Abstract
TGF-β-dependent activation of lung fibroblasts is a hallmark of Idiopathic Pulmonary Fibrosis (IPF) which results in excessive collagen deposition and progressive scarring. Collagen production by lung fibroblasts is supported by de novo synthesis of glycine, the most abundant amino acid in collagen protein. SHMT2 produces glycine by transferring a one-carbon (1 C) unit from serine to tetrahydrofolate (THF), producing 5,10-methylene-THF (meTHF). meTHF is then converted back to THF in the mitochondrial 1 C pathway. It is unknown how 1 C metabolism contributes to collagen protein production and fibrosis. Here, we demonstrate that TGF-β induces the expression of mitochondrial 1 C pathway enzymes, including MTHFD2, in human lung fibroblasts. MTHFD2 was required for TGF-β-induced cellular glycine accumulation and collagen protein production in lung fibroblasts. Pharmacologic inhibition of MTHFD2 ameliorated fibrotic responses after intratracheal bleomycin instillation in vivo. Our findings suggest that mitochondrial 1 C metabolism is a therapeutic target for IPF and other fibrotic diseases.
Similar content being viewed by others
Introduction
Idiopathic pulmonary fibrosis (IPF) is a progressive, fatal disease, which has a median survival of 3.5 years and affects approximately 150,000 people in the United States1,2. The etiology of IPF is unknown; however, the primary effector cell population is lung mesenchymal cells, particularly alveolar fibroblasts, which respond to signals from epithelial and immune cells by secreting extracellular matrix, including collagens, leading to progressive scarring3,4,5,6. Among the signals responsible for fibroblast activation, transforming growth factor-β (TGF-β) is the most potent inducer of matrix production and loss of TGF-β signaling in lung fibroblasts prevents development of fibrosis downstream of lung injury7,8. Replacement of healthy lung tissue with matrix proteins leads to progressive loss of lung function and thus, lung fibroblasts, and the production of collagens by lung fibroblasts represents an important target for the development of novel therapies for IPF6,9,10.
Metabolic reprogramming during the process of fibroblast activation is an emerging hallmark of IPF and an increasingly-investigated area for therapeutic intervention11,12,13. Lung fibroblasts activated with TGF-β exhibit increased levels of glycolysis and mitochondrial respiration which is required for pathologic phenotypes14,15. We and others have demonstrated that this metabolic reprogramming contributes to the de novo synthesis of glycine and proline, which together constitute over 50% of the amino-acid content of collagen proteins16. Inhibition of the de novo synthesis of these amino acids prevents collagen protein production downstream of TGF-β17,18,19,20,21.
Upregulation of glycine synthesis by TGF-β requires the mTOR and ATF4-dependent induction of the enzymes of the de novo serine/glycine synthesis pathway, including PHGDH (phosphoglycerate dehydrogenase), PSAT1 (phosphoserine aminotransferase), PSPH (phosphoserine phosphatase), and SHMT2 (serine hydroxymethyltransferase 2)19,22,23. SHMT2 produces glycine from serine and is the entry point for carbon into the mitochondrial one-carbon (1 C) pathway24,25,26. SHMT2 catalyzes the transfer of a 1 C group from serine to tetrahydrofolate (THF), producing glycine and 5,10-methylene-THF (meTHF). In order for the SHMT2 reaction to proceed in the direction of glycine synthesis, mitochondrial meTHF must be recycled back to THF by the enzymes of the mitochondrial 1 C pathway. First, the bifunctional dehydrogenase/cyclohydrolase, MTHFD2 (meTHF Dehydrogenase 2) catalyzes the conversion of meTHF to 10-formyl-THF. 10-formyl-THF is then converted back to THF by either MTHFD1L (meTHF Dehydrogenase 1 Like), producing formate, or by ALDH1L2 (Aldehyde Dehydrogenase Family 1 Member L2), producing CO2 and reducing NADP+ to NADPH. It is unknown how 1 C metabolism contributes to glycine synthesis and collagen protein production downstream of TGF-β signaling.
Here, we demonstrate that TGF-β induces the mRNA and protein expression of MTHFD2, MTHFD1L, and ALDH1L2. We find that MTHFD2 is required for intracellular glycine accumulation downstream of TGF-β and for collagen matrix protein production in lung fibroblasts. The combined knockdown of MTHFD1L and ALDH1L2 recapitulated the effects of MTHFD2 knockdown; however, the individual knockdown of either enzyme did not affect glycine or collagen accumulation, suggesting that lung fibroblasts have flexibility to utilize either enzyme to regenerate THF. Chemical inhibition of MTHFD2 using DS18561882 had a similar effect as MTHFD2 knockdown on glycine and collagen production downstream of TGF-β. Furthermore, treatment of mice with DS18561882 ameliorated fibrotic responses after intratracheal instillation of bleomycin. Our results demonstrate that mitochondrial one carbon metabolism is required for TGF-β-induced collagen synthesis and suggest that MTHFD2 may be a viable therapeutic target for IPF and other fibrotic diseases.
Results
TGF-β induces the expression of mitochondrial one carbon pathway enzymes through mTORC1 and ATF4
We have previously demonstrated that lung fibroblasts depend on de novo synthesis of glycine to support production of collagen protein downstream of TGF-β17,18. Synthesis of glycine from serine produces 1 C units which are transferred to THF, producing 5,10-methylene-THF (meTHF), which must be recycled back to THF to support glycine production by SHMT2 (Fig. 1a)26. To determine whether TGF-β regulates the expression of 1 C enzymes, we treated human lung fibroblasts (HLFs) with TGF-β (1 ng/mL) and measured mRNA and protein expression of the mitochondrial 1 C enzymes MTHFD2, ALDH1L2, and MTHFD1L. TGF-β significantly increased the mRNA and protein expression of all three enzymes (Figs. 1b, c and S1A–C, E). TGF-β induced the expression of these enzymes to a similar extent as stimulation with 10% fetal bovine serum (FBS, Fig. S1F).
a Schematic representation of metabolite flux through the mitochondrial and cytosolic one-carbon pathways. b qRT-PCR analysis of 1 C enzyme mRNA expression in human lung fibroblasts (HLFs) cultured in the presence or absence of TGF-β for 24 hours. c Western blot analysis of 1 C enzyme protein expression in HLFs treated with TGF-β for the indicated intervals. d qRT-PCR analysis of 1 C enzyme mRNA expression in HLFs transfected with siRNA targeting ATF4 or nontargeting siRNA. Cells were treated with TGF-β or left untreated for 24 hours. e Western blot analysis of 1 C enzyme protein expression in HLFs transfected with siRNA targeting ATF4 or nontargeting siRNA. Cells were treated with TGF-β for the indicated intervals. f qRT-PCR analysis of 1 C enzyme mRNA expression in HLFs either left untreated or treated with TGF-β in the presence or absence of the mTORC1 inhibitor Rapalink-1. g Western blot analysis of 1 C enzyme protein expression in HLFs treated with TGF-β for the indicated intervals in the presence or absence of Rapalink-1. Bar graphs represent mean ± SD, n = 3 biological replicates. Significance was calculated by unpaired two-tailed Student’s t-test (b), 2-way ANOVA with Tukey’s post-test (d) and 1-way ANOVA with Tukey’s post-test (f). d, f indicated P values are for comparisons with TGF-β-treated control samples. Source data are provided as a Source Data file.
Flux through the mitochondrial 1 C pathway supplies 1 C units to the cytosolic 1 C pathway through formate production and mitochondrial export. Formate overflow from mitochondria supplies 1 C units for the generation of purines, thymidylate, and methionine (Fig. 1a)26. In contrast to the mitochondrial pathway, which uses multiple enzymes, the cytoplasmic 1 C pathway is catalyzed only by MTHFD1. We found that while MTHFD1 transcript levels were increased by TGF-β, its protein expression was reduced compared with untreated cells (Figs. 1b, c and S1D).
Glycine synthesis and 1 C metabolism have been shown to be promoted by activation of ATF4 downstream of mTORC127,28,29,30,31. Thus, we sought to determine the requirement of these factors for the regulation of 1 C enzyme expression downstream of TGF-β. We found that TGF-β-mediated induction of MTHFD2 and ALDH1L2 was inhibited by ATF4 knockdown, while MTHFD1L and MTHFD1 were unaffected (Figs. 1d, e and S2A–D). Consistent with a role of mTORC1 in promoting ATF4 activation, inhibition of mTORC1 with the selective inhibitor Rapalink-1 resulted in similarly reduced expression of MTHFD2 and ALDH1L2, with a partial inhibition of MTHFD1L induction downstream of TGF-β (Figs. 1f, g and S2E–H). Similar findings were found when mTOR was inhibited with TORIN1, which is an inhibitor of both mTORC1 and mTORC2 (Fig. S3).
Mitochondrial one carbon metabolism is required for TGF-β-induced collagen protein production in HLFs
We have previously shown that SHMT2-mediated production of glycine is required to support collagen protein production in lung fibroblasts17. Because the SHMT reaction requires mitochondrial THF regeneration (Fig. 1a), we sought to determine whether mitochondrial 1 C metabolism is required for collagen protein production downstream of TGF-β. We thus knocked down MTHFD2 in HLFs and treated them with TGF-β. Consistent with a critical role of mitochondrial 1 C metabolism in promoting collagen protein production, we found that Collagen 1 protein accumulation downstream of TGF-β was inhibited by MTHFD2 knockdown in HLFs (Fig. 2a, b). Levels of α-smooth muscle actin protein were not significantly affected by MTHFD2 knockdown (Fig. 2a, b). We assessed the requirement of MTHFD2 for TGF-β-induced collagen production in multiple clones of lung fibroblasts from control donor lungs and from patients with IPF (Fig. S4A–E). We found that MTHFD2 was required for the production of collagen 1 in all clones. TGF-β-induced production of collagen 3 protein was similarly inhibited by MTHFD2 knockdown (Fig. S4A–E). The sensitivity of collagen production to MTHFD2 knockdown was not affected by the presence of 10% FBS in the media (Fig. S4F). We also measured the secretion of collagens into the culture media. Consistent with reduced collagen protein production, levels of collagen 1 and collagen 3 proteins were reduced in the media of MTHFD2 knockdown HLFs (Fig. S4G). By contrast, levels of fibronectin in the media of MTHFD2 knockdowns were not significantly changed compared with media from control cells.
a Western blot analysis of collagen and α-smooth muscle actin (α-SMA) protein expression in human lung fibroblasts (HLFs) transfected with siRNA targeting MTHFD2 or nontargeting siRNA. Cells were treated with TGF-β for the indicated intervals. b Quantification of Collagen 1 and α-SMA levels in MTHFD2 knockdown HLFs relative to control cells at the indicated intervals. c Western blot analysis of GCN2 and S6-kinase phosphorylation and Collagen 1 expression in control and MTHFD2 knockdown HLFs. Cells were cultured in the presence or absence of extracellular glycine and were treated with TGF-β for the indicated intervals. Blots are representative of 4 separate experiments. d Western blot analysis of puromycin incorporation into newly translated proteins. Control and MTHFD2 knockdown HLFs were treated with TGF-β for 48 hours or left untreated. Cells were pulse labeled with puromycin for the indicated intervals. e qRT-PCR analysis of COL1A1, ACTA2, and COL3A1 mRNA expression in control and MTHFD2 knockdown HLFs either left untreated or treated with TGF-β for 48 hours. f Heatmap analysis of TGF-β-induced gene expression (differentially expressed genes between control knockdown untreated and control knockdown TGF-β, P < 0.01) in control and MTHFD2 knockdown HLFs treated with TGF-β for 48 hours. g Volcano plot showing differentially expressed genes between control knockdown TGF-β and combined MTHFD2 knockdown TGF-β-treated HLFs. h Proliferation analysis of control and MTHFD2 knockdown HLFs. i, j Analysis of cellular migration of control and MTHFD2 knockdown HLFs. k, l Analysis of contraction of control and MTHFD2 knockdown HLFs embedded in agarose and treated with TGF-β for 24 hours before gel release. All plots are presented as mean ± SD. Significance was calculated by 1-way ANOVA with Dunnett’s post-test (b, n = 3 independent experiments), (h, n = 4 biological replicates), (j, n = 16 biological replicates), (l, n = 4 biological replicates), 2-way ANOVA with Tukey’s post-test (e, n = 3 biological replicates), and quasi-likelihood F-test (f, n = 3 biological replicates). Source data are provided as a Source Data file.
MTHFD2 knockdown did not affect TGF-β-induced phosphorylation of the mTOR target S6-kinase or induce phosphorylation of GCN2, indicating that MTHFD2 inhibition does not cause stress that leads to inhibition of general protein translation (Fig. 2c). As we have previously shown17, removal of glycine from the culture media did not inhibit collagen protein production downstream of TGF-β in control cells (Fig. 2c); however, removal of glycine from the media of MTHFD2 knockdown cells resulted in activation of GCN2, and greater inhibition of collagen production than MTHFD2 knockdown alone. Phosphorylation of S6-kinase was not affected by MTHFD2 knockdown in glycine deficient conditions, suggesting that mTORC1 is not sensitive to cellular glycine levels.
To confirm that MTHFD2 knockdown does not inhibit general protein translation, we pulse-labeled cells with puromycin for the final 5, 10, 15, or 20 minutes of TGF-β treatment to label actively translating proteins. As shown in Fig. 2d, TGF-β greatly increased the rate of incorporation of puromycin into polypeptide chains as indicated by western blot for puromycin. MTHFD2 knockdown did not decrease the incorporation of puromycin into newly translated peptides, suggesting specificity for the effect of MTHFD2 knockdown on collagens.
Consistent with a post-transcriptional role of mitochondrial 1 C metabolism in regulating collagen production, knockdown of MTHFD2 did not affect TGF-β-induced expression COL1A1 and COL3A1 mRNA as measured by qPCR (Fig. 2e). We also sequenced RNA from control and MTHFD2 knockdown HLFs treated with TGF-β or left untreated. Figure 2f shows heatmap analysis of all significant differentially expressed genes between untreated and TGF-β-treated control knockdown HLFs. Figure 2g shows volcano plot analysis of all differentially expressed genes between TGF-β-treated control and MTHFD2 knockdown HLFs. We found no significant differences in TGF-β-induced gene expression between control and MTHFD2 knockdown cells.
Finally, we assessed the effect of MTHFD2 knockdown on other fibroblast functions. Consistent with lack of a stress response, MTHFD2 knockdown did not induce cell death in HLFs (Fig. S4H). When cells were cultured in growth medium, we observed decreased proliferation in MTHFD2 knockdown cells, consistent with previous reports showing a role of mitochondrial 1 C metabolism in promoting cell growth27,32 (Fig. 2h). Wound closure of confluent layers of HLFs was not affected by MTHFD2 knockdown (Fig. 2i, j). We then embedded control and MTHFD2 knockdown cells in collagen gels and stimulated with TGF-β for 24 hours before releasing the gels from the side of the wells. Consistent with a lack of effect of MTHFD2 knockdown on TGF-β-induced expression of α-smooth muscle actin, we did not observe a significant effect of MTHFD2 knockdown on contraction of HLFs (Fig. 2k, l). Together, these findings suggest that the major role of MTHFD2 in lung fibroblasts is to promote collagen production.
Mitochondrial one carbon metabolism is required for TGF-β-induced cellular glycine accumulation
As we have previously demonstrated the crucial role of de novo glycine synthesis for collagen production17,18, we assessed glycine synthesis in control and MTHFD2 knockdown HLFs using gas chromatography mass spectrometry (GC-MS). Cells were cultured in media containing 13C2-glycine (glycine labeled with 13C on both carbon atoms) to allow us to determine the relative contribution of extracellular (M + 2) glycine as well as glycine synthesized de novo from either extracellular glucose or serine (M + 0) to total cellular glycine pools (Fig. S5A). As shown in Fig. S5B, unlabeled glycine, derived from either extracellular glucose or extracellular serine constituted over half of intracellular glycine in untreated cells. TGF-β increased both total cellular glycine and levels of de novo-synthesized (M + 0) glycine in HLFs (Fig. S5C, D). Both total glycine and de novo-synthesized glycine were reduced in MTHFD2 knockdown cells (Fig. S5C, D).
MTHFD2 knockdown was also associated with a significant increase in cellular serine levels as well as increased levels of cellular serine labeled from extracellular glycine (Fig. S5E–G). This suggests that in the absence of MTHFD2, catabolism of serine is inhibited and reverse flux through SHMT2 is increased, leading to increased serine synthesis from glycine. This was confirmed in SHMT2 knockdown cells, which also displayed reduced total glycine and de novo synthesized glycine levels (Fig. S6A–D). While SHMT2 knockdown cells had elevated total serine levels compared with control knockdown cells, labeling of serine from extracellular glycine was inhibited in SHMT2 knockdown cells (Fig. S6E–G). These findings are consistent with recent reports on the reversibility of SHMT233,34.
While the percentage of intracellular glycine from de novo production did not differ between control and MTHFD2 knockdown HLFs, intracellular serine and glycine are known to exchange extensively with extracellular serine and glycine at steady state, even when there is no net uptake or release of these amino acids35,36,37 (Fig. S7A). Indeed, we observed accumulation of de novo synthesized glycine in the culture medium of HLFs over the experimental time course, which was reduced in MTHFD2 knockdown HLFs consistent with a reduced rate of glycine synthesis in these cells (Fig. S7B, C). HLFs cultured in the absence of extracellular glycine exhibited increased production of glycine from extracellular 13C3-serine (Fig. S7D–G); however, MTHFD2 knockdown HLFs exhibited greatly reduced levels of intracellular glycine when cultured in the absence of extracellular glycine (Fig. S7H–J). Thus, MTHFD2 is an important determinant of intracellular glycine levels in HLFs and is required for TGF-β-mediated increases in intracellular glycine concentration.
To better parse the relative contributions of the individual mitochondrial 1 C enzymes to production of glycine and other downstream metabolites, we labeled control and MTHFD2 knockdown HLFs with 2,3,3-D3-Serine (serine labeled with one deuterium atom on the 2-carbon, and 2 deuterium atoms on the 3-carbon), which has been used to demonstrate the contribution of 1 C metabolism to downstream metabolic pathways38,39 (Fig. 3a). Consistent with our findings in Fig. S5, cellular glycine levels, and glycine synthesized from serine were increased in TGF-β-treated cells (Figs. 3b, c and S8A). Total cellular serine levels were elevated in MTHFD2 knockdown cells, as were M + 1 and M + 2 serine resulting from reverse flux through SHMT (Figs. 3d, e and S8B).
a Schematic representation of metabolite labeling downstream of 2,3,3-D3-Serine. Human lung fibroblasts (HLFs) were transfected with siRNA targeting MTHFD2 or nontargeting siRNA. Cells were labeled with 2,3,3-D3-Serine and treated with TGF-β for 48 hours or left untreated. b–g Gas chromatography/mass spectrometry analysis of cellular metabolites. b Analysis of cellular glycine labeling from 2,3,3-D3-Serine in control and MTHFD2 knockdown HLFs. c Relative levels of M + 1 glycine from (b). d Analysis of cellular serine after labeling with 2,3,3-D3-Serine in control and MTHFD2 knockdown HLFs. e Relative levels of M + 1 and M + 2 serine from (d). f Analysis of cellular proline labeling from 2,3,3-D3-Serine in control and MTHFD2 knockdown HLFs. g Relative levels of M + 1 and proline from (f). h Relative levels of media formate content in NHLFs treated with TGF-β or left untreated. i–k Liquid chromatography/mass spectrometry analysis of cellular metabolites. i Analysis of cellular GTP labeling from 2,3,3-D3-Serine in control and MTHFD2 knockdown HLFs. j Analysis of cellular ATP labeling from 2,3,3-D3-Serine in control and MTHFD2 knockdown HLFs. k Analysis of cellular SAM labeling from 2,3,3-D3-Serine in control and MTHFD2 knockdown HLFs. Bar graphs represent mean ± SD, n = 3 biological replicates (b–g, i–k), n = 4 biological replicates (h). Significance was calculated by 2-way ANOVA with Tukey’s post-test. Source data are provided as a Source Data file.
The reduction of NADP+ by ALDH1L2 leads to labeling of mitochondrial NADPH with deuterium, which can then be transferred to proline during its de novo synthesis (Fig. 3a)38,39. We thus tested the hypothesis that TGF-β-induced glycine and proline biosynthesis may be linked by an NADPH/NADP+ redox cycle in which NADPH produced during glycine synthesis is used to support proline synthesis. Indeed, we found that TGF-β increased cellular proline levels and labeling of cellular proline (M + 1) with deuterium from serine (Fig. 3f, g and S8C). While labeling of proline was abolished in MTHFD2 knockdown cells, TGF-β-induced proline accumulation was not inhibited. Thus, we conclude that while mitochondrial 1 C metabolism contributes to mitochondrial NADPH pools, it is not required to support cellular proline production.
Formate, which is exported from mitochondria and can be released from the cell, was elevated in the media of control cells after TGF-β treatment, consistent with increased MTHFD1L activity (Fig. 3h). MTHFD2 knockdown reduced formate accumulation in the media of TGF-β-treated HLFs. Together, our findings demonstrate that both ALDH1L2 and MTHFD1L are active in HLFs after treatment with TGF-β
To better understand the effects of MTHFD2 knockdown on HLFs, we used liquid chromatography mass spectrometry (LC-MS) to assess the effects of MTHFD2 knockdown on nucleotides and redox cofactors. We found that MTHFD2 knockdown did not significantly reduce nucleotide levels in HLFs (Fig. S9A). Tracing of 2,3,3-D3-Serine into the purine synthesis pathway showed that 10-formyl-THF-dependent labeling of IMP, ATP, and GTP was increased by TGF-β treatment (Figs. 3i, j and S9B–D). While labeling was reduced in MTHFD2 knockdown cells, total levels of these metabolites were not affected (Fig. S9B–D). Labeling of S-adenosylmethionine (SAM) by 2,3,3-D3-Serine was similar to that of ATP, consistent with a role for 1C-dependent de novo ATP synthesis in promoting production of SAM, without a major role for 1C-dependent remethylation of homocysteine (Fig. 3k and S9E)40. Total levels of SAM were not affected by MTHFD2 knockdown (Fig. S9E).
Metabolism of serine also contributed to cellular NADH pools in TGF-β-treated HLFs as low levels of labeling from 2,3,3-D3-serine were detected on cellular malate (<2%), aspartate (<1%), and lactate (<1%) (Fig. S10A–C). Consistent with this low level of MTHFD2-dependent labeling, knockdown of MTHFD2 did not significantly affect total cellular levels of NADH or NAD+ (Fig. S10D). Similarly, MTHFD2 knockdown did not affect total cellular levels of NADPH, NADP+ , or glutathione (GSH, GSSG) (Fig. S10E, F).
Lung fibroblasts can use either ALDH1L2 or MTHFD1L to support glycine production
Because our findings suggest that both ALDH1L2 and MTHFD1L are active downstream of MTHFD2 in lung fibroblasts, we sought to determine whether either pathway was required for TGF-β-induced glycine and collagen production. Knockdown of either ALDH1L2 or MTHFD1L did not prevent TGF-β-induced increases in cellular glycine levels or lead to a significant decrease in M + 1 glycine derived from 2,3,3-D3-Serine (Figs. 4a, e and S11A, B and S12A, B). An increase in total serine levels and in M + 2 serine (Figs. 4b, f and S11C, D, S12C, D) was detected after either ALDH1L2 or MTHFD1L knockdown, suggesting a level of inhibition in the mitochondrial 1C pathway that was insufficient to prevent TGF-β-induced glycine accumulation. Consistent with NADP+ reduction by ALDH1L2, TGF-β-induced proline labeling was abolished in ALDH1L2 knockdowns (Figs. 4c and S11F) while MTHFD1L knockdown had no effect (Figs. 4g and S12F). Total proline accumulation was unaffected by either knockdown, consistent with our findings from MTHFD2 knockdowns (Figs. S11E and S12E). Conversely, and consistent with the role of MTHFD1L in producing formate downstream of MTHFD2, MTHFD1L knockdown inhibited formate accumulation in the media after TGF-β treatment (Fig. 4h), while ALDH1L2 knockdown had no effect (Fig. 4d).
Human lung fibroblasts (HLFs) were transfected with the indicated siRNAs targeting ALDH1L2, MTHFD1L, or both enzymes. a–c Analysis of cellular glycine (a) serine (b) and proline (c) in ALDH1L2 knockdown NHLFs after labeling with 2,3,3-D3-Serine in the presence or absence of TGF-β for 48 hours. d Relative levels of media formate in ALDH1L2 knockdown HLFs treated with TGF-β or left untreated for 48 hours. e–g Analysis of cellular glycine (e), serine (f), and proline (g) in MTHFD1L knockdown HLFs after labeling with 2,3,3-D3-Serine in the presence or absence of TGF-β for 48 hours. h Relative levels of media formate in MTHFD1L knockdown HLFs treated with TGF-β or left untreated for 48 hours. i–k Analysis of cellular glycine (i) serine (j) and proline (k) in ALDH1L2 + MTHFD1L knockdown HLFs after labeling with 2,3,3-D3-Serine in the presence or absence of TGF-β for 48 hours. l Relative levels of media formate in ALDH1L2 + MTHFD1L knockdown HLFs treated with TGF-β or left untreated for 48 hours. m, n Western blot analysis of Collagen 1 and α-SMA protein expression in control and ALDH1L2 knockdown HLFs. Cells were treated with TGF-β for the indicated intervals. o, p Western blot analysis of Collagen 1 and α-SMA protein expression in control and MTHFD1L knockdown HLFs. Cells were treated with TGF-β for the indicated intervals. q, r Western blot analysis of Collagen 1 and α-SMA protein expression in control and ALDH1L2 + MTHFD1L knockdown HLFs. Cells were treated with TGF-β for the indicated intervals. Bar graphs represent mean ± SD, n = 3 biological replicates (a–c, e–g, i–k), n = 3 independent experiments (n, p, r), n = 4 biological replicates (d, h, l). Significance was calculated by two-way ANOVA with Tukey’s post-test (d, h, l) or 1-way ANOVA with Dunnett’s post-test (n, p, r). Source data are provided as a Source Data file.
Consistent with a lack of effect of ALDH1L2 or MTHFD1L knockdown on TGF-β-induced glycine accumulation, knockdown of either gene did not affect collagen induction in HLFs treated with TGF-β (Fig. 4m–p). We thus knocked down both genes to determine whether HLFs have the flexibility to use either ALDH1L2 or MTHFD1L to regenerate mitochondrial THF. We found that combined knockdown of both MTHFD1L and ALDH1L2, inhibited TGF-β-induced production of glycine from serine (Figs. 4i and S13A, B) and mimicked the effect of MTHFD2 knockdown on serine and proline labeling as well as on media formate accumulation (Figs. 4j–l and S13C–F). Consistent with the reduction of TGF-β-induced glycine synthesis with knockdown of both ALDH1L2 and MTHFD1L, the combination knockdown also reduced collagen production downstream of TGF-β (Fig. 4q, r).
Our results suggest that mitochondrial formate production and cytosolic 1 C metabolism are dispensable for collagen protein production downstream of TGF-β. Consistent with this, knockdown of the cytosolic enzyme MTHFD1 did not affect glycine or collagen production downstream of TGF-β (Fig. S14). Moreover, formate supplementation, which rescues cytosolic 1 C metabolism when mitochondrial 1 C metabolism is inhibited41, was insufficient to rescue the effects of MTHFD2 knockdown on collagen production by HLFs (Fig. S15A). Treatment of HLFs with methotrexate, which reduces cytosolic THF by inhibiting dihydrofolate reductase similarly had no effect on collagen protein production downstream of TGF-β (Fig. S15B). Finally, to determine whether folate supplementation into the media could rescue MTHFD2 knockdown, we added increasing doses of 5-methyl-THF to the media of MTHFD2 knockdown HLFs. Consistent with an absolute requirement of mitochondrial THF regeneration for collagen protein production by lung fibroblasts, 5-methyl-THF was unable to rescue the effects of MTHFD2 knockdown (Fig. S15C). This finding is consistent with previous reports demonstrating high levels of compartmentalization of cellular folate pools with limited transport42,43.
MTHFD2 is upregulated by fibrotic responses in vivo
Our results demonstrate that mitochondrial 1 C metabolism is critical for TGF-β-induced elevations in glycine content and collagen production in cultured lung fibroblasts. Recent studies using single-cell RNA sequencing (scRNAseq) have demonstrated heterogeneity in fibroblast populations from both healthy and fibrotic lungs and have shed light on the changes that occur during disease pathogenesis. Tsukui and colleagues demonstrated that mouse alveolar fibroblasts give rise to fibroblasts with inflammatory and fibrotic phenotypes after bleomycin-induced lung injury (Fig. 5a)8. Moreover, the development of fibrotic fibroblasts and lung fibrosis after bleomycin was shown to be completely dependent on TGF-β signaling in alveolar fibroblasts. We thus analyzed expression of 1 C metabolic enzymes in lineage-labeled alveolar fibroblasts from mice 0, 7, 14, and 21 days after bleomycin instillation as published and annotated by Tsukui et al.8. We found that the expression of Mthfd2, Aldh1l2, and Mthd1l was elevated in alveolar fibroblasts after bleomycin instillation, peaking at day 7 after bleomycin and remained elevated until day 21 (Figs. 5b, c and S16A). We then examined the expression of these enzymes in fibroblast subpopulations in fibrotic lungs 21 days after bleomycin instillation. Mthfd2, Aldh1l2, and Mthfd1l exhibited the highest expression in fibrotic fibroblasts compared with alveolar and inflammatory fibroblasts (Figs. 5d and S16B). Expression of Mthfd1 was highest in alveolar fibroblasts (Figs. 5d and S16B).
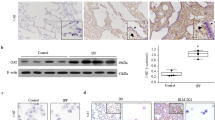
a Uniform manifold approximation and projection (UMAP) clustering of lineage labeled alveolar fibroblast populations from mice as annotated by Tsukui et al.8. b UMAP representation of the expression of Mthfd2 in lineage labeled alveolar fibroblasts at the indicated day after bleomycin instillation. c Dot plot representation of the expression of 1 C metabolic enzymes in total lineage labeled alveolar fibroblasts at the indicated day after bleomycin instillation. d Dot plot representation of the expression of 1 C metabolic enzymes in fibroblast subpopulations as in (a) on Day 21 after bleomycin instillation. e UMAP clustering of lung fibroblast populations from 10 control lungs and 20 lungs from patients with pulmonary fibrosis as defined by Habermann et al.44. f UMAP representation of the expression of MTHFD2 in human lung fibroblasts as in (e). g Dot plot representation of the expression of 1 C metabolic enzymes in fibroblast subpopulations as in (e). h UMAP representation of subclustered alveolar fibroblasts and myofibroblasts from (e) showing alveolar, inflammatory, and fibrotic groups. i UMAP representation of the expression of MTHFD2 in human lung fibroblasts as in (h). j Dot plot representation of the expression of 1 C metabolic enzymes in fibroblast subpopulations as in (h). k Histological analysis of Mthfd2 protein expression in mouse lung tissue 21 days after instillation of either bleomycin or saline (vehicle). l Histological analysis of MTHFD2 protein expression in lung tissue from a control donor, or a patient with IPF. m Pearson correlation of MTHFD2 gene expression with two independent measures of lung function (FVC-pre-BD and DLCO) in patients with IPF. Chart represents best fit linear regression with 95% confidence interval. Data is from GSE32537.
scRNAseq studies on tissue from human patients with pulmonary fibrosis have similarly highlighted a diversity of fibroblast populations. Lung fibroblasts from control donor lungs consist primarily of fibroblast and myofibroblast populations (Fig. S17A)44. In lungs from patients with pulmonary fibrosis, the myofibroblast population is expanded and disease-specific PLIN2+ and HAS1High populations emerge (Fig. S17A)44. We recently found that TGF-β-regulated metabolic enzymes, including PHGDH and SHMT2 are upregulated in these disease-specific fibroblast populations23. We thus analyzed expression of 1 C pathway enzymes in scRNAseq data from 10 control subjects and 20 patients with pulmonary fibrosis published by Habermann and colleagues44. We found that expression of MTHFD2 was increased in lung fibroblasts of patients with pulmonary fibrosis compared with control subjects (Fig. S17B, C). Expression of MTHFD2 was highest in disease-specific PLIN2+ and HAS1High populations but was also elevated in myofibroblast populations compared with fibroblast populations (Fig. 5e–g).
Tsukui and colleagues demonstrated that the fibroblast and myofibroblast populations from human data sets are themselves heterogeneous populations and identified alveolar, inflammatory, and fibrotic fibroblast populations within the fibroblast and myofibroblast populations of Habermann’s dataset8. We thus reclustered these populations and categorized subpopulations based on alveolar, inflammatory, and fibrotic markers as we have previously (Figs. 5h and S18A)23. We found that MTHFD2 and ALDH1L2 were most highly expressed in fibrotic fibroblasts compared with alveolar fibroblasts (Figs. 5i, j, and S18B). We found that MTHFD1L was expressed most highly in alveolar fibroblasts, suggesting that in vivo, fibroblasts may be more dependent on ALDH1L2 than MTHFD1L (Figs. 5j and S18B).
We found that Mthfd2 protein was highly upregulated in mouse lung tissue after bleomycin instillation (Fig. S19A). Immunohistochemical analysis showed that Mthfd2 protein is expressed most highly in fibrotic areas of mouse lung on day 21 after bleomycin instillation (Figs. 5k and S19B). Analysis of tissue from patients with IPF showed that MTHFD2 is expressed in fibroblastic foci of IPF patient lungs with very little staining detected in lung tissue from control donors (Figs. 5l and S19C–H). Finally, we examined a publicly available data set in which lung tissue gene expression from patients with IPF was associated with DLCO (diffusing capacity for carbon monoxide) and FVC-pre-BD (forced vital capacity prebronchodilator), two independent measurements of lung function45. We found that MTHFD2 expression correlated negatively with both measures of lung function in IPF patients (Fig. 5m). Together, our findings suggest that mitochondrial 1 C metabolism is upregulated in lung fibroblasts during fibrosis in vivo, and that MTHFD2 expression correlates negatively with patient outcomes.
MTHFD2 promotes fibrotic responses in vivo
DS18561882 is a small molecule inhibitor of MTHFD2 that has been shown to inhibit cancer cell growth and T-cell-mediated inflammation in vivo46,47,48. We found that treatment of HLFs with DS18561882 inhibited TGF-β-induced collagen protein production post-transcriptionally (Figs. 6a and S20A, B). Furthermore, DS18561882 treatment mimicked the effects of MTHFD2 knockdown on TGF-β-induced glycine accumulation and labeling from 2,3,3-D3-Serine (Figs. 6b and S20C, D). DS18561882 treatment also led to elevated cellular levels of serine, loss of labeling on proline from 2,3,3-D3-serine, and inhibition of media formate accumulation, consistent with inhibition of MTHFD2 (Figs. 6c–e and S20E–H). As with MTHFD2 knockdown, DS18561882 treatment did not significantly affect cellular levels of nucleotides; however, cellular proliferation under growth conditions was inhibited (Fig. S20I, J).
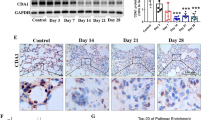
a Western blot analysis of collagen and α-smooth muscle actin protein expression in human lung fibroblasts (HLFs) treated with TGF-β for the indicated intervals in the presence or absence of DS18561882 (5 μM). b–d Analysis of cellular (b) glycine, (c) serine, and (d) proline in HLFs after labeling with 2,3,3-D3-Serine in the presence or absence of TGF-β and DS18561882. e Relative levels of media formate in HLFs treated with TGF-β or left untreated in the presence or absence of DS18561882. f–i Analysis of fibrotic responses in mice to a single dose bleomycin instillation. f Representative coronal microCT images from DS18561882 or vehicle treated mice prior to bleomycin instillation (Day 0), and 7 or 14 days after bleomycin instillation. g Quantification of aerated lung volumes from microCT-scanned mice as in (f). h Histological analysis of mouse lung tissue from DS18561882 or vehicle treated mice on Day 21 after bleomycin instillation. i Hydroxyproline content of lungs from DS18561882 or vehicle treated mice on Day 21 after bleomycin or vehicle (saline) instillation. j–m Analysis of fibrotic responses in mice after multiple bleomycin instillations. j Representative coronal microCT images from DS18561882 or vehicle treated mice on Day 70 after the first bleomycin instillation. k Quantification of aerated lung volumes from microCT-scanned mice treated as in (j). l Histological analysis of mouse lung tissue on Day 70 after the first bleomycin instillation. m Hydroxyproline content of lungs from DS18561882 or vehicle treated mice on Day 70 after the first bleomycin instillation. All plots represent mean ± SD. n = 3 biological replicates (b–d), n = 4 biological replicates (e), n = 4 mice per treatment group (g), n = 5 control mice, 10 bleomycin-treated mice per treatment group (i), n = 5 mice per treatment group (k), n = 7 mice per treatment group (m). Significance was calculated by 1-way ANOVA with Tukey’s post-test (b–e, g, i) or by unpaired two-tailed Student’s t-test (k, m). Source data are provided as a Source Data file.
We thus treated mice with DS18561882 to determine if MTHFD2 inhibition can therapeutically inhibit progression of lung fibrosis after bleomycin instillation. Mice were intratracheally instilled with bleomycin, which induces an acute lung injury/inflammatory state lasting approximately one week, followed by a fibrotic phase which lasts 2-3 weeks. To determine the therapeutic efficacy of MTHFD2 inhibition on active fibrotic processes, mice were treated with DS18561882 or vehicle beginning on day 8 after bleomycin instillation (Fig. S21A). Using micro computed tomography (microCT), we measured lung aeration at baseline and at day 7 and 14 after bleomycin instillation. All mice had similar levels of lung aeration at day 7 after bleomycin. Mice receiving vehicle displayed decreased aeration on day 14, concomitant with the induction of fibrosis. Mice receiving DS18561882 beginning on day 8 did not exhibit a significant drop in lung aeration on day 14 (Figs. 6f, g and S21C). Endpoint analysis at day 21 post-bleomycin revealed a greatly reduced degree of fibrosis in DS18561882-treated mice as assessed histologically (Fig. 6h). Lung hydroxyproline levels, an indicator of collagen content, were also significantly reduced in mice treated with DS18561882 (Fig. 6i). Similar numbers of mice from both treatment groups died during the time course of the experiment (Fig. S21D).
While initiation of fibrosis with a single instillation of bleomycin is the most commonly used model of pulmonary fibrosis, this model does not completely replicate all of the histological hallmarks of IPF and resolves spontaneously over the course of several weeks. More recently, dosing regimens involving multiple instillations of bleomycin have been shown to more faithfully replicate certain features of IPF, including development of a progressive fibrotic response that does not resolve49,50. We thus employed a multidose model in which bleomycin was instilled into the lungs of mice 3 times with each instillation separated by 14 days (Days 0, 14, and 28) (Fig. S21E). Treatment with DS18561882 or vehicle began 8 days after the last bleomycin instillation (Day 36) and endpoint analysis was conducted 42 days after the last bleomycin instillation (Day 70). Fibrosis development after multidose bleomycin was histologically distinct from that induced by single dose, including evidence of septal thickening and formation of epithelium-lined cystic structures, consistent with previous reports (Fig. S21B, F)49,50. Mice treated with DS18561882 exhibited reduced fibrosis in this model, including increased aerated lung volume, reduced fibrotic area, and decreased lung hydroxyproline content (Figs. 6j–m and S21G). Similar numbers of mice from both treatment groups died during the time course of the experiment (Fig. S21H). Together, our results suggest a critical role for MTHFD2 in promoting fibrotic responses both in vitro and in vivo.
Discussion
Metabolic reprogramming in lung fibroblasts is an emerging mechanism required to support matrix production and the development of fibrotic disease. We have previously demonstrated that TGF-β induces metabolic reprogramming in lung fibroblasts characterized by increased levels of de novo serine and glycine synthesis17,18,22. Glycine constitutes one third of the primary structure of collagens, and we and others have found that de novo glycine synthesis supports collagen synthesis downstream of TGF-β17,18,19,20,21,22. Because glycine synthesis by SHMT2 is linked with the mitochondrial 1 C pathway, we sought to determine the roles and requirements of 1 C metabolism in glycine synthesis and collagen protein production downstream of TGF-β.
We found that TGF-β significantly increases the expression of mitochondrial 1 C pathway enzymes in lung fibroblasts. Consistent with our previous findings that the enzymes of the serine/glycine synthesis pathway are regulated by mTORC1 and ATF4, we found that MTHFD2 and ALDH1L2 are induced by TGF-β through these same mechanisms. This is consistent with previous reports of regulation of these enzymes by mTORC1 and ATF4 downstream of insulin signaling27,28,30. MTHFD1L has been shown to be regulated by transcription factors including NRF2 and c-myc51,52,53. While the mechanism of MTHFD1L regulation by TGF-β remains to be determined, it is clear that metabolic pathways that support amino acid biosynthesis are among the primary targets of TGF-β-mediated transcriptional responses.
Our results demonstrate that MTHFD2 is a central regulator of glycine synthesis and collagen protein production in lung fibroblasts. Our findings suggest that loss of MTHFD2 function not only inhibits de novo glycine synthesis, but promotes the reversal of the SHMT2 reaction, potentially making MTHFD2 a more effective therapeutic target for fibrotic disease than SHMT2. The cytoplasmic 1 C pathway has been shown to reverse flux after inhibition of the mitochondrial 1 C pathway38, and may contribute to some of the de novo glycine synthesis that we observed after MTHFD2 or SHMT2 knockdown; however, this is not sufficient to maintain elevated cellular glycine levels after TGF-β treatment. Our current and previous findings show that SHMT1 and MTHFD1 expression are decreased by TGF-β treatment17, which may limit the ability of lung fibroblasts to utilize the reverse cytoplasmic 1 C pathway in the absence of MTHFD2.
MTHFD2 knockdown did not affect global protein translation, suggesting that collagen protein production is particularly sensitive to inhibition of mitochondrial 1 C metabolism. MTHFD2 knockdown slowed proliferation of lung fibroblasts but had no effect on contraction or migration. Our metabolomic assessments showed that MTHFD2 knockdown does not significantly affect levels of nucleotides in HLFs treated with TGF-β even though the contribution of serine to de novo purine synthesis is reduced. Whether nucleotides become limiting during growth conditions will need to be assessed to determine how MTHFD2 knockdown affects proliferation of HLFs. Our findings also showed that the TGF-β-induced transcriptome was largely unaffected by MTHFD2 knockdown. While MTHFD2-dependent 1 C metabolism may regulate DNA, RNA, and histone methylation in HLFs, potentially altering chromatin conformation and RNA stability, we found that total SAM levels were not affected by MTHFD2 knockdown in HLFs despite reduced labeling from serine. Whether this reduced contribution of 1 C metabolism to SAM synthesis affects the methylome of HLFs will require further study.
While MTHFD2 is essential, we find that both MTHFD1L and ALDH1L2 are dispensable for collagen protein production by HLFs, supporting our hypothesis that it is the regeneration of mitochondrial THF and synthesis of glycine that are the essential functions of MTHFD2 for collagen synthesis, and not the downstream functions of either pathway. The effect of combined knockdown of MTHFD1L and ALDH1L2 was not as strong as MTHFD2 knockdown, likely due to the closer proximity of MTHFD2 to production of meTHF at SHMT2. Residual MTHFD2 activity in this double knockdown may cause dual MTHFD1L and ALDH1L2 inhibition to not be as effective as direct inhibition of MTHFD2.
We originally hypothesized that glycine and proline synthesis may be linked via an NADPH/NADP+ redox cycle in which glycine synthesis (which produces NADPH) supports proline synthesis (which consumes NADPH). Our findings do not support this possibility as loss of either MTHFD2 or ALDH1L2 abolishes NADPH-dependent labeling on proline without affecting cellular proline levels. Thus, other sources of mitochondrial NADPH are sufficient to support proline production in the absence of one-carbon flux. NAD kinase 2 (NADK2) has recently been shown be required for proline synthesis54,55, and thus, phosphorylation of mitochondrial NAD(H) may be the primary mechanism by which NADPH is produced to support proline synthesis. While total cellular levels of NADPH/NADP+ and NADH/NAD+ were not affected by MTHFD2 knockdown, the effect on mitochondrial levels of these cofactors will need to be determined.
Recent advances in single cell transcriptomics have highlighted the diversity and etiology of multiple fibroblast populations in fibrotic lungs, particularly fibrotic fibroblasts, which are the primary source of pathologic matrix production. Tsukui and colleagues recently demonstrated that alveolar fibroblasts are the source of fibrotic fibroblasts after bleomycin-induced lung injury8. Moreover, the development of a fibrotic fibroblast signature was dependent on TGF-β signaling in alveolar fibroblasts. Our findings demonstrate that, similar to cultured fibroblasts, TGF-β-dependent processes in vivo lead to increased mitochondrial 1 C enzyme expression, with Mthfd2 expressed at the highest level in fibrotic fibroblasts. We similarly found that MTHFD2 expression was elevated in fibroblasts from patients with pulmonary fibrosis, and that expression of MTHFD2 was elevated in fibrotic fibroblasts compared with alveolar fibroblasts. We also found that MTHFD2 was highly expressed in the pathologic PLIN2+ and HAS1High populations identified by Habermann and colleagues44. While the role and etiology of these populations has not been fully elucidated, our findings suggest that their phenotype may be dependent on mitochondrial 1 C metabolism.
We found that Mthfd2 is highly induced in mouse lung tissue after bleomycin and was most highly expressed in fibrotic areas of the lung. In human tissue, MTHFD2 was expressed in fibroblastic foci of patients with IPF. Expression was also detected in the epithelium overlaying the foci, suggesting that local TGF-β signaling promotes MTHFD2 expression in both cellular compartments. While the role of 1 C metabolism in lung epithelium is beyond the scope of this manuscript, our findings using DS18561882 in vivo suggest that MTHFD2 is a viable therapeutic target for treatment of pulmonary fibrosis. We found that DS18561882 mimics the effect of MTHFD2 knockdown on glycine accumulation and collagen production after TGF-β exposure in vitro. In vivo, DS18561882 was effective at inhibiting the development of fibrosis in two models of bleomycin-induced lung fibrosis. Effects of DS18561882 on cells other than lung fibroblasts cannot be ruled out, and experiments using conditional knockout mice will be required to show the extent to which Mthfd2 inhibition specifically in lung fibroblasts is responsible for ameliorating fibrotic responses. Nevertheless, we conclude that mitochondrial 1 C metabolism is required for collagen deposition during lung fibrosis. MTHFD2 inhibition did not appear to have toxic effects on mice, even after 5 weeks of treatment, suggesting that there is a therapeutic index for targeting this pathway. This is consistent with previous studies showing beneficial effects of DS18561882 and other MTHFD2 inhibitors in multiple cancer and inflammatory disease models46,47,48,56,57. With the continued development of drugs targeting the mitochondrial 1 C pathway58,59,60, our findings suggest that these drugs may have efficacy in the setting of fibrotic disease.
Methods
Ethics statement
The collection and use of human lung specimens were approved by the University of Chicago Institutional Review Board. Deidentified lung tissue from 3 patents with IPF who underwent lung transplantation and lung tissue from 3 healthy donor lungs were used for immunohistochemistry. All participants gave written informed consent to participate in the research. The protocol for the use of animals, including bleomycin instillation and use of DS18561882 was approved by the University of Chicago Institutional Animal Care and Use Committee.
Fibroblast culture
Normal (catalog number CC-2512) and IPF (catalog number CC-7231) human lung fibroblasts were purchased from Lonza and maintained in Fibroblast Growth Medium 2 (PromoCell, C023020). Cells were plated at 1 × 105 on 12 well plates for experiments. For experiments not involving metabolic labeling, 24 hours after plating, cells were serum starved in DMEM (Gibco, 11054020) containing 0.1% bovine serum albumin (BSA), 5.5 mM glucose, 2 mM glutamine, and 1 mM pyruvate for 24 hours prior to treatment with 1 ng/mL TGF-β (Peprotech, 100-21 C). DS18561882 (5 μM, MedChemExpress, HY-130251), TORIN1 (125 nM, MedChemExpress, HY-13003), or Rapalink-1 (2 nM, MedChemExpress, HY-111373) were added at the time of TGF-β treatment. For serine and glycine labeling experiments, MEM (Gibco, 11095080) was supplemented with MEM vitamin solution (Gibco, 1120052). For glycine labeling, 400 μM 13C2-glycine (Cambridge Isotope Laboratories, CLM-1017) and 400 μM unlabeled L-Serine (Sigma, 84959) were added. For serine labeling, 400 μM 2,3,3-D3-L-Serine or 13C3-L-Serine (Cambridge Isotope Laboratories, DLM-582 or CLM-1574) and 400 μM unlabeled Glycine (Sigma, 50046).
siRNA knockdowns
For siRNA knockdowns, 1×106 HLFs were transfected with 250 pmol ON-TARGETplus siRNA (Dharmacon) using an Amaxa Nucleofector 2b set to program A-024. Cells were plated on 10 cm dishes for 24 hours and then replated for experiments as above. Dharmacon product numbers: ATF4 (si1: J-005125-10, si2: J-005125-11), MTHFD2 (si1: J-009340-09, si2: J-009349-11), MTHFD1L (si1: J-009949-09, si2: J-009949-12), ALDH1L2 (si1: J-026918-09, si2: J-026918-12), MTHFD1 (si1: J-009577-05, si2: J-009577-06), SHMT2 (si1: J-004906-05, si2: J-004906-06).
Western blotting
Cells were lysed, and electrophoresis was performed as we previously described61. Wells were lysed in 100 μL Urea Sample Buffer (8 M deionized urea, 1% SDS, 10% Glycerol, 60 mM Tris pH 6.8, 0.1% pyronin-Y, 5% β-mercaptoethanol). Lysates were run through a 28 gauge needle and were electrophoresed on Criterion gels (Bio-Rad) and transferred to nitrocellulose using a Trans-Blot Turbo (Bio-Rad) set to the Mixed MW program. Primary antibodies used were: Collagen 1 (Abcam, ab138492), Collagen 3 (Proteintech, 22734-1-AP), α-SMA (Sigma, A2547), MTHFD2 (Proteintech, 12270-1-AP), ALDH1L2 (Proteintech, 21391-1-AP), MTHFD1L (Proteintech, 18112-1-AP), MTHFD1 (Proteintech, 10794-1-AP), ATF4 (Proteintech, 10835-1-AP), Phospho-GCN2 (Cell Signaling, 94668), GCN2 (Cell Signaling, 3302), Phospho-P70S6K (Cell Signaling, 9234), P70S6K (Cell Signaling, 9202), Puromycin (Millipore, MABE343), Fibronectin (Proteintech, 66042-1-AP), GAPDH (Cell Signaling, 2118), Vinculin (Cell Signaling, 13901). Secondary antibodies were HRP-linked anti-rabbit and anti-mouse (Cell Signaling, 7074 and 7076, respectively). Blots were imaged using a ChemiDoc Touch (Bio-Rad). Densitometry was performed using ImageJ.
Quantitative PCR
RNA as isolated using GenElute Mammalian Total RNA Miniprep Kit (Sigma RTN350) and reverse transcribed using iScript Reverse Transcription Supermix (Bio-Rad, 1708841). Quantitative mRNA expression was determined by quantitative PCR using iTaq Universal SYBR Green Supermix (Bio-Rad, 172-5124). Primers used were: MTHFD2 (F: 5’-AAACACATCTGTCTGGTATGGT-3’, R: 5’-TGGTTAGGTCACAACTAGGAGTC-3’), ALDH1L2 (F: 5’-GCACTAATTGGCCAGAGCCT-3’, R: 5’-AGCCAGAGGGTCAGCTTTTC-3’), MTHFD1L (F: 5’-TTTGGTCGGAACGATGAGCA-3’, R: 5’-GTCCTGTGAGAGCCTTGTCC-3’), MTHFD1 (F: 5’-ACCCGGCCCTGTTTTTATGA-3’, R: 5’-TCCCAGTGGGCCTGAATAGA-3’), Col1A1 (F: 5’-GGTCAGATGGGCCCCCG-3’, R: 5’-GCACCATCATTTCCACGAGC-3)’, Col3A1 (F: 5’-TGGAGGATGGTTGCACGAAA-3’, R: 5’-ACAGCCTTGCGTGTTCGATA-3’), ACTA2 (F: 5’-AGCCAAGCACTGTCAGGAAT-3’, R: 5’-TTGTCACACACCAAGGCAGT-3’), RPL13 (F: 5’-GTCGTACGCTGTGAAGGCAT-3’, R: 5’-GGAAAGCCAGGTACTTCAACTT-3’).
RNA sequencing
Total RNA isolated as above was sequenced on an Illumina NovaSEQ6000 at the University of Chicago Genomics Core Facility (100 bp paired end). Sequence qualities of generated FASTQ files were assessed using FastQC. Transcript expression was quantified using Kallisto v. 0.46.162. The Kallisto index was created with the GENCODE (Human Release 45, GRCh38.14) quantification was performed in quant mode using default parameters. Gene abundances were computed using the R package txiimport v. 1.34.063. Differential gene expression analysis was performed using the edgeR package v. 4.4.2 quasi-likelihood F model64. Differential gene expression was considered significant for genes with an p-value ≤ 0.01. All packages were run in RStudio (2024.12.0 + 467) with R version 4.4.2.
Growth, viability, migration, and contraction assays
For cellular proliferation assays, 2 × 104 HLFs were plated on 6 well dishes in FGM2. Cells were trypsinized and counted by hemocytometer each day. Viability was assessed by trypan blue exclusion 72 hours after transfection with MTHFD2 or control siRNAs. For migration assays, cells were plated at 2.5×105 on 6 well TPP dishes. After 24 hours, wounds were made using 1 mm wounding tips. Wounds were imaged using a Zeiss Axiocam 105 Color at the indicated timepoints. For contraction assays cells were plated at 2×105 on 24 well plates in 0.5 mL of a 1.85 mg/mL bovine Type 1 collagen gel (Cell Biolabs, CBA201). After polymerization, 1 mL of media containing TGF-β was added on top of the gels. 24 hours later, gels were released from the side of the wells to initiate contraction.
Gas chromatography-mass spectrometry
HLFs grown for 48 hours in the presence or absence of TGF-β were washed in blood bank saline (Thermo, 23-293-184) and metabolites were extracted in 600 μL ice cold 80% Methanol (Fisher, A456 (MeOH), W6 (H2O)). The solution was vortexed and centrifuged at 21,000 × g for 20 minutes. 400 μL of each extract was transferred to a new tube and dried under nitrogen. Dried metabolites were derivatized in 16 μL Methoxamine reagent (Thermo, TS-45950) for 1 hr at 37 °C followed by 20 μL of 1% N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide (Sigma, 394882) for 1 hr at 60 °C. Derivatized samples were analyzed with an 8890 gas chromatograph with an HP-5MS column (Agilent) coupled with a 5977B Mass Selective Detector mass spectrometer (Agilent). Helium was used as the carrier gas at a flow rate of 1.2 ml/min. One microliter of each sample was injected in split mode (1:8) at 280 °C. After injection, the GC oven was held at 100 °C for 1 min and increased to 300 °C at 3.5 °C/min. The oven was then ramped to 320 °C at 20 °C/min and held for 5 minutes. The MS system was operated under electron impact ionization at 70 eV and the MS source was operated at 230 °C and quadrupole at 150 °C. The detector was used in scanning mode, and the scanned ion range was 100–650 m/z. Peak ion chromatograms for metabolites of interest were extracted at their specific m/z with Mass Hunter Quantitative Analysis software (Agilent Technologies). Ions used for quantification of metabolite levels were as follows: Glycine m/z 246, Serine m/z 390, Proline m/z 258. Mass isotopomer distributions were determined by integrating appropriate ion fragments for each metabolite using IsoCor to correct for natural abundance65. Peak areas were normalized to cell number counted from parallel wells.
Liquid chromatography-mass spectrometry
For LC/MS analysis, cells were plated on six-well dishes at 240,000 per well and treated as above. For nucleotide analyses, cells were quenched in ice-cold 300 μL of 4:4:2 acetonitrile:methanol:water containing 0.1 M formic acid. Cells were scraped and transferred to a conical tube within 3 minutes. After 3 minutes, samples were neutralized with 15% ammonium bicarbonate, incubated on ice for 20 minutes, and spun at 18,000 × g for 20 minutes at 4 °C. The supernatant was transferred to an LC-MS vial and data was immediately acquired. For stable isotope tracing, cells were quenched with dry ice cold 80% methanol, sonicated for 3 minutes, vortexed for 5 minutes at 4 °C, spun at 18,000 × g at 4 °C for 20 minutes, and the supernatant was dried using a vacuum dryer. On the day of analysis, dried pellets were resuspended in 60:40 acetonitrile:water before analysis.
Metabolite separation was performed using a Vanquish Horizon UHPLC system (Thermo) and iHILIC-(P) Classic (2.1 × 150mm, 5 μm, HILICON AB part 160.152.0520). A high resolution Orbitrap ID-X (nucleotides) or IQ-X (stable isotope tracing) Tribrid mass spectrometer (Thermo) with an H-ESI probe operating in negative mode was used to detect and quantify metabolites. The mobile phase A (MPA) was 20 mM ammonium bicarbonate at pH 9.6, adjusted by ammonium hydroxide addition and the MPB was acetonitrile. The column temperature, injection volume, and the flow rate were 40 °C, 2 μL, and 0.2 mL/minute, respectively. The chromatographic gradient was 0 minutes: 85% B, 0.5 minutes: 85% B, 18 minutes: 20% B, 20 minutes: 20% B, 20.5 minutes: 85% B, and 28 minutes: 85% B. MS parameters were as follows: Acquisition range 300–800 m/z at 60 K resolution for nucleotides, 70–800 m/z at 120 K resolution for stable isotope tracing. Separate injections were made for each ionization mode. Spray voltage was 3600 for positive ionization mode, 2800 for negative ionization mode. Sheath gas: 35, auxiliary gas: 5, sweep gas: 1, ion transfer tube temperature: 250 °C, vaporizer temperature: 350 °C, AGC target: 100%, and a maximum injection time of 118 ms. Poole QC samples were injected intermittently to determine the reproducibility of the detection and metabolite stability over time. Data acquisition was done using Xcalibur software (Thermo) and data analysis was performed using Compound Discoverer 3.3 ( ± 5 ppm) and Skyline software66. Metabolite identification was done by matching the retention time and MS/MS fragmentation to the in-house database generated using commercially available reference standards. Natural abundance correction was performed using Accucor67.
Single cell RNAseq analysis
For mouse data, pre-analyzed data from Scube2-CreER; ROSA26-tdTomato lineage-labeled alveolar fibroblasts published by Tsukui et al.8, were downloaded from Gene Expression Omnibus (GEO) (GSE210341). Cell-type annotations of fibroblasts were as defined by Tsukui et al. with the exception that Alveolar1 and Alveolar2 populations were combined into one Alveolar Fibroblast population.
For human data, preanalyzed data from 10 control subjects and 20 patients with pulmonary fibrosis published by Habermann et al.44 were downloaded from GitHub (https://github.com/tgen/banovichlab/tree/master/pulmonary_fibrosis/10x_scRNA-Seq_2019). This dataset is also publicly available at GEO: GSE135893. We excluded cells with fewer than 250 detected genes or larger than 20% mitochondrial genes. Cell-type annotation of fibroblasts was performed based on markers defined by Habermann or Tsukui as we have previously described23.
Seurat v5.3.0 package in R version 4.4.3 within RStudio (2024.12.1 + 563) was used to process and analyze scRNA-seq data, including normalization, scaling, and visualization68. Visualization of the cells and clusters on a 2D map was performed with uniform manifold approximation and projection (UMAP). Dot plots and UMAP plots overlaid with gene expression levels were generated using Seurat and the scCustomize package v. 3.0.1 (https://doi.org/10.5281/zenodo.14529706).
Analysis of gene expression and patient data
Processed gene expression datasets from GSE32537 were downloaded from Gene Expression Omnibus. Pearson correlation coefficients between gene expression levels and clinical data were calculated using the cor.test function in R, with pairwise complete observations.
Bleomycin-induced pulmonary fibrosis
Male C57Bl/6 J mice (8–12 weeks old; Jackson Laboratory) were intubated and bleomycin (0.7U/kg single dose, 0.5U/kg multi dose, TEVA Pharmaceuticals) was instilled intratracheally. Mice were sacrificed on the indicated days and perfused lung tissue was either snap frozen for hydroxyproline measurement or processed for histology. Mice receiving MTHFD2 inhibitor were IP injected with DS18561882 (MedChemExpress, HY-130251) beginning on day 8 after final bleomycin instillation. Drug was administered at 100 mg/kg in 150 μL of 10% DMSO, 40% PEG300, 5% Tween-80, 45% saline.
Micro computed tomography
MicroCT was performed on an X-Cube CT imager (Molecubes, Ghent, Belgium). Animals were anesthetized via inhalation of 2% isoflurane. MicroCT scan was performed using a respiratory-gated protocol to reduce motion artifacts. A window delay of 30% and window width of 50% was used to capture the end-expiration phase of the breathing cycle. Voltage was set to 50 kVp and current at 350 mAS. For quantitative 3D assessment of lung volume, HU range [-1000, -450] was considered as normally-aerated lung tissue. Images were processed using VivoQuant 4 patch 1 (InviCRO).
Immunohistochemistry
After deparaffinization and rehydration, tissue sections were treated with antigen retrieval buffer (DAKO, S1699) in a steamer for 20 min. Rabbit polyclonal anti-MTHFD2 antibody (Proteintech, 12270-1-AP, 1:400) was applied on tissue sections for a 1-h incubation at room temperature in a humidity chamber. Following a TBS wash, tissue sections were incubated with biotinylated anti-rabbit IgG (1:200, Vector Laboratories, BA-1000) for 30 min at room temperature. The antigen-antibody binding was detected by Elite kit (PK-6100, Vector Laboratories) and DAB (DAKO, K3468) system. Tissue sections were briefly immersed in hematoxylin for counterstaining and were covered with cover glasses.
Formate and hydroxyproline measurement
Formate content of media was measured using a Formate Assay Kit (Abcam, ab111748) on 50 μL of culture media following manufacturer’s instructions. For hydroxyproline measurements, 50 mg of cryogenically pulverized lung tissue was homogenized in 500 μL ultrapure H2O and hydroxy proline content was measured using a Hydroxyproline Assay Kit (Sigma-Aldrich, MAK357) following manufacturer’s instructions.
Statistics and reproducibility
Statistical analysis was performed using Prism 10 (GraphPad Software, Inc) v10.5.0. All data plots are presented as mean ± deviation (SD). Significance was determined using tests indicated in the figure legends. No statistical method was used to predetermine sample size. No data was excluded from the analyses. Mice were randomized to treatment groups before bleomycin instillation. Fibrosis assessment was conducted by core laboratories or by technicians with no knowledge of sample identity.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Publicly available transcriptomic data analyzed in this manuscript are available from GEO under the following accession numbers: Mouse lineage labeled alveolar fibroblast scRNAseq data from Tsukui et al.8—GSE210341, Human control and pulmonary fibrosis scRNAseq data from Habermann et al.44—GSE135893, Human lung tissue microarray and associated clinical readouts from Yang et al.45—GSE32537. RNA-seq data generated from MTHFD2 knockdown HLFs were deposited into GEO under accession number GSE302114. All other data supporting the findings of this study are found within the article, supplementary information, or source data file. Source data are provided with this paper.
Code availability
All code used in this manuscript is available at https://github.com/MutluHamanakaLab/MTHFD2 and has been linked to Zenodo under the https://doi.org/10.5281/zenodo.17049760.
References
Glassberg, M. K. Overview of idiopathic pulmonary fibrosis, evidence-based guidelines, and recent developments in the treatment landscape. Am. J. Manag Care 25, S195–S203 (2019).
Martinez, F. J. et al. Idiopathic pulmonary fibrosis. Nat. Rev. Dis. Prim. 3, 17074 (2017).
Ligresti, G. et al. Mesenchymal cells in the lung: evolving concepts and their role in fibrosis. Gene 859, 147142 (2023).
Wynn, T. A. & Ramalingam, T. R. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat. Med. 18, 1028–1040 (2012).
Wolters, P. J., Collard, H. R. & Jones, K. D. Pathogenesis of idiopathic pulmonary fibrosis. Annu. Rev. Pathol. 9, 157–179 (2014).
Scotton, C. J. & Chambers, R. C. Molecular targets in pulmonary fibrosis: the myofibroblast in focus. Chest 132, 1311–1321 (2007).
Sheppard, D. Transforming growth factor beta: a central modulator of pulmonary and airway inflammation and fibrosis. Proc. Am. Thorac. Soc. 3, 413–417 (2006).
Tsukui, T., Wolters, P. J. & Sheppard, D. Alveolar fibroblast lineage orchestrates lung inflammation and fibrosis. Nature 631, 627–634 (2024).
Staab-Weijnitz, C. A. Fighting the fiber: targeting collagen in lung fibrosis. Am. J. Respir. Cell Mol. Biol. 66, 363–381 (2022).
Sivakumar, P., Ntolios, P., Jenkins, G. & Laurent, G. Into the matrix: targeting fibroblasts in pulmonary fibrosis. Curr. Opin. Pulm. Med. 18, 462–469 (2012).
Hamanaka, R. B. & Mutlu, G. M. Metabolic requirements of pulmonary fibrosis: role of fibroblast metabolism. FEBS J. 288, 6331–6352 (2021).
Selvarajah B., Azuelos I., Anastasiou D., Chambers R. C. Fibrometabolism-an emerging therapeutic frontier in pulmonary fibrosis. Sci. Signal. 14, eaay1027 (2021).
Liu, G. & Summer, R. Cellular metabolism in lung health and disease. Annu. Rev. Physiol. 81, 403–428 (2019).
Bernard, K. et al. Metabolic reprogramming is required for myofibroblast contractility and differentiation. J. Biol. Chem. 290, 25427–25438 (2015).
Xie, N. et al. Glycolytic reprogramming in myofibroblast differentiation and lung fibrosis. Am. J. Respir. Crit. Care Med. 192, 1462–1474 (2015).
Hamanaka, R. B. & Mutlu, G. M. The role of metabolic reprogramming and de novo amino acid synthesis in collagen protein production by myofibroblasts: implications for organ fibrosis and cancer. Amino Acids 53, 1851–1862 (2021).
Nigdelioglu, R. et al. Transforming growth factor (TGF)-beta promotes de novo serine synthesis for collagen production. J. Biol. Chem. 291, 27239–27251 (2016).
Hamanaka, R. B. et al. Inhibition of phosphoglycerate dehydrogenase attenuates bleomycin-induced pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 58, 585–593 (2018).
Selvarajah B. et al. mTORC1 amplifies the ATF4-dependent de novo serine-glycine pathway to supply glycine during TGF-beta(1)-induced collagen biosynthesis. Sci. Signal. 12, eaav3048 (2019).
Schworer, S. et al. Proline biosynthesis is a vent for TGFbeta-induced mitochondrial redox stress. EMBO J. 39, e103334 (2020).
Hamanaka, R. B. et al. Glutamine metabolism is required for collagen protein synthesis in lung fibroblasts. Am. J. Respir. Cell Mol. Biol. 61, 597–606 (2019).
O’Leary, E. M. et al. TGF-beta Promotes Metabolic Reprogramming in Lung Fibroblasts via mTORC1-dependent ATF4 Activation. Am. J. Respir. Cell Mol. Biol. 63, 601–612 (2020).
Shin, K. W. D. et al. mTOR signaling regulates multiple metabolic pathways in human lung fibroblasts after TGF-beta and in pulmonary fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 328, L215–L228 (2025).
Ducker, G. S. & Rabinowitz, J. D. One-carbon metabolism in health and disease. Cell Metab. 25, 27–42 (2017).
Locasale, J. W. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer 13, 572–583 (2013).
Tibbetts, A. S. & Appling, D. R. Compartmentalization of mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 30, 57–81 (2010).
Ben-Sahra, I., Hoxhaj, G., Ricoult, S. J. H., Asara, J. M. & Manning, B. D. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733 (2016).
Park, Y., Reyna-Neyra, A., Philippe, L. & Thoreen, C. C. mTORC1 balances cellular amino acid supply with demand for protein synthesis through post-transcriptional control of ATF4. Cell Rep. 19, 1083–1090 (2017).
Reina-Campos, M. et al. Increased serine and one-carbon pathway metabolism by PKClambda/iota deficiency promotes neuroendocrine prostate cancer. Cancer Cell 35, 385–400 e389 (2019).
Torrence M. E. et al. The mTORC1-mediated activation of ATF4 promotes protein and glutathione synthesis downstream of growth signals. Elife 10, e63326 (2021).
Byles, V. et al. Hepatic mTORC1 signaling activates ATF4 as part of its metabolic response to feeding and insulin. Mol. Metab. 53, 101309 (2021).
Ye, J. et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 4, 1406–1417 (2014).
McBride, M. J. et al. Glycine homeostasis requires reverse SHMT flux. Cell Metab. 36, 103–115 e104 (2024).
Ghrayeb, A. et al. Serine synthesis via reversed SHMT2 activity drives glycine depletion and acetaminophen hepatotoxicity in MASLD. Cell Metab. 36, 116–129 e117 (2024).
Heinz, E. & Walsh, P. M. Exchange diffusion, transport, and intracellular level of amino acids in Ehrlich carcinoma cells. J. Biol. Chem. 233, 1488–1493 (1958).
Heinz, E. & Mariani, H. A. Concentration work and energy dissipation in active transport of glycine into carcinoma cells. J. Biol. Chem. 228, 97–111 (1957).
DeNicola, G. M. et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 47, 1475–1481 (2015).
Ducker, G. S. et al. Reversal of cytosolic one-carbon flux compensates for loss of the mitochondrial folate pathway. Cell Metab. 23, 1140–1153 (2016).
Gao, X. et al. Serine availability influences mitochondrial dynamics and function through lipid metabolism. Cell Rep. 22, 3507–3520 (2018).
Maddocks, O. D., Labuschagne, C. F., Adams, P. D. & Vousden, K. H. Serine metabolism supports the methionine cycle and DNA/RNA methylation through de novo ATP synthesis in cancer cells. Mol. Cell 61, 210–221 (2016).
Labuschagne, C. F., van den Broek, N. J., Mackay, G. M., Vousden, K. H. & Maddocks, O. D. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 7, 1248–1258 (2014).
Trent, D. F., Seither, R. L. & Goldman, I. D. Compartmentation of intracellular folates. Failure to interconvert tetrahydrofolate cofactors to dihydrofolate in mitochondria of L1210 leukemia cells treated with trimetrexate. Biochem. Pharm. 42, 1015–1019 (1991).
Horne, D. W., Patterson, D. & Cook, R. J. Effect of nitrous oxide inactivation of vitamin B12-dependent methionine synthetase on the subcellular distribution of folate coenzymes in rat liver. Arch. Biochem. Biophys. 270, 729–733 (1989).
Habermann, A. C. et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 6, eaba1972 (2020).
Yang, I. V. et al. Expression of cilium-associated genes defines novel molecular subtypes of idiopathic pulmonary fibrosis. Thorax 68, 1114–1121 (2013).
Kawai, J. et al. Discovery of a potent, selective, and orally available MTHFD2 inhibitor (DS18561882) with in vivo antitumor activity. J. Med. Chem. 62, 10204–10220 (2019).
Sugiura, A. et al. MTHFD2 is a metabolic checkpoint controlling effector and regulatory T cell fate and function. Immunity 55, 65–81 e69 (2022).
Zhu, Z. et al. Folate enzyme MTHFD2 links one-carbon metabolism to unfolded protein response in glioblastoma. Cancer Lett. 549, 215903 (2022).
Degryse, A. L. et al. Repetitive intratracheal bleomycin models several features of idiopathic pulmonary fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 299, L442–L452 (2010).
Redente, E. F. et al. Persistent, progressive pulmonary fibrosis and epithelial remodeling in mice. Am. J. Respir. Cell Mol. Biol. 64, 669–676 (2021).
Lee, D. et al. Folate cycle enzyme MTHFD1L confers metabolic advantages in hepatocellular carcinoma. J. Clin. Invest. 127, 1856–1872 (2017).
Pikman, Y. et al. Targeting MTHFD2 in acute myeloid leukemia. J. Exp. Med. 213, 1285–1306 (2016).
Satoh, K. et al. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc. Natl. Acad. Sci. USA 114, E7697–E7706 (2017).
Zhu, J. et al. Mitochondrial NADP(H) generation is essential for proline biosynthesis. Science 372, 968–972 (2021).
Tran, D. H. et al. Mitochondrial NADP(+) is essential for proline biosynthesis during cell growth. Nat. Metab. 3, 571–585 (2021).
Bonagas, N. et al. Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress. Nat. Cancer 3, 156–172 (2022).
Ju, H. Q. et al. Modulation of redox homeostasis by inhibition of MTHFD2 in colorectal cancer: mechanisms and therapeutic implications. J. Natl. Cancer Inst. 111, 584–596 (2019).
Liu, S., Wang, X., Zhao, L., Zhang, L. & Song, Y. MTHFD2: A significant mitochondrial metabolic enzyme and a novel target for anticancer therapy. Drug Discov. Today 30, 104372 (2025).
Dekhne, A. S., Hou, Z., Gangjee, A. & Matherly, L. H. Therapeutic targeting of mitochondrial one-carbon metabolism in cancer. Mol. Cancer Ther. 19, 2245–2255 (2020).
Cuthbertson, C. R. et al. A review of small-molecule inhibitors of one-carbon enzymes: SHMT2 and MTHFD2 in the spotlight. ACS Pharm. Transl. Sci. 4, 624–646 (2021).
Hamanaka, R. B. & Mutlu, G. M. PFKFB3, a direct target of p63, is required for proliferation and inhibits differentiation in epidermal keratinocytes. J. Invest. Dermatol. 137, 1267–1276 (2017).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Millard, P. et al. IsoCor: isotope correction for high-resolution MS labeling experiments. Bioinformatics 35, 4484–4487 (2019).
Pino, L. K. et al. The Skyline ecosystem: informatics for quantitative mass spectrometry proteomics. Mass Spectrom. Rev. 39, 229–244 (2020).
Su, X., Lu, W. & Rabinowitz, J. D. Metabolite spectral accuracy on orbitraps. Anal. Chem. 89, 5940–5948 (2017).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Acknowledgements
R01ES010524 and W81XWH-22-1-0387 (G.M.M.), and R01HL151680 (R.B.H.).
Author information
Authors and Affiliations
Contributions
Conception and design: A.Y.M., R.C.A., A.M., G.M.M., R.B.H. Acquisition of data: A.Y.M., K.D.S., Y.T., J.C.H.S., T.C., K.A.S., P.S.W., O.R.S., B.C., H.S., R.B.H. Resources: N.O.D., A.N.H. Analysis and interpretation of data: A.Y.M., R.C.A., M.V.A., N.O.D., A.N.H., J.C.H.S., A.M., H.S., G.M.M., R.B.H. Manuscript writing: A.Y.M., R.C.A., G.M.M., R.B.H. Final approval of manuscript: A.Y.M., K.D.S., R.C.A., M.V.A., Y.T., J.C.H.S., T.C., K.A.S., P.S.W., O.R.S., B.C., N.O.D., A.N.H., A.M., H.S., G.M.M., R.B.H.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Kim Kampen, Mark Jones and Victor Thannickal for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Meliton, A.Y., Shin, K.W.D., Cetin-Atalay, R. et al. Mitochondrial one-carbon metabolism is required for TGF-β-induced glycine synthesis and fibrotic responses. Nat Commun 16, 9250 (2025). https://doi.org/10.1038/s41467-025-64320-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-64320-2