Abstract
The circumvallate papillae and foliate papillae of the posterior tongue contain taste buds in close proximity to specialized salivary glands, known as von Ebner glands. The developmental relationship between taste buds and these salivary glands has been suggested but remains largely unexplored at postnatal and adult stages. Lineage tracing studies in mice have revealed that Lgr5 marks taste bud stem cells. Here, we report single-cell RNA sequencing of the entire circumvallate and foliate papillae of mice, providing a transcriptional atlas of cells from tongue surface epithelium, taste buds, and the associated and non-associated salivary glands. We unveil a developmental trajectory in which taste buds, the associated salivary glands, and the non-taste tongue surface epithelium originate from a common Lgr5 cell. We confirm this tripotency at the clonal level in vitro and with multicolor lineage tracing in vivo. Thus, the circumvallate and foliate papillae harbor chemosensory units composed of taste bud and salivary gland cells derived from the same parental Lgr5-positive stem cell.
Similar content being viewed by others
Introduction
Taste buds occur in distinct structures, each with its unique location on the tongue: circumvallate papillae (CVP) reside on the posterior tongue, foliate papillae (FoP) on the bilateral sides of the tongue, and the fungiform papillae (FP) on the anterior tongue. The CVP and FoP are of endodermal origin1, while the FP are of ectodermal origin2,3,4. Regardless of their location, taste buds consist of clusters of specialized post-mitotic epithelial cells (types I, II, III & IV), of which type II and type III cells are primarily responsible for detecting tastants (sweet, sour, salty, bitter, and umami), while type IV cells represent their precursors. Type I cells are glial-like cells and have a more supportive role5, whilst type IV cells represent the precursor of the continuously self-renewing taste buds6,7. FP papillae protrude from the surface and harbor individual taste buds, whereas in CVP and FoP, the taste buds are embedded within epithelial trenches. CVP and FoP uniquely appear in close proximity to small salivary glands. These minor salivary glands, the so-called von Ebner glands (VEG) are located below the taste bud proper. VEG produce serous mucus to support taste sensitivity, and in addition, excrete digestive enzymes (e.g., lipase, amylase)8,9,10. Dysgeusia (altered taste perception) and ageusia (complete loss of taste) are frequently observed in conditions where salivary function is impaired, implying the dependence of taste perception on saliva. These sensory disturbances arise from diverse physiological causes. For instance, up to 80% of head and neck cancer patients develop radiation-induced xerostomia, which is often accompanied by dysgeusia or ageusia11,12,13,14. These taste disturbances may result not only from reduced salivary function but also from direct damage to taste bud progenitors15. Similarly, autoimmune diseases such as Sjögren’s syndrome can lead to chronic dry mouth and associated taste dysfunctions16. These concurrent processes not only compromise oral health but also significantly affect quality of life by reducing nutrient intake and contributing to depression due to the loss of taste enjoyment17,18,19.
Developmentally, the tongue surface epithelium is formed from basal progenitors (Krt5+ and Krt14 + ) that are also known to give rise to the taste buds20. Mapping early development of the CVP showed the formation of the trenches followed by an invagination forming the von Ebner gland21,22,23. Postnatally, Lgr5 stem cells maintain taste buds and give rise to precursors marked by Sox2, Shh, and Lgr524,25, known as Type IV cells. Sox2, a key player in the development of multiple adult epithelia26,27, is critical for taste bud development24,28. Despite being broadly expressed across the tongue epithelium, Sox2 is most highly expressed near taste papillae24.
Lgr5, a member of the Wnt-signaling pathway, marks actively dividing stem cells in many tissues, such as the intestine, stomach, hair follicles, and liver29,30,31,32. Comparable to Lgr5+ stem cell populations observed in other tissues and as first shown for the gut33, isolated Lgr5+ cells from taste buds can form self-organizing three-dimensional (3D) organoids in vitro25. An alternative approach for generating taste bud organoids involves the isolation of the entire CVP/FoP34. Both approaches yield functional taste bud cells (TBCs). In CVP and FoP, Lgr5 expression is enriched in the trench epithelium and sporadically present in adjacent VEG cells located beneath the trenches35 and subsequent lineage tracing from Lgr5+ cells in the tongue marks their taste bud progeny36. Cell lineage principles of the von Ebner glands remain unknown.
Here, we demonstrate that the CVP and FoP harbor a chemosensory unit that includes taste bud and salivary gland cells derived from the same parental Lgr5+ stem cell, while the same stem cell can also generate tongue surface epithelium.
Results
Single-cell atlas of the posterior tongue
To provide a comprehensive atlas of the taste buds of the posterior tongue, we dissected 20 CVP and 40 FoP from 20 adult female mice and subjected these to single-cell RNA sequencing (scRNA-seq) (Fig. 1a). Our initial dataset captured a total of 20,320 high-quality cell profiles (6389 CVP and 13,931 FOP) (Supplementary Fig. 1a). We filtered out immune (Ptprc) and connective tissue (Vim, Pecam1) clusters by subsetting the dataset (Supplementary Fig. 1b), leaving us with 4921 CVP and FoP 11,570 cells, divided over 10 clusters (Fig. 1b).
a Schematic overview of the single-cell data collection. Briefly, CVPs (n = 20) and FoPs areas (n = 40) were picked, dissociated and subjected to scRNA-seq. Partly created in BioRender. Van de wetering, M. (2025) https://BioRender.com/pcvss09. b UMAP of the annotated clusters from mouse CVP (n = 4921) and FoP (n = 11570). Descriptive cluster labels are projected. c Dotplot showing relative expression of markers used to annotate the clusters. Basal Basal progenitor, LEC non-taste Lingual epithelial cell, End Endothelial, L-Pr Lgr5+ Stem cell, Myo Myoepithelial, Aci Acinar, Duct Ductal, CVP circumvallate papillae, FoP Foliate papillae.
Annotation with established markers identified the following cell types: non-cycling basal progenitors (Krt5, Krt1420), cycling basal progenitors (Krt5, Krt14, Mki67), non-taste lingual epithelial cells (non-taste LEC#1; Sprr1b, Krt4, Krt1337,38), Lgr5 stem cell (Lgr5, Lgr625), Type I, III(Krt8, Krt19, Kcnq1, Entpd2, Snap25, Car45,39,40,41,42) & IV TBCs (Lgr5, Shh, Sox2high )24, and Type II TBCs (Krt8, Krt19, Kcnq1, Gnat3, Trpm543,44)(Fig. 1c). A cluster displaying intermediate-like signatures—marked by low expression of basal markers (Krt5 and Krt14) alongside Krt4 and Krt13, typically associated with non-taste lingual epithelial cells (LECs)—was characterized as 'non-taste LEC#2'. Gene Ontology analysis of its top marker genes revealed strong enrichment for immune-related pathways (Supplementary Fig. 1e), a feature commonly observed in non-taste lingual populations of the oral cavity45,46,47.
Interestingly, our dataset captured the presence of ductal (Dcpp148), myoepithelial (Acta249), and acinar (Muc1950) cells, as well as a Krt14 basal progenitor population, characteristic of salivary gland49. Thus, VEG cells were represented in our dataset, in addition to basal keratinocytes, non-taste LEC, and TBCs, exemplifying the completeness of our dataset.
As the complement of taste buds and associated salivary glands has not been extensively studied, we then aimed to provide an overview of both compartments to identify cell-specific markers. We generated a separate data analysis for taste bud cells and for salivary glands and annotated these using established markers. For the taste compartment, the previously described type I (Entpd2), II (Gnat3, Gna14, Trpm5), III (Car4, Snap25) & IV (Lgr5, Shh, Sox2high) taste bud cells (TBCs) were represented in the dataset (Fig. 2a). Compared to the more general TBC marker Krt8 and Kcnq1, Krt19 appeared to mark differentiated TBC. Within the type II clusters, there was a distinct difference between sweet/umami- and bitter-designated clusters. In line with previous research, sweet/umami TBCs uniquely expressed taste receptor type 1 (T1r) receptors in combination with the downstream G-protein Gna1451 (Fig. 2c), whilst bitter TBCs expressed a variety of T2rs as well as the G-protein Gnat3. Furthermore, we identified a set of immature type II cells expressing both T1rs, T2rs and taste progenitor markers such as Sox2 and Pou2f352 (Fig. 2c). Type III TBCs expressed high levels of markers Car4, Otop1(Sour taste), and Snap25. Interestingly, although Otop1 was proposed to be solely expressed by type III TBCs53,54, we also observed its expression in type I TBCs (Supplementary Fig. 2a).
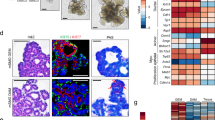
a UMAP projection of a separate Seurat object of the taste bud compartment (n = 532) showing Type I, II, III and IV taste cells over six clusters. For the Type II cells, we identify three clusters for Bitter, Sweet/Umami, and progenitor-like cells (Type II only cluster). b UMAP projection of the salivary gland compartment (n = 2139) projecting differentiated cell types in the ductal, ductal/serous mix, acinar (Serous and Serous/Mucous) clusters. Progenitor compartments were subdivided into multiple basal, Krtr5+Krt14+, and Krt14+/Kit+/ Krt7low, and Myoepithelial. c Dot plot representing all classical taste markers over the Type I, II & III clusters. d Violin Plot representing all module scores from the SMG/SLG murine dataset utilized for the annotation of the eight clusters in the Salivary gland compartment. SMG Submandibular gland, SLG Sublingual gland, GCT granular convoluted tubule. X-axis: 0; Basal (Krt5/Krt14), 1; Basal (Krt14), 2; Ductal, 3; Ductal/Serous, 4; Myoepithelial, 5; Acinar (Serous), 6; Acinar (Mucus), 7; Prolif. Endbud, 8; Ductal Keratinocyte.
To characterize the salivary compartment, we annotated clusters using previously conducted scRNA-seq studies on mouse salivary glands (submandibular (SMG), sublingual (SLG), and parotid (PG))49,55. We calculated module scores for all compartments and projected these on our salivary gland dataset (Fig. 2b, d). Generally, large salivary glands exhibit a distinct architecture consisting of intercalated ducts, striated ducts, granular convoluted ducts, high Krt19+ ducts, and acini56,57. Myoepithelial cells surround the ducts and acini of the gland to facilitate the propulsion of saliva.
In our VEG dataset, we discovered an acinar compartment harboring mixed serous (Bhla15, Amy1, Lipf 58) and mucous (Aqp5, Muc19, Muc5b) cells (Fig. 2b, d, Supplementary Fig. 2b). Intriguingly, we also noted a serous-like cluster with a ductal signature, which we annotated as ductal/serous. Of note, in the CVP we identified both serous and mucous cells, whilst in the FoP we exclusively identified mucous cells (Supplementary Fig. 2d). The CVP phenotype more closely resembled the SMG, whilst the FoP phenotype was more similar to the SLG. Our salivary dataset lacked the expression of PG-specific Amy2, further indicating a more SLG/SMG-like origin49.
Upon further analysis, we discerned two distinct populations of basal duct cells, one characterized by Krt14 and Krt5 expression, the other expressing Krt14 to some higher degree than Krt5, accompanied by Kit 59 expression and low levels of Krt7 + (Supplementary Fig. 2b, c). This second population predominantly occurred in the FoP (Supplementary Fig. 2d). Otherwise, the make-up of the VEG of the FoP closely mirrored that of the CVP. Additionally, we identified myo-epithelial cells, as indicated by Acta2 and Myl9 expression, and ductal cells (Ltf, Krt19, Krt7) (Supplementary Fig 2b). Notably, the ductal cluster appeared more mixed, consisting of intercalated duct (marked by Dcpp1, Dmbt1 and Kit expression), striated duct (marked by Dmbt1 and Cftr expression), and more differentiated duct cells (Krt19high, Krt7high) (Fig. 2d, Supplementary Fig 2b). Furthermore, we observed a minor ductal population (Krt19high, Krt7high) with keratinocyte-like features, characterized by the expression of Psca and Cnfn. Taken together, our dataset appeared to capture the cellular diversity of the posterior tongue epithelium plus its appendages.
Sox9 and Lgr5 co-expressing stem cells in the tongue
The transition from Lgr5-expressing progenitor cells, which eventually acquire epithelial characteristics through the expression of keratins Krt8, Krt19, and Krt7, represents a tightly regulated process in taste bud homeostasis25,60. Critical for the taste lineage are progenitors that express high levels of Lgr5, Shh, and Sox2. Indeed, such progenitors were present in our dataset (Fig. 3a). While Sox2 was expressed across the entire dataset, its expression distinctively increased in taste lineage cells. These taste bud-committed Sox2high/Lgr5 progenitors are known to be key to efficient taste generation25.
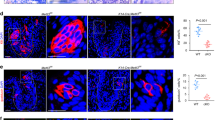
a Feature plot projecting relative expression of Sox9, Sox2, Lgr5 over the UMAP. Note the overlap in Lgr5 and Sox9 expression. b, c Immunofluorescent staining on mouse primary CVP tissue for Sox9 (green; marks the saliva compartment), Sox2 (magenta; marks the taste compartment), and Krt8 (yellow; the mature compartment for both taste bud and VEG entities). DAPI (blue) marks all nuclei. Arrows highlight Sox2-, Sox9- and Krt8- regions. Scale bars, 200 µm (b), 50 µm (c). d Whole CVP immunofluorescent images of the trench area of a Lgr5-EGFP-IRES-creERT2 mouse. GFP antibody was used for Lgr5 (green), Sox9 (yellow) and Sox2 (magenta). DAPI was used to stain nucleus (blue) of the cells. Highlighted is the Lgr5 niche. Arrows indicate Sox2/Lgr5 and Sox9/Lgr5 cells. Zoomed-in image shows this in greater detail. Scale bar: 50 µm, zoom: 10 µm. e Schematic overview of the proposed stem cell niche in the posterior tongue. Created in BioRender. Van de wetering, M. (2025) https://BioRender.com/xpednf4. f Schematic representation of the mouse tongue and brightfield images of the organoid cultures derived from the CVP and FoP. g RT-qPCR data reveal expression of ductal (Dmbt1, Cldn2) and Acini (Aqp5, Cldn2) markers in the taste bud organoids derived from CVP and FoP. Data are shown as mean ± SEM. n = 4 organoids, n = 3 for tissues. No statistical test was performed. CVP circumvallate papillae, FoP Foliate papillae, VEG von Ebner Gland.
The VEG has remained relatively understudied. As in related salivary tissues61,62, Sox9 expression was observed throughout the salivary lineage hierarchy (Fig. 3a, Supplementary Fig. 2b), with ductal and acinar cell populations expressing Krt8, Krt19, and Krt7 63. We noted that a subpopulation of the Lgr5 stem cell cluster also expressed Sox9 (Fig. 3a). Interestingly, it was reported in the initial taste bud organoid protocol that taste bud organoids34 contain Sox9+ cells. Histologically, we observed a region in the CVP where Sox2low Sox9- and Krt8- cells reside (Fig. 3b, c). Analysis of Lgr5-EGFP-IRES-creERT2 mice32 revealed Lgr5-driven GFP expression in the trenches where the taste cell compartment ‘touches’ the VEG compartment (Fig. 3d). Interestingly, we also discovered double positive Lgr5/Sox9 cells, besides the known double positive Lgr5/Sox2 cells (Fig. 3d). Together, these observations prompted us to investigate the possibility that taste buds (Lgr5, Sox2) and salivary glands (Lgr5, Sox9) originate from the same bi-potent Lgr5 stem cell. Our dataset contained taste bud-committed Lgr5 precursors, often referred as Type IV cells. These cells expressed Lgr5 in combination with Krt8 and Sox2high (Supplementary Fig. 3a). Previous research on progenitors in the posterior tongue particularly focused on taste, by looking either in Krt8-labeled38 or in Sox2high expressing cells20. Our more comprehensive dataset appeared to contain an earlier Lgr5 stem cell, not yet uniquely fated to generate taste bud cells only. The characterization of this Lgr5 cluster showed increased expression levels of Ptch1, Cldn10, Krt17, Il33, and Krt75 (Supplementary Fig. 3b, c).
It has previously been shown that taste bud organoids can be grown from single Lgr5+ cells25,64. If taste buds and the associated salivary glands are derived from the same Lgr5+ stem cell, this raised the possibility that organoids derived from progenitor cells of the CVP and FoP contain salivary cells as well as taste cells. We therefore generated organoids derived from mouse FoP and CVP tissues, following the protocol of Aihara et al.34 (Fig. 3f). Briefly, CVP and FoP were placed in culture, with optimizations to the medium composition (i.e the removal of Wnt from the medium). After five passages, we indeed detected expression of ductal markers (Dmbt1, Cldn2) and acinar (Aqp5, Cldn2) in taste bud organoids derived from the CVP and FoP (Fig. 3g). Next, we investigated a public dataset of scRNA-seq of taste bud organoids (Supplementary Fig. 4a). The authors did not mention the presence of VEG cells but refer to an ‘undefined’ cell population in their dataset65. We then calculated a module score of the top 100 genes from the acinar and ductal clusters. Indeed, module scores of the acinar and ductal compartments of primary tissue CVP and FoP showed a strong correlation with clusters in organoids derived from CVP and FoP (Supplementary Fig. 4b, c). Cluster 10 especially showed a ductal signature with Cldn2, Dmbt1, Dcpp1, Dcpp3 and Sox9 being highly expressed (Supplementary Fig. 4d). Intriguingly, cluster 11 demonstrated high expression of Krt19, Krt7 and Psca, the ductal keratinocyte-like population we found in the salivary gland compartment (see above). The acinar compartment signature was less apparent, but we observed cells expressing acinar mucins (Muc5b, Muc16) and Aqp5. Taken together, this confirmed our notion that both taste bud and salivary gland cells were present in posterior tongue-derived taste bud organoids.
Tripotent Lgr5 stem cells form lingual, taste and salivary lineages
Lgr5+ stem cells are known to reside in the trenches of the CVP to give rise to the taste compartment35,36. Earlier studies have demonstrated that Lgr5+ stem cell-derived organoids contain non-taste LEC besides TBCs24,25.
To further study a possible tri-potent role of the Lgr5 stem cell in taste/saliva and LEC biology, we generated CVP organoids from a Lgr5-EGFP-IRES-creERT2 mouse32 (Fig. 4a, b). Single Lgr5-GFP positive cells, sorted from CVP organoids, were clonally expanded into organoids, following previously established protocols33,66,67,68 (Supplementary Fig. 5a, b). These organoids (n = 7) again expressed stem cell (Lgr5) and differentiation markers (Krt8) for both entities (taste bud and salivary gland))(Supplementary Fig. 6a). Examination with more mature markers revealed salivary gland (Dmbt1 and Aqp5), taste (Enptd2 and Snap25), and non-taste LEC markers (Krt13, Krt4) cells, implying that Lgr5+ cells represent the tri-potent precursor for all epithelial elements of the posterior tongue tissues (Supplementary Fig. 6b–d). We next performed staining on clone 1,3, and 7 for Krt13 (LEC), Aqp5 (Saliva) and Entpd2 (Taste), confirming the presence of all three lineages in the clonal lines (Supplementary Fig 7a–d), even though clone 3 portrayed minimal Krt13 staining, similar to the mRNA levels we detected.
a Experimental set-up to generate single-cell clones from Lgr5-eGFP cells. n = 7 clones. b A representative brightfield and fluorescent image of dissected mouse tongue showing GFP (green) expression in the CVP. Scale bar = 500 µm. c Integrated UMAP of the annotated clusters from mouse Lgr5-eGFP-CreERT2 clones 1 (n = 2643 cells), 3 (n = 3248 cells), and 7 (n = 4144 cells). UMAP is split by its origin. (n = 10,035 cells). d Dot plot presenting all markers utilized for the annotation of taste bud, salivary gland, and non-taste lingual epithelium. e Integrated UMAP of clonal organoid cultures and primary tissue (CVP and FoP). Highlighted are non-taste LEC, Salivary Gland, and Taste buds. f A bar plot showing cell proportions of clone 1, 3, 7, primary CVP, and FoP tissue datasets. CVP circumvallate papillae, FoP Folliate papillae, LEC Lingual epithelial cell.
To accurately confirm the presence of all lineages, we subjected Clones 1, 3, and 7 to scRNA-seq. Initially, we captured a total of 12,494 cells. Subsequent filtering left us with 10 clusters equally distributed over each clone, totaling 10,035 high quality cells: clone 1 (2643 cells), clone 3 (3248 cells) and clone 7 (4144 cells) (Fig. 4c). Annotation of these clusters led to the identification of progenitor-like and differentiated cell types (Fig. 4d). As in the primary dataset, we detected a cycling basal progenitor population (Krt14, Krt5, Mki67) and a Lgr5/Lgr6 stem cell-like cluster. Notably, we identified a myoepithelial-like population characterized by Acta2 and Myl9 expression. We also discovered an ‘Intermediate #2’ population, characterized by expression of progenitor markers (Sox2low, Sox9, and Lgr6). This was further supported when we generated a module score of the top 100 genes and projected this over our primary dataset (Supplementary Fig. 7e). In the differentiated clusters, we observed non-taste LEC (Krt13, Krt4, Sprr1b)- and mixed saliva- and taste clusters (Krt7, Krt19, Krt8). Within this mixed population, we noted ductal markers (Sox9, Cldn10, Ltf, Kit) and acinar markers (Aqp5, Muc16, Lipf). For the taste lineage, we detected the previously mentioned classical taste markers Sox2, Hes6, Foxa1, Gna14, Entpd2, and Kcnq1. Of note, we did not observe mature taste cells expressing T1rs or T2rs, likely due to the medium composition, which supports expansion over terminal differentiation.
Next, we integrated the organoid dataset with the CVP and FoP datasets (Fig. 4e, f, Supplementary Fig. 7f). Here, we noted the presence of ‘non-taste LEC’, ‘Taste buds’ and ‘Salivary Gland’ cells in the clonal organoids (Fig. 4f, Supplementary Fig. 7e). All major cell populations were represented across all clones and all tissue samples. However, Clone 3 contained a lower proportion of taste cells, while Clone 7 exhibited an enrichment of Sox9⁺ progenitors, suggestive of increased salivary gland lineage contribution.
To investigate if single Lgr5+ stem cells can generate all lineages in vivo, we performed multicolor lineage tracing in Lgr5-eGFP-IRES-CreERT2/ R26-Confetti mouse (n = 3)69. The tamoxifen induction protocol was designed to yield sporadic tracing of nuclear (n) GFP, membrane-bound (m) CFP, YFP and RFP, while cytoplasmic GFP+ signal indicated expression of the Lgr5-GFP knock-in allele by stem cells. Sporadic tracing is critical to observe if single-Lgr5 stem cells have tripotent capacity32,69,70,71,72. We included a 1-week timepoint to visualize the initial tracing event and tracked tracing over 2 months to visualize the ‘steady-state’ dynamics of the Lrg5 stem cell population. We included three mice per timepoint and analyzed consecutive slices (approx. 8 slices per CVP, 60 μm). After imaging of the Confetti-signal we performed whole-mount immunostaining with differentiation markers (Dapi, Sox2, Sox9, GFP, and Krt8) to reveal the contribution to the three lineages. We used DAPI as a reference to determine CVP orientation, facilitating the 3D rendering/visualization of endogenous Confetti signals and interpreting the whole-mount stainings. Non-induced CVP showed widespread cytoplasmic GFP expression, revealing cells that express Lgr5. GFP expression reaches deeper down in the VEG accompanied by Sox9 (Fig. 5a). After one week, we observed sporadic stochastic induction of the fluorescent confetti markers throughout the CVP (Fig. 5b), in particular towards taste buds and to a lesser extent the VEG.
a 3D whole-mount imaging of the CVP without tamoxifen induction. Cytoplasmic GFP (green) labels Lgr5⁺ stem cells. Krt8 (yellow) marks both differentiated taste buds and the salivary compartment. Sox2 (magenta, gradient intensity) labels the tongue epithelium with the highest expression in taste buds. Sox9 (red) marks VEG ductal progenitors. DAPI (cyan) stains nuclei. Scale bar = 50 µm (3D) and 20 µm (ZOOM). b Endogenous Confetti signal one week after induction, capturing the initiation of lineage tracing in Lgr5-eGFP-IRES-CreERT2/R26-Confetti mice. Tracing is visualized via expression of mCFP, nGFP, YFP, and RFP. Re-staining with differentiation markers reveals early YFP⁺ clones in the VEG and trench area. GFP labels both Lgr5⁺ stem cells and traced populations (nGFP, YFP). DAPI (cyan) marks nuclei. Scale bars = 50 µm and 20 µm. c Two-month lineage tracing in Lgr5-eGFP-IRES-CreERT2/R26-Confetti mice reveals long-term persistence of labeled clones (mCFP, nGFP, YFP, RFP). A 3D reconstructed image highlights a nGFP⁺ clone contributing to taste buds, non-taste lingual epithelium (LEC), and salivary gland from the same mouse (m30). This clone spans multiple consecutive sections, identified in Regions A and B. Re-staining reveals the anatomical compartments and directionality of lineage contribution. Cytoplasmic GFP marks Lgr5⁺ stem cells. Scale bar = 20 µm. d Z-projection of Region A showing nGFP⁺ lineage tracing within a taste bud. Markers include Krt8 (yellow), Sox2 (magenta), and GFP (green). Scale bar = 10 µm. e Z-projection of Region B showing nGFP⁺ lineage tracing extending into the taste bud, non-taste LEC, and VEG compartments. Stained with Krt8 (yellow), Sox2 (magenta), and GFP (green). Scale bar = 10 µm. f Quantification of lineage tracing events in Taste bud, VEG or LEC in the two-month mice (m30, m33 and m62). CVP Circumvallate Papillae, VEG von Ebner’s gland, LEC Lingual epithelial cell.
In the 2-month tracing samples, we observed fewer tracings (Fig. 5c–f). Intriguingly, 2-month after tamoxifen induction we observed nGFP+ tracing in the trench area in mouse 30 (m30). This nGFP trace showed progeny in the taste buds (Region A; Fig. 5c, d, see arrows), von Ebner ducts and the non-taste lingual epithelium (Krt8-) of the CVP when reviewed in consecutive slices (Region B; Fig. 5c–e, see arrows). Quantifying 2-month tracings led to the discovery that most of the two-month tracings contributed to taste buds (Fig. 5f).
We observed that most lineage tracings within taste buds remained confined to individual taste buds and did not extend across multiple structures. Interestingly, in the m30 nGFP tracings initiated from the trench area, we detected labeling of multiple adjacent taste buds. This suggested a hierarchical organization within the Lgr5+ stem cell pool, where some cells exhibit lineage restriction, while others possess broader regenerative potential. Collectively, these findings implied that non-taste LEC, taste, and saliva cells can derive from a common Lgr5 stem cell.
Transcriptional networks linked to Lgr5 stem cells
Having ascertained that the Lgr5 stem cell is the common precursor for non-taste LEC, taste, and saliva lineages, we analyzed the transcriptional pathways that regulate cell type specification. For this, we focused on the cell fate decisions towards the taste and saliva gland compartments and conducted pseudotime analysis on the trajectory from the Lgr5 stem cell (starting node) to the taste and saliva clusters (ending node) in the primary tissue dataset (Fig. 6a–c). Original UMAP clustering (Fig. 6a) captures static transcriptional states. We performed the pseudotime analysis73,74 (Fig. 6b) to infer differentiation trajectory from progenitors transition toward either taste bud or VEG fates. This analysis allowed us to follow the sequential transcriptional changes over lineage trajectories. Based on the observations in Figs. 4 and 5, we chose the Lgr5 stem cell as the starting point in these trajectories and designated the salivary gland and the taste buds as the endpoints.
a UMAP visualization of original clusters, excluding the acinar population. b Pseudotime values projected on the UMAP primary FoP and CVP dataset. c Schematic overview of cell fate decisions from Lgr5 stem cell towards taste and salivary gland. Created in BioRender. Van de wetering, M. (2025) https://BioRender.com/3ouqtcg. d Schematic overview of cell fate decisions Lgr5 stem cell towards taste. e Trajectory analysis of Lgr5 stem cell towards taste. Feature plot projecting the pseudotime over the subsetted Lgr5 stem cell towards taste dataset. f ‘Gene switches’ reveal top 100 up- and downregulated genes, with several known genes being highlighted (UP:Krt8, Krt7,Scnn1a, Entpd2;Down: Trp63, Krt5, Foxe1). Geneswitch analysis revealing switched-on and -off genes over the pseudotime (x-axis) of cell fate decision towards taste. Surface proteins (green), Transcription Factors(blue), other genes (red). g Schematic overview cell fate decisions Lgr5 stem cell towards saliva. h Trajectory analysis of Lgr5 stem cell towards to salivary glands. Feature plot projecting the pseudotime over the subsetted Lgr5 stem cell towards salivary gland dataset. i ‘Gene switches’ reveal top 100 up- and downregulated genes, with several known genes being highlighted (UP:Krt8, Krt7, Dmbt1;Down: Trp63, Krt5, Sox2). Geneswitch analysis revealing switched-on and -off genes over the pseudotime (x-axis) of cell fate decision towards saliva. Surface proteins (green), Transcription Factors(blue), other genes (red). j Relative expression of TFs (DOWN) over the pseudotime from Lgr5 stem cell to taste lineage (x-axis). k Relative expression of TFs (DOWN) over the pseudotime from Lgr5 stem cell to saliva lineage (x-axis). c, d and g were created with BioRender.com under an academic license.
Pseudotime computation is limited to clusters within the primary cluster. Because the primary clustering (Fig. 1b) failed to include the acinar cluster, we created a new Seurat object excluding the acinar cluster by subsetting the cells, followed by re-clustering (Fig. 6a). Next, we subsetted the following branches: Lgr5 stem cell to TBCs and the Lgr5 stem cell to salivary gland cells and we analyzed the transcriptional program over the pseudotime by focusing on genes that are switched on and off, using the ‘Geneswitches’ algorithm. We initially focused on transcription factors (TFs) that were differentially expressed in the Lgr5 stem cell cluster (LFC > 0.5) (Supplementary Fig. 8a). Among these TFs were Xbp1, Runx1, Foxe1, Hes6, Tcf4, Etv1 and Pax1, the latter four previously shown to participate in CVP/FoP development38,75 confirming our approach.
We then focused on the differentiation paths from Lgr5 stem cell to TBC (Fig. 6d, e) and salivary gland cells (Fig. 6g, h). For both paths, we observed a loss of basal progenitor markers such as Krt5 and Trp63, and a gain of markers such as Krt7 and Krt8 (Fig. 6f, i). Regarding the taste trajectory, we noted the taste-specific genes Entpd2 (Type I) and Scnn1a (Type III) upregulated in TBCs compared to Lgr5 stem cells (Fig. 6f). As expected, some previously reported critical TFs that are switched on along the trajectory showed increased expression levels in TBCs: Foxa2, Hes6, Sox2 and Prox176,77 (Supplementary Fig. 8b).
For the differentiation towards salivary cells, we observed upregulation of salivary-associated genes Krt23, Dmbt1, Ltf and Krt19 (Fig. 6i). In addition, the salivary regulators Tfcp2l1, Ehf, Barx2, Sox9, and Foxc149,55,78,79 increased in expression (Supplementary Fig. 8c). Other TFs that decreased over both trajectories included Nfe2l2, Mafb, Sox15, Hlf, and Foxe1 (Fig. 6j, k). We specifically noted TFs with changing expression along the differentiation trajectory belonging to the Forkhead family, known regulators of development80. Among these, Foxc1 is associated with salivary gland development79, Foxa2 has been implicated in taste bud formation77, while the role of Foxe1 in this lineage context has remained unexplored.
Foxe1 regulates posterior tongue differentiation
To identify potential regulators involved in the maintenance of the Lgr5 stem cell population, we investigated which TFs are increased in expression in the Lgr5 stem cell (LFC > 0.5) and then decrease in expression from Lgr5 stem cell to both taste and salivary gland. This left us with one TF: Foxe1 (Fig. 7a, Supplementary Fig. 8b–e). Foxe1 is involved in the development of diverse tissues, such as tongue, palate, epiglottis, pharynx, and esophagus81. In hair follicles, where Lgr5 marks the tissue-specific stem cells30, Foxe1 is found to be critical for proper hair formation82. In our dataset, Foxe1 was expressed in Lgr5 stem cells and in type II TBCs (Supplementary Fig. 9a, b). Subsequent IHC shows indeed that Foxe1+ nuclei sit in the trench area and taste buds in vivo (Supplementary Fig. 9c).
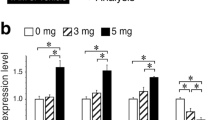
a Venn diagram showcasing all TFs going down after differentiation (taste and saliva) and up from basal to Lgr5 lineage. b Representative immunostaining images of WT and Foxe1-/- organoids. Krt8 (yellow) marker for both differentiated VEG and TBCs. DAPI (blue). Scale bar = 25 µm. c Quantification of proportion of Krt8+ cells in CVP and FoP Foxe1-/- versus WT organoids as determined. Box plots show the median (centre line), 25th–75th percentiles (bounds of box), and whiskers extending to the minimum and maximum values; all individual data points are shown. n = Organoids: CVP WT; n = 11, CVP Foxe1 -/-; n = 11, FoP WT n = 10, FoP Foxe1 -/-; n = 18. **P < 0.002, ***P < 0.0002. Two-tailed unpaired t-test. d Scatter PCA plot of bulk RNA-seq WT (red) vs Foxe1-/- (blue). n = 2 clones, with different mutations for each origin, with technical duplicates. e Heatmap of selected genes WT vs Foxe1-/-. f Projections of ‘WT-score’ (top 100 genes upregulated in WT organoids) displayed on the UMAP. Regions with a high score are highlighted. Projections of ‘WT-score’ and ‘Knockout-score’ on VEG (g, h) and taste (I, j) Seurat objects. We included the Fig. 2a, b to better visualize the affected populations. CVP Circumvallate papillae, FoP Folliate papillae, VEG von Ebner gland.
We generated CRISPR-mediated homozygous Foxe1 knockout (Foxe1-/-) organoids from CVP-Lgr5-EGFP and FoP-Lgr5-EGFP organoids (n = 2 clones for each origin) (Supplementary Fig. 10a). Interestingly, we observed a dramatic decrease of Krt8+ cells in Foxe1-/- organoids (CVP:2.4%; FoP: 3.5%) compared to WT organoids (CVP:28.2%; FoP: 17.2%) (Fig. 7b, c). Given this robust phenotype, we further delineated the effects of Foxe1 loss by bulk RNA-seq (Fig. 7d). In the PCA plot, CVP Foxe1-/- and FoP Foxe1-/- samples appear separated along PC2, this component accounts for only 16% of the total variance, compared to 58% for PC1, this difference likely reflects limited biological variability. GO analysis of the top 100 genes contributing to PC2 revealed enrichment for DNA replication-related processes, suggesting cell cycle variation underlie the observed distribution (Supplementary Fig. 10b).
CVPFoxe1-/- and FoPFoxe1-/- organoids displayed elevated levels of progenitor markers (Sox9, Lgr5, and Krt14), while differentiated markers such as Krt19, Krt7 (taste and saliva), Krt20 (taste83), Cldn6 and Dmbt1 (saliva) were decreased, in comparison to WT organoids (Fig. 7e). Foxe1-/- organoids show decreased expression of Tfc2pl1 (saliva) and Elf3 (taste), alongside an increase in Foxa2 (taste), Hes6 (taste), and Sox9 (saliva) expression. Examination of the top 50 differentially expressed genes (DEGs; Foldchange >1, p < 0.01; 1171 DEGs; Table S2) indicated a depletion of salivary gland-associated genes in both CVP and FoP Foxe1-/- organoids (Supplementary Fig. 10c). Next, we derived a top 100 DEGs signature (module score) for both WT organoids (‘WT score’) and Foxe1-/- organoids (‘Knockout score’), enabling the visualization of affected populations (Table S2). Indeed, detailed analysis revealed that within the scRNA-seq UMAP of the CVP and FoP, the ‘WT score’ showed the highest expression levels in the ductal and acinar salivary gland populations (Fig. 7f, g). Conversely, the ‘Knockout score’ was highest in a subpopulation of the progenitor compartment of the salivary gland (Fig. 7h). In the TBC clusters, the ‘WT score’ appeared to be highest in the type I branch (Fig. 7f), whereas the ‘Knockout score’ was enriched in the type II cell cluster (Fig. 7i, j). This was supported by a significant increase in expression of type II markers in Foxe1-/- organoids (Fig. 7e) (Gna14, Tas1r1). In addition, Otop1 expression, present in type I and III TBCs (Supplementary Fig. 2a), was lost in Foxe1-/- organoids. This coincided with the absence of the classical taste marker Krt20, which -in our scRNA-seq data- was exclusively expressed in type I and III TBCs but completely absent in type II TBCs (Supplementary Fig. 2a). Both suggested a shift from type I and III toward a more type II-like phenotype in the TBCs of Foxe1-/- organoids. In summary, these results illustrated a regulatory role of Foxe1 in determining salivary gland differentiation and in the makeup of TBC subtypes. Following Foxe1 loss, salivary gland cells obtain a more progenitor-like phenotype, while TBCs shift from type I and III to type II cells.
Discussion
In this study, we present evidence that CVP and FoP taste buds harbor a Lgr5 stem cell that can give rise to non-taste LEC, TBC, and salivary gland cells. Single-cell analysis predicted such a lineage design, which was subsequently confirmed by analysis of organoids grown from single Lgr5 stem cells. We also show that -in this combined chemosensory unit- Foxe1 plays a regulatory role in cell fate decision of the Lgr5+ stem cell along the lineage trajectory: loss of Foxe1 results in an increase of progenitor markers and a dramatic ablation of the salivary compartment, combined with a shift in the make-up of the TBC compartment.
Through our single-cell transcriptome analysis, we characterize the posterior tongue epithelium and its appendages in detail. While our study was under review, two scRNA-seq studies84,85 were the first to describe taste and salivary gland cell types in the posterior tongue. We propose a stem cell niche where the Lgr5/Foxe1/Sox2low/Sox9- progenitor resides prior to being recruited for differentiation. Progeny of this stem cell (1) generate non-taste lingual epithelium, (2) ascends towards the taste bud structure to generate TBCs (Sox2high 24), or (3) descend to generate salivary gland cells (Sox9). Consistent with earlier research, we note that in Lgr5-EGFP knock-in mice, the GFP signal is elevated near the trenches where the taste compartment touches the salivary compartment35 (Fig. 3d). We propose that this marks the location where the uncommitted tripotent Lgr5 stem cell resides. While previous studies have shown that stem cells marked by Lgr5 generate taste bud and non-taste epithelium35 or von Ebner’s gland (VEG) ducts86, these studies did not address if a single Lgr5+ stem cell could generate all three tissues. Our findings provide evidence for a single tripotent Lgr5+ stem cell capable of generating taste bud cells, non-taste lingual epithelium, and VEG structures. This supports that the taste epithelium and VEG, previously described as a single chemosensory unit87, developmentally appear as a single unit, even in the adult mouse, deriving from a shared stem cell. We have reported very recently that a similar situation exists in the developing human pancreas, where a single Lgr5+ stem cell can be expanded over long periods of time in organoid culture and remains capable of generating ductal, acinar, and endocrine lineage cells88.
Single cell analysis of organoids, clonally derived from single Lgr5+ stem cells, revealed cells from all three lineages. We used an unbiased media composition without favoring a specific lineage, likely underlying the lack of mature taste bud markers in clonal organoids. Future studies could refine media composition to direct lineage-specific outcomes. Next, as a proof of principle, we conducted Rosa-confetti lineage tracing to observe this phenomenon in vivo. We observe clonal tracing from single Lgr5+ stem cells into the salivary ducts, non-taste lingual epithelium (LEC), and taste buds, revealing tripotent potential of these cells within the CVP. Quantification of our lineage tracing data shows that the majority of clones contribute to taste buds, which is consistent with the high turnover rate of taste bud cells (10–14 days)89,90 in contrast to the relatively slow turnover of salivary glands (>60 days)91. The low frequency of tripotent tracing events is likely a result of the divergent renewal kinetics across tissues. Our clonal analysis reveals that the majority of labeled cells remain confined within individual taste buds, suggesting localized stem cell activity. Intriguingly, tracings originating from the trench region occasionally span multiple taste buds, besides the salivary gland, pointing to a hierarchical structure within the Lgr5+ stem cell population.
The lineage tracing data further supports that Lgr5 stem cells contribute to tongue epithelium near the CVP, which physiologically will likely only make a small contribution to homeostatic self-renewal of the surface epithelium: it is widely recognized that Krt14+ basal progenitors maintain the tongue surface20,92. In the skin, hair follicles contain Lgr5+ stem cells. While these normally only generate hair, they will drive to repair of the intrafollicular surface epithelium, functioning as a secondary stem cell hub69,93,94. A putative similar role may exist for the Lgr5 stem cell in the posterior tongue with regard to tissue regeneration upon damage.
In humans, FOXE1 is known to be key for craniofacial development, as biallelic loss results in a rare autosomal recessive disease called Bamforth-Lazarus syndrome, manifesting with athyreosis, cleft palate, bilateral choanal atresia, bifid epiglottis, and spiky hair95,96. Similar phenotypes have been observed in mice, where Foxe1 has been identified as a downstream target of the Shh-pathway82. In the hair follicle, Foxe1 and Lgr5 are co-expressed in the outer root sheath30,82,97, and a Foxe1-/- leads to disturbed hair formation. Our computational analysis suggested a role for Foxe1 along the differentiation axis of Lgr5⁺ stem cells in the posterior tongue, which led us to interrogate its function experimentally. Functionally, we show that Foxe1 regulates cell fate decisions in both CVP and FoP regions, as its loss alters lineage differentiation in the corresponding organoid models. The Shh pathway is well-established as being essential for maintaining the posterior tongue epithelium21,77,98, while Foxe1 may act downstream of Shh to mediate this effect. The observed shift toward a more Type II-dominant taste bud phenotype in Foxe1-deficient states implies impaired specification or maintenance of Type I cells. As Type I cells retain Shh expression (Fig. 2c), Foxe1 may act downstream in their differentiation pathway. The resulting reduction in Type I cells would then contribute to a compensatory increase in the proportion of Type II cells. We cannot exclude the possibility that Foxe1 influences the survival of specific taste cell types rather than their initial specification. However, Foxe1 loss may also directly impact Lgr5 stem cells in a lineage-specific manner. Several putative mechanisms may explain how loss of Foxe1 perturbs the differentiation axis.
Taste sensation is strongly affected by reduced saliva production in dry-mouth disease (e.g., radiotherapy-induced or in the context of Sjögren’s syndrome). Anterior TBCs, not studied in detail in this study, do not appear to rely on associated minor salivary glands. These taste buds lie embedded in tongue epithelium directly facing the tastants with a constant saliva flow produced by the main salivary glands (SMG, SLG, and PG), whereas the CVP and FoP taste buds -given their location in clefts/trenches- appear to rely on a constant flow of saliva produced within their own anatomical structure. This suggests that the salivary gland and taste buds have co-evolved on the posterior tongue. Beyond these developmental considerations, the taste buds and the associated salivary glands represent unified chemosensory units. While VEG involvement is often overlooked in xerostomia-related studies, its anatomical proximity to taste buds implies a significant role in modulating taste perception. This integrated perspective may improve our understanding of ageusia and dysgeusia in the context of xerostomia. In summary, we report the existence of a Lgr5 stem cell in the posterior tongue capable of generating TBC, salivary gland cells, as well as the cells of the tongue surface.
Methods
Animals
All animal experiments were carried out under the guidelines and approval of the animal welfare committees of the Netherlands Cancer Institute and Hubrecht Institute (KNAW). Single-cell experiments were performed using female adult mice (n = 20). Transgenic Lgr5-EGFP-IRES-creERT2 were first described in Barker et al.32. Lineage tracing experiments were performed with male and female Lgr5-EGFP-IRES-creERT2 /R26-confetti mouse strains described in Snippert et al.69. All experiments were performed in adult mice maintained under specific-pathogen-free conditions, with a 12 h light/12 h dark cycle and ad libitum access to food and water.
Organoid culture
Organoid cultures were established using either C57BL/6 adult mice WT or Lgr5-EGFP-IRES-creERT2 mouse strains generated by the Barker paper's methods32. Targeted regions, identified under a stereomicroscope, were picked for organoid isolation. The Aihara protocol34 was used for CVP and FoP organoid culture, and the media composition, referred to as RNE medium, including Advanced DMEM/F12 (Gibco), 1x B27, 1x N2, 1x Glutamax, 10 mmol/L Hepes, 100 μU/ml penicillin–streptomycin, 50 ng/ml EGF(Peprotech), 10% Rspondin1(in-house), and 10% Noggin conditioned media (in-house), supported optimal growth. Media refreshment every 2–3 days and routine splitting every 7–10 days maintained organoid viability.
Multi-color Lineage Tracing in mice
For lineage tracing, Lgr5-EGFP-IRES-creERT2 /R26- confetti mouse strains were used described in Snippert et al. (2010). Induction was achieved following procedures by Yee et al. (2013). Briefly, creERT2 activity was induced by injecting intraperitoneally into mice five times, once per day, 0.22 mg/g Tamoxifen (Sigma Aldrich T5648) in peanut oil (8002-03-7) per injection in adult mice, as described in Yee et al.35. Mice were sacrificed and tongues were left overnight at 4 °C in PLP (periodate, lysine, paraformaldehyde) buffer. Tongues were embedded in low-melting agarose (3%) and sectioned using a vibratome (Leica; settings: 1 mm, 0.8 mm/s, 60 µm). Next, samples were embedded in FunGi99,100 clearing solution overnight to visualize endogenous nGFP, mCFP, YFP, and RFP tracing.
Clonal culture
Clonal cultures were generated as described in the protocol of Pleguezuelos-Manzano et al.66,67,68. Briefly, organoids were treated 1 hour before with 10 µM ROCK inhibitor and trypsinized with TrypLE Express for 15 min at 37 °C. Next, trypsinization was halted, and the cell suspension was filtered stepwise with a 70 µm and 40 µm filter. Organoids were washed and resuspended in FACS buffer (1% FBS, 1 mM EDTA, PBS). Gates were set on the Sony SH800 Cell Sorter for single-cells, and sorted fractions were checked if sorting of actual single cells was achieved. GFP was gated based on the negative control (Wild-type organoids; Supplementary Fig. 5a, b). Lgr5-EGFP cells were gated based on single-cell and GFP. Data was analyzed using FlowJo (10.8.:1). Low-density cell suspension was seeded in a Matrigel dome in RNE medium supplied with Wnt-surrogate (1:1000) and 10 µM ROCK inhibitor. Organoids were picked after 7-10 days and clonally expanded. Clonal cultures 1,3, and 7 were send for scRNA-seq.
CRISPR-cas9 gene editing
Gene editing was performed on CVP and FoP organoids derived from the Lgr5-EGFP-IRES-creERT2 mouse following a brief expansion period. For Foxe1 sgRNA construction, sgRNA’s were designed using CHOPCHOP (https://chopchop.cbu.uib.no/; Table S1) and ligated into the pSPgRNA plasmid (Addgene #47108) by Ran et al.101 was utilized. A plasmid encoding both SpCas9 and mCherry for the visualization of transfected cells (Addgene #66940) was employed, along with a piggyBac carrying a hygromycin B cassette and piggyBac transposase-expressing plasmid. Electroporation was carried out using a Neon electroporator (Nepagene) with 2 mm cuvettes (Bio-Rad). Poring pulses were set at a voltage of 175 V, pulse length of 7.5 msec, pulse interval of 50 msec, and a total of 2 pulses; transfer pulses were configured with a voltage of 20 V, pulse length of 50 msec, pulse interval of 50 msec, and a total of 5 pulses. Selection with 300 µg/ml Hygromycin B commenced after 5 days and was maintained for 7-10 days. Fifteen clones were manually picked using a P20 pipette. WT and knock-out organoids were expanded in parallel. For genomic DNA (gDNA) extraction, cells were lysed at 56 °C overnight using a lysis buffer (100 mM Tris.HCl, pH 8.5, 5 mM EDTA, 0.2% SDS, and 200 mM NaCl) containing 60 µg/ml proteinase K. DNA was precipitated by adding 1x volume isopropanol, washed in 70% ethanol, and then resuspended in sterile MilliQ water. The DNA concentration, determined using a NanoDropTM spectrophotometer, was further diluted to a concentration of 100-200 ng/µl. Genotype was conducted using Sanger Sequencing (Macrogen) to confirm successful knock-out (Primers in Supplementary Table 2).
Immunohistochemistry
The organoids or primary tissues were fixed in 4% paraformaldehyde (PFA) and left overnight. Tissues were either cryopreserved or paraffin-embedded. Cryoprotection was achieved with sucrose involving successive incubations in 15% and 30% sucrose, followed by embedding in cryomolds with Tissue tek(Takara bio). Sections of 20 μm were prepared using a CryoStar (NX70 cryostat). For blocking and permeabilization, a mixture of 1x phosphate-buffered saline (PBS), 5% bovine serum albumin (BSA), and 0.3% Triton X-100 was applied for 1 hour at room temperature. Subsequently, primary antibody incubation occurred overnight at 4 °C, and secondary staining was conducted for 1 hour at room temperature using a solution of 1x PBS, 2.5% BSA, and 0.2% Triton X-100. The specific antibodies used are detailed in Supplementary Table 1. Slides were mounted with ProLong™ Gold Antifade Mountant. Tissue slices for whole-mount staining were incubated in primary antibody for 3 days, then incubated overnight with the appropriate secondary antibody. Next, samples were embedded in FunGi clearing solution overnight prior to visualization.
Imaging
Tissue sections with fluorescent markers were subjected to imaging using a Leica SP8 Confocal microscope. Following the imaging process, the fluorescent images obtained underwent processing using ImageJ software (version 2.16.0/1.54p) to create a maximum projection of all z-stacks. For confetti lineage tracing, images were acquired with the Zeiss LSM880, utilizing online fingerprinting to optimize for CFP, GFP, YFP and RFP signals. After imaging the endogenous signal, whole-mount stainings were done and imaged on the Leica Stellaris, which allows >4 color imaging. Sox9 staining did not work for 2-month lineage tracing. However, Sox2 and Krt8 were sufficient to distinguish compartments. Images were analyzed in Imaris Image Analysis Software (Bitplane). Tracing events directions was quantified based on whole-mount staining with differentiation markers. Regarding Foxe1, for each condition (wild-type and Foxe1 knockout), Krt8-positive cells were quantified manually using ImageJ across ≥10 organoids per group in both the circumvallate papilla (CVP) and foliate papilla (FoP) cultures. The proportion of Krt8+ cells was calculated relative to the total number of nuclei per organoid based on DAPI staining. Statistical analysis was performed using two-tailed unpaired t-tests and Mann–Whitney U tests, error bars represent standard deviation.
RNA extraction and qPCR
Cell pellets for RNA extraction were collected and snap frozen in liquid nitrogen. RNA-easy kit (Qiagen) was used for RNA-isolation, and concentration and quality were checked using a Nanodrop. Subsequent cDNA(input: 500 ng RNA) was generated using the SuperScript III kit (Thermo Fisher) using Oligo dt or random primers. Prior to qPCR, cDNA was 1:5 diluted and 1 µl was used per qPCR reaction, iQSYBRGreen mix (Bio-Rad) with a final volume of 10 µl. For qPCR, Actb(b-actin) and L19 were picked as housekeeping genes and
RNA sequencing
WT and mutant Foxe1-/- organoids were cultured separately for 8 days in RNE (Rspo, Noggin, Egf) medium. Of each origin (CVP and FoP), two clones were cultured. RNA was extracted from 12 samples(duplicates) as mentioned above, and concentration was determined using a Qubit RNA HS Assay. Quality was checked using an Agilent RNA 6000 nano kit and run on an Agilent 2100 Bioanalyzer. Samples were sent for library preparation and subsequent sequencing to USEQ (Utrecht Sequencing Facility). The R-package DESeq2 was used for analysis102. First, a combined analysis of both origins was performed to WT vs Foxe1-KO. Next, we made separate DESeq objects, and significant genes were selected with log2FoldChange > = 0.5 & baseMean > 30 & padj<0.05. Heatmaps were generated with the package pheatmap and were scaled by ‘row’. A module score of the TOP 100 DEGs in WT organoids (‘WT score’) and Foxe1-/- organoids (‘Knockout score’) (Supplementary Data 1) was generated and added to the metadata and main Seurat object of Fig. 1 with AddModuleScore. Featureplots projected the score on the scRNA-seq UMAP. Dataset is publicly available under GEO-accesion number GSE274014.
Single-cell RNA sequencing
Freshly dissected adult mouse tongues served as the primary source for scRNA-seq of taste bud tissue. Mouse tongues were initially cut, and under a stereomicroscope, the circumvallate papillae (CvP) and foliate papillae (FoP) were surgically removed. Subsequently, CVPs and FoPs were incubated in collagenase 1A at 37 °C for 20–25 minutes. Concurrently, CVPs and FoPs were transferred to medium (Advanced DMEM +++ with Glutamax, PenStrep, and HEPES), spun down, and treated with TryPLE, including 10 µM ROCK inhibitor. For Lgr5-eGFP CVP clonally cultured organoids, organoids were harvested in ice-cold DMEM, spun down and treated with TrypLE. The cells underwent incubation at 37 °C, with periodic pipetting and supernatant collection. Examination under a light microscope confirmed successful detachment of taste bud cells. Following incubation, cells were centrifuged at ~300 g for 5 minutes.
Cell pellets underwent two washes with cold FACS buffer (1% FBS, 0.1 mM EDTA, PBS) and filtration through a 70 µm filter. The resulting cell pellets were resuspended in FACS buffer and filtered through a 40 µm filter FACS tube.
Cells were stained with Draq5 for 10 minutes at room temperature and DAPI just before FACS sorting. Dead (DAPI+Draq5-) and live (DAPI-Draq5+) cells were sorted, and the collected cells were placed in SC-buffer (1% BSA, PBS) with 10 mM ROCK inhibitor in an Eppendorf tube. The cell suspension was then transferred to a 1.5 mL Eppendorf tube, spun down, and resuspended in SC buffer. Live cells, manually counted using Trypan blue, were resuspended in 1X PBS containing 0.1% BSA at a concentration of 700–1200 cells/µl. For clonally cultured organoids, 8000 cells per sample, and for primary tissues, approximately 10,000–18,000 cells per sample were loaded onto a Chromium Single Cell B chip for library preparation using a Chromium Next GEM Automated Single Cell 3’ Library and Gel Bead Kit v3 (10X Genomics), according to the manufacturer’s instructions. The libraries were subsequently sequenced using the NovaSeq6000 1092 (Illumina) flowcell. Datasets (clonal and tissue) are publicly available under GEO accession number GSE274015.
Single-cell RNA seq analysis
Data preprocessing involved the use of the CellRanger count function. Output files were read into R (v4.1.0) using the Read10X function from the Seurat package (v5.0.X)103 (CVP = 6389 cells, FoP = 13931). Mitochondrial transcripts were filtered, and the expression matrices were merged. Genes expressed in fewer than 3 cells and with a minimum of 200 features per cell were filtered. The preprocessed expression matrix was used to create a Seurat object, excluding cells with nFeature_RNA > 8000 or >30% mitochondrial transcript percentage for further analysis (CVP = 6094, FoP=12926) (Supplementary Fig. 1c-d).
To ensure robust and comparable analysis, the scRNA-seq data underwent preprocessing steps. Initially, the data were normalized using the LogNormalize method with a factor of 10,000. Following normalization, variable features were identified using the FindVariableFeatures function. Next, an Elbowplot was generated to inform the selection of dimensions crucial for capturing the dataset's intrinsic structure. The dimensionality assessment guided subsequent clustering analyses (pcs = 35). We regressed out cell cycle markers (CellCycleScores,) and Mitochondrial genes were regressed out. Clusters were identified using the FindNeighbors and FindClusters functions, followed by RunUMAP. In general, to characterize cell types globally, cell signatures were defined based on module scores (AddModuleScore) or by multiple well-established marker genes in the literature. These markers, depicted in Fig. 1 & S2 and detailed in Supplementary Data 2, served as a reliable reference for assigning cell types.
For further analysis, exclusion of clusters "15", "14", "13", and "17" based on Vim, Ptprc, and Pecam1 expression was performed. The Seurat object was reformatted following the above-mentioned pipeline. Through subsetting, we narrowed down our investigation to the salivary gland and taste papillae for in-depth exploration. Data visualization, carried out in the RStudio environment using Seurat plotting functions, leveraged UMAP, Violin Plots, and Dot Plots for a detailed and expressive representation of the analytical outcomes.
The pseudotime analysis was executed using monocle373,74, which requires careful cluster management by excluding outlier clusters, notably the acinar cluster. Establishing a starting node at the Lgr5 stem cell cluster, we delineated distinct trajectories: Lgr5 to taste and Lgr5 to salivary gland, achieved through subsetting. A dedicated function was constructed to generate a SingleCellExperiment class object for the identified cluster from a Seurat class, an object that was compatible with the Geneswitches package104. Geneswitches allow visualization of genes along the pseudotime that are switched on /off. Implementation of the geneswitch package involved binarizing gene expression with a set cutoff of 0.1, providing binary values for gene states (on/off). Logistic regression was then employed to estimate switching times. Visualization of the ordered switching genes involved a rigorous filtering process based on zero-expression percentage (>90%), FDR (>0.05), and McFadden’s Pseudo R^2 (<0.03). The analysis honed in on top genes with high McFadden’s Pseudo R^2. Next, all mouse transcription factors (TFs)105 and surface proteins(Cell Surface Protein Atlas)106 were systematically extracted from gene lists to highlight for precise plotting. Finally, we plotted the top 150 over each trajectory over the Pseudotime (x-axis) (plot_timeline_ggplot). Heatmaps of TFs switched-off/switched-on for Lgr5 stem cells towards both taste and saliva over pseudotime were generated utilizing smoothspline. All other TFs were plotted over the pseudotime to visualize per cell the expression of each TF over the pseudotime (Fig. 6, Supplementary Fig. 8).
For clonal cultured Lgr5-eGFP derivatives, clones 1, 3 and 7 were sent for scRNA-seq according to earlier earlier-mentioned procedures. A total of 12,494 cells (Clone1:3472, Clone3: 4119, Clone7: 4903) were captured. We filtered the dataset with parameters nFeature_RNA > 8000 or <30% mitochondrial transcript percentage for further analysis. Next, we integrated the dataset using the RPCA-based method. Cluster ‘5’ had abnormally high mitochondrial transcript and had a very low nfeature_RNA so we excluded it for further analysis. After filtering, we were left with 10 clusters equally distributed over each clone, totaling 10,035 high-quality cells; clone 1 (2643 cells), clone 3 (3248 cells), clone 7 (4144 cells) (Supplementary Fig. 7f). For integration with CVP and FoP tissue datasets, we used the ‘cca’ based method. First layers were split, and datasets were merged. Next, layer integration was performed. This resulted in 13 clusters. Annotation was performed using the same markers as in Fig. 1.
Statistics & reproducibility
Information regarding sample sizes and statistical analyses is provided in the relevant methods sections and figure legends. No data were excluded from the analysis. No statistical method was used to predetermine sample size. For Foxe1 knock-outs, differential expression analysis was performed with DESeq2 using the Wald test. Default parameters were applied, except that the significance threshold was set to α = 0.05. Reported p-values are two-sided and adjusted for multiple comparisons using the Benjamini–Hochberg method. GraphPad Prism (version 10.0.3.) was used for the generation of quantification bar graphs and qPCR boxplots. All corresponding data is distributed in the Source Data file.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Processed bulk and single-cell RNA sequencing data are deposited to the GEO repository (GSE274014 (Foxe1 knock-out RNA-seq), GSE274015 (scRNA-seq clonal and tissue) and are available in a database with open access. Source data are provided with this paper.
Code availability
Code is made available on https://github.com/PMC-Clevers/Tripotent-Lgr5/ and has been released https://doi.org/10.5281/zenodo.17152273.
References
Rothova, M., Thompson, H., Lickert, H. & Tucker, A. S. Lineage tracing of the endoderm during oral development. Dev. Dyn. 241, 1183–1191 (2012).
Petersen, C. I. et al. FGF signaling regulates the number of posterior taste papillae by controlling progenitor field size. PLoS Genet. 7, e1002098 (2011).
Zhang, C. & Oakley, B. The distribution and origin of keratin 20-containing taste buds in rat and human. Differentiation 61, 121–127 (1996).
Barlow, L. A. & Northcutt, R. G. Embryonic origin of amphibian taste buds. Dev. Biol. 169, 273–285 (1995).
Bartel, D. L., Sullivan, S. L., Lavoie, ÉG., Sévigny, J. & Finger, T. E. Nucleoside Triphosphate Diphosphohydrolase-2 (NTPDase2) is the ecto-ATPase of Type I cells in taste buds. J. Comp. Neurol. 497, 1 (2006).
Miura, H., Scott, J. K., Harada, S. & Barlow, L. A. Sonic hedgehog-expressing basal cells are general post-mitotic precursors of functional taste receptor cells. Dev. Dyn. 243, 1286–1297 (2014).
Sullivan, J. M., Borecki, A. A. & Oleskevich, S. Stem and progenitor cell compartments within adult mouse taste buds. Eur. J. Neurosci. 31, 1549–1560 (2010).
Harada, K., Miki, K., Tanaka, S., Kogo, M. & Wakisaka, S. Lectin histochemistry of posterior lingual glands of developing rats. Sci. Rep. 13, 1–11 (2023).
Field, R. B., Dromy, R. & Hand, A. R. Regulation of Secretion of Enzymes from von Ebner’s of Rat Tongue. 66, 586–587 https://doi.org/10.1177/00220345870660023501 (1987).
Proctor, G. B. The physiology of salivary secretion. Periodontol 2000 70, 11–25 (2016).
Deshpande, T. S. et al. Radiation-related alterations of taste function in patients with head and neck cancer: a systematic review. Curr. Treat. Options Oncol. 19, 72 (2018).
Ruo Redda, M. G. & Allis, S. Radiotherapy-induced taste impairment. Cancer Treat. Rev. 32, 541–547 (2006).
Wang, X. & Eisbruch, A. IMRT for head and neck cancer: reducing xerostomia and dysphagia. J. Radiat. Res. 57, i69–i75 (2016).
Nathan, C. A. O. et al. Current management of xerostomia in head and neck cancer patients. Am. J. Otolaryngol. 44, 103867 (2023).
Asif, M., Moore, A., Yarom, N. & Popovtzer, A. The effect of radiotherapy on taste sensation in head and neck cancer patients- A prospective study. Radiat. Oncol. 15, 1–9 (2020).
Brito-Zerón, P. et al. Sjögren syndrome. Nat. Rev. Dis. Prim. 2, 1–20 (2016).
Hutton, J. L., Baracos, V. E. & Wismer, W. V. Chemosensory dysfunction is a primary factor in the evolution of declining nutritional status and quality of life in patients with advanced cancer. J. Pain. Symptom Manag. 33, 156–165 (2007).
Ogama, N. et al. Appetite and adverse effects associated with radiation therapy in patients with head and neck cancer. Eur. J. Oncol. Nurs. 14, 3–10 (2010).
Ravasco, P., Monteiro-Grillo, I., Vidal, P. M. & Camilo, M. E. Impact of nutrition on outcome: A prospective randomized controlled trial in patients with head and neck cancer undergoing radiotherapy. Head. Neck 27, 659–668 (2005).
Okubo, T., Clark, C. & Hogan, B. L. M. Cell lineage mapping of taste bud cells and Keratinocytes in the mouse tongue and soft palate. Stem Cells 27, 442–450 (2009).
Lee, M. J. et al. Association of Shh and Ptc with keratin localization in the initiation of the formation of the circumvallate papilla and von Ebner’s gland. Cell Tissue Res. 325, 253–261 (2006).
Sohn, W.-J. et al. Morphological evidences in circumvallate papilla and von Ebners’ gland development in mice. Anat. Cell Biol. 44, 274 (2011).
Kim, J. Y. et al. Shh and ROCK1 modulate the dynamic epithelial morphogenesis in circumvallate papilla development. Dev. Biol. 325, 273–280 (2009).
Shechtman, L. A. et al. High Sox2 expression predicts taste lineage competency of lingual progenitors in vitro. Development 150, (2023).
Ren, W. et al. Single Lgr5- or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proc. Natl. Acad. Sci. USA 111, 16401–16406 (2014).
Arnold, K. et al. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell 9, 317–329 (2011).
Novak, D. et al. SOX2 in development and cancer biology. Semin Cancer Biol. 67, 74–82 (2020).
Ohmoto, M. et al. SOX2 regulates homeostasis of taste bud cells and lingual epithelial cells in posterior tongue. PLoS One 15, e0240848 (2020).
Barker, N. et al. Article in the stomach and build long-lived gastric units. Vitr. Stem Cell 6, 25–36 (2010).
Jaks, V. et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet 40, 1291–1299 (2008).
Huch, M. et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494, 247–250 (2013).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Aihara, E. et al. Characterization of stem/progenitor cell cycle using murine circumvallate papilla taste bud organoid. Sci. Rep. 5, 1–15 (2015).
Yee, K. K. et al. Lgr5-EGFP marks taste bud stem/progenitor cells in posterior tongue. Stem Cells 31, 992–1000 (2013).
Takeda, N. et al. Lgr5 Identifies Progenitor Cells Capable of Taste Bud Regeneration after Injury. PLoS One 8, e66314 (2013).
Winter, H., Rentrop, M., Nischt, R. & Schweizer, J. Tissue-specific expression of murine keratin K13 in internal stratified squamous epithclia and its aberrant expression during two-stage mouse skin carcinogenesis is associated with the methylation state of a distinct CpG site in the remote 5′-flanking region of the gene. Differentiation 43, 105–114 (1990).
Vercauteren Drubbel, A. & Beck, B. Single-cell transcriptomics uncovers the differentiation of a subset of murine esophageal progenitors into taste buds in vivo. Sci. Adv. 9, eadd9135 (2023).
Yang, R., Crowley, H. H., Rock, M. E. & Kinnamon, J. C. Taste Cells With Synapses in Rat Circumvallate Papillae Display SNAP-25-Like Immunoreactivity. J. Comp. Neurol. 424, 205–215 (2000).
Lossow, K., Hermans-Borgmeyer, I., Behrens, M. & Meyerhof, W. Genetic labeling of Car4-expressing cells reveals subpopulations of type III taste cells. Chem. Senses 42, 747–758 (2017).
Knapp, L., Lawton, A., Oakley, B., Wong, L. & Zhang, C. Keratins as markers of differentiated taste cells of the rat. Differentiation 58, 341–349 (1995).
Wang, H. et al. Expression of the voltage-gated potassium channel KCNQ1 in mammalian taste bud cells and the effect of its null-mutation on taste preferences. J. Comp. Neurol. 512, 384–398 (2009).
Wong, G. T., Gannon, K. S. & Margolskee, R. F. Transduction of bitter and sweet taste by gustducin. Nature 381, 796–800 (1996).
Zhang, Y. et al. Coding of sweet, bitter, and umami tastes: Different receptor cells sharing similar signaling pathways. Cell 112, 293–301 (2003).
Iglesias-Bartolome, R. et al. Transcriptional signature primes human oral mucosa for rapid wound healing. Sci. Transl. Med. 10, 1–14 (2018).
Wu, T. et al. Genome-wide analysis reveals the active roles of keratinocytes in oral mucosal adaptive immune response. Exp. Biol. Med. 236, 832–843 (2011).
Formanek, M., Knerer, B. & Kornfehl, J. Cytokine Expression of Human Oral Keratinocytes. ORL 61, 103–107 (1999).
Bekhor, I. et al. cDNA cloning, sequencing and in situ localization of a transcript specific to both sublingual demilune cells and parotid intercalated duct cells in mouse salivary glands. Arch. Oral. Biol. 39, 1011–1022 (1994).
Yoon, Y.-J. et al. Salivary gland organoid culture maintains distinct glandular properties of murine and human major salivary glands. Nat. Commun. 13, 1–16 (2022).
Culp, D. J. et al. The gene encoding mouse Muc19: cDNA, genomic organization and relationship to Smgc. Physiol. Genomics 19, 303–318 (2005).
Shindo, Y. et al. Gα14 is a candidate mediator of sweet/umami signal transduction in the posterior region of the mouse tongue. Biochem. Biophys. Res. Commun. 376, 504–508 (2008).
Matsumoto, I., Ohmoto, M., Narukawa, M., Yoshihara, Y. & Abe, K. Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nat. Neurosci. 14, 685–687 (2011).
Zhang, J. et al. Sour sensing from the tongue to the brain. Cell 179, 392–402.e15 (2019).
Tu, Y. H. et al. An evolutionarily conserved gene family encodes proton-selective ion channels. Science (1979) 359, 1047–1050 (2018).
Hauser, B. R., Aure, M. H., Kelly, M. C., Hoffman, M. P. & Chibly, A. M. Generation of a single-cell RNAseq Atlas of Murine Salivary Gland Development. iScience 23, 101838 (2020).
de Paula, F. et al. Overview of human salivary glands: highlights of morphology and developing processes. Anat. Rec. 300, 1180–1188 (2017).
Humphrey, S. P. & Williamson, R. T. A review of saliva: normal composition, flow, and function. J. Prosthet. Dent. 85, 162–169 (2001).
Hamosh, M. & Scow, R. O. Lingual lipase and its role in the digestion of dietary lipid. J. Clin. Invest 52, 88–95 (1973).
May, A. J. et al. STEM CELLS AND REGENERATION Diverse progenitor cells preserve salivary gland ductal architecture after radiation-induced damage. https://doi.org/10.1242/dev.166363 (2018).
Asano-Miyoshi, M., Hamamichi, R. & Emori, Y. Cytokeratin 14 is expressed in immature cells in rat taste buds. J. Mol. Histol. 39, 193–199 (2008).
Chatzeli, L., Gaete, M. & Tucker, A. S. Fgf10 and Sox9 are essential for the establishment of distal progenitor cells during mouse salivary gland. Dev. Dev. 144, 2294–2305 (2017).
Lombaert, I. M. A. & Hoffman, M. P. Epithelial stem/progenitor cells in the embryonic mouse submandibular gland. Front. Oral. Biol. 14, 90–106 (2010).
Knox, S. M. et al. Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science (1979) 329, 1645–1647 (2010).
Shechtman, L. A. et al. Generation and culture of lingual organoids derived from adult mouse taste stem cells. J. Vis. Exp. 170, https://doi.org/10.3791/62300 (2021).
Adpaikar, A. A. et al. Fine-tuning of epithelial taste bud organoid to promote functional recapitulation of taste reactivity. Cell. Mol. Life Sci. 79, 211 (2022).
Blokzijl, F. et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016).
Roerink, S. F. et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 556, 457–462 (2018).
Pleguezuelos-Manzano, C. et al. Establishment and culture of human intestinal organoids derived from adult stem cells. Curr. Protoc. Immunol. 130, 1–24 (2020).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Wuidart, A. et al. Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 30, 1261–1277 (2016).
Baggiolini, A. et al. Premigratory and migratory neural crest cells are multipotent in vivo. Cell Stem Cell 16, 314–322 (2015).
VanHorn, S. & Morris, S. A. Next-generation lineage tracing and fate mapping to interrogate development. Dev. Cell 56, 7–21 (2021).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nat. 566, 496–502 (2019).
Kist, R. et al. The formation of endoderm-derived taste sensory organs requires a pax9-dependent expansion of embryonic taste bud progenitor cells. PLoS Genet 10, e1004709 (2014).
Nakayama, A. et al. Expression of the basal cell markers of taste buds in the anterior tongue and soft palate of the mouse embryo. J. Comp. Neurol. 509, 211–224 (2008).
Golden, E. J. et al. Onset of taste bud cell renewal starts at birth and coincides with a shift in shh function. Elife 10, 1–28 (2021).
Song, E. A. C. et al. Genetic Study of Elf5 and Ehf in the Mouse Salivary Gland. J. Dent. Res. 102, 340–348 (2023).
Tanaka, J. et al. Generation of orthotopically functional salivary gland from embryonic stem cells. Nat. Commun. 9, 4216 (2018).
Golson, M. L. & Kaestner, K. H. Fox transcription factors: from development to disease. Development 143, 4558–4570 (2016).
Dathan, N., Parlato, R., Rosica, A., De Felice, M. & Di Lauro, R. Distribution of the titf2/foxe1 gene product is consistent with an important role in the development of foregut endoderm, palate, and hair. Dev. Dyn. 224, 450–456 (2002).
Brancaccio, A. et al. Requirement of the forkhead gene Foxe1, a target of sonic hedgehog signaling, in hair follicle morphogenesis. Hum. Mol. Genet. 13, 2595–2606 (2004).
Ren, W. et al. Transcriptome analyses of taste organoids reveal multiple pathways involved in taste cell generation. Sci. Rep. 7, 1–13 (2017).
Yu, W. et al. The duct of von Ebner’s glands is a source of Sox10 + taste bud progenitors and susceptible to pathogen infections. Front Cell Dev. Biol. 12, 1460669 (2024).
Ren, W. et al. Single-cell transcriptomic atlas of taste papilla aging. Aging Cell 23, e14308 (2024).
Harrison, T. A. et al. Lgr5+ ductal cells of von Ebner’s glands are stem cells for turnover of posterior tongue taste buds. bioRxiv 2024.04.18.589316 https://doi.org/10.1101/2024.04.18.589316 (2024).
Sbarbati, A., Crescimanno, C. & Osculati, F. The anatomy and functional role of the circumvallate papilla/von Ebner gland complex. Med. Hypotheses 53, 40–44 (1999).
Andersson-Rolf, A. et al. Long-term in vitro expansion of a human fetal pancreas stem cell that generates all three pancreatic cell lineages. Cell 187, 7394–7413.e22 (2024).
Beidler, L. M. & Smallman, R. L. Renewal of cells within taste buds. J. Cell Biol. 27, 263–272 (1965).
Hamamichi, R., Asano-Miyoshi, M. & Emori, Y. Taste bud contains both short-lived and long-lived cell populations. Neuroscience 141, 2129–2138 (2006).
Zajicek, G., Schwartz-Arad, D., Arber, N. & Michaeli, Y. The streaming of the submandibular gland II: parenchyma and stroma advance at the same velocity. Cell Prolif. 22, 343–348 (1989).
Tanaka, T. et al. Identification of stem cells that maintain and regenerate lingual keratinized epithelial cells. Nat. Cell Biol. 15, 511–518 (2013).
Joost, S. et al. Single-cell transcriptomics of traced epidermal and hair follicle stem cells reveals rapid adaptations during wound healing. Cell Rep. 25, 585–597.e7 (2018).
Ito, M. et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med. 11, 1351–1354 (2005).
Clifton-Bligh, R. J. et al. Mutation of the gene encoding human TTF-2 associated with thyroid agenesis, cleft palate and choanal atresia. Nat. Genet. 19, 399–401 (1998).
Moreno, L. M. et al. FOXE1 association with both isolated cleft lip with or without cleft palate, and isolated cleft palate. Hum. Mol. Genet 18, 4879–4896 (2009).
Joost, S. et al. The Molecular Anatomy of Mouse Skin during Hair Growth and Rest. Cell Stem Cell 26, 441–457.e7 (2020).
Castillo, D. et al. Induction of ectopic taste buds by SHH reveals the competency and plasticity of adult lingual epithelium. Development 141, 2993–3002 (2014).
van Ineveld, R. L., Ariese, H. C. R., Wehrens, E. J., Dekkers, J. F. & Rios, A. C. Single-cell resolution three-dimensional imaging of intact organoids. J. Vis. Exp. 2020, 1–8 (2020).
Rios, A. C. et al. Intraclonal plasticity in mammary tumors revealed through large-scale single-cell resolution 3D Imaging. Cancer Cell 35, 618–632.e6 (2019).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
Stuart, T. et al. Comprehensive integration of single-. Cell Data. Cell 177, 1888–1902.e21 (2019).
Cao, E. Y., Ouyang, J. F. & Rackham, O. J. L. GeneSwitches: ordering gene expression and functional events in single-cell experiments. Bioinformatics 36, 3273–3275 (2020).
Lambert, S. A. et al. The human transcription factors. Cell 172, 650–665 (2018).
Bausch-Fluck, D. et al. A mass spectrometric-derived cell surface protein atlas. PLoS One 10, 1–22 (2015).
Acknowledgements
We say our gratitude towards the Single cell facility, especially to Aleksandra Balwierz and Thanasis Margaritis, the Flow facility, and the Organoid facility at the Prinses Maxima Centrum. Furthermore, we would like to thank the animal facility at the Hubrecht Institute (Utrecht), in particular Simona Lafirenze (Lgr5-eGFP-IRES-CreERT2) and Jeroen Korving (Wildtype). This work was funded by Givaudan (Geneva, Switzerland). Illustrations as a whole or parts in Figs. 1a, 3e and 6c, d, g were created using BioRender.com.
Author information
Authors and Affiliations
Contributions
Conceptualization: L.V., S.Y.K., M.v.W., H.C.; Data curation: L.V., S.Y.K., G.v.S.; Formal Analysis: L.V., S.Y.K., G.v.S.; Investigation: L.V., S.Y.K., D.L., L.L., F.R., H.B.; Methodology: L.V., S.Y.K., D.L.; Project administration: L.V., S.Y.K., K.S., M.v.W., H.C.; Resources: L.V., S.Y.K., D.L., L.L., G.v.S; Supervision: C.W., J.S., A.R., K.S., J.v.R., M.v.W., H.C.; Visualization: L.V., S.Y.K., D.L., G.v.S., R.v.I.; Writing – original draft: L.V., S.Y.K., K.S., M.v.W., H.C.; Writing – review & editing: L.V., S.Y.K., K.S., M.v.W., H.C.; Funding acquisition: K.S., M.v.W., H.C.
Corresponding author
Ethics declarations
Competing interests
J.S. is an employee and shareholder of GIVAUDAN – C.W. is an employee of GIVAUDAN. H.C. is listed as an inventor on patents related to organoid technology held by the Royal Netherlands Academy of Arts and Sciences. He is currently Head of Pharma Research and Early Development at Roche, Basel, Switzerland. A full disclosure is available at https://uu.nl/staff/JCClevers/. All other authors declare that they have no competing interests.
Peer review
Peer review information
Nature Communications thanks Marit H. Aure and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Verweij, L.H.G., Kim, SY., Laskaris, D. et al. Tripotent Lgr5 stem cells in the posterior tongue generate lingual, taste, and salivary gland lineages. Nat Commun 16, 10266 (2025). https://doi.org/10.1038/s41467-025-65140-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65140-0