Abstract
Self-aggregation into biofilms is a bacterial stress response that promotes antimicrobial resistance because biofilms comprise viscous extracellular polymeric matrices that impede antimicrobial diffusion. Extracellular DNA (eDNA) is typically a principal component of the biofilm matrix. Here we show that persistent R-loops, which are three-stranded nucleic acid structures consisting of single DNA and a DNA:RNA hybrid, contribute to the viscoelastic behaviour of eDNA in Pseudomonas aeruginosa biofilms. The RNA strands are inserted throughout the genome by the strand exchange protein RecA, at locations in the genome distant from the site of their own transcription i.e. in trans. R-loop formation creates genomic instability in bacterial cells that subsequently die and release R-loops. These events appear to occur as part of a programmed cell death pathway, which is activated by the stringent stress response. The released R-loops become building blocks of the viscoelastic extracellular matrix, for the benefit of the remaining population. Our results indicate that R-loops facilitate the formation of the viscoelastic eDNA matrix in the context of bacterial stress responses, and that interfering with the R-loops may provide a broadly effective strategy for biofilm control.
Similar content being viewed by others
Introduction
The importance of extracellular DNA (eDNA) as a key structural component of Pseudomonas aeruginosa biofilm matrices, alongside extracellular polysaccharides and proteins, was first reported more than twenty years ago1,2. It was subsequently demonstrated that eDNA is the principal viscoelastic component of the extracellular matrix of P. aeruginosa as well as other pathogens, including Staphylococcus epidermidis, Streptococcus mutans, uropathogenic Escherichia coli (UPEC) and non-typeable Haemophilus influenzae (NTHI)3,4,5,6. Bacteria within biofilms express different phenotypes than their planktonic counterparts, including increased stress tolerance, and the presence of eDNA is a major contributor to increased antimicrobial resistance.7,8,9,10,11. Understanding the origins of viscoelastic eDNA networks in bacteria is therefore critically important for developing biofilm control technologies.
While sequentially very similar to chromosomal DNA (cDNA), there are several characteristic higher-order structural features that differentiate eDNA from cDNA and possibly suggest a mechanism by which DNA strand exchange and subsequent crosslinking into viscoelastic eDNA matrix occurs. In NTHI, UPEC and S. epidermidis biofilms, binding of eDNA to DNABII proteins, integration host factor (IHF) and histone-like proteins promotes stable Holliday Junction-like eDNA crossed-strands, which contribute to eDNA assembling an extracellular network12. The DNABII proteins also stabilise eDNA in the rare Z form, rather than the B form, in which the Z form of eDNA promotes biofilm rather than planktonic growth6. The ability of P. aeruginosa biofilm eDNA to form viscoelastic cross-linked structures is due to G-quadruplex DNA3, which is also a critical component of the eDNA matrix of S. epidermidis biofilms13. Binding of eDNA to other molecules, such as the exopolysaccharide Pel, can also increase the mechanical stability of P. aeruginosa biofilms14. Moreover, extracellular RNA (eRNA), which binds to eDNA as extracellular RNA:DNA hybrids, contribute to the primary biofilm structural matrix of model P. aeruginosa biofilms, and the sputum of chronic lung disease patients infected with P. aeruginosa15. It is not clear, however, how extracellular nucleic acid packaging is regulated to accomplish its characteristic structures, including RNA:DNA hybrids, nor is it known whether eDNA assembly is determined by exogenous physicochemical factors16,17,18,19.
RNA:DNA hybrids are intermediates in many biological processes (e.g., replication and gene expression). They are also produced by homologous recombination and DNA double-strand break repairs, as for Holliday Junction intermediates, which were previously described as viscoelastic eDNA matrix constituents12. If left unresolved, however, RNA:DNA hybrids can disrupt DNA repair processes and cause genomic instability20. We aimed to establish here whether extracellular RNA:DNA hybrids were the result or cause of homologous recombination, which we hypothesised would inform on how bacteria assemble their viscoelastic eDNA networks. We subsequently show that the RNA:DNA hybrids are present in biofilms as extracellular R-loops. R-loops are three-stranded nucleic acid structures comprising RNA:DNA hybrids and a displaced DNA strand that can threaten genomic stability if left unresolved.
In trans R-loops are formed when RNA molecules, either transcribed from distant loci or introduced externally, hybridize with complementary DNA at separate genomic or extracellular sites. This distinguishes them from transient transcriptional intermediates such as in cis R-loops (those R-loops formed co-transcriptionally, where the RNA hybridises to its template DNA strand at the same genomic locus)21. In contrast to in cis R-loops, the mechanism for in trans R-loop formation is poorly understood22. Nonetheless, in trans R-loops were previously shown to be promoted in yeast by eukaryotic strand exchange protein Rad51p, which led to genomic instability23,24. We find here that the bacterial equivalent RecA performs a similar function in P. aeruginosa upon induction of the stringent stress response (SSR). This led us to develop a model for viscoelastic biofilm matrix formation, where genomic instability occurs in a subset of suicide cells that subsequently release R-loops as building blocks for extracellular matrix assembly upon lysis. Thus, the stringent stress response, long understood to lead to biofilm formation25,26, is intrinsically linked to viscoelastic eDNA matrix production through R-loop production. These findings will likely inform on and deliver new strategies for managing biofilm growth.
Results
Extracellular R-loops impart viscoelasticity to biofilms
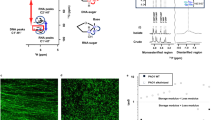
R-loops are a common source of RNA:DNA hybrids in cells. The displaced single strand of R-loops is additionally characterised by the presence of G-quadruplexes, which are also features of viscoelastic biofilm networks27. We thus sought to ascertain whether extracellular R-loops account for the previous observation of eRNA:DNA hybrids in P. aeruginosa biofilms15. We performed immunofluorescence microscopy on the biofilms using an R-loop-specific S9.6 antibody that detects RNA:DNA hybrids regardless of sequence28. R-loops co-localised with eDNA fibres in the matrix of mature (5 d) P. aeruginosa biofilms (Fig. 1a). The R-loop content in eDNA fibres increased with the period of biofilm growth (Supplementary Fig. 1a), and there was no non-specific binding of R-loops to other regions of the matrix. 26 ± 5.7 % of eDNA fibres overlapped with an R-loop signal in mature P. aeruginosa biofilms (Supplementary Fig. 1b), which is the same biofilm model used previously to describe the presence of eRNA:DNA hybrids15. 70 ± 0.07% of R-loops colocalised with eDNA fibres, and the remainder colocalised with cells (Supplementary Fig. 1b). No extracellular R-loop signals were observed in planktonic cells (Supplementary Fig. 1c). Secondary fluorophore staining of biofilms without the S9.6 antibody showed no signal throughout the biofilms, indicating that the matrix signal resulted from binding of primary antibody (Supplementary Fig. 1d). Therefore, the eDNA matrix of the model P. aeruginosa biofilm is characterized by the presence of R-loops.
Confocal micrographs of mature (5 d) P. aeruginosa PAO1 biofilms (n = 4) represented in two dimensional (2D) images showing eDNA fibres (TOTO-1, green; left) and extracellular R-loops (S9.6 antibody, red; middle) as well as merged images (yellow; right) in PAO1 wildtype (a) without enzymatic treatment, (b) after RNase H digestion (0.3 mg/mL, 37 °C, 30 min) (c) after Nuclease P1 digestion (0.3 mg/ml, 37 °C, 30 min) and (d) after RNase H (0.3 mg/ml, 37 °C, 30 min) followed by nuclease P1 (0.3 mg/mL, 37 °C, 30 min). The Manders coefficient for the proportion of eDNA fibres containing R-loops was 26.8 ± 3.4 %. e Fluorescence intensity quantification (n = 4) of eDNA (green bars) and R-loops (red bars) expression in mature P. aeruginosa wildtype biofilms treated with and without enzymatic digestion. f Rheogram for frequency sweep of mature P. aeruginosa PAO1 wildtype biofilm with and without enzymatic treatment, (25 °C, 0.026 mm gap, 0.3 amplitude). Biological triplicates (n = 3) were averaged for each condition and plotted against frequency. g, h Confocal micrographs of mature P. aeruginosa ΔtopA and ΔrnhB PAO1 transposon mutant biofilms (n = 6) represented in 2D micrographs showing extracellular R-loops (S9.6 antibody, red), eDNA fibres (TOTO-1, green) and merged images (yellow). i Quantification of biofilm biomass of PAO1 wildtype, ΔtopA and ΔrnhB mutants from days 2 to 5 using the crystal violet assay (biological triplicate, n = 3). j Fluorescence intensity quantification (n = 6) of percentage eDNA fibres containing R-loops (red bars) in mature (5 d) PAO1 wildtype, ΔtopA and ΔrnhB biofilms. k Fluorescence intensity quantification (n = 5) of percentage of eDNA fibres containing R-loops in mature untreated (red bars) and RNase H + nuclease P1-treated (yellow bars) S. epidermidis biofilms and COPD infected lung sputum highly enriched with P. aeruginosa. All three-dimensional (3D) biofilms were imaged with a Z-stack thickness of 10 µm. For Fig. 1F, error bars represent mean ± standard deviation (SD). The centre point indicates the mean of the data. n represents the number of biological replicates, with the exception of microscopy images and data analysis, where it is number of representative microscopy images collected. For microscopy data analysis, all images were acquired across biological triplicates, and the statistics were determined based on the number of representative images collected. All scale bars represent 10 µm. P < 0.05 for differences between and within treatment groups. Source data are provided as a source data file.
There was a reduction in eDNA fluorescence by 19.6 ± 5.7% and 28.8 ± 12.0%, respectively, following treatment of mature P. aeruginosa biofilms with RNase H, which specifically removes RNA:DNA hybrids, and with Nuclease P1, which digests the displaced single-stranded DNA in R-loops (Fig. 1b–e, Supplementary Fig. 1e,f). Correspondingly, the percentage of eDNA fibres overlapping with an R-loop signal decreased by 26.0 ± 4.3% with RNase H and 24.9 ± 15.1% with Nuclease P1 (Fig. 1d,e, Supplementary Fig. 1e–g). However, when RNase H digestion was coupled with Nuclease P1 digestion, eDNA fluorescence intensity and R-loop percentage were reduced by 99.7 ± 0.1% and 99.9 ± 0.1, respectively (Fig. 1d,e, Supplementary Fig. 1e-g). An increase in dissipation factor tan δ from 0.1 to 0.4 was observed for RNase H treated biofilms relative to untreated biofilms in the rheology frequency sweep, indicating a limited decrease in biofilm viscoelastic property rather than complete dissolution (i.e., tan δ < 1) (Fig. 1f). tan δ remained unchanged following nuclease P1 treatment. However, with combined treatments of RNase H and nuclease P1, the biofilm viscoelasticity was completely removed (tan δ > 1) (Fig. 1f). Thus, the eDNA fibres of P. aeruginosa were only fully degraded following digestion of both single stranded DNA (ssDNA) and hybrid RNA:DNA, which demonstrates the importance of R-loops for imparting viscoelastic behaviour to the mature biofilm.
Mutants lacking topoisomerase (topA) and ribonuclease HII (rnhB) were tested to assess the effect of disrupting endogenous R-loop removal. As for ribonuclease HII, topoisomerase I (topA) resolves R-loops structures in the genome during replication and transcription22,29,30. Mature biofilms of topoisomerase (ΔtopA) and ribonuclease HII (ΔrnhB) transposon mutants displayed two and three times more biofilm biomass, respectively, relative to wildtype biofilms, along with a greater percentage of eDNA fibres containing R-loops (30 ± 3.7 % and 35 ± 2.8 % respectively, compared to 25 ± 1.3 % for the wildtype) (Fig. 1g–j, Supplementary Fig. 2a–d). This pattern of increased biofilm biomass was consistent across days 2–5 of biofilm growth (Fig. 1i, Supplementary Fig. 2c). These results further suggest that R-loops contribute to increasing the biomass of P. aeruginosa biofilms.
R-loops were also detected along the eDNA fibres in S. epidermidis wildtype biofilms and in clinical bronchiectasis airway sputum specimens with high loads of P. aeruginosa (SG0003, TBE0001, TBE0005) (Fig. 1k and Supplementary Fig. 3a–d). Combined RNase H and Nuclease P1 enzymatic treatment reduced the percentage of eDNA fibres with R-loops in both S. epidermidis biofilms and P. aeruginosa–infected sputum from 27 ± 5.9% to 7.5 ± 1.9% and from 15 ± 2.9% to 5 ± 1.1%, respectively, compared to untreated controls. Importantly, this R-loop depletion also eliminated the viscoelastic properties of the biofilm matrices (Supplementary Fig. 3c), indicating that their R-loops are structurally and functionally similar to those observed in P. aeruginosa biofilms. Additionally, sputum from patients with chronic P. aeruginosa infection exhibited a 70-fold higher R-loop fraction in eDNA compared to sputum from individuals (SUD0004, Nano0015, and Nano0016) without chronic lung disease (Supplementary Fig. 3e,f). These findings suggest that extracellular R-loops are not species-specific but could represent a widespread mechanism contributing to the formation and mechanical properties of eDNA networks across diverse biofilms, including those relevant to human disease.
P. aeruginosa biofilm extracellular R-loop content is independent of transcription (i.e., in trans)
The RNA profile of extracellular R-loops from the model P. aeruginosa biofilm following RNA:DNA immunoprecipitation (DRIP) sequencing was distinct from intracellular RNA from P. aeruginosa planktonic cells, with a p-value and log 2- fold change distribution closer to zero for most R-loop RNA genes compared to a broader range of values for the planktonic RNA (Fig. 2a). This pattern was also observed when comparing R-loop RNA to both biofilm intracellular (i.e. genomic) and extracellular RNA (Supplementary Fig. 4a–h), indicating that the RNA content of the R-loops was different from those actively transcribed. Notably, transcripts such as ssrA, crcZ, ssrS, and lasB, which were previously shown to be highly expressed in P. aeruginosa planktonic, biofilm intracellular, and extracellular RNA samples15, were evenly expressed in R-loop RNA (Fig. 2b, Supplementary Fig. 5a). Furthermore, the top 100 expressed genes accounted for only 9% of R-loop RNA, compared to 60% in biofilm intracellular RNA (Supplementary Fig. 5b), highlighting the distinct and less biased composition of R-loop–associated RNA. The greater evenness in the gene profile for the R-loop RNA relative to planktonic, biofilm intracellular and extracellular RNA across different days of growth is further highlighted by its more consolidated log 10 standardised mRNA count (Fig. 2c). A flatter gene expression was also observed for non-coding DNA sequences (non-CDS) in R-loop RNA, compared to biofilm intracellular (Fig. 2d), extracellular RNA and planktonic RNA (Supplementary Fig. 6a-b). Furthermore, the RNA: DNA hybrid recovery yield from the mature P. aeruginosa biofilm by immunoprecipitation was more than seventy times greater than following RNase H treatment and more than forty times greater than that of planktonic cells (Supplementary Table 1). This validates the specificity of the DRIP sequencing for R-loops and demonstrates that the biofilm matrix is characterized by a higher R-loop content relative to intracellular RNA. The extracellular matrix of the mature biofilms, therefore, contains high amounts of R-loops with a gene profile that is independent of total RNA expression.
a Volcano plot of immunoprecipitated R-loop RNA sequenced from the biofilm matrix of mature (5 d) P. aeruginosa PAO1 biofilms (R-loop RNA; blue square, right) and 16 h planktonic P. aeruginosa cells (16 h) (Planktonic RNA; blue rectangle, left). mRNA transcripts with a positive log 2-fold change and higher negative log 10 p-value such as PA2424 and PA0041 are more abundant in the extracellular R-loop sample relative to planktonic cells. b Box plot representation (n = 3) showing normalised relative abundance (%) of highly expressed mRNA transcripts such as (i) ssrA, (ii) crcZ, (iii) lasB and (iv) ssrS in RNA extracted from 16 h PAO1 planktonic cells, PAO1 biofilm intracellular and biofilm extracellular (1, 2, 3 and 5 d), and 5 d PAO1 biofilm R-loop samples. c Violin plot showing log 10 standardised mRNA count for each gene and differential expression profile across 16 h planktonic, mature biofilm intracellular, extracellular, and R-loop RNA samples. d mRNA transcript mapping along PAO1 genome plot showing both distribution of coding DNA sequences (CDS) and non-CDS from R-loop and mature biofilm intracellular RNA samples presented in reads per kilobase per million mapped reads (RPKM). All extractions and sequencing were performed in biological triplicate. For (b), error bars represent mean ± standard deviation (SD) and box plots display the 25th and 75th percentiles (lower and upper edges of the box). Whiskers extend to the minimum and maximum values within 1.5 times the interquartile range. n represents the number of biological replicates in (b). Significance was determined using DeSeq2 to identify differentially expressed genes which incorporates two-tailed Wald test with a P < 0.01 (a–d). Source data are provided as a source data file.
These results indicate that the extracellular R-loops of P. aeruginosa are decoupled from the transcription bubble22,31,32, form in genomic regions other than the transcription site, and are therefore in trans R-loops33,34. In contrast, R-loops appearing at double stranded breakage (DSB) sites and contributing to DSB repair, are coupled to transcription and therefore in cis35 where the nascent RNA hybridises back to the template DNA strand36,37,38,39. Other common sources of R-loops, including those derived from transcriptional regulation, pre-mRNA hybridisation, or polymerase backtracking, all lead to in cis R-loops35. The finding that the extracellular R-loops are in trans, therefore, removes the possibility that these extracellular R-loops of P. aeruginosa, and hence its eDNA matrix formation, are aberrant by-products of transcription, or transcription-coupled DNA repair processes.
Extracellular R-loops are produced by RecA as part of the SOS response in P. aeruginosa biofilms
In trans R-loops have been shown to be produced by bacterial strand exchange protein RecA, forming filaments with single stranded DNA and recruiting nucleoid mRNA to bind to the complementary DNA strand24. Accordingly, we found that the SOS response gene recA was upregulated in P. aeruginosa biofilm intracellular RNA compared to planktonic RNA (Fig. 3a). The amount of biofilm biomass in P. aeruginosa ΔrecA mutant biofilms was approximately half that of the wildtype across days 2–5 (Fig. 3b,c, Supplementary Fig. 7a–d). This decrease in biomass occurred coincident with a 64 ± 9 % decrease in the percentage of eDNA fibres with R-loops (Supplementary Fig. 7e), and a reduction in viscoelasticity (Supplementary Fig. 7f) in mature ΔrecA biofilms compared to the wildtype. Inactivating the recA gene therefore impairs biofilm production, and resultant biofilms are characterised by fewer R-loops. In contrast, in a mutant strain targeting an alternative EPS production mechanism in P. aeruginosa (i.e., pslF), biofilm biomass was reduced but not R-loop abundance (Supplementary Fig. 7g). These changes to the biofilm matrix occurred despite no difference in the growth rates of ΔrecA mutant and wildtype PAO1 cells (Supplementary Fig. 7h), suggesting that these observations are biofilm phenotype-specific. The correlation between R-loop coverage and biofilm biomass observed here and throughout this study is therefore likely due to an R-loop effect on biofilm structure.
a Box plot representation (n = 3) showing normalised relative abundance (%) of upregulated SOS gene recA in RNA extracted from 16 h P. aeruginosa planktonic cells, 1, 2, 3 and 5 d P. aeruginosa biofilm samples, both intracellular and extracellular. b Three-dimensional (3D) confocal micrographs (n = 5) of mature (5 d) P. aeruginosa wildtype and ΔrecA transposon mutant biofilms showing eDNA fibres in top panel stained with TOTO-1(green), and R-loop specific s9.6 antibody in bottom panel (red). All 3D biofilms were imaged with a Z-stack thickness of 10 µm. c Quantification of biofilm biomass of P. aeruginosa wildtype and ΔrecA transposon mutants across days 2–5 using crystal violet assay (n = 3). d Quantification of biofilm biomass of mature P. aeruginosa (n = 3) PAO1 wildtype and ΔrecA transposon mutants after treatment with different concentrations of SOS response inducing chemical methyl methane sulfonate (MMS). e Two-dimensional (2D) confocal micrographs (n = 4) of mature P. aeruginosa wildtype and ΔrecA biofilms showing RecA specific antibody (1 mg/ml; red) across eDNA fibre (green). All scale bars represent 10 µm. n represents the number of biological replicates, with the exception of microscopy images, where it is the number of representative microscopy images collected. For (a), error bars represent mean ± standard deviation (SD) and box plots display the 25th and 75th percentiles (lower and upper edges of the box). Whiskers extend to the minimum and maximum values within 1.5 times the interquartile range. The centre point indicates the mean of the data. For (a), significance was determined using DeSeq2 to identify differentially expressed genes which incorporates two-tailed Wald test with a P < 0.01 and for (c, d), significance between and within different treatment groups was determined using both one and two factor ANOVA with replication with P < 0.05. Source data are provided as a source data file.
Both wildtype and ΔrecA P. aeruginosa biofilms were then treated with methyl methane sulfonate (MMS), which activates the SOS response by generating abnormal DNA bases through methylation40. Biofilm biomass and the fraction of eDNA fibres containing R-loops increased two-fold with increasing MMS concentration for the mature wildtype biofilm but not the ΔrecA mutant strain (Fig. 3d, Supplementary Fig. 8a–f). Finally, the RecA protein was also observed along eDNA fibres of mature P. aeruginosa biofilms following immunofluorescence microscopy with the RecA antibody (Fig. 3e, Supplementary Fig. 8g–h). RecA protein was not observed in the ΔrecA mutant strain biofilm, confirming antibody specificity and the inability of the mutant strain to produce RecA protein (Fig. 3e, Supplementary Fig. 8g–i). Thus, recA expression correlates with in trans R-loop abundance and biofilm biomass, and the appearance of RecA protein in eDNA fibres directly links recA with the viscoelastic eDNA matrix of P. aeruginosa. A correlation between RecA and increased eDNA has been described previously41. Given the structural importance of R-loops we have demonstrated here, and previous work also implicating RecA in R-loop formation24,42,43, it is therefore likely that extracellular R-loops are an important link between RecA and viscoelastic eDNA production in P. aeruginosa biofilms. Specifically, extracellular R-loop production could be a mechanism by which RecA promotes biofilm formation.
Stringent stress response and amino acid starvation trigger production of extracellular R-loops and biofilm formation
Confoundingly, RecA is also a key component of the SOS response that rescues damaged DNA43,44. The SOS response has many functions in addition to DNA repair, and it is not clear how these seemingly divergent functions of RecA (i.e., to destabilise and rescue damaged genomes) are regulated for inserting in trans R-loops into genomic DNA for release into the extracellular matrix of P. aeruginosa biofilms. We thus sought to identify the source of the stress contributing to activation of the SOS response. Similar to Zhai et al. (2023) and Pribis et al. (2022), who described the stringent stress response as a regulator of the SOS response in Escherichia coli45,46, we observed here that the concentrations of guanosine pentaphosphate and tetraphosphate (ppGpp) were elevated in mature biofilms (17.5 ± 0.3 nmol per mg dry biomass) compared to planktonic cells (4.2 ± 0.3 nmol per mg) (Supplementary Fig. 9a–b). This suggests that the stringent stress response (SSR) was also activated, by which ppGpp is produced by combined activities of RelA and SpoT in bacteria under conditions of nutrient stress47. Initiation of the SSR in mature P. aeruginosa biofilms, as observed in this study, is further supported by the consistent upregulation of the SSR genes relA, spoT and dksA across several days of P. aeruginosa biofilm growth, relative to planktonic cells (Fig. 4a), and the suppression of ribosome and RNA degradation pathways with P. aeruginosa biofilm growth (Supplementary Fig. 9c).
a Box plot representation showing normalised relative abundance (%) (n = 3) of upregulated SSR genes relA, spoT and dksA in RNA extracted from 16 h planktonic cells and 1, 2, 3 and 5 d biofilm intracellular samples of P. aeruginosa. b Quantification of biofilm biomass of mature P. aeruginosa wildtype, ΔrelA and ΔspoT transposon mutants using crystal violet assay (n = 3) and fluorescence intensity quantification (n = 5) of percentage of eDNA fibres containing R-loops (red bars) in mature (5 d) P. aeruginosa wildtype, ΔrelA, and ΔspoT transposon mutants. c Quantification of biofilm biomass of untreated, 8 mM SHT and 100 mM AAmix (glycine, threonine and methionine) treated mature P. aeruginosa wildtype (n = 3) (green bars), and fluorescence intensity quantification (n = 5) of percentage of eDNA fibres containing R-loops (red bars) in both treated and untreated mature P. aeruginosa wildtype biofilms. d Three-dimensional (3D) confocal micrographs of mature P. aeruginosa wildtype biofilms (n = 5) with and without treatment showing eDNA stained with TOTO-1 (green) and R-loop specific s9.6 antibody (red). e Quantification of biofilm biomass of untreated and 8 mM SHT treated P. aeruginosa wildtype and mutants from days 2–5 biofilms (n = 3). f Fluorescence intensity quantification (n = 5) of percentage of eDNA fibres containing R-loops (red bars) in both untreated and 8 mM SHT treated mature P. aeruginosa biofilms. All 3D biofilms were imaged with a Z-stack thickness of 10 µm. All scale bars represent 10 µm. For (a), significance was determined using DeSeq2 to identify differentially expressed genes which incorporates two-tailed Wald test with a P < 0.01 and for (b, c, e and f), significance between and within different treatment groups was determined using both one and two factor ANOVA with replication with P < 0.05. n represents the number of biological replicates, with the exception of microscopy images and data analysis, where it is number of representative microscopy images collected. All images were acquired across biological triplicates, and the statistics were determined based on the number of representative images collected. Source data are provided as a source data file.
Simultaneous reduction of biofilm biomass and R-loop coverage was also observed in mature P. aeruginosa SSR mutants ΔrelA, and ΔspoT (Fig. 4b, Supplementary Fig. 9d–f). Both mutants produced one half the amount of biofilm biomass, relative to mature wildtype P. aeruginosa. The percent coverage of R-loops across eDNA fibres was also reduced in mature ΔrelA, and ΔspoT biofilms to 7.0 ± 0.8% and 11.5 ± 1.7% respectively, compared to 25 ± 1.3% for the mature wildtype biofilm (Fig. 4b, Supplementary Fig. 9d–g).
To further verify the link between SSR, R-loop formation and viscoelastic eDNA fibre formation, P. aeruginosa biofilms were grown in the presence of serine hydroxamate (SHT), which activates SSR by promoting ppGpp accumulation48. SHT added following commencement of P. aeruginosa biofilm growth (i.e. 1 day), increased subsequent biomass production and the percentage of eDNA fibres with R-loops in the mature biofilm by 2.3 and 1.5 times, respectively, relative to the non-treated wildtype biofilm (Fig. 4c-d). In contrast, no increase in biofilm growth (Fig. 4e) or the percentage of eDNA fibres with R-loops in mature biofilm (Fig. 4f) was observed following SHT treatment of the ΔrelA P. aeruginosa biofilm, i.e. similar to the untreated mutant biofilm. RelA is therefore required for this stringent stress effect on R-loop abundance and biofilm biomass.
Furthermore, the same addition of SHT had no effect on the biofilm matrix of ΔrecA P. aeruginosa biofilms, with the biomass and R-loop content similar to the ΔrecA mutant biofilm without SHT treatment (Fig. 4e,f, Supplementary Fig. 10a-c). This indicates that RecA also plays a role in mediating the SSR effect on viscoelastic eDNA matrix formation. This trend in biofilm biomass following SHT addition was consistent for biofilms from days 2–5 (Supplementary Fig. 10d,e).
Amino acid deficiency leads to SSR activation in bacteria49 and we hypothesised that preventing the onset of SSR by amino acid addback would reduce R-loop production and delay biofilm formation. Glycine, threonine and methionine reduce SSR48,50, and when added together here (i.e., AAmix, 100 mM) resulted in a three-fold decrease in P. aeruginosa biofilm biomass relative to non-treated cells (Fig. 4c,d, Supplementary Fig. 10b) despite eliciting no change in growth rate relative to untreated planktonic cells (Supplementary Fig.10f). This decrease in biomass with the addition of AAmix was concentration-dependent (Supplementary Fig.10g). This pattern was consistent across days 2–5 of biofilm growth (Supplementary Fig. 10h). The delay in biofilm formation also coincided with a reduction in the percentage of eDNA fibres containing extracellular R-loops in AAmix-treated biofilms by 55 ± 6 %, relative to non-treated biofilms (Fig. 4c,d). AAmix treatment reduced the extent of stringent stress response in biofilm cells relative to the untreated control, as indicated by activation of RNA degradation and ribosome pathways (i.e. increased translation rates; Supplementary Fig. 11a). Finally, AAmix-treatment downregulated SOS response gene recA relative to non-treated and SHT-treated mature biofilms (Supplementary Fig. 11b). This clearly demonstrates that removing amino acid starvation reduces recA expression and subsequently R-loop production, and P. aeruginosa biofilm formation.
R-loops are a regulated agent of death but also enable dying cells to contribute to matrix assembly
Adding antibodies specific to extracellular proteins contributing to matrix structural networks has been shown previously to inhibit biofilm formation (e.g., DNA binding proteins and integration host factor)51,52. Similarly, providing the R-loop specific S9.6 antibody reduced biofilm biomass here (Fig. 5a, Supplementary Fig. 12a–b). Viscoelasticity continued to decrease from days 2–5 after S9.6 addition, despite no further antibody addition, with tan delta increasing to 0.8 after adding 20 µg/ml of S9.6 antibody compared to 0.1 for untreated control (Supplementary Fig. 12c). There was no additional increase in percentage of eDNA fibres containing R-loop fraction after the second round of S9.6 immunofluorescence on mature biofilms (Supplementary Fig. 12d). Adding increasing concentrations of S9.6 antibody to wildtype biofilm reduced the average biovolume and length of eDNA fibres by 30 and 10 times, respectively, relative to untreated wildtype P. aeruginosa (Fig. 5b). Thus S9.6 antibody inhibits biofilm formation and the S9.6 effect persists during biofilm growth. The IgG antibody had no effect on the biofilms, demonstrating that this effect was specific to the S9.6 antibody (Fig. 5a). In contrast, no biofilm inhibition was observed following addition of increasing concentrations of RecA antibodies to P. aeruginosa biofilms (Fig. 5a, Supplementary Fig. 12a), despite the presence of RecA in eDNA fibres (Fig. 3f). This could reflect that RecA binding to ssDNA to form nucleoprotein filaments and extracellular R-loops occurs intracellularly and prior to their release.
a Quantification of biofilm biomass (n = 3) of untreated, RecA, S9.6 and IgG antibody-treated mature (5 d) P. aeruginosa wildtype. b Quantification of biovolume and ellipticity of each eDNA fibre (n = 5) from 1, 5, 10 and 20 µg/ml of R-loop specific S9.6 antibody treated mature P. aeruginosa biofilms. c Quantification of biofilm biomass (n = 3) of both untreated and 8 mM serine hydroxamate (SHT) treated mature P. aeruginosa wildtype, ΔalpA, Δlys and Δhol PAO1 transposon mutants. d Fluorescence intensity quantification (n = 5) of percentage of eDNA fibres containing R-loops (red bars) in mature P. aeruginosa wildtype, ΔalpA, Δlys and Δhol PAO1 transposon mutants. All experiments were performed in biological triplicate. Significance between and within different treatment groups was determined using both one and two factor ANOVA with replication with P < 0.05 (a–d). n represents the number of biological replicates, with the exception of microscopy data analysis, where it is number of representative microscopy images collected. All images were acquired across biological triplicates, and the statistics were determined based on the number of representative images collected. Source data are provided as a source data file.
Finally, increased ppGpp levels can also promote virulence gene expression and cell death in P. aeruginosa through the AlpA protein, a positive regulator of the alpBCDE cell lysis genes53. Accordingly, reduced biofilm growth and R-loop coverage were observed here for the alpA mutant, relative to the wildtype (Fig. 5c-d, Supplementary Fig. 12e–f). In contrast, SHT treatment did not lead to changes in biofilm formation in the ΔalpA PAO1 strain (Fig. 5c, Supplementary Fig. 12e–f), unlike the wildtype. Activating the SSR, therefore, only increases biofilm biomass and R-loop coverage when alpA is present. This suggests that the insertion of in trans R-loops by RecA, which are understood to kill cells31, involves the programmed cell death (PCD) pathway as well. We previously described the presence of particular mRNA in the matrix e.g., lasB15. We also observed here the upregulation of various DNA rescue genes such as ruvA and sulA in wildtype biofilms relative to planktonic cells (Supplementary Fig. 13). RecA’s contribution to toxic R-loop formation and genomic instability at the same time that DNA rescue genes are activated could be attributed to SSR activating PCD for a subset of suicidal cells through the PCD regulator gene alpA53. A high increase in cell death upon maturation as described previously for the biofilm model used in this study, supports this claim15.
eDNA release in P. aeruginosa has been independently attributed to cell lysis54 and activation of the SOS response41. These studies, albeit with the observation of extracellular vesicles, which was not the case here, therefore support our assertion that the in trans R-loops are also released by cell lysis. This is demonstrated by our findings that inactivation of alpA reduced both R-loop coverage and biofilm biomass, and that SHT treatment failed to enhance these phenotypes in the absence of alpA (Fig. 5c,d, Supplementary Fig. 12e) i.e., AlpA-dependent lysis plays a key role in enabling extracellular export of these nucleic acid structures. Accordingly, knockouts for cell lysis genes lys and hol also displayed reduced biofilm formation and extracellular R-loop coverage relative to the wildtype (Fig. 5d). Increased biofilm formation upon SHT exposure was similarly not observed in either of these knockouts relative to the wildtype and SHT treatment inhibited growth of planktonic cells (Fig. 5c, Supplementary Figs. 12e, 14a).
Discussion
R-loops were once considered unscheduled intermediate by-products of biological processes; however, this study demonstrates for the first time a biological function for extracellular R-loops in promoting matrix formation in biofilms. Our findings also suggest that extracellular R-loop formation is initiated by RecA within cells prior to their release. This is illustrated by the ineffectiveness of the RecA antibody at inhibiting biofilm formation (Fig. 5a). Meanwhile, R-loops act extracellularly to promote biofilm growth upon release, as evidenced by the inhibition of biofilm growth when S9.6 antibody bind to the R-loops. R-loops thus have the dual function of population control, presumably to a number that can survive the given amino acid availability and promoting biofilm formation. For a planktonic population the transition to a biofilm mode of life, would enable surviving cells to colonise host tissue and access less bioavailable amino acid sources (i.e., insoluble proteins) through proteolysis (i.e., pathogenic virulence and elastase)15,55,56.
Biofilm growth and other phenotypes associated with biofilm formation (e.g., virulence, antibiotic resistance) have been described separately for both the SOS response and stringent stress response (SSR)57,58. SOS response genes recA, sulA and uvrA are additionally triggered by nutrient starvation and SSR59. Similarly, in our study, activation of SSR in SHT-treated P. aeruginosa biofilms was verified by lower rates of translation, ribosome suppression, higher rates of transcription and upregulation of SOS response genes such as sulA, recA and ruvA, relative to non-treated biofilms (days 1 and 5) and 16 h planktonic cells (Supplementary Fig. 14b–e). Thus, the findings presented here provide further evidence for a link between the bacterial SOS and SSR responses and enable us to describe a mechanism by which activation of the SOS response and SSR produces viscoelastic biofilms. Accordingly, activation of the stringent stress response (SSR) by amino acid starvation induces the SOS response to insert toxic R-loops into genomic DNA. These R-loops are subsequently released by cell lysis and become key viscoelastic matrix structures. This all occurs coincident with upregulation of DNA repair genes, and the appearance of specific stress-related transcripts in the eDNA fibres of P. aeruginosa biofilms (Supplementary Figs. 11b and 13). This apparent contradiction, between simultaneous activation of DNA repair or degradation, likely occurs because AlpA is coordinated according to ppGpp levels to implement either DNA repair or PCD processes. RecA is then engaged to recover DNA or achieve cell suicide, respectively. For killing cells, RecA inserts the toxic R-loops into the genome (see model in Fig. 6).
Amino acid starvation in biofilms activates stringent stress response (SSR) and ppGpp alarmone overproduction. ppGpp binds to RNA polymerase, slows down mRNA degradation and only activates transcription of stress response genes. ppGpp activates programmed cell death (PCD) in a subpopulation of suicidal cells using the AlpA master regulator. In suicidal cells, the RecA protein binds to single-stranded DNA (ssDNA) at sites of damage and facilitates the recruitment of excess mRNA that has accumulated within the cell to these damaged regions. In trans R-loop formation is generated by RecA across the genome, which creates widespread stalling of replication forks and subsequently genomic instability. The SOS response additionally induces explosive cell lysis through endolysin overproduction. R-loop structures are released into the biofilm matrix from suicidal cells. In contrast, RecA activates DNA repair machinery in survivor cells. Stress response genes such as lasB are released into the matrix and interact with R-loops to act as building blocks for biofilm formation. Created in BioRender. Group, S. (2025) https://BioRender.com/y95q1n6.
One possible explanation for R-loop-mediated biofilm stability is that they promote crosslinking in biofilm matrices. Adding S9.6 antibodies neutralises R-loop released from the cells and prevents the establishment of biofilm matrix. This could reflect the binding of single stranded DNA R-loops to mRNA from healthy cells, or that they bind indirectly through an intermediary (such as a protein). R-loops crosslinking DNA from the survivor cells could either be present in genomic DNA where the chain structure is preserved or present as small fragments after the chain DNA has been digested. It is additionally unknown how the R-loops are inserted into the eDNA matrix upon release. Nonetheless, these results validate our earlier observation that R-loops are key factors in providing structural stability and increasing P. aeruginosa biofilm biomass upon activation of SSR and SOS responses.
Amino acid starvation is widely implicated in biofilm formation50,60. SSR and SOS responses are highly conserved mechanisms in bacterial systems59,61. Additionally, antimicrobial strategies that rely on stressing bacteria often have unforeseen consequences, such as increased biofilm formation, mutations, and resistance resulting from, among other factors, hyperactivation of the SOS response62,63, which our study shows could result in R-loop release. While demonstrated here for P. aeruginosa, it is therefore possible that R-loops provide a broadly applicable bacterial strategy to build viscoelastic higher-order nucleic acid structures under conditions of stress, and facilitate the biofilm mode of growth across a range of ecosystems50,60.
Methods
This study complies with all relevant ethical regulations, operating under the ethical framework of the National Research Foundation, Singapore
Bacterial strains and growth conditions
Pseudomonas aeruginosa PAO1 wildtype (WT), topA::ISphoA/hah, rnhB::ISphoA/hah, relA::ISphoA/hah, spoT::IsphoA/hah, rpoS::ISphoA/hah and recA::ISphoA/hah, alpA::ISphoA/hah, lys::ISphoA/hah, and hol::ISphoA/hah transposon mutants were acquired from the PAO1 Manoil transposon library64. Complementation of all genes resulted in the reversal of the phenotypes described here for the knockouts (Supplementary Fig. 14f, detailed protocol is shared below). These strains were grown on Luria-Bertani (LB) agar plates (5 g/l NaCl, 5 g/l yeast extract, 10 g/l tryptone, 15 g/l agar) at 37 °C for 16 h. Staphylococcus epidermidis wildtype was grown on MHB agar at 37 °C for 16 h. Overnight pre-cultures of P. aeruginosa strains and S. epidermidis WT were grown at 37 °C, 200 rpm for 16 h in either 10 ml of Luria-Bertani (LB) broth (5 g/l NaCl, 5 g/l yeast extract, 10 g/l tryptone) or Mueller Hinton broth (MHB), respectively.
Biofilm growth
Biofilm cultures were grown under static conditions with no media replenishment for 5 d at 37 °C throughout the study. Briefly, 10 ml of overnight culture was diluted 20 times with respective broth (i.e., either LB or MHB) depending on the bacteria used for biofilm growth. All biofilm growth experiments were performed with three biological replicates. For extracellular nucleic acid isolation, DRIP, RNA sequencing and rheology property measurements, viscoelastic biofilms grown in conical flasks were concentrated by centrifugation at 10,000 * g for 10 min in 50 ml falcon tubes3,15. For confocal microscopy and biofilm quantification, biofilms were grown in 24-well glass bottom plates in shaking conditions at 37 °C, 150 rpm for stipulated time of growth and gently collected using a cut-off 1 ml pipette to minimise the disturbance of biofilms and to maintain the native biofilm structure. For experiments involving the crystal violet assay for biofilm quantification, excess supernatant from non-static biofilms grown using 24-well glass bottom plate was removed before measuring absorbance.
Enzymatic digestions
The digestion was performed according to the previously published protocol3,15. Briefly 20 mg (wet weight) of P. aeruginosa, S. epidermidis, clinical sputum sample from conical flasks, were resuspended in 1 ml of either (i) RNase H buffer (20 mM HEPES-KOH buffer (pH 8.0), 50 mM KCl, 4 mM MgCl 2, 1 mM DTT, 50 µg/ml BSA and 2 units Ribonuclease H (R6501, Sigma Aldrich) or (ii) Nuclease P1 buffer (25 mM Tris-HCl, 50 mM NaCl, 1 mM ZnCl2, 50% glycerol) and 5 units of Nuclease P1 (M0660S, New England Biolabs). All digestions were performed in biological triplicate at 37 °C for 30 min. For viscoelasticity measurements, 200 µl of both untreated and digested biofilm samples were collected and pipetted gently onto a HAAKE MARS 60 Rheometer (Thermofisher Scientific, catalogue number: 3,790,600) with parallel plates and measured according to the published protocol3,15. For confocal microscopy, 100 µl of untreated and digested biofilm samples were washed in situ with distilled water, followed by immunolabelling using S9.6 antibody, as described below.
RNA extraction for sequencing
Overnight planktonic cultures (16 h) of P. aeruginosa grown at 37 °C grown in conical flasks were harvested by centrifugation (10,621 * g for 10 min at 4 °C). For biofilm intracellular RNA, P. aeruginosa biofilms grown in 500 ml conical flasks were lyophilised. Both pelleted planktonic cells and biofilms were further processed for RNA extraction according to our published protocol15. Briefly, both sets of samples were washed with ice-cold 1x PBS, resuspended in TE buffer, probe sonicated and centrifuged separately to collect supernatant. Both planktonic and biofilm RNA across different days in the supernatant were extracted using Trizol method65 and purified according to our published protocol15.
For biofilm extracellular RNA, biofilms collected from different days of growth in conical flasks were solubilised in ionic liquid 40% (v/v) 1-ethyl-3-methylimidazolium acetate (EMIM Ac):60% (v/v) dimethyl acetamide (DMAc) at 55 °C for 1 h. The supernatant was collected and subjected to perchloric acid precipitation (70%) on ice for 15–30 min. The recovered pellet was resolubilised in Ionic liquid solvent, precipitated using isopropanol and further downstream processed and purified according to the detailed protocol published in our previous study15. All biofilms were grown in biological triplicate, and sampled on days 1, 2, 3 and 5 for extraction of intracellular RNA (iRNA) and extracellular RNA (eRNA).
For serine hydroxamate (SHT) and amino acid (AA) addback experiments, 1 d static P. aeruginosa PAO1 biofilms were either treated with 8 mM of SHT or 100 mM of glycine, methionine and threonine. The biofilms were collected post 0, 15 and 30 min of both SHT and AA treatment and processed for biofilm intracellular RNA extraction as described above. Biofilm intracellular RNA was also extracted from untreated 1 d (BF day 1) and mature 5 d (BF day 5) static biofilms and used as a reference for this sample batch. All biofilms were grown in biological triplicate and sampled for iRNA.
DNA-RNA immunoprecipitation (DRIP) sequencing
One to two micrograms of purified biofilm matrix nucleic acid were extracted from mature (5 d) P. aeruginosa biofilms grown in conical flasks in biological triplicate, according to our previously published protocol15. The extracted samples were not treated with turbo DNase but directly fragmented using a cocktail of restriction enzymes (New England BioLabs) including 30 units of each of the following: BsrGI (cat. No. R0575S), EcoRI (cat. No. R0101S), HindIII (cat. No. R0104S), SspI (cat. No. R0132S), XbaI (cat. No. R0145S) at 37 °C for 1 h66. This was followed by immunoprecipitation (IP) using 1 µg/ml of Anti-DNA-RNA Hybrid, clone S9.6 antibody, (Cat. No. MABE1095, Merck Millipore), incubated overnight at 4 °C. S9.6 antibody is a mouse monoclonal antibody that was generated against a ΦX174 bacteriophage-derived synthetic DNA–RNA antigen and recognizes RNA-DNA hybrids of various lengths28. Prior to IP, antibodies were crosslinked to dynabead protein A (Cat. No. 10002D, ThermoFisher Scientific, Life Technologies) using crosslinker bis (sulfosuccinimidyl) suberate (BS3) (Cat. No. 21,580, ThermoFisher Scientific, Life Technologies). Crosslinked antibody was used to pull down R-loops from fragmented extracellular nucleic acid extract, following the manufacturer’s protocol (Cat. No. 10002D, ThermoFisher Scientific, Life Technologies). R-loops were eluted using elution buffer (10 mM Tris, pH 8.5, 0.1 mM EDTA). The sample purity and concentration were measured using Qubit high sensitivity ssDNA and RNA fluorometry kit (Thermofisher Scientific, Cat No: Q32852, Q33230, respectively) and NanoDrop 2000 Spectrophotometer, respectively. Library preparation was performed using the modified ribozero method according to the published protocol66.
RNA-seq analysis
Illumina sequencing raw reads first underwent a quality check, filtering, and adaptor trimming with BBDuk67. This was followed by ribosomal RNA read removal with SortMeRNA68. The remaining non-rRNA aligned reads were mapped to a P. aeruginosa PA01 reference genome15 (accession number NC_002516.2) using Bowtie269. SAM files generated from Bowtie2 alignment were converted to BAM files and sorted with SAMtools70. The read mapping information was quantified with HTSeq count, followed by differential gene expression analysis using DESeq271,72. RNA-seq was performed in biological triplicate. Raw reads used in this study have been uploaded to Sequence Read Archive (SRA) with accession numbers.
Statistics
Differential gene expression analysis was performed using DESeq2 package in R. A negative binomial model was employed to estimate size factors and dispersion. Statistical significance was assessed using two-tailed tests, with adjusted p-values corrected for false discovery rate and unadjusted p-values (P < 0.05) evaluated against a threshold for log 2-fold change. Volcano plot, box plot of gene expression level and violin plot of overall gene expression profile were generated with ggplot2. KEGG pathway enrichment analysis was performed with clusterProfiler using gene set enrichment function73. Significance of difference in treatment groups was determined using one and two factor ANOVA.
Staining and confocal microscopy
Biofilms were gently collected from 24-well glass bottom plate using 1 ml cut-off pipette tip, as described above. R-loop distribution in biofilms were visualised by immunofluorescence (IF) using S9.6 antibody (Cat. No. MABE1095, host-mouse, Merck Millipore). Briefly, 1 μg/ml of either S9.6, IgG or RecA specific antibody (Abcam, Cat no: 63797) resuspended in 1X PBS was added to 1 ml of biofilm and incubated for 1 h at 37 °C. Biofilms were washed twice with 1x PBS for 1 min each. Anti-R-loop and RecA antibodies were detected with Alexa Fluor 568 and 647 tagged goat anti-mouse IgG (2 mg/mL stock solution, 1 µg/mL) in 1× PBS for 1 h at 37 °C, respectively, followed by a washing step, as described earlier. Post IF, the distribution of extracellular DNA (eDNA) fibres in biofilms was visualised using eDNA specific TOTO-1 stain (3 µg/ml in DI water; Thermofisher Scientific, Cat No: T3600) for 15 min in the dark3. For biomass fluorescence quantification, biofilms were stained with SYTO9 (Cat No:S34854, Thermofisher Scientific) according to manufacturer’s protocol and quantified by calculating raw integrated intensity of green channel using Image J Fiji software. Antibody validation data is available in the relevant manufacturer’s website.
All microscopy imaging was conducted on a Zeiss LSM 780 confocal microscope with a 63x objective. 100 µl of stained biofilms were deposited on poly-L lysine coated slide (Cat No: P0425-72EA, Sigma Aldrich) with a coverslip for imaging. All biofilms were grown in biological triplicate. Due to low sample availability, clinical sputum samples were imaged from technical triplicates.
Image quantification
For quantification of differential expression levels of eDNA fibres and percentage of eDNA fibres with R-loops, Image J Fiji software was used to set a threshold option to eliminate the bright cell signal based on the fluorescence intensity difference between cells and eDNA fibres. The total fluorescence intensity was also obtained from images by calculating the raw integrated intensity using Image J. Mander’s coefficient of colocalisation was also measured to calculate the percentage of R-loops in eDNA fibres post enzymatic treatment and 3D images were processed using Imaris x64 software. Biovolume and ellipticity of eDNA fibres post S9.6 antibody treatment was measured using Imaris x64 software by thresholding both eDNA and R-loop channels to absolute intensity. Biovolume surfaces over 10 µm³ were used as a threshold for calculation. Representative images were chosen by taking biological triplicates into account.
Biofilm biomass quantification
One milliliter of biofilm was grown at 37 °C, 150 rpm in 24-well glass bottom plate (Cat.no: P24-0-N, Cellvis) for stipulated time of growth in biological triplicate, as described above in biofilm growth section. After the stipulated time of growth, supernatant was gently removed, and the surface attached biofilms were washed twice with 1x PBS. This is followed by incubation of 1 ml of 0.1 % crystal violet (CV) to biofilms for 15 min at room temperature, washing twice with 1X PBS to remove excess CV and finally resuspending the biofilms in 1 ml of 95% methanol for 10 min. The biofilm biomass was quantified by measuring absorbance at 600 nm.
Antibody growth assay
One millilitre of P. aeruginosa biofilms was grown at 37 °C, 150 rpm in 24-well glass bottom plate in biological triplicate as described above in biofilm biomass quantification section. Different concentrations of both RecA and S9.6 antibody such as 1, 5, 10 and 20 µg/ml were either added separately or mixed following one day of biofilm growth. Biofilms were allowed to grow for 4 d, and the CV assay was performed according to the protocol described above. For the rheology assay, biofilms were collected daily for 5 d post S9.6 antibody addition and viscoelasticity were measured according to the protocol described above.
To verify whether S9.6 antibody binds to and neutralises R-loops, two rounds of immunofluorescence (IF) were performed on biofilms using S9.6 antibody. The percentage of R-loops in eDNA fibres was measured after each round of IF to quantify the amount of unbound free R-loops in the matrix. The experiment was performed in biological triplicate.
Stress and SOS response inducer assays
For serine hydroxamate (SHT) and amino acid addback experiments, one-day static biofilms grown in 24-well glass bottom plates were treated with 8 mM of SHT and 100 mM of glycine, methionine, and threonine, respectively and biofilms were allowed to grow for 4 d and processed as described above for S9.6 antibody staining and microscopy imaging. For biofilm biomass quantification, 8 mM of SHT and 100 mM of glycine, methionine and threonine were added to 1 d biofilms grown at 37 °C, 150 rpm in a 24-well glass bottom plate in biological triplicate. Biofilms were grown for 4 d, and the CV assay was performed according to the protocol described above.
For the methyl methane sulfonate (MMS, Sigma Aldrich, Cat no: 129925) assay to induce SOS response, 1 d biofilms of wildtype and recA mutant P. aeruginosa biofilms grown at 37 °C, 150 rpm in 24-well glass bottom plate in biological triplicate were treated with different concentrations of MMS: 0.1%, 0.5%, 1%, 5%, 10% v/v. Biofilms were collected four days post MMS treatment for biofilm quantification using CV assay. S9.6 antibody staining was performed on MMS treated mature static biofilms to quantify R-loops in eDNA fibres respectively.
ppGpp extraction and quantification
Different concentrations of ppGpp standard (Merck, Cat no: 7415-69-2) were prepared from stock and a standard curve was established to calculate the area of each standard. For ppGpp extraction, 40 mg of dry biofilms grown in conical flasks were treated with 1 ml of 100% methanol and left at room temperature for 5 min followed by vortexing for 60 s. The sample was frozen using liquid nitrogen and thawed on ice followed by centrifugation at 4000 rpm for 5 min at 4 °C. The supernatant was collected and filtered using 0.22 µm filter. Another cycle of extraction was performed on the remaining pellet and supernatants were pooled for HPLC quantification of ppGpp74. Samples were injected onto Shimpack Bio IEX-Q, 5 µm, 4.6 * 30 anion exchange column and ppGpp was eluted by comparing the retention time of the ppGpp standard75. The experiment was conducted in biological triplicate.
Clinical sample collection
We obtained three lung sputum samples (SG0003, TBE0001, and TBE0005) from patients with severe chronic obstructive pulmonary disease (COPD). In addition, sputum samples from healthy individuals (Nano0015 and Nano0016) were collected as controls. This study was approved by the Institutional Review Boards (IRBs) and ethical approval for sample collection was obtained from Singapore General Hospital (CIRB 2017/2109) and Nanyang Technological University (NTU IRB-2017-05-035). Written informed consent was obtained from all participants prior to sample collection. The specimens were transported on ice and reached the NTU laboratory within 4 h post-collection.
Following analysis, all sputum samples were disposed in accordance with institutional biosafety and ethical regulations. Experiments involving clinical samples were performed in technical triplicate due to the limited quantity of material available.
Detection and identification of Pseudomonas aeruginosa in clinical sputum samples
Pseudomonas aeruginosa was identified in all three sputum samples mentioned above using microbiological detection methods. Sputum was cultured on blood and MacConkey agar (BD-BBL) and incubated at 37 °C. Colourless colonies on MacConkey agar were tested with Gram staining, oxidase tests, and BD BBL™ Crystal™ microbial ID strips (N/H) and confirmed with MALDI-TOF (Bruker MALDI Biotyper).
Further confirmation was achieved through metagenomic sequencing of SG0003, TBE0001, TBE0005, Nano0015 and Nano0016. Sputum DNA was extracted with the Roche High-pure PCR Template Preparation Kit and quantified using the Qubit dsDNA HS Assay Kit76,77,78. Sequencing was performed on a HiSeq 2500 platform (Illumina) at SCELSE-NTU. Libraries were prepared with the Swift Biosciences Accel-NGS 2S Plus DNA Kit, and DNA was sheared to ~450 bp. Library size and validation were performed using Agilent Bioanalyzer and qPCR, respectively79,80. Raw reads were trimmed and mapped against the GRCh38 human genome with Bowtie269. Non-host reads were aligned to the NCBI non-redundant protein database using Diamond, and microbial taxonomy was classified with MEGAN81,82. The relative abundance of P. aeruginosa was calculated based on non-human reads.
Gene complementation protocol
To confirm the role of specific genes in the observed phenotypes, we performed gene complementation by introducing wildtype copies of relA, recA, alpA, topA, and lys into their respective PAO1 transposon mutant strains. Genomic DNA was extracted from 16 h planktonic PAO1 wildtype cultures using the PureLink Genomic DNA Mini Kit (ThermoFisher Scientific, Cat. No. K182001). PCR primers were designed to include gene-specific restriction sites and ribosome binding sites, with melting temperatures optimised for amplification. Primers were ordered from Integrated DNA Technologies (IDT), and sequences are provided in following link: template-paste-entry_Sudarsan NTU.xlsx. PCR amplification was carried out using standard protocols, and amplicons were purified using the PureLink™ Quick Gel Extraction and PCR Purification Combo Kit (ThermoFisher Scientific, Cat. No. K220001). PCR product size was confirmed by 1% agarose gel electrophoresis (TAE buffer, 90 V, 45 min).
The purified gene inserts and the expression vector pBBRMCS4 (ampicillin resistant gene) were digested using EcoRI and XbaI (New England Biolabs, Cat. Nos. R0145S and R0101S), based on compatibility and the absence of internal restriction sites (determined using NEB Cutter). Ligation was performed using T4 DNA ligase (NEB, Cat. No. M0202), and the ligation product was transformed into NEB® 5-alpha Competent E. coli (High Efficiency, Cat. No. C2987H) following the manufacturer’s protocol. Transformants were selected on LB agar plates containing 100 µg/mL ampicillin, and successful insertion was verified by colony PCR using GoTaq® Green Master Mix (Promega, Cat. No. M712). Plasmids were purified from overnight cultures using the PureLink™ HiPure Plasmid Miniprep Kit (ThermoFisher Scientific, Cat. No. K210002). The confirmed plasmids were electroporated into P. aeruginosa mutant strains using the Gene Pulser Xcell Microbial System (Bio-Rad, Cat. No. 1652662). Transformants were selected on carbenicillin-containing agar plates (typically effective against P. aeruginosa). Phenotypic assays, including biofilm biomass quantification and R-loop coverage in eDNA fibres, were then performed to assess whether complementation restored the wildtype phenotype.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA sequencing data analysis from PAO1 and metagenomic data of clinical sputum sample have been deposited to Sequence Read Archive (SRA) with a BioProject accession number PRJNA1207270, PRJNA595703, PRJNA590225 and PRJNA982297. Other data supporting the findings of this study are available within the paper, its Supplementary Information, and Source Data file. Source data are provided with this paper.
References
Whitchurch, C. B., Tolker-Nielsen, T., Ragas, P. C. & Mattick, J. S. Extracellular DNA required for bacterial biofilm formation. Science 295, 1487 (2002).
Böckelmann, U. et al. Bacterial extracellular DNA forming a defined network-like structure. FEMS Microbiol. Lett. 262, 31–38 (2006).
Seviour, T. et al. The biofilm matrix scaffold of Pseudomonas aeruginosa contains G-quadruplex extracellular DNA structures. NPJ Biofilms Microbiomes 7, 27 (2021).
Peterson, B. W., van der Mei, H. C., Sjollema, J., Busscher, H. J. & Sharma, P. K. A distinguishable role of eDNA in the viscoelastic relaxation of biofilms. mBio 4, e00497–00413 (2013).
Wells, M. et al. Perspective: The viscoelastic properties of biofilm infections and mechanical interactions with phagocytic immune cells. Front. Cell Infect. Microbiol. 13, https://doi.org/10.3389/fcimb.2023.1102199 (2023).
Buzzo, J. R. et al. Z-form extracellular DNA is a structural component of the bacterial biofilm matrix. Cell 184, 5740–5758.e5717 (2021).
Sharma, S. et al. Microbial biofilm: a review on formation, infection, antibiotic resistance, control measures, and innovative treatment. Microorganisms 11, https://doi.org/10.3390/microorganisms11061614 (2023).
Flemming, H.-C. et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575 (2016).
Fux, C. A., Wilson, S. & Stoodley, P. Detachment characteristics and oxacillin resistance of Staphyloccocus aureus biofilm emboli in an in vitro catheter infection model. J. Bacteriol. 186, 4486–4491 (2004).
Dal Co, A., van Vliet, S. & Ackermann, M. Emergent microscale gradients give rise to metabolic cross-feeding and antibiotic tolerance in clonal bacterial populations. Philos. Trans. R. Soc. B: Biol. Sci. 374, 20190080 (2019).
Sharma, D., Misba, L. & Khan, A. U. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob. Resist Infect. Control 8, 76 (2019).
Devaraj, A. et al. The extracellular DNA lattice of bacterial biofilms is structurally related to Holliday junction recombination intermediates. Proc. Natl. Acad. Sci. USA 116, 25068–25077 (2019).
Minero, G. et al. Extracellular G-quadruplexes and Z-DNA protect biofilms from DNase I, and G-quadruplexes form a DNAzyme with peroxidase activity. Nucleic Acids Res. 52, 1575–1590 (2024).
Secchi, E. et al. The structural role of bacterial eDNA in the formation of biofilm streamers. Proc. Natl. Acad. Sci. USA 119, e2113723119 (2022).
Mugunthan, S. et al. RNA is a key component of extracellular DNA networks in Pseudomonas aeruginosa biofilms. Nat. Commun. 14, 7772 (2023).
Li Wong, L. et al. Surface-layer protein is a public-good matrix exopolymer for microbial community organisation in environmental anammox biofilms. ISME J. 17, 803–812 (2023).
Wong, L. L. et al. Microbial biofilms are shaped by the constant dialogue between biological and physical forces in the extracellular matrix. Environ. Microbiol 25, 199–208 (2023).
Kundukad, B. et al. Viewing biofilm formation through a multifocal lens of physics and biology. Microbiol Aust. 44, 69–74 (2023).
Seviour, T. et al. Phase transitions by an abundant protein in the anammox extracellular matrix mediate cell-to-cell aggregation and biofilm formation. mBio 11, https://doi.org/10.1128/mbio.02052-02020, (2020).
McLean, E. K., Nye, T. M., Lowder, F. C. & Simmons, L. A. The impact of RNA-DNA hybrids on genome integrity in bacteria. Annu. Rev. Microbiol. 76, 461–480 (2022).
Brambati, A., Zardoni, L., Nardini, E., Pellicioli, A. & Liberi, G. The dark side of RNA:DNA hybrids. Mutat. Res./Rev. Mutat. Res. 784, 108300 (2020).
Hegazy, Y. A., Fernando, C. M. & Tran, E. J. The balancing act of R-loop biology: The good, the bad, and the ugly. J. Biol. Chem. 295, 905–913 (2020).
Wahba, L., Amon, J. eremyD., Koshland, D. & Vuica-Ross, M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell 44, 978–988 (2011).
Wahba, L., Gore, S. K. & Koshland, D. The homologous recombination machinery modulates the formation of RNA–DNA hybrids and associated chromosome instability. eLife 2, e00505 (2013).
He, H., Cooper, J. N., Mishra, A. & Raskin, D. M. Stringent response regulation of biofilm formation in Vibrio cholerae. J. Bacteriol. 194, 2962–2972 (2012).
Strugeon, E., Tilloy, V., Ploy, M. C. & Da Re, S. The stringent response promotes antibiotic resistance dissemination by regulating integron integrase expression in biofilms. mBio 7, https://doi.org/10.1128/mBio.00868-16 (2016).
Petermann, E., Lan, L. & Zou, L. Sources, resolution and physiological relevance of R-loops and RNA–DNA hybrids. Nat. Rev. Mol. Cell Biol. 23, 521–540 (2022).
Boguslawski, S. J. et al. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods 89, 123–130 (1986).
Brochu, J., Breton É, V. & Drolet, M. Supercoiling, R-loops, replication and the functions of bacterial type 1A topoisomerases. Genes 11, https://doi.org/10.3390/genes11030249 (2020).
Manzo, S. G. et al. DNA Topoisomerase I differentially modulates R-loops across the human genome. Genome Biol. 19, 100 (2018).
Niehrs, C. & Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 21, 167–178 (2020).
Bou-Nader, C., Bothra, A., Garboczi, D. N., Leppla, S. H. & Zhang, J. Structural basis of R-loop recognition by the S9.6 monoclonal antibody. Nat. Commun. 13, 1641 (2022).
Belotserkovskii, B. P., Tornaletti, S., D’Souza, A. D. & Hanawalt, P. C. R-loop generation during transcription: formation, processing and cellular outcomes. DNA Repair 71, 69–81 (2018).
Brickner, J. R., Garzon, J. L. & Cimprich, K. A. Walking a tightrope: The complex balancing act of R-loops in genome stability. Mol. Cell 82, 2267–2297 (2022).
Marnef, A. & Legube, G. R-loops as Janus-faced modulators of DNA repair. Nat. Cell Biol. 23, 305–313 (2021).
Ginno, P. A., Lott, P. L., Christensen, H. C., Korf, I. & Chédin, F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 45, 814–825 (2012).
Aguilera, A. & García-Muse, T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46, 115–124 (2012).
Merrikh, H., Machón, C., Grainger, W. H., Grossman, A. D. & Soultanas, P. Co-directional replication-transcription conflicts lead to replication restart. Nature 470, 554–557 (2011).
Sollier, J. et al. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell 56, 777–785 (2014).
Janion, C. Inducible SOS response system of DNA repair and mutagenesis in Escherichia coli. Int J. Biol. Sci. 4, 338–344 (2008).
Demjanenko, P., Zheng, S. & Crane, J. K. SOS-inducing drugs trigger nucleic acid release and biofilm formation in Gram-negative bacteria. Biomolecules 14, 321 (2024).
Mazin, A. V. & Kowalczykowski, S. C. A novel property of the RecA nucleoprotein filament: activation of double-stranded DNA for strand exchange in trans. Genes Dev. 13, 2005–2016 (1999).
Zaitsev, E. N. & Kowalczykowski, S. C. A novel pairing process promoted by Escherichia coli RecA protein: inverse DNA and RNA strand exchange. Genes Dev. 14, 740–749 (2000).
Kasahara, M., Clikeman, J. A., Bates, D. B. & Kogoma, T. RecA protein-dependent R-loop formation in vitro. Genes Dev. 14, 360–365 (2000).
Pribis, J. P., Zhai, Y., Hastings, P. J. & Rosenberg, S. M. Stress-induced mutagenesis, gambler cells, and stealth targeting antibiotic-induced evolution. mBio 13, e01074–01022 (2022).
Zhai, Y. et al. Drugging evolution of antibiotic resistance at a regulatory network hub. Sci. Adv. 9, eadg0188 (2023).
Boes, N., Schreiber, K. & Schobert, M. SpoT-triggered stringent response controls usp gene expression in Pseudomonas aeruginosa. J. Bacteriol. 190, 7189–7199 (2008).
Tosa, T. & Pizer, L. I. Effect of serine hydroxamate on the growth of Escherichia coli. J. Bacteriol. 106, 966–971 (1971).
Winther, K. S., Roghanian, M. & Gerdes, K. Activation of the stringent response by loading of RelA-tRNA complexes at the ribosomal A-site. Mol. Cell 70, 95–105.e104 (2018).
Traxler, M. F. et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68, 1128–1148 (2008).
Novotny, L. A., Jurcisek, J. A., Goodman, S. D. & Bakaletz, L. O. Monoclonal antibodies against DNA-binding tips of DNABII proteins disrupt biofilms in vitro and induce bacterial clearance in vivo. EBioMedicine 10, 33–44 (2016).
Martyn, L. et al. Antibodies against the DNABII protein integration host factor (IHF) inhibit sinus implant biofilms. Laryngoscope 130, 1364–1371 (2020).
Peña, J. M. et al. Control of a programmed cell death pathway in Pseudomonas aeruginosa by an antiterminator. Nat. Commun. 12, 1702 (2021).
Turnbull, L. et al. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat. Commun. 7, 11220 (2016).
Yu, H. et al. Elastase LasB of Pseudomonas aeruginosa promotes biofilm formation partly through rhamnolipid-mediated regulation. Can. J. Microbiol. 60, 227–235 (2014).
Kuang, Z. et al. Pseudomonas aeruginosa elastase provides an escape from phagocytosis by degrading the pulmonary surfactant protein-A. PLoS One 6, e27091 (2011).
Krishnan, S., Petchiappan, A., Singh, A., Bhatt, A. & Chatterji, D. R-loop induced stress response by second (p)ppGpp synthetase in Mycobacterium smegmatis: functional and domain interdependence. Mol. Microbiol 102, 168–182 (2016).
Podlesek, Z. & Žgur Bertok, D. The DNA damage inducible SOS response is a key player in the generation of bacterial persister cells and population wide tolerance. Front Microbiol 11, 1785 (2020).
Maslowska, K. H., Makiela-Dzbenska, K. & Fijalkowska, I. J. The SOS system: A complex and tightly regulated response to DNA damage. Environ. Mol. Mutagen 60, 368–384 (2019).
Reffuveille, F. et al. Bone environment influences irreversible adhesion of a methicillin-susceptible Staphylococcus aureus Strain. Front. Microbiol. 9, 2865 (2018).
Urwin, L., Savva, O. & Corrigan, R. M. Microbial Primer: What is the stringent response and how does it allow bacteria to survive stress? Microbiology 170, https://doi.org/10.1099/mic.0.001483 (2024).
Yakimov, A., Bakhlanova, I. & Baitin, D. Targeting evolution of antibiotic resistance by SOS response inhibition. Comput. Struct. Biotechnol. J. 19, 777–783 (2021).
Mo, C. Y. et al. Systematically altering bacterial SOS activity under stress reveals therapeutic strategies for potentiating antibiotics. mSphere 1, https://doi.org/10.1128/mSphere.00163-16 (2016).
Held, K., Ramage, E., Jacobs, M., Gallagher, L. & Manoil, C. Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J. Bacteriol. 194, 6387–6389 (2012).
Rio, D. C., Ares, M. Jr., Hannon, G. J. & Nilsen, T. W. Purification of RNA using TRIzol (TRI reagent). Nat. Protoc. 2010, pdb.prot5439 (2010).
Sanz, L. A. & Chédin, F. High-resolution, strand-specific R-loop mapping via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc. 14, 1734–1755 (2019).
Bushnell, B., Rood, J. & Singer, E. BBMerge–accurate paired shotgun read merging via overlap. PLoS One 12, e0185056 (2017).
Kopylova, E., Noé, L. & Touzet, H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinform 28, 3211–3217 (2012).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinform 25, 2078–2079 (2009).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinform 31, 166–169 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Wu, T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Bai, K. et al. Extraction and detection of guanosine 5’-diphosphate-3’-diphosphate in amino acid starvation cells of Clavibacter michiganensis. Braz. J. Microbiol. 52, 1573–1580 (2021).
Ochi, K., Kandala, J. C. & Freese, E. Initiation of Bacillus subtilis sporulation by the stringent response to partial amino acid deprivation. J. Biol. Chem. 256, 6866–6875 (1981).
Chotirmall, S. H. et al. Sputum Candida albicans presages FEV₁ decline and hospital-treated exacerbations in cystic fibrosis. Chest 138, 1186–1195 (2010).
Mac Aogáin, M. et al. Immunological corollary of the pulmonary mycobiome in bronchiectasis: the CAMEB study. Eur. Respir. J. 52, 1800766 (2018).
Ali, N. A. T. B. M., Mac Aogáin, M., Morales, R. F., Tiew, P. Y. & Chotirmall, S. H. Optimisation and benchmarking of targeted amplicon sequencing for mycobiome analysis of respiratory specimens. Int. J. Mol. Sci. 20, 4991 (2019).
Mac Aogáin, M. et al. Metagenomics reveals a core macrolide resistome related to microbiota in chronic respiratory disease. Am. J. Respir. Crit. Care Med. 202, 433–447 (2020).
Mac Aogáin, M. et al. Integrative microbiomics in bronchiectasis exacerbations. Nat. Med. 27, 688–699 (2021).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Huson, D. H., Auch, A. F., Qi, J. & Schuster, S. C. MEGAN analysis of metagenomic data. Genome Res. 17, 377–386 (2007).
Acknowledgements
We thank Professor Anders Løbner-Olesen from Copenhagen University for his discussions on possible sources of RNA: DNA hybrids. We acknowledge Dr Sujatha Subramoni for providing transposon mutants and help with molecular cloning work, Dr Yong Hwee Foo for discussions regarding microscopy imaging and analysis, and Dr Sharon Longford for proofreading. The Singapore Centre for Environmental Life Sciences Engineering (SCELSE) is funded by the Ministry of Education, Singapore, the National Research Foundation of Singapore, Nanyang Technological University Singapore (NTU) and National University of Singapore (NUS) and hosted by NTU in partnership with NUS (S.K.).
Author information
Authors and Affiliations
Contributions
S.M. performed all experiments; S.M., T.S., and Z.D. designed experiments. Z.D. performed sequencing and analysis. S.H.C. helped with providing clinical sputum sample and metagenome analysis. S.M., T.S., Z.D., and S.K. wrote the manuscript. T.S. and S.K. (project management).
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Erin Gloag, John Crane, and the other anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mugunthan, S., Dong, Z., Chotirmall, S.H. et al. Stress-induced toxic genomic R-loops support biofilm extracellular matrix formation. Nat Commun 16, 11451 (2025). https://doi.org/10.1038/s41467-025-66315-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66315-5