Abstract
Broadly neutralizing antibodies (bNAbs) against HIV have demonstrated robust protective efficacy against sensitive viruses in humans and nonhuman primates. However, the potential of rare resistant viral variants to abrogate bNAb-based protection remains to be determined. Here we show that simian-human immunodeficiency virus (SHIV) challenge containing minor resistant variants can compromise protective efficacy of the V2-specific bNAb PGDM1400 in rhesus macaques. Macaques were infused with PGDM1400 and challenged intrarectally with either 500 monkey infectious dose 50 (MID50) of the PGDM1400-sensitive virus SHIV-325C or with 500 MID50 of SHIV-325C mixed with a sub-infectious 0.5 or 0.05 MID50 dose of the PGDM1400-resistant virus SHIV-SF162P3. All PGDM1400-treated animals challenged with SHIV-325C were protected as expected, but animals that received the mixed SHIV challenge were productively infected either with the resistant virus SHIV-SF162P3 or with rare resistant clones in the SHIV-325C challenge stock. These data demonstrate that minor resistant viral variants comprising just 0.01-0.1% of the challenge stock, at sub-infectious doses when administered alone, effectively abrogated bNAb protection against SHIV challenge in rhesus macaques. These findings suggest rare resistant variants may be more important than previously appreciated as a mechanism of virologic failure, emphasizing the importance of utilizing bNAb cocktails for HIV prevention.
Similar content being viewed by others
Introduction
Pre-exposure prophylaxis (PrEP) with antiretroviral drugs, such as emtricitabine and tenofovir1, cabotegravir2, and lenacapavir3, have been shown to be highly effective at preventing HIV infection. The broadly neutralizing antibody (bNAb) VRC01 has also been shown to block HIV infection with sensitive viruses4, and clinical studies are currently underway to explore bNAb cocktails5,6,7. Studies in macaques have similarly shown that bNAbs are highly protective against SHIV challenge with sensitive viruses8,9,10.
We previously reported that a cocktail of the V2-specific bNAb PGDM1400 and the V3-specific bNAb PGT121 was required to protect against a mixed SHIV challenge containing a 1:1 mixture of SHIV-SF162P3, which is neutralized by PGT121 but not PGDM1400, and SHIV-325C, which is neutralized by PGDM1400 but not PGT12110. In this study, we sought to determine if rare resistant viral variants in the challenge stock at levels that are sub-infectious when administered alone could compromise bNAb based protection against SHIV challenge in rhesus macaques.
Results
Baseline resistance variants abrogate bNAb therapeutic efficacy following ART discontinuation in persons with HIV
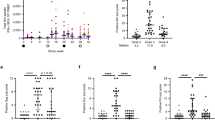
In a recently published clinical study7, we reported the therapeutic efficacy of the combination of PGT121, PGDM1400, and VRC07-523LS to control viral replication after discontinuation of antiretroviral therapy (ART) in persons with HIV. In this study, 10 of 12 participants demonstrated virologic suppression for the duration of bNAb therapy, but 2 individuals showed early viral rebound during bNAb treatment7. In these 2 participants, we amplified Env sequences from CD4+ T cells prior to ART discontinuation and observed that both had proviruses that were partially resistant to both PGT121 and PGDM1400 at baseline (Fig. 1). These data show that partially resistant variants can abrogate bNAb-based therapeutic efficacy in humans. To explore whether a defined frequency of rare resistant variants can abrogate bNAb protective efficacy, we utilized the model of bNAb protection against SHIV challenge in rhesus macaques in which we spiked a sensitive SHIV challenge stock with low amount of a resistant SHIV challenge stock (Fig. 2a).
As described in a previous study7, PWH on ART were given a bNAb cocktail of PGT121, PGDM1400, and VRC07-523LS and then discontinued ART. Env sequences were cloned from CD4 T cells prior to ART discontinuation and from plasma virus post-rebound. Pseudoviruses with these Env sequences were tested for sensitivity to these three bNAbs. Viral loads of individuals a 65-021, and c 98-059, with times of sampling for Env sequencing indicated by a red arrow and ART by gray shading. Neutralization results from b 65-021, and d 98-059, are shown at baseline and post-rebound. Source data are provided as a Source Data file.
Macaques received a single intrarectal challenge with SHIV-325C or SHIV-SF162P3, and viral loads were monitored for 70–84 days. Median values are shown in red. a Experimental design. b Animals (n = 3) intrarectally challenged with 500 MID50 of SHIV-325C were productively infected. c Animals (n = 4) intrarectally challenged with 500 MID50 of SHIV-SF162P3 were productively infected. d Animals (n = 4) intrarectally challenged with 0.5 MID50 of SHIV-SF162P3 were not productively infected. Source data are provided as a Source Data file.
Rare resistant viral variants abrogate protective efficacy of PGDM1400 against SHIV challenge in rhesus macaques
SHIV-325C has a subtype C HIV envelope and is sensitive to PGDM1400, whereas SHIV-SF162P3 has a subtype B HIV envelope and is resistant to PGDM140010. In the absence of bNAb treatment, a high-dose intrarectal challenge of either 500 MID50 SHIV-325C or 500 MID50 SHIV-SF162P3 infected 4 of 4 (100%) rhesus macaques each, as expected10,11,12 (Fig. 2b, c). In contrast, a 1000-fold lower intrarectal dose of 0.5 MID50 SHIV-SF162P3 infected 0 of 4 (0%) animals (Fig. 2d).
To evaluate the potential impact of a minor resistant viral variant on bNAb based protective efficacy, 8 Indian-origin rhesus macaques received an intravenous infusion with 10 mg/kg PGDM1400 one day prior to intrarectal SHIV challenge. Animals were then challenged with either 500 MID50 SHIV-325C (n = 4) or 500 MID50 SHIV-325C mixed with 0.5 MID50 SHIV-SF162P3 (n = 4). As we previously reported10, 0 of 4 (0%) of PGDM1400-treated macaques challenged with 500 MID50 SHIV-325C were productively infected and showed no detectable plasma viremia (<50 copies/ml) for 70 days following challenge (Fig. 3a). In contrast, 4 of 4 (100%) of PGDM1400-treated macaques that received the mixed SHIV challenge were productively infected with peak viral loads of 4.8–7.2 log RNA copies/mL by days 10-63 following challenge (Fig. 3b), despite the fact that the low dose of 0.5 MID50 SHIV-SF162P3 did not result in productive infection when given alone (Fig. 2d). Plasma PGDM1400 pharmacokinetics were comparable in both groups, except for one animal in the SHIV-325C only group that showed more rapid clearance kinetics (Fig. 3c, d). These data demonstrate that a 0.1% minor resistant viral variant spiked into a sensitive SHIV challenge stock effectively abrogated the protective efficacy of PGDM1400.
8 rhesus macaques received 10 mg/kg PGDM1400 one day prior to challenge with either 500 MID50 SHIV-325C (n = 4) or 500 MID50 SHIV-325C mixed with 0.5 SHIV-SF162P3 (n = 4). Viral loads and PGDM1400 pharmacokinetics were monitored. a SHIV-325C challenge alone did not result in productive infection, whereas b the mixed SHIV challenge resulted in productive infection in all animals. c, d PGDM1400 serum concentrations in the respective challenge groups. Source data are provided as a Source Data file.
Viral env sequencing of breakthrough viruses following mixed SHIV challenge
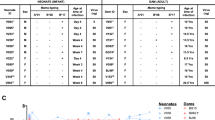
To determine the identity of the breakthrough viruses, we performed single genome amplification (SGA) sequencing from plasma samples from the 4 productively infected animals following the mixed SHIV challenge in the experiment described above. Animals BA58 and BA73 had sequences derived from the SHIV-SF162P3 challenge stock as expected, but animals AP82 and BA93 had sequences that surprisingly clustered with SHIV-325C (Fig. 4a; Supplementary Fig. 1). All env sequences isolated from AP82 and BA93, however, exhibited the amino acid mutation S380H (A1138C, G1139A) compared with the consensus SHIV-325C Env sequence (Fig. 4b). In addition, in all clones that were sequenced, AP82 also had a K115E mutation (A343G), and BA93 also had a N193D mutation (A577G) (Fig. 4b). None of these mutations were present in the SHIV-SF162P3 sequence (Fig. 4c; Supplementary Fig. 1), suggesting that they did not originate from recombination between the two challenge stocks. In contrast, deep sequencing of the SHIV-325C challenge stock showed the presence of a low frequency of the S380H mutation, with A1138C at a frequency of 0.05% and G1139A at a frequency of 0.2% (p = 1.9 ×10−148; Supplementary Table 1). The K115E (A343G; p = 1.5 ×10−7) and K193D (A577G; p = 4.7 ×10−52) mutations were also detected at a frequency of 0.1% in the SHIV-325C challenge stock (Supplementary Table 1). However, in rhesus macaques infected with SHIV-325C in a prior study without PGDM140010, none of the animals showed the S380H mutation in plasma viral sequences (Fig. 4d), suggesting that the emergence of these mutations in AP82 and BA93 likely reflected selection pressure from PGDM1400.
Plasma Env sequences from the animals that received the mixed SHIV challenge were determined by single genome amplification. Genetic analysis was performed with Geneious. a Phylogenetic tree if Env sequences from BA58 (blue), BA73 (black), BA93 (green), and AP82 (purple), along with reference Env sequences of HIV HXB2, SHIV-SF162P3, and SHIV-325C in red. b Highlighter plot of the Env sequences from AP82 and BA93 in relation to SHIV-325C Env. Sequences from both animals share the amino acid mutation S380H (relative to SHIV-325C). AP82 sequences all share the mutation K115E, and BA93 sequences all share the mutation N193D. c The common amino acid mutations are not a result of SHIV-325C recombining with SHIV-SF162P3, as the mutations are not present in the latter viral Env. d Highlighter plot comparing Env sequences obtained from five macaques infected with SHIV-325C in the absence of PGDM1400 to AP82 and BA93 around S380H. None of the five macaques had the mutation shared by AP82 and BA93.
To evaluate the neutralization properties of these SHIV-325C variants, we generated pseudoviruses with env clones from AP82 and BA93 as well as the consensus SHIV-325C sequence and assessed for resistance to PGDM1400 in TZM-bl pseudovirus neutralization assays. The AP82 and BA93 variants showed only modest resistance to PGDM1400 by IC50 and IC80 measurements, but they showed substantial resistance by IC90 and IC99 measurements (Fig. 5), suggesting that IC90 and IC99 may be more relevant than IC50 and IC80 measurements for predicting viral breakthrough in vivo. One BA93 clone that had only S380H and N193D mutations showed only mild resistance, suggesting that a third mutation P122S or A226V is likely required for high level IC99 resistance (Fig. 5). Moreover, the SHIV-325C consensus sequence was more sensitive than the SHIV-325C challenge stock, likely reflecting the approximate 0.1% rare resistant viral clones within the stock (Supplementary Table 1). These data demonstrate that BA58 and BA73 were productively infected by the resistant virus SHIV-SF162P3, which was experimentally spiked at levels of 0.1% into the SHIV-325C challenge stock, and AP82 and BA93 were productively infected by rare resistant clones that were present at baseline at levels of 0.05–0.2% in the SHIV-325C challenge stock.
TZM-bl luciferase neutralization assays were performed on pseudoviruses constructed from Env sequences isolated from AP82 and BA93 as well as the SHIV-325C consensus sequence and the SHIV-SF162P3 and SHIV-325C challenge stocks. Inhibitory concentrations 50, 80, 90, and 99 are shown. Source data are provided as a Source Data file.
Serum cytokines in PGDM1400-treated animals following SHIV-325C challenge
We next investigated potential mechanisms why the PGDM1400-treated animals challenged with the mixed SHIV challenge demonstrated viral breakthrough with rare resistant variants derived from both the SHIV-SF162P3 and the SHIV-325C challenge stocks, whereas the PGDM1400-treated animals challenged with only 500 MID50 SHIV-325C were fully protected10. We assessed serum cytokines on samples from days 3 and 7 following challenge in the two groups of animals, as well as in a control group of 5 rhesus macaques that were not productively infected following challenge with 0.5 MID50 SHIV-SF162P3 in the presence (n = 3; Supplementary Fig. 2) or absence (n = 2) of PGDM1400. The day 3 timepoint was prior to detectable plasma viremia. Both the group that received only 500 MID50 SHIV-325C and the group that received the mixed SHIV challenge demonstrated substantially higher levels of IL-8, IL-15, IP-10, and TARC than the controls that received 0.5 MID50 SHIV-SF162P3 (p = 0.02 for all comparisons; Supplementary Fig. 3), suggesting that the high dose challenge led to inflammatory cytokine responses even if the virus was fully neutralized.
In addition, we also observed higher levels of proinflammatory cytokines in animals that received the mixed SHIV challenge compared with the animals that received only SHIV-325C (Supplementary Fig. 3). A partial least squares discriminant analysis showed different cytokine patterns in these two groups of animals (Supplementary Fig. 4a), and a Random Forest analysis showed higher MCP-1, IP-10, and VEGF levels in animals that received the mixed SHIV challenge compared with animals that received only SHIV-325C on day 3 post-challenge (p = 0.02; Supplementary Fig. 4b). Moreover, a k-means clustering analysis using the cytokines selected by the Random Forest model showed clustering accuracy of 87% (Supplementary Fig. 4c). We speculate that inflammatory differences in the local tissue microenvironments may be greater than serum cytokines, although exploration of this hypothesis will require further studies. It is possible that the fully resistant SHIV-SF162P3 virus in the mixed SHIV challenge may have increased inflammatory responses that facilitated viral replication of not only the resistant SHIV-SF162P3 clones but also the resistant SHIV-325C clones. No differences were observed between these two groups of animals in terms of lymphocyte subsets (Supplementary Fig. 5a) or activation markers (Supplementary Figs. 5b, 6) by flow cytometry.
Very rare resistant viral variants abrogate protective efficacy of PGDM1400 against repeated SHIV challenges
We next sought to determine the protective efficacy of PGDM1400 against repeated challenge with 500 MID50 SHIV-325C containing an even lower amount of SHIV-SF162P3 (Fig. 6a). To titrate the dose of SHIV-SF162P3 that would be non-infectious after repeated challenges, we challenged 8 macaques intrarectally 6 times with 5 or 0.05 MID50 SHIV-SF162P3 (n = 4 per group). All animals in the 5 MID50 dose group but none of the animals in the 0.05 MID50 dose group became productively infected by this repeated challenge protocol (Fig. 6b, c), and animals that received repeated infusions of PGDM1400 and repeated challenges with 500 MID50 SHIV-325C were not infected (Fig. 6d).
a Experimental design for repeat challenges. b Macaques (n = 4) that received 6 intrarectal challenges with 5 MID50 SHIV-SF162P3 were productively infected. c Macaques (n = 4) that received 6 intrarectal challenges with 0.05 MID50 SHIV-SF162P3 were not productively infected. d Macaques received 10 mg/kg of PGDM1400 and then 6 intrarectal challenges with 500 MID50 SHIV-325C. e Macaques received 10 mg/kg of PGDM1400 and then 5 intrarectal challenges with 500 MID50 SHIV-325C mixed with 0.05 MID50 SHIV-SF162P3. f PGDM1400 serum concentrations. Red arrows indicate time of each antibody administration and challenge. Source data are provided as a Source Data file.
To assess the impact of a 0.01% minor resistant variant on PGDM1400 mediated protective efficacy, 5 rhesus macaques received intravenous infusions with 10 mg/kg PGDM1400 on days −1, 27, 41, 55, and 76 to maintain therapeutic bNAb levels. Animals were challenged intrarectally with 500 MID50 SHIV-325C mixed with 0.05 MID50 SHIV-SF162P3 on days 0, 28, 42, 56, and 77. By the end of this repeat dose challenge protocol, 4 of 5 (80%) macaques were productively infected, and 1 animal (BM34) had a confirmed blip of viremia but did not develop sustained viremia (Fig. 6e). Plasma PGDM1400 pharmacokinetics showed similar antibody levels after each infusion (Fig. 6f). One animal showed rapid PGDM1400 decay kinetics and was infected after the first SHIV challenge (BC41, purple; Fig. 6e, f).
To assess the sequences of the breakthrough viruses, we performed SGA sequencing on plasma virus from the four animals with productive infection (BM18, BM20, BM22, BC41). BM18 showed primarily but not exclusively SHIV-SF162P3 clones (Fig. 7a), whereas BM20, BM22, and BC41 showed SHIV-325C variants with the same S380H mutation that was observed in the prior experiment but with different secondary mutations (Fig. 7b, c). The SHIV-325C variants found in BM18 also showed these mutations. These mutations were present at low levels in the SHIV-325C challenge stock (Supplementary Table 1). These results confirm that rare resistant viral variants in both challenge stocks abrogated PGDM1400 protective efficacy against repeated SHIV challenges in macaques.
Plasma Env sequences from the animals that received the repeated mixed SHIV challenge were determined by single genome amplification. Genetic analysis was performed with Geneious. a Phylogenetic tree of Env sequences from BM18 (blue), BM20 (black), BM22 (green), and BC41 (purple), along with reference Env sequences of HIV HXB2, SHIV-SF162P3, and SHIV-325C in red. b Highlighter plot with selected Env sequences from BM18, BM20, BM22, and BC41 in relation to SHIV-325C Env. Sequences from all four animals share the amino acid mutation S380H (relative to SHIV-325C). c Mutations common to all SHIV-325C variant sequences in each animal were not present in SHIV-SF162P3 and were thus not a result of recombination.
Discussion
In this study, we demonstrate that rare resistant viral variants at frequencies of 0.01–0.1% in the challenge stock effectively abrogated PGDM1400 protective efficacy against SHIV challenge in rhesus macaques, even though these doses of the resistant variants were sub-infectious when given alone. Our data are consistent with a model in which the high dose of the sensitive SHIV-325C challenge virus, even if fully neutralized by PGDM1400, led to increased proinflammatory and antiviral cytokines that facilitated viral breakthrough by resistant SHIV-SF162P3 clones as well as rare resistant SHIV-325C variants in the context of a mixed SHIV challenge. Our findings also suggest that IC90 and particularly IC99 measurements may be more informative than IC50 and IC80 measurements for predicting viral breakthrough. Taken together, our findings demonstrate the capacity of rare resistant variants to undermine bNAb protective efficacy against SHIV challenge in macaques, which has important potential clinical implications.
Unexpectedly, we observed viral breakthrough with not only the resistant virus SHIV-SF162P3 that was experimentally spiked into the SHIV-325C challenge stock, but also with rare resistant SHIV-325C clones that were present at baseline in the SHIV-325C challenge stock in the context of the mixed SHIV challenge. Since we did not observe routine emergence of the resistant SHIV-325C clones following SHIV-325C challenge in the absence of SHIV-SF162P3, we speculate that the resistant SHIV-SF162P3 virus may have increased the inflammatory milieu in the local tissue microenvironment that could have facilitated replication of both the resistant SHIV-SF162P3 and the rare resistant SHIV-325C clones, although further studies are required to evaluate this hypothesis. SHIV-SF162P3 is fully resistant to PGDM1400 by IC50 measurements, whereas the rare resistant clones of SHIV-325C are only resistant to PGDM1400 by IC90 and IC99 measurements, and thus it is possible that initial replication with SHIV-SF162P3 in the mixed SHIV challenge may have led to increased inflammatory and antiviral cytokines that facilitated the emergence of both resistant viruses. Consistent with this model is that observation that the mixed SHIV challenge that included 0.01–0.1% SHIV-SF162P3 led to higher levels of MCP-1, IP-10, VEGF, and other cytokines as compared with the SHIV-325C only challenge.
All the animals that received the mixed SHIV challenge and that were productively infected with the rare resistant SHIV-325C variants had the S380H mutation as well as at least one additional mutation. Since the S380H mutation alone did not result in resistance to PGDM1400 (IC99 of 0.322 ug/mL), it appears that at least two mutations are required for SHIV-325C to develop resistance to PGDM1400. Both N193D and N204D would be predicted to result in loss of a glycosylation site and introduction of a new negative charge in the V2 loop, although these changes have not individually been shown to confer resistance to PGDM140013. In contrast, K176E has been reported to confer resistance to PGDM140013.
We previously reported that the combination of PGDM1400 and PGT121 was required to protect against a 1:1 mixed challenge of SHIV-SF162P3 and SHIV-325C10. In this prior study, bNAb monotherapy led to breakthrough with the resistant SHIV as expected10. Our results extend this study by showing that low frequency resistant variants, either spiked experimentally into a sensitive challenge stock or naturally occurring in the challenge stock, can similarly lead to viral breakthrough to bNAb monotherapy. A recent study also showed that macaques given SIV-specific bNAbs developed subclinical occult infections despite therapeutic levels of protective bNAbs14. Taken together, these data suggest the importance of developing bNAb cocktails for clinical development to increase neutralization coverage including coverage of minor resistant variants.
It is sobering that a low sub-infectious dose of a minor resistant variant in the context of an otherwise sensitive challenge stock consistently abrogated bNAb protective efficacy against SHIV challenge in rhesus macaques. Neutralized sensitive virus may even facilitate the emergence of rare resistant variants by increasing systemic or local inflammation. These data suggest that minor resistant variants may be more important than previously appreciated and should be evaluated in clinical trials. Moreover, future studies could investigate the mechanism of how neutralized sensitive virus induces inflammatory responses and facilitates productive infection with resistant variants.
Methods
Animals
Animal studies were conducted with the approval of the appropriate Institutional Animal Care and Use Committee (IACUC) at Beth Israel Deaconess Medical Center, AlphaGenesis (19-04, 18-14), and Bioqual (17-014.2, 17-015.2). All animals used in these studies were outbred, young adult (3–10 years old) Indian-origin rhesus macaques (Macaca mulatta) of both sexes housed at either AlphaGenesis (Yemasee, SC) or Bioqual (Rockville, MD). Animals were randomly allocated for each experiment. For the single challenge experiments, animals were intravenously infused with PGDM1400 (10 mg/kg) one day prior to challenge. The animals were then intrarectally challenged with 500 MID50 of PGDM1400-sensitive SHIV-325C (n = 4) or 500 MID50 SHIV-325C combined with a sub-infectious dose of 0.5 MID50 PGDM1400-resistant SHIV-SF162P3 (n = 4). Control animals were intrarectally challenged with either 500 MID50 SHIV-325C (n = 3), 500 MID50 SHIV-SF162P3 (n = 4), or 0.5 SHIV-SF162P3 (n = 4) to show infectivity of the virus stocks at those doses.
In the repeat challenge experiments, macaques were infused with PGDM1400 one day prior to each challenge. These animals (n = 5) were challenged five times with 500 MID50 SHIV-325C combined with a lower sub-infectious dose of 0.05 MID 50 SHIV-SF162P3. Control animals were challenged intrarectally every two weeks until the animals became viremic with either 500 MID50 SHIV-325C (n = 4), 5 MID50 SHIV-SF162P3 (n = 4), or the sub-infectious dose of 0.05 MID50 SHIV-325C (n = 4). In both studies, immunological and virological assays were performed blinded.
PGDM1400 pharmacokinetics
Serum levels of human PGDM1400 mAb were monitored using a previously described human IgG specific enzyme-linked immunosorbent assay (ELISA)15. In brief, ELISA plates were coated overnight at 4 °C with 1 μg/mL of goat anti-human IgG (H + L) secondary antibody (monkey pre-adsorbed) (Novus Biologicals) and then blocked for 2 h. Serum samples were assayed at 3-fold dilutions starting at a 1:3 dilution in Blocker Casein in PBS (Thermo Fisher Scientific) diluent. Samples were incubated for 1 h at ambient temperature and then removed, and plates were washed. Wells then were incubated for 1 h with horseradish peroxidase (HRP)-conjugated goat anti-human IgG (monkey pre-adsorbed) (Southern Biotech) at a 1:4,000 dilution. Wells were washed and then incubated with SureBlue Reserve TMB Microwell Peroxidase Substrate (100 μL/well; Seracare) for 3 min followed by TMB Stop Solution (100 μL/well; Seracare) to stop the reaction. Microplates were read at 450 nm. The concentration of PGDM1400 mAb was interpolated from the linear range of concurrently run purified human IgG (Sigma) standard curves using Prism software, version 10.3.1 (GraphPad).
Viral RNA analyses
Viral RNA was extracted from plasma with the QIAmp Viral RNA Kit (Qiagen) and then converted to cDNA with Superscript III VILO (Invitrogen). The cDNA was quantified via qPCR with QuantStudio 1.7.1 as previously described16 using the following primers (Integrated DNA Technologies) and probe (Applied Biosystems) with TaqMan Fast Advanced Master Mix (Applied Biosystems): Fwd, 5′- GTCTGCGTCATCTGGTGCATTC -3′; Rev, 5′- CACTAGGTGTCTCTGCACTATCTGTTTTG -3′; and Probe 5′-(FAM) CTTCCTCAGTGTGTTTCACTTTCTCTTCTGCG-(BHQ)-3′. Human viral loads were run either the Hologic Aptima HIV-1 Quant or Abbott Real-Time HIV-1 assays as described previously7.
Serum cytokine analysis
Serum cytokines and chemokines were measured via multiplex ELISA U-PLEX assays on Multiplex ELISA (Meso Scale Discovery) according to manufacturer’s instructions by the Metabolism and Mitochondrial Research Core (Beth Israel Deaconess Medical Center, Boston, MA).
Flow cytometry
PBMCs were isolated from blood at day 42, frozen, and stored in a liquid nitrogen freezer. Cells were later defrosted, washed, and then stained with monoclonal antibodies at concentrations suggested by the manufacturer (Becton Dickinson unless noted) against CD3 (SP34; Alexa Fluor 700), CD4 (OKT4; BV510, Biolegend), CD8 (SK1; APC-Cy7), CD14 (M5E2; BUV737), CD16 (3G8; BV650), CD25 (PE-Cy7; M-A251), CD28 (L293; PerCP-Cy5.5), CD38 (APC; HB-7), CD56 (NCAM16; BV786), CD69 (TP1.55.3; PE-TexasRed; Beckman Coulter), CD95 (DX2; BV711), CCR5 (3A9; PE), CCR7 (3D12; BV421), HLA-DR (BUV-395; G46-6), Ki67 (B56; FITC), and PD-1 (EH21.1; BV605). Samples were run on an LSR II and analyzed in FlowJo (see Supplementary Fig. 6 for gating strategy).
Single genome amplification (SGA) and pseudovirus construction
SGA was performed on macaque samples as previously described17. Briefly, RNA isolated from plasma samples at one to two timepoints within detection of positive viral loads with the QIAmp Viral RNA Mini Kit (QIAgen) was reverse transcribed with Superscript IV reverse transcriptase (Invitrogen) and the primer 5’- TGTAATAAATCCCTTCCAGTCCCCCC-3’. All primers for this assay were sourced from Integrated DNA Technologies (IDT). Limiting dilution PCR with the outer primers (Fwd: 5’- CCTCCCCCTCCAGGACTAGC-3’; Rev: 5’- TGTAATAAATCCCTTCCAGTCCCCCC-3’) was then performed with Platinum SuperFi II PCR Master Mix (Invitrogen) and the following PCR conditions: 1’ at 98 °C; 35 cycles of 10” of 98 °C, 10” of 60 °C, and 2’15” of 72 °C; 5’ at 72 °C; hold at 4 °C. The inner PCR reaction, using 1.5 uL of the outer PCR product, was then performed with Platinum SuperFi II PCR Master Mix (Invitrogen), a second set of primers (Fwd: 5’- ATAGACATGGAGACACCCTTGAGGGAGC-3’; Rev: 5’- ATGAGACATRTCTATTGCCAATTTGTA-3’), and slightly altered PCR conditions (45 cycles and a 55 °C annealing temperature). Amplicons were considered to be the result of a single cDNA molecule according to the Poisson distribution were processed for sequencing if one third or fewer wells were positive (p < 0.05). Unique Env sequences with complete open reading frames were reamplified with Platinum PCR SuperMix High Fidelity (Invitrogen) to generate 3’-A overhangs with an altered set of inner PCR primers (Fwd: 5’- CACCTTAGGCATCTCCTATGGCAGGAAGAAG-3’; Rev: same as previous) and the following conditions: 1’ at 94 °C; 45 cycles of 30” at 94 °C, 30” at 55°, and 3’30” at 68 °C; 5’ at 68 °C; hold at 4 °C. HIV-specific primers and PCR conditions were used as previously described for the human samples7. The resultant sequences were inserted into CMV-based plasmid expression vectors with the pcDNA 3.1/V5-His TOPO TA Expression Kit (Invitrogen) and used for pseudovirus generation as described previously18. Briefly, 293T cells were transfected with 3.3 ug of the Env expression plasmid and 10 ug of env-deficient HIV-1 backbone plasmid pSG3ΔEnv using Fugene 6 (Promega). Pseudovirus was collected after two days and underwent 0.45 μm filtration prior to storage at −80 °C.
Sequence analysis
Whole envelope sequencing was performed for the viruses from the single-challenge experiment by the MGH CCIB DNA Core. Partial envelope sequencing was performed for relevant regions of virus from the macaques infected with SHIV-325C in the absence of PGDM1400 and the macaques from the repeat challenge experiments. Alignments and neighbor-joining consensus trees were generated with Geneious Prime (Version 2024.0.2). Highlighter plots were generated using the Los Alamos National Laboratories (LANL) Highlighter webtool (https://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html). Potential N-linked glycosylation sites (PNGS) were detected using the LANL N-GlycoSite webtool (https://www.hiv.lanl.gov/content/sequence/GLYCOSITE/glycosite.html) via any amino acid sequence matches to the N-linked glycosylation recognition sequence N-X-[ST], where X may be any amino acid except proline.
Neutralization assay
Pseudoviruses and the SHIV-SF162P3 and SHIV-325C stocks were analyzed for sensitivity to PGDM1400 neutralization via the TZM-bl luciferase reporter neutralization as previously described19. Briefly, reduction in Tat-regulated luciferase (Luc) reporter gene expression in TZM-bl cells was used to measure the neutralization of Env pseudoviruses by PGDM1400 using a fivefold dilution series starting with a concentration of 50 μg/mL in duplicate wells. IC50, IC80, IC90, and IC99 were then calculated.
Deep sequencing of SHIV-325C Env
RNA isolated from our SHIV-325C stock was reverse transcribed with Superscript IV reverse transcriptase (Invitrogen), and the resultant cDNA was amplified with Platnium SuperFi II PCR Master Mix (Invitrogen) and our Env-specific outer primers as described above. Specific regions of Env were then amplified with Platnium SuperFi II PCR Master Mix and sent out for library preparation and Illumina MiSeq 150 bp paired-end sequencing at the MGH CCIB DNA Core. Primer sets (Integrated DNA Technologies) are described in Supplementary Table 2. Pre-processing of deep sequencing data to remove adaptor sequences and low-quality bases was performed by the MGH CCIB DNA Core. Using Geneious Prime (Version 2024.0.2), the processed Illumina sequences were aligned to the SHIV-325C Env consensus sequence, and single-nucleotide polymorphisms were detected using the program’s built-in tool to detect single-nucleotide polymorphisms and variants and calculate approximate p values for those variants.
Statistical analyses
Virological and immunological data analysis (excluding modeling) was performed using GraphPad Prism Version 10 (GraphPad Software). Two-sided Mann–Whitney tests were used to compare groups.
Partial least squares discriminant analysis (PLS-DA)
To investigate the association between predictor variables (cytokines) and the two challenge groups, we performed Partial Least Squares Discriminant Analysis (PLS-DA) using R (version R.4.4.2). PLS-DA is a supervised dimensionality reduction technique that models the relationship between the feature matrix (cytokines) and a categorical response variable (groups), aiming to find latent components that maximize the separation between groups.
The analysis was conducted using the mixOmics 6.28.0 package. To visualize the results, we generated a biplot using the plotIndiv() and plotVar() functions. The biplot displays both the samples and the features in the space defined by the first two latent components. Each sample is represented as a point, colored by its group membership, while each feature is represented as an arrow. The direction of each arrow indicates the orientation of the variable in the component space, and the length of the arrow reflects the strength of the association between the feature and the components. Features with longer arrows are more strongly associated with the variation that separates the groups. Arrows pointing in the same direction as a group cluster suggest a positive association between that feature and the group. All visualizations were refined using base R plotting or ggplot2 to ensure clear interpretation and high-quality presentation.
Machine learning analysis of serum cytokines
To identify markers that significantly contribute to the classification of samples into “Mixed” and “NotMixed” groups at the D3 and D7 timepoints, a machine learning-based Random Forest approach was employed to determine the relative importance of each protein marker and to evaluate their association with the respective groups. The dataset underwent a series of preprocessing steps to ensure its suitability for machine learning analysis. First, samples corresponding to the D3 or D7 timepoint were filtered for analysis. Non-numeric columns, such as sample identifiers and time labels, were removed as they were not relevant to the classification task. Rows containing missing values were excluded to maintain the completeness and integrity of the data used for model training.
To evaluate the discriminative power of protein markers, a Random Forest classifier was used to classify samples into the “Mixed” and “NotMixed” groups. The Category column, which indicated group membership, served as the dependent variable, while the protein marker measurements acted as independent variables. The model was configured to generate 500 decision trees to ensure robust predictions and reliable feature importance metrics. Feature importance was assessed using the Mean Decrease in Gini impurity, which measures the contribution of each protein marker to reducing classification uncertainty.
To further interpret the importance of individual protein markers, their levels were compared between the “Mixed” and “NotMixed” groups. The mean expression difference for each protein marker was calculated, and features were categorized based on whether their higher expression was associated with the “Mixed” or “NotMixed” group. This allowed for the identification of proteins with stronger ties to either group.
The feature importance scores were visualized using a dot plot. In the plot, protein markers were arranged along the y-axis in descending order of their importance scores. The x-axis displayed the feature importance values derived from the Random Forest model. A red gradient color scale was used to highlight the magnitude of importance, with darker shades representing higher importance. The dot size was proportional to the feature importance score, further emphasizing the most critical markers.
To assess the statistical significance of the association of a group of cytokines instead of individual cytokines with the mixed challenge, we applied the Rotation Gene Set Testing (ROAST) method from the limma R package. Specifically, we are interested in determining whether a group of cytokines, exhibit significant changes in expression associated with the Mixed challenge compared to the Not Mixed challenge groups. ROAST works by applying a rotation-based resampling technique to simulate the null distribution, rather than relying on traditional permutation tests, which makes it particularly powerful for smaller sample sizes. It adjusts for correlations between genes, providing a more accurate assessment of whether the cytokines in the set are collectively upregulated, downregulated, or exhibit mixed regulation.
K-means clustering analysis
To assess how well the cytokine profiles naturally group the samples, independent of group labels, we used K-means clustering on the log-transformed cytokine data after filtering out cytokines with zero variance and removing highly collinear features. The clustering results were compared to the actual group labels to evaluate accuracy. The confusion matrix showed the distribution of samples across predicted clusters versus true groups, and the overall clustering accuracy was calculated as the proportion of correctly assigned samples. The results were visualized with a cluster plot, showing separation of samples colored by their assigned cluster. The following R packages were used for data analysis and visualization: “ggplot2”, “randomForest”, “mixOmics”, “FactoMineR”, “factoextra”, and “cluster”.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw data for individual monkeys is shown in the figures. Source data are provided with this paper. The sequencing data generated in this study have been deposited in GenBank under accession numbers: PX498814-PX498958. The deep sequencing data generated in this study have been deposited in the Sequence Read Archive under BioProject ID PRJNA1348589 and accession number SAMN52904265. The remaining data generated in this study are provided in the Source Data file. Source data are provided with this paper.
References
Baeten, J. M. et al. Antiretroviral prophylaxis for HIV prevention in heterosexual men and women. N. Engl. J. Med. 367, 399–410 (2012).
Marzinke, M. A. et al. Characterization of Human Immunodeficiency Virus (HIV) Infection in Cisgender Men and Transgender Women Who Have Sex With Men Receiving Injectable Cabotegravir for HIV Prevention: HPTN 083. J. Infect. Dis. 224, 1581–1592 (2021).
Bekker, L. G. et al. Twice-Yearly Lenacapavir or Daily F/TAF for HIV Prevention in Cisgender Women. N. Engl. J. Med. 391, 1179–1192 (2024).
Corey, L. et al. Two Randomized Trials of Neutralizing Antibodies to Prevent HIV-1 Acquisition. N. Engl. J. Med. 384, 1003–1014 (2021).
Gaebler, C. et al. Prolonged viral suppression with anti-HIV-1 antibody therapy. Nature 606, 368–374 (2022).
Sneller, M. C. et al. Combination anti-HIV antibodies provide sustained virological suppression. Nature 606, 375–381 (2022).
Julg, B. et al. Safety and antiviral effect of a triple combination of HIV-1 broadly neutralizing antibodies: a phase 1/2a trial. Nat. Med. 30, 3534–3543 (2024).
Ferrantelli, F. et al. Complete protection of neonatal rhesus macaques against oral exposure to pathogenic simian-human immunodeficiency virus by human anti-HIV monoclonal antibodies. J. Infect. Dis. 189, 2167–2173 (2004).
Hessell, A. J. et al. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 5, e1000433 (2009).
Julg, B. et al. Protection against a mixed SHIV challenge by a broadly neutralizing antibody cocktail. Sci. Transl. Med. 9, eaao4235 (2017).
Borducchi, E. N. et al. Antibody and TLR7 agonist delay viral rebound in SHIV-infected monkeys. Nature 563, 360–364 (2018).
Julg, B. et al. Broadly neutralizing antibodies targeting the HIV-1 envelope V2 apex confer protection against a clade C SHIV challenge. Sci. Transl. Med. 9, eaal1321 (2017).
Bricault, C. A. et al. HIV-1 Neutralizing Antibody Signatures and Application to Epitope-Targeted Vaccine Design. Cell Host Microbe 25, 59–72 e58 (2019).
Gonelli, C. A. et al. Antibody prophylaxis may mask subclinical SIV infections in macaques. Nature 639, 205–213 (2025).
Zost, S. J. et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449 (2020).
Whitney, J. B. et al. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 512, 74–77 (2014).
Julg, B. et al. Safety and antiviral activity of triple combination broadly neutralizing monoclonal antibody therapy against HIV-1: a phase 1 clinical trial. Nat. Med. 28, 1288–1296 (2022).
Seaman, M. S. et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84, 1439–1452 (2010).
Montefiori, D. C. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 485, 395–405 (2009).
Acknowledgements
We acknowledge support from NIH grants (AI128751, AI145801, AI149670, AI164556, AI169615, AI177687; D.H.B.) and the Ragon Institute of MGH, MIT, and Harvard (D.H.B.). We thank Michelle Lifton and Orion Barrett for technical assistance.
Author information
Authors and Affiliations
Contributions
D.H.B. designed the study. V.W.S, J.N., J.L., B.J., and M.S.S. performed the immunological and virological assays. M.A. conducted the machine learning analysis. V.W.S. and D.H.B. wrote the paper with the assistance of all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Walker-Sperling, V.E.K., Nkolola, J., Liu, J. et al. Minor SHIV variants abrogate protective efficacy of broadly neutralizing antibodies in rhesus macaques. Nat Commun 16, 11226 (2025). https://doi.org/10.1038/s41467-025-66321-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66321-7