Abstract
Disrupted in Schizophrenia 1 (DISC1) is essential for neuronal development and has been implicated in various psychiatric disorders. Our transcriptomic and proteomic analyses identified Zika virus (ZIKV) infection enhanced DISC1 expression, however, its functional role in ZIKV infection and caused congenital Zika syndrome (CZS) and ZIKV-induced long-term neurodevelopmental defects remain unexplored. In this study, we demonstrate that DISC1 attenuates ZIKV infection in human placental and neuroglia cells, as well as in murine macrophages and primary cortical cells. DISC1 also decreases ZIKV dissemination from peripheral tissues to key organs of mice, including the uterus, testis, and brain, thereby reducing fetal abortion rates and intrauterine growth restriction. Notably, DISC1 is associated with brain damage and long-term ZIKV effects, including memory loss, reduced anxiety and depression, declines in sociability and social novelty. Mechanistically, DISC1 activates autophagy by enhancing AMPKα phosphorylation and reducing mTOR phosphorylation, protecting against ZIKV infection. Additionally, DISC1 interacts with LC3 to further activate autophagy, partially contributing to reduce ZIKV infection. In conclusion, DISC1 plays a critical factor in controlling ZIKV infection and mitigating CZS and ZIKV-induced neurocognitive decline.
Similar content being viewed by others
Introduction
Zika virus (ZIKV), a neurotropic flavivirus, poses significant risks to pregnant women, potentially leading to a variety of developmental defects in fetuses and infants. These defects, which include microcephaly, brain development issues, and sensory impairments such as vision and hearing loss, are collectively referred to as congenital Zika syndrome (CZS)1,2. In addition to these immediate defects, long-term neurological developmental disorders associated with vertical ZIKV transmission—such as neurodevelopmental dysfunction, behavioral and sociability deficits, and memory impairments—can emerge over time3,4,5. These effects may manifest even in infants with a normal head circumference at birth or in the absence of overt symptoms at birth6,7,8. A recent follow-up study of 216 children with prenatal ZIKV exposure found that microcephaly was present in only 3.7% of cases. However, despite the absence of obvious signs of CZS, 40.4% of the children scored below average on the Bayley-III neurodevelopmental evaluation, which assessed cognitive, language, and motor domains9. These findings underscore the need for a better understanding of host-ZIKV interactions, both during acute infection and after apparent recovery, to identify potential interventions that could reduce the developmental impact and minimize the long-term damage caused by ZIKV. However, the host factors critical for ZIKV pathogenesis and long-term neurological outcomes remain largely unidentified. Through transcriptomic and proteomic analyses in ZIKV-infected human first-trimester extravillous trophoblast cells (HTR8), we have identified Disrupted in Schizophrenia 1 (DISC1) as a potential molecular regulator of ZIKV infection.
Although the DISC1 gene has been proposed as a risk factor for psychiatric disorders such as schizophrenia, major depression, bipolar disorder, and potentially autism spectrum disorders, most of the supporting genetic evidence originates from rare familial cases. Large-scale genome-wide association studies (GWAS) have not replicated these findings10,11,12, suggesting that the genetic association between DISC1 gene and common psychiatric disorders is limited and may not generalize to broader populations. Despite the lack of strong genetic evidence, DISC1 protein has been functionally implicated in the pathogenesis of neuropsychiatric diseases. The DISC1 protein is a critical regulator of neuronal development, notably through its interaction with the LIS1/NDEL1/dynein complex, which governs microtubule dynamics and neuronal migration. C-terminal truncation of DISC1 disrupts its ability to support neurite outgrowth and leads to cortical development abnormalities—phenotypes linked to increased schizophrenia risk13,14,15. Furthermore, aberrant DISC1 aggregation can impair dopamine signaling, disrupt interactions with key partners such as NDEL1, and interfere with neurodevelopmental processes, thereby contributing to behavioral deficits and disease susceptibility16,17. DISC1 also directly regulates new neuron development in the adult brain by modulating the AKT-mTOR signaling pathway through interaction with KIAA121218. Additionally, DISC1 stabilizes β-catenin by inhibiting GSK3β activity via direct interaction, promoting neural progenitor proliferation19. Importantly, a 4-bp mutation in DISC1 can lead to a global depression of serine/threonine kinase activity, affecting kinases such as AMP-activated protein kinase (AMPK), extracellular signal-regulated kinases (ERK), and thousand-and-one amino acid (TAO) kinases, highlighting DISC1’s critical role in the kinase network20. Furthermore, DISC1 directly binds to LC3 to promote mitophagy, thereby protecting synaptic plasticity from Aβ accumulation-induced toxicity21. Despite its diverse functions, the role of DISC1 in viral infections remains underexplored. One study suggests that Influenza virus (H1N1) infection may lead to DISC1 aggregation, potentially contributing to the pathogenesis of synucleinopathies or certain mental disorders, positioning DISC1 as a potential risk factor22. However, the specific role of DISC1 in viral infections, particularly neurotropic viruses, is not well understood.
In this work, we uncover a previously unrecognized role of DISC1 as a host factor that restricts ZIKV infection. We demonstrate that DISC1 suppresses viral replication in placental and neuronal cells, limits viral dissemination to critical organs, and mitigates adverse pregnancy outcomes. Furthermore, DISC1 is implicated in ZIKV-associated behavior alterations, including memory deficits, reduced anxiety- and depression-like behaviors, and impairments in sociability and social novelty. Mechanistically, DISC1 enhances host antiviral defense by promoting autophagy through activation of the AMPK–mTOR signaling pathway and interaction with LC3. Collectively, these findings identify DISC1 as a key regulator of both acute infection and long-term neurological outcomes, offering new insights into host–virus interactions.
Results
DISC1 reduces ZIKV infection in placental cells and neuroglia cells
In our previous studies, we identified differentially expressed genes (DEGs) in HTR8 in response to ZIKV infection by transcriptomics23. Building on this work, we performed proteomic analyses to investigate molecular alterations induced by ZIKV (Asian lineage strain GZ01, GenBank: KU820898) infection in HTR8 cells. Among the four upregulated genes identified at both the transcriptional and translational levels, DISC1 gene stood out due to its established role in mental illness (Fig. 1a). We first validated the increased expression of DISC1 using quantitative real-time PCR (qRT-PCR) and Western blot in both HTR8 cells and the ZIKV-susceptible human glioblastoma cell line U251. The mRNA levels of DISC1 were significantly elevated on days 1 and 2 post-infection in ZIKV-infected HTR8 (Fig. S1a) and U251 (Fig. S1b) cells, compared to non-infected controls. Correspondingly, an increase in DISC1 protein levels was observed on day 1 post-infection in HTR8 cells (Fig. S1c), and on both days 1 and 2 post-infection in U251 cells (Fig. S1d), relative to non-infected controls. These findings indicate that ZIKV infection induces upregulation of DISC1 in both placental and neuroglia cells. Unlike H1N1 infection in lund human mesencephalic cells and human neuroblastoma NLF cells22, we did not observe DISC1 aggregation in HTR8 and U251 cells upon ZIKV infection (Fig. S1e, f). Next, we investigated the potential role of DISC1 in modulating ZIKV infection in vitro. Overexpression of DISC1 significantly reduced ZIKV infection, as evidenced by decreased viral mRNA, protein expression, and viral titers in both HTR8 (Fig. 1b–d) and U251 (Fig. 1e–g) cells on days 1 and 2 post-infection. Additionally, we performed siRNA-mediated knockdown experiments to further validate the role of DISC1 in ZIKV infection. Treatment with siDISC1 significantly reduced DISC1 protein expression, achieving a knockdown efficiency of at least 50% (Fig. 1h, i, and Fig. S1g, h). Specifically, cells treated with siDISC1 exhibited an ~0.5- to 1-fold increase in ZIKV E protein expression relative to control groups (Fig. 1h, i, and Fig. S1i, j). Furthermore, DISC1 knockdown resulted in a significant elevation of viral mRNA levels and viral titers in both cell lines on days 1 and 2 post-infection, relative to negative controls (Fig. 1j–m). To clarify, neither the transfection of the DISC1 plasmid nor the DISC1 siRNA affected cell viability compared to the controls (Fig. S1k–S1n). Taken together, these results suggest that DISC1 acts to reduce ZIKV infection in HTR8 and U251 cells.
a Venn diagram showing the overlap of differentially expressed genes (DEGs) identified through transcriptomic and proteomic analyses of ZIKV-infected HTR8 cells compared to non-infected controls. The overlapping area indicates shared DEGs between the two datasets. b−d HTR8 cells were transfected with pcDNA or HA-DISC1 (1 μg) plasmids for 24 hours (h) and then infected with ZIKV at a multiplicity of infection (MOI) of 1. Viral mRNA levels, protein expression and titers were assessed on day 1 (D1) and day 2 (D2) post-infection using qRT-PCR (b), Western blot (c), and plaque assay (d). e−g U251 cells were transfected with pcDNA or HA-DISC1 (1 μg) plasmids for 24 h and infected with ZIKV at a MOI of 1. Viral mRNA levels, protein expression and titers were measured on D1 and D2 post-infection using qRT-PCR (e), Western blot (f), and plaque assay (g). h, i HTR8 cells (h) or U251 cells (i) were transfected with siRNA targeting DISC1 for 24 h prior to ZIKV infection at a MOI of 1. Viral protein was analyzed on D1 and D2 post-infection using Western blot. j−m HTR8 cells (j, k) or U251 cells (l, m) were transfected with siRNA targeting DISC1 for 24 h prior to ZIKV infection at a MOI of 1. Viral mRNA levels and viral loads were analyzed on D1 and D2 post-infection using qRT-PCR ( j, l) and plaque assay (k, m). Data shown in (b, d–e, g, j–m) are from one representative experiment out of three independent replicates with similar results, each including 3 biological replicates per group (n = 3). Images in (c−i) are from one representative experiment out of three independent replicates with similar results (n = 3). All the data were confirmed to follow a normal distribution using the Shapiro-Wilk test, and statistical analysis was performed using Student’s unpaired two-tailed t test. Data are presented as means ± SD. Source data are provided as a Source Data file.
DISC1 reduces ZIKV infection in murine models
To further investigate the role of DISC1 in ZIKV infection, we generated a Disc1-deficient mouse model using CRISPR/Cas9 technology to delete exon 2 of the Disc1 gene (Fig. S2a and Supplementary Note 1). Genotyping was confirmed by PCR analysis of genomic DNA (Fig. S2b). Western blot analysis revealed a significant reduction in DISC1 protein levels across multiple tissues—including the spleen, brain, uterus and testis—confirming at least a 50% decrease in DISC1 expression (Fig. S2c–f). These findings indicate that deletion of exon 2 significantly reduces—but does not completely eliminate—DISC1 expression. Therefore, this model should be considered a Disc1 knockdown (KD) rather than a complete knockout. Throughout this study, we refer to it as the Disc1 KD model.
As shown in Fig. 2a, both wild-type (WT) and Disc1 KD mice were pretreated with 2 mg of anti-type I interferon receptor (IFNR) antibody (MAR1-5A3) and subsequently challenged with 1 × 106 plaque-forming units (PFU) of the ZIKV strain (Asian lineage strain GZ01, GenBank: KU820898) one day later. Viral burdens in blood, spleen, brain, testis, and uterus were measured at days 2 and 6 post-infection. Disc1 KD mice exhibited significantly higher viremia (Fig. 2b, c) and increased viral mRNA levels in the spleen on both day 2 and day 6 post-infection compared to WT mice (Fig. 2d). On day 6 post-infection, viral mRNA levels in the brains of Disc1 KD mice were significantly higher than those in WT mice, whereas no such difference was observed on day 2 (Fig. 2e). In the testis, viral mRNA levels were also lower in WT mice than in Disc1 KD mice on day 2 post-infection (Fig. 2f). In addition, viral mRNA levels in the uterus were lower on day 6 post-infection (Fig. 2g), and viral loads on both days 2 and 6 post-infection were reduced in WT mice compared to Disc1 KD mice (Fig. 2h). These findings indicate that DISC1 reduces ZIKV replication in multiple organs of mice. Additionally, we assessed viral mRNA levels and viral loads in ZIKV-infected macrophages isolated from both WT and Disc1 KD mice. Prior to infection, macrophages differentiation was confirmed by flow cytometry, with a differentiation efficiency of at least 90% and the representative showed here is 92.3% (Fig. S3). We found that both viral mRNA levels (Fig. 2i) and viral titers (Fig. 2j) were significantly reduced in macrophages from WT mice on days 1 and 3 post-infection. These results suggest that DISC1 reduces ZIKV replication in murine tissues and cells.
a Schematic representation of the experiment set up. 6-8-week-old WT or Disc1 KD mice were treated with 2 mg MAR1-5A3 on the day prior to infection and then intraperitoneally (i.p.) inoculated with PBS or 1 × 106 PFU of ZIKV. Mouse tissues were collected on D2 and D6 post-infection. b−h Viral titers in plasma (b) and uterus (h) were determined by plaque assay, and mRNA levels in blood cell (c), spleen (d), brain (e), testis (f) and uterus (g) were measured by qRT-PCR on D2 and D6 post-infection. i, j Primary cells isolated from WT or Disc1 KD mice were induced to differentiate into macrophages using mouse macrophage colony-stimulating factor. Cells were pre-incubated with 20 μg/mL anti-IFNAR1 antibody MAR1-5A3 for 6 h, followed by infection with ZIKV at a MOI of 1. Viral mRNA levels and titers were measured on D1 and D3 post-infection using qRT-PCR (i) and plaque assay (j). Data for plasma (b), blood cells (c), spleen (d), and brain (e) are one representative experiment out of two independent replicates with similar results, each including 6 mice per group (n = 6), consisting of 3 males and 3 females. Data for the testis (f) are one representative experiment out of two independent replicates with similar results, each including 6 male mice per group (n = 6). Data for the uterus (g, h) are one representative experiment out of two independent replicates with similar results, each including 6 female mice per group (n = 6). Data shown in (i, j) are one representative experiment out of two independent replicates with similar results, each including 4 biological replicates per group (n = 4). Data shown in (b, e, j) did not follow a normal distribution according to the Shapiro-Wilk test, and statistical analysis was performed using two-tailed Mann-Whitney test. Data shown in (c–d, f–i) were confirmed to follow a normal distribution using the Shapiro-Wilk test, and statistical analysis was performed using Student’s unpaired two-tailed t test. Data are presented as means ± SD. Source data are provided as a Source Data file. Figure 2a was created in BioRender. Hl, Z. (2025) https://BioRender.com/bh2c5ad.
DISC1 attenuates ZIKV-induced congenital abnormalities in murine fetuses
We subsequently explored the impact of Disc1 knockdown on CZS. Both WT and Disc1 KD pregnant dams received treatment with 2 mg MAR1-5A3 on embryonic day 5.5 (E5.5) to enhance ZIKV permissiveness, followed by challenge with 1 × 107 PFU ZIKV one day later (Fig. 3a). On E13.5, we observed no significant differences in survival rates between non-infected WT and Disc1 KD dams. However, we observed fetal demise and subsequent absorption in fetuses from ZIKV-infected dams, leaving only placental remnants in the uterus (Fig. 3b, red arrows and Fig. 3c). Additionally, the fetuses from ZIKV-infected dams exhibited marked pallor (Fig. 3b, green arrows, left panel). Notably, fetuses from ZIKV-infected Disc1 KD group developed with more evident pallor (Fig. 3b, green arrows, right panel), and exhibited more severe miscarriage or absorption (Fig. 3b, red arrows, right panel and Fig. 3c). Next, we assessed individual fetuses isolated from the uterus by measuring their weight, crown-rump length (CRL), and occipito-frontal diameter (OFD) of the fetal head. As controls, no significant differences in weight (Fig. 3d), CRL (Fig. 3e), OFD (Fig. 3f), or overall fetal size (CRL × OFD) (Fig. 3g) were observed between WT and Disc1 KD fetuses in the non-infected groups. However, ZIKV infection induced intrauterine growth restriction (IUGR), as evidenced by reductions in all four parameters. More pronounced reductions were observed in the ZIKV-infected Disc1 KD group compared to the infected WT group (Fig. 3d–g). Correspondingly, the viral mRNA levels in the placentas and fetal heads from the ZIKV-infected Disc1 KD fetuses were significantly higher compared to that in WT fetuses (Fig. 3h, i). Additionally, we observed a significant increase in viral mRNA levels in the spleen of Disc1 KD dams compared to WT dams (Fig. 3j). These findings suggest that DISC1 reduces ZIKV infection and contributes to the mitigation of IUGR in fetuses.
a Schematic representation of the experimental setup. One day prior to infection, 2 mg of the anti-IFNAR1 antibody MAR1-5A3 was administered to 8-10-week-old WT dams and Disc1 KD dams on E5.5. On E6.5, dams were i.p. inoculated with either PBS or 1 × 107 PFU of ZIKV. Placenta and fetal head were harvested on E13.5. b Representative images of E13.5 uteri (upper panel) and fetuses (lower panel). Partial demise and growth restriction was shown in ZIKV-infected WT and Disc1 KD pregnant dams. Red arrows indicate the placental residues, and green arrows show the growth restriction of fetuses. c Resorption rates were analyzed on E13.5. d−g The weight (d), CRL (e), OFD (f) and size (g, CRL × OFD) of fetuses were measured on E13.5. h, i ZIKV mRNA levels in placentas (h) and fetal heads (i) of WT and Disc1 KD mice was measured by qRT-PCR. j ZIKV mRNA levels in the spleen of in WT or Disc1 KD dams were measured by qRT-PCR. Data shown in (d−i) are one representative experiment out of two independent replicates with similar results, each including 4 pregnant dams per group (n = 4). The offspring analyzed were NF-WT (n = 36), NF-KD (n = 37), ZIKV-WT (n = 28), and ZIKV-KD (n = 20). Data shown in j are one representative experiment out of two independent replicates with similar results, each including 4 pregnant dams per group (n = 4). Data shown in (d−i) did not follow a normal distribution according to the Shapiro-Wilk test, and statistical analysis was performed using two-tailed Kruskal-Wallis test followed by Dunn’s multiple comparison test (d−g) or two-tailed Mann-Whitney test (h, i). Data shown in e and j were confirmed to follow a normal distribution using the Shapiro-Wilk test, and statistical analysis was performed using two-tailed one-way ANOVA followed by Tukey’s multiple comparison test (e) or Student’s unpaired two-tailed t test (j). Data are presented as means ± SD. Source data are provided as a Source Data file. Figure 3a was created in BioRender. Hl, Z. (2025) https://BioRender.com/dvzm7vs.
DISC1 is associated with the brain damage and long-term behaviors in ZIKV infected-neonatal mice
Given the critical role of DISC1 in mental illness, we investigated its involvement in ZIKV-induced brain damage in newborns. Pups were intracranially injected with 200 PFU of ZIKV or an equal volume of PBS on day 2 after birth. Their brains were collected on days 3, 6 and 9 post-infection, as well as at 6 weeks of age (Fig. 4a). ZIKV NS5 mRNA was detected in the brain on days 3, 6, and 9 post-infection, peaking on day 6, and was undetectable at 6 weeks post-infection, notably, Disc1 KD enhanced ZIKV replication on days 3, 6, and 9 post-infection (Fig. 4b). Similarly, ZIKV-infected Disc1 KD mice exhibited a more significant reduction in body weight compared to ZIKV-infected WT mice on days 3, 6, and 9 post-infection, as well as at 6 weeks, relative to non-infected controls (Fig. 4c). Additionally, decreased brain weight was observed in both WT and Disc1 KD groups on days 3, 6, 9, and at 6 weeks post-infection, compared to non-infected groups, with the ZIKV-infected Disc1 KD group showing a more significant reduction than the ZIKV-infected WT group (Fig. 4d). Notably, when we measured the brain size of the pups, a similar trend of reduction was observed (Fig. 4e–g). While no significant differences were observed between WT and Disc1 KD mice in the non-infected groups, these findings suggest that Disc1 KD in mice leads to more severe impairments in brain development following ZIKV infection.
a Schematic representation of the experiment set up. On the second day after birth, WT and Disc1 KD neonatal mice were intracranially injected with 200 PFU of ZIKV or an equal volume of PBS. Brains were collected on D3, D6 and D9 post-infection, as well as at 6 weeks of age. b ZIKV mRNA levels in WT and Disc1 KD brains were assessed by qRT-PCR on D3, D6 and D9 post-infection, as well as at 6 weeks of age. c, d Body weight (c) and brain weight (d) of WT and Disc1 KD mice were measured on D3, D6, D9 post-infection, as well as at 6 weeks of age. e, f Representative images of WT and Disc1 KD mice brains on D3, D6, D9 post-infection (e), as well as at 6 weeks of age (f). The yellow scale bar indicates consistent size within the same age group. g The cerebral size of WT and Disc1 KD mice was measured on D3, D6, D9 post-infection, as well as at 6 weeks of age. Data shown in (b−d, g) are collected from two independent replicates, including 8 mice per group (n = 8), consisting of 4 males and 4 females. Images in (e, f) are one representative experiment out of two independent replicates with similar results. Data shown in (b) did not follow a normal distribution according to the Shapiro-Wilk test, and statistical analysis was performed using two-tailed Mann-Whitney test. Data shown in (c–d, g) were confirmed to follow a normal distribution using the Shapiro-Wilk test, and statistical analysis was performed using two-tailed one-way ANOVA followed by Tukey’s multiple comparison test. Data are presented as means ± SD. Source data are provided as a Source Data file. Figure 4a was created in BioRender. Hl, Z. (2025) https://BioRender.com/s3be69y.
To investigate whether DISC1 influences long-term behavior following ZIKV infection, we assessed neurological and behavioral changes at 6 weeks of age using various tests: the Y-maze for working memory, the balance beam for balance and coordination, the elevated plus maze for anxiety and depression, and the three-chamber test for sociability and social novelty. While Disc1 KD alone did not impair working memory, ZIKV infection significantly affected performance in WT mice, and this deficit was further exacerbated in Disc1 KD mice (Fig. 5a), suggesting a protective role for DISC1 in cognitive resilience against ZIKV exposure. In contrast, Disc1 KD alone impaired motor coordination, while ZIKV infection had no additional effect on this domain (Fig. 5b). We also observed that both Disc1 KD and ZIKV infection reduced anxiety-like behaviors, but this effect was absent in ZIKV-infected Disc1 KD mice, suggesting that DISC1 may be required for the anxiety-related effects of ZIKV (Fig. 5c, d). Interestingly, ZIKV infection decreased sociability in WT mice, whereas this effect was attenuated in Disc1 KD mice (Fig. 5e), further implicating DISC1 as a modulator of ZIKV-induced social behavior changes. Finally, both Disc1 KD and ZIKV infection independently impaired social novelty recognition. However, the ZIKV-induced deficit was not observed in the context of Disc1 knockdown, highlighting the critical role of DISC1 in mediating this behavioral effect (Fig. 5f). To summarize the overall behavioral alterations, a detailed overview is presented in Table S1.
a On the second day after birth, WT and Disc1 KD neonatal mice were intracranially injected with 200 PFU of ZIKV or an equal volume of PBS, and behavioral tests were conducted when the mice reached 6 weeks of age. The spontaneous alternation rate of WT and Disc1 KD mice in the Y-maze test. b The passing time of WT and Disc1 KD mice in the balance beam test. c, d Proportion of entries into the open arms (c) and the percentage of time spent in the open arms (d) by WT and Disc1 KD mice in the elevated plus maze test. e, f The time of WT and Disc1 KD mice spent on socializing with Stranger 1 (e) and Stranger 2 (f) during the 3-chamber test. Data shown in (a−d) are collected from one independent experiment, each including 8 mice per group (n = 8), consisting of 4 males and 4 females. Data shown in (e, f) are collected from one independent experiment, each including 10 mice per group (n = 10), consisting of 5 males and 5 females. Data shown in (a-b, d–f) were confirmed to follow a normal distribution using the Shapiro-Wilk test, and statistical analysis was performed using two-tailed one-way ANOVA followed by Tukey’s multiple comparison test. Data shown in c did not follow a normal distribution according to the Shapiro-Wilk test, statistical analysis was performed using two-tailed Kruskal-Wallis test followed by Dunn’s multiple comparison test. Data are presented as means ± SD. Source data are provided as a Source Data file.
DISC1 reduces ZIKV infection through autophagy mediated by AMPK-mTOR signaling pathway
To investigate the mechanism by which DISC1 reduces ZIKV infection, we analyzed the DEGs in ZIKV-infected U251 cells overexpressing DISC1 compared to a control group transfected with pcDNA. Among the top 20 significantly enriched KEGG pathways associated with these DEGs, we identified a notable enrichment of genes related to the lysosome pathway and AMPK signaling pathway, specifically in the context of autophagy (Fig. 6a). To further investigate the molecular mechanisms underlying DISC1-mediated antiviral activity, we performed immunoprecipitation followed by mass spectrometry (IP/MS) analysis in DISC1-overexpressing U251 cells to identify DISC1-interacting proteins (Supplementary Data 1). The IP/MS results revealed that these proteins are enriched in the autophagic AMPK-mTOR signaling pathway compared to the control group (Fig. 6b, c). Notably, DISC1 overexpression led to increased phosphorylation of AMPKα (pAMPKα) at 12, 24, and 36 h post-ZIKV infection (Fig. 6d and Fig. S4a). Furthermore, DISC1 overexpression resulted in decreased phosphorylation of mTOR (p-mTOR) at 12 and 24 h compared to controls (Fig. 6d and Fig. S4b). We also observed a reduction in P62 protein levels at 12, 24, and 36 h (Fig. 6d and Fig. S4c), along with an enhanced conversion of LC3A/B Ⅰ to LC3A/B Ⅱ at 0, 12, and 24 h (Fig. 6d and Fig. S4d), indicating that DISC1 activates AMPK-mTOR-mediated autophagy. Based on these findings, we hypothesized that DISC1 may reduce ZIKV replication through the AMPK-mTOR-mediated autophagy. We then treated cells with rapamycin (an mTOR inhibitor and autophagy activator) or MHY1485 (an mTOR activator and autophagy inhibitor). Our results showed that MHY1485 reversed the ZIKV reduction caused by DISC1, while rapamycin failed to prevent this reduction (Fig. 6e and Fig. S4e). We further evaluated chloroquine (CQ), which impairs the fusion of autophagosomes with lysosomes at later stages, and 3-MA, which blocks autophagosome formation at earlier stages. The results demonstrated that CQ, but not 3-MA, effectively blocked the DISC1-mediated reduction in ZIKV protein expression, suggesting that DISC1 decreases ZIKV infection by promoting autophagic flux at a late stage (Fig. 6f and Fig. S4f).
a KEGG pathway enrichment of DEGs in ZIKV-infected U251 cells transfected with HA-DISC1 (1 μg) plasmid compared with the control group transfected with pcDNA. b KEGG pathway enrichment of differentially expressed proteins identified by IP/MS analysis in U251 cells overexpressing HA-DISC1 (5 μg) plasmid compared with the control group transfected with pcDNA. c Chord diagram illustrates the proteins identified in the IP/MS analysis. d U251 cells were transfected with pcDNA or HA-DISC1 (1 μg) plasmids for 24 h followed by ZIKV infection. The protein levels of AMPKα, pAMPKα, mTOR, pmTOR, P62 and LC3A/B were assessed by Western blot at 0, 12, 24, and 36 h post-infection. e U251 cells were transfected with pcDNA or HA-DISC1 (1 μg) plasmids for 24 h, followed by ZIKV infection for 6 h and treated with DMSO, rapamycin (25 μM), and MHY1485 (10 μM) for 24 h. ZIKV E expression was assessed by Western blot. f U251 cells were transfected with pcDNA or HA-DISC1 (1 μg) plasmids for 24 h, followed by ZIKV infection for 6 h and treated with DMSO, autophagosome inhibitor 3-MA (1 mg/ml), and lysosome inhibitor CQ (100 μM) for 24 h. ZIKV E expression was assessed by Western blot. KEGG pathway enrichment analyses in (a, b) were performed using a one-tailed Fisher’s exact test. Images in (d−f) are one representative experiment out of three independent replicates with similar results (n = 3). Source data are provided as a Source Data file.
Besides, previous studies have shown that DISC1 regulates neuronal development through its interaction with the LIS1/NDEL1/dynein complex13,14,15, and that the ZIKV envelope (E) protein can interact with dynein24, raising the possibility that viral proteins may interfere with host intracellular transport machinery. To investigate whether DISC1 plays a role in stabilizing the LIS1/NDEL1/dynein complex during ZIKV infection, we performed co-immunoprecipitation (Co-IP) assays in 293T cells co-transfected with HA-tagged DISC1 and Flag-tagged ZIKV E protein expression plasmids. As shown in Fig. S4g, our Co-IP experiments did not detect a direct physical interaction between DISC1 and the ZIKV E protein. However, overexpression of the E protein markedly weakened the interaction between DISC1 and endogenous NDEL1. In contrast, we did not observe detectable interactions between DISC1 and either endogenous DYNC1I2 or LIS1 under our experimental conditions, which may reflect limitations of the cell type used.
DISC1 can directly interact with LC3 to partially inhibit ZIKV replication
As we observed that autophagy plays a role in DISC1-mediated regulation of ZIKV infection, and given that DISC1 is known to interact with LC3 through its LIR motif to induce mitophagy-a type of autophagy21, we constructed a DISC1 mutant plasmid based on the HA-DISC1 construct with a specific point mutation (highlighted in yellow) in the LIR motif (amino acids 210-213, FSFI mutated to AAAA)21 to assess whether this interaction with LC3 is essential for decreasing ZIKV infection (Fig. 7a). We first confirmed the interaction between DISC1 and LC3 in 293 T cells, and mutation of DISC1 abolished this interaction (Fig. 7b). We then compared the inhibitory effects of WT DISC1 and the DISC1 mutant during ZIKV infection. Although we observed significant reductions in ZIKV mRNA levels (Fig. 7c), protein expression (Fig. 7d), and viral loads (Fig. 7e) on both days 1 and 2 post-infection in cells transfected with WT DISC1 compared to those transfected with the DISC1 mutant or negative control, the reduction was not completely reversed by DISC1 mutant. This suggests that the interaction between DISC1 and LC3 plays a partial role in reducing ZIKV infection.
a Conserved LIR motifs (amino acids 210 FSFI 213) in DISC1 were highlighted in yellow, which were mutated into AAAA in DISC1 mutant. b 293T cells were co-transfected with HA-DISC1 (5 μg) or HA-DISC1 mutant (5 μg) and GFP-LC3 (5 μg) plasmids for 48 h. Cellular lysates were subjected to immunoprecipitation with anti-Flag or anti-HA magnetic beads and Western blot assays using the indicated antibodies. c−e U251 cells were transfected with HA-DISC1 (1 μg) or HA-DISC1 mutant (1 μg) plasmids for 24 h, followed by ZIKV infection for 24 and 48 h. Viral mRNA levels, protein expression and titers were measured on D1 and D2 post-infection by qRT-PCR (c), Western blot (d), and plaque assays (e). Images in (b, d) are from one representative experiment out of three independent replicates with similar results (n = 3). Data shown in (c, e) are from one representative experiment out of three independent replicates with similar results, each including 3 biological replicates per group (n = 3). Data shown in (c, e) were confirmed to follow a normal distribution using the Shapiro-Wilk test, and statistical analysis was performed using two-tailed one-way ANOVA followed by Tukey’s multiple comparison test. Data are presented as means ± SD. Source data are provided as a Source Data file.
DISC1 reduces ZIKV infection in the primary cortical neurons, placenta and fetal head by activating autophagy via the AMPK–mTOR signaling pathway
To gain further mechanistic insight, we isolated and differentiated primary cortical neurons from WT and Disc1 KD mice, achieving a differentiation efficiency of at least 90% with a representative efficiency of 92.1% shown in Fig. 8a and Fig. S5a, and approximately a 50% reduction in DISC1 protein expression (Fig. S5b). Following ZIKV infection on day 1 and day 2, Disc1 KD neurons displayed significantly higher viral RNA levels (Fig. 8b), viral loads (Fig. 8c), and ZIKV E protein expression (Fig. 8d) than WT neurons, indicating enhanced viral infection in the context of Disc1 knockdown. To explore the role of autophagy in this context, we examined key markers of the AMPK-mTOR signaling pathway. In non-infected neurons, Disc1 KD neurons exhibited no significant difference in the pAMPKα/AMPKα ratio (Fig. 8d, e) but a higher pmTOR/mTOR ratio (Fig. 8d and f) and elevated P62 levels (Fig. 8d and g), along with reduced expression of LC3A/B Ⅰ (Fig. 8d and h) and LC3A/B Ⅱ (Fig. 8d and i), indicating impaired autophagic activity under basal conditions. Upon ZIKV infection, the AMPK-mTOR signaling pathway was further activated in WT neurons, as evidenced by an increase in the pAMPKα/AMPKα ratio (Fig. 8d, e), a decrease in the pmTOR/mTOR ratio (Fig. 8d and f), reduced P62 levels (Fig. 8d and g), and elevated expression of LC3A/B Ⅰ (Fig. 8d and h) and LC3A/B Ⅱ (Fig. 8d and i), consistent with enhanced autophagic flux. In contrast, these changes were attenuated in ZIKV-infected Disc1 KD neurons (Fig. 8d–i), suggesting an impaired to activate autophagy in response to infection. Together, these findings support the conclusion that DISC1 reduces ZIKV infection, at least in part, by promoting AMPK/mTOR-mediated autophagy signaling.
a Primary cortical neurons were isolated from neonatal mice (P0–P1) and induced to differentiate in vitro. On day 7 of differentiation, neurons exhibiting neurite outgrowth were observed under bright-field microscopy. Confocal imaging showed neuronal nuclei stained with DAPI (blue), and endogenous Neun labeled with an anti-Neun antibody (red). Scale bar = 50 μm. b, c WT and Disc1 KD primary cortical neurons were pre-incubated with 20 μg/mL MAR1-5A3 for 6 h, followed by infection with ZIKV at a MOI of 1. Viral mRNA levels and titers were measured on D1 and D2 post-infection using qRT-PCR (b) and plaque assay (c). d WT and Disc1 KD primary cortical neurons were pre-incubated with 20 μg/mL MAR1-5A3 for 6 h, followed by infection with ZIKV at a MOI of 1. The protein levels of ZIKV E protein, DISC1, AMPKα, pAMPKα, mTOR, pmTOR, P62 and LC3A/B were assessed by Western blot on D1 and D2 post-infection. e, f The ratios of pAMPKα to AMPKα (e) and pmTOR to mTOR (f) were determined by comparing the corresponding protein band intensities at each time point in Fig. 8d. g−i Quantification of P62 (g), LC3A/BⅠ (h) and LC3A/BⅡ (i) protein levels relative to GAPDH at each time point in Fig. 8d. j, k Placentas (j) and fetal heads (k) from non-infected and ZIKV-infected WT and Disc1 KD mice were collected on E13.5. The protein levels of DISC1, AMPKα, pAMPKα, mTOR, pmTOR, P62 and LC3A/B were assessed by Western blot. l−o The ratios of pAMPKα to AMPKα (l, m) and pmTOR to mTOR (n, o) were determined by comparing the corresponding protein band intensities at each time point in Fig. 8j (l, n) and Fig. 8k (m, o). p−s Quantification of P62 (p, q) and LC3A/BⅡ (r, s) protein levels relative to GAPDH at each time point in Fig. 8j (p, r) and Fig. 8k (q, s). Data shown in (b, c) are from one representative experiment out of three independent replicates with similar results, each including 3 biological replicates per group (n = 3). Images in (a, d, j–k) are from one representative experiment out of three independent replicates with similar results (n = 3). Data shown in (e−i, l−s) are collected from three independent experiments (n = 3). Data shown in (b−c, e−i, l−s) were confirmed to follow a normal distribution using the Shapiro-Wilk test, and statistical analysis was performed using Student’s unpaired two-tailed t test (b, c) or two-tailed one-way ANOVA followed by Tukey’s multiple comparison test (e−i, l−s). Data are presented as means ± SD. Source data are provided as a Source Data file.
To provide in vivo validation, we examined autophagy-related protein levels in placentas (Fig. 8j) and fetal heads (Fig. 8k) from both non-infected and ZIKV-infected WT and Disc1 KD mice. As expected, DISC1 protein levels were significantly reduced in Disc1 KD placentas (Fig. 8j and Fig. S5c) and fetal heads (Fig. 8f and Fig. S5d). Unlike the clear impairment of autophagy observed in neurons, we detected only elevated P62 levels under non-infected conditions in placentas and fetal heads with Disc1 knockdown (Fig. 8j, k and p, q). Upon ZIKV infection, WT placentas and fetal heads exhibited a marked increase in the pAMPKα/AMPKα ratio (Fig. 8j–m), a decrease in the pmTOR/mTOR ratio (Fig. 8j, k and n, o), reduced P62 levels (Fig. 8j, k and p, q), and elevated expression of LC3A/BⅡ (Fig. 8j, k and r, s). However, these changes were notably attenuated in the ZIKV-infected Disc1 KD group. Notably, LC3A/BⅠ was undetectable under our experimental conditions. Together, these in vivo data demonstrate that DISC1 promotes autophagy in the placenta and fetal head by modulating AMPK/mTOR signaling, thereby contributing to reduced ZIKV replication.
DISC1 reduces ZIKV-induced brain injury and long-term neurobehavioral impairments in mice via autophagy
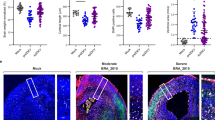
To assess whether DISC1 modulates ZIKV-induced brain injury through autophagy, we performed hematoxylin and eosin (H&E) staining (Fig. 9a) and immunofluorescence analysis (Fig. 9b, Fig. S6a–S6b and S7) on brain tissues collected from WT and Disc1 KD adult mice 6 days after intraperitoneal ZIKV infection. As shown in Fig. 9a, Disc1 KD mice exhibited neuronal abnormalities in the hippocampal dentate gyrus (DG) and cornu ammonis 3 (CA3) regions, characterized by shrunken, hyperchromatic neurons with indistinct boundaries (black arrows), compared to WT controls. ZIKV infection exacerbated these pathological changes, resulting in more severe neuronal damage in both the DG and CA3 of Disc1 KD mice than in infected WT mice. Autophagy levels, visualized as yellow puncta labeled with antibody against LC3A/B, were markedly elevated in ZIKV-infected brains relative to non-infected controls. However, the autophagy signal was notably stronger in WT mice than in Disc1 KD mice, particularly in the DG (Fig. 9b), CA3 (Fig. S6a), cortex (Fig. S6b), and hypothalamus (Fig. S7). These findings further support the conclusion that DISC1 promotes autophagy in response to ZIKV infection, thereby mitigating virus-induced neuronal injury.
a 6-8-weeks-old WT or Disc1 KD mice were treated with 2 mg MAR1-5A3 on the day prior to infection and then intraperitoneally (i.p.) inoculated with PBS or 1 × 106 PFU of ZIKV. Mouse brains were collected on D6 post-infection. Representative H&E-stained sections of the hippocampal DG and CA3 regions are shown. Black arrows indicate shrunken, hyperchromatic neurons with indistinct boundaries Scale bar = 100 μm. b Representative immunofluorescence-stained sections of the hippocampal DG region. Cell nuclei were stained using DAPI (blue). Endogenous Neun was labeled with anti-Neun antibody (red). Endogenous LC3A/B was labeled with anti-LC3A/B antibody (yellow). Scale bar = 100 μm. Representative images in (a, b) are from one independent experiment, including 6 mice per group (n = 6), consisting of 3 males and 3 females.
To further support our behavioral findings and address the need for detailed neuropathological and molecular characterization, we performed H&E staining and immunofluorescence analysis on brain tissues collected from WT and Disc1 KD neonatal mice 6 days or 6 weeks after intracranial ZIKV infection. As shown in Fig. 10a, both WT and Disc1 KD mice displayed normal neuronal morphology in the CA1 region under non-infected conditions. Following ZIKV infection, WT mice still exhibited no apparent neuronal abnormalities in this region. However, Disc1 KD mice displayed neuronal abnormalities in the CA1 region, characterized by shrunken, hyperchromatic neurons with indistinct boundaries (black arrows), along with nuclear pyknosis and fragmentation (orange arrows), compared to WT controls. Immunofluorescence analysis revealed elevated autophagy levels and viral protein expression in ZIKV-infected brains compared to non-infected controls. Autophagy was visualized as yellow puncta (anti-LC3A/B), and ZIKV E protein as green puncta. Notably, the autophagy signal was significantly stronger in ZIKV-infected WT mice than in ZIKV-infected Disc1 KD mice, particularly in the DG and CA3 regions (Fig. 10b), as well as in the cortex (Fig. S8a), thalamus (Fig. S8b), hypothalamus (Fig. S9a), and cerebellum (Fig. S9b), whereas ZIKV E protein levels were correspondingly lower in WT mice. By 6 weeks of age, Disc1 KD mice exhibited neuronal abnormalities in the DG region, characterized by shrunken, hyperchromatic neurons with indistinct boundaries (black arrows), compared to WT controls. ZIKV infection exacerbated these pathological changes, resulting in more severe neuronal damage in the DG of Disc1 KD mice than in infected WT mice (Fig. S10). Collectively, these results indicate that DISC1 alleviates ZIKV-induced neuropathology and associated behavioral deficits primarily by enhancing autophagic responses.
a Neonatal mice were intracranially injected with 200 PFU of ZIKV GZ01 or an equal volume of PBS into the λ point of the brain on postnatal day 2 (P2). Mouse brains were collected on D6 post-infection. Representative H&E-stained sections of the hippocampal CA1 region are shown. Black arrows indicate shrunken, hyperchromatic neurons with indistinct boundaries. Orange arrows indicate nuclear pyknosis and fragmentation. Scale bar = 100 μm. b Representative immunofluorescence-stained sections of the hippocampal DG region. Cell nuclei were stained using DAPI (blue). Endogenous Neun was labeled with anti-Neun antibody (red). ZIKV E protein was labeled with anti-E antibody (green). Endogenous LC3A/B was labeled with anti-LC3A/B antibody (yellow). Scale bar = 100 μm. Representative images in (a, b) are from one representative experiment out of two independent experiments with similar results, each including 4 mice per group (n = 4), consisting of 2 males and 2 females.
Discussion
Although DISC1 is a crucial scaffolding protein involved in neuronal migration and neural stem cell renewal25,26, its role during viral infections, particularly in neurotropic virus infections, remains largely unexplored. In this study, we demonstrate that DISC1 plays a protective role against ZIKV infection in both in vitro and in vivo models. Specifically, we show that DISC1 reduces ZIKV replication through AMPK-mTOR signaling-mediated autophagy. Furthermore, we identify a novel function of DISC1 in modulating behavioral alterations during ZIKV infection, which may help explain the long-term neurological effects observed in some infants born to ZIKV-infected mothers.
While most previous studies have focused on the biological functions of DISC1, a recent report has shown that acute H1N1 infection can induce DISC1 aggregation, thereby disrupting proteostasis of proteins associated with neurodegenerative diseases22. However, the role of DISC1 in viral infections remains largely unclear. Notably, our study demonstrates that DISC1 reduces ZIKV infection without evidence of DISC1 aggregation, suggesting a distinct antiviral role that does not involve protein misfolding or aggregation. Our findings further suggest that the AMPK-mTOR signaling pathway mediates the protective effect of DISC1 against ZIKV infection. The AMPK-mTOR pathway, known for regulating various cellular processes such as growth, survival, and proliferation27,28,29, has been widely implicated in flavivirus infections. For example, dengue virus infection activates the AMPK-mTOR axis and reduces viral replication30, while mTOR promotes West Nile virus replication31, highlighting the complex and context-dependent role of AMPK-mTOR in viral infections. In the context of ZIKV infection, the role of mTOR-related autophagy remains controversial. Some studies have reported that inhibition of mTOR reduces ZIKV replication, suggesting that mTOR-driven autophagy may support viral propagation32,33. Conversely, our findings show that DISC1 functions as an upstream regulator of the AMPK-mTOR pathway, promoting autophagy that reduces ZIKV replication. This is in line with other studies reporting an antiviral role of autophagy in various cell types34,35. Together, these observations underscore the complex, context-dependent nature of autophagy in ZIKV pathogenesis, warranting further investigation to clarify its precise role.
Although we were unable to generate a completely Disc1-deficient mouse model, previous studies have shown that Disc1 KD or point mutation models can effectively reveal functional roles of DISC1 in neurodevelopment and disease25,36,37, and our findings are consistent with these literature. Specifically, our data suggest that DISC1 contributes to ZIKV-induced behavioral abnormalities, including impaired working memory, reduced anxiety-like behavior, impaired sociability, and diminished social novelty recognition. Importantly, we observed no significant differences in body size or weight between Disc1 KD and control pups or adult mice, indicating that the knockdown did not interfere with our investigation of ZIKV dissemination and vertical transmission. The current model, which achieves approximately a 50% reduction in DISC1 expression, enabled us to uncover the role of DISC1 in modulating ZIKV pathogenesis. Notably, we found that DISC1 attenuated ZIKV infection in key target tissues such as the brain, uterus, and testes, supporting a protective role for DISC1 in host defense. Most critically, partial DISC1 knockdown led to increased ZIKV replication in the placenta, fetal brain, and primary neurons, thereby promoting key pathological features associated with CZS. These findings highlight DISC1 as a potential genetic modifier of susceptibility to ZIKV and provide new insights into gene–virus interactions in neurodevelopmental disorders.
This study has several limitations. First, we acknowledge that impaired motor coordination may confound the interpretation of certain behavioral outcomes, particularly in the elevated plus maze and social novelty recognition tests. Despite the widespread acceptance of the current experimental design, we were unable to fully disentangle motor-related impairments from domain-specific behavioral deficits. Therefore, the behavioral impact of ZIKV must be considered in the context of an existing genetic vulnerability. Further in-depth investigations are needed to clarify the mechanisms underlying these behavioral changes. Second, we used neonatal intracranial injection of ZIKV as an alternative model for studying ZIKV-associated behavioral changes in offspring, based on previous studies38,39,40,41. However, this model does not allow us to directly determine the role of DISC1 in vertical transmission in relation to behavioral changes, which will need to be investigated in future studies. Third, although we identified that DISC1 promotes autophagy through the AMPK–mTOR signaling axis, thereby reducing ZIKV replication and viral titers in both cellular and animal models, additional pathways—such as those involving the phagosome and proteasome—were also enriched in our transcriptomic analysis and warrant further investigation.
In conclusion, our study provides novel insights into the role of DISC1 in ZIKV infection, particularly its involvement in the development of CZS. We show that DISC1 not only modulates viral replication through the AMPK-mTOR signaling pathway but also contributes to long-term behavioral alterations, which may underlie some of the lasting neurological effects observed in affected individuals. These findings highlight DISC1 as a potential target for therapeutic strategies aimed at mitigating the long-term consequences of ZIKV infection and further underscore the complex interplay between viral infections and host cellular processes.
Methods
Ethics statement
The experiments with infectious ZIKV were conducted under biosafety level 2 (BSL-2) facilities at School of Public Health, Shenzhen Campus of Sun Yat-sen University, Sun Yat-sen University Laboratory Animal Center and Beijing Laboratory Animal Research Center. All animal experiments were approved by the Animal Care and Use Committee of Sun Yat-sen University (Ethics File Approval No.: SYSU-IACUC-2025-000887) and Beijing Laboratory Animal Research Center (Ethics File Approval No.:BLARC-SSYYDW/013-JL/001).
Cell culture and virus
The trophoblast cell line (HTR-8/SVneo) was purchased from the American Type Culture Collection (ATCC, CRL-3271) and cultured in RPMI-1640 medium (ATCC, 30–2001) with 5% fetal bovine serum (FBS; CLARK, FB25015) and 100 units/ml Penicillin-Streptomycin (PS; Gibco, 15140–122). The human glioblastoma cell line (U251) was obtained from the Procell Life Science & Technology Co., Ltd (Procell; Wuhan, China) and maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Cienry, CR-12800) containing 10% FBS and 100 units/ml PS. The African green monkey (Vero) cell line (ATCC, CCR-81) was provided by Prof. Caijun Sun from Sun Yat-sen University and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, C11995500BT) containing 10% FBS and 100 units/ml PS. All cells were cultured at 37 °C in a 5% CO2 incubator. DMSO (Sigma, D2650) was purchased from Sigma-Aldrich. 3-MA (3-Methyladenine, HY-19312), CQ (Chloroquine, HY-17589A), Rapamycin (HY-10219) and MHY1485 (HY-B0795) were obtained from MedChemExpress.
The Asian lineage ZIKV GZ01 (GenBank accession number: KU820898) used in this study was provided by Prof. Jincun Zhao from Guangzhou Medical University.
Viral infection
HTR8 cells and U251 cells were infected with ZIKV GZ01 at a multiplicity of infection (MOI) of 1 PFU or non-infected group in the culture medium. The culture medium used for infection was the respective complete culture medium for HTR8 cells or U251 cells, without PS. The cells were incubated with the virus for 1 h with intermittent shaking every 20 min. After incubation, the inoculum was removed, and the cells were washed twice with phosphate-buffered saline (PBS; Biosharp, BL601A) to remove any unbound virus. A complete culture medium was added to each well, and the cells were further incubated at 37 °C with 5% CO2. Cell samples were collected at different time points for analysis.
Plaque assay
Vero cells were seeded in 24-well plates and grown to 80%-90% confluence. Viral samples were ten-fold serially diluted six times in DMEM containing 2% FBS and added to each well. The plates were incubated at 37 °C for 1 h with gentle rocking every 15 min. The viral solution was then replaced with 1 mL of 0.8% methylcellulose overlay containing 3.7 g/L NaHCO3 (Sigma, S5761), 0.025 M HEPES buffer (Gibco, 15630-080), 2% FBS, 1% PS and 0.5% DMSO, and plates were incubated at 37 °C for 4 days. After incubation, cells were fixed with 3.7% formaldehyde (Aladdin, F111936) for 30 min and stained with 0.1% crystal violet (Sigma, C0775) for plaque visualization. Viral titers were calculated as plaque-forming units (PFU) per milliliter based on the number of plaques and the dilution factor.
Animal experiments
The Disc1 knockdown mice (C57BL/6 background) were generated by Cyagen Biosciences (Guangzhou, China) using CRISPR/Cas9-mediated deletion of exon 2 of the Disc1 gene. Four guide RNAs (gRNAs) were designed to target exon 2:
gRNA-A1 (AGGCTAGCTCGAACTCCCTCTGG)
gRNA-A2 (CGGGACCAGCACTCAGTCGGTGG)
gRNA-B1 (TTTCACGCTGTGTCTTGGATAGG)
gRNA-B2 (AAGTATGGGCCCCATCAGAGAGG).
All mice were maintained on a 12 h light/12 h dark cycle, at 18-23 °C with 40%−60% humidity. To establish an adult viral infection model, the 6-8-week-old mice (both male and female, n = 6 per group) were treated with 2 mg of anti-IFNAR antibody (MAR1-5A3; Leinco Technologies, 0622L335) on the day before infection and then intraperitoneally inoculated with 1 × 106 plaque-forming units (PFU) of ZIKV GZ01 or an equivalent volume of PBS. Tissues were harvested on day 2 and day 6 upon ZIKV infection. For the prenatal microcephaly model, 8-10-week-old mice (female, n = 4 per group) were set up for timed-mating and were intraperitoneally inoculated with 1 × 107 PFU of ZIKV GZ01 or PBS on E6.5. Pregnant mice were injected intraperitoneally with 2 mg MAR1-5A3 on E5.5 and euthanized on E13.5. On E13.5, placentas and fetal heads were harvested. Fetal head weight was measured using a high-precision analytical balance, while occipito-frontal diameter (OFD) and crown-rump length (CRL) were measured using a digital caliper. Collected placentas and fetal heads were used for viral mRNA levels quantification and protein expression analysis. For the neonatal infection model, 200 PFU of ZIKV GZ01 or an equal volume of PBS was stereotactically injected into the λ point of the brain in P2 mice (both male and female, n = 8 per group). The injection was performed at a depth of 2–3 mm using a 10 μL Hamilton syringe (Hamilton, 81216) equipped with a 33-gauge needle. A total volume of 5 μL was delivered per mouse, and the needle was held in place for 1 min post-injection to minimize back flow. Based on visual assessment of reflux and our experience, nearly all of the inoculum was retained in the brain. For the behavioral experiments, neonatal mice at P2 were intracranially injected with 200 PFU of ZIKV GZ01 or an equal volume of PBS, and behavioral tests were conducted at 6 weeks of age (both male and female, n = 8 or 10, per group).
Behavioral tests
Y maze: During the training phase, the 6-week-old WT or Disc1 KD mice were placed into the start arm facing the wall and were allowed to explore the start and trained arm for 5 min, while the entry to the 3rd arm (novel arm) was blocked. The maze was cleaned between each mouse to remove odor cues, and the trained arm was alternated between mice. After training, the mouse was returned to its home cage. After 1 h, the mouse was returned to the start arm and was allowed to explore all three arms for 5 min. The number of entries and the time spent in each arm was quantified using the Smart Video Tracking Software. The percentage of spontaneous alternation was calculated using the following formula: Alternation (%) = (Number of actual alternations/Total number of possible alternations) × 100%.
Balance beam test: Motor coordination and balance were evaluated using the balance beam test. The apparatus consisted of a 1 meter-long metal beam with a square cross-section of 10 mm, positioned horizontally 50 cm above the bench surface. One end of the beam was mounted on a narrow support, while the opposite end was connected to an enclosed escape box. The beam was positioned such that the distance from the starting point to the entrance of the escape box was 100 cm. Two permanent markers were placed on the beam: one located 10 cm from the starting end and the other 10 cm from the escape box entrance. The 80 cm segment between these two markers served as the timing zone. The traversal time was recorded from the moment the mouse crossed the first marker (10 cm from the start) to the point it reached the second marker (10 cm from the goal). Prior to testing, mice underwent two days of training to familiarize them with traversing the beam and entering the escape box. After each successful traversal during training, mice were allowed to stay in the escape box for 15 s before being gently removed. Following the training period, each mouse performed three test trials. The average traversal time across the three trials was calculated for analysis. To minimize fatigue and potential stress-induced variability, a minimum interval of 15 min was maintained between trials for each mouse.
Elevated plus maze: The elevated plus maze test was used to assess anxiety- and depression-like behaviors in mice. The apparatus consisted of four arms arranged in a cross shape: two open arms (30 × 5 cm) without walls and two closed arms (30 × 5 × 15 cm) with walls, elevated 50 cm above the floor. The arms extended from a central platform (5 × 5 cm), and the maze was placed in a dimly lit, quiet testing room. Each mouse was placed individually on the central platform facing an open arm and allowed to explore the maze freely for 5 min. The number of entries and the time spent in each arm were recorded using a video tracking system. An arm entry was defined as all four paws entering an arm. Two anxiety-related parameters were calculated: Entries to open arms (%) = (Number of open arm entries / Total number of entries) × 100%. Time in open arms (%) = (Time spent in open arms / Total time in all arms) × 100%.
Three-chamber test: The three-chamber apparatus consists of a rectangular transparent acrylic box (60 cm × 45 cm × 30 cm) evenly divided into three connected compartments by two transparent partitions with small openings (5 cm × 5 cm) allowing free movement between chambers. The test is performed under dim lighting in a quiet room. The test includes three consecutive 10-min phases: (i) Habituation phase: The test mouse is placed in the central chamber and allowed to freely explore all three chambers without any stimuli. This allows the animal to habituate to the environment. (ii) Sociability phase: An unfamiliar mouse (Stranger 1, age- and sex-matched) is placed in a ventilated wire cage (10 cm diameter) in one of the side chambers, while the opposite chamber contains an empty but identical wire cage. The test mouse is then allowed to explore all three chambers freely for 10 min. The position of the social target is alternated between tests to avoid side bias. (iii) Social novelty phase: A second unfamiliar mouse (Stranger 2) is placed in the previously empty cage, while Stranger 1 remains in place. The test mouse is again allowed to explore freely for another 10 min to assess preference for novel versus familiar social targets. Sociability was evaluated by measuring the time the test mouse spent interacting with Stranger 1 compared to the empty wire cage during the sociability phase. Social novelty preference was assessed by comparing the interaction time with the familiar mouse (Stranger 1) and the novel mouse (Stranger 2) during the social novelty phase. Interaction was defined as direct sniffing or remaining within 2 cm of the wire cage.
Plasmids and siRNA
The HA-tagged DISC1 expression plasmid was constructed by inserting the HA-tagged DISC1 coding sequence (NCBI accession number: NM_001012957.2) into the pcDNA vector in Sangon Biotechnology (Shanghai, China). The Flag-tagged E expression plasmid was constructed by inserting the Flag-tagged E coding sequence (NCBI accession number: YP 009430300.1) into the pcDNA vector in Sangon Biotechnology (Shanghai, China). The HA-tagged DISC1 mutant expression plasmid was generated by site-directed mutagenesis using the HA-DISC1 wild-type construct as a template. Four primers were designed to introduce targeted substitutions (FSFI to AAAA) within the LC3-interacting region (LIR) motif of the DISC1 gene:
DISC1 mutant Forward-1: GCTCGGATCCACTAGTCCAGTGTGGTGGAATTCGCCACCATGCCAGGC
DISC1 mutant Reverse-1: GAGCCAAGCGAGAGCCGCGCTGCAGCGGCGCTTGAGGTAAAGGCACTGTGAG
DISC1 mutant Forward-2: CAGTGCCTTTACCTCAAGCGCCGCTGCAGCGCGGCTCTCGCTTGGCTCTGCCG
DISC1 mutant Reverse-2: GAGGCTGATCAGCGGGTTAAACGGGCCCTCTAGATTAGGCGTAATCGGGCAC.
PCR amplification was performed using the HA-DISC1 plasmid as a template and T8 high-fidelity DNA polymerase (TSINGKE, TSE111). The PCR program was: initial denaturation at 95 °C for 3 min; followed by 25 cycles of 95 °C for 25 s, 62 °C for 20 s, and 72 °C for 40 s; with a final extension at 72 °C for 1 min. The two overlapping fragments were fused by overlap extension PCR using DISC1 mutant Forward-1 and Reverse-2 under the same PCR conditions. The full-length PCR product was purified using an agarose gel extraction kit (Axygen, AP-GX-500). The pcDNA3.1(+) vector was digested with EcoRI (NEB, R3101) and XbaI (NEB, R0145V), gel-purified, and recombined with the purified mutant insert using a recombinase-based cloning system. The ligation mix was incubated at 50 °C for 25 min and transformed into E. coli competent cells (TSINGKE, TSC-C14). Transformants were selected on ampicillin-containing agar plates and incubated overnight at 37 °C. Positive colonies were screened by colony PCR using primers: SEQ1: TGGCACCAAAATCAACGGGACT, SEQ2: TGACACCTACTCAGACAATG. Colonies showing the correct band size were cultured overnight, and plasmids were extracted for sequencing validation using the following primers: SEQ1: TGGCACCAAAATCAACGGGACT, SEQ2: TGACACCTACTCAGACAATG, SEQ3: CTCGGCGCACTTTGGGATTC, SEQ4: CACAGCCAGGGCTGCAGCTG, SEQ5: CTTGGCTACACGGGTCTCTGC, SEQ6: GAAAAGCAGCAGCTACAGAAAG. Sanger sequencing confirmed the presence of the intended mutation and overall sequence integrity of the HA-DISC1 mutant plasmid.
The GFP-tagged LC3 expression plasmid was constructed by inserting the GFP-tagged LC3 coding sequence (NCBI accession number: NM_022818.5) into the pcDNA vector in Sangon Biotechnology (Shanghai, China). Endogenous DISC1 expression was knocked down using siRNA (siDISC1: 5′-GGCAGAUGGAUGACUUAGATT-3′). JetPRIME transfection reagent (Polyplus, 101000046) was used to transfect the DNA plasmids and the siRNAs.
Quantitative real-time PCR (qRT-PCR)
Total intracellular RNA was isolated using the TRIzol reagent (Invitrogen, 15596018). The isolated RNA was reverse-transcribed into complementary DNA (cDNA) using the HiScript IV RT SuperMix kit (Vazyme, R423-01), which contains both oligo (dT) and random hexamer primers, according to the manufacturer’s instructions. The qPCR was performed in the CFX96 real-time PCR system (Bio-Rad) with ChamQ Universal SYBR qPCR Master Mix Kit (Vazyme, Q711-02). All primers were synthesized by Sangon Biotechnology (Shanghai, China). Relative mRNA expression was determined using GAPDH as the internal control and calculated using the 2^(-∆∆Ct) method. The primers used for qPCR were as follows:
ZIKV NS5 Forward: 5ʹ-GGTCAGCGTCCTCTCTAATAAACG-3ʹ
ZIKV NS5 Reverse: 5ʹ-GCACCCTAGTGTCCACTTTTTCC-3ʹ
Human DISC1 Forward: 5ʹ-AGACTCACCTCATCCCCTCTC-3ʹ
Human DISC1 Reverse: 5ʹ-TCCAAGTACAATAGCTTCTTGCC-3ʹ
Human GAPDH Forward: 5ʹ-CTGACTTCAACAGCGACACC-3ʹ
Human GAPDH Reverse: 5ʹ-TAGCCAAATTCGTTGTCATACC-3ʹ
Mouse GAPDH Forward: 5ʹ-AGGTCGGTGTGAACGGATTTG-3ʹ
Mouse GAPDH Reverse: 5ʹ-TGTAGACCATGTAGTTGAGGTCA-3ʹ.
Genotyping
Extraction of genomic DNA from mouse tails and the PCR amplification system were accomplished with a One Step Mouse Genotyping Kit (Vazyme, PD101). Two primer PCR strategies were employed for genotyping, primer set 1 (Forward: 5’-TATGTATCATCAGCACTAGGACAG-3’, Reverse: 5’-TATGGGGCAAGGAGAAGTAATG-3’, product size 1817 bp and 484 bp from WT and Disc1 knockdown alleles, respectively. Primer set 2 (Forward: 5’-TGGATGGCATACATTGCTCAG-3’, Reverse: 5’-TATGGGGCAAGGAGAAGTAATG-3’), produced 410 bp fragments from the Disc1 knockdown allele.
Cell viability assay
HTR8 and U251 cells were seeded in each well of the 96-well plate (NEST, 701011), and a series of concentrations of plasmids or siRNA were added. After incubating for 24 h, 10 μl CCK8 (Biosharp, CCK8-BS350B) solution was added into each well, followed by 2 h in the dark at 37 °C. The OD at 450 nm was then detected by a microplate reader (Biotek, Synergy HTX, USA).
Western blot
At different time points, proteins were harvested from cells and animal tissues and lysed in RIPA lysis buffer (Beyotime, P0013B) containing 1% PMSF (Selleck, S3025). Protein samples were prepared and separated on 12% SDS-PAGE (Vazyme, E304), followed by transfer to PVDF membranes (Merck Millipore, IPVH00010). After blocking with 5% bovine serum albumin (BSA; Sigma, V900933) in PBS containing 0.05% Tween-20 (Beyotime, ST1726) for 1 h at room temperature (RT), the membranes were washed three times with PBS containing 0.05% Tween-20 and then incubated with the indicated primary antibodies overnight at 4 °C. The next day, the blots were incubated with secondary antibodies. Protein bands were then visualized using Tanon™ High-sig ECL Western Blotting Substrate (Tanon) and imaged using a Chemiluminescence imaging system (SAGECREATION, MiniChemi610). The following primary antibodies were used: anti-DISC1 antibody (1:1000, Invitrogen, 40-6800), anti-DISC1 antibody (1:500, Santa Cruz Biotechnology, sc-365591), anti-DISC1 antibody (1:500, GeneTex, GTX33154), anti-Zika virus E antibody (1:000, Genetex, GTX133314), anti-HA tag antibody (1:2000, Genetex, GTX115044), anti-AMPKα antibody (1:1000, Beyotime, AF1627), anti-pAMPKα antibody (1:1000, Cell Signaling Technology, 2535S), anti-mTOR antibody (1:500, Beyotime, AF1648), anti-pmTOR antibody (1:1000, Cell Signaling Technology, 5536), anti-P62 antibody (1:1000, Cell Signaling Technology, 39749), anti-LC3A/B antibody (1:1000, Cell Signaling Technology, 4108), anti-GFP antibody (1:2000, Genetex, GTX113617), anti-Flag antibody (1:2000, Sigma, F1804), anti-DYNC1I2 antibody (1:1000, Proteintech, 12219-1-AP), anti-LIS1 antibody (1:1000, Proteintech, 20678-1-AP), anti-NDEL1 antibody (1:1000, Proteintech, 17262-1-AP), anti-GAPDH antibody (1:20000, Fude Biotech, FD0063), HRP conjugated goat anti-mouse antibody (1:10000, Fude Biotech, FDM007), HRP conjugated goat anti-rabbit antibody (1:10000, Fude Biotech, FDR007).
Co-immunoprecipitation (Co-IP)
HEK293T cells containing HA-DISC1 or GFP-LC3 plasmids were lysed in RIPA lysis buffer (weak) (Beyotime, P0013D) containing 1% PMSF. Cell lysates were centrifuged at 12000 × g for 20 min at 4 °C, and the supernatant was incubated with HA-tagged magnetic beads (ThermoFisher, 88836) at 4 °C for 12−14 h. Then the beads were washed with TBST (Biosharp, BL1423A) containing 1% NP40 (Solarbio, N8030) and 0.5% Tween-20 for three times and resolved with protein loading buffer. The target proteins were detected by Western blot.
Isolation and culture of primary mouse macrophages and cortical neurons
Primary macrophages were derived from the bone marrow of 3–4 week-old WT or Disc1 KD mice. Briefly, femurs and tibias were aseptically harvested and flushed with sterile DMEM/F-12 medium (Gibco, 10565) using a 1 mL sterile syringe (Prosperich, HC-ZSQ-20) to extract bone marrow cells. The cell suspension was filtered through a 70 μm cell strainer (Biologix, 15−1070), centrifuged at 300 × g for 5 min at 4 °C, and resuspended in complete macrophage differentiation medium containing DMEM/F-12 supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin (PS), 5% L-glutamine (ThermoFisher, 25030081), and 20 ng/mL recombinant murine macrophage colony-stimulating factor (M-CSF, PeproTech). Cells were plated in Petri dishes (NEST, CM400039-704001) at a density of 2 × 106 cells/mL and cultured at 37 °C in a humidified incubator with 5% CO2. On day 3, half of the medium was replaced with fresh differentiation medium. By day 7, adherent cells were considered fully differentiated macrophages. For ZIKV infection, macrophages were detached using cell dissociation buffer (ThermoFisher, 13151014) at 37 °C for 5 min and then replated as needed. Prior to infection, macrophages were pretreated with 20 μg/mL MAR1-5A3 for 6 h. Macrophages were then infected with ZIKV (MOI of 1) in DMEM/F-12 medium supplemented with 20 μg/mL MAR1-5A3 for 1 h. After incubation, cells were washed twice with DMEM/F-12 medium and cultured in complete DMEM/F-12 medium containing 20 μg/mL MAR1-5A3 for subsequent assays.
Primary cortical neurons were isolated from neonatal (postnatal day 0–1) WT or Disc1 KD mice. Cerebral cortices were carefully dissected in ice-cold PBS, followed by enzymatic digestion with 0.5 mg/mL papain (Sigma, P-4762) and 0.01 mg/mL DNase I (Sigma, 10104159001) at 37 °C for 20 min. The tissue was then gently triturated using a flame-polished Pasteur pipette to obtain a single-cell suspension. Digestion was terminated by adding Neurobasal Plus medium (Gibco, A3582901) supplemented with 2% B27 (Thermo Fisher, 17504-044), 1% L-glutamine, and 1% penicillin-streptomycin-amphotericin B (Biosharp, P7630). The cell suspension was filtered through a 70 μm cell strainer, centrifuged at 300 × g for 5 min at 4 °C, and resuspended in the complete neuronal culture medium. For plating, 6-well culture plates (NEST, 703011) and confocal dishes (Sorfa, 201200) were pre-coated with 0.1 mg/mL poly-L-lysine (Sigma, P-6282) for 12 h at 37 °C in a humidified incubator with 5% CO₂. Prior to seeding, the coating solution was aspirated, and the surfaces were washed twice with PBS and air-dried in the incubator. Primary cortical neurons were maintained at 37 °C in a humidified atmosphere containing 5% CO₂, with one-third of the culture medium replaced every 2–3 days. On day 7 in vitro, neurons were pretreated with 20 μg/mL MAR1-5A3 antibody for 6 h before infection. Neurons were then infected with ZIKV (MOI of 1) in complete neuronal medium containing 20 μg/mL MAR1-5A3 for 1 h. After infection, cells were washed twice with fresh complete medium and maintained in medium containing 20 μg/mL MAR1-5A3 for subsequent assays.
Flow cytometry
To assess macrophage differentiation, cells were analyzed by flow cytometry on day 7 of induction. Adherent macrophages were gently detached using Cell Dissociation Buffer (Thermo Fisher, 13151014) at 37 °C for 5 min. The cells were then washed twice with FACS buffer (PBS supplemented with 2% fetal bovine serum, FBS). To block Fc receptors and minimize nonspecific antibody binding, cells were incubated with anti-mouse CD16/32 antibody (1:200, BioLegend, 101320) on ice for 30 min. After blocking, cells were washed twice with FACS buffer and subsequently stained with fluorochrome-conjugated anti-CD11b antibody (1:200, BioLegend, 101211) for 1 h on ice in the dark. Following staining, cells were again washed twice with FACS buffer and resuspended in the same buffer for acquisition. Samples were acquired using a CytoFLEX S flow cytometer (Beckman Coulter, USA), and data were analyzed with FlowJo software (version 10.8.1). The proportion of CD11b+ cells was used as an indicator of macrophage differentiation efficiency.
Neuronal differentiation efficiency was assessed by flow cytometry on day 7 in vitro following primary cortical neuron culture. Cells were gently dissociated using 0.25% trypsin-EDTA (Gibco, 25200072) at 37 °C for 3−5 min, followed by two washes with FACS buffer. For intracellular staining, cells were fixed and permeabilized using Cytofix/Cytoperm solution (BD Biosciences, 554722) for 30 min at room temperature. After permeabilization, cells were incubated with Perm/Wash buffer (BD Biosciences, 554723) for 30 min on ice to block nonspecific binding. Cells were then incubated with anti-MAP2 primary antibody (1:200, Proteintech, 67015) for 1 h at room temperature. After washing, they were stained with Alexa Fluor 488-conjugated goat anti-mouse IgG H&L secondary antibody (1:500, Abcam, ab150113) for 1 h at room temperature in the dark. Following two final washes with FACS buffer, cells were resuspended in FACS buffer for analysis. Flow cytometric acquisition was performed using a CytoFLEX S flow cytometer (Beckman Coulter, USA), and data were analyzed using FlowJo software (version 10.8.1). Neuronal differentiation efficiency was quantified as the percentage of MAP2⁺ cells within the total cell population.
Hematoxylin and Eosin (H&E) staining
To assess histopathological changes in mouse brains, hematoxylin and eosin (H&E) staining was performed on paraffin-embedded brain sections. Mice were euthanized, and brains were carefully harvested and post-fixed in 4% paraformaldehyde (PFA; Prosperich, PL0059) at 4 °C overnight. The fixed tissues were dehydrated through a graded ethanol series (Macklin, E809061), cleared in xylene (Macklin, X820563), and embedded in paraffin. Coronal or sagittal brain sections (4−6 μm thickness) were prepared using a rotary microtome (Leica, RM2016) and mounted onto glass microscope slides. Paraffin sections were deparaffinized in xylene, rehydrated through a descending ethanol gradient, and stained with hematoxylin (Servicebio, G1076) for 5−10 min. After rinsing with running tap water, the sections were counterstained with eosin (Servicebio, G1076) for 1−3 min. Finally, the slides were dehydrated through graded ethanol, cleared in xylene, and mounted with a resin-based mounting medium (Macklin, N861409). Stained brain sections were examined under a bright-field optical microscope (Nikon, ECLIPSE E100), and images were captured using a digital imaging system (Nikon, DS-U3). For the adult viral infection model, 6−8 weeks-old mice (both male and female, n = 6 per group) were used. For the neonatal infection model, P2 mice (both male and female, n = 4 per group) were used.
Immunofluorescence assay
The cells were fixed with 4% formaldehyde (Beyotime, P0099) for 15 min and then permeabilized with Triton X-100 (Beyotime, P0096) for 5 min at RT. Next, cells were blocked in Immunol Staining Blocking Buffer (Beyotime, P0102) for 1 h at RT, followed by overnight incubation at 4 °C with primary antibodies diluted in PBS containing 3% BSA. The following primary antibodies were used: anti-DISC1 antibody (1:500, Invitrogen, 40-6800), anti-Neun antibody (1:500, Servicebio, GB15138). After three washing steps with PBS containing 0.5% Tween-20, the cells were incubated for 1 h with secondary antibodies: Goat Anti-Mouse IgG H&L tagged with Alexa Fluor 488 (1:500) and Goat Anti-Rabbit IgG H&L tagged with Alexa Fluor 594 (1:500, Abcam, ab150080). Thereafter, cells were washed with PBS containing 0.5% Tween-20 and mounted in Antifade Mounting Medium with DAPI (Beyotime, P013). Images were captured using the All-in-One Fluorescence Microscope BZ-X800 (KEYENCE).
Immunofluorescence staining was performed on paraffin-embedded brain sections from both 6-8-week-old WT and Disc1 KD mice on day 6 post-intraperitoneal ZIKV infection (both male and female, n = 6 per group), as well as from neonatal mice at day 6 and 6 weeks of age following intracranial ZIKV injection (both male and female, n = 4 per group). Two different staining approaches were employed depending on the sample origin.
For adult mice, sections were deparaffinized and subjected to antigen retrieval in citrate buffer (pH 6.0; Servicebio, G1202) using a water bath at 95 °C for 40 min. After natural cooling to room temperature, sections were washed three times in PBS (pH 7.4; Servicebio, G0002) using a pendulum shaker. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide for 25 min in the dark. Sections were then blocked with 3% bovine serum albumin (BSA; Servicebio, GC305006) for 30 min at room temperature. Primary antibodies—anti-NeuN antibody (1:300) and anti-LC3A/B antibody (1:300)—were mixed and applied to the tissue sections, followed by overnight incubation at 4 °C in a light-protected humidified chamber. After three PBS washes, sections were incubated for 1 h at room temperature with a mixture of Alexa Fluor 488-conjugated goat anti-mouse IgG (1:400; Servicebio, GB25301) and Alexa Fluor 594-conjugated goat anti-rabbit IgG (1:400; Jackson ImmunoResearch, 111-585-003). Slides were washed again in PBS, treated with an autofluorescence quencher (Servicebio, G1221) for 5 min, and rinsed in running water for 10 min. Finally, sections were mounted with antifade mounting medium (Servicebio, G1401) and imaged using a fluorescence microscope (Nikon, Eclipse C1).
For neonatal mouse brain sections, triple immunofluorescence staining was carried out using tyramide signal amplification (TSA) to enable high-sensitivity multiplex detection. After deparaffinization and antigen retrieval as described above, the first primary antibody, anti-LC3A/B (1:300), was applied and incubated overnight at 4 °C in the dark. The following day, sections were washed and incubated with an HRP-conjugated secondary antibody, followed by tyramide signal amplification using iF488-Tyramide (Servicebio, G1232) for 10 min at room temperature. Slides were then washed in TBST (Servicebio, G0004). Bound antibodies were removed using an antibody elution buffer (Servicebio, G1266), first for 5 min at room temperature, then for 30 min at 37 °C. The same steps were repeated sequentially for the second and third primary antibodies—anti-ZIKV E (1:500) and anti-NeuN (1:300, mouse monoclonal)—with TSA fluorophore development using iF488-Tyramide (Servicebio, G1231) and iF555-Tyramide (Servicebio, G1233), respectively. Nuclei were counterstained with DAPI (Servicebio, G1012) for 10 min at room temperature in the dark. Autofluorescence was quenched (Servicebio, G1221), followed by a 10-min rinse in running water. Slides were sealed with antifade mounting medium (Servicebio, G1401) and imaged using the fluorescence microscope (Nikon, Eclipse C1). Multichannel images were acquired for subsequent analysis.
RNA sequencing
The RNA-sequencing data from ZIKV-infected HTR8 cells, as reported in a previous study23, have been deposited in NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE234128, and raw sequence data deposited at the NCBI Sequence Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA883202. U251 cells were infected with ZIKV GZ01 (MOI of 1) post pcDNA or HA-DISC1 transfection for 24 h. Total RNA was isolated using TRIZOL reagent. The purity and quantity of the total RNA were assessed using the NanoDrop ND-1000 spectrophotometer (NanoDrop, Wilmington, DE, USA). The RNA sequencing was performed on an Illumina NovaseqTM 6000 platform by Tsingke Biotechnology Co., Ltd. (Beijing, China). The resulting sequencing data were analyzed using the “edgeR” package in R software to identify differentially expressed genes (DEGs). Genes with fold change >2 and a P-value < 0.05 were considered significantly differentially expressed. Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs was performed using the cluster-Profiler R package.
Proteomic analysis
HTR8 cells were infected with ZIKV GZ01 (MOI of 1) for 24 h. Total cellular proteins were extracted from both infected and non-infected cells using RIPA lysis buffer supplemented with 1% PMSF. The lysates were incubated on ice for 30 min and then centrifuged at 12,000 × g for 20 min at 4 °C. A total of 100 µg of protein from each sample was reduced with 10 mM dithiothreitol (Macklin, 3483-12-3) at 56 °C for 30 min, followed by alkylation with 20 mM iodoacetamide (Aladdin, 144-48-9) at room temperature for 30 min in the dark. The samples were then digested with sequencing-grade trypsin (Beyotime, P4209) at 37 °C for 20 h. Digested peptides were acidified with formic acid (Macklin, 64-18-6) to a pH of <3 and purified using C18 Spin Columns (Thermo Fisher Scientific, 89870). Peptides were analyzed using high-resolution data-independent acquisition (DIA) mass spectrometry on a Thermo Fisher Q Exactive™ HF-X instrument coupled with an EASY-nLC 1200 system, performed by BGI Genomics (Guangdong, China). The raw DIA data were processed using Spectronaut Pulsar X software (Biognosys) with reference to the human UniProt protein database. Differentially expressed proteins were defined as those with fold change >2 and a P-value < 0.05.
Immunoprecipitation and mass spectrometry (IP-MS)
U251 cells containing HA-DISC1 or pcDNA plasmids were lysed in RIPA lysis buffer (weak) containing 1% PMSF. Cell lysates were centrifuged at 12000 × g for 20 min at 4 °C, and the supernatant was incubated with HA-tagged magnetic beads at 4 °C for 12−14 h. Then the beads were washed with TBST containing 1% NP40 and 0.5% Tween-20 three times and resolved with protein loading buffer. Protein samples were subjected to reduction with 10 mM dithiothreitol at 56 °C for 30 min and alkylation with 20 mM iodoacetamide at room temperature for 30 min in the dark. The treated proteins were digested with sequencing-grade trypsin at 37 °C for 20 h. Digested peptides were desalted with C18 solid-phase extraction columns (ThermoFisher, 60108-303) lyophilized, and reconstituted in 0.1% formic acid for LC-MS/MS analysis. Peptide separation was conducted on a nanoElute high-performance liquid chromatography (HPLC) system (Bruker) coupled with C18 Spin Columns. After chromatographic separation, the peptides were analyzed using a timsTOF Pro mass spectrometer (Bruker) for mass spectrometry analysis. Raw mass spectrometry data files (.d folder) were processed using MaxQuant software (version 1.6.14). Database searches were performed against the UniProt human reference proteome. A false discovery rate (FDR) of 1% was applied at both the peptide and protein levels. Proteomics sequencing was performed at Applied Protein Technology Company. Candidate proteins were selected based on the following criteria: (1) the candidates should be presented in the group of anti-HA tagged DISC1 immunoprecipitation but absent in the control pcDNA group; (2) the number of unique peptides should be ≥2. Candidate proteins are shown in Supplementary Data 1.
Data analysis
All data were analyzed using GraphPad Prism software (version 8.0.2) and R (v 4.1.0). The quantification of the Western blot was conducted using ImageJ (1.53t/Java 1.8.0 345). The results are presented as means ± standard deviation (SD). Normality of the data was assessed using the Shapiro-Wilk test. For data following a normal distribution, statistical analyses were performed using an unpaired two-tailed Student’s t test or a one-way ANOVA followed by Tukey’s multiple comparison test. For data not following a normal distribution, analyses were performed using the two-tailed Mann-Whitney test or the Kruskal-Wallis test followed by Dunn’s multiple comparison test. P-values <0.05 were considered significant. ns, not significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-sequencing data generated in this study have been deposited in NCBI Gene Expression Omnibus (GEO) database under accession number GSE289385. The proteomic data and mass spectrometry proteomics generated in this study have been deposited via the ProteomeXchange Consortium with the dataset identifier PXD060751. Source data are provided with this paper.
References
Melo, A. S.dO. et al. Congenital Zika virus infection: beyond neonatal microcephaly. JAMA Neurol. 73, 1407–1416 (2016).
Johansson, M. A., Mier-y-Teran-Romero, L., Reefhuis, J., Gilboa, S. M. & Hills, S. L. Zika and the risk of microcephaly. N. Engl. J. Med. 375, 1–4 (2016).
Lopes Moreira, M. E. et al. Neurodevelopment in Infants Exposed to Zika Virus In Utero. N. Engl. J. Med 379, 2377–2379 (2018).
Cardoso, T. F. Jr. et al. Congenital Zika infection: neurology can occur without microcephaly. Arch. Dis. Child 104, 199–200 (2019).
Ireland, D. D. C. et al. Long-term persistence of infectious Zika virus: Inflammation and behavioral sequela in mice. PLoS Pathog. 16, e1008689 (2020).
van der Linden, V. et al. Description of 13 infants born during october 2015-January 2016 with congenital Zika virus infection without microcephaly at birth - Brazil. MMWR-Morbidity Mortal. Wkly. Rep. 65, 1343–1348 (2016).
Mulkey, S. B. et al. Sequential neuroimaging of the fetus and newborn with in utero Zika virus exposure. JAMA Pediatrics 173, 52–59 (2019).
Rice, M. E. et al. Vital signs: Zika-associated birth defects and neurodevelopmental abnormalities possibly associated with congenital zika virus infection—US territories and freely associated states, 2018. MMWR-Morbidity Mortal. Wkly. Rep. 67, 858–867 (2018).
Nielsen-Saines, K. et al. Delayed childhood neurodevelopment and neurosensory alterations in the second year of life in a prospective cohort of ZIKV-exposed children. Nat. Med. 25, 1213–1217 (2019).
Hamshere, M. L. et al. Genomewide linkage scan in schizoaffective disorder: significant evidence for linkage at 1q42 close to DISC1, and suggestive evidence at 22q11 and 19p13. Arch. Gen. Psychiatry 62, 1081–1088 (2005).
Kilpinen, H. et al. Association of DISC1 with autism and asperger syndrome. Mol. Psychiatry 13, 187–196 (2008).
Hodgkinson, C. A. et al. Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am. J. Hum. Genet 75, 862–872 (2004).
Ozeki, Y. et al. Disrupted-in-schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc. Natl. Acad. Sci. USA 100, 289–294 (2003).
Morris, J. A., Kandpal, G., Ma, L. & Austin, C. P. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum. Mol. Genet. 12, 1591–1608 (2003).
Kamiya, A. et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat. Cell Biol. 7, 1167–1178 (2005).
Leliveld, S. R. et al. Insolubility of disrupted-in-schizophrenia 1 disrupts oligomer-dependent interactions with nuclear distribution element 1 and is associated with sporadic mental disease. J. Neurosci. 28, 3839–3845 (2008).
Trossbach, S. V. et al. Misassembly of full-length Disrupted-in-Schizophrenia 1 protein is linked to altered dopamine homeostasis and behavioral deficits. Mol. Psychiatry 21, 1561–1572 (2016).
Kim, J. Y. et al. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron 63, 761–773 (2009).
Mao, Y. et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell 136, 1017–1031 (2009).
Bentea, E. et al. Kinase network dysregulation in a human induced pluripotent stem cell model of DISC1 schizophrenia. Mol. Omics 15, 173–188 (2019).
Wang, Z.-T. et al. Disrupted-in-schizophrenia-1 protects synaptic plasticity in a transgenic mouse model of Alzheimer’s disease as a mitophagy receptor. Aging Cell 18, e12860 (2019).
Marreiros, R. et al. Disruption of cellular proteostasis by H1N1 influenza A virus causes α-synuclein aggregation. Proc. Natl. Acad. Sci. USA 117, 6741–6751 (2020).
Chen, Q. et al. ZIKV infection differentially affects the transcriptional profiles in HTR8 and U251 cells. Virus Res. 334, 199166 (2023).
Zavala-Vargas, D. I. et al. Interaction of the Zika virus with the cytoplasmic dynein-1. Virol. J. 20, 43 (2023).
Kim, N.-S. et al. Pharmacological rescue in patient iPSC and mouse models with a rare DISC1 mutation. Nat. Commun. 12, 1398 (2021).
Wu, Q. et al. DISC1 Regulates the proliferation and igration of mouse neural stem/progenitor cells through Pax5, Sox2, Dll1 and Neurog2. Front. Cell. Neurosci. 11, 261 (2017).
Hindupur, S. K., Gonzalez A., Hall M. N. The opposing actions of target of rapamycin and AMP-activated protein kinase in cell growth control. Cold Spring Harbor Perspect. Biol. 7, a019141 (2015).
Chun, Y., Kim, J. AMPK-mTOR signaling and cellular adaptations in hypoxia. Int. J. Molecul. Sci. 22, 9765 (2021).
Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017).
Jordan, T. X., Randall G. Dengue virus activates the AMP kinase-mTOR axis to stimulate a proviral lipophagy. J. Virol. 91, e02020−16 (2017).
Shives, K. D. et al. West nile virus-induced activation of mammalian target of rapamycin complex 1 supports viral growth and viral protein expression. J. Virol. 88, 9458–9471 (2014).
Sahoo, B. R., Pattnaik A., Annamalai A. S., Franco R., Pattnaik A. K. Mechanistic target of rapamycin signaling activation antagonizes autophagy to facilitate Zika virus replication. J. Virol. 94, e01575−20 (2020).
Liang, Q. et al. Zika virus NS4A and NS4B Proteins deregulate akt-mTOR signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell 19, 663–671 (2016).
Cao, B., Parnell, L. A., Diamond, M. S. & Mysorekar, I. U. Inhibition of autophagy limits vertical transmission of Zika virus in pregnant mice. J. Exp. Med. 214, 2303–2313 (2017).
Lennemann, N. J. & Coyne, C. B. Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy 13, 322–332 (2017).
Clapcote, S. J. et al. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron 54, 387–402 (2007).
Kuroda, K. et al. Behavioral alterations associated with targeted disruption of exons 2 and 3 of the Disc1 gene in the mouse. Hum. Mol. Genet. 20, 4666–4683 (2011).
Zhang, F. et al. American strain of Zika virus causes more severe microcephaly than an old asian strain in neonatal mice. Ebiomedicine 25, 95–105 (2017).
Manet, C. et al. Zika virus infection of mature neurons from immunocompetent mice generates a disease-associated microglia and a tauopathy-like phenotype in link with a delayed interferon beta response. J. Neuroinflamm. 19, 307 (2022).
de Oliveira Souza, I. N. et al. Acute and chronic neurological consequences of early-life Zika virus infection in mice. Sci. Trans. Med. 10, eaar2749 (2018).
Snyder-Keller, A., Kramer, L. D., Zink, S. & Bolivar, V. J. Mouse strain and sex-dependent differences in long-term behavioral abnormalities and neuropathologies after developmental Zika infection. J. Neurosci. 39, 5393–5403 (2019).
Acknowledgements
This work was supported by National Natural Science Foundation of China (32270147, 32000116, to H.L.L.), Shenzhen Science and Technology Program (JCYJ20240813151010014 to H.L.L., KQTD20200820145822023 to H.L.L., KCXFZ20211020172545006 to X.Z., ZDSYS20230626091203007 to H.L.L.), High-Level Project of Medicine in Nanshan, Shenzhen; Sanming Project of Medicine in Shenzhen (SZSM202103008 to Y.L.S.), Science and Technology Planning Project of Guangdong Province in China (2021B1212040017 to C.J.S.).
Author information
Authors and Affiliations
Contributions
H.L.L. and S.Z.Z. conceptualized the study and designed the experiments. S.Z.Z. conducted most of the experiments. H.L.Z., Q.Q.C., N.N.L., C.Y.Z., T.X., P.J.M., W.J.T., S.H.B., C.M.L., N.W. (Ning Wang), X.Z. and S.S.F. assisted with experiments. Y.J., J.H.Y., N.W. (Nan Wu), C.J.S., Y.L.S., and H.L.L. generated resources. H.L.L., S.Z.Z., H.L.Z., Q.Q.C., N.N.L., N.W. (Nan Wu) and Y.L.S. contributed to data analyses and manuscript writing. All authors discussed the results and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Indira Mysorekar, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, S., Zha, H., Chen, Q. et al. DISC1 Protects Against Zika Virus Infection and Long-Term Neurological Damage Through AMPK-mTOR-Mediated Autophagy. Nat Commun 16, 9841 (2025). https://doi.org/10.1038/s41467-025-64809-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-64809-w