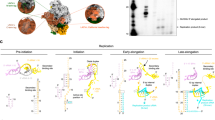
Abstract
Enteroviral replication-linked cloverleaf RNAs recruit the viral 3CD protein, a fusion of 3C protease and 3D RNA-dependent RNA-polymerase, for negative-strand synthesis during genome replication. However, the structures and mechanisms of this virological process remain unclear. Using the coxsackievirus B3 model, we determine the crystal structures of both intact cloverleaf-3C and isolated sD-3C complexes at 2.69 Å and 2.41 Å resolutions, respectively. Our structures reveal that the sD stem-loop is the sole determinant for binding two 3C monomers, with each monomer recognizing the lateral surface of the sD stem either upstream (toward the apical tetraloop) or downstream (near the dinucleotide bulge) of the Py•Py helix. Binding studies with structure-guided cloverleaf and 3C mutants further clarify the roles of specific nucleotides and residues involved in the interactions between cloverleaf and 3C, explaining earlier virological observations. Through comparative structural and binding studies of 3C, 3D, and 3CD with cloverleafs from seven different enteroviral species, we demonstrate that while the 3D domain does not contribute to cloverleaf binding, the sD sequence and its structural pattern govern 3CD-cloverleaf interactions through the 3C domain. Our work establishes a high-resolution structural framework for understanding enteroviral replication mechanisms, which will aid in developing antivirals targeting this platform.
Similar content being viewed by others
Introduction
The enterovirus genus comprises a group of RNA viruses responsible for various human diseases, including the common cold, poliomyelitis, acute flaccid paralysis, aseptic meningitis, and myocarditis1,2,3. These viruses contain a (+)-strand RNA genome with modular RNA structures within the 5′ and 3′ untranslated regions (UTRs), which promote and regulate the viral protein translation and genome replication. When the viral genome is released into the cytoplasm during infection, multiple RNA domains within the 5′ UTR, known as internal ribosome entry sites (IRESs), recruit the host cell machinery to translate the genome-encoded proteins4,5,6,7. Once sufficient viral proteins are available, viral proteases cleave the host protein factors bound to the IRES domains, such as PCBP2, to halt translation and trigger genome replication8,9. To initiate the replication, the viral 3CD protein (precursor of 3C protease and 3D RNA-dependent RNA-polymerase, RdRp) and host PCBP2 bind to a replication-linked cloverleaf-like RNA domain (CL) at the extreme 5′ end of the viral genome10,11,12,13,14,15,16,17. The host poly(A)-binding protein (PABP) binds at the 3′ UTR and subsequently interacts with the CL-3CD-PCBP2 ternary complex to facilitate genome circularization10,11,12,13,14,18,19. This assembly recruits a uridylated viral protein VPg as the primer, forming a replication-competent initiation complex20,21,22,23 (Fig. 1a). Finally, 3CD self-cleaves in trans to yield an active 3D polymerase21,24, which elongates the primer to synthesize (−)-strand RNA21,22,24. The (−)-strand RNA then acts as a template for synthesizing back the (+)-strand genomic RNA20,22, which can be translated, replicated, or packaged into new virions. Notably, since the actively translating genomic RNA is inhibited from replication and vice versa, the CL domain is also considered a molecular switch that modulates viral genome translation and replication11,25. However, the structures and mechanisms of these critical virological processes remain largely unknown.
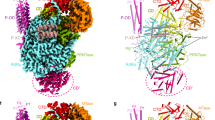
a Schematics showing the CL-mediated assembly of the replication initiation complex during (−)-strand RNA synthesis of enteroviral genome replication. b The crystal structure of intact CVB3 CL in complex with the viral 3C was solved at 2.69 Å resolution, which showed two copies of 3C monomers (designated 3C1 and 3C2) bound per RNA molecule. c The crystal structure of the sD-3C complex solved at 2.41 Å resolution shows two copies of 3C monomers bound per sD molecule, identical to that in the intact CL-3C complex. d The secondary structure of the CL showing the key features of CL-3C interactions within the sD stem-loop structure. e The superposition of the sD-3C complex with the corresponding region of the CL-3C complex (all atoms RMSD = 0.64 Å). f Two copies of 3C bind the sD stem-loop as monomers with no significant interactions between themselves except the K12 residue within the hydrogen-bonding distance ( ~ 3 Å) closer to the protein-RNA interface. g Representative chromatograms for the 21.4 kDa 3C protein (red) superimposed with a molecular size marker (blue), suggesting no dimerization of the 3C in solution. h Similar chromatograms of 3C-RNA complexes for the wild-type CL (magenta) and a sD loop mutant (C63G, C65G, G66A) CL (green). The elution peak labels for these complexes are based on the elution profile of the protein marker (blue) and 3C alone (red). i The alignment of the previously determined protein-free CL structure26,27 with the 3C-bound CL (all atoms RMSD = 1.86 Å) shows no significant change in the overall structure of the RNA upon 3C binding to the sD stem-loop. j Similarly, the alignment of the previous 3C-only structure33 with the CL-bound 3C (all atoms RMSD = 0.33 Å) shows no change in the overall structure of the 3C upon binding to the RNA. The figures in each panel and corresponding labels have been colored analogously for facile comparisons.
Using Fab-assisted RNA crystallography, we recently determined the high-resolution crystal structures of the protein-unbound CLs from three enteroviral species: coxsackievirus B3 (CVB3), rhinovirus B14 (RVB14), and rhinovirus C15 (RVC15)26,27. Our findings revealed that the enteroviral CLs adopt a conserved H-type four-way junction topology composed of four subdomains: sA, sB, sC, and sD, featuring co-axially stacked sA-sD and sB-sC helices26,27 (Fig. 1a). Notably, in the crystal structures, we also identified conserved A•C•U base-triple formation through tertiary interactions between an adenosine within the sC loop and a pyrimidine-pyrimidine (Py•Py) helix within the sD stem (Fig. 1a), which organizes the sA-sB and sC-sD helices in near-parallel orientations26,27. These conserved structural features observed in our crystal structures enabled us to generate 3-dimensional CL models for four additional enteroviral species, EV71, PV1, EVD68, and RVA2, using a homology modeling approach26. These homology models demonstrated that their sequences were fully compatible with the solved crystal structures26, indicating that enteroviral CLs maintain a conserved H-type topology, consistent with a high degree of sequence conservation across enteroviral genomes and their functionality, even in CL-swapped chimeras26,27.
While the sB-loop and a C-rich spacer sequence (3′SP) located between the CL and IRES are known to bind to PCBP225,26, the sD-loop and a dinucleotide bulge in the sD stem interact with 3CD through its 3C domain27 (Fig. 1a). Our previous studies revealed that the adjacent positioning of the two PCBP2 binding sites, the sB-loop and the 3′SP, facilitates a cooperative binding of PCBP2 at these sites (Fig. 1a). In this configuration, disrupting the sC-sD tertiary interaction by destabilizing the A•C•U base-triple reduces PCBP2 binding, even when the sB-loop and 3′SP remain intact26, leading us to propose that the spatial arrangement of the sB-loop and the spacer is crucial for the effective high-affinity binding of PCBP2, as the stability of the A•C•U base-triple allosterically modulates PCBP2’s interaction with the enteroviral CLs26. Conversely, disrupting the sC-sD tertiary interaction did not affect the affinity of CL-3C binding; the isolated sD stem-loop maintained a similar affinity for 3C as the intact CLs27, aligning with previous biochemical and virological studies that identified 3C as the RNA binding domain within 3CD17,28,29,30,31.
Nevertheless, previous gel electrophoresis-based studies have proposed that 3CD binds to the CL with about ten-fold greater affinity than the isolated 3C domain19, indicating that the CL-3CD interactions might involve additional contacts from the 3D domain19,32. Moreover, while the positioning of the sD stem-loop, sB-loop, and 3′SP suggests discrete binding of 3CD and PCBP2 proteins at opposite ends of the H-shaped CL scaffold26,27, PCBP2 showed around two orders of magnitude greater affinity to the CL in the presence of 3CD11. Although PCBP2 can bind to CLs independently of 3CD and interactions between 3CD and PCBP2 have yet to be established, 3CD binding appears to stabilize the interactions between the CL and PCBP2. Such stabilization could occur by promoting a CL conformation conducive to PCBP2 binding or providing additional surface area for PCBP2 interactions within the preformed CL-3CD complex. Either way, 3CD is crucial in modulating the interactions between CL and PCBP2, influencing negative-strand synthesis during viral genome replication. However, the structural basis for the interactions between these proteins and enteroviral CLs remains unknown, mainly due to the lack of high-resolution structures for any of these CL-protein complexes.
Here, we crystallized and determined the intact CL and isolated sD structures in complex with 3C protein at 2.69 Å and 2.41 Å resolution, respectively, using CVB3 as a model system. Our crystallographic data confirmed that the sD stem-loop alone suffices to bind 3C, with the sD-loop and dinucleotide bulge identified as the primary interaction sites. Consistent with previous molecular virology and biochemical studies11,17,27,28,29,30, we observed that each crystallized complex exhibited two 3C molecules bound per RNA, suggesting the recruitment of two 3CD molecules to the CL site during viral genome replication. Neither the CL nor the 3C structure showed a noticeable conformational change upon binding. Although the functional significance of recruiting two copies warrants further investigation, 3C and 3CD appear to exist as monomers in solution. The structure-guided binding studies with CL and 3C mutant constructs using isothermal titration calorimetry (ITC) elucidated the roles of key nucleotides and residues for specific interactions between 3C and enteroviral CLs in solution, aligning well with earlier biochemical and virological findings17,27,28,29,30. Moreover, comparing the binding interactions of CVB3, RVB14, and EV71 3Cs with CLs from seven different enteroviral species underscored that both the sequence and structure of the sD stem-loop are critical for 3CD recognition, with specificity in interactions arising from subtle variations within the sD stem-loop or 3C structures. Further binding studies with recombinantly expressed CVB3 3D and 3CD using ITC and biolayer interferometry (BLI) assays showed no significant interactions between 3D and CL and illuminated additional potential interactions of the 3CD linker region with CL, which explained why 3CD exhibited a higher affinity for the CL than 3C alone in both previous and our current binding studies. Although subtle variations in 3CD-CL interactions may occur, the high conservation of both 3CD and CL structures among enteroviruses suggests that our findings apply to a general recruitment mechanism of 3CD to the replication initiation site in enteroviral genomes and offer a structural basis to explain previous biochemical, virological, and cellular observations. Since 3C and 3D have been extensively investigated as potential therapeutic targets against enteroviruses, our structural studies pave the way for the rational design of drugs targeting the CL-3CD platform. Given the high conservation of the CL and 3CD protein three-dimensional structures among enteroviruses26,27, developing universal drugs to target multiple species that cause different diseases might be possible.
Results
Crystallization and structure determination of CVB3 CL-3C complex
Our previous binding studies have shown that 3C recognizes both the loop and a dinucleotide bulge within the sD stem-loop27. Our findings also suggested that 3C exhibits an affinity for an isolated sD stem-loop construct comparable to the intact 5′ CL27. To elucidate the structural basis of the 3C-CL interactions, we aimed to crystallize and determine the structures of both the intact 5′ CL and the isolated sD RNA construct in complex with the 3C protein, using CVB3 as our model system. The intact 5′ CL construct consists of 90 nucleotides (nts 2 to 88 from the CVB3 genome) and includes two additional G-C pairs at the start of the sA helix to improve transcription and RNA stability (Supplementary Fig. 1). Likewise, the isolated sD construct (36 nts) includes nts G47-C80 of the sD stem-loop with an additional G-C pair to close the stem (Supplementary Fig. 1). We purified recombinant CVB3 3C (see methods for details) and prepared the CL-3C and sD-3C complexes for crystallization trials. Among more than 600 conditions screened for each complex, we observed crystals for the CL-3C and sD-3C complexes, which diffracted to 2.69 Å and 2.41 Å, respectively. To solve these structures, we used the previously determined crystal structure of CVB3 3C (PDB: 2ZU1)33 as a search model for molecular replacement, which provided a robust electron density map after initial phasing and allowed us to model the RNA nucleotides unambiguously. The final structures, following iterative rounds of model building and refinement for the CL-3C (Rwork/Rfree = 19.8/23.2%) and sD-3C (Rwork/Rfree = 19.4/23.8%) complexes, were determined at resolutions of 2.69 Å and 2.41 Å, respectively. Supplementary Figs. 2 and 3 show these structures with color-coding based on the crystallographic B-factors and with the 2|Fo | -|Fc | electron density maps. Supplementary Table 1 provides details of the data collection and refinement statistics.
Composition of CL-3C and sD-3C complexes
Our crystallographic results indicate that the CVB3 CL-3C and sD-3C complexes crystallized in C121 and P212121 lattice space groups, respectively. However, the overall structures of both complexes reveal two 3C molecules (designated 3C1 and 3C2) bound to a single RNA molecule (Fig. 1b–d), and the sD-3C structure is almost identical to the corresponding region of the CL-3C complex (root mean square deviation, RMSD = 0.64 Å, Fig. 1e). Notably, no 3C interactions were identified with other areas of CL within the CL-3C complex structure, suggesting that sD is sufficient to recruit 3C, consistent with our previous binding studies27. Whereas the analysis of the macromolecular interfacial areas within the crystal lattice using PDBePISA34 for both complexes revealed crystal contacts involving protein-protein (Supplementary Fig. 4), two 3C molecules bound to the RNA in both complexes exhibited no apparent interactions (except side-chains K12s from two 3C molecules appeared within ~3 Å, Fig. 1f), suggesting that two copies of 3C bind to the sD stem-loop of the CL likely as monomers rather than as a stable dimer. Notably, 3C was previously crystallized in two lattice space groups (2.0 Å resolution; space group P21, and 1.72 Å resolution; space group C2)33. While the former contained two 3C molecules per crystallographic asymmetric unit, the latter contained only one 3C molecule. The 3C molecules in the 3C-CL complex structure face each other using different surfaces than those observed in the unbound 3C structure within the crystallographic asymmetric unit (Supplementary Fig. 5). This implies that the proximity of 3C molecules in the complex or the RNA-free structure does not indicate any specific dimerization interface within the 3C. To further confirm that 3C exists as a monomer in solution, we performed a size-exclusion chromatography (SEC) assay with the 3C and sD-3C complex to understand these crystallographic findings in solution. For the 3C protein alone (molecular weight, MW = 21.4 kDa), we observed a single peak between 13.7 and 44.3 kDa, compared to a standard protein marker, suggesting that 3C exists as a monomer in solution (Fig. 1g), which aligns with an earlier in vivo study that reported no evidence of 3C dimerization35. Additionally, the CL-3C complex exhibited a peak and a shoulder between the 150 and 44.3 kDa markers, consistent with the 1:2 CL:3C complex (MW = 71.7 kDa) and the 1:1 CL:3C complex (MW = 50.3 kDa, Fig. 1h), respectively. Moreover, a mutant CL with C63G, C65G, and G66A mutations in the sD loop that complexed with 3C under the same conditions produced a single peak corresponding to the 1:1 CL:3C complex (Fig. 1g). These mutations likely affected only one 3C binding site (i.e., upstream of the Py•Py) without impacting the other site (i.e., downstream), suggesting that 3C likely binds to the CL as two monomers rather than a dimer.
Structural features of the CL-3C complex
Since the sD-3C complex crystal structure was nearly identical to the corresponding region of the CL-3C complex (RMSD = 0.64 Å), we used the latter structure for additional structural analysis. Except for minor local variations, the previous crystal structures of protein-free CL26,27 and RNA-free 3C protein33 superimpose well with the CL-3C complex structures (RMSDs = 1.86 and 0.34 Å, respectively), suggesting that no significant conformational changes occur in either RNA or 3C upon complex formation and that 3C binds to a pre-organized RNA scaffold (Fig. 1i, j). Subtle variations, such as around the sB and sD loops, and bulge regions, reflect differences caused by the Fab-binding loop and random sampling of the dynamic bulge conformation in the 3C-free CL structure27. Although the two 3C molecules bound to the CL exhibit identical folds and interact with the RNA using the same protein surface, our structure revealed that one primarily recognizes the sD structural features toward the loop and the other toward the sD bulge, consistent with our findings from previous binding studies in solution27. However, each 3C molecule resides along the helical axis on nearly opposite sides of the sD stem-loop structure, allowing some residues of each 3C to interact with both the sD loop and bulge regions.
Like other previously reported enteroviral 3C structures33,35,36,37,38,39, our crystal structure of the CL-3C complex revealed a chymotrypsin-like fold of 3C, which consists of two topologically equivalent β-barrels (residues S15-K76 and V101-F179) that pack together to form a substrate-binding pocket for protease activity. The RNA binding region, which is located far from the catalytic site, primarily consists of an α-helix formed by N-terminal residues G1-N14, a long unstructured loop (L1, residues L77-A100) in the first β-barrel domain, and a short loop (L2, residues S153-K156) in the second β-barrel domain, creating a binding surface that aligns parallel to the sD helix (Fig. 2a). While one of the 3Cs places the α-helix and L2 close to the bulge area and L1 toward the sD loop, the other 3C mirrors the same arrangement, positioning the α-helix near the sD loop (Fig. 2b). With this sD-3C configuration at the 3C-CL interface, the 3C residues specifically interact with the RNA involving minor- and major-groove nucleotides along the sD stem (Fig. 2c). Some key interactions are located in the major groove near the Py•Py region, suggesting a crucial role for this Py•Py helix in tailoring the major groove and facilitating access for 3C residues, even though this helix does not directly interact with 3C (Fig. 2c). Specifically, nucleotides A53, G77, and C78 located downstream of Py•Py and near the bulge interact with R79, N80, E81, and K82 of 3C1 through direct hydrogen bonding and electrostatic interactions (Fig. 2d). Similar interactions of the residues R13, R84, E7, and T154 in the same 3C molecule also occur with the nucleotides C70, A69, U68, G67, and G66 located upstream of the Py•Py helix near the sD loop region (Fig. 2e). In contrast, while the residues N80, E81, K82 and D85 of the second 3C (3C2) interact with the nucleotides C71, A69, U68, U62, A61 near the sD loop region (Fig. 2f), the K156, E5, R84 and R13 residues of this 3C molecule interact with the nucleotides U49, A50, G51, C52, A53, and C54 positioned downstream of the Py•Py helix and near the bulge region (Fig. 2g). While some 3C contacts occur with the nucleobases, most of the 3C interactions happen through the phosphodiester backbone of the sD stem-loop RNA. Although the sD loop and bulge were proposed previously to dictate the 3C binding, 3C mainly recognizes the lateral surface of the sD stem, where bulge and loop define the precise gauge for organizing such a 3C binding site within the sD RNA. The interactions of two 3C monomers thus establish a consolidated electrostatically favorable positively charged binding surface to recognize a specific structural pattern of the negatively charged surface of the sD stem-loop within the enteroviral CLs (Fig. 2h).
a Structure of CL-bound 3C showing the RNA binding interface that includes N-terminal α-helix and L1 and L2 loops (colored yellow). b The crystal structure of the CL-3C complex elucidates the positions of each 3C molecule interacting with the sD stem-loop region, which mainly interacts with nucleotides upstream of the Py•Py helix (cyan) and 3C2 with nucleotides downstream of the Py•Py helix. Each 3C molecule uses the same RNA-binding surface (colored wheat for 3C1 and colored yellow for 3C2 for clarity). c The specific residues of the 3C1 (left) and 3C2 (right) involved in RNA binding interactions. The sD stem-loop has been represented as a molecular surface (wheat) with a Py•Py helix (cyan). The 3C1 residues interact with nucleotides (d) downstream and (e) upstream of the Py•Py helix. f Similarly, the 3C2 residues interact with nucleotides upstream and (g) downstream of the Py•Py helix. h The molecular surface of 3C molecules according to the electrostatics of each residue (blue = positive; red = negative; white = neutral charge) shows how two 3C molecules establish a positively charged surface for binding sD RNA (ribbon). The pale cyan meshes in d–g represent the simulated annealing composite omit 2|Fo| -|Fc| electron density map at contour level 1σ and carve radius 1.8 Å, the dashed lines represent hydrogen bonding distances, and an orange sphere in g depicts a water molecule. Each panel and corresponding labels have been colored analogously for facile comparisons.
Binding studies with CL and 3C mutants
To investigate the nature of CL-3C interactions and the roles of specific residues and nucleotides, we prepared several rationally designed, structure-guided CL and 3C mutant constructs and conducted binding assays using ITC. First, we examined 3C binding to the CL constructs with mutations at or near the sD loop (Fig. 3a). We found that a single nucleotide G66C mutation within the sD loop reduced the 3C binding affinity by more than 15-fold (apparent dissociation constant, Kd = 17.0 ± 8.0 µM; mean ± standard deviation, s.d.; n = 3) compared to wild-type RNA (apparent Kd = 0.998 ± 0.096 µM, Fig. 3b and Table 1). Although we observed only a single hydrogen bonding interaction of G66 with 3C T154 (Fig. 2e), this nucleotide appeared critical for maintaining a UNCG-type tetraloop conformation of the sD loop, which agrees well with a GNRA-type tetraloop formation in a G66C mutant crystal structure27. When comparing the structures of this mutant with 3C-bound wild-type sD, the GNRA-tetraloop appeared to tilt away from the 3C-binding surface (Fig. 3c), possibly interfering with the specific interactions between other nearby RNA and 3C residues. Notably, both 3C-bound (CVB3) and 3C-unbound PV1 and RVC15 CL structures formed UNCG-type sD tetraloops26,40, suggesting that 3C docks into the preorganized sD tetraloop structure and emphasizing the significance of the high conservation of the UNCG-type sD tetraloop among enteroviral CLs. Similarly, CL constructs with a single C63A and triple C63G, C65G, and G66A mutations to install a GAGA tetraloop exhibited comparably lower binding affinities with 3C (apparent Kd = 24.4 ± 1.1 and 24.4 ± 1.7 µM, respectively), indicating that specific sD-loop sequence and an appropriate conformation are essential for adequate recognition of the CL by 3C (Fig. 3b, Table 1). However, the A64U mutation only reduced the 3C binding affinity by approximately 3-fold (apparent Kd = 3.42 ± 0.15 µM) compared to the wild-type, which is consistent with the nucleotide being flipped out in both UNCG- and GNRA-type sD tetraloops and has no apparent interactions with the 3C (Fig. 3b, Table 1). The consensus secondary structure of enteroviral CLs indicates that most sD tetraloops are closed by a wobble U•G pair, with some exceptions, such as the EV71 and RVB14 CLs, where a C-G pair closes the sD loop. Interestingly, the U62C mutation to replace this loop-closing wobble pair with the C62-G67 pair and swapping the positions of these nucleotides with U62G and G67C double mutations resulted in no detectable binding affinity with 3C (Fig. 3b, Table 1). This aligns with the disruption of a network of hydrogen bonding and electrostatic interactions observed between this U•G pair and the 3C residues within the binding interface (Fig. 2e, f) and agrees well with previous NMR results41.
a Representations of all CVB3 CL constructs used for ITC measurements showing the positions of mutations or insertions within the sD loop. The nucleotides colored in black indicate point mutations or insertions. The red, green, and blue colored nucleotides represent the corresponding simultaneous mutations or insertions. b The apparent Kds for binding wild-type CVB3 3C with CL mutants as measured by ITC. The data with * were adopted from Das et al.27 for facile comparisons. c Alignment of 3C-bound sD and protein-free sD crystal structures showing altered conformations of the loop and bulge regions. The 3C-free sD loop forms a GNRA-type tetraloop due to the G66C mutation27, but the protein-bound WT loop forms a UNCG-type tetraloop. d Superposition of the 3C-bound sD and a hypothetical sD model (green) with the non-canonical Py•Py region replaced by canonical pairs via U72A, U73G, and U74A mutations. e The apparent Kds for binding wild-type CVB3 CL with 3C mutant constructs. The horizontal dotted line in b and d indicates the Kd for wild-type CL binding with wild-type 3C for facile comparison. The apparent Kds for cross-binding of (f) CVB3 3C, g RVB14 3C, and (h) EV71 3C with seven different enteroviral species. The reported Kds (depicted as bars) in b, e and f are the mean ± s.d. from three biological replicates, and those in g and h are the mean from two biological replicates. The filled circle indicates the Kd from each replicate, and the error bars in b, e, and f represent the s.d. (see Supplementary Figs. 6–10 for representative ITC profiles). The N.B., in b and e–h, depicts no detectable binding observed by ITC at the given conditions. Source data are provided as a Source Data file.
Second, we investigated the binding of 3C with CL constructs containing sD bulge and stem mutations. In line with the specific interactions of 3C with the dinucleotide bulge near the foot of the sD stem observed in the crystal structures, our prior 3C binding studies have demonstrated that mutations or the removal of the bulge through complementary base insertions reduce the CL binding affinity for 3C27. The comparative reinterpretation of these data in the context of the CL-3C complex crystal structures indicates that separate mutations in either the sD loop or bulge do not entirely abolish the 3C binding (Fig. 3b, Table 1), implying that each of these regions could independently bind one molecule of 3C. In alignment with these findings, a CL construct lacking both the bulge and the G66C mutation in the sD loop exhibited no detectable binding to 3C (Fig. 3b, Table 1). Additionally, although the sD loop and bulge were previously proposed as key binding sites19,31,32, most 3C residues in the complex crystal structures interacted with the RNA backbone (phosphodiester-sugar) of the sD stem upstream or downstream of the Py•Py base pairs (Fig. 2c). However, despite not directly interacting with the 3C, substituting three noncanonical base pairs in the sD (55U•74U, 56 C•73U, and 57U•72U) with their canonical counterparts (55U-74A, 56C-73G, and 57U-72A) resulted in an about 5-fold decrease in the CL binding affinity for 3C (apparent Kd = 4.88 ± 1.15 µM, Fig. 3b, Table 1). As shown previously27, the Py•Py stem in 3C-unbound CL is slightly narrower (diameter = 16.8 ± 0.3 Å) compared to the stem flanking this region (diameter = 18.6 ± 0.6 Å). While a narrower Py•Py stem caused no significant deformation of the sD stem from a standard A-form helix27, a hypothetical sD with canonical base pairs replacing the Py•Py stem exhibited a wider sD stem, which perhaps causes steric clashes with the 3C residues compared to the 3C-bound native sD (Fig. 3d). The decreased 3C affinity for these mutants thus suggests that the disruption of the narrower Py•Py helix may either sterically hinder 3C for binding or allosterically affect the interactions of specific residues in both the upstream (near the sD loop) and downstream (near the dinucleotide bulge) regions of the sD helix, thereby reducing the accessibility of the 3C residues by altering the shape-complementarity pattern in the sD structure (see Fig. 2c, h). Moreover, consistent with observed direct hydrogen bonding interactions of R13 and K82 residues from both 3C monomers (Fig. 2e, g), the A69U mutant showed about 12-fold decreased binding affinity with 3C (Fig. 3b, Table 1), explaining why this nucleotide is highly conserved across enteroviral CLs26,27.
Third, we purified structure-guided 3C mutants and performed binding studies with the wild-type CL. The ITC measurements (see Supplementary Fig. 10 and Source Data for detailed data and results) indicated that while the K156A mutant binds to CL with approximately 50-fold lower affinity (apparent Kd = 54.0 ± 2.9 µM) compared to the wild-type, the R13A mutant exhibited no detectable binding affinity (Fig. 3d, Table 1), suggesting that R13 may be a key determinant for the CL-3C binding. Structurally, K156 in one of the 3C molecules (3C2) recognizes the bulge nucleotides (U49 and A50) via electrostatic interactions and a network of hydrogen bonds. We also observed that the R13s from both 3C molecules are positioned near each other, recognizing the RNA backbone near the Py•Py helical region (A69, C70, A53, and C54). Notably, although most of the CL-3C interactions in crystal structures occurred through the RNA backbone, R13 recognizes the major groove of the sD stem just downstream and upstream of the Py•Py helix (see Fig. 2c). This suggests that the Py•Py configuration plays a prominent role in widening the major groove to facilitate 3C interactions via electrostatics and a network of hydrogen bonds. Since the structures of 3Cs appear highly conserved among enteroviruses, these findings agree well with previously observed inhibition of (-)-strand synthesis caused by the K12N and R13N mutations in PV142.
Comparison of 3C interactions among enteroviral CLs
Since the CL and 3C structures are conserved across enteroviral species26,43, we hypothesized that CVB3 3C might bind promiscuously to the sD stem-loop of various enteroviral CLs. To gain further insight into sD-3C recognition among enteroviruses, we first performed binding studies of CVB3 3C with CLs from seven different enteroviral species (see Supplementary Fig. 6 and Table 2 for detailed data and results). Our ITC measurements revealed that CVB3 3C indeed binds to PV1, EVD68, RVA2, and RVC15 CLs with a similar affinity (apparent Kds = 0.86 ± 0.04, 0.61 ± 0.01, 1.85 ± 0.16, and 0.57 ± 0.04 µM, respectively) as the CVB3 CL (Fig. 3e, Table 2), consistent with conserved structural features of the sD stem-loop among enteroviruses. However, CVB3 3C did not exhibit any binding to RVB14 and EV71 CLs (Fig. 3e, Table 2). Interestingly, consistent with our prior crystallographic structures of CVB3, RVB14, and RVC15 CLs26,27, most enteroviral CLs showed a tetranucleotide sD loop in mFold-predicted secondary structures, except a trinucleotide loop observed in RVB14 (Supplementary Fig. 11). This implies that the structural attributes of the sD loop, rather than sequence, are critical for defining the specificity of 3C interactions, consistent with earlier biochemical studies27.
Surprisingly, recombinantly purified RVB14 3C bound to all seven CLs (Fig. 3f, Table 2). While the structure of the RVB14-3C complex has yet to be determined to clarify the roles of specific nucleotides or 3C residues, RVB14 3C seems more promiscuous than CVB3 3C in binding enteroviral CLs (Fig. 3f and Table 2). Nevertheless, the superposition of the previous RVB14 3C structure39 with each copy in the CL-3C complex structure (all atoms RMSD = 0.993 Å) indicates that subtle variations in the sD loop and bulge regions may synergistically contribute to the promiscuity of RVB14 3C interactions with enteroviral CLs (Supplementary Fig. 12). On the other hand, while the EV71 sD features a tetranucleotide loop like that of CVB3 (Supplementary Fig. 11), it also did not bind to CVB3 3C (Fig. 3e, Table 2). First, the EV71 sD loop has a C-G loop closing pair, unlike the U•G wobble pair in other CLs (Supplementary Fig. 11). Second, the superposition of the EV71 3C structure36 with the CVB3 3C within the CL-3C complex structure implies that an additional motif in EV71 3C could play a crucial role in modulating the sD-3C interaction (Supplementary Fig. 12). This suggests that the 3C specificity for CL binding arises from subtle structural variations in both sD and 3C binding sites. Intriguingly, recombinantly purified EV71 3C bound to all CLs except RVA2 and RVB14 (Fig. 3g, Table 2). Despite both containing a tetraloop, RVA2 sD is characterized by a single-nucleotide sD bulge, while EV71 sD features a dinucleotide bulge (Supplementary Fig. 11). Although further studies are warranted, these observations imply that enteroviral 3C proteins may attain specificity through subtle structural variations in both sD and 3C binding interfaces. Nonetheless, enteroviral 3Cs maintain high promiscuity when binding to various CLs with structurally similar features.
Interactions of 3CD with enteroviral CLs
The 3CD protein is a multifunctional precursor that includes the 3C proteinase and 3D RNA-dependent RNA-polymerase domains linked by a seven-residue polypeptide44. It shows full protease functionality with distinct specificity but lacks polymerase activity44. Although 3C displays robust RNA-binding activity, prior gel electrophoresis-based studies have proposed that 3CD may bind enteroviral CL with approximately ten times greater affinity than 3C alone19, indicating that 3CD is a better RNA binder than 3C alone. This suggested that 3D may exhibit RNA-binding activity or induce a conformational change in the 3C domain to enhance its affinity for the CL. To further explore the interactions between CL and 3CD, we recombinantly purified CVB3 3CD and 3D proteins and performed binding assays using the CVB3 CL. First, our gel electrophoresis assay did not reveal any discrete CL-3D complex band, even at elevated concentrations of 3D, indicating that 3D failed to form a kinetically stable complex with the CL. In agreement with these findings, the ITC assay showed no measurable affinity of 3D for the CL, suggesting that 3D may lack any specific RNA-binding activity toward the CL (Supplementary Fig. 10). However, due to the low yield of recombinant 3CD, we could not measure the binding interactions by ITC, which requires a large amount of the protein, so we turned our efforts to biolayer interferometry (BLI) measurements. We immobilized a biotinylated CL construct (Fig. 4a) into the streptavidin biosensors, recorded the BLI responses upon incubation of the sensor-immobilized CL with various concentrations of the proteins, and used steady-state parameters to calculate the equilibrium Kd for the binding interactions between the CL and 3C or 3CD (see supplementary Fig. 13 for representative data). Interestingly, while 3C showed a similar affinity as measured by ITC (apparent Kd = 1.03 ± 0.3 µM, Fig. 4b) and 3D no detectable binding, 3CD exhibited an affinity to the CL approximately two times greater (apparent Kd = 0.67 ± 0.1 µM) than 3C alone (Fig. 4c), which aligns with prior research indicating that 3CD may be the protein recruited to the CL in vivo45.
a Schematics of a CVB3 CL construct used for BLI measurements of 3C, 3D, and 3CD proteins. The 3' overhang sequence in the CL construct hybridizes with a 5' biotinylated complementary DNA oligo, allowing the construct to immobilize in streptavidin biosensors. The saturated BLI responses (wavelength shift) generated by the interactions of sensor-immobilized CL for various concentrations of (b) CVB3 3C and (c) 3CD (filled circles) were fitted with a binding isotherm (dotted curve). The reported error in Kds represents the standard error of the fitting. See Supplementary Fig. 13 and the methods for details. d A model of CVB3 3CD predicted by the AlphFold3, showing 3C and 3D domains connected by a flexible linker. e Superposition of the AlphaFold3-predicted 3CD model with each 3C molecule of the CL-3C complex crystal structure, showing no apparent 3D domain interaction with the CL. f The alignment of the previously determined PV1 3CD crystal structure44 with each 3C molecule of the CVB3 CL-3C complex crystal structure shows potential contacts of the linker region in both 3CD molecules with the CL. g The apparent Kd for a mutant CVB3 3CD with E190A and K193A mutations in the linker region decreased significantly compared to the wild-type 3CD. h A proposed model for binding interactions of two 3C domains with CVB3 cre involved in VPg uridylation. The structure of the cre was predicted by AlphaFold 3 and superimposed with the sD region of the CVB3 CL. Each panel and corresponding labels, where appropriate, have been colored analogously for facile comparisons. Source data are provided as a Source Data file.
To further understand the interactions between 3CD and the CL, we generated a structural model of CVB3 3CD (Fig. 4d, see Supplementary Fig. 14 for the modeling statistics) using AlphaFold346 and superimposed it with the 3C domain of the 3C-CL complex structure (Fig. 4e). The 3C and 3D domains of this predicted 3CD closely resemble previously solved structures of isolated 3C and 3D (RMSDs = 0.31 and 0.38 Å, respectively); however, the flexible and less structured linker between them appears to be responsible for adjusting their relative orientations (Supplementary Figs. 15 and 16). Previous crystal structures of both isolated CVB3 3C and 3D also aligned well with the crystal structure of PV1 3CD (RMSDs = 0.67 and 0.93 Å, respectively; Supplementary Fig. 17), consistent with our observations for similar binding of CVB3 3C to PV1 CL (see Fig. 4f and Table 2). When superimposed with the 3C within the CL-3C complex crystal structure, the CVB3 3CD model exhibited no significant structural variations in the 3C domain (RMSD = 0.43 Å), suggesting that the presence of 3D does not induce conformational changes in the 3C’s RNA binding site (Fig. 4e). Notably, the 3D domain shows no substantial contact with the CL in this computed model. However, while the PV1 3CD similarly aligned well with the 3C domain of the CL-3C complex (RMSD = 0.68 Å), its linker region is positioned near the CL’s sD helix, indicating potential interactions with the RNA (Fig. 4f). Although we had this sequence in our isolated 3D, the high dynamicity of this N-terminus in isolated 3D may account for no detectable binding between the 3D and the CL. Consistent with CL-3CD linker interactions, a mutant CVB3 3CD (3CD-Mut) with E190A and K193A mutations near the linker region showed about a 3-fold decrease in CL binding affinity (apparent Kd = 2.03 ± 0.83 µM, Fig. 4g) compared to the wild-type 3CD (apparent Kd = 0.67 ± 0.1 µM). While the conformational changes or differences in the relative positioning of 3C or 3CD due to these mutations cannot be ruled out, such a 3CD-CL complex model underscores the importance of the linker region in the CL binding. Because of the flexible linker, 3CD likely adopts multiple conformations with varying relative positions of the 3C and 3D domains, and the CL RNA perhaps recognizes one of these sampled 3CD conformations. Although further investigation and structural determination of the 3CD-CL complex are necessary to elucidate this recognition mechanism, our binding and modeling results indicate that additional interactions occurring in the 3C-3D linker region may account for the higher affinity of 3CD for CLs compared to the isolated 3C.
Discussion
During enteroviral infections, 5′ CLs recruit various viral and host proteins, including a multifunctional viral protein called 3CD, to form the functional ribonucleoprotein (RNP) complex essential for genome replication. The 3CD consists of a 3C proteinase and a 3D RdRp component, where 3C shows RNA-binding activity and interacts with the sD stem-loop of CL to recruit intact 3CD. Since 3CD functions as a proteinase but lacks polymerase activity, cleavage of 3C releases the active polymerase required for negative-strand RNA synthesis. While separate crystal structures of 3C, 3CD, and CLs from various enteroviruses have been determined previously26,27,33,35,36,37,38,39,44,47,48, the role of 3CD in recognizing CL remained ambiguous due to the lack of high-resolution structures of the CL-3C complex. Here, we solved the crystal structures of the intact CL-3C and sD-3C complexes and characterized their binding interactions through CL and 3C mutagenesis. Our findings enhanced the understanding of how enteroviral CLs interact with 3CD during genome replication and explained previous biochemical and cellular observations at both molecular and atomic scales. Although structures of ternary complexes involving RNA and proteins would provide deeper insights into the roles of each component, our study lays out a structural framework for such future investigations.
Previous studies have demonstrated that 3C interacts with the isolated sD and the entire CL in yeast three-hybrid and mammalian cell systems45. Our structural analysis supported these findings, confirming that the sD is the sole determinant for 3C binding to enteroviral CLs. While the structures of the CL-3CD complex remain undetermined, computational models for the CL-3CD complex structure and model-guided mutagenesis studies suggest additional potential interactions of 3CD, explaining the higher affinity of 3CD-CL binding compared to isolated 3C. Our analysis has shown that the bound and unbound CL and 3C structures are nearly identical, with only minor deviations in flexible regions such as unpaired nucleotides and loops. This suggests a relatively simple docking of 3C into prefolded sD without significant conformational changes on either side of the binding interface. However, the precise roles of the 3CD linker region in the CL RNA binding remain elusive, and possible roles of other protein factors, such as PCBP2, in the ternary complex cannot be ruled out.
Notably, while crystallographic results revealed two 3C molecules bound per CL, our SEC and ITC results are consistent with the monomeric form of 3C in solution. Since there are no specific interactions between 3C molecules within the CL-3C complex structure, and each molecule preferentially recognizes the sD loop or dinucleotide bulge region, we ruled out the stable homodimerization of 3C within the CL, which aligns with the stoichiometric analysis from ITC measurements. However, our data conflict with recent claims made by Dias-Solange et al.49 that suggested dimerization of 3C with or without the CL RNA. We identified several differences in their experimental conditions and flaws in data interpretation while exploring potential reasons for this discrepancy. First, based on a previous study, they introduced multiple mutations in the 3C protein (G55A, D58A, and V63A) to reduce aggregation or dimerization44. Although it remains unclear how these mutations affected dimerization, interpreting the 3C dimerization using such a construct is self-contradictory. Second, some conditions, such as the K+ concentration used in their measurements, differ from ours. While K+ seems to influence 3C oligomerization based on their mass-photometry (MP) results, their report does not clarify its role49. Third, the limitations of the MP technique in analyzing the molecular mass of a small protein with an MW less than 50 kDa make their data interpretations questionable. Although our MP results for 3C alone are somewhat similar to theirs, the data are inconclusive in confirming whether 3C exists as a monomer or dimer, likely due to the limitations of the MP method (see Supplementary Fig. 18 for details). Fourth, enteroviral 3C dimerization, as Dias-Solange et al.49 proposed, also conflicts with a previous study that found no evidence of 3C dimerization in cells35. Although dimeric assemblies were also observed in the crystals of other enteroviral 3Cs (EVD68 and EV93), gel filtration and dynamic light scattering (DLS) showed no evidence of dimerization in solution37,50, indicating that the dimeric assemblies observed in the crystals are not biologically relevant. These findings align with what has been observed for enteroviral 3Cs that lack a dimerization domain, unlike the related dimer-forming proteases from coronaviruses51,52.
In addition to the CLs, a stem-loop structure found within the coding region of enteroviral genomes, known as cre, also interacts with 3CD to facilitate the uridylylation of the viral VPg21,23,53. The uridylylated VPg is a primer for synthesizing negative-strand RNA during genome replication23. Although the precise mechanisms and functions of 3CD in VPg uridylation and its relationships with the CL-3CD complex formation remain elusive, it has been proposed that the sequence and structure of CL may influence VPg uridylation, viral genome stability, and perhaps assist the replication machinery in recognizing the enteroviral genome in a milieu of myriad cellular RNAs21,22,23,53. Interestingly, previous studies suggested that two 3D or 3CD molecules bind to the cre during VPg uridylylation21,22,23,53, implying that two copies could be necessary for the cooperative function. Given the predicted stem-loop secondary structure of the CVB3 cre, we hypothesized that the recognition of the cre by 3CD would be similar to that by the CL sD stem-loop (Fig. 4h). Although the functional significance of requiring two 3CD molecules warrants further investigation, they likely have distinct roles, potentially with one binding RNA and the other performing catalytic functions. Notably, 3CD exhibits both RNA binding and protease activities but lacks polymerase function. Conceivably, two copies within the replication complex could promote intermolecular cleavage of 3CD, releasing the active polymerase essential for RNA synthesis. The stoichiometry and positioning of two 3CDs within the enteroviral CLs may play roles in the context of the CL-3CD-PCBP2 ternary complex. Since 3D activity is vital for viral replication and needs strict regulation, the polymerase-inactive form, 3CD, may play a regulatory role.
Consistent with our structural findings, several molecular virology and cell-based studies involving mutagenesis have shown that disrupting the dinucleotide bulge, Py•Py mismatches, or sD loop impacts both positive- and negative-strand synthesis27,30,54,55. However, removing one nucleotide from the bulge or swapping their positions minimally impacted the negative-strand synthesis17,31,32, consistent with our structural analyses showing that only a single bulge nucleotide extensively interacts with the 3C protein. This observation helps explain why certain enteroviruses, such as RVB14 and RVC15, possess a single-nucleotide bulge yet continue to bind 3C from species with a dinucleotide bulge, providing a mechanistic rationale for the conservation of a bulge structure in the sD stem across many enteroviruses. Additionally, our results indicate that conformational changes disrupting the GNRA-type tetraloop formation by the sD loop hinder 3C binding, further supporting previous findings that mutations in the sD loop inhibit negative-strand synthesis22. Furthermore, removing the Py•Py helix by introducing canonical base pairs enhanced negative-strand synthesis while reducing positive-strand synthesis17,31,32. However, these mutations only moderately affected 3C’s binding affinity for CL27. Since 3C interactions primarily recognize the RNA backbone pattern, this slight reduction in binding affinity may result from the disruption of a narrower Py•Py helix, which could allosterically alter the interactions of specific residues in the sD loop and bulge, modifying RNA accessibility for 3C residues. Although the effects of Py•Py mismatch pairs go beyond positive-strand synthesis, our findings partially explain the conservation of these Py•Py pairs in enteroviral CLs. Overall, our cross-binding studies of 3Cs and CLs from seven different enteroviruses suggest that 3C-CL recognition depends more on structural pattern recognition than specific sequences, and specificity for a particular enterovirus arises from subtle differences in the three-dimensional structures of the 3C or CL binding interfaces.
The 3CD precursor protein is multifunctional, playing several critical roles in the life cycle of enteroviruses as 3C and 3CD cleave various host factors to downregulate cellular activities and promote viral genome replication. The 3D protein is essential for synthesizing new strands of the viral genome, and its interactions with CLs and cre during this process are crucial for enteroviral proliferation in the host cells. Due to the significant conservation of amino acid sequences and three-dimensional structures of these proteins across various enteroviral species, they have become attractive targets for antiviral drug development56,57,58. The crystallographic structures of the isolated 3C, 3D, and 3CD proteins from various enteroviral species33,35,36,37,38,39,44,47,48 have improved our understanding of their functional mechanisms and protein-drug interactions. We have recently identified a remarkable structural conservation of CLs among several human disease-causing enteroviruses. This research has highlighted a conserved recognition mechanism between the enteroviral CLs, 3Cs, and 3CDs, paving the way for developing antiviral drugs that target this RNP platform to disrupt viral replication. Considering the high conservation of this RNP platform among enteroviruses, developing universal therapeutics that could treat various enteroviral infections may be feasible.
Methods
Materials
The sequences of the CL, 3C, 3D, and 3CD constructs used in this study are listed in Supplementary Table 2. Unless stated otherwise, all ssDNA templates and PCR primers for the CL construct synthesis were purchased from Integrated DNA Technologies (IDT) Inc.
RNA synthesis and purification
The RNA constructs for this study were synthesized by in vitro transcription using protocols as previously described26,27. The DNA template with a T7 promoter sequence for transcription reaction was produced by PCR amplification of ssDNA purchased from IDT. The first two nucleotides of the reverse primer were 2′ OMe modified to reduce the 3′ end heterogeneity of the transcript59. The transcription reaction was conducted for 3 h at 37 °C in a buffer containing 40 mM Tris-HCl, pH 8.0, 2 mM spermidine, 10 mM NaCl, 25 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 40 U/ml RNase inhibitor, 5 U/ml TIPPase, 5 mM of each NTP, 50 pmol/ml DNA template, and 50 μg/ml homemade T7 RNA polymerase60. The reaction was then quenched by adding 10 U/ml DNase I (Promega) and incubating at 37 °C for 1 h. All RNA samples were purified by denaturing polyacrylamide gel electrophoresis (dPAGE). The RNA band was visualized by UV shadowing, excised from the gel, crushed, and eluted overnight at 4 °C in 10 mM Tris, pH 8.0, 2 mM EDTA, and 300 mM NaCl. The buffer of eluted RNA was exchanged with pure water three times using a 10 kDa cut-off Amicon column (Millipore Sigma). RNA was collected, aliquoted into 300 μl fractions, and stored at −80 °C until further use.
3C, 3D, and 3CD expression and purification
The recombinant 3C, 3D, and 3CD proteins were expressed and purified using previously described protocols with some modifications17,27,28,33,36,39,44. The codon-optimized DNA sequences encoding the CVB3 3C, 3D, 3CD, RVB14 3C, and EV71 3C were cloned into the pET-22b (+) vector between NdeI and XhoI restriction sites by GenScript (https://www.genscript.com). The final protein sequence for CVB3 3C, 3D, 3CD, and EV71 3C contained a C-terminal 6× His-tag, and RVB14 3C contained an N-terminal GB1 domain with a 6× His-tag. All purified proteins were used for all experiments without removing the 6× His-tag or the GB1domain. The protein purification was done using a Bio-Rad NGC FPLC system (Bio-Rad, https://www.bio-rad.com). The expression plasmid was transformed into BL21 (DE3) E. coli, and the cells were cultured in a 2×YT medium supplemented with 100 μg/ml Ampicillin at 37 °C with 220 rpm shaking until the OD of ~0.6. The cells were then induced for protein expression by using IPTG (isopropylthio-β-galactoside) to a final concentration of 1 mM for all 3C and 3D proteins, grown for 6 h at 25 °C, and harvested by centrifugation at 6000 × g. For CVB3 3CD, cells were induced with 0.5 mM IPTG and grown for 6 h at 30 °C. For 3C and 3D proteins, the cell pellet was resuspended in a lysis buffer containing 50 mM Tris-HCl, pH 7.5 (pH 6.5 for RVB14 3C), 300 mM NaCl, and 20 mM imidazole and was lysed using sonication. The lysate was centrifuged at 18,000 rpm at 4 °C, and the supernatant was passed through a 0.45-micron filter. The clarified lysate was then applied to a HisTrap™ column (Cytiva), and the protein was eluted from the column with a buffer (50 mM Tris HCl, 300 mM NaCl, and 250 mM imidazole, pH 7.5 (pH 6.5 for RVB14 3C) after washing the column with 5 column volumes of the lysis buffer. The eluted fractions were collected, dialyzed against a buffer containing 50 mM Tris HCl, 100 mM KCl, 1 mM EDTA, and 5 % Glycerol, pH 7.5 (pH 6.5 for RVB14 3C), and purified further by size-exclusion chromatography (HiLoad® 26/600 Superdex® 200 PG column, Cytiva). For the CVB3 3D, 400 mM NaCl was used instead of 100 mM KCl. The 3CD lysis buffer contained 100 mM potassium phosphate (pH 8.0), 350 mM NaCl, 20 mM imidazole, and 5% glycerol elution buffer containing 250 mM imidazole. The final protein was further purified, similar to the other 3C and 3D proteins, using a buffer containing (50 mM Tris-HCl, 350 mM NaCl, and 5 % Glycerol, pH 8.0). The single-peak protein fractions were pooled and concentrated using the Amicon centrifugal filters (molecular weight cut-off 10-30 kDa, Millipore Sigma), flash frozen with liquid N2 in small aliquots, and stored at −80 °C.
Crystallization
The purified RNA in water was heated at 90 °C for 2 min, and an appropriate volume of 10× refolding buffer (500 mM Tris-HCl, pH 7.4, 100 mM MgCl₂, 1 M NaCl, and 1 mM EDTA) was added. It was then incubated at 50 °C for 10 min, followed by incubation on ice for 5 min. The refolded RNA was then incubated for 30 min at 25 °C with an excess of 3C (~4 equivalents), and the complex was purified using SEC with a Superdex® 200 Increase 10/300 GL column. The peak corresponding to the RNA-3C complex was collected and concentrated using appropriate molecular weight cutoff filters (Amicon Ultra-1, Millipore-Sigma). The 3C-RNA complexes were passed through 0.2 μm cutoff Millipore centrifugal filter units to remove any preformed crystal seeds. The resulting complex for crystallization trials contained ~3.5 mg/ml of sD for the sD-3C complex and ~5.5 mg/ml of CL for the CL-3C complex. The crystallization screening was performed in a Mosquito Xtal3 liquid-handling robot (TTP Labtech, ttplabtech.com) as described previously26,27. Using commercially available screening kits from Hampton Research (https://hamptonresearch.com), Molecular Dimensions (https://moleculardimensions.com), and Jena Bioscience (www.jenabioscience.com), the sitting-drop vapor-diffusion method was employed to set up crystallization trials at 22 °C and 80% relative humidity. The best diffracting crystals for the CL-3C and sD-3C complexes were observed within a week under several conditions. Drops containing suitable crystals were brought up to 40% glycerol for cryoprotection without changing other compositions. The crystals were immediately flash-frozen in liquid nitrogen after being fished in the loops from the drops and shipped to the Brookhaven National Laboratory (BNL) for X-ray diffraction screening and data collection.
Crystallographic data collection, processing, and analysis
The X-ray diffraction data sets were collected at the Brookhaven National Laboratory, NSLSII beamlines 17-ID-1 (AMX) and 17-ID-2 (FMX). Datasets were collected for several single crystals at a single wavelength, 0.97934 Å, under a jet of liquid nitrogen (77 K or −196 °C). The crystal grown in a condition with 0.05 M Bis-Tris, 1.4 M ammonium sulfate, and 25% glycerol provided the best resolution of 2.69 Å for the CL-3C complex. Similarly, the best diffracting crystal (2.41 Å resolution) for the sD-3C complex was obtained in a condition with 0.2 M DL-malic Acid, 20% PEG 3350. All the datasets were then integrated and scaled using the NSLSII on-site automated programs (AutoProc). Some datasets were also processed and analyzed using the Xia2/Dials platform on CCP424,61. The crystallographic data suggested that the CL-3C complex was crystallized in the C121 space group and contained two 3C molecules per RNA within the crystallographic asymmetric unit (a = 106.41 Å, b = 95.25 Å, c = 113.64 Å, α = 90°, β = 90.63°, and γ = 90°) with the 41.1% solvent content (see Supplementary Table 1 for details of the data collection statistics). Unlike the CL-3C complex, the sD-3C complex crystallized in the P 21 21 21 space group, with two sets of complexes, each containing two 3C molecules per RNA within the crystallographic asymmetric unit (a = 52.30 Å, b = 78.87 Å, c = 106.51 Å, α = 90°, β = 90°, and γ = 90°). Details of data collection statistics are provided in Supplementary Table 1. For solving both structures, the initial phases were obtained by molecular replacement with the previously reported structure of CVB3 3C (PDB: 2ZU1)33 as the search model using Phaser on Phenix62. Iterative model building and refinement were performed using the COOT63 and the Phenix package62. The RNA structure was built unambiguously by modeling the individual nucleotides into the electron density map obtained from the molecular replacement. The refinement used default NCS options and auto-selected TLS parameters in Phenix. The Ramachandran plot showed 98.3% of the protein residues in the preferred regions and 1.66% in the allowed regions for the sD-3C complex, while 96.41% of the residues were in the preferred regions and 3.59% in the allowed regions for the CL-3C complex. The structure-related figures were made in PyMOL (The PyMOL Molecular Graphics System, Version 2.0, Schrödinger, LLC), UCSF ChimeraX-1.7.1, and the figure labels were edited in CorelDRAW (Corel Corporation, http://www.corel.com).
Isothermal titration calorimetry
The isothermal titration calorimetry (ITC) experiments were carried out using MicroCal PEAQ-ITC Automated (Malvern Panalytical) equipment and freshly prepared RNA and protein samples. The RNA and protein samples were dialyzed overnight into a buffer containing 50 mM Tris-HCl, pH 7.5, 100 mM KCl, 1 mM EDTA, and 5 % glycerol. The injection syringe contained 150 μl of ~150 to 800 µM of 3C protein, and the calorimetry cell was loaded with 400 μl of ~5 to 10 µM RNA. After thermal equilibration at 25 °C and an initial 60-second delay, a single injection of 200 nl, followed by 19 serial injections of 2 μl 3C protein, was made into the calorimetry cell. The ITC data were analyzed using the integrated MicroCal PEAQ-ITC software.
Biolayer interferometry (BLI)
The experiments were performed in the Octet R2 system (Sartorius) using the Octet® Streptavidin (SA) biosensors (Sartorius). The CL RNA containing a 3′ spacer sequence was refolded and hybridized with a 5′ biotinylated DNA oligo (see Fig. 4a), and the complex was purified by SEC (Superdex® 200 Increase 10/300 GL, 200 PG column, Cytiva). Then, the purified biotinylated RNA construct (10 ng/µl concentration) was immobilized on the SA biosensor surface after hydrating the biosensor for 10 min in the assay buffer (PBS pH 7.4). These RNA-immobilized biosensors were then dipped into varying concentrations of the protein solution and a reference sample (assay buffer) for the background correction. The BLI responses as a function of time data were recorded and then processed by aligning the baselines using the BLI system-integrated software. To determine the Kd from the BLI data, the responses from the plateau region of the association curves (i.e., average responses from the 110 to 115 seconds region for each concentration were plotted against the protein concentration and fitted using the following binding isotherm.
\({Response}(Y)=\frac{{R}_{\max }\left[{Protein}\right]}{\left(\right.{K}_{d}+\left[{Protein}\right]}\), where Rmax refers to the maximum response due to the protein binding to the sensor-immobilized RNA.
AlphaFold prediction
The AlphaFold3 Server46 was used to predict the three-dimensional structures of the CVB3 3CD. The AlphaFold server (https://alphafoldserver.com) was run using default parameters for the 3CD sequence (Supplementary Table 2), generating 5 predicted models using a random seed. The models for analysis were selected based on the predicted local distance difference test (pLDDT) score, which provides a per-atom confidence estimate on a 0 to 100 scale, where higher values indicate greater confidence (see Supplementary Fig. 14). Moreover, the predicted template modeling (pTM) scores64 and the interface template modeling (ipTM) scores64 were also considered for evaluating a predicted structure. All five lowest-energy predicted models were analysed and compared with the crystal structures (Supplementary Figs. 15 and 16). The predicted structures were viewed and analyzed in PyMOL (The PyMOL Molecular Graphics System, Version 3.1.3.1, Schrödinger, LLC).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The atomic coordinates and structure factors for the reported crystal structures of the cloverleaf-3C and sD-3C complexes have been deposited with the Protein Data Bank (PDB) under accession codes 9D9P and 9D9O, respectively. The atomic coordinates of CVB3 3C147A used in this study for molecular replacement are available in the PDB under accession code 2ZU1. All other relevant data are available in the main text or the supplementary materials. The authors will provide plasmids for the recombinant expression of 3C, 3D, and 3CD proteins upon request in compliance with the material transfer agreements. Requests should be directed to D.K. Source data are provided as a Source Data file. Source data are provided with this paper.
References
Palacios, G. & Oberste, M. S. Enteroviruses as agents of emerging infectious diseases. J. Neurovirol. 11, 424–433 (2005).
Tapparel, C., Siegrist, F., Petty, T. J. & Kaiser, L. Picornavirus and enterovirus diversity with associated human diseases. Infect. Genet Evol. 14, 282–293 (2013).
Royston, L. & Tapparel, C. Rhinoviruses and respiratory enteroviruses: not as simple as ABC. Viruses 8, 16 (2016).
Rohll, J. B. et al. The 5’-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J. Virol. 68, 4384–4391 (1994).
Jackson, R. J., Hellen, C. U. & Pestova, T. V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. cell Biol. 11, 113–127 (2010).
Jan, E., Mohr, I. & Walsh, D. A cap-to-tail guide to mRNA translation strategies in virus-infected cells. Annu. Rev. Virol. 3, 283–307 (2016).
Jaafar, Z. A. & Kieft, J. S. Viral RNA structure-based strategies to manipulate translation. Nat. Rev. Microbiol. 17, 110–123 (2019).
Perera, R., Daijogo, S., Walter, B. L., Nguyen, J. H. C. & Semler, B. L. Cellular protein modification by poliovirus: the two faces of Poly(rC)-binding protein. J. Virol. 81, 8919–8932 (2007).
Chase, A. J., Daijogo, S. & Semler, B. L. Inhibition of poliovirus-induced cleavage of cellular protein PCBP2 reduces the levels of viral RNA replication. J. Virol. 88, 3192–3201 (2014).
Zell, R. et al. Poly (rC)-binding protein 2 interacts with the oligo (rC) tract of coxsackievirus B3. Biochem. Biophys. Res. Commun. 366, 917–921 (2008).
Gamarnik, A. V. & Andino, R. Interactions of viral protein 3CD and poly (rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74, 2219–2226 (2000).
Parsley, T., Towner, J., Blyn, L., Ehrenfeld, E. & Semler, B. Poly (rC) binding protein 2 forms a ternary complex with the 5’-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3, 1124–1134 (1997).
Spear, A., Sharma, N. & Flanegan, J. B. Protein–RNA tethering: the role of poly (C) binding protein 2 in poliovirus RNA replication. Virology 374, 280–291 (2008).
Toyoda, H., Franco, D., Fujita, K., Paul, A. V. & Wimmer, E. Replication of poliovirus requires binding of the poly (rC) binding protein to the cloverleaf as well as to the adjacent C-rich spacer sequence between the cloverleaf and the internal ribosomal entry site. J. Virol. 81, 10017–10028 (2007).
Zell, R. et al. Interaction of poly (rC)-binding protein 2 domains KH1 and KH3 with coxsackievirus RNA. Biochem. Biophys. Res. Commun. 377, 500–503 (2008).
Xiang, W., Harris, K. S., Alexander, L. & Wimmer, E. Interaction between the 5’-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 69, 3658–3667 (1995).
Zell, R., Sidigi, K., Bucci, E., Stelzner, A. & Görlach, M. Determinants of the recognition of enteroviral cloverleaf RNA by coxsackievirus B3 proteinase 3C. RNA 8, 188–201 (2002).
Herold, J. & Andino, R. Poliovirus RNA replication requires genome circularization through a protein–protein bridge. Mol. Cell 7, 581–591 (2001).
Andino, R., Rieckhof, G. E., Achacoso, P. L. & Baltimore, D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5’-end of viral RNA. EMBO J. 12, 3587 (1993).
Goodfellow, I. et al. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74, 4590–4600 (2000).
Rieder, E., Paul, A. V., Kim, D. W., van Boom, J. H. & Wimmer, E. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74, 10371–10380 (2000).
van Ooij, M. J. et al. Structural and functional characterization of the coxsackievirus B3 CRE (2C): role of CRE (2C) in negative-and positive-strand RNA synthesis. J. Gen. Virol. 87, 103–113 (2006).
Murray, K. E. & Barton, D. J. Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J. Virol. 77, 4739–4750 (2003).
Agirre, J. et al. The CCP4 suite: integrative software for macromolecular crystallography. Acta Crystallogr. D. Struct. Biol. 79, 449–461 (2023).
Beckham, S. A. et al. Structure of the PCBP2/stem–loop IV complex underlying translation initiation mediated by the poliovirus type I IRES. Nucleic Acids Res. 48, 8006–8021 (2020).
Das, N. K., Vogt, J., Patel, A., Banna, H. A. & Koirala, D. Structural basis for a highly conserved RNA-mediated enteroviral genome replication. Nucleic Acids Res. 52, 11218–11233 (2024).
Das, N. K. et al. Crystal structure of a highly conserved enteroviral 5′ cloverleaf RNA replication element. Nat. Commun. 14, 1955 (2023).
Ohlenschläger, O. et al. The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure 12, 237–248 (2004).
Yang, Y., Rijnbrand, R., Watowich, S. & Lemon, S. M. Genetic evidence for an interaction between a picornaviral cis-acting RNA replication element and 3CD protein. J. Biol. Chem. 279, 12659–12667 (2004).
Sharma, N. et al. Functional role of the 5′ terminal cloverleaf in Coxsackievirus RNA replication. Virology 393, 238–249 (2009).
Du, Z., Yu, J., Ulyanov, N. B., Andino, R. & James, T. L. Solution structure of a consensus stem-loop D RNA domain that plays important roles in regulating translation and replication in enteroviruses and rhinoviruses. Biochemistry 43, 11959–11972 (2004).
Andino, R., Rieckhof, G. E., Trono, D. & Baltimore, D. Substitutions in the protease (3Cpro) gene of poliovirus can suppress a mutation in the 5’ noncoding region. J. Virol. 64, 607–612 (1990).
Lee, C.-C. et al. Structural Basis of Inhibition Specificities of 3C and 3C-like Proteases by Zinc-coordinating and Peptidomimetic Compounds*. J. Biol. Chem. 284, 7646–7655 (2009).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Mosimann, S. C., Cherney, M. M., Sia, S., Plotch, S. & James, M. N. G. Refined X-ray crystallographic structure of the poliovirus 3C gene product11Edited By D. Rees. J. Mol. Biol. 273, 1032–1047 (1997).
Cui, S. et al. Crystal structure of human enterovirus 71 3C protease. J. Mol. Biol. 408, 449–461 (2011).
Tan, J. et al. 3C protease of enterovirus 68: structure-based design of michael acceptor inhibitors and their broad-spectrum antiviral effects against picornaviruses. J. Virol. 87, 4339–4351 (2013).
Kawatkar, S. P. et al. Design and structure–activity relationships of novel inhibitors of human rhinovirus 3C protease. Bioorg. Med. Chem. Lett. 26, 3248–3252 (2016).
Bjorndahl, T. C., Andrew, L. C., Semenchenko, V. & Wishart, D. S. NMR solution structures of the Apo and peptide-inhibited human rhinovirus 3C protease (Serotype 14): structural and dynamic comparison. Biochemistry 46, 12945–12958 (2007).
Gottipati, K., McNeme, S. C., Tipo, J., White, M. A. & Choi, K. H. Structural basis for cloverleaf RNA-initiated viral genome replication. Nucleic Acids Res. 51, 8850–8863 (2023).
Zoll, J., Tessari, M., Van Kuppeveld, F. J., Melchers, W. J. & Heus, H. A. Breaking pseudo-twofold symmetry in the poliovirus 3’-UTR Y-stem by restoring Watson-Crick base pairs. Rna 13, 781–792 (2007).
Spear, A., Ogram, S. A., Morasco, B. J., Smerage, L. E. & Flanegan, J. B. Viral precursor protein P3 and its processed products perform discrete and essential functions in the poliovirus RNA replication complex. Virology 485, 492–501 (2015).
Wang, J. et al. Crystal structures of enterovirus 71 3C protease complexed with rupintrivir reveal the roles of catalytically important residues. J. Virol. 85, 10021–10030 (2011).
Marcotte, L. L. et al. Crystal structure of poliovirus 3CD protein: virally encoded protease and precursor to the RNA-dependent RNA polymerase. J. Virol. 81, 3583–3596 (2007).
Blair, W. S. et al. Utilization of a mammalian cell-based RNA binding assay to characterize the RNA binding properties of picornavirus 3C proteinases. RNA 4, 215–225 (1998).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Campagnola, G., Weygandt, M., Scoggin, K. & Peersen, O. Crystal structure of coxsackievirus B3 3Dpol highlights the functional importance of residue 5 in picornavirus polymerases. J. Virol. 82, 9458–9464 (2008).
Love, R. A. et al. The crystal structure of the RNA-dependent RNA polymerase from human rhinovirus: a dual function target for common cold antiviral therapy. Structure 12, 1533–1544 (2004).
Dias-Solange, D., Le, M. T., Gottipati, K. & Choi, K. H. Structure of coxsackievirus cloverleaf RNA and 3Cpro dimer establishes the RNA-binding mechanism of enterovirus protease 3Cpro. Sci. Adv. 11, eads6862 (2025).
Costenaro, L. et al. StrucTural Basis For Antiviral Inhibition Of The Main Protease, 3C, from human enterovirus 93. J. Virol. 85, 10764–10773 (2011).
Shi, J., Wei, Z. & Song, J. Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme: defining the extra domain as a new target for design of highly specific protease inhibitors. J. Biol. Chem. 279, 24765–24773 (2004).
Chen, S. et al. Residues on the dimer interface of SARS coronavirus 3C-like protease: dimer stability characterization and enzyme catalytic activity analysis. J. Biochem. 143, 525–536 (2008).
Steil, B. P. & Barton, D. J. Cis-active RNA elements (CREs) and picornavirus RNA replication. Virus Res. 139, 240–252 (2009).
von Eije, K. J., ter Brake, O. & Berkhout, B. Human immunodeficiency virus type 1 escape is restricted when conserved genome sequences are targeted by RNA interference. J. Virol. 82, 2895–2903 (2008).
Nagy, P. D., Zhang, C. & Simon, A. E. Dissecting RNA recombination in vitro: role of RNA sequences and the viral replicase. EMBO J. 17, 2392–2403 (1998).
van der Linden, L. et al. The RNA template channel of the RNA-dependent RNA polymerase as a target for development of antiviral therapy of multiple genera within a virus family. PLOS Pathog. 11, e1004733 (2015).
Zhai, Y. et al. Cyanohydrin as an anchoring group for potent and selective inhibitors of enterovirus 71 3C protease. J. Med. Chem. 58, 9414–9420 (2015).
Matthews, D. A. et al. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Natl. Acad. Sci. USA 96, 11000–11007 (1999).
Kao, C., Rüdisser, S. & Zheng, M. A simple and efficient method to transcribe RNAs with reduced 3′ heterogeneity. Methods 23, 201–205 (2001).
Rio, D. C. Expression and purification of active recombinant T7 RNA polymerase from E. coli. Cold Spring Harb. Protoc. 2013, pdb.prot078527 (2013).
Winter, G. et al. DIALS: implementation and evaluation of a new integration package. Acta Crystallogr. D. Struct. Biol. 74, 85–97 (2018).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 (2010).
Zhang, Y. & Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins Struct. Funct. Bioinforma. 57, 702–710 (2004).
Acknowledgements
This work was primarily supported by the NSF CAREER award (MCB 2236996) to D.K. and partially by the NIH T32 grant GM066706 to N.K.D. The crystallographic data were collected at the NSLS-II beamlines (17-ID-1 and 17-ID-2) at Brookhaven National Laboratory (BNL) using the beamtime obtained through the NECAT BAG proposal # 311950. The NIH-NIGMS primarily supports the Center for Bio-Molecular Structure (CBMS) at BNL through a Center Core P30 Grant (P30GM133893) and by the DOE Office of Biological and Environmental Research (KP1607011). NSLS-II is a U.S. DOE Office of Science user facility operated under Contract No. DE-SC0012704. The authors also thank Prof. Michael F. Summers’s laboratory at the University of Maryland, Baltimore County, for providing the ITC facilities.
Author information
Authors and Affiliations
Contributions
D.K. and N.K.D. conceived and designed the experiments. N.K.D. prepared the samples, conducted most crystallographic and binding experiments, and solved the crystal structures with assistance from D.K. Undergraduate A.P. and high school student R.A. helped N.K.D. with sample preparation, analysis, and crystallization trials. N.K.D. and D.K. analyzed most of the biochemical and structural data and interpreted the results. N.K.D. and D.K. wrote the manuscript, and all authors reviewed it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Quentin Vicens and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Das, N.K., Patel, A., Abdelghani, R. et al. Structural basis for 3C and 3CD recruitment by enteroviral genomes during negative-strand RNA synthesis. Nat Commun 16, 9293 (2025). https://doi.org/10.1038/s41467-025-64376-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-64376-0