C., Keeble, A. H., Khan, Z., Rhodes, D. A. & Trowsdale, J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc. Natl Acad. Sci. USA 104, 6200–6205 (2007).
Jones, E. L., Laidlaw, S. M. & Dustin, L. B. TRIM21/Ro52 - roles in innate immunity and autoimmune disease. Front Immunol. 12, 738473 (2021).
Mallery, D. L. et al. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21). Proc. Natl Acad. Sci. USA 107, 19985–19990 (2010).
Clift, D. et al. A method for the acute and rapid degradation of endogenous proteins. Cell 171, 1692–1706 e1618 (2017).
Kiss, L. et al. Trim-Away ubiquitinates and degrades lysine-less and N-terminally acetylated substrates. Nat. Commun. 14, 2160 (2023).
Benn, J. et al. Aggregate-selective removal of pathological tau by clustering-activated degraders. Science 385, 1009–1016 (2024).
Miller, L. V. C. et al. Co-opting templated aggregation to degrade pathogenic tau assemblies and improve motor function. Cell 187, 5967–5980 e5917 (2024).
Lu, P. et al. Selective degradation of multimeric proteins by TRIM21-based molecular glue and PROTAC degraders. Cell https://doi.org/10.1016/j.cell.2024.10.015 (2024).
Li, X. et al. Chemically Induced Nuclear Pore Complex Protein Degradation via TRIM21. ACS Chem Biol https://doi.org/10.1021/acschembio.4c00833 (2025).
Cheng, Y. et al. TRIM21-NUP98 Interface Accommodates Structurally Diverse Molecular Glue Degraders. ACS Chem. Biol. 20, 953–959 (2025).
Li, X. et al. Chemically induced nuclear pore complex protein degradation via E3 Ligase TRIM21. Biorxiv. https://doi.org/10.1101/2024.12.03.626577 (2024).
Huang, W. et al. Synthesis and evaluation of quinazolin-4-ones as hypoxia-inducible factor-1alpha inhibitors. Bioorg. Med Chem. Lett. 21, 5239–5243 (2011).
Sahasrbudhe, S. R. et al. Selective in vitro and in vivo anti-tumor activity of PRLX 93936 in biological models of melanoma and ovarian cancer. J. Clin. Oncol. 26, 14586 (2008).
Hunt, J. T. et al. Discovery of (R)-7-cyano-2,3,4, 5-tetrahydro-1-(1H-imidazol-4-ylmethyl)-3- (phenylmethyl)-4-(2-thienylsulfonyl)-1H-1,4-benzodiazepine (BMS-214662), a farnesyltransferase inhibitor with potent preclinical antitumor activity. J. Med Chem. 43, 3587–3595 (2000).
Hyer, M. L. et al. A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat. Med 24, 186–193 (2018).
Jin, J., Li, X., Gygi, S. P. & Harper, J. W. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 447, 1135–1138 (2007).
Rees, M. G. et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 12, 109–116 (2016).
Manne, V. et al. Apoptotic and cytostatic farnesyltransferase inhibitors have distinct pharmacology and efficacy profiles in tumor models. Cancer Res. 64, 3974–3980 (2004).
Balabanov, S. et al. Quantitative proteomics analysis of BMS-214662 effects on CD34 positive cells from chronic myeloid leukaemia patients. Proteomics 13, 153–168 (2013).
Wada, K. & Kamitani, T. Autoantigen Ro52 is an E3 ubiquitin ligase. Biochem. Biophys. Res. Commun. 339, 415–421 (2006).
Pan, J. A. et al. TRIM21 ubiquitylates SQSTM1/p62 and suppresses protein sequestration to regulate redox homeostasis. Mol. Cell 61, 720–733 (2016).
Tolvanen, T. A. Current sdvances in CETSA. Front. Mol. Biosci. 9, 866764 (2022).
Foss, S. et al. TRIM21 immune signaling is more sensitive to antibody affinity than its neutralization activity. J. Immunol. 196, 3452–3459 (2016).
Bernad, R., Engelsma, D., Sanderson, H., Pickersgill, H. & Fornerod, M. Nup214-Nup88 nucleoporin subcomplex is required for CRM1-mediated 60 S preribosomal nuclear export. J. Biol. Chem. 281, 19378–19386 (2006).
Hutten, S. & Kehlenbach, R. H. Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol. Cell Biol. 26, 6772–6785 (2006).
Laplante, S. R. et al. Assessing atropisomer axial chirality in drug discovery and development. J. Med. Chem. 54, 7005–7022 (2011).
Kumarasamy, E., Raghunathan, R., Sibi, M. P. & Sivaguru, J. Nonbiaryl and heterobiaryl atropisomers: molecular templates with promise for atropselective chemical transformations. Chem. Rev. 115, 11239–11300 (2015).
Voorhees, P. eterM. et al. An open-label, dose escalation, multi-center phase 1 study of PRLX 93936, an agent synthetically active against the activated ras pathway, in the treatment of relapsed or relapsed and refractory multiple myeloma. Blood 124, 2140 (2014).
Ramanathan, R. K. R. et al. A phase I pharmacodynamic and pharmacokinetic study of a Ras inhibitor, PRLX 93936, in patients with advanced solid tumors. J. Clin. Oncol. 28, https://doi.org/10.1200/jco.2010.28.15_suppl.e13042 (2010).
Cortes, J. et al. Phase I study of BMS-214662, a farnesyl transferase inhibitor in patients with acute leukemias and high-risk myelodysplastic syndromes. J. Clin. Oncol. 23, 2805–2812 (2005).
Acknowledgements
This work was supported by the CWRU School of Medicine, Thomas F. Peterson, Jr., and John and Virginia Johnson. M.A.S.dG. and P.Z. were supported by the CWRU Medical Scientist Training Program (NIH T32GM152319). Additional support was provided by the Small-Molecule Drug Development and Proteomics Shared Resources of the Case Comprehensive Cancer Center (P30 CA043703, Y.F., D.J.A). The TimsTof Pro2 instrument used for LC-MS/MS was purchased via an NIH shared instrument grant, S10 OD030398 (B. Willard, Cleveland Clinic Lerner College of Medicine, Case Western Reserve University). Fig 1A was made in Biorender. We thank B. Willard, L. Li, Y.F. Chen, D. Taylor, W. Huang, E. Schick, B. Clayton, and F. Najm for technical assistance and discussion. We also thank Y. Ze, Y. Li, J. Hao, and B. Wu (WuXi Apptec) for discussion and support of chemical synthesis.
Author information
Authors and Affiliations
Contributions
M.A.S.dG. and D.J.A conceived the project. M.A.S.dG., Y.F., and D.J.A. performed screening and analysis of bioactive small molecules. M.A.S.dG. led experimental efforts to characterize PRLX-93936 and BMS-214662. E.H., A.R.A., P.Z., R.B.G., and A.N.F. performed cell-based assays. M.A.S.dG and D.J.A. wrote the manuscript. All authors participated in data analysis and manuscript editing and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
D.J.A. and M.A.S.dG. are inventors on a provisional patent application (63/735,365) filed by Case Western Reserve University on the synthesis of PRLX-93936 analogs and their applications towards degrading protein aggregates. The other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Mikołaj Słabicki and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Scemama de Gialluly, M.A., Allen, A.R., Hayes, E.H. et al. Elaboration of molecular glues that target TRIM21 into TRIMTACs that degrade protein aggregates. Nat Commun 16, 6548 (2025). https://doi.org/10.1038/s41467-025-61818-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-61818-7
This article is cited by
-
State-selective small molecule degraders that preferentially remove aggregates and oligomers
Nature Communications (2025)