Abstract
The limited proliferative capacity of erythroid precursors is a major obstacle to generate sufficient in vitro-derived red blood cells for clinical purposes. While BMI1, a Polycomb Repressive Complex 1 member, is both necessary and sufficient to drive extensive proliferation of self-renewing erythroblasts, its mechanism of action remains poorly understood. Here we report that BMI1 overexpression leads to 10 billion-fold increase in self-renewal of human erythroblasts, which can terminally mature and agglutinate with typing reagent monoclonal antibodies. BMI1 and RING1B occupancy, along with repressive histone marks, are present at known BMI1 target genes, including the INK-ARF locus, consistent with altered cell cycle kinetics following BMI1 inhibition. Upregulation of BMI1 target genes with low repressive histone modifications, including key regulators of cholesterol homeostasis, along with functional studies, suggest that both cholesterol import and synthesis are essential for BMI1-associated self-renewal. We conclude that BMI1 regulates erythroid self-renewal not only through gene repression but also through gene activation and offer a strategy to expand immature erythroid precursors for eventual clinical uses.
Similar content being viewed by others
Introduction
Self-renewal, the generation of daughter cells with the same developmental potential as the parent cell, has been investigated within the hematopoietic system as a specific characteristic of hematopoietic stem cells (HSCs)1. However, hematopoietic progenitors and even some mature blood cells, such as subpopulations of tissue-resident macrophages and B1 lymphocytes, can also self-renew to maintain and expand their cellular compartments2,3. In the erythroid lineage, ex vivo culture of adult immature erythroid precursors leads to their limited, approximately 200–300-fold, expansion in the presence of erythropoietin (EPO), Stem Cell Factor (SCF), and the synthetic glucocorticoid dexamethasone (Dex)4,5. These self-renewing erythroblasts (SREs) have a proerythroblast morphology and spontaneously mature upon more prolonged in vitro culture. However, we previously discovered that these culture conditions enabled the essentially unlimited (>1060-fold) self-renewal of immature murine erythroblasts derived from transient hematopoietic progenitors in the early mouse embryo6. This acquisition of an extensive self-renewal state was accompanied by increased expression of Bmi1 and Ring1a, members of the Polycomb Repressive Complex 1 (PRC1)7. Importantly, we determined that increasing Bmi1 expression in adult murine erythroid cells with limited self-renewal capacity was sufficient to induce the essentially unlimited self-renewal capacity of their embryonic counterparts7. These Bmi1-induced self-renewing erythroblasts, which we termed iSREs, maintained their capacity to mature not only in vitro but also in vivo, where they generated a wave of circulating red blood cells (RBCs) with a normal lifespan7. The overexpression of BMI1 can also expand the self-renewal capacity of adult human-derived erythroblasts8.
Bmi1 was originally identified as a target of Moloney virus insertion, leading to accelerated B lymphoid tumors in mice9. It was subsequently recognized that Bmi1 regulates the self-renewal of normal and malignant stem cells, as well as more mature cell types, including fetal-derived B1a lymphocytes3,10. BMI1 is a member of PRC1, which classically functions as a transcriptional repressor through the mono ubiquitination of histone H2A at Lysine 119 (H2AK119Ub) via the E3 ubiquitin ligase, RING1(A/B)11,12. In this way, BMI1 can impact multiple cellular functions/pathways, including the cell cycle, survival, senescence, and DNA repair13. Interestingly, ribosome biogenesis was recognized as a target of Bmi1 in maturing murine erythroblasts14. Additionally, BMI1 was found to directly bind multiple ribosomal protein genes in K562 cells, which was associated surprisingly with gene activation14. Recent CUT&RUN studies in erythroid-lineage cells revealed that IGF2BP1, IGF2BP3, and Lin28B are direct targets of BMI1 repression, which in turn regulate fetal hemoglobin levels15. However, the broader potential functional targets of BMI1 in the erythroid lineage otherwise remain poorly understood.
Here, we investigated the function of BMI1 in the regulation of human erythroid self-renewal. Using CUT&RUN as a global and unbiased approach, multiple potential targets of BMI1 were identified, including the INK/ARF locus, where BMI1 acts as a repressor to silence the expression of cell cycle inhibitors. Importantly, we also identified a group of upregulated BMI1 target genes not marked by repressive histone modifications following BMI1 overexpression, including key genes encoding regulators of cholesterol homeostasis such as HMGCR, the rate-limiting enzyme in cholesterol synthesis. Despite prolonged culture and BMI1 overexpression, iSREs retain the ability to terminally mature with ~50% rate of enucleation and the ability to agglutinate with typing reagent monoclonal antibodies against Rh, Glycophorin, Kell, and Duffy system antigens. Our findings support the concept that BMI1 acts not only as a transcriptional repressor but also as a transcriptional activator to enhance the in vitro self-renewal of erythroid precursors. Expanding the pool of immature erythroid precursors will ultimately help meet the increasing need for standardized reagent RBCs and for cultured RBCs for transfusion of alloimmunized patients.
Results
Increased BMI1 expression drives extensive in vitro expansion of immature human erythroblasts
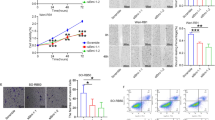
Adult human erythroblasts normally have a restricted proliferative capacity when cultured in vitro4,5. Lentiviral transduction of BMI1 on day 5 of cultured peripheral blood mononuclear cells (PBMCs) (Fig. 1A) resulted in an approximately 4-fold increase in both BMI1 transcript and protein levels (Fig. 1B, C and Supplementary Fig. 1C). While untransduced (UT) and empty vector (EV)-transduced control self-renewing erythroblasts (SREs) underwent limited (~200-400-fold) expansion, BMI1-transduced self-renewing erythroblasts (iSREs) underwent approximately 104-fold expansion (Fig. 1D). Analysis of these cultures at later timepoints of expansion revealed a significant subpopulation of small, maturing cells with condensed nuclei that had downregulated CD36 surface expression, as well as BMI1 expression as evidenced by decreased GFP levels (Fig. 1E, population 3). Since the presence of late-stage erythroid precursors can negatively impact the self-renewal of immature erythroblasts16, we instituted weekly purification of FSChi GFPhi immature erythroblasts (Supplementary Fig. 1B) by FACS. This approach resulted in approximately 1012-fold total expansion of immature human erythroblasts over a 2-3 month period (Fig. 1F). We found similar overall expansion of iSREs when initiated from CD34+ cells (Supplementary Fig. 1A, D).
A Schema of BMI1-induced SREs (iSRE) establishment and culture from PBMCs. B Lentiviral transduction leads to an approximately four-fold increase in BMI1 transcript levels in iSREs compared to compared to untransduced (UT) SREs and empty vector transduced (EV) SREs at day 13 of expansion, which have, as expected, similar levels of BMI1. 4 independent iSRE cultures derived from 3 donors, mean ± SEM. p-value was calculated based on paired two-tailed Student's t-test. ***p < 0.0001. Source data are provided as a Source Data file. C Lentiviral transduction leads to an approximately four-fold increase in BMI1 protein levels compared to compared to UT and EV SREs at day 13 of expansion. 4 independent iSRE cultures derived from 3 donors, mean ± SEM. p-value was calculated based on paired two-tailed Student's t-test **p < 0.01. Source data are provided as a Source Data file. D iSREs have approximately a 104-fold expansion, compared to untransduced (UT) SREs and empty vector transduced (EV) SREs, which have similar limited (200-300-fold) expansion. 4 independent iSRE cultures derived from 3 donors, mean ± SEM. E FACS-based analysis of iSRE cultures reveals 3 subpopulations of cells. Population 1 is CD36+CD235lo and consist of cells with an immature erythroblast morphology. Population 2 is CD36 + CD235+ and consists of cells with a slightly more mature morphology. Population 3 is CD36loCD235+ and consist of smaller hemoglobinizing cells with condensed nuclei. Cells are colored based on GFP expression, which is highest in Population 1 and lowest in Population 3. Size bar = 10um. A representative example of three independent experiments from three donors is shown. F Enrichment of immature erythroblasts by weekly fluorescence-activated cell sorting (FACS) leads to a 1012-fold expansion of iSREs, compared to untransduced (UT) and empty vector transduced (EV) control SREs. 3 independent iSRE cultures derived from 3 donors.
iSRE are poised at an immature erythroblast stage of maturation
iSREs have an immature erythroblast morphology, characterized by a large nuclear:cytoplasmic ratio and nuclear staining suggesting open chromatin (Fig. 2A). Consistent with an immature erythroid phenotype, iSREs are not only CD36+ , CD235mid but also CD71+, CD44+, CD49dlo, and CD34- (Fig. 2B). The average cell area and nuclear area of self-renewing iSREs remain consistent throughout extensive culture (Fig. 2C). Importantly, despite prolonged culture and increased BMI1 expression, iSREs maintain a normal karyotype and lack evidence of clonal aberrations based on G-banding analysis (Fig. 2D and Supplementary Fig. 2).
A Morphology of self-renewing iSREs at expansion day 34. Size bar = 10 um. Representative example of 6 independent cultures derived from 3 donors. B Surface expression of CD71, CD44, CD49d, and CD34 on iSREs at expansion day 30. Representative plots from one of three independent iSRE cultures. C Consistency of cell and nuclear area of iSREs on days 15, 30, and 55 of culture, as analyzed by imaging flow cytometry. Data from 3 independent cultures derived from 3 donors, mean ± SEM. D Karyotype of iSREs at day 45 of culture. Data from 1 of 3 independent cultures are shown (see Supplementary Fig. 2 for additional two donors). E Principal Component Analysis (PCA) of global transcriptomes of CD34-derived erythroid populations (GEO GSE53983 and GSE128268)17,18 and of iSREs (Supplementary Fig. 6C, D) indicate that iSREs are closest in gene expression identity to proerythroblasts (ProE). HSPC = hematopoietic stem and progenitor cells, BFU-E = burst forming units erythroid, CFU-E = colony forming units erythroid, EBasoE = early basophilic erythroblasts, LBasoE = late basophilic erythroblasts, PolyE = polychromatophilic erythroblasts, and OrthoE = orthochromatic erythroblasts. F BMI1 and EEF1A1 transcript levels during the differentiation trajectory of CD34+ HSPCs to OrthoE (GEO GSE53983 and GSE128268)17,18.
We compared the transcriptomes of iSREs with those of progressive stages of the human erythron differentiated from CD34+ cells17,18. As shown in Fig. 2E, principal component analysis (PCA) revealed that 3 independent (population 1) iSRE samples cluster together and are found closest to proerythroblasts (ProE). These data, together with the cellular morphology and cell surface phenotype, suggest that iSREs are poised at a ProE-like stage between upstream erythroid progenitors and downstream terminally maturing erythroid precursors. BMI1 transcript levels are highly expressed in ProE and rapidly decline during terminal maturation of erythroid precursors (Fig. 2F, red bars)17,18.
iSREs retain the ability to terminally mature in vitro
To test if iSREs remain capable of terminal maturation despite prolonged expansion, cells were transferred into maturation media and cultured for 7 days. Morphological analysis revealed a reduction in cell and nuclear size, with acidification of the cytoplasm, and approximately 47% rates of enucleation (Fig. 3A, B; Supplementary Fig. 3A). Maturing iSREs express adult (HBB, HBA) and fetal (HBG) globin genes (Fig. 3C). The terminal maturation of SREs revealed similar morphologic changes, expression of adult and fetal globin genes, and rates of enucleation (Supplementary Fig. 3B,C, D). BMI1 transcript expression decreases in murine erythroblasts as they terminally mature14. Consistent with these data and the marked downregulation of BMI1 transcript levels seen in CD34 + -derived late erythroblasts (Fig. 2F), we found that iSREs rapidly downregulate BMI1 transcript and protein levels (Fig. 3D; Supplementary Fig. 3E). The EEF1A1 promoter, which drives the overexpression of BMI1, is also normally downregulated during terminal erythropoiesis in a pattern very similar to that of BMI1 (Fig. 2F, grey bars)17,18, supporting the concept that lentiviral-derived BMI1/GFP silencing is a property of the vector. This concept is also consistent with the loss of GFP as iSREs spontaneously mature in expansion media (Fig. 1E, population 3).
A Morphology of terminally maturing iSREs after 8 days of maturation culture. Representative example of 6 independent cultures derived from 3 donors. B Flow-based assay of enucleation of iSREs after 8 days of maturation. Representative example of 4 independent iSRE cultures derived from 3 donors. Percent gated is the mean ± SEM of the replicates. C Globin gene expression of iSREs matured for 3 days and quantified by qPCR. 4 independent iSRE cultures derived from 3 donors, mean ± SEM. Source data are provided as a Source Data file. D BMI1 transcript levels rapidly decrease during in vitro maturation of iSREs. 4 independent iSRE cultures derived from 3 donors. Paired two-tailed Student's t-test *p = 0.027. Source data are provided as a Source Data file. E Flow-based surface expression of CD235a, Band3, Kell, Glycophorin C (GPC), and Rh on day 8 matured iSREs compared to FMO (fluorescence minus one) controls. F Gel column assay of 1.5 million terminally maturing iSREs after 7 days of maturation culture18. Absence of antibody served as a negative control. G Gel column assay of expression of Rh antigens on D + C + c + E-e+ terminally maturing iSREs. H Gel column assay of Glycophorin B (S+, s-), Kell (K-, k+), and Duffy Fy(a-b+) blood groups on terminally maturing iSREs. I Gel column assay of type A terminally maturing iSREs using plasma from a type O and a type A donor. J Ficin-treated terminally maturing iSREs agglutinated with plasma from type A patients containing anti-D, anti-C, or anti-e, respectively.
We next asked if these iSRE-derived cells could be used as an alternative source of test RBCs for blood bank assays to identify antibodies that guide consent selection and RBC compatibility19. Terminally maturing iSREs are characterized by surface expression of glycophorin A (CD235a), Band3, glycophorin C (GPC), Rh, and Kell (Fig. 3E). Gel column agglutination is a common method that is used to assess direct or indirect agglutination for RBC antigen phenotyping or for RBC antibody detection. Increased agglutination prevents RBCs from passing through the gel matrix after centrifugation. To test if iSRE-derived cultured RBCs have properties of mature erythrocytes (e.g., small size) for testing by laboratory transfusion service gel column assay, we first added 1.5 million cultured RBCs to gel columns in the absence of reagent antibodies to ensure that the cells could pass through the gel matrix upon centrifugation to form a pellet (Fig. 3F). iSRE-derived cells agglutinated with typing reagent monoclonal antibodies against Rh antigens D, C, c, and e, demonstrating Rh expression on the cell surface (Fig. 3G). These cultured RBCs also agglutinated as expected with typing reagent antibodies against the Glycophorin B (S,s), Kell (K,k), and Duffy (Fya,Fyb) systems (Fig. 3H). iSRE-derived cells (from a Group A donor) agglutinated with plasma from an O donor with expected naturally occurring anti-A, shown to lack antibodies to other RBC antigens, and did not agglutinate with plasma from type A donors who lack anti-A (Fig. 3I). Ficin acts as a proteolytic agent to enhance antigen reactivity with antibodies in the ABO, Rh, Kidd, Lewis, I, and P blood group systems20. Ficin-treated iSRE-derived cells agglutinated with plasma containing anti-D, -C, or -e, respectively (Fig. 3J). Taken together, these data support the concept that iSRE-derived cultured RBCs could potentially serve as an in vitro source of reagent RBCs for pre-transfusion antibody testing.
iSRE proliferation remains dependent on BMI1 and exogenous cytokines
Bmi1 expression is necessary for the continued self-renewal of adult murine self-renewing erythroblasts7. We asked if human iSRE self-renewal remains dependent on BMI1 expression. shRNA-mediated knock-down of BMI1 transcript levels markedly reduced iSRE proliferation compared to control (Fig. 4A, B). As an alternative approach, we treated iSREs with PTC-209, a small molecule inhibitor of BMI1, which caused dose-dependent reductions in BMI1 transcript and protein levels (Fig. 4C; Supplementary Fig. 4A, B). PTC-209 treatment resulted in a rapid loss of iSRE, as well as SRE, proliferation (Fig. 4D and Supplementary Fig. 4C). Taken together, these data indicate that BMI1 is necessary for the ex vivo self-renewal of both SREs and of iSREs. Despite overexpression of BMI1, human iSREs, like their murine counterparts7, remain dependent on the continued presence of EPO, SCF, and dexamethasone for continued self-renewal (Fig. 4E). While overexpression of BMI1 does not obviate the requirement for dexamethasone, we asked if dexamethasone might regulate iSRE self-renewal by upregulating BMI1. However, removal of dexamethasone from iSRE cultures with concomitant addition of RU486 did not impact BMI1 levels (Supplementary Fig. 4D). Additionally, matches for the GR binding motif, AGAACANNNTGTTCT, were not present near the BMI1 gene, suggesting that BMI is not directly regulated by glucocorticoids. Taken together, these data suggest that glucocorticoid signaling does not impact BMI1 expression.
A shRNA knockdown of BMI1 decreases BMI1 transcript levels compared to control luciferase shRNA culture. 3 independent iSRE cultures derived from 3 donors, mean ± SEM. p-value was calculated based on one-tailed Student's t-test. **p < 0.01. B shRNA knockdown of BMI1 leads to rapid collapse of iSRE cultures. Representative example shown, 3 independent iSRE cultures derived from 3 donors. C PTC-209 inhibition of BMI1 for 24 hours decreases BMI1 transcript levels compared to vehicle-treated controls. 3 independent iSRE cultures derived from 3 donors, mean ± SEM. p-value was calculated based on a one-tailed Student's t-test. **p = 0.010. ***p = 7.8e-6. D PTC-209 treatment leads to a dose-dependent inhibition of iSRE self-renewal. Representative example shown of 3 independent iSRE cultures, derived from 3 donors. E iSRE self-renewal remains dependent on the continued presence of exogenous erythropoietin (EPO), stem cell factor (SCF), and Dexamethasone (Dex). 6 independent iSRE cultures from 3 donors, mean ± SEM. All experiments were conducted with iSREs between expansion days 25 and 35. Source data are provided as a Source Data file.
Identification of potential downstream targets of BMI1
To gain a better understanding of BMI1 function in erythroid self-renewal, we sought to identify direct downstream targets using CUT&RUN to determine genome-wide BMI1 binding sites in both untransduced SREs and BMI1-transduced iSREs. RING1B, a core E3 ubiquitin ligase in PRC1, and its associated histone modification H2A119ub were also included in the analysis because shRNA-mediated knock-down of RING1B impaired expansion of iSREs (Fig. 5A), suggesting a critical role of RING1B in erythroid self-renewal. As dynamic interplay between PRC1 and PRC2 can regulate gene expression, genomic patterns of the PRC2-asociated histone modification H3K27me3 were also determined. A minimum of four replicates, including at least 2 donors, were analyzed for each condition, and Spearman correlation revealed high agreement between replicates (Supplementary Fig. 5A).
See Supplementary Fig. 5A for samples used in CUT& RUN. A shRNA knockdown of RING1B leads to rapid collapse of iSRE cultures. p-value was calculated based on two-way ANOVA of all RING1B shRNA vs Luciferase. 2 independent iSRE cultures from 1 donor, mean ± range, expansion day 30. B Heatmaps of BMI1 enrichment in untransduced (UT) SREs and iSREs over union peaks (±2 kb). The color scale represents the RPKM of each sample using merged replicates. Ranked based on mean RPKMs. C Volcano plot showing differential occupancy of BMI1 in iSRE versus untransduced SREs. Significantly increased and decreased peaks are shown in red and blue, respectively, with FDR < 0.05. D Heatmaps of BMI1 occupancy, RING1B occupancy, H2A119ub, and H3K27me3 over BMI1 differentially increased (top) and decreased (bottom) peaks in untransduced SREs and in iSREs. The color scale represents RPKM of each sample using merged replicates. Ranking was sorted based on BMI1 binding in iSREs. Blue line- increased BMI1 peaks. Orange line- decreased BMI1 peaks. E Heatmaps of BMI1, RING1B, H2A119ub, and H3K27me3 over 2 clusters of BMI1 differentially increased peaks (kmeans=2, clustering based on H3K27me3 in iSREs) in untransduced SREs and in iSREs. The color scale represents RPKM of each sample using merged replicates. Ranking was based on BMI1 binding in iSREs. Blue line- Cluster 1. Orange line- Cluster 2. F Table of selected published gene sets that contain significant overlap with CUT&RUN gene sets (Enrichr). See Supplementary Data 1 for details.
Our CUT&RUN analysis revealed extensive overlap of BMI1 occupancy in SREs and iSREs (Fig. 5B). Consistent with BMI1 overexpression in iSREs, BMI1 enrichment was differentially increased at 1,061 regions compared to SREs (Fig. 5C, D, BMI1 blue lines). The pattern of RING1B binding overall paralleled BMI1 binding both in SREs and in iSREs (Fig. 5D; Supplementary Fig. 5C, D). H2AK119Ub and H3K27me3 increased to a lesser extent at the same regions of differentially increased BMI1 enrichment (Fig. 5D, Supplementary Fig. 5B). We also found BMI1 binding differentially decreased at 681 regions (Fig. 5C,D BMI1, orange lines). These regions had significantly more enrichment for H2A119ub and H3K27me3 in both SREs and iSREs than regions of BMI1 gain (Fig. 5D, Supplementary Fig. 5B). The mechanism causing decreased BMI1 binding at sites in iSREs despite increased BMI1 expression is unclear. However, we speculate that the higher H3K27me3 levels in SREs compared to iSREs in these regions might promote the recruitment of BMI1. Examination of regions with increased BMI1 occupancy in iSREs revealed one subset where H3K27me3 was concordantly increased based on Kmeans clustering (Fig. 5E, Cluster 1, Supplementary Fig. 5E). These regions also revealed higher levels of RING1B and H2AK119Ub (Supplementary Fig. 5E). Thus, both Cluster 1 and regions with decreased BMI1 occupancy (Fig. 5D, bottom) display the expected association of histone modifications with canonical BMI1 function. Consistent with this, gene enrichment analysis (Enrichr) of genes associated with these regions revealed a high correlation with genes bound by BMI1 and other components of PRC1, as well as PRC2 components, in published datasets (Fig. 5F; Supplementary Data 1).
In contrast, the larger subset of sites with increased BMI1 occupancy in iSREs was associated with minimal H3K27me3 and modest levels of H2AK119Ub in both SRE and iSRE (Fig. 5E, Cluster 2; Supplementary Fig. 5E). Gene enrichment analysis of Cluster 2 did not reveal strong correlations with known PRC1/2 data sets but did show enrichment of genes bound by proteins identified in other cell systems to be regulated by BMI1 (e.g., CCND1, p53) and, interestingly, processes associated with erythropoiesis (e.g., heme metabolism, MCH GWAS) (Fig. 5F, Supplementary Data 1). RING1B increased in Cluster 2 iSREs without an increase in H2AK119ub (Fig. 5E, orange line; Supplementary Fig. 5E), consistent with the ability of RING1B to function in some contexts independently of its ubiquitin ligase activity21,22,23,24. Taken together, Cluster 2 findings suggest that acquired BMI1 is acting through mechanisms other than H2AK119Ub deposition.
Approximately half (53%) of all differentially increased BMI1-occupied regions are located at promoters, while the differentially decreased BMI1-occupied sites are located at promoters (27%), introns (33%), and intergenic regions (19%) (Fig. 6A; Supplementary Fig. 6A). We found that BMI1 was enriched near (i.e., within 2 kb of) the transcription start sites (TSS) of 6,538 genes in iSREs (Fig. 6B, Supplementary Data 2). Paralleling our genome-wide BMI1 occupancy findings, increases in BMI1 enrichment at TSSs were associated with increases in RING1B, whereas the associations between BMI1 and H2AK119Ub or H3K27me3 were not as strong (Fig. 6B; Supplementary Fig. 6B). The weak correlation between changes in ubiquitin ligase RING1B occupancy and enrichment of H2A119ub at BMI1-bound promoters in iSRE further suggests that BMI1 overexpression can alter gene expression via mechanisms other than H2A119ub deposition (Supplementary Fig. 6B).
See Supplementary Fig. 5A for samples used in CUT&RUN and Supplementary Fig. 6C, D for samples used in RNA-Seq. A Genomic annotations of BMI1 differentially increased peaks (left) and decreased peaks (right). B Heatmaps of BMI1, RING1B, H2A119ub, and H3K27me3 over genes (scaled TSS to TES ± 2 kb) associated with iSRE BMI1 peaks in untransduced SREs and iSREs. The color scale represents RPKM of each sample using merged replicates ranked based on the binding of BMI1 in iSREs. C Heatmaps of BMI1, RING1B, H2A119ub, and H3K27me3 over promoters (TSS ± 2 kb) of differentially expressed, upregulated (top) and downregulated (bottom), genes in untransduced SREs and in iSREs as determined by DeSeq2 analysis of RNA-Seq. The color scale represents RPKM of each sample using merged replicates ranked based on BMI1 binding in iSREs. Blue line- upregulated genes. Orange line- downregulated genes. D Quantitation of log2-fold change (iSRE/SRE) for BMI1, RING1B, H2A119ub, and H3K27me3 over promoters (TSS ± 500 bp) of the same differentially expressed genes shown in Fig. 6C. Box is Q1-Q3 with bar at the median and whiskers represent Q1-1.5x IQR and Q3 + 1.5x IQR. Statistics for log2 -old change is not equal to 0 based on a two-tailed Wilcoxon signed rank test with continuity correction. ****p < 2.2e−16, ns not significant. E Table of published gene sets with significant overlap with differentially upregulated and downregulated genes that have BMI1 associated with their promoters indicated on the left (Enrichr). See Supplementary Data 2 for details.
To further examine potential targets modulated by BMI1 in iSREs, we compared the global transcriptome of iSREs with that of empty vector transduced SREs. There was high concordance of all samples with the highest concordance among replicates (Supplementary Fig. 6C), and we identified 2147 up- and 1957 down-regulated genes (Supplementary Fig. 6D; Supplementary Data 2). Consistent with the well-established role of BMI1 as a transcriptional repressor, nearly half of the differentially downregulated genes were bound by BMI1 (Supplementary Fig. 6E). Of note, 43% (927 of 2,147) of upregulated genes were also bound by BMI1 (Supplementary Fig. 6E). Further analyses revealed that the subset of BMI1 promoter peaks with significant increases in iSRE were associated with similar numbers of up and down regulated genes (Supplementary Fig.6E). Consistent with these findings, BETA analysis, which integrates transcriptomic and protein binding/epigenetic data, strongly predicted that BMI functions to activate, as well as repress, gene expression (Supplementary Fig. 6F)25. The levels of BMI1 and RING1B occupancy in iSREs were also increased both at upregulated and at downregulated genes (Fig. 6C, D). Intriguingly, the upregulated genes had decreased levels of both H2AK119uB and H3K27me3, while the downregulated maintained H2AK119Ub levels and gained H3K27me3 (Fig. 6C, D; Supplementary Fig. 6G). We found similar transcription factor motifs, such as KLF and SP motifs, enriched at promoters of both upregulated and downregulated genes, without evidence for distinct BMI1 cofactors at activating versus repressive activity (Supplementary Fig. 6H).
PRC1 and PRC2 occupancy, as well as H2A119ub deposition, have been associated with CpG islands26,27. In erythroid lineage cells, BMI1 occupancy at CpG islands was identified at the LIN28B, IGF2BP1, and IGF2BP3 genes, where it was associated with gene repression15. Consistent with this report, we found that BMI1, RING1B, as well as H2A119ub and H3K27me3 were localized at the CpG islands of these 3 genes, whose expression was repressed, particularly in iSREs (Supplementary Fig. 6I, J). To further investigate the relationship of BMI1 and CpG island occupancy in iSREs, we undertook a global analysis and found that BMI1 and RING1B occupancy were higher at genes whose promoter contained CpG islands than those that did not contain CpG islands (Supplementary Fig. 6K, L). However, as seen with BMI1 and RING1B occupancy, BMI1 binding at CpG islands was associated equivalently with upregulated and with downregulated genes (Supplementary Fig. 6K, L).
Gene set enrichment analysis of BMI1-bound genes that are downregulated in iSREs compared to SREs have significant overlap with genes bound by transcription factors associated with myeloid and megakaryocyte lineages (FLI1, RUNX1, SPI1), as well as with regulators of cell cycle and apoptosis (Fig. 6E, Supplementary Data 2). Downregulated genes also overlapped with gene sets upregulated following BMI1 loss of function in other cell types (e.g., TP53, CCND1). In contrast, upregulated genes were correlated with gene sets bound by known regulators of erythropoiesis (e.g., GATA1, EKLF) and of cholesterol and lipid homeostasis (e.g., SREBF1, SREBF2) (Fig. 6E; Supplementary Data 2). Additionally, they were correlated with gene sets associated with known BMI1 functions, including positive regulation of the cell cycle and ribosomal proteins. Taken together, our CUT&RUN data suggest roles for BMI1 in regulating self-renewal of erythroblasts that extend beyond the classical repressive function ascribed to PRC1 and reveal potentially novel pathways upregulated by BMI in iSRE, including cholesterol metabolism.
BMI1 occupies the INK/ARF locus and regulates the cell cycle of SREs and iSREs
To begin investigating direct targets of BMI1 in erythroid self-renewal, we focused first on the INK/ARF locus, a known repressive target of BMI1 in multiple cell types28,29. We confirmed BMI1 and RING1B occupancy and H2A119ub deposition specifically at CDKN2A, as well as CDKN2B, which encodes the pl61NK4a and pl4ARF, as well as p15INK4B (Fig. 7A). Our CUT&RUN studies also revealed that iSRE had an increase in RING1B occupancy and H2A119ub compared to SRE at these loci. Consistent with the repressive function of PRC1, the increased BMI1 expression in iSREs was associated with reduced expression of p16, p14, and p15 in iSREs compared to SREs (Fig. 7B). Additionally, treatment of iSREs with the BMI1 inhibitor PTC-209 caused an increase in the expression of p15 and p16 but not p27 (Fig. 7C), and siRNA-mediated knockdown of BMI1 increased p16 gene expression (Fig. 7D).
A Occupancy of indicated factors and presence of CpG islands (purple) in SRE (black) and iSRE (green) at the INK/ARF locus. Data represent merged replicates for all CUT&RUN studies. B The INK/ARF genes are expressed at lower levels in iSREs compared to SREs at day 13 of expansion. 6 independent cultures derived from 3 donors. C Relative expression of INK/ARF genes in day 25-35 iSRE following 24 hours of PTC-209 treatment relative to DMSO (Veh) control. N = 3 for p16. N = 5 for CDKN1B and CDKN2B, each derived from 2 donors. D Expression of BMI1 and p16 relative to luciferase (Luc) at 24 hours after siRNA treatment of expansion day 41-46 CD34-derived iSREs. N = 7 for Luc and BMI1-1 and N = 4 for BMI1-2, each derived from 2 donors. N = 3 for p16 with iSREs derived from 2 donors. E PTC-209 for 12 hours decreases the percentage of iSREs in S-phase in a dose-dependent manner compared to vehicle (Veh)-treated controls. 4 independent iSRE cultures expanded for 25-35 days, derived from 2 donors. F PTC-209 treatment of iSREs for 48 hours leads to dose-dependent increases of G0 cells. 4 independent iSRE cultures expanded for 25-35 days, derived from 2 donors. G PTC-209 treatment of iSRE for 12 hours leads to dose-dependent increases in early apoptotic cells. 4 independent iSRE cultures expanded for 25-35 days, derived from 2 donors. H Venn Diagram showing overlap in BMI1 occupied ribosomal protein genes (yellow) and ribosomal protein genes differentially upregulated in iSREs vs. SREs (blue). I Occupancy of indicated factors in SRE (black) and iSRE (green), and CpG islands (purple) at the Ribosomal Protein S19 (RPS19) locus. Data represent merged replicates for all CUT&RUN studies. J Occupancy of indicated factors in SRE (black) and iSRE (green) and compared with RNA-seq reads at the BMI1 locus. Boxed region is not present in the viral overexpression construct and thus represents endogenous gene expression. All graphs plot mean ± SEM. p values were calculated using a two-tailed Student's t-test. *p < 0.05 **p < 0.01 ***p < 0.001. Source data are provided for as a Source Data file.
Having identified BMI1 binding and regulation of gene expression of the INK/ARF locus, we more directly tested the function of BMI1 in the regulation of the cell cycle. Treatment of SREs and of iSREs with the BMI1 inhibitor PTC-209 caused a dose-dependent loss of S-phase (BrdU+) cells at 12 h post treatment (Fig. 7E; Supplementary Fig. 7A, D). Cell cycle analysis of iSREs also revealed a significant increase of cells in G0 (Ki67-) at 48 h of PTC-209 treatment (Fig. 7F, Supplementary Fig. 7B). These studies also revealed an increase in sub-G0 cells, consistent with an increase in cell death of iSREs (Supplementary Fig. 7B). p14ARF, which is bound by BMI1, regulates apoptosis, and p14 expression is downregulated in iSREs compared to SREs (Fig. 7B). We therefore analyzed rates of apoptosis in SREs and iSREs following BMI1 inhibition. As shown in Fig. 7G and Supplementary Fig. 7E, PTC-209 induced a dose-dependent increase in early apoptotic (PI-, Annexin5 + ) cells in iSREs and to a lesser extent in SREs. Taken together, these findings support the hypothesis that erythroid self-renewal is controlled in part by BMI1-mediated regulation of cell cycle and cell survival via the INK/ARF locus.
BMI1 positively regulates expression of ribosomal protein genes in iSREs
Gene Enrichment analyses revealed positive regulation of ribosomal proteins (Fig. 6E). Given that ribosomal protein genes have been reported to be bound by BMI1 in K562 cells14, we next examined BMI1 occupancy of these genes in human iSREs. BMI1 and RING1B were bound to the promoters of 49 of 79 ribosomal protein genes (Fig. 7H), as exemplified by the RPS19 gene (Fig. 7I). However, in contrast to the INK/ARF locus, there was minimal H2A119ub or H3K27me3 deposition at these loci. Consistent with the lack of repressive marks, 23 of the 49 ribosomal protein genes bound by BMI1 were upregulated in iSREs compared to SREs (Fig. 7H; Supplementary Fig. 7F). These data, taken together with our findings at the INK/ARF locus above, highlight contrasting roles for BMI1 and RING1b- both the classical repressive function ascribed to PRC1, as well as binding of positively regulated genes in the absence of repressive marks.
BMI1 overexpression represses the endogenous BMI1 locus
Our CUT&RUN datasets demonstrate that BMI1 binds the BMI1 locus (Fig. 7J). Further, the BMI1 locus is one of the most significant differentially bound regions between SRE and iSRE (Log2-fold change 4.95, p = 1.42E−14). BMI1 occupancy at the BMI1 locus is accompanied by gain of H2AK119Ub and H3K27Me3 in iSRE (Fig. 7J). Consistent with the gain of repressive histone modifications, RNA-Seq data reveal that the increase in BMI1 binding correlates with a downregulation of BMI1 endogenous gene expression in iSREs compared to SREs. Together, these data suggest that BMI1 regulates its own expression, with BMI1 overexpression repressing expression of the endogenous BMI1 locus.
BMI1 regulates cholesterol homeostasis in iSREs
Our CUT&RUN studies also revealed that BMI1 binds the gene promoter regions of the rate limiting enzymes of cholesterol biosynthesis (HMGCR, SQLE), of other enzymes in the cholesterol biosynthesis pathway (HMGCS1), of the primary cholesterol importer (LDLR), of the enzymes driving cholesterol esterification (ACAT1, ACAT2), as well as of the primary transcriptional regulator of cholesterol synthesis (SREBP2) (Fig. 8A, 8B; Supplementary Fig. 8). Similar to our findings of RPS19 and other ribosomal protein genes (Fig. 7H, I), CUT&RUN studies also revealed minimal H2A119ub and H3K27me3 marks despite BMI1 and RING1B occupancy at these loci, suggesting a lack of PRC1-mediated repression (Fig. 8B, Supplementary Fig. 8). Consistent with this notion, treatment of iSRE with the BMI1 inhibitor PTC-209 resulted in decreased expression of HMGCR and HMGCS1, while the upregulation of p16 in the same iSREs served as a control for the activity of the BMI1 inhibitor (Fig. 8C).
A Cholesterol homeostasis depends on cholesterol synthesis, import, export and storage. B CUT&RUN studies reveal BMI1 and RING1B bind the HMGCR locus at higher levels in iSRE than SRE. Data represent merged replicates for all CUT&RUN studies. C The BMI1 inhibitor PTC-209 decreases the expression of BMI1, HMGCR, and HMGCS1 in iSREs at 25 to 35 days of expansion. In contrast, PTC-209 treatment increases expression of p16. 6 independent iSRE cultures expanded for 25 to 35 day and derived from 2 donors, mean. ± SEM. p-value was calculated using a two-tailed Student's t-test. *p < 0.05, **p < 0.01. Source data are provided as a Source Data file. D iSREs contain cytoplasmic droplets that stain with filipin (cholesterol). Representative example of one of two cultures shown. E iSREs contain cytoplasmic droplets that stain with Nile red (lipids). Representative example of one of two cultures shown. F iSREs contain higher levels of cholesterol (filipin stain) compared to primary human proerythroblast (ProE). Staining of both cell populations normalized to primary orthochromatic erythroblasts (OrthoE). Human marrow samples derived from 3 donors. 4 independent iSRE cultures expanded for 25 to 35 day and derived from 2 donors, mean ± SEM. Two-tailed Student's t-test. **p < 0.01. Source data are provided as a Source Data file. G iSREs contain higher levels of lipid rafts (CT-B staining) compared to primary human ProE. Staining of both cell populations normalized to primary OrthoE. Human marrow samples derived from 3 donors. 2 independent iSRE cultures expanded for 25 to 35 day and derived from 2 donors. Paired two-tailed Student's t-test. *p < 0.05 mean ± range. Source data are provided as a Source Data file. H While short-term removal of exogenous lipids does not alter iSRE proliferation, concomitant block of cholesterol synthesis with Ro 48 obviates iSRE proliferation, which can be partially rescued by addition of exogenous lipids. 3 independent iSRE cultures expanded for 25 to 35 day and derived from 2 donors. p-value was calculated based on two-tailed Student's t-test, mean ± SEM. **p < 0.01, ***p < 0.001. Source data are provided as a Source Data file.
We next examined the accumulation of cholesterol in proliferating iSREs using an immunohistochemical approach with filipin and Nile red staining of cholesterol and lipids, respectively. Both stains revealed accumulation of lipids not only in the cell membrane but also in lipid droplets (Fig. 8D, E), consistent with the vacuoles present in the cytoplasm of iSRE (Fig. 2A). Since primary human erythroblasts lack such vacuoles, we compared the total levels of filipin staining in iSREs to primary human erythroid cells isolated from the bone marrow. Filipin staining was approximately 1.7-fold higher in ProE than orthochromatic erythroblasts (OrthoE), while iSRE contained approximately 2.8-fold higher levels than marrow ProE (Fig. 8F). Since membrane cholesterol can be localized to lipid rafts, we used Cholera toxin subunit B (CT-B) staining to quantify lipid rafts and found approximately 3-fold higher levels of staining in iSREs compared to normal marrow ProE (Fig. 8G). Taken together, these data indicate that iSREs accumulate cholesterol, which localizes in lipid droplets and in lipid rafts. These data also indicate that cholesterol content and lipid raft levels normally decrease as ProE undergo terminal differentiation, which also parallels the decrease seen in BMI1 transcript and protein levels.
To test the function of cholesterol synthesis and import on iSRE self-renewal, iSREs were cultured for 48 hours in the absence of cholesterol-rich lipids normally added to our cultures. This short-term removal of exogenous lipids did not affect iSRE proliferation (Fig. 8H, vehicle). However, concomitant treatment of the cultures with increasing concentrations of the cholesterol synthesis inhibitor, Ro 48, led to near complete cessation of proliferation (Fig. 8H). Addition of exogenous cholesterol-rich lipids partially rescued iSRE self-renewal. The incomplete rescue suggests that iSREs depend both on cholesterol import and on endogenous cholesterol synthesis to maintain their proliferative capability.
Discussion
RBCs are the most common form of cell-based therapy with approximately 10 million units transfused annually in the US and over 110 million worldwide. Cultured RBCs could provide standardized reagent red cells for pre-transfusion testing and ultimately supplement donor-derived units19, particularly for alloimmunized patients requiring chronic transfusions for whom compatible units are difficult to obtain. However, the limited proliferative capacity of erythroid progenitors and the fixed number of cell divisions associated with terminal erythroid precursor maturation is a major obstacle to generate sufficient numbers of cRBCs for clinical purposes. Increasing the self-renewal capacity of an erythroid progenitor would facilitate the production of greater numbers of cultured RBCs from the limited number of starting hematopoietic stem and progenitor cells available in the peripheral blood, even after mobilization.
Here, we confirmed that increasing the expression of BMI1 in erythroid progenitors leads to an expansion in the self-renewal capacity of human erythroid cells8. Four-fold overexpression of BMI1 was sufficient to increase the ex vivo self-renewal capacity of human erythroblasts by 10 billion-fold compared to untransduced and empty vector control cultures. Morphology, immunophenotyping, and global transcriptomic studies indicate that human iSREs share most similarity with proerythroblasts, which is the stage of erythropoiesis where BMI1 levels normally peak (Fig. 2E).
To better understand the function of BMI1 in erythroid self-renewal, we used a non-biased approach to screen for potential downstream targets of BMI1 in human iSREs. Consistent with BMI1 regulation of the cell cycle in multiple cell types3,30,31, we found that BMI1 and RING1B each occupy the INK/ARF locus in SREs and iSREs, which was associated with H2A119ub and H3K27Me3 deposition. These data exemplify the canonical repressive activity associated with BMI1-containing polycomb repressive complexes. Functional studies provide data consistent with the notion that BMI1 enhances the cell cycle and inhibits apoptosis in iSREs, potentially expanding their self-renewal capacity. Additionally, BMI1 and other cell cycle regulators, including E2F1 and CCND1, share downstream targets, suggesting that cell cycle control of erythroid precursors is multifaceted.
While BMI1, as a member of PRC1 along with RING proteins, is thought to act predominantly in gene repression, our CUT&RUN and transcriptomic studies surprisingly revealed nearly as many upregulated as downregulated genes associated with BMI1 occupancy in iSREs. A role for BMI1 in gene upregulation is consistent with studies in murine erythroblasts14 demonstrating that ribosomal protein genes, such as RPS19, were both bound and upregulated by BMI1, as well as a recent study in spermatogonia where BMI1 was also found to act as a transcriptional activator32. Despite extensive RING1B – BMI1 co-occupancy, the upregulated genes were in general, not associated with H2A119ub and H3K27me3 deposition. RING1B is a relatively weak ubiquitin ligase, particularly when in a complex with BMI133, and can regulate gene expression independently of its ubiquitin ligase activity21,22,23,24. Together, these data suggest that BMI1 positively regulates gene expression via mechanisms other than canonical PRC1 activity. Our data are consistent with growing evidence that Polycomb Group Proteins can bind regions of active transcription, including promoters and enhancers, which are not marked with repressive histone modifications34,35,36,37.
Our CUT&RUN studies also revealed cholesterol homeostasis as a novel and direct target of BMI1. The levels of intracellular cholesterol in cells are dynamically balanced by several processes including 1) synthesis, regulated by the master transcription factor SREBP2, 2) import primarily from LDL via LDLR also regulated by SREBP2, 3) export via ABC transporters, and 4) storage via esterification by ACAT1 and ACAT2 and transport into and out of lipid droplets. Recently, downregulation of cholesterol homeostasis was shown to regulate terminal erythropoiesis38. However, highly proliferating cells, including many cancer cell types, require high levels of cholesterol (reviewed by ref. 39). In addition, high erythroid output disorders, such as hereditary spherocytosis and thalassemia intermedia, as well as polycythemia vera are associated with hypocholesterolemia, suggesting that expansion of erythropoiesis leads to increased utilization of serum cholesterol40,41. Dexamethasone has been shown to alter lipid metabolism in human erythroblasts making them dependent on import of exogenous lipids via LDL and VLDL42. Consistent with these findings, we found that removal of exogenous lipids from the culture media reduced human erythroblast self-renewal. However, this effect only occurred after several days of culture, suggesting that iSREs can compensate through the use of stored cholesterol and/or by de novo synthesis. Our studies furthermore suggest that iSREs depend both on cholesterol import and on cholesterol synthesis to support their proliferation (Fig. 8H).
We found that BMI1 occupied multiple gene loci associated with cholesterol synthesis, import, and esterification, suggesting extensive regulation of cholesterol homeostasis. Furthermore, upregulated genes bound by BMI1 in iSREs were enriched for genes associated with lipid metabolism and SREBP2, a master regulator of cholesterol homeostasis. BMI1 has previously been implicated in regulating cholesterol homeostasis in glioblastoma cells, however, in these tumor cells BMI1 was considered to be a negative regulator of cholesterol synthesis via an indirect and undefined pathway43. In contrast, BMI1 appears to act in iSREs as a positive regulator of cholesterol biosynthesis. This includes directly binding to the promoters and increasing the expression of the regulators of the first 2 steps of cholesterol biosynthesis, including the rate-limiting HMGCR. Consistent with these findings, iSREs accumulate large amounts of cholesterol, which is localized not only in the cell membrane associated with increased lipid rafts but also in an extensive network of lipid droplets. The in vitro maturation of iSREs is associated with the loss of lipid droplets. The addition of exogenous cholesterol to the in vitro maturation of erythroblasts can affect enucleation rates and reticulocyte stability44,45. Since cholesterol content regulates multiple parameters of the RBC membrane, including gas transport and cell deformability, it will be important to further analyze the cholesterol content of iSRE-derived reticulocytes before considering their use in transfusion therapy.
Despite overexpression of BMI1, which can contribute to immortalization of multiple different cell types46,47,48, iSREs maintain a normal karyotype and a strong tendency to terminally differentiate, necessitating weekly FACS-based enrichment to maintain a population of self-renewing cells. This is in marked contrast to erythroid cell lines such as HUDEP2, BEL-A, and iEPC, which are immortalized using HPV16 E6/E7. These immortalized cell lines, unlike iSREs, have marked chromosomal abnormalities and generally lower (2–40%) rates of enucleation49,50,51,52. Our transcriptomic analysis indicates that iSREs appear poised between upstream proliferating erythroid progenitors and downstream terminally maturing erythroid precursors. Indeed, transfer of iSREs into maturation media leads to the rapid downregulation of BMI1 and their terminal differentiation into orthochromatic erythroblasts and reticulocytes, with approximately 50% rates of enucleation. We tested the ability of these differentiated cells to express common blood group antigens. Using standard gel card-based assays, Rh, S/s, Kell and Duffy antigens were detected on iSRE-derived cells. BMI1-driven expansion of the pool of immature erythroid precursors, coupled with future improvements in terminal maturation, provide a basis for the eventual generation of sufficient numbers of cultured RBCs to study erythroid intrinsic disorders, for laboratory use for RBC antibody identification, and clinically for drug delivery and transfusion therapy.
Methods
Human biological samples
All human samples used in this study were obtained after receiving informed consent. Peripheral blood mononuclear cells (PBMCs) were isolated from 40 mls of peripheral blood collected from consented individuals under the University of Rochester School of Medicine IRB approved protocol RSRB00003723. The University of Rochester Institutional Biosafety Committee approved all experiments involved with human peripheral blood. PBMCs, isolated by Ficoll-Paque Premium (Cytiva), were seeded at 0.5–1 × 107 cells/ml in “phase 1 expansion medium” consisting of Stemspan SFEM (Stem Cell Technologies) supplemented with Stem Cell Factor (SCF; 100 ng/ml, Peprotech), Erythropoietin (EPO; 2 U/ml), water-soluble dexamethasone (1uM, Sigma), IGF-1 (40 ng/ml, Peprotech), IL-3 (5 ng/ml, Peprotech), and cholesterol-rich lipids (40 mg/mL, Sigma), and cultured from expansion day 0 to day 5 (Fig. 1A).
Human CD34+ cells, purchased from the Yale Cooperative Center of Excellence in Hematology, served as an alternate source of hematopoietic stem and progenitor cells. Cells were cultured for 4 days in IMDM with Penicillin / Streptomycin, 2% PB plasma, 3% AB serum, insulin (10 ug/ml), heparin (3 U/ml), transferrin (200 ug/ml), SCF (10 ng/ml), EPO (3 U/ml), IL-3 (1 ng/ml), and Glutamax (2 mM)53. The cells were then transferred for 2 days of culture in “phase 1 expansion medium” (Supplementary Fig. 1A).
Human bone marrow cells were isolated from hip surgery samples collected through the Tissue Donation Program at Northwell Health and hip aspirates from the Hospital for Special Surgery, as IRB-approved at Northwell Health, the New York Blood Center, and the Hospital for Special Surgery. Primary human proerythroblasts and orthochromatic erythroblasts cells were identified by flow cytometry, according to the immunophenotype described54.
Lentiviral constructs and lentiviral transduction
The pReceiver-Lv165 human BMI1 overexpression plasmid (EX-B0015-Lv165, GeneCopoeia) and the pReceiver-Lv165 empty vector plasmid (EX- NEG-Lv165, GeneCopoeia) contain an EEF1A1 promoter with an IRES2 followed by an eGFP fluorescent marker. Site-directed mutagenesis was used to create an additional Lv165 BMI1 overexpression vector with a P2A sequence instead of the IRES by designing primers using the New England Biolabs (NEB)-base changer website (https://nebasechanger.neb.com) in combination with the Q5 Site-Directed Mutagenesis kit (NEB, E0554S). Lentiviral particles were harvested from HEK293T cells 72 hours post transduction, stored at −80 °C, and added to cultured PBMCs or CD34+ cells at day 5 or day 6, respectively (Fig. 1A and Supplementary Fig. 1A). Following overnight culture, cells were pelleted and resuspended in fresh “phase 1 expansion media” at a concentration of 5 × 105 cells/ml for 24 h.
SRE/iSRE expansion cultures
Two or three days following lentiviral transduction, PBMC- or CD34+ -derived cells, respectively, were reseeded at 5 × 105 cells/ml in “phase 2 expansion media”, which consists of the same components as “phase 1 expansion media”, but without IL-3 (Fig. 1A and Supplementary Fig. 1A). Following a further two days of culture, DAPI- GFP+ cells were enriched by fluorescence-activated cell sorting (FACS) and resuspended in “phase 2 expansion media” at a concentration of 5 × 105 cells/ml (Fig. 1A). The DAPI-, FSChi, GFPhi self-renewing iSREs were enriched weekly by FACS (Supplementary Fig. 1B). Total live cells were counted daily and the cell concentration was brought to 5 × 105 total cells/mL with at least half volume medium changes. The daily fold expansion was calculated by the number of live cells counted on that day, normalized to the number of live cells that seeded the culture. Most experiments were performed with PBMC-derived iSREs and we note in the figure legends when CD34-derived iSREs were used.
To test the function of exogenous factors, iSREs were cultured in “phase 2 expansion media” with the exclusion of either EPO, SCF, or dexamethasone. The minus-dexamethasone condition also included the glucocorticoid receptor antagonist, RU486 (1 μM; Sigma). Total live cells were counted daily.
iSRE maturation culture
iSREs were transferred at a concentration of 1 × 106 cells/ml into “maturation media” consisting of 1X Iscove’s Modified Dulbecco’s Medium (IMDM; Gibco), SCF (100 ng/ml), EPO (2 U/mL), 5% plasma-derived serum (PDS; Animal Technologies), 10% serum replacement (Invitrogen), 10% Protein-Free Hybridoma Media (PFHM-II; Invitrogen), 1:10 MTG (12.7 ul/100 ml; Sigma), and 20% BIT (Stem Cell Technologies). The SCF was removed after day 3 of maturation, and cells were cultured at 2 × 106 cells/ml from maturation day 4 to 7.
Knockdown of BMI1 expression
Three different lentiviral BMI1 shRNAs were generated by site-directed mutagenesis. iSREs were cocultured overnight with lentiviral particles containing BMI1 shRNA or a Luciferase control shRNA and resuspended in fresh “phase 2 expansion media” at a concentration of 5 × 105 cells/ml. Four days post lentiviral shRNA transduction, the transduced cells were enriched 4 days later by FACS for single, live (DAPI-negative), mCherry+ cells.
30 nM BMI1 siRNAs (Bmi1#1 SASI_Hs01-0017-5764/Bmi1 GAAUCAAGAUCACUGAGCAU; Bmi1#2 SASI_Hs01-0017-5768/Bmi1 CACUAAUGGAUAUUGCCU; Luciferase EHURLUC Lot: VKY) were transfected via electroporation (Lonza Nucleofector) into iSREs in Solution SF plus supplement in strip well format with Program DS-120. iSREs were subsequently diluted with pre-warmed expansion media.
BMI1 inhibition by PTC-209
The BMI1 inhibitor PTC-209 (5191, Tocris Bioscience) was suspended in DMSO and added to iSRE cultures at final concentrations ranging from 0.5 – 12.5 μM. DMSO equivalent to that used at the highest PTC-209 dose served as vehicle control.
RBC blood group antigen expression
Cryopreserved iSREs were thawed into “phase 1 expansion media”, cultured for 3 days, and resuspended in “maturation media” for 7 days19. Matured iSREs were serologically phenotyped for ABO; Rh (D, C, c, E, e); Kell (K, k); Duffy (Fya, Fyb); and Glycophorin B (S, s) using a gel card method (Ortho Diagnostics). Rh antigen typing was performed using commercial monoclonal IgM typing reagents anti-D (Alba Alpha), anti-C, anti-c, anti-E, and anti-e (Immucor Gammaclone) on buffered gel cards (Ortho Clinical Diagnostics). Kell, Duffy, and Glycophorin B antigen typing was performed using commercial anti-K, anti-k, anti-Fya, anti-Fyb, anti-S, and anti-s, (Ortho Sera) on anti-IgG gel cards (Ortho Clinical Diagnostics). Matured iSREs treated with 0.1% ficin solution (Immucor) for 10 minutes at 37 °C were washed with PBS before testing with Rh antibodies. 1 to 1.5 million mature iSREs resuspended in 50 μL of Micro Typing Systems 2 diluent (MTS2, Ortho Clinical Diagnostics) were added to each gel column then incubated with 25 μL of each typing reagent antibody for 30 min at room temperature for buffered gel cards or at 37 °C for anti-IgG gel cards before centrifugation on the MTS Workstation (Ortho Clinical Diagnostics) at 1032 rpm for 10 min.
Plasma from type O and type A patients with antibody testing performed for clinical care was collected after informed consent under a protocol approved by the IRB at the Children’s Hospital of Philadelphia. 1 to 1.5 million iSREs were added to anti-IgG cards (Ortho Clinical Diagnostics) and incubated with 25 μL plasma at 37 °C for 30 min followed by centrifugation on the MTS Workstation (Ortho Clinical Diagnostics).
Analysis of iSREs
Morphologic analysis
50,000 iSREs in PB2 (Dulbecco phosphate-buffered saline, 0.1% glucose, and 0.3% bovine serum albumin [Gemini Bioproducts]) were cytospun (Shandon II; Thermo-Scientific) onto a glass slide and air dried. Slides were stained with Wright-Giemsa and analyzed using the Nikon Eclipse 80i microscope, DS-Fi1 camera, and NIS Elements Software (Nikon).
Cell cycle analysis
BrdU incorporation was performed using the BrdU Flow kit (Supplementary Table 1) according to manufacturer’s instructions. iSREs were stained with BrdU for one hour. Ki67 staining was performed on ethanol-fixed and permeabilized iSRE stained with Ki67-APC antibody (Supplementary 1). BrdU and Ki67 data were collected using an LSR-II (BD Biosciences) flow cytometer and analyzed using FlowJo software (version 10; BD Biosciences).
Apoptosis assay
500,000 iSREs were resuspended in Annexin V binding buffer (Fisher, #NC1267456) and stained with Annexin V-AF647 and propidium iodide (Supplementary Table 1) according to manufacturer’s instructions. Data were collected on an LSR-II and analyzed using FlowJo software.
Karyotype analysis
iSREs (5 × 106 cells) from three donors were cultured for 24 hours in expansion media and then harvested using a standard cytogenetic method in the URMC Cytogenetics laboratory55. Briefly, cells were incubated with Colcemid (100 ng/ul) at 37 °C for 20 minutes, then treated with a hypotonic solution (0.075 M KCl) at 37 °C for 20 min, and washed three times in fresh Carnoy’s fixative. Chromosomes were analyzed by G-banding using trypsin digestion and Wright’s staining (GTW). Twenty metaphase spreads were analyzed for each cell line at approximately the 425-band level.
Flow cytometry
Surface immunophenotyping was carried out on cells blocked in 10% normal mouse serum (Invitrogen) in PB2 and then stained with the antibodies (Supplementary Table 1). DRAQ5 (Ebioscience) was used as a DNA stain when indicated. For lipid analysis, cells were stained with anti-cholera toxin beta unit antibody (CT-B, Sigma) according to manufacturer’s instructions, washed in PB2, and stained with surface antibodies as above. For filipin staining, surface-stained cells were fixed in 4% formaldehyde for 15 min, washed in PB2, and stained with 100 ug/ml filipin (Sigma) for 3 h. Standard flow cytometric analysis and sorting were carried out utilizing the LSR-II and Aria-II flow cytometers, respectively, using DIVA (v.8) acquisition software (BD Biosciences) and analyzed with FlowJo software (v.10, BD Biosciences). At least 100,000 events were collected for each sample.
Imaging flow cytometry was carried out using the ImageStream GenX with INSPIRE (v3.0) acquisition software (Amnis/Cytek), and analyzed with the IDEAS software (v.6.2, Amnis/Cytek). At least 50,000 events were collected for each sample. Cytoplasmic area was determined using a combined Morphology mask based on staining of CD44, CD235a, CD71, CD36, and Adaptive Erode at 81% of the brightfield. These were used alone or in combination with each other, depending on the stain used. The nuclear area was determined using the Morphology mask of DNA stain (DRAQ5) mask as previously described56.
Histochemistry
Nile red staining (Sigma) was performed in solution on paraformaldehyde-fixed iSREs at a concentration of 2.5 ug/ml for 30 min at room temperature. Filipin staining (Sigma) also occurred in paraformaldehyde fixed cells at a concentration of 50ug/ml for 2 h at room temperature after cells were pre-blocked in 1.5 mg/ml glycine in PBS. All staining was performed in the dark, and cells were then cytospun on slides, counterstained with DAPI (2ug/ml, Sigma), mounted, and viewed with widefield fluorescence microscopy.
qPCR
RNA was harvested using the Qiagen RNeasy kit following manufacturer’s guidelines. Quantitative reverse-transcription polymerase chain reaction was performed as described57 using Taqman Gene Expression Assays (Thermofisher, Supplementary Table 2) with an annealing temperature of 60 °C. Transcript expression was calculated relative to the 18S control gene.
Western blotting
Cells were resuspended in RIPA buffer (Cell Signaling Technology), vortexed, sonicated, and frozen. Following 4–20% gradient SDS page gel (Biorad) electrophoresis, proteins were transferred to a 0.2 um nitrocellulose membrane and stained with primary antibodies including BMI1 (6964S, Cell Signaling Technology), H4 (13919, Cell Signaling Technology), Beta globin (sc-21757, Santa Cruz), Gamma globin (sc-21756, Santa Cruz), Alpha globin (sc-514378. Santa Cruz), or Epsilon globin (ab156041, Abcam). The HRP-conjugated secondary antibodies were detected with WesternSure PREMIUM Chemiluminescent Substrate and quantitated on a LICOR C-DiGit Blot Scanner.
RNA-Seq
Sequencing of poly A-selected mRNA from three replicates of SRE and iSRE cells was performed at the Yale Center for Genome Analysis. For comparison of iSRE data to stage-selected RNA-sequencing data in GEO GSE53983 and GSE128268, reads were mapped to the human genome using STAR58, and reads in genes were counted using featureCounts59. For comparison of iSRE and SRE data, reads in genes were quantified using kallisto60. Differential expression between different sample types was identified using DESeq261.
CUT&RUN
CUT&RUN was performed as outlined in ref. 62. Proliferating iSREs in “phase 2 expansion media” were cultured past day 30 and collected for analysis and compared to SRE cultures at expansion day 13. CUT&RUN was performed using the CUTANA ChIC/CUT&RUN Kit (Epicypher) using 260,000 to 500,000 cells/sample and the following antibodies: BMI1 (D20B7, Cell Signaling), RING1B (D22F2, Cell Signaling), H2AK119Ub (D27C4, Cell Signaling), and H3K27me3 (9733, Cell Signaling). Libraries were generated using the NEBNext Ultra II DNA Library Prep kit (NEB). For transcription factors (BMI1 and RING1B), size selection was optimized for small fragments (end prep for 20 °C for 30 min followed by 50 °C for 60 min, ligation AMPure XP cleanup at 1.75X, annealing temperature of 65 °C for library amplification, and library AMPure XP cleanup using 1st round negative selection at 0.8X followed by a 2nd round positive selection at 1.2X). Libraries were sequenced with illumina HiSeq, paired end 150 bp length reads.
Fastq files were aligned to hg38 using Bowtie2, and duplicates were removed using Picard. Read count normalization (RPKM) was performed on alignment files. Replicate variability was assessed by Spearman correlation plots (see Supplementary Data) and genome track visualization, resulting in exclusion of one outlier BMI1 iSRE replicate for downstream analyses. Peaks were called on individual replicates using macs2 callpeak (narrowPeak), and union peak sets were used for DiffBind differential analysis in R (BMI1 CUT&RUN iSRE N = 5, SRE N = 4). Volcano plots were generated in R. Replicate merged bigwig files were generated using bigWigMerge (deepTools) for heatmap and genome track visualization. Heatmaps were generated using plotHeatmap (deepTools). Kmeans clustering was performed with plotHeatmap --kmeans and --outFileSortedRegions. Genomic regions were annotated using Homer annotatePeaks.pl. Region gene associations were determined as the single nearest gene within 2 kb of TSSs. Enrichr was used to identify gene associations63.
Statistics
Differences between groups were compared with an unpaired one-tailed or two-tailed Student’s t-test. A two-tailed paired Student’s t-test and two-way ANOVA analyses with Bonferroni correction were used when appropriate and p-values are indicated in the figure legends. Statistical analyses were performed using Excel (Microsoft Corp., Redmond, WA) or Prism software (GraphPad Software, San Diego, CA).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
RNA sequencing data and primary CUT&RUN data generated in this study have been deposited in GEO under accession code GSE253291 and GSE253289, respectively, under superseries GSE253292. Source data are provided with this paper.
References
Bryder, D., Rossi, D. J. & Weissman, I. L. Hematopoietic stem cells: The paradigmatic tissue-specific stem cell. Am. J. Pathol. 169, 338–346 (2006).
Sieweke, M. H. & Allen, J. E. Beyond stem cells: Self-renewal of differentiated macrophages. Science 342, 1242974 (2013).
Kobayashi, M. et al. Bmi1 maintains the self-renewal property of innate-like B lymphocytes. J. Immunol. 204, 3262–3272 (2020).
Panzenbock, B., Bartunek, P., Mapara, M. Y. & Zenke, M. Growth and differentiation of human stem cell factor/erythropoietin-dependent erythroid progenitor cells in vitro. Blood 92, 3658–3668 (1998).
von Lindern, M. et al. The glucocorticoid receptor cooperates with the erythropoietin receptor and c-kit to enhance and sustain proliferation of erythroid progenitors in vitro. Blood 94, 550–559 (1999).
England, S. J., McGrath, K. E., Frame, J. M. & Palis, J. Immature erythroblasts with extensive ex vivo self-renewal capacity emerge from the early mammalian fetus. Blood 117, 2708–2717 (2011).
Kim, A. R. et al. Bmi-1 regulates extensive erythroid self-renewal. Stem Cell Rep. 4, 995–1003 (2015).
Liu, S. et al. Bmi1 enables extensive expansion of functional erythroblasts from human peripheral blood mononuclear cells. Mol. Ther. 29, 1918–1932 (2021).
van Lohuizen, M. et al. Identification of cooperating oncogenes in e mu-myc transgenic mice by provirus tagging. Cell 65, 737–752 (1991).
Park, I. K. et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423, 302–305 (2003).
Wang, H. et al. Role of histone H2A ubiquitination in polycomb silencing. Nature 431, 873–878 (2004).
Cao, R., Tsukada, Y. & Zhang, Y. Role of bmi-1 and ring1a in h2a ubiquitylation and hox gene silencing. Mol. Cell 20, 845–854 (2005).
Bhattacharya, R. et al. Bmi-1: At the crossroads of physiological and pathological biology. Genes Dis. 2, 225–239 (2015).
Gao, R. et al. Bmi1 promotes erythroid development through regulating ribosome biogenesis. Stem Cells 33, 925–938 (2015).
Qin, K. et al. Molecular basis of polycomb group protein-mediated fetal hemoglobin repression. Blood 141, 2756–2770 (2023).
Migliaccio, G. et al. Under hema conditions, self-replication of human erythroblasts is limited by autophagic death. Blood Cells Mol. Dis. 47, 182–197 (2011).
An, X. et al. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood 123, 3466–3477 (2014).
Li, J. et al. Isolation and transcriptome analyses of human erythroid progenitors: Bfu-e and cfu-e. Blood 124, 3636–3645 (2014).
An, H. H. et al. The use of pluripotent stem cells to generate diagnostic tools for transfusion medicine. Blood 140, 1723–1734 (2022).
Hill, B. C. et al. Ficin-treated red cells help identify clinically significant alloantibodies masked as reactions of undetermined specificity in gel microtubes. Lab Med. 48, 24–28 (2017).
Boyle, S. et al. A central role for canonical prc1 in shaping the 3d nuclear landscape. Genes Dev. 34, 931–949 (2020).
Eskeland, R. et al. Ring1b compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell 38, 452–464 (2010).
Francis, N. J., Kingston, R. E. & Woodcock, C. L. Chromatin compaction by a polycomb group protein complex. Science 306, 1574–1577 (2004).
Zhang, Y. et al. The polycomb protein ring1b enables estrogen-mediated gene expression by promoting enhancer-promoter interaction and r-loop formation. Nucleic Acids Res. 49, 9768–9782 (2021).
Wang, S. et al. Target analysis by integration of transcriptome and chip-seq data with beta. Nat. Protoc. 8, 2502–2515 (2013).
Blackledge, N. P. & Klose, R. J. The molecular principles of gene regulation by polycomb repressive complexes. Nat. Rev. Mol. Cell Biol. 22, 815–833 (2021).
Kallin, E. M. et al. Genome-wide uh2a localization analysis highlights bmi1-dependent deposition of the mark at repressed genes. PLoS Genet 5, e1000506 (2009).
Jacobs, J. J. et al. The oncogene and polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397, 164–168 (1999).
Dhawan, S., Tschen, S. I. & Bhushan, A. Bmi-1 regulates the ink4a/arf locus to control pancreatic beta-cell proliferation. Genes Dev. 23, 906–911 (2009).
Cales, C. et al. Inactivation of the polycomb group protein ring1b unveils an antiproliferative role in hematopoietic cell expansion and cooperation with tumorigenesis associated with ink4a deletion. Mol. Cell Biol. 28, 1018–1028 (2008).
Rizo, A. et al. Repression of bmi1 in normal and leukemic human CD34 (+) cells impairs self-renewal and induces apoptosis. Blood 114, 1498–1505 (2009).
Liu, R. et al. Bmi1 fine-tunes gene repression and activation to safeguard undifferentiated spermatogonia fate. Front Cell Dev. Biol. 11, 1146849 (2023).
Taherbhoy, A. M., Huang, O. W. & Cochran, A. G. Bmi1-ring1b is an autoinhibited ring e3 ubiquitin ligase. Nat. Commun. 6, 7621 (2015).
Giner-Laguarda, N. & Vidal, M. Functions of polycomb proteins on active targets. Epigenomes 4, 17 (2020).
Zhao, W. et al. Polycomb group ring finger proteins 3/5 activate transcription via an interaction with the pluripotency factor tex10 in embryonic stem cells. J. Biol. Chem. 292, 21527–21537 (2017).
Xu, J. et al. Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol. Cell 57, 304–316 (2015).
Loubiere, V. et al. Coordinate redeployment of prc1 proteins suppresses tumor formation during drosophila development. Nat. Genet. 48, 1436–1442 (2016).
Lu, Z. et al. Fine-tuning of cholesterol homeostasis controls erythroid differentiation. Adv. Sci.9, e2102669 (2022).
Huang, B., Song, B. L. & Xu, C. Cholesterol metabolism in cancer: Mechanisms and therapeutic opportunities. Nat. Metab. 2, 132–141 (2020).
Gilbert, H. S., Ginsberg, H., Fagerstrom, R. & Brown, W. V. Characterization of hypocholesterolemia in myeloproliferative disease. Relation to disease manifestations and activity. Am. J. Med. 71, 595–602 (1981).
Shalev, H. et al. Hypocholesterolemia in chronic anemias with increased erythropoietic activity. Am. J. Hematol. 82, 199–202 (2007).
Zingariello, M. et al. Dexamethasone predisposes human erythroblasts toward impaired lipid metabolism and renders their ex vivo expansion highly dependent on plasma lipoproteins. Front Physiol. 10, 281 (2019).
Freire-Beneitez, V. et al. Elucidation of the bmi1 interactome identifies novel regulatory roles in glioblastoma. NAR Cancer 3, zcab009 (2021).
Claessen, M. et al. Production and stability of cultured red blood cells depends on the concentration of cholesterol in culture medium. Sci. Rep. 14, 15592 (2024).
Bernecker, C. et al. Cholesterol deficiency causes impaired osmotic stability of cultured red blood cells. Front Physiol. 10, 1529 (2019).
Dimri, G. P. et al. The bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer Res. 62, 4736–4745 (2002).
Song, L. B. et al. Bmi-1 is a novel molecular marker of nasopharyngeal carcinoma progression and immortalizes primary human nasopharyngeal epithelial cells. Cancer Res. 66, 6225–6232 (2006).
Tan, J. J. et al. Establishment of immortalized laryngeal epithelial cells transfected with bmi1. Cell Transpl. 29, 963689720908198 (2020).
Kurita, R. et al. Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS One 8, e59890 (2013).
Trakarnsanga, K. et al. An immortalized adult human erythroid line facilitates sustainable and scalable generation of functional red cells. Nat. Commun. 8, 14750 (2017).
Daniels, D. E. et al. Comparing the two leading erythroid lines bel-a and hudep-2. Haematologica 105, e389–e394 (2020).
Bagchi, A. et al. Direct generation of immortalized erythroid progenitor cell lines from peripheral blood mononuclear cells. Cells 10, 523 (2021).
Hu, J. et al. Isolation and functional characterization of human erythroblasts at distinct stages: Implications for understanding of normal and disordered erythropoiesis in vivo. Blood 121, 3246–3253 (2013).
Yan, H. et al. Comprehensive phenotyping of erythropoiesis in human bone marrow: Evaluation of normal and ineffective erythropoiesis. Am. J. Hematol. 96, 1064–1076 (2021).
Craig, C. P. et al. Diagnosis of Foxg1 syndrome caused by recurrent balanced chromosomal rearrangements: Case study and literature review. Mol. Cytogenet 13, 40 (2020).
McGrath, K. E., Catherman, S. C. & Palis, J. Delineating stages of erythropoiesis using imaging flow cytometry. Methods 112, 68–74 (2017).
Kingsley, P. D. et al. “Maturational” globin switching in primary primitive erythroid cells. Blood 107, 1665–1672 (2006).
Dobin, A. et al. Star: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. Featurecounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with deseq2. Genome Biol. 15, 550 (2014).
Skene, P. J. & Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6, e21856 (2017).
Chen, E. Y. et al. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinforma. 14, 128 (2013).
Acknowledgements
We acknowledge the assistance of the University of Rochester Flow Cytometry Shared Resource. Funding sources include NIH U01 HL134696 (S.T.C, J.P., C.M.W.), NIH R01 HL130670 (J.P.), NIH R01 DK124777 (L.A.S.), NIH R01 DK104920 (L.A.S.), NIH R35 HL171168 (L.B.), and NIH UL1TR002001 University of Rochester CTSI pilot funding (J.L.O.). We also acknowledge the Yale Cooperative Center of Excellence in Hematology (NIH U54 DK106857) from which we purchased frozen CD34+ cell aliquots.
Author information
Authors and Affiliations
Contributions
Ka.E.M., Kr.E.M., J.L.O., V.P.S., and H.H.A. designed and conducted experiments, analyzed data, and wrote the manuscript. Ju.P. provided human bone marrow cells. A.D.K., M.G., A.R.K., B.Z., T.L.C., P.D.K., and C.M.W. designed and conducted experiments. L.B. and P.G.G. interpreted data and wrote the manuscript. S.T.C., L.A.S., and J.P. designed experiments, analyzed data, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
McGrath, K.E., Olsen, J.L., Koniski, A.D. et al. BMI1 regulates human erythroid self-renewal through both gene repression and gene activation. Nat Commun 16, 7619 (2025). https://doi.org/10.1038/s41467-025-62993-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-62993-3