Abstract
Multidrug transport by ATP binding cassette (ABC) exporters entails a mechanism to modulate drug affinity across the transport cycle. Here, we combine cryo-EM and molecular dynamics (MD) simulations to illuminate a lipid-competition mechanism to drive substrate translocation by ABC exporters. We determine cryo-EM structures of the ABC transporter BmrCD in drug-loaded inward-facing (IF) and outward-facing (OF) conformations in lipid nanodiscs to reveal the structural basis of alternating access, details of drug-transporter interactions, and the scale of drug movement between the two conformations. Remarkably, the structures uncover lipid molecules bound in or near the transporter vestibule along with the drugs. MD trajectories from the IF structure show that these lipids stimulate drug disorder and translocation towards the innermost constricted region of the vestibule. Similarly, bound lipids enter the OF vestibule and weaken drug-transporter interactions facilitating drug release. Our results complete a near-atomic model of BmrCD’s conformational cycle and suggest the modulation of substrate-transporter interactions by lipids.
Similar content being viewed by others
Introduction
Ubiquitous across all kingdoms of life, ABC transporters traffic molecules across cell membranes in both directions by directly consuming ATP1,2,3. A subclass of these transporters, ABC exporters, specializes in exporting structurally and chemically diverse substrates, often cytotoxic molecules, thereby imparting a multidrug resistance phenotype3,4,5. The canonical alternating access mechanism of the ABC exporter class invokes the population of a suite of conformations powered by ATP binding and hydrolysis in the dedicated nucleotide-binding domains (NBDs)6,7,8. Through the underlying conformational changes, substrate molecules bind an inward-facing (IF) vestibule cradled by the transmembrane domains (TMDs) and are then extruded following a switch of the vestibule orientation to face the extracellular side. For multidrug ABC exporters, amphipathic substrates can bind from either the inner leaflet of the bilayer or the cytoplasm and are released to the outer leaflet of the bilayer or the extracellular milieu9,10.
Insight into the determinants of polyspecific binding has been gleaned from multiple drug-bound IF conformations of multidrug ABC transporters11,12. However, there is a paucity of drug-bound outward-facing (OF) structures7,10,13,14, although a low-resolution OF structure has been reported for substrate-bound BmrA15. In addition, P-glycoprotein (ABCB1) was captured in IF and purportedly OF structures bound to a synthetic substrate that was trapped via disulfide bonds in cysteine mutant backgrounds. The covalent trapping, the unexpected location of the synthetic substrate and the occluded nature of the conformation raise questions regarding its mechanistic identity16. Moreover, neither Pgp structures were determined in a wild-type background and in a lipid bilayer environment15,16. The dogma in the field associates the release of substrates by ABC transporters to a reduction in affinity in the OF conformation. However, addressing the unanswered and central question of how interactions with the transporter are weakened to drive release, following the power stroke, requires high-resolution structures of drug-bound transporters in a native-like environment.
Here, we illuminate a lipid-driven mechanism of substrate release through an integrated investigation combining single particle cryo-EM and MD simulations of the heterodimeric multidrug ABC exporter BmrCD from B. subtilis. BmrCD has been the subject of extensive functional, biochemical, spectroscopic and cryo-EM investigations in lipid bilayers, yielding a profound understanding of multiple facets of its mechanism8,17,18,19,20. We selected Hoechst as a model substrate because it was shown to be transported in inside-out vesicles and to cooperatively stimulate the ATP turnover in detergent micelles, liposomes and lipid nanodiscs8,17,21,22. The work presented in this paper describes the interactions of Hoechst with the transporter in the OF conformation. Observation of bound lipid molecules in or near the vestibule suggests a mechanism of how drugs are released to the extracellular side and proposes a general model of multidrug export.
Results
Cryo-EM structure of Hoechst-bound BmrCD in an OF conformation
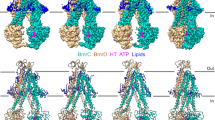
We have previously reported a series of BmrCD conformations in lipid nanodiscs, including multiple ATP- and drug-bound IF intermediates and an occluded (OC) conformation8. Driven by the evidence from double electron-electron resonance (DEER) spectroscopy that an OF conformation is populated8,17, we identified 2D images that profile such a structure featuring an open extracellular side (Supplementary Fig. 1). Subsequent 3D classification, based on OC particles, from a dataset of the WT transporter in an ADP-Vi trapped biochemical preparation yielded a small subset of OF particles (Supplementary Fig. 2). Biochemical, spectroscopic and structural data established that under vanadate trapping following ATP hydrolysis, ABC exporters populate a high-energy, post-hydrolysis state, often in a predominantly OF conformation8,17,22. A final map with a resolution of 3.1 Å of an OF conformation was obtained (Fig. 1a and Supplementary Fig. 2) following non-uniform and local refinement. The OF model (Supplementary Table 1, and Supplementary Figs. 2 and 3) features a V-shaped vestibule open to the extracellular side and the outer leaflet of the bilayer. In this OF vestibule, two Hoechst molecules pack in an antiparallel conformation (Figs. 1a and 2). In addition to either 2 molecules of ATP or an ATP/ADP-Vi in the NBDs, twenty lipid molecules are observed to decorate the intracellular and extracellular halves of the transporter. Lipids were conspicuously bound to each side of the V-shaped interface of BmrC and BmrD at the extracellular side (Fig. 1a and Supplementary Fig. 2g). Previous DEER analysis17 demonstrated that ATP hydrolysis is required for population of the OF conformation, thus, we have modeled the consensus site as bound to ADP-Vi in the cryo-EM structure.
a Cryo-EM map and structure model of BmrCD OF conformation bound to Hoechst. The extracellular domain (ECD), transmembrane domain (TMD), and nucleotide-binding domain (NBD) are indicated. BmrC and BmrD are shown in light sea green and tan. Hoechst (HT) and ATP/ADPVi are shown as sticks in magenta, and lipids are blue, hereafter if not indicated. b Conformational changes of the TMD intracellular (left panels) and extracellular sides (right panels) between BmrCD_OF, BmrCD_IF (PDB 8FMV), and BmrCD_OC (PDB 8T1P). All alignments are based on BmrD. BmrCD_IF and BmrCD_OC are shown in gray and slate, respectively. Large movements are indicated by red arrows. c DEER distance distributions for spin-label pairs on the intracellular (96BmrC/188BmrD) and extracellular (55BmrC/146BmrD) sides in BmrCD. Upper panels: close-up views of BmrCD_OF highlighting spin label pairs. TMD helices are shown as cylinders, ECD and NBD are shown as transparent surfaces. The spin-labeled locations are shown by raspberry spheres. Lower panels: experimental distance distributions are shown in solid lines. Simulate distance distributions derived from cryo-EM structures by MDDS are shown in dashed lines.
a Close-up view of the two bound Hoechsts represented by magenta sticks and colored by heteroatom. The cryo-EM density of Hoechsts are shown in the inset. b Electrostatic potential surface of the Hoechst-binding chamber of BmrCD_OF. c Perspective on Hoechst movement from the IF to the OF conformation. BmrC and BmrD of IF are shown in slate and gray, and Hoechsts of IF are represented by gray sticks colored by heteroatom. Left panel: BmrCD OF and IF are superimposed based on BmrD. The shift in distances and angles of the Hoechsts are noted. Transmembrane helices TM5 and TM6 from both BmrC and BmrD are shown in a transparent cartoon. Middle and right panels: key residues that stabilize Hoechsts in the OF and IF conformations are labeled, residues in common are boxed. d Radius measurement of the vestibule in the OF and IF structures reveals that WH-latch (W290BmrCH338BmrD-latch)8 is located at the bottleneck of the chamber.
Superposition of Hoechst-bound BmrCD OF (BmrCD_OF) and IF (PDB 8FMV hereafter referred to as BmrCD_IF)8 conformations reveals that the closing of the intracellular side is mainly contributed by BmrC rearrangement, whereas opening the extracellular side is executed by BmrD reconfiguration. Specifically, TM1, TM2, TM3, and TM6 of BmrC, along with TM4 and TM5 of BmrD, exhibit substantial displacement, resulting in the closure of the intracellular vestibule, with distance changes ranging from 8 to 12 Å. Concomitantly, the extracellular side of the TM helices move outward, facilitating the formation of an OF vestibule. Opening of the extracellular side is primarily driven by TM1 and TM2 of BmrC and TM3, TM4, TM5, and TM6 of BmrD, with distance changes ranging from 6 to 15 Å (Fig. 1b, upper panels). These distance changes are validated by DEER distance measurements in the corresponding lipid nanodiscs8,17,22. A minor population at the larger distance of the extracellular pair 55/146, observed in the vanadate-trapped post-hydrolysis state8 (Fig. 1c, solid red line), overlaps the predicted distance distribution from the OF structure (Fig. 1c, dashed red line), further reinforcing the mechanistic identity of this structure. Superposition of BmrCD_OF and OC (PDB 8T1P hereafter referred to as BmrCD_OC)8 conformations reveals that the closing of the extracellular side at this step is mainly contributed by BmrD rearrangement (Fig. 1b, lower panels). In addition, the asymmetry of TMD between BmrC and BmrD is reduced in the OF compared to the IF and OC structures (Supplementary Fig. 4).
The conformational changes propagate to the extracellular domain (ECD) that blocks the TMD in the IF conformation. Four loops, residues 92–96, 103–106, 111–116, and 120–124, are displaced ~4-5 Å in the IF to OF transition, while they move back 4-7 Å in the transition from OF to OC (Supplementary Fig. 5a). These conformational changes indicate that ECD movement is coupled to Hoechst extrusion. The local structural changes of the ECD have a profound effect on the electrostatic surface, which could play a role in funneling the Hoechst molecules out of the transporter. Specifically, the positive central patch (black circle) of the ECD in OF can repel the positively charged Hoechst, whereas the two negative side patches (blue and yellow circles) attract it, which may guide the Hoechst extrusion. Notably, these charge distributions are reversed in the IF and OC conformations (Supplementary Fig. 5b). Similar to the TMD, distance distributions for spin label pairs in the ECD, predicted from the OF structure, superimposes on the experimental data, further confirming the conformational changes at these sites8 (Supplementary Fig. 5c).
Interactions of Hoechst with the outward-facing vestibule of BmrCD
Clear density of two Hoechst substrates (labeled HT-1 and HT-2) enabled a detailed analysis of their interactions with the transporter in the OF conformation (Fig. 2a, b). The two Hoechst molecules are located between TM1, TM3, and TM6 of BmrC and TM5 and TM6 of BmrD. Specifically, D141 of BmrC and D377 of BmrD hydrogen-bond with the piperazine group of HT-1 and the benzimidazole group of HT-2, respectively. The pivotal role of D141 is consistent with previous results showing that its alanine substitution abrogates Hoechst stimulation of ATP hydrolysis in BmrCD22. Y284 of BmrC interacts with HT-1 via a π-π interaction, and V283 of BmrC forms a hydrophobic interaction with HT-2. M34 and M287 of BmrC hydrophobically interact with the two Hoechsts separately (Fig. 2c, middle panel). Similar to the IF conformation, the major residues contributing to Hoechst binding are from BmrC, suggesting that BmrC interactions guide Hoechst during transport (Fig. 2c). W290 of BmrC, H338, D377 and Q384 of BmrD appear to interact with Hoechst in both conformations (Fig. 2c).
Comparison of the IF and OF conformations (Fig. 2c) captures the translocation of the two Hoechst molecules by around 15 Å to 21 Å from the lower part to the upper part of the TMD. In the IF conformation, the two Hoechsts form an angle of 43°, while in the OF conformation, they become almost antiparallel. The alternating access of the vestibule is contributed by the intracellular and the extracellular sides of BmrD TM5 and BmrC TM6 which move to close the entrance and open the exit simultaneously. The substrate binding vestibule’s volume is reduced from 7882 Å3 to 4995 Å3 during the IF to OF transition. The fulcrum of this transition (Fig. 2d) appears to be located in the vicinity of a previously8 identified ionic interaction between W290BmrC and H338BmrD (referred to as an ionic latch), both critical residues for binding of Hoechsts (Fig. 2c, middle and right panels). Interestingly, inspection of the putative OF conformation of mPgp16 shows that TM6 and TM11 (equivalent to TM5 in the second half of Pgp) contribute to substrate binding in both OF and IF conformations with the equivalent residue to W290 (F339) in mPgp contacting the substrate (Supplementary Fig. 6). This residue is also conserved in hPgp, and the bacterial heterodimers TmrAB and TM287/28815,23 (Supplementary Fig. 6).
MD simulations uncover the pivotal role of lipids in Hoechst release
We simulated the dynamics of the lipid molecules visualized in the cryo-EM structure with particular focus on the lipid molecule, a POPA molecule labeled POV15, which is poised to enter the extracellular cleft of the protein. Our simulations show a rapid descent of this lipid into the cleft. Its entrance is captured in kernel density estimation analysis, which shows regions of high occupancy or density, revealing locations accessible to membrane phospholipids (Fig. 3a, b). In one simulation replica, the lipid-binding event is aborted, while in the other 3 replicas it continues until the lipid has fully entered the transporter OF cavity.
a Kernel Density Estimation analysis. Lipid phosphorous atoms in the outer leaflet have been binned in their XY projections, with high density appearing yellow and low density appearing dark purple. b Molecular rendering of POV15 entering into the cleft and attracting Hoechst molecules. Gray and color models are the MD simulation results at 0 ns and 140 ns, respectively. Loops are omitted for clarity. c Time series of fraction of native contacts. Each color in the trace belongs to an independent replica simulation. Native contacts are defined as any protein residue with heavy atoms within 4.5 Å of the Hoechst molecules in the initial structure. d Distribution of Hoechst tilt angles as measured with respect to the positive Z-axis. This was measured by defining the vector going from the piperazine to the ethoxy group of Hoechst and measuring the angle formed with the vector (0, 0, 1). e Time series for the salt-bridge distance between E33 and R345 of BmrC and BmrD, respectively. Each trace represents an independent replica. f Distribution of E33BmrC-R345BmrD salt-bridge distances over all the simulations.
One consequence of the lipid entrance into the vestibule is the perturbation of the bound Hoechst molecules, highlighted in native contact analysis (Fig. 3c). While in the cryo-EM density the Hoechst molecules adopt roughly vertical, antiparallel conformations, the presence of the negatively charged lipid induces a strong tilt for both HT molecules (Fig. 3d). This tilting motion of the drug molecules breaks several of the stabilizing interactions formed between the two chains of BmrCD and Hoechst molecules. One such critical residueinvolved in substrate binding, D141 of BmrC22, is also implicated in the observed lipid-mediated substrate dislodging. As the lipid weakens protein substrate contacts, D141 continues to hold tightly onto the charged end of HT-1, heavily contributing to the tilting behavior observed as lipids tug on the opposite end of HT-2. As POV15 begins pulling on HT-2, we also observe a change in the electrostatic interactions between BmrCD and Hoechst (Supplementary Fig. 7a). This energetic perturbation occurs one substrate at a time and correlates with structural rearrangement of TM1 of BmrD (Supplementary Fig. 7b, c).
In another simulation replica, we observed HT-1 beginning to depart the original binding site into the membrane, opposite of POV15 (Supplementary Fig. 7d, e). This lipid-substrate interaction helps to break the D141-HT-1 salt bridge as well as the nearby E33BmrC-R345BmrD salt bridge, allowing E33 to now form a new salt bridge with the exposed charged nitrogen of HT-1 (Fig. 3e, f, and Supplementary Fig. 8a). Concurrently, HT-2 terminal piperazine group forms a salt bridge to E51 of BmrD, causing the substrate to rapidly translocate up towards the ECD domain (Supplementary Fig. 8b). The piperazine group then is passed off to the nearby E75 of the ECD, while E51 goes on to form a salt bridge with the benzimidazole ring in the Hoechst molecule.
While HT-2 is traveling up towards the ECD domain, the ethoxy group of HT-1 glides out of the extracellular cleft, an event accompanied with a correlated opening of the ECD domain (Supplementary Fig. 8c, d). In summary, we observe an unbinding motion which is initiated by the entry of POV15 into the transporter chamber. Shortly after lipid entry, HT-2 is engaged in a pattern of electrostatic and hydrophobic interactions with the lipid, allowing HT-1 to dislodge and presumably diffuse into the nearby membrane. The opening of the ECD domain provides a potential role for this appended domain in substrate unbinding and may explain why BmrCD contains this structural domain relative to other type IV ABC transporters.
In contrast, simulations in which lipids were restrained from entering the binding pocket confirm their disruptive effect on ligand binding. The kernel density estimation of the non-restrained (referred to as wild-type (WT) in supplementary Fig. 9) and lipid- restrained (RS) systems (Supplementary Fig. 9a, b) shows the density of lipids dramatically decreased near the binding pocket compared to the WT, indicating almost no lipids entered and interacted with HT-1 and HT-2 in the RS system (Supplementary 1–4). The lower RMSDs of HT-1 and HT-2 (Supplementary Fig. 9 and Supplementary Table 2) show that the RS system (orange) maintains a more stable conformation (HT-1: 2.9 ± 0.9 Å, HT-2: 3.1 ± 0.9 Å), whereas WT (blue) shows higher and more variable RMSD values, indicating reduced conformational stability in the absence of lipid restraints.
MD simulations reveal decreased Hoechst binding stability in D141A BmrCD
In light of the previous experimental data and the above simulations of Hoechst dynamics in the OF conformation, we further probed the role of D141 in the release mechanism by MD simulations in a background where this residue is substituted by alanine. We tracked the distance between the center of mass (COM) of HT‑1 or HT‑2 and the COM of residue 141 over WT and D141A simulations (2 µs). As shown in Supplementary Table 3 and Supplementary Fig. 10a and b, HT‑1 maintained a short distance of 4.0 ± 1.7 Å to D141, in the WT with transient fluctuations generally under 2.0 Å (n = 3 replicates). In contrast, in the D141A mutant, the distance to A141 drifted to 7.5 ± 2.5 Å, occasionally even reaching above 14 Å. While less considerable, a similar pattern arose for HT‑2: compared to a distance of 8.4 ± 2.7 Å in the WT, we observed a distance of 12.0 ± 1.8 Å for D141A. The RMSD of each ligand relative to its initial bound conformation (Supplementary Fig. 10c–f) further substantiates the impact of D141 removal. In WT, the average RMSD of HT‑1 plateaued to 3.8 ± 1.0 Å (n = 3 replicates), while the mutant showed larger fluctuations, reaching up to 12.2 Å, with an average RMSD of 6.2 ± 3.1 Å (n = 3 replicates). Likewise, HT‑2 in WT settled to an RMSD of 3.9 ± 1.8 Å, whereas in the D141A mutant it reached 4.8 ± 1.4 Å during the same period. These data demonstrate that, once D141 is removed, both HT‑1 and HT‑2 lose a key electrostatic anchor and adopt more mobile binding poses that deviate from the original binding pose more wildly. Consistent with these shifts, ionic bond analysis (Supplementary Fig. 10g, h) shows that D141 in the WT established recurring salt‑bridge interactions with HT‑1, but after mutation, these contacts disappeared, prompting the ligands to form compensatory electrostatic interactions with other negatively charged residues (e.g., E33 in BmrC).
Overall, analysis of the distances, RMSD and ionic interactions strongly indicates that D141 is a pivotal anchor for HT‑1 and HT‑2, and when it is replaced by a non‑charged residue (D141A), the ligands experience increased fluctuations and less stability in the binding pocket. These collectively drive HT‑1 and HT‑2 to shift away from the canonical binding region. While we did not observe full unbinding of the ligands from their binding pocket over the simulation timescales, the observed increased fluctuations after introducing the mutation clearly reflect an increased unbinding probability.
Lipid binding to the IF conformation
The discovery of the role of lipids in the release of Hoechst from the OF conformation prompted us to investigate if lipids play a role in Hoechst binding to the IF conformation. Searching the same ADP/Vi-trapped dataset identified two IF conformations at a resolution of 3.4 Å, both with lipids bound in proximity to the two Hoechst molecules (Fig. 4a, Supplementary Figs. 11, 12, and 13, and Supplementary Table 1). In one IF conformation, hereafter referred to as BmrCD_IF1, a POPA molecule (labeled as POV4), directly interacts with H184 of BmrC (Fig. 4b, c, middle panels), which is part of an ionic “latch” that involves its interaction with R304 side chain, as we previously noted8. The latch closes upon the binding of the two Hoechst molecules manifested by the distance between the two side chains8. In BmrCD_IF1, the latch is in the open state as the lipid engages H184. In the second identified IF conformation, BmrCD_IF2, a lipid POPC molecule (POV23) is bound in the substrate binding vestibule with the latch in the closed position. In addition, another lipid (POV4) is poised nearby (Fig. 4b, c).
a The structure models and density maps of BmrCD_IF1 and BmrCD_IF2. Density maps are shown as a transparent surface, and the color scheme is the same as Fig. 1a. b The RH-latch (R304BmrC-H184BmrC latch) densities for BmrCD_IF1 (middle panel, tan), and BmrCD_IF2 (right panel, light sea green) are shown with side-chain distance indicated. BmrCD_IF (PDB 8FMV, left panel, gray) is shown for comparison. c The lipid molecules close to the RH-latch are shown in blue sticks colored by heteroatom with the distance highlighted. The lipid code is from the corresponding structure. d RH-latch monitored by MD simulations. Time series (left panel) and probability distributions (right panel) are shown. In the IF2 without lipid replica 1 (IF2_nolipid_rep1) simulation, a lipid molecule interacts at the TM4/TM6 interface after 400 ns. e Stability of the substrates in all MD simulated conditions.
Bound lipids loosen drug-transporter interactions in the IF conformation
To explore the functional role of these lipids in the transport mechanism, we carried out MD simulations of BmrCD_IF2 in the presence and absence of bound lipids, monitoring the dynamics of bound lipids, the stability of the substrates under all conditions, and the distance between TM4 and TM6, where the H184-R304 latch is located. Most of the 27 lipid molecules identified from the cryo-EM density were flexible, showing atomic fluctuations larger than 3.5 Å for the phosphorous atom of the headgroups, and even larger tail deviations. Only 6 molecules showed atomic fluctuations between 2.7 and 3.5 Å (Supplementary Fig. 14), and these do not include the two lipid molecules near the latch. Indeed, lipids 4 and 23 appeared quite flexible.
We monitored the Cα-Cα distance for H184-R304 under all simulated conditions as illustrated by the time series and its probability distribution (Fig. 4d and Supplementary Table 4). The analysis of the RH latch revealed that the presence of a single substrate molecule (Fig. 4d, gray) stabilizes shorter distances, while introducing a second substrate (silver) triggers the opening of the TM4/TM6 interface (shoulder in the silver distribution). The presence of two lipid molecules keeps the interface more open (extremely pronounced in replica 1 of the lipid bound simulation set, red curve), although we observed that it began to close as the lipids started to depart from the site during the simulation (red curve). Simulations after removal of the lipid bound molecules (green) show two peaks in the distance distributions. Remarkably, the interface begins to close until a new lipid molecule, from the bulk lipids, comes in after 400 ns (snapshots from the simulations are shown in Supplementary Fig. 15a). The implied role of the TM4/TM6 interface as a gate is in agreement with previous structural studies of mammalian Pgp and MD simulations of the ABC heterodimer TM287/TM28824,25.
To monitor the substrate stability, we calculated the atomic fluctuations of all HT heavy atoms as time series for all simulated conditions (Fig. 4e and Supplementary Fig. 15). Furthermore, to map the movement of the substrates during the simulations, we represented the HT molecules as vectors connecting the nitrogen of the piperazine to the ethoxy group (reported every 100 frames, Fig. 4e and Supplementary Fig. 15). When only one HT molecule is present, it is free to explore the wide BmrCD cavity (Fig. 4e, gray curve). In contrast, when two HT molecules are bound (Fig. 4e, silver), the first one becomes more flexible and starts packing against the second one (as shown by the blue and magenta vectors which tend to cluster together in Supplementary Fig. 15c). The presence of lipid molecules prevents substrate packing and pushes the first substrate towards the extracellular half of the protein at the innermost region of the vestibule, its apex (Supplementary Fig. 15c). Once it pushes the substrate, the lipid molecule leaves the interface presumably allowing the transporter to switch from inward-open to inward-occluded prior to transitioning to the outward open conformation to extrude the substrates.
Discussion
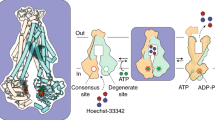
The structures presented here, in conjunction with previous structures, paint an almost complete model of the conformational landscape of BmrCD at near atomic resolution in a lipid bilayer (Fig. 5). In this model, the substrate-coupled cycle includes previously8 described stepwise binding of Hoechst and Mg2+. cryo-EM structures of BmrCD IF without and with Hoechst, along with corresponding DEER data, demonstrate the lack of major conformational changes upon binding of Hoechst (steps 1 to 3).
BmrCD is shown in a cartoon with helices represented by cylinders. Hoechsts, lipids: lipids POV23 in IF(2HT/2ATP) and POV15 in OF(2HT), and ATP/ADPVi are shown by ball-and-stick model. The rest of the lipids are shown in sticks. Mg2+ ions are shown in green ball. Red arrows indicate the conformational changes. Four steps of the transport cycle are indicated by circled numbers. The structure models are as follows: IF(1ATP), AlphaFold3 generated model (See details in “Methods”); IF(2ATP), PDB 8FPF; IF(1HT/2ATP), PDB 8SZC; IF(2HT/2ATP), BmrCD_IF2; OF(2HT), BmrCD_OF; OC, PDB 8T1P.
ATP hydrolysis, primarily in the consensus NBS, triggers transition to the OF conformation. B-factor analysis (Supplementary Fig. 16) shows that the NBDs are dynamic in the IF state prior to formation of the catalytic NBSs, whereas the ECD becomes dynamic in the OC and OF states. Following dissociation of Hoechst, the transporter transitions to the OC conformation (step 4). Previous DEER analysis demonstrates that the population of the OF conformation in lipid nanodiscs requires the binding of Hoechst8,17. The stability of the drug-bound OF conformation in BmrCD as opposed to other ABC exporters can be attributed, among other factors to the ECD, which may increase the energetic cost of drug release, and the angle of extracellular opening, which is smaller than other ABC exporters, possibly promoting stronger substrate interactions with the transporter (Supplementary Fig. 17 and Supplementary Table 5). Consistent with the notion of a role for the ECD, a BmrCD construct lacking this domain loses the stimulation of ATP turnover by Hoechst22.
Unique to our model is the profiling of interactions that stabilize bound substrates to both IF and OF intermediates. Contacts between Hoechst and the two protomers are asymmetric, with BmrC contributing the majority of interacting residues. This finding echoes the asymmetry that shapes the conformational cycle previously noted8,22 and reinforced here by the structure of the OF conformation. The BmrC side of the V-shaped extracellular vestibule is closer to the substrate (3.4 Å), display a more flexible TM1, and an opening near the exit. In contrast, the BmrD side is further from the substrate (8.9 Å), has a less flexible TM1, and an electrostatic interaction between K40BmrD and R381BmrD that may obstruct the exit (Supplementary Fig. 18).
Starting from lipid molecules bound at the entrance of the OF vestibule, the MD simulations delineate a potential mechanism by which lipids can act to disrupt the interactions of the Hoechst molecules with the transporter. While the driving force for movement of the substrate from the IF to the OF vestibule is contributed overwhelmingly by the energy of ATP binding and hydrolysis that power conformational changes, a mechanism is needed for the release from the OF conformation. Functional and spectroscopic data of BmrCD harmonize with our model8,17,22. Formation of the OF conformation for BmrCD in lipids requires Hoechst binding. In addition, the OF stability is reduced upon transition from detergent micelles to lipid bilayers8,17,22. Although MD simulations did not show Hoechst release, presumably due to the limited time scale of simulations, they did reveal that Hoechst binding is destabilized by the mutation of D141, a residue implicated in Hoechst binding to the IF and OF conformations.
The energetic cost of substrate release is dependent on the strength of its interactions with the OF conformation. It is possible that despite the higher concentration of substrate on the release side of the membrane, its intrinsic affinity is weak enough such that its dissociation is spontaneous. Such spontaneous dissociation was previously observed in MD simulations of TM287/TM28824 and was ascribed to changes in water hydration in the OF cavity. Alternatively, for those transporters wherein ATP hydrolysis occurs after the formation of the OF conformation, the energetic drive to reset to the IF conformation could also overcome low-affinity binding of the substrate to the OF state.
These models, however, do not explicitly include the energetic contribution of lipids. The amphipathic substrates of multidrug ABC transporters tend to be soluble in the lipid phase. Indeed, early models of ABC transporters posited a hydrophobic “vacuum cleaner” model where substrate molecules are bound from the inner leaflet of the membrane and extruded into the outer leaflet26. Similarly, a lipid bound in the substrate cavity of the multidrug transporter LmrP was interpreted to reflect a mechanism for polyspecificity27. Lipids are amphipathic, and their high concentrations can overcome their intrinsic low affinity to the transporters, thereby driving binding to the transporter. The MD simulations establish that lipids loosen stabilizing interactions between the transporter and substrate prior to release.
Thus, we propose a dynamic “competition” model between substrates and lipids for multidrug ABC exporters. We posit that this competition results from the modest affinity of the substrates to these polyspecific transporters and the high concentration of lipids, which have access to the rather wide vestibules in both IF and OF conformations. The constant entry and exit of lipid molecules disrupt interactions with bound substrates, although the weak lipid affinity leads to short dwell times in the vestibule of the transporter. This model is supported by previous MD simulations for other ABC transporters in the IF conformations, where lipids spontaneously enter the vestibule25,28. This dynamic binding, along with disorder in the bound state, may hinder the ability to visualize lipids in the vestibule by structural techniques. Rigorous testing of this model requires a systematic study of BmrCD structure and dynamics in bilayers of different lipid composition. Finally, such a lipid-induced modulation of substrate affinity may be a general mechanism for other ABC exporters that are specific to amphipathic substrates.
Methods
BmrCD cloning, expression, and purification
Wild-type BmrCD was cloned in the pET21b(+) plasmid and then transformed into Escherichia coli BL21(DE3) cells to be expressed in minimal media supplemented with 0.5% v/v glycerol, 2.5 μg/ml thiamin, 100 μg/ml ampicillin, 1 mM MgSO4, and 50× MEM amino acids. Cell cultures were grown at 37 °C until the optical density at 600 nm (OD600) reached 1.2. Subsequently, induction of BmrCD expression at 25 °C was initiated by adding 0.7 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Cells were harvested after 5 h of expression and stored at −80 °C till purification.
For BmrCD purification, cell pellets were resuspended in a lysis buffer containing 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 1 mM PMSF, and a complete EDTA-free protease inhibitor cocktail tablet (Roche) before undergoing sonication. The lysate was centrifuged at 9000 × g for 10 min at 4 °C, yielding the supernatant utilized for high-speed centrifugation at 185,000 × g for 1 hat 4 °C to precipitate membrane pellets. To solubilize the membrane pellets, a resuspension buffer comprising 50 mM Tris-HCl (pH 8.0), 0.1 M NaCl, 15% v/v glycerol, 1 mM DTT, and 1.25% w/v n-dodecyl-β-D-maltopyranoside (β-DDM) was applied for 1 h on ice. Following this, centrifugation at 185,000 × g for 1 h at 4 °C separated insoluble material, and the supernatant was combined with pre-equilibrated Ni-NTA beads for a 2 h incubation in binding buffer (50 mM Tris-HCl, pH 8.0, 0.1 M NaCl, 15% v/v glycerol, 0.05% β-DDM). After resin washing with binding buffer supplemented with 20 mM imidazole, protein elution was performed using binding buffer containing 250 mM imidazole.
Subsequently, protein purification involved size exclusion chromatography (SEC) with a Superdex 200 Increase 10/300 column (Cytiva). The SEC buffer 1, which consisted of 50 mM Tris-HCl (pH 7.5), 0.15 M NaCl, 10% v/v glycerol, and 0.05% β-DDM, was used for equilibration.
BmrCD nanodiscs preparation
The preparation of BmrCD nanodiscs followed the procedure outlined previously8. In summary, lipids consisting of PC (L-α phosphatidylcholine) (Cat #: 840051, Avanti Polar Lipids, Alabaster, USA) and PA (L-α phosphatidic acid) (Cat #: 840101, Avanti Polar Lipids, Alabaster, USA) were combined in a molar ratio of 9:1. The BmrCD nanodiscs were then created by mixing lipids, MSP1D1, and BmrCD micelles at a molar ratio of 1800:360:3:1 (β-DDM:lipid:MSP1D1:BmrCD). The mixtures were incubated for 30 minutes before adding biobeads (0.8-1 g/ml) and rotating at 4 °C overnight. Following the removal of biobeads by centrifugation, BmrCD in nanodiscs was filtered through a 0.45 μm filter and subsequently purified via size exclusion chromatography using a Superdex 200 Increase 10/300 column (Cytiva) with SEC buffer 2 (50 mM Tris-HCl, pH 8.0, 0.15 M NaCl).
Cryo-EM sample preparation and data acquisition
Reconstituted BmrCD at a concentration of approximately 3 mg/ml was incubated at 37 °C for 15 min with a mixture containing 10 mM ATP, 4 mM MgSO4, 4 mM vanadate, and 1.2 mM Hoechst-33342 (Thermo Fisher Scientific) before plunging. Cryo-EM grids were prepared by applying 2.5 μl of the sample to a Quantifoil UltrAuFoil grid (R1.2/1.3, 300 mesh, Electron Microscopy Sciences) using a Vitrobot Mark IV (Thermo Fisher), pre-equilibrated at 4 °C with 100% humidity. Grids suitable for cryo-EM data collection were selected through screening by the Glacios system (Thermo Fisher Scientific).
Cryo-EM data collection was conducted using a 300 keV Titan Krios G4 microscope (Thermo Fisher Scientific) equipped with a Gatan K3 direct electron camera. Movies containing 50 frames were recorded with three exposures per hole using EPU in normal mode. The magnification was set at 130,000 with a physical pixel size of 0.647 Å/pixel, and defocus values were within the range of − 0.9 to − 2.0 μm. The total electron dose over the sample during data collection was 56 e-/Ų.
Cryo-EM data processing
The cryo-EM dataset was processed in cryoSPARC29, implemented with MotionCor230 for beam-induced motion correction and Gtcf31 for CTF estimations. For BmrCD_OF, BmrCD_OC was used as a template for the Template picker to identify approximately 6.6 million particles, followed by 2D classification, heterogeneous refinement, and Ab initio to obtain high-quality particles. Followed by obtaining the particle images for occluded conformation, one round of 3D classification was performed to get 27,802 out of 185,953 particles to generate the outward facing conformation map (Supplementary Fig. 2). Subsequently, non-uniform32 and local refinement were applied, resulting in a final map with a resolution of 3.11 Å. For BmrCD_IF1 and IF2, similar strategies were applied (See details in Supplementary Figs. 11, 12 and 13).
All structural models refinement and validation were performed using Real-space refinement33 and MolProbity34 implemented in Phenix 1.21.1-528635. Coot36 was applied to build the models. The predicted structure model of BmrCD_IF(1ATP) was generated by AlphaFold 337 with input files of the sequences of BmrC and BmrD, as well as 1 copy of ATP in autoseed mode. Visualization of the results and preparation of figures were carried out using UCSF Chimera 1.1838, ChimeraX 1.439,40,41, and PyMol 2.4.1. In addition, the B-factor was calculated using the B-factor putty tool implemented in PyMol 2.4.1 with default settings. Solvent-accessible volume calculations were conducted using CASTp42. Radius measurement of the vestibule was performed using the software HOLE43.
Molecular dynamics simulations
MD simulations of the OF state (BmrCD_OF)
Constructing the initial model
Starting with the cryo-EM model, we first completed partially resolved lipids (Supplementary Data 1). To do this, we used the PSFGEN utility in VMD44 with the CHARMM36 forcefield45,46. Protein-lipid clashes with the built lipid moieties were resolved by a brief energy minimization of 2000 steps in NAMD347. The D141A mutation was modeled using mutate.tcl in VMD44. The resulting wild-type (WT) and mutant models were then embedded in membranes generated with CHARMM-GUI48. Each membrane consisted of a 9:1 ratio of POPC:POPA, in accordance with the nanodisc composition used in the experiments, and was solvated with TIP3P49 water and ionized to 150 mM NaCl in VMD44.
Equilibrium molecular dynamics
All molecular dynamics (MD) simulations were performed using NAMD347. The equilibration protocol included the following: (i) 10,000 steps of energy minimization; (ii) Gentle heating increasing the temperature by 25 K at a time for 1000 steps of MD until the final temperature of 310 K was reached; (iii) 2.5 ns of equilibration with harmonic restraints (k = 1.5 kcal mol−1 Å−2) on the heavy atoms of the lipid headgroups, the protein backbone, Hoechst (HT) molecules, and ATP and coordinating Mg2+ ions; (iv) 2.5 ns of equilibration with harmonic restraints (k = 1.0 kcal mol−1 Å−2) on the heavy atoms of the protein backbone, HT molecules, and the ATP and coordinating Mg2+ ions; (v) 2.5 ns of equilibration with harmonic restraints (k = 0.5 kcal mol−1 Å−2) on the protein backbone and HT molecules; and, (vi) 5.0 ns of unrestrained equilibration. After equilibration, four replicates for the WT and three replicates for the mutant were simulated, each for 2 µs, resulting in an aggregate of 14 µs simulation data. Simulation parameters included a 12-Å cutoff distance for long-range interactions, a 10-Å switching distance, a 2-fs timestep, the particle mesh Ewald (PME) method for electrostatics50, Nosé-Hoover Langevin piston for pressure coupling with a target of 1 atm, oscillation period of 100 fs and decay of 50 fs, and Langevin dynamics with a damping coefficient of 1 ps−1 for controlling the temperature51.
Analysis
To investigate the influence of lipids on HT binding and release, we first applied kernel density estimation (KDE) to map the density distribution of lipid molecules, thereby identifying high-occupancy regions of phospholipids. Building on this initial analysis, we then measured native contacts and non-bonded interaction energies between HT and BmrCD using the NAMD Interaction Energy module in VMD44,47, which allowed us to assess lipid-induced perturbations in substrate binding. Furthermore, we analyzed the distribution of HT tilt angle and changes in salt bridges to elucidate lipid-induced conformational shifts. Next, we evaluated the structural dynamics by measuring the backbone root-mean-square deviation (RMSD) of BmrCD transmembrane helices over time, revealing rearrangements associated with lipid binding and substrate displacement. In addition, we quantified lipid–substrate interactions by tracking the number of lipid heavy atoms within 3.5 Å of HT. Finally, we calculated the minimum distance between E51 and E75 of BmrD and HT over time, as well as the distribution of extracellular domain (ECD) domain angles relative to the negative z axis (0, 0, − 1).
To determine the binding stability of HT-1 and HT-2 molecules in the binding pocket of BmrCD, we calculated the RMSD of each ligand relative to its initial binding pose over time. Prior to RMSD calculation, each frame of the trajectory was aligned to the first frame using the backbone atoms of the binding pocket residues (defined as all amino acid residues within 8 Å of the ligand’s center of mass: residues 29–45, 48, 78–83, 107, 134–149, 237, 241, 279–298, 338, 341–345, 349, 370, 372–385, 387–388, and 391). This alignment removes global translational and rotational motions of the protein and its binding region, ensuring that RMSD reflects only the movement of the ligand within the binding site. The ligand’s RMSDs were then averaged over three replicate simulations for each system. To quantify ionic interactions, ion pairs between positive or negative residues and the ligands within 5.0 Å, were considered.
In order to assess the behavior of the HT molecules when not in contact with lipids, in some simulations, the lipids were prevented from entering the protein binding pocket while allowing natural membrane fluctuations. To do so, positional restraints were applied to all lipid atoms within 30 Å of the pocket. These restraints were defined using the distanceXY colvars in NAMD, with reference to the center of mass (COM) of the binding pocket. A flat-bottom harmonic potential was applied with a narrow restraint window between 21.0 Å and 39.0 Å, corresponding to the effective pocket boundary and its immediate surroundings. The lower and upper wall force constants were set to 7.0 and 1.0 kcal mol−1Å−2, respectively, to gently confine lipids within this annular region while avoiding excessive forces that may perturb the membrane structure. The restraints were applied in the XY plane (forceNoPBC yes in NAMD) to maintain flexibility along the membrane normal (z-axis). A harmonic potential with a force constant of 100 kcal mol-1Å−2 was applied to restrain the COM of the protein relative to a fixed reference point at (0.0, 0.0, 0.0), in order to prevent lateral drift and maintain proper alignment of the protein within the membrane. To account for lipid diffusion during simulation, the list of restrained lipid atoms was updated every 50 ns using a custom script. This adaptive strategy ensures that only lipids adjacent to the pocket are restrained. The analyses were performed using custom scripts either in VMD44 or with Python 3.9 using the following packages: SciPy52, MDAnalysis 2.8.053, and Seaborn54. Plots were generated with Matplotlib. Analysis codes can be accessed through https://github.com/YYYYYunsen/Analysis-code-of-BmrCD.
Molecular dynamics simulations for BmrCD_IF1 and BmrCD_IF2
Protein structure construction
Three distinct BmrCD protein structures were employed here, specifically pdb IDs 8FMV (BmrCD_IF) and 8SZC (BmrCD_1HT/ATP)8, alongside a solved structure, BmrCD_IF2. These structures, chosen for their relevance to the BmrCD transporter mechanism, included variations in ligand binding—two structures with dual Hoechst (HT) molecules and one with a single HT molecule (Supplementary Data 2). The determined BmrCD_IF2 revealed some of the bound lipid molecules, especially at the interface between transmembrane helices 4 and 6 (TM4/6). To elucidate the role of these lipids, two different simulation setups were prepared: one including the 27 bound lipids (BmrCD_IF2) and one without them (BmrCD_IF2_nolipid) (Supplementary Table 4).
The Molecular Operating Environment (MOE) software, v2022.0255 was utilized to model missing loops for the protein and missing heavy atoms for the solved lipid molecule. Protonation of the side chains were determined according to the physiological pH (7.4).
System preparation and molecular dynamics simulation
The Amber22 software56 was used to prepare each system. The packmol-memgen tool facilitated membrane embedding in a lipid bilayer with the same lipid composition as in the OF system. The systems were solvated in a 150 mM NaCl solution, with topologies and initial coordinates generated via tleap56 using the amber19SB force field for the protein, lipid21 for the lipid, and the OPC water model. All structures were simulated with ATP and Mg2+ bound at the consensus site, and ATP alone at the degenerate site. The ATP topologies were sourced from the Bryce group’s website, utilizing parameter sets detailed by Meagher et al.57.
Ligand preparation
The Hoechst molecule was computationally refined using Gaussian 1658 to ensure accurate molecular interactions in subsequent simulations. Geometry optimization was first performed with the B3LYP functional combined with the 6-31 G* basis set59,60, enforcing a stringent (tight) convergence criterion on the self-consistent field (SCF) calculations. Following this step, the electrostatic potential (ESP) was calculated using the Hartree-Fock method with a 6-31 G* basis set, coupled with Merz-Kollman population analysis to accurately determine the electrostatic potential surface. Subsequently, the Amber22 Antechamber tool56 was employed to fit RESP charges to the optimized structure, which were then utilized in molecular dynamics simulations to accurately model HT’s behavior.
Moleculardynamics protocols
Each prepared system was subjected to an initial energy minimization to relieve any steric clashes or unfavorable geometries, followed by a canonical ensemble (NVT) heating step where the system was gradually heated from 0 to 310 K over 200 ps, and then sustained at this target temperature for an additional 50 ps. The NVT phase employed a 1 fs timestep with positional restraints applied to all heavy atoms, excluding water oxygens and ions in solution. Upon reaching the desired temperature, the systems were subjected to the isothermal-isobaric (NPT) equilibration phase to stabilize pressure and density, using semi-isotropic pressure scaling. The positional restraints on proteins, ligands (inclusive of ATP, Mg2+, and HT molecules), and lipid head groups were gradually released over a period of 10 ns. This stepwise easing of restraints—from 10.0 kcal/mol/Å2 applied to the heavy atoms of the protein and ligands and 2.5 kcal/mol/Å2 applied to the lipid head groups—culminated in the complete removal of restraints for the last equilibration step prior to the NPT production run of ~ 4μs. Such a gradual approach facilitated orderly water diffusion, membrane equilibration, and stabilization of protein binding sites and ligands. Temperature control was done using Langevin dynamics61, stabilizing the system at 310 K. The pressure was controlled at 1 bar using the Monte Carlo Barostat62, which operates with a friction coefficient of 1 ps−1. The timestep for the simulations was set at 1 fs during the initial NVT and first NPT equilibration steps and was increased to 2 fs for the remainder of the equilibration stages and the production phase. The SHAKE algorithm63 was used to constrain hydrogen atoms, ensuring the stability of each simulation, and allowing for a larger timestep. Long-range electrostatic interactions were calculated via the particle mesh Ewald method, while the cutoff for van der Waals and Coulombic interactions was set between 10 and 12 Å to ensure accurate force field representation.
Analysis were performed using cpptraj from the AmberTools23 package suite or custom scripts in VMD44 using the tcl scripting language. Plots were generated using custom Python scripts and matplotlib.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The cryo-EM maps of BmrCD_OF, BmrCD_IF1, and BmrCD_IF2 have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-45940, EMD-45938, EMD-45939, respectively. The atomic coordinates of BmrCD_OF, BmrCD_IF1, and BmrCD_IF2 have been deposited in the Protein Data Bank (PDB) under accession codes 9CUS, 9CUP, and 9CUR, respectively. For MD simulation, initial coordinates, simulation input files and coordinate files of the final output are provided as Supplementary Data 1 and 2. Code is available from GitHub [https://github.com/YYYYYunsen/Analysiscode-of-BmrCD]. Source data are provided in this paper.
References
Beek, J., Guskov, A. & Slotboom, D. J. Structural diversity of ABC transporters. J. Gen. Physiol. 143, 419–435 (2014).
Rees, D. C., Johnson, E. & Lewinson, O. ABC transporters: the power to change. Nat. Rev. Mol. Cell Biol. 10, 218–227 (2009).
Locher, K. P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016).
Thomas, C. & Tampe, R. Structural and mechanistic principles of ABC transporters. Annu Rev. Biochem 89, 605–636 (2020).
Higgins, C. F. & Linton, K. J. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 11, 918–926 (2004).
Shvarev, D., Januliene, D. & Moeller, A. Frozen motion: how cryo-EM changes the way we look at ABC transporters. Trends Biochem. Sci. 47, 136–148 (2022).
Hofmann, S. et al. Conformation space of a heterodimeric ABC exporter under turnover conditions. Nature 571, 580–583 (2019).
Tang, Q. et al. Asymmetric conformations and lipid interactions shape the ATP-coupled cycle of a heterodimeric ABC transporter. Nat. Commun. 14, 7184 (2023).
Seeger, M. A. & van Veen, H. W. Molecular basis of multidrug transport by ABC transporters. Biochim. Biophys. Acta 1794, 725–737 (2009).
Kim, Y. & Chen, J. Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation. Science 359, 915–919 (2018).
Johnson, Z. L. & Chen, J. Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell 168, 1075–1085 (2017).
Alam, A., Kowal, J., Broude, E., Roninson, I. & Locher, K. P. Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science 363, 753 (2019).
Johnson, Z. L. & Chen, J. ATP Binding enables substrate release from multidrug resistance protein 1. Cell 172, 81–89 (2018).
Dawson, R. J. & Locher, K. P. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 581, 935–938 (2007).
Chaptal, V. et al. Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter. Sci. Adv. 8, eabg9215 (2022).
Gewering, T. et al. Tracing the substrate translocation mechanism in P-glycoprotein. Elife 12, https://doi.org/10.7554/elife.90174 (2024).
Mishra, S. et al. Conformational dynamics of the nucleotide binding domains and the power stroke of a heterodimeric ABC transporter. Elife 3, e02740 (2014).
Galian, C. et al. Optimized purification of a heterodimeric ABC transporter in a highly stable form amenable to 2-D crystallization. PLoS ONE 6, e19677 (2011).
Reilman, E., Mars, R. A., van Dijl, J. M. & Denham, E. L. The multidrug ABC transporter BmrC/BmrD of Bacillus subtilis is regulated via a ribosome-mediated transcriptional attenuation mechanism. Nucleic Acids Res. 42, 11393–11407 (2014).
Dezi, M., Di Cicco, A., Bassereau, P. & Levy, D. Detergent-mediated incorporation of transmembrane proteins in giant unilamellar vesicles with controlled physiological contents. Proc. Natl. Acad. Sci. USA 110, 7276–7281 (2013).
Torres, C., Galian, C., Freiberg, C., Fantino, J. R. & Jault, J. M. The YheI/YheH heterodimer from Bacillus subtilis is a multidrug ABC transporter. Biochim. Biophys. Acta 1788, 615–622 (2009).
Thaker, T. M. et al. Asymmetric drug binding in an ATP-loaded inward-facing state of an ABC transporter. Nat. Chem. Biol. 18, 226–235 (2022).
Di Cesare, M. et al. The transport activity of the multidrug ABC transporter BmrA does not require a wide separation of the nucleotide-binding domains. J. Biol. Chem. 300, 105546 (2024).
Goddeke, H. & Schafer, L. V. Capturing substrate translocation in an ABC exporter at the atomic level. J. Am. Chem. Soc. 142, 12791–12801 (2020).
Wen, P. C., Verhalen, B., Wilkens, S., McHaourab, H. S. & Tajkhorshid, E. On the origin of large flexibility of P-glycoprotein in the inward-facing state. J. Biol. Chem. 288, 19211–19220 (2013).
Jones, P. M. & George, A. M. The switch and reciprocating models for the function of ABC multidrug exporters: perspectives on recent research. Int. J. Mol. Sci. 24, https://doi.org/10.3390/ijms24032624 (2023).
Debruycker, V. et al. An embedded lipid in the multidrug transporter LmrP suggests a mechanism for polyspecificity. Nat. Struct. Mol. Biol. 27, 829–835 (2020).
Rasouli, A. et al. Differential dynamics and direct interaction of bound ligands with lipids in multidrug transporter ABCG2. Proc. Natl. Acad. Sci. USA 120, e2213437120 (2023).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D. Struct. Biol. 74, 531–544 (2018).
Williams, C. J. et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Meng, E. C. et al. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Tian, W., Chen, C., Lei, X., Zhao, J. & Liang, J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 46, W363–W367 (2018).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph 14, 354–376 (1996).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph 14, https://doi.org/10.1016/0263-7855(96)00018-5 (1996).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010).
Phillips, J. C. et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 153, 044130 (2020).
Wu, E. L. et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 (2014).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995).
Martyna, G. J., Tobias, D. J. & Klein, M. L. Constant-pressure molecular-dynamics algorithms. J. Chem. Phys. 101, 4177–4189 (1994).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Michaud-Agrawal, N., Denning, E. J., Woolf, T. B. & Beckstein, O. Software news and updates MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 32, 2319–2327 (2011).
Waskom, M. L. seaborn: statistical data visualization. J. Open Source Softw. 6, 4 (2021).
Molecular Operating Environment (MOE), C. C. G. U., 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7 (2022).
Case, D. A. et al. Amber 2023, University of California, San Francisco, San Francisco, CA. (2023).
Meagher, K. L., Redman, L. T. & Carlson, H. A. Development of polyphosphate parameters for use with the AMBER force field. J. Comput. Chem. 24, 1016–1025 (2003).
Frisch, M. J. T. et al. Gaussian 16, Revision A.03, Gaussian, Inc., Wallingford, CT. (2016).
Stephens, P. J., Devlin, F. J., Chabalowski, C. F. & Frisch, M. J. Ab-initio calculation of vibrational absorption and circular-dichroism spectra using density-functional force-fields. J. Phys. Chem. 98, 11623–11627 (1994).
Hehre, W. J., Ditchfield, R. & Pople, J. A. Self-consistent molecular-orbital methods. XII. further extensions of Gaussian-type basis sets for use in molecular-orbital studies of organic-molecules. J. Chem. Phys. 56, 2257 (1972).
Loncharich, R. J., Brooks, B. R. & Pastor, R. W. Langevin dynamics of peptides: the frictional dependence of isomerization rates of N-acetylalanyl-N’-methylamide. Biopolymers 32, 523–535 (1992).
Åqvist, J., Wennerström, P., Nervall, M., Bjelic, S. & Brandsdal, B. O. Molecular dynamics simulations of water and biomolecules with a Monte Carlo constant pressure algorithm. Chem. Phys. Lett. 384, 288–294 (2004).
Kräutler, V., Van Gunsteren, W. F. & Hünenberger, P. H. A fast SHAKE:: Algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 22, 501–508 (2001).
Acknowledgements
We acknowledge the use of the Glacios cryo-TEM for grids screening, which was acquired by NIH S10 award OD030292-01 and the cryo-EM data were collected at the Center for the Structural Biology Cryo-EM Facility at Vanderbilt University. We appreciate Erkan Karakas for his help in structure analysis and Richard A. Stein for discussions. This work was supported by National Institutes of Health grant 3R35GM152382 to H.S.M. and P41-GM104601 and R24-GM145965 to E.T.
Author information
Authors and Affiliations
Contributions
Q.T. performed all the experiments. M.S. and Y.Z. performed the MD simulations for BmrCD_OF under the supervision of E.T. P.B. performed the MD simulations for BmrCD_IF1 and BmrCD_IF2. Q.T. and H.S.M. designed the research, analyzed and interpreted the structures and wrote the manuscript with input from all the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tang, Q., Sinclair, M., Bisignano, P. et al. Drug-bound outward-facing conformation of a heterodimeric ABC exporter suggests a putative mechanism of drug translocation. Nat Commun 16, 10403 (2025). https://doi.org/10.1038/s41467-025-65318-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65318-6