Abstract
Over the past decade, non-viral DNA template delivery has been used with engineered nucleases to target single-stranded DNA sequences in hematopoietic stem and progenitor cells. While effective for gene therapy, this method is limited to short DNA donor templates, restricting its applications to gene corrections. To expand its scope, we developed an editing process using kilobase-long circular single-stranded DNA donor templates and TALEN technology. Our results show that the CssDNA editing process achieves high gene insertion frequency in HSPCs. Compared to AAV-edited HSPCs, CssDNA-edited HSPCs show a higher propensity to engraft and maintain gene edits in a female NCG murine model. This positive outcome is partly due to higher levels of primitive edited HSPCs, a more quiescent metabolic state, and elevated expression of bone marrow niche adhesion markers. Our findings highlight the strong potential of CssDNA as a universal, scalable and efficient non-viral DNA template for gene therapy applications.
Similar content being viewed by others
Introduction
Gene therapy strategies utilizing hematopoietic stem and progenitor cells (HSPCs) hold the promise of delivering a lifelong supply of therapeutic agents to patients. Gene editing techniques expand these therapeutic prospects by employing engineered nucleases to introduce sequence-specific double-strand breaks (DSBs) and promote the targeted insertion of therapeutic genes vectorized as DNA donor templates. While viral vectors, particularly Adeno-associated viruses (AAVs), are the most prevalent and efficient carriers of DNA donor templates in vitro1,2,3,4,5,6, they have been recently associated with genotoxicity, impairment of HSPCs engraftment in murine models as well as loss of gene insertion events7,8,9,10,11,12. In this context, non-viral DNA templates recently emerged as promising alternatives to AAV6. Non-viral DNA templates come in different formats. The first format explored was linear double-stranded DNA (LdsDNA). When vectorized with engineered nuclease, LdsDNA enables gene correction and gene insertion in multiple cell types. But its recognition by intracellular DNA-sensing pathways triggers cellular toxicity and impairs the overall efficiency of gene editing, especially in HSPCs13,14. To overcome this hurdle, linear single-stranded DNA (LssDNA) was considered. LssDNA, mostly used as short linear oligonucleotides (LssODNs, <300b), promotes efficient gene correction in Long-Term Hematopoietic Stem cells (LT-HSCs) without apparent cellular toxicity8,9,11,15. This correlated with an efficient engraftment of edited HSPCs in murine models and to the maintenance of their corrective events. Despite this, the limitation of short LssODNs to single-point mutation corrections led the field to explore longer LssDNA templates (>1 kb) for full gene insertion in primary cells. Although highly efficient in T cells, LssDNA showed a dose-dependent cellular toxicity and displayed low levels of gene insertion efficacy in HSPCs16, likely due to its degradation by exonucleases in the cytoplasm17.
Circular single-stranded DNA (CssDNA) offers a promising solution to the aforementioned challenges observed with different DNA donor template formats. Indeed, it may mitigate the activation of intracellular DNA-sensing pathways that evolved to specifically recognize foreign dsDNA and is inherently resistant to exonuclease attacks. Recent proof-of-concept studies on CRISPR-CAS9/CssDNA-mediated gene insertion in human cell lines have demonstrated up to 25% gene insertion at multiple endogenous loci18. These promising results were very recently confirmed in HSPCs, although the editing frequency and viability obtained seemed suboptimal (~60 % cell viability and ~20% KI efficiency). Most importantly, the functionality of the resulting edited HSPCs was not assessed in vitro and in vivo19. Furthermore, there has been no direct in vitro and in vivo comparison with AAV6, the standard DNA template vectorization method for targeted gene insertion. Therefore, a thorough investigation of CssDNA’s potential for gene insertion in HSPCs and comparison with other DNA vectorization methods, including linear single-stranded DNA (LssDNA) and AAV6, is essential for gauging its absolute and relative potential for the field of cell and gene therapy.
Here we developed a gene insertion process incorporating non-viral DNA donor templates (LssDNA or CssDNA) and TALEN technology for HSPC editing. Our findings indicate that the CssDNA editing process yields a 3- to 5-fold higher gene knock-in (KI) frequency than LssDNA process, with efficiencies surpassing 40%. Notably, this correlates with increased cell viability and a reduction in the frequency of gene knock-out (KO, via the promotion of insertions and deletions) in CssDNA-edited cells compared to those edited with LssDNA. These outcomes are consistent across various DNA donor template lengths (from 0.6 kb to 2.2 kb) inserted at multiple loci and are transportable to primary T cells. Further comparative analysis of CssDNA and AAV gene insertion processes using a combination of Flow cytometry, CITE-seq transcriptomics, and long-term engraftment in NCG mice, suggest the superiority of CssDNA over AAV6 gene editing process. Our data also unveiled key transcriptomic and phenotypic distinctions associated with the efficacy of both gene insertion processes, including Long-Term HSC-enriched subpopulation (LT-HSCe) number, editing frequency, and bioenergetic status. This work underscores the strong potential of combining CssDNA delivery with an engineered nuclease for gene insertion in LT-HSCs and marks a crucial step towards advancing next-generation cell and gene therapies.
Results
Circular single-stranded DNA molecules can be used as DNA donor template for efficient TALEN-mediated gene insertion in HSPCs
To assess the potential of ssDNA molecules and TALEN editing to promote gene insertion in HSPCs, we chose the exon 1 of the B2M gene as an endogenous genomic target. We used a B2M exon 1-specific TALEN (TALENB2M) and designed promoterless DNA templates encoding cell surface reporters (CSR1 and CSR2) flanked by 300 bases of homology arms specific for the B2M locus. These specific template designs allowed to seamlessly rewire the B2M gene in a disruptive or non-disruptive manner and to re-express the CSRs (Fig. 1a and Supplementary Fig. 1a). Because B2M is uniformly expressed in HSPCs and all their progenitors, this experimental system allows high-content, multiparametric single-cell level characterization of edited HSPCs and progenitor cells using either flow cytometry (FCM) and/or CITE-seq analysis.
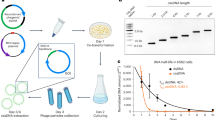
a HSPCs editing protocol using an mRNA-encoded TALEN targeting the B2M locus and LssDNA or CssDNA as DNA donor templates to insert a tag (0.6 kb) or a reported gene (2.2 kb) via non-disruptive and disruptive insertions, respectively (LHA and RHA stand for right and left homology arm, respectively). Created in BioRender. Valton, J. (2025) https://BioRender.com/7qzeb34. mRNAs encoding a viability enhancer and a HDR enhancer (Via-Enh01 and HDR-Enh01, respectively) were also incorporated in the first transfection. The timing is indicated in days (D0-D7). Edited HSPCs obtained 7 days post-thawing (D7) were characterized by flow cytometry to assess the level of phenotypic knock-in (KI) of DNA donor templates and phenotypic knock-out (KO) of B2M as well as their viability. Their differentiation capacity into erythroid and myeloid progenitors was also assessed by colony forming unit (CFU) assay. b Flow cytometry results illustrating the frequency of cells harboring KI events, the ratio KI/KO, the viability and plating efficiency of HSPCs either untreated, electroporated (Mock EP), edited with TALEN only (TALEN), or edited with TALEN and LssDNA or CssDNA donor templates (LssDNA or CssDNA, respectively). Top and bottom panels illustrate results obtained with 0.6 kb and 2.2 kb DNA donor templates, respectively. On each box plot, the central mark indicates the median, the bottom and top edges of the box indicate the interquartile range (IQR), and the whiskers represent the maximum and minimum data point. Each dot represents data obtained from one HSPC donor (numbers of independent donor, i.e., biological replicate, are indicated underneath of each plot). Mann–Whitney two-tailed non-parametric unpaired test with a confidence interval of 95%. P-values are indicated. Source data are provided as a Source data file.
Based on our former work11, we set up a four-day-long gene insertion process consisting of HSPCs thawing (D0), followed by an expansion phase (D0-D2), a transfection of 3 mRNA molecules encoding the TALENB2M, the HDR-Enh01 and the Via-Enh0111 (D2) followed by a second transfection (D3) of the ssDNA donor template. These steps were followed by one (D4) or multiple days of culture (D7 or D14) depending on the read out to be performed (Fig. 1a). Using this experimental setup, we first assessed the influence of the format of ssDNA donor template (Linear or Circular) on the frequency of gene editing (phenotypic gene knock-in, KI, and gene knock-out, KO) as well as on the viability and differentiation capacity of edited HSPCs (Fig. 1 and Supplementary Fig. 1). Three negative controls, consisting in untreated cells and electroporated cells with and without TALEN (“Untreated”, “TALEN” and “Mock EP” groups, respectively) were performed as references. Our results obtained with the 0.6 kb ssDNA molecule encoding the CSR1 showed that both ssDNA formats elicited precise and non-disruptive KI at the B2M locus as demonstrated by the detection of a CSR1( + ) HSPC population by FCM (Supplementary Fig. 1b, c). Interestingly, CssDNA promoted 5-fold higher KI than LssDNA, (Fig. 1b top panel, 45.2% ± 5.0% and 8.8% ± 3.6%, mean ± SD, respectively and Supplementary Fig. 1c) with up to 51% of KI. This correlated with a 10-fold higher KI/KO ratio obtained with the CssDNA compared to the LssDNA (Fig. 1b top panel, 1.30 × 1.18±1 gMean x gSD±1, giving a dispersion from 1.10 to 1.52 and 0.11 × 1.82±1 gMean x gSD±1, giving a dispersion from 0.06 to 0.21, respectively), indicating that CssDNA was more efficiently integrated at the B2M locus than the LssDNA. Although the electroporation process significantly impacted HSPC viability with respect to untreated cells (Fig. 1b top panel, median cell viability 81.3% and 94.5%, respectively, p-value = 0.01), addition of TALENB2M, LssDNA, or CssDNA did not further impact it significantly (Fig. 1b top panel, median cell viability 71%, 71% and 78%, respectively). This indicated that the electroporation process was mainly responsible for the slight loss of HSPC viability observed after gene editing. A similar pattern was observed by colony-forming unit (CFU) assay. Indeed, HSPCs electroporated alone or edited in the presence of TALENB2M, LssDNA, or CssDNA were able to differentiate in a similar fashion (Fig. 1b top panel, median plating efficiency of 17%, 14%, 14% and 17%, respectively).
To assess if our editing process could enable longer gene insertion in HSPCs, we performed similar experiments with a 2.2 kb ssDNA template encoding CSR2, designed for disruptive insertion at the B2M locus. Our results showed that both ssDNA formats elicited precise and disruptive KI at the B2M locus as demonstrated by the detection of a CSR2( + ) HSPC population by FCM. CssDNA promoted higher KI and KI/KO ratio than LssDNA (Fig. 1b, bottom panel, 3.6-fold and 6.2-fold respectively, Supplementary Fig. 1b and c) with up to 49% of KI. HSPC viability and differentiation capacity observed after 2.2 kb ssDNA editing followed the same trend as the one observed with the 0.6 kb ssDNA. Together, these results indicate that CssDNA and TALENB2M editing process promoted significantly higher gene insertion than LssDNA in HSPCs without severely impacting HSPC viability and differentiation capacity.
Circular ssDNA-mediated gene insertion is applicable to different endogenous Loci
To gauge the applicability of CssDNA and TALEN editing for gene insertion at other loci in HSPCs, we designed 3 additional CssDNA donor templates specific for AAVS1, CD11b and S100A9 loci, demonstrated earlier to allow pan-lineage or myeloid-specific expression of therapeutic transgenes20 (Supplementary Fig. 2). Using the aforementioned gene editing protocol, loci-specific TALEN and promoterless matrices encoding the reported gene CSR3, we showed that AAVS1, CD11b and S100A9 loci were efficiently modified, reaching gene insertion frequency and viability similar to those obtained with TALENB2M and CssDNA encoding CSR2 (Supplementary Fig. 2b, 34% ± 9.6%, 26% ± 6.6% and 39% ± 10.5%, mean KI frequency ± SD, respectively). This additional dataset indicates that CssDNA could be used to promote efficient gene insertion at a variety of loci in HSPCs.
Circular ssDNA-edited HSPCs efficiently engraft in NCG mice
To verify the suitability of CssDNA for HSPC-based ex vivo gene therapy applications, we assessed the ability of HSPCs edited by the TALENB2M and the CssDNA encoding CSR2, to engraft in an NCG murine model, to differentiate and maintain their gene insertion events, 16 weeks post injection onset (Fig. 2a). A control experiment was performed with HSPCs edited with an adeno-associated virus editing process (AAV process, TALENB2M and AAV6 encoding the CSR2), since AAV is considered as the conventional and therapeutically relevant DNA donor template vectorization method for targeted gene insertion in human HSPCs1,2,3,4,5,6. We employed a transduction protocol akin to the most advanced one, which has been optimized, vetted and employed by multiple teams in combination with mRNA encoding Zinc Finger nuclease, TALEN and CRISPR-CAS9 delivered as mRNA or with CRISPR-Cas9 delivered as RNPs10,11,12,21,22,23. This 4-days process, incorporating an AAV transduction step right after RNP or mRNA transfection, was also adapted to GMP-compliant conditions to generate CRISPR-CAS9 and AAV6-edited CD34 + HSPCs1 evaluated in the clinical trial NCT0481984. In addition, to allow for rigorous side-by-side comparisons of both processes and mitigate the occurrence of the confounding side effects usually associated with AAV612,24, we defined the minimal MOI of AAV6 (350 vg/cells) to reach similar gene insertion efficiency as the one obtained with the CssDNA editing process. By design, both CssDNA and AAV editing processes elicited similar KI frequency (Supplementary Fig. 3a left panel, 30.7% ± 2.1% and 26.1% ± 5.4%, mean ± SD, respectively), KI/KO ratio (Supplementary Fig. 3a left panel, 0.6 × 1.14±1 gMean x gSD±1, giving a dispersion from 0.53 to 0.69 and 0.52 ×1.33±1 gMean x gSD±1, giving a dispersion from 0.39 to 0.69, respectively) and cell viability (Supplementary Fig. 3a middle panel, 81.0% ± 1.0% and 82.5% ± 4.3%, mean ± SD, respectively) using 3 HSPC donors. As demonstrated earlier, edited HSPCs were able to differentiate in vitro as observed by CFU assay (Supplementary Fig. 3a, right panel). Interestingly, although not significant, the AAV editing process led to a 1.7-fold and 1.5 to 2-fold increase of plating efficiency and proliferation capacity, respectively, compared to its CssDNA counterparts (Supplementary Fig. 3a right panel, 31.3% ± 5.2% and 19.2% ± 4.5%, mean plating efficiency ± SD, respectively, Supplementary Fig. 3a lower panel, 1.6 ± 0.2 and 1.1 ± 0.1 mean proliferation ± SD between D0 and D4, respectively, 6.7 ± 1.0 and 3.5 ± 0.2 mean proliferation ± SD between D0/D7, respectively).
a HSPC editing protocol. Created in BioRender. Valton, J. (2025) https://BioRender.com/hjz2iqm. b left panel, Frequencies of human CD45(+) cells (hCD45(+)) engraftment. b middle panel, Frequencies of KI and KI/KO ratio determined either before mice injection (input), or in hCD45(+) cells engrafted in the bone marrow (BM) of NCG mice, 16 weeks after cells injection onset (output). Two-way ANOVA followed by Bonferroni multi-comparison test. P-values are indicated (n = 2 donors). b right panel, Number of KI(+) hCD45(+) cell detected in the bone marrow. Mann–Whitney two-tailed non-parametric unpaired test with a confidence interval of 95%. P-value is indicated. On each box plot, the central mark indicates the median, the bottom and top edges of the box indicate the interquartile range (IQR), and the whiskers represent the maximum and minimum data point. Each dot represents data obtained from one HSPC donor and one mouse. c UMAP plots showing aggregated CITE-seq analysis obtained at the time of NCG mice injection (D4). The different cell subpopulations identified are illustrated by a color code indicated underneath the plot. The definition of each subpopulation is documented in the methods section. d UMAP plots showing the position of LT-HSCe subpopulation. e, f UMAP plots showing the cell cycle phases (G0/G1, G2M and S). Number of biological replicates (n) used to obtain UMAPs are indicated underneath the plots. g left panel, Mean frequency ± SD of LT-HSCe within all subpopulations. Two-way ANOVA with Bonferroni post-tests. P-values are indicated. g middle and right panels, Mean frequency ± SD of KI and KO within the LT-HSCe subpopulation, respectively. Two tailed paired t-test. P-values are indicated (n = 3 donors). h gMean x gSD±1 of KI/KO ratio and mean frequency ± SD of KI or KO computed from each subpopulation in top and bottom panels, respectively (n = 3 donors; subpopulations identified with fewer than 100 cells are not displayed). i Gene Set Enrichment Analysis (GSEA) obtained in unedited and edited LT-HSCe. Normalized Enrichment Score (NES) and Log P value obtained for each donor computed as two-sided p-value corrected for multiple comparisons, are illustrated (n = 3 donors). Source data are provided as a Source data file.
HSPCs donors 1 and 2, showing the most similar editing outcomes in vitro between CssDNA and AAV6 processes, were used for further side-by-side comparison in vivo. Our results obtained 16 weeks post-injection onset, showed that both processes led to similar HSPC engraftment frequencies, although HSPC edited by the AAV process showed the lowest one (Fig. 2b left panel, 65.0% ± 13.6% and 46.0% ± 17.0%, mean ± SD, for CssDNA and AAV6 processes, respectively). They also elicited similar levels of HSPC differentiation into Lymphocyte T and B, myeloid cells, monocytes, macrophages, and neutrophils with respect to untreated cells (Supplementary Fig. 3b). Notably, the CssDNA editing process allowed for preservation of a significantly higher frequency of KI events than the AAV editing process. Indeed, we found a 3-fold higher KI frequency in hCD45 cells engrafted in animals injected with CssDNA-edited HSPCs compared to those injected with AAV-edited HSPCs (Fig. 2b middle panel, 14.9% ± 4.1% and 5.5% ± 3.2% mean ± SD, respectively). This trend was consistent among the different cell subpopulations studied (Supplementary Fig. 3c). In addition, the CssDNA editing process allowed to keep a KI/KO ratio constant between the input and output, in contrast with the drop observed with the AAV6 editing process (Fig. 2b middle panel, 0.59 versus 0.50, and 0.58 versus 0.14, gMean KI/KO ratio obtained at input and output for CssDNA and AAV processes, respectively). Ultimately, the overall engraftment efficiency of edited HSPCs, calculated by multiplying the bone marrow engraftment and output KI frequencies, was 5-fold higher for HSPCs edited using the CssDNA process compared to those edited using the AAV process (Fig. 2b right panel, 9.20 × 104 versus 1.85 × 104, mean number of KI(+) HSPCs per 106 bone marrow cells). Collectively, our findings indicate that the CssDNA editing process enables efficient targeted gene insertion in HSPCs, producing cells capable of engrafting in the bone marrow of the NCG murine model at significantly higher levels than those edited with the AAV6 process.
Circular ssDNA is efficiently inserted in LT-HSCe
To understand the significant differences observed between CssDNA- and AAV6-edited HSPC engraftment in the murine model, an in-depth characterization of cells gathered right before their injection in NCG mice (D4), was performed using single-cell CITE-seq analysis. A total of 22,359 encapsulated cells coming from 3 different donors (including those used for in vivo engraftment analysis), enabled us to analyze a median of 2414 cells per experimental group and per donor, with a median of 6318 genes analyzed per cell (Supplementary table 1). This comprehensive dataset enabled us to first decipher the different cell populations present in the cellular mixture using as reference, the comprehensive dataset documented in ref. 25 (Fig. 2c and Supplementary Fig. 4). Among them, we identified the most primitive subpopulation harboring, among others, the markers commonly expressed by Long-Term HSCs (AVP, HLF, ITGA3, CRHBP, THY1, and PROM1, Fig. 2d and Supplementary Fig. 4)11,25,26,27. This subpopulation, displaying a majority of cells in G0/G1 (Fig. 2e,f and Supplementary Fig. 4), will be called Long-Term HSC enriched subpopulation (LT-HSCe) in the following, for the sake of simplicity.
Interestingly, the frequency of LT-HSCe was found to be 5-fold higher in the CssDNA-edited HSPCs compared to the AAV-edited or untreated HSPC groups (Fig. 2g left panel, 7.7% ± 3.0%, 1.4% ± 0,5%, and 2.0% ± 1%, median ± SD, respectively). Further analysis of the sequence of B2M transcripts as well as surface expression of the CSR2, enabled us to confidently identify cells displaying phenotypic and genotypic KI as well as genotypic KO, in the following referred to as insertion and deletion (indels) events, within each subpopulation of edited HSPCs (Supplementary Fig. 5a). Using this additional layer of information, we determined that the frequency of LT-HSCe harboring KI events (KI( + ) LT-HSCe) obtained using the CssDNA process was 10-fold higher than that obtained using the AAV process (Fig. 2g middle panel, 10.7% ± 4.0% and 0.7% ± 1.2%, mean ± SD, respectively). In stark contrast, the frequency of LT-HSCe harboring indel events (Indels(+) LT-HSCe) was surprisingly similar from one process to another (Fig. 2g right panel, 14.7% ± 2.5% and 12.0% ± 7.2%, mean ± SD, respectively). This phenomenon was also observed in other most primitive subpopulations, including Late Multipotent Progenitor and Macrophage Dendritic Progenitor (LMPP and MDP, respectively), but not in differentiated subpopulations including Progenitor Megakaryocytes, Erythroid and Myeloid intermediates (Prog_MK, Ery, Myeloid Int, respectively) as non-limiting examples, Fig. 2h). Deeper investigation of the KI events within each cell cycle phase of LT-HSCe subpopulation, showed that CssDNA-edited KI( + ) LT-HSCe were mainly set in G0/G1 phase (80%, Supplementary Fig. 5b, left panel). Unfortunately, such analysis could not be rigorously performed on AAV-edited LT-HSCe due to the low number of KI( + ) LT-HSCe cells identified in this group (1 LT-HSCe KI(+) cell, Supplementary Fig. 5b, right panel).
Nevertheless, we could compare the overall transcriptomics status of LT-HSCe detected in each experimental group using Gene Set Enrichment Analysis (GSEA, Reactome, Fig. 2i). Our results showed that pathways associated with DNA metabolism, cell cycle, DNA repair, and mitochondrial bioenergetics were significantly downregulated in CssDNA-edited LT-HSCe compared to its AAV-edited and untreated counterparts. This indicates a lower overall proliferative status of the former group, in agreement with the conclusions made earlier from the cell cycle analysis and cellular fold expansion (Supplementary Fig. 3a). It also suggests that the double electroporation may slow down LT-HSCe proliferation and differentiation into early and late progenitor cells. Interestingly, the opposite phenomenon was observed with respect to inflammation pathways (upregulation of IFNα/β and γ pathways in CssDNA-edited LT-HSCe compared to AAV-edited and untreated LT-HSCe, Fig. 2i), indicating that CssDNA was more likely to be recognized by DNA-sensing pathways than the AAV particles in the cytoplasm, in agreement with a former study11. Furthermore, we found no evidence of P53 pathway upregulation between the AAV-edited and CssDNA-edited or untreated cells, as expected from an AAV-editing process performed at low MOI10,21. This allowed us to rule out the potential contribution of the P53-dependent toxicity to the difference of engraftment and KI stability observed in vivo between the different experimental groups. Finally, we found that the CXCR4, CD44, and F11R, three surface markers involved in cellular adhesion to the bone marrow niche, were significantly upregulated in the LT-HSCe subpopulation of CssDNA-edited cells compared to their AAV-edited counterparts or untreated cells (Supplementary Fig. 5c, Log fold change of expression ≥1).
Circular ssDNA-mediated gene insertion is applicable to T cells
While CssDNA bears a strong potential for ex vivo HSPC-based gene therapy, it may also find applicability in other fields, including CAR T-cell therapy and immuno-oncology. We thus also assessed the transposability of CssDNA and TALEN editing to T cell engineering. To investigate that aspect, we established a T-cell-centric TALEN engineering protocol in the absence of Via-Enh01 and HDR-Enh01 encoding mRNA and used it to assess the efficiency of gene insertion mediated by 2.2-kb CssDNA and LssDNA templates encoding the CSR2 specific for the B2M locus (Supplementary Fig. 6a). Our results show that CssDNA promoted higher frequency of gene insertion than the LssDNA, whatever the dose being used, with up to about 40% of KI (Supplementary Fig. 6b and c). This process did not significantly impact the viability of edited T cells at D7 (Supplementary Fig. 6d, mean viable cell frequency >90% for both CssDNA-, LssDNA-, and TALEN-edited T cells) and allowed them to proliferate, although to a lower extent than T cells edited by TALENB2M alone (~3.7-fold lower proliferation rate of CssDNA-treated T cells compared to TALENB2M alone-treated T cells, Supplementary Fig. 6e).
Discussion
The goal of this study was to evaluate the potential of linear and circular ssDNA donor templates (LssDNA and CssDNA) to enable TALEN-mediated gene insertion in HSPCs. Our findings demonstrate that both DNA donor templates enable precise gene insertion at endogenous loci. Circularization of ssDNA significantly and markedly increases the efficiency of gene insertion frequency by up to 5-fold, with absolute values surpassing 40% using a 2.2 kb CssDNA template. This unprecedented efficiency of CssDNA insertion correlates with a significant decrease of Indel events, considered as by-products of the gene insertion process. Our CssDNA-editing process can be used to insert kilobase-long genes at multiple independent loci and is applicable to other cell types, including primary T cells used for therapeutic applications. Comparative analysis with an optimized AAV-editing process reveals that HSPCs edited by CssDNA display a higher propensity to engraft and maintain their gene insertion edits in a murine model compared to HSPCs edited by AAV. This difference could be explained in part, by significantly higher levels of KI( + ) LT-HSCe subpopulation, a more quiescent metabolic state, and an elevated expression of bone marrow niche adhesion markers, in CssDNA- compared to AAV6-edited HSPCs. Our findings underscore the strong potential of CssDNA as a universal and efficient non-viral DNA template for cell and gene therapy applications.
We found that the efficiency of TALEN-mediated gene insertion was up to 5-fold higher with CssDNA than LssDNA donor template. This significant difference, observed in HSPCs and primary T cells, using two lengths of ssDNA templates and across several donors, is consistent with results obtained in K562 and HEK293T cell lines edited with CRISPR-Cas9 nuclease18. It also aligned with a very recent study published just before the submission of this work, showing that circularization of ssDNA increases gene insertion efficiency by 5-fold in CRISPR-CAS9 edited K562 cells19. In that study, CssDNA was also used as DNA donor template in combination with CRISPR-CAS9 nuclease or nickase (vectorized as RNP or mRNA) to insert, in a targeted fashion, kilobase-long DNA in primary T cells with KI efficiency ranging from 15% to 40% and a cell viability around 80% (depending on the locus edited and T cell donor). Targeted insertion of CssDNA was also demonstrated in other cell types with variable cell viability and KI efficiency outcomes including in iPSCs (~30% cell viability and 40–60% KI efficiency), NK cells (~80% cell viability and 20–40% KI efficiency), B-cells (~80% cell viability and 25% KI efficiency), and HSPCs (~60% cell viability and ~20% KI efficiency). Our work thus corroborates and extends these former investigations by demonstrating that circularization of ssDNA improves KI efficiency with up to 49% of multi-kilobase-long gene insertion efficiency in HSPCs. It also shows that edited HSPCs are viable (>80% viability, Fig. 1) and endowed with long-term engraftment capacity in an immunodeficient murine model (Fig. 2). Furthermore, taking into account former work, our study implies that a variety of engineered nuclease platforms, including CRISPR-Cas9 (SpyCas9), D10A CRISPR-Cas9 nickase19, CRISPR-Cas12a (AspCas12, LbaCas12a, FnoCas12a)18 and TALEN, could be combined with CssDNA donors to promote non-viral gene insertion in different primary cells. The similar gene insertion efficiency obtained with these different nuclease platforms suggests that the insertion mechanism of CssDNA is independent from the pattern of DNA cleavage ranging from near blunt ends for CRISPR-CAS928, single-strand nicking for D10A CRISPR-CAS929, to 5’ overhangs for CRISPR-CAS12a30 and TALEN31,32. However, a direct comparison of these platforms for corresponding parameters, such as different homology arms, strandedness, and chemistry of CssDNA donors, would be needed for a definitive statement and detection of any platform-specific preferences and mechanisms.
The difference in gene insertion efficiency observed in the presence of LssDNA and CssDNA could be due to multiple factors. One factor could be the hydrodynamic volume and shape of ssDNA, which may affect its penetration of the nuclear membrane during electroporation. Another more likely factor could be its stability and susceptibility, respectively, to Trex1 exonuclease-dependent hydrolysis, as recently demonstrated in the context of CRISPR-CAS9-mediated insertion of short LssODN17. Adding the widely used and extensively characterized phosphorothioate bonds at the ends of LssDNA sequence could be beneficial by preventing its hydrolysis, increasing its stability, and by extension, its insertion efficiency17,33,34,35,36. Although the incorporation of such modifications in long LssDNA molecules is not streamlined yet with conventional synthesis technics, the enzymatic in vitro technology used in this study and derived from37 to assemble LssDNA, could be suitable for the incorporation of chemically modified dNTPs by natural or engineered DNA polymerases. Thus, further research should be conducted to investigate this avenue, assess its suitability for therapeutic applications, and monitor its potential cellular toxicity as suggested earlier38.
Nevertheless, we show that adapting the format of the ssDNA donor template through simple circularization is sufficient to significantly improve the efficacy of gene insertion in primary cells. This improvement was demonstrated by rigorously comparing LssDNA- and CssDNA-mediated gene insertion in the presence of an mRNA encoding HDR-Enh01, a genetically encoded inhibitor of 53BP1, known to increase the efficiency of gene correction in HSPCs without generating additional and detectable adverse events11. The systematic incorporation of HDR-Enh01 in our gene editing process thus contributes to the high levels of gene insertion obtained in this study. Nonetheless, achieving these high levels of gene insertion did not require Cas9 targeting sequences (CTS)39, HDR-boosting modular sequences40 or chemical modifications17,33,34,35,36, although their incorporation in the CssDNA-mediated editing process could be beneficial. In addition, it did not need small molecule-based inhibition of DNA-PK and Polθ, an elegant approach known to dramatically increase the efficiency of gene insertion/correction in multiple cell types41,42 but likely to promote large-scale genomic alterations in HSPCs when used in the presence of short ssDNA donor templates43.
Our CssDNA-mediated editing process enables targeted insertion of a multi-kilobase gene in HSPCs, achieving unprecedented efficiency with respect to recent studies16,19. However, beyond editing efficiency, the functionality of edited cells is of paramount importance for their effective translation into HSPC-based gene therapy products. In this regard, the choice of the DNA donor template format as well as the editing process appear to be critical8,9,11. With these considerations in mind, we assessed the functionality of CssDNA-edited HSPCs in vivo using an NCG murine model and analyzed their transcriptomic status at a single-cell level. This in-depth characterization was performed alongside AAV-edited HSPCs, since AAV is regarded as a reference of DNA donor template format for ex vivo HSPC-based gene therapy applications1,2,3,4,5,6 and was the first to be used in the clinic in the context of sickle cell disease targeted gene correction therapy (CEDAR trial, NCT04819841). Our results demonstrate that both groups of edited HSPCs were able to engraft and differentiate in NCG mice, suggesting they contain a fraction of LT HSCs. However, the level of engrafted HSPCs harboring gene insertion events (KI(+) HSPCs) was 5-fold higher in animals injected with CssDNA-edited HSPCs compared to those injected with AAV-edited HSPCs (Fig. 2b right panel). This differential outcome was obtained despite similar levels of KI observed at input for both groups and the extremely low AAV6 MOI used to mitigate the well-known toxicity of this DNA vectorization approach in HSPCs (Supplementary Fig. 3a left panel and Fig. 2i, respectively)12,24. These results are reminiscent of former works8,9,11 and suggest that the CssDNA editing process enables the generation of higher levels of LT HSCs harboring KI events compared to its AAV counterpart. Of note, a SCID repopulating cell assay and multiple serial transplantations would be needed to fully confirm this claim44,45,46. In agreement with this hypothesis, transcriptomic analysis performed on HSPCs just before their injection in mice showed significantly higher numbers of LT-HSCe in CssDNA-edited HSPCs than in AAV-edited HSPCs (Fig. 2g, left panel, 5-fold). In addition, CssDNA-edited LT-HSCe showed a significantly higher frequency of KI and a higher expression of bone marrow niche adhesion markers than their AAV counterparts (Fig. 2g middle panel, 10-fold and Supplementary Fig. 5c, > 1 log fold change of CXCR4, CD47, and F11R expression, respectively). We hypothesize that the combination of these different features contributes to an efficient engraftment of CssDNA-edited LT-HSCe and to the maintenance of their KI events in our murine model. Notably, this pattern was observed despite a pronounced and significant upregulation of interferon α, β, and γ inflammatory pathways in the CssDNA-edited LT-HSCs compared to their AAV and untreated counterparts. This suggests that CssDNA activates the cGAS-STING pathway-dependent DNA sensing response in the cytoplasm, whereas AAV6 effectively evades this response by delivering its DNA payload directly into the nucleus47. These results are consistent with previous studies11,14. We hypothesize that the CssDNA editing process elicits a transient and reversible inflammatory response that does not greatly affect the engraftment capacity of edited HSPCs. It is however, noteworthy that upregulation of inflammation pathways has recently been associated with negative therapeutic outcomes of engineered HSPC-mediated gene therapies of sickle cell disease48 and chronic granulomatous disease49. Thus, preventing CssDNA-mediated inflammation could potentially be beneficial to optimizing the therapeutic potency of CssDNA-edited HSPCs. In that respect, incorporation of cGAS-STING inhibitory chemical modifications or DNA gapmer sequences34,50,51 within the ssDNA donor template appears to be relevant avenues to explore.
Interestingly, while the phenotypic KI/KO ratio remained stable after long-term engraftment of CssDNA-edited HSPCs (Fig. 2b, middle panel), it significantly decreased in their AAV-edited counterparts. This suggests that the CssDNA editing process generates similar levels of phenotypic KI and KO editing events within LT-HSCs, whereas its AAV counterpart may favor phenotypic KO over KI events in LT HSCs. In accordance with this finding, our transcriptomic analysis showed that while the CssDNA-editing process elicited similar levels of genotypic KI and indel events within LT-HSCe, the AAV-editing process rather favored genotypic indels over KI events, specifically in LT-HSCe (Fig. 2g, h). This imbalance of KI over indel events was expected in AAV-edited LT-HSCe7,24,52,53. Indeed, genotypic KI events mainly occur in the S/G2M phases through the homology-directed repair (HDR) pathway. These events are thus less favorable in quiescent and primitive cells compared to indel events, which can happen in any cell cycle phase through the non-homologous end joining (NHEJ) pathway7,24,52,53. In agreement with this, such imbalance was also observed in other primitive subpopulations but was found normalized (KI/indels ratio ~1), if not inverted (KI/indels ratio >1), in most of the differentiated subpopulations, except for B cells, showing a KI/indels ratio <1 for 2 out 3 donors (Fig. 2g and h, AAV experimental group). It is important to note that this imbalance may also be attributed to the higher toxicity of AAV particles on primitive subpopulations compared to differentiated subpopulations, despite the absence of any observed evidence of P53 pathway upregulation, commonly observed as a toxic side effect of the AAV-based delivery of DNA editing components (Fig. 2i, compare AAV versus Untreated and CssDNA groups)10,11,21. Further work is now needed to fully investigate this point.
Surprisingly, in contrast to the observation described above for the AAV-editing process, the CssDNA-editing process elicits similar levels of KI and indel events in the most primitive subpopulations including MPP, LMPP, MDP, and LT-HSCe (Fig. 2g and h). Moreover, when focusing on the LT-HSCe subpopulation, this process was also found to skew the KI events toward cells set in G0/G1 phases over those set in G2M or S phases (Supplementary Fig. 5b). This unexpected pattern could be due to multiple parameters including the overall engineering process incorporating 2 electroporations and/or to alternative mechanisms of ssDNA template insertion (via synthesis-dependent strand annealing, SDSA, single-stranded DNA incorporation, ssDI or single-strand annealing, SSA)54,55,56,57. These alternative mechanisms differ from the standard RAD51-dependent HR-mediated dsDNA insertion promoted in the presence of AAV6 and could be cell cycle independent, as described for SSA57. Further work is now needed to fully investigate this point, which lies outside the scope of this study.
The success of HSPC-based ex vivo gene therapy relies on multiple factors ranging from the efficiency of gene modification/insertions, the stemness of edited HSPC subpopulation, the viability, differentiation capacity and long-term engraftment potential of the resulting edited HSPCs. Multiple avenues have been explored in the past decade to improve the efficiency of targeted gene insertion in mobilized HSPCs, a cell population relevant for ex vivo gene therapy. Utilization of engineered nuclease and AAV6 as DNA donor template showed highly efficient gene targeting outcomes without affecting the overall HSPC viability and differentiation capacity in vitro. These outcomes were obtained with different engineered nuclease platforms, were reproducible across multiple loci, and obtained by several teams across the world1,2,3,4,5,6. The vivid enthusiasm for this approach was however dampened by the suboptimal functionality of edited HSPCs including their engraftment capacity in murine models as well as their poor ability to maintain their gene insertion edits after long-term engraftment in vivo7,8,9. Further in-depth investigations shed light onto these phenomena by demonstrating the propensity of AAV6 to activate P53 pathways in a dose-dependent manner, and reduce the clonal repertoire of engrafted HSPCs10,11,12. These converging evidences led the field to explore other strategies, including those aimed at reducing the confounding toxicity of AAV624 and those using alternative DNA donor template vectorization8,9,10,11,15,58. Our work further explores and expands the latter avenue by showing that CssDNA can be used as DNA donor template along TALEN gene editing to efficiently promote non-viral full gene insertion in HSPCs and more specifically, in the LT-HSCe subpopulation. We demonstrate that CssDNA-edited HSPCs are viable, functional, and can engraft and differentiate in vivo in a xenograft murine model, 16 weeks post-injection onset. We also demonstrate that it can be used to insert kilobase-long genes at multiple loci and in different cell types, including primary T cells used for therapeutic applications. CssDNA templates can be generated with lengths of up to 9 kilobases, with yields varying based on the methodologies employed19,37,59. In this study, the CssDNA was synthesized using in vitro enzymatic DNA synthesis via rolling-circle amplification37. This production process is highly scalable due to its reliance on isothermal enzymatic reactions and is also potentially safer than fermentation-based DNA production methods19,59, which are often susceptible to endotoxin contamination. As a result, it could be seamlessly implemented as a GMP-compliant process. We thus believe that CssDNA bears strong potential and will be crucial in guiding the next generation of HSPCs-based gene therapies for the benefit of patients.
Methods
All procedures and animal housing for NCG xenotransplantations were performed at TRANSCURE bioServices (Archamps, France) and were reviewed and approved by the local ethics committee (CELEAG). Animal experiments abode by the ARRIVE guidelines. HPSCs and the T cells used in this study were purchased from AllCells (Almeda). The experiments presented in this study that used human samples (HSPC and primary T-cells) were approved by the GMO Department of the French Ministry of Education and Research on September 20, 2023, is documented under the DUO file number 10,849 and is valid for five years.
CD34 + HSPC sourcing and culture
Frozen CD34+ HSPCs purified from healthy donor G-CSF-mobilized and Plerixafor-mobilized peripheral blood were purchased from AllCells (Almeda). After thawing, CD34+ HSPCs were cultured at a concentration of 0.4 × 106 cells/mL in complete medium: StemSpan II (Stemcell, #09655), 1X CD34 expansion supplement (Stemcell, #02691) and 1X penicillin-streptomycin (Gibco, #15140-122) at 37 °C, 5% CO2. HSPCs were assessed for viability by Nucleocounter or by the expression of CD34 and viability marker by flow cytometry 2 or 5 days after gene editing. Flow cytometry staining was performed in Annexin V Binding Buffer 1X (BD Pharmingen, #556454) with antibody panel documented in Supplementary Table 2. The flow cytometry gate used to identify CD34(+) viable subpopulations were set using unstained and stained samples obtained 2 days after HSPC thawing, leaving <1 % of false positive with respect to the unstained control and <3 % of false negative with respect to the stained sample obtained at D2. The same gate was then used to analyze, in batch, the samples from the same donor, gathered at later time points. The overall gating strategy used to analyse cellular suspension is documented in Supplementary Fig. 1.
TALENs and DNA donor templates
Plasmids of the TALEN arms, HDR-Enh0111 and Via-Enh0111, containing a T7 promoter and a polyA sequence (120 residues), were produced using a proprietary and automated Golden Gate Assembly method derived from60 and linearized for mRNA in vitro transcription. TALEN, HDR-Enh0111, and Via-Enh0111mRNAs were produced in-house, using a proprietary in vitro transcription (IVT) process.
For non-viral mediated gene insertion, a DNA donor template sequence containing the coding sequences of CSR1 (HA-Tag), CSR2 (HLAE trimeric construct61) or CSR3 (ΔLNGFR) flanked by 300-nt left and right homology arm sequences specific for the locus to be modified (B2M, AAVS1, CD11b or S100A9) was designed. The corresponding LssDNA and CssDNA were produced by Moligo Technologies, using an enzymatic in vitro technology, using a proprietary protocol, partially based on the RCA approach documented in ref. 37. All sequences documenting the left, right homology arms and the coding sequences are available in Supplementary Table 3. For viral-mediated gene insertion, the DNA donor template containing CSR2 and documented in ref. 61 was used to produce the corresponding AAV6 particles.
HSPC engineering
The procedures used to transfect and/or transduce HSPCs were adapted from our former work documented in refs. 11,62. Briefly, two days after thawing, the cells were washed twice in BTXpress buffer and resuspended at a final concentration of 40 × 106 cells/mL in the same solution. The cellular suspension (4 x 106 cells) was mixed with 5 µg mRNA encoding each TALEN arm in the presence of 4 µg mRNA and 1 µg mRNA encoding for HDR-Enh01 and Via-Enh01, respectively, in a final volume of 100 µl. The cellular suspension was transfected in 4 mm cuvette gap size using PulseAgile technology. The electroporation program consisted of two 0.1 ms pulses at 1000 V/cm followed by four 0.2 ms pulses at 130 V/cm. Immediately after electroporation, the HSPCs were transferred to a new plate containing prewarmed medium at a concentration of 2 × 106 cells/mL and incubated for 15 min at 37 °C.
For AAV-mediated transduction experiments, 15 minutes after TALEN electroporation, HSPCs were cultured at a concentration of 2 × 106 cells/mL in the presence or absence of AAV6 particles (MOI = 350 viral genome/cell) and incubated for 15 minutes at 37 °C. HSPCs were then incubated at 30 °C overnight. For ssDNA transfection experiments, HSPCs were kept in culture at 30 °C for 16 h after TALEN electroporation, recovered and washed 2 times in PBS (Gibco, 10010056). A second electroporation was performed, using 1 × 106 cells in presence or absence of 0.02 nmol of ssDNA in a final volume of 100 µl using Nucleofector 2b apparatus (Lonza, AAB-1001) and human CD34+ Cell Nucleofector™ Kit (Lonza, #VPA-10003) using U-08 program. After ssDNA transfection, HSPCs were seeded at a concentration of 2 × 106 cells/mL and incubated at 30 °C overnight. The following day, cells were seeded at a density of 0.3 × 106 cells/mL in complete medium and cultured at 37 °C in the presence of 5% CO2.
T cells engineering
The procedure used to transfect T cells were adapted from our former work documented in refs. 11,62. Briefly, one day after thawing, T cells were activated using Transact (Miltenyi#200-076-204, 60 µL/106 CD3(+) cells) and allowed to grow for 3 days. Activated T cells were then recovered, washed twice in Cytoporation T buffer and resuspended at a final concentration of 50 × 106 cells/mL in the same solution. The cellular suspension (5 x 106 cells) was mixed with 5 µg mRNA encoding each TALENB2M arm in a final volume of 100 µl. The cellular suspension was transfected in 2 mm cuvette gap size using PulseAgile technology. The electroporation program consisted of two 0.1 ms pulses at 400 V/cm followed by four 0.2 ms pulses at 100 V/cm. Immediately after electroporation, the transfered were transferred to a new plate containing prewarmed medium at a concentration of 2 × 106 cells/mL and incubated at 30 °C. T cells were kept in culture at 30 °C for 6 hours after TALEN electroporation, recovered and washed 2 times in PBS (Gibco, 10010056). A second electroporation was performed using 2.5 x 106 cells in absence or presence different quantities of ssDNA in a final volume of 100 µl using Nucleofector 2b apparatus (Lonza, AAB-1001) and human T-Cell Nucleofector™ Kit (lonza, #VPA-10002) using T-023 program. After ssDNA transfection, T cells were seeded at a concentration of 2.5 x 106 cells/mL and incubated at 30 °C overnight. The following day, cells were transferred to 37 °C and cultivated in the presence of 5% CO2 for further assessment of gene editing efficiency and rate of proliferation.
Colony forming unit (CFU) assays
The procedure used to perform CFU assay were adapted from our former work documented in ref. 11. Briefly, CD34+ HSPCs recovered two days after electroporation, were plated in methylcellulose (Stemcell, #04435) for Colony Forming Unit (CFU) assays. A total of 200-500 cells (CD34 + ) were resuspended in 100 µL of Stemspan II and transferred to an aliquot of 1 mL of methylcellulose, mixed, and plated in a Smartdish well (Stemcell, #27371). Cells were cultured for 12–14 days in methylcellulose according to the manufacturer’s instructions. At the end of the culture, colonies were automatically counted using a Stemvision (StemCell) automated colony counter to assess plating efficiency (number of colonies counted at day 14/ number of cells plated at day 0).
Single-cell CITE-seq procedure and bioinformatic analysis of sequencing results
Single-cell mRNA barcoding and library generation were performed following the 10X Genomics protocol from the Chromium Next GEM Single-cell 5ʹ Kit v2 (#1000263) as described in ref. 11. Samples from different donors were multiplexed following a previously described cell-hashing protocol. Briefly, 200,000 edited or non-edited HSPCs (D4) were thawed and incubated with human TruStain FcX™ Fc Blocking reagent (Biolegend, #422301) for 10 minutes at 4 °C. Cells were stained with TotalSeq-C Hashtag antibodies diluted at 1/50 in Cell Staining buffer (Biolegend, #420201) for 30 min at 4 °C. After washing, different conditions bearing different hashtags were pooled. 500,000 cells were stained for 30 min at 4 °C with TotalSeq-C Universal Cocktail V1.0 (Biolegend, #399905) complemented with 18 additional TotalSeq-C antibodies (Supplementary table 2) at 0.25 µg per 500,000 cells. The list of antibody-derived tag (ADT) used for CITE-seq procedure is documented in Supplementary table 2. After washing, cells were loaded into Chromium Single-Cell Chip (10X Genomics) at a target capture rate of ~10,000 individual cells per sample. Gene Expression and Cell Surface libraries were checked using a Bioanalyzer High Sensitivity DNA kit (Agilent, #5067-4626) according to the manufacturer’s recommendations. Libraries generated were sequenced by Institut du Cerveau (ICM) IGenSeq plateformusing a NovaSeq X Plus sequencer from Illumina following 10X recommendations.
Raw demultiplexed sequences were processed by Cell Ranger multi v8 using human reference genome GRCh38 to generate the count matrices. Cells with less than 1000 genes or more than 9000 genes were filtered out, as well as cells with more than 10% mitochondrial genes. Data were processed using Seurat v5. Briefly, data were normalized, the 2000 most variable mRNA features were retained, before scaling and doing a Principal Component Analysis (PCA). Protein and mRNA features were integrated using Seurat WNN function FindMultiModalNeighbors. Cell cycle was predicted using CellCycleScoring function from Seurat. A dataset of CD34(+) high cells from 8 donors (about 190,000 cells) obtained from11 was used as a reference to map our cells using Symphony63. Cell type acronyms were used in Fig. 2 and Supplementary Fig. 4 to name the different HSPC subpopulations identified. We used LT-HSCe for LT HSC enriched, the most primitive Hematopoietic Stem Cells identified in this dataset; MPP for Multipotent Progenitors; LMPP for Lymphoid Primed Multipotent Progenitors; CLP for Common Lymphoid Progenitors; MEP for Megakaryocytic Erythroid Progenitors; MDP for Monocyte-macrophage and Dendritic cells Progenitors; Prog MK for Progenitor Megakaryocytes; Ery for Erythroid Progenitors; Myeloid_Int for Myeloid Intermediate Progenitors; Neu for Neutrophil Progenitors; Mono for Monocyte Progenitors; BaEoMa for Basophil Eosinophil Mast Progenitors; Prog DC for Progenitor Dendritic Cells and Bcells for Progenitors and Precursor B cells. KI resulting from AAV or CssDNA produced a large increase of the expression of HLA-E at the surface which could be detected by one of the antibodies present in the CITE-seq. Taking as a reference the untreated samples, a threshold on HLA-E ADT signal was determined above which a cell could safely (false positive rate estimated at 1%) be evaluated as having KI from its phenotype (called phenotypic KI). Differential gene expression was assessed using Seurat FindMarkers with default parameters, but setting no minimum for the log fold change and min.pct. Gene Set Enrichment Analysis (GSEA) was carried out using using Reactome pathways retrieved from MSigDB with the R package msigdbr. Sankey diagrams were generated using Excel and UDT macro add-in.
Transplantation of HSPCs into NCG mice
The transplantation of edited and non-edited HSPCs into mice was adapted from the procedure documented in ref. 11. Briefly, frozen aliquots of mobilized peripheral blood CD34+ cells from healthy donors obtained 2 days after editing (D4) were sent to TRANSCURE bioServices (Archamps, France) for xenotransplantation into 5-weeks-old female NODPrkdcem26Cd52 Il2rgem26Cd22/NjuCrl (NCG) mice (TRANSCURE bioServices). Pre-transplant conditioning was based on Busulfan (Sigma, #B1170000) according to the TRANSCURE bioServices protocol. Thirty-two NCG mice followed an acclimatation period of 7 days prior to being used in experiments and were housed at a constant temperature T = 22 ± 2 °C, at a constant relative humidity RH = 55 ± 10% and a photoperiod of 12:12-hour light-dark cycle 7am:7 pm with water and food (VRF1 food pellets, Safe Diets, # DS861912G10R) available ad libitum. Mice were monitored daily for unexpected signs of distress. Body weight was measured once a week. Mice with a cumulative clinical score ≥7 were euthanized. For a mouse with a body weight loss >20% associated to a clinical score <6, the veterinarian was consulted. The decision to euthanize an animal in pain or having reached the ethical limit was at the sole decision of the veterinarian. Mice showing a body weight loss >10 and <20%, were fed with nutritionally fortified food until recovery to 10% body weight loss. A total of 0.75 ×106 edited or non-edited control HSPCs were transplanted via tail vein injection. Sixteen weeks after transplantation, mice were sacrificed, and peripheral blood and bone marrow were recovered. One hundred thousand cells from each organ were harvested, and chimerism was assessed by flow cytometry using the following antibodies: mouse CD45 perCPvio700, Clone# REA747(Miltenyi, #130-110-636), human CD45 BV650, Clone# HI30 (BD, #563717), and viability dye FVS780, (BD, #565388). Editing was also assessed by cytometry using the following antibodies: HLA-E-ABC Vioblue, Clone#REA230 (Miltenyi, #130-120-435), HLA-E APC, Clone1031 (Miltenyi, #130-117-402. The gating strategy used to analyze cellular suspension is documented in Supplementary Fig. 7. All procedures and animal housing for NCG xenotransplantations were performed at TRANSCURE bioServices (Archamps, France) and were reviewed and approved by the local ethics committee (CELEAG). Animal experiments abode by the ARRIVE guidelines.
Statistical analysis
Comparisons of numerical variables between two groups were carried out using Mann–Whitney U tests as specified in the figure legends. Two-way ANOVA with Bonferroni post-tests was used to analyse experiments comparing two variables as specified in the figure legends. P-values are indicated in the plots. All statistical analyses were performed using GraphPad Prism v.9.4 (GraphPad).
Instrument and software used for data acquisition and analysis
The instruments used for this study were similar to the ones used in ref. 11. Cell viability was acquired using NucleoCounter® NC-250 (ChemoMetec) and analyzed using NucleoView™ software v4.3. CFU colonies were detected using a STEMvision apparatus (STEMCELL Technologies) and analyzed using STEMvision Colony marker (STEMCELL Technologies) v2.0.3.0. DNA quantification was performed using a NanodropOne device (ThermoScientific) and analyzed using NanoDrop QC software v1.6.198. Flow cytometry was performed using Novocyte Quanteon 4025 (Agilent) or Attune NxT (Life Technologies) flow cytometers. BD FACS DIVA software v9.0 (BD), FlowJo v10.8.1 (Treestar), were used to analyze flow cytometry dataset. Single-cell formulations were generated using Chromium single-cell system (10X Genomics), high throughput DNA sequencing was performed using NovaSeq systems (Illumina). Sequence read were demultiplexed and aligned to the human reference genome (GRCh38), using the CellRanger pipeline v6.1.2 (10X Genomics). GraphPad Prism software v.9.4. was used to plot most of the datasets illustrated in this study.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that the data supporting the findings of this study are available in the article and in the Supplementary information files. The CITE-seq dataset generated in this study has been deposited in the NCBI SRA under the BioProject accession code PRJNA1313799 [https://www.ncbi.nlm.nih.gov/bioproject/1313799]. Source data are provided with this paper.
References
Lattanzi, A. et al. Development of β-globin gene correction in human hematopoietic stem cells as a potential durable treatment for sickle cell disease. Sci. Transl. Med. 13, eabf2444 (2021).
Dever, D. P. et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature 539, 384–389 (2016).
Wang, J. et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 33, 1256–1263 (2015).
Pavel-Dinu, M. et al. Gene correction for SCID-X1 in long-term hematopoietic stem cells. Nat. Commun. 10, 1634 (2019).
Rai, R. et al. Targeted gene correction of human hematopoietic stem cells for the treatment of Wiskott-Aldrich Syndrome. Nat. Commun. 11, 4034 (2020).
Scharenberg, S. G. et al. Engineering monocyte/macrophage−specific glucocerebrosidase expression in human hematopoietic stem cells using genome editing. Nat. Commun. 11, 3327 (2020).
Lomova, A. et al. Improving Gene Editing Outcomes in Human Hematopoietic Stem and Progenitor Cells by Temporal Control of DNA Repair. Stem Cells 37, 284–294 (2019).
Pattabhi, S. et al. In Vivo Outcome of Homology-Directed Repair at the HBB Gene in HSC Using Alternative Donor Template Delivery Methods. Mol. Ther. Nucleic Acids 17, 277–288 (2019).
Romero, Z. et al. Editing the Sickle Cell Disease Mutation in Human Hematopoietic Stem Cells: Comparison of Endonucleases and Homologous Donor Templates. Mol. Ther. 27, 1389–1406 (2019).
Ferrari, S. et al. Choice of template delivery mitigates the genotoxic risk and adverse impact of editing in human hematopoietic stem cells. Cell Stem Cell 29, 1428–1444.e9 (2022).
Moiani, A. et al. Non-viral DNA delivery and TALEN editing correct the sickle cell mutation in hematopoietic stem cells Check for updates. Nat. Commun. 15, 4965 (2024).
Schiroli, G. et al. Precise Gene Editing Preserves Hematopoietic Stem Cell Function following Transient p53-Mediated DNA Damage Response. Cell Stem Cell 24, 551–565.e8 (2019).
Vu, G. T., Awad, V., Norberto, M. F., Bowman, T. V. & Trompouki, E. Nucleic acid-induced inflammation on hematopoietic stem cells. Exp. Hematol. 131, 104148 (2024).
Liao, W., Du, C. & Wang, J. The cGAS-STING Pathway in Hematopoiesis and Its Physiopathological Significance. Front. Immunol. 11, 1–11 (2020).
Magis, W. et al. High-level correction of the sickle mutation is amplified in vivo during erythroid differentiation. iScience 25, 104374 (2022).
Shy, B. R. et al. High-yield genome engineering in primary cells using a hybrid ssDNA repair template and small-molecule cocktails. Nat. Biotechnol. 41, 521–531 (2023).
Karasu, M. E. et al. Removal of TREX1 activity enhances CRISPR–Cas9-mediated homologous recombination. Nat. Biotechnol. 43, 1168–1176 (2025).
Iyer, S. et al. Efficient Homology-Directed Repair with Circular Single-Stranded DNA Donors. Cris. J. 5, 685–701 (2022).
Xie, K. et al. Efficient non-viral immune cell engineering using circular single-stranded DNA-mediated genomic integration. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02504-9 (2024).
Seclen, E. et al. TALEN-mediated intron editing of HSPCs enables transgene expression restricted to the myeloid lineage. Mol. Ther. 32, 1643–1657 (2024).
Baik, R. et al. Transient inhibition of 53BP1 increases the frequency of targeted integration in human hematopoietic stem and progenitor cells. Nat. Commun. 15, 111 (2024).
Wallace, K. A. et al. A differentiated β-globin gene replacement strategy uses heterologous introns to restore physiological expression. Mol. Ther. 33, 1407–1419 (2025).
Dudek, A. M. et al. A simultaneous knockout knockin genome editing strategy in HSPCs potently inhibits CCR5- and CXCR4-tropic HIV-1 infection. Cell Stem Cell 31, 499–518.e6 (2024).
Ferrari, S. et al. Efficient gene editing of human long-term hematopoietic stem cells validated by clonal tracking. Nat. Biotechnol. 38, 1298–1308 (2020).
Zhang, X. et al. An immunophenotype-coupled transcriptomic atlas of human hematopoietic progenitors. Nat. Immunol. 25, 703–715 (2024).
Lehnertz, B. et al. HLF expression defines the human hematopoietic stem cell state. Blood 138, 2642–2654 (2021).
Tomellini, E. et al. Integrin-α3 Is a Functional Marker of Ex Vivo Expanded Human Long-Term Hematopoietic Stem Cells. Cell Rep. 28, 1063–1073.e5 (2019).
Longo, G. M. C. et al. Linking CRISPR–Cas9 double-strand break profiles to gene editing precision with BreakTag. Nat. Biotechnol. 43, 608–622 (2025).
Jinek, M. et al. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821 (2012).
Fonfara, I., Richter, H., Bratovič, M., Le Rhun, A. & Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521 (2016).
Liang, Z., Sunder, S., Nallasivam, S. & Wilson, T. E. Overhang polarity of chromosomal double-strand breaks impacts kinetics and fidelity of yeast non-homologous end joining. Nucleic Acids Res. 44, 2769–2781 (2016).
Hiroyuki, S. & Susumu, K. New restriction endonucleases from Flavobacterium okeanokoites (FokI) and Micrococcus luteus (MluI). Gene 16, 73–78 (1981).
Renaud, J.-B. et al. Improved Genome Editing Efficiency and Flexibility Using Modified Oligonucleotides with TALEN and CRISPR-Cas9 Nucleases. Cell Rep. 14, 2263–2272 (2016).
Kanke, K. L. et al. Single-stranded DNA with internal base modifications mediates highly efficient knock-in in primary cells using CRISPR-Cas9. Nucleic Acids Res. 52, 13561–13576 (2024).
Ghanta, K. S. et al. 5′-Modifications improve potency and efficacy of DNA donors for precision genome editing. Elife 10, e72216 (2021).
Yu, Y. et al. An efficient gene knock-in strategy using 5′-modified double-stranded DNA donors with short homology arms. Nat. Chem. Biol. 16, 387–390 (2020).
Ducani, C., Kaul, C., Moche, M., Shih, W. M. & Högberg, B. Enzymatic production of ‘monoclonal stoichiometric’ single-stranded DNA oligonucleotides. Nat. Methods 10, 647–652 (2013).
Croft, L. V. et al. Sequence- and Structure-Dependent Cytotoxicity of Phosphorothioate and 2′-O-Methyl Modified Single-Stranded Oligonucleotides. Nucleic Acid Ther. 34, 143–155 (2024).
Improving non-viral knock-in with modified single-stranded DNAs and small molecules. Nat. Biotechnol. 41, 478–479 (2023).
Jin, Y.-Y. et al. Enhancing homology-directed repair efficiency with HDR-boosting modular ssDNA donor. Nat. Commun. 15, 6843 (2024).
Wimberger, S. et al. Simultaneous inhibition of DNA-PK and Polϴ improves integration efficiency and precision of genome editing. Nat. Commun. 14, 4761 (2023).
Selvaraj, S. et al. High-efficiency transgene integration by homology-directed repair in human primary cells using DNA-PKcs inhibition. Nat. Biotechnol. 42, 731–744 (2024).
Cullot, G. et al. Genome editing with the HDR-enhancing DNA-PKcs inhibitor AZD7648 causes large-scale genomic alterations. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02488-6 (2024).
Yahata, T. et al. A highly sensitive strategy for SCID-repopulating cell assay by direct injection of primitive human hematopoietic cells into NOD/SCID mice bone marrow. Blood 101, 2905–2913 (2003).
Larochelle, A. et al. Identification of primitive human hematopoietic cells capable of repopulating NOD/SCID mouse bone marrow: Implications for gene therapy. Nat. Med. 2, 1329–1337 (1996).
Anjos-Afonso, F. & Bonnet, D. Human CD34+ hematopoietic stem cell hierarchy: how far are we with its delineation at the most primitive level? Blood 142, 509–518 (2023).
Wang, D., Tai, P. W. L. & Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 185, 358–378 (2019).
Sobrino, S. et al. Severe inflammation and lineage skewing are associated with poor engraftment of engineered hematopoietic stem cells in patients with sickle cell disease. Nat. Commun. 16, 3137 (2025).
Sobrino, S. et al. Severe hematopoietic stem cell inflammation compromises chronic granulomatous disease gene therapy. Cell Rep. Med. 4, 100919 (2023).
Valentin, R. et al. Sequence-dependent inhibition of cGAS and TLR9 DNA sensing by 2′-O-methyl gapmer oligonucleotides. Nucleic Acids Res. 49, 6082–6099 (2021).
Alharbi, A. S. et al. Rational design of antisense oligonucleotides modulating the activity of TLR7/8 agonists. Nucleic Acids Res. 48, 7052–7065 (2020).
Shin, J. J. et al. Controlled Cycling and Quiescence Enables Efficient HDR in Engraftment-Enriched Adult Hematopoietic Stem and Progenitor Cells. Cell Rep. 32, 108093 (2020).
Karanam, K., Kafri, R., Loewer, A. & Lahav, G. Quantitative Live Cell Imaging Reveals a Gradual Shift between DNA Repair Mechanisms and a Maximal Use of HR in Mid S Phase. Mol. Cell 47, 320–329 (2012).
Hussmann, J. A. et al. Mapping the genetic landscape of DNA double-strand break repair. Cell 184, 5653–5669.e25 (2021).
Kan, Y., Ruis, B., Takasugi, T. & Hendrickson, E. A. Mechanisms of precise genome editing using oligonucleotide donors. Genome Res. 27, 1099–1111 (2017).
Gallagher, D. N. et al. A Rad51-independent pathway promotes single-strand template repair in gene editing. PLoS Genet 16, (2020).
Saito, S. & Adachi, N. Characterization and regulation of cell cycle-independent noncanonical gene targeting. Nat. Commun. 15, 5044 (2024).
Park, S. H. et al. Highly efficient editing of the β-globin gene in patient-derived hematopoietic stem and progenitor cells to treat sickle cell disease. Nucleic Acids Res. 47, 7955–7972 (2019).
Shepherd, T. R., Du, R. R., Huang, H., Wamhoff, E.-C. & Bathe, M. Bioproduction of pure, kilobase-scale single-stranded DNA. Sci. Rep. 9, 6121 (2019).
Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39, e82 (2011).
Jo, S. et al. Endowing universal CAR T-cell with immune-evasive properties using TALEN-gene editing. Nat. Commun. 13, 3453 (2022).
Yang, M. et al. Optimized two-step electroporation process to achieve efficient nonviral-mediated gene insertion into primary T cells. FEBS Open Bio 12, 38–50 (2022).
Kang, J. B. et al. Efficient and precise single-cell reference atlas mapping with Symphony. Nat. Commun. 12, 5890 (2021).
Acknowledgements
Figures include some illustrations created using Biorender.com. Gil Letort, Aymeric Duclert, Diane Le Clerre, Isabelle Chion-Sotinel, Emilie Dessez, Margaux Sevin, Marco Rotondi, Philippe Duchateau & Julien Valton perceived salary from Cellectis S.A. Cellectis S.A. funded this research project. Cosimo Ducani & Roger Salvatori received Salary from Moligo Technologies. This paper is dedicated to the memory of our friend and colleague Arianna Moiani, a talented scientist who was part of the early design of this study and unfortunately deceased in 2024 after her courageous battle against cancer.
Author information
Authors and Affiliations
Contributions
P.D. and J.V. conceived the study. G.L. designed, performed and analyzed most of the experiments using HSPCs as starting biological material including the preparation of cellular sample for CITE-seq analysis. A.D. analyzed all CITE-seq datasets with the help of G.L. and J.V., who participated in curating and plotting datasets. DL.C., I.C-S. and M.S. participated to the establishment of the optimal experimental conditions of HSPCs engineering. E.D. performed and analyzed most of the experiments using T cells as starting biological material and M.R, supervised E.D., experimental work. R.S. produced the LssDNA and CssDNA used in this study and C.D. supervised R.S., experimental work and the collaboration between Moligo Technologies and Cellectis. SA. G.L., DL.C., I.C-S., M.S., and J.V. designed the in vivo study performed with NCG mice, and G.L., I.C-S. analyzed the dataset. J.V. supervised the study and wrote the manuscript with the help of all authors. J.V. and C.D., established and supervised the collaboration between Cellectis. SA and Moligo Technologies which was coordinated with the help of M.S.
Corresponding author
Ethics declarations
Competing interests
Gil Letort, Aymeric Duclert, Diane Le Clerre, Isabelle Chion-Sotinel, Emilie Dessez, Margaux Sevin, Marco Rotondi, Philippe Duchateau and Julien Valton are current employees and equity holders at Cellectis S.A. Roger Salvatori and Cosimo Ducani are current employees and equity holders at Moligo Technologies. The authors declare no other competing interests. TALEN® is a Cellectis patented technology.
Peer review
Peer review information
Nature Communications thanks Carsten Lederer, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Letort, G., Duclert, A., Le Clerre, D. et al. Circular single stranded DNA potentiates non-viral gene insertion in hematopoietic stem and progenitor cells. Nat Commun 16, 10125 (2025). https://doi.org/10.1038/s41467-025-66318-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66318-2