Abstract
Our understanding of epigenetic processes is based on the hypothesis that individual posttranslational modifications of DNA and histones, or combinations thereof, function to direct unique downstream effects on transcription. Still, histone modifications are broadly categorized as repressive or activating, raising the question of potential functional redundancy. Here, we present an approach for addressing this question by substituting the genome-wide H3K27me3 pattern with other histone modifications. By taking advantage of the modular organization of PRC2, we direct de novo recruitment of H3K9me3 and H3K36me3 to PRC2 target genes in H3K27me3 null mouse embryonic stem cells (mESCs). We show that despite accurate genome-wide re-establishment of H3K36me3 at PRC2 target genes, which leads to significant reduction in H3K4me3 levels, the remaining H3K4me3 prevents H3K36me3 from recruiting sufficient DNA methylation to substitute for H3K27me3-mediated repression. In contrast, we demonstrate that H3K9me3 is more efficient in repressing H3K27me3 regulated genes, however this repression is also contingent on H3K4me3 status. Taken together, these results highlight the unique repressive functions of H3K27me3 and suggest that the functional effects of individual posttranslational modifications are highly dependent on the interplay with the existing chromatin environment.
Similar content being viewed by others
Introduction
It is widely accepted that individual histone posttranslational modifications (PTMs) and combinations thereof are essential for regulating chromatin structure and transcription by directing unique gene expression states1,2. H3K9me3, H3K27me3, and H3K36me3 are three of the most extensively studied histone PTMs, which all have been implicated in transcriptional repression, albeit in different genomic contexts. H3K9me3, enriched in gene poor regions and heterochromatin, is considered a strong transcriptional repressor acting through a self-reinforcing feed-forward loop involving HP1 and DNA methyltransferases (DNMTs)3. H3K36me3, which is enriched across actively transcribed genes and associated with RNA processing and RNA splicing4,5, has been suggested to have a transcription-coupled repressive role that safeguards transcriptional fidelity by repressing cryptic transcriptional initiation, in part, through the recruitment of DNMT3B6,7,8. Finally, the Polycomb modification H3K27me3, together with H2AK119ub1, is essential for maintaining the transcriptional repression of several thousand Polycomb target genes9,10,11,12,13. Because the genome-wide enrichment patterns for these PTMs are generally mutually exclusive, a large-scale, direct comparison of their repressive capacities has not been possible.
The Polycomb Repressive Complex 2 (PRC2) is responsible for catalyzing all levels of methylation of lysine 27 on histone H314. PRC2 is primarily found enriched at CpG island (CGI) H3K27me3-positive promoters of transcriptionally silent genes – here forward described as PRC2 target genes9,15,16. In mESCs, many PRC2 target genes are additionally co-occupied by H3K4me3 at their CpG promoters and are referred to as bivalent genes17,18. Previously, we discovered that H3K27me3 could be accurately established at PRC2 target genes independent of self-autonomous epigenetic inheritance by SUZ12-directed de novo recruitment of PRC214. Further, we demonstrated that PRC2 has a unique modular organization that can be functionally divided into a catalytic module and a chromatin-binding module. The SUZ12 C-terminal VEFS domain stabilizes the core subunits EZH1/2 and EED, forming a minimal catalytically competent complex, while the N-terminal fragment of SUZ12, lacking the C-terminal VEFS domain (S12N), interacts with all non-core PRC2 subunits and specifies the binding of the complex to PRC2 target sites across the genome (Fig. 1a)14,19. The independent chromatin binding ability of S12N thus makes it suitable as a specific recruiting domain for tethering of other activities to PRC2 target genes. Here, we have taken advantage of this unique modular organization of PRC2 by directing de novo recruitment of the catalytic activity of SUV39H2 and SETD2 to PRC2 target genes in H3K27me3-depleted mESCs. This robust system allows for genome-wide substitution of the H3K27me3 pattern with the repressive modifications H3K9me3 and H3K36me3 to directly study and compare their functional effects on gene expression of thousands of Polycomb target genes.
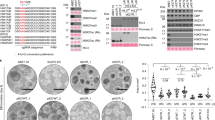
a Schematic illustration of PRC2 composition. SUZ12 acts as a scaffold for the complex, containing a C-terminal VEFS domain binding to core-subunits EZH1/2 and EED and an N-terminal region (S12N) interacting with the PRC2 non-core subunits. The VEFS domain of SUZ12 is required for catalytic activity, while chromatin binding depends on the S12N domain and the associated non-core subunits. b Mean SUZ12 ChIP-seq signals (RPKM) for WT mESCs and Suz12 KO mESCs with ectopic expression of S12N or WT SUZ12 within a 40 kb window centered on 7732 H3K27me3 positive promoter peaks identified in WT mESCs (n = 1). c Representative western blot probing for global H3K27me3 and the expression of fusion constructs using an anti-SUZ12 antibody (n = 3). d Pearson correlation for the expression phenotypes of the indicated cell lines in three independent biological replicates. e Schematic drawing of the experimental strategy to recruit H3K9me3 and H3K36me3 to PRC2 target genes, using the S12N-PRC2 as a recruiter. f Mean SUZ12 ChIP-seq signals (RPKM) (n = 1) of extracts prepared from Suz12 KO mESCs with ectopic expression of fusion constructs within a 40 kb window centered on 7732 H3K27me3 positive promoter peaks identified in WT mESCs.
Results
Establishment of Polycomb targeting chimeric epigenetic complexes
To establish a system for genome-wide replacement of H3K27me3 with H3K36me3 or H3K9me3, we first deleted Suz12 in mESCs using CRISPR/Cas9. Knockout of Suz12 abolishes H3K27 methylation and Suz12 KO cells, therefore provides a suitable system for a functional comparison between de novo establishment of H3K27me3, H3K9me3 and H3K36me314. Following confirmation of H3K27me3 depletion in Suz12 KO mESCs, we assessed genome-wide localization patterns of the S12N (aa 1–544) upon re-expression in Suz12 KO mESCs. Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) analysis revealed that S12N re-expression led to its proper localization at consensus PRC2-binding regions, similar to the re-expressed full-length SUZ12 (Fig. 1b). In both cases, enrichment at these sites was reduced as compared to wild type SUZ12 (Fig. 1b), consistent with lower total protein levels (Supplementary Fig. 1a, b and Fig. 1c). However, binding site fidelity and peak enrichments were maintained (Supplementary Fig. 1c, d). In agreement with the loss of EZH1/2 and EED interaction, we found no detectable levels of H3K27 methylation in the S12N expressing mESCs (Fig. 1c), and the cells displayed similar gene expression phenotypes as Suz12 KO mESCs (Fig. 1d). Thus, despite recapitulating the normal PRC2 binding pattern, binding of S12N alone has no significant repressive effect on gene transcription.
Next, we generated new Polycomb targeting chimeric epigenetic complexes with activity towards H3K36 and H3K9 by expressing S12N fusion proteins containing either the catalytic SET domain of the H3K36 histone methyltransferase SETD2 (S12N:SD2), or a fragment including the chromodomain and SET domain of the H3K9 histone methyltransferase SUV39H2 (S12N:SUV2) in Suz12 KO mESCs (Fig. 1e, f and Supplementary Fig. 1a). Both chimeric complexes localized to PRC2 target genes, although at a reduced level, which was more pronounced for S12N:SUV2 (Fig. 1f and Supplementary Fig. 1e). Thus, these results show that the N-terminal of SUZ12 is sufficient to drive the proper localization of the chimeric proteins to Polycomb target genes genome-wide.
Ectopic H3K36me3 deposition resembles endogenous H3K27me3 genome-wide
To assess whether S12N:SD2 could deposit H3K36me3 to mimic the genome-wide H3K27me3 pattern, we performed Cleavage Under Targets & Release Using Nuclease (CUT&RUN) for H3K36me3. To control for the specificity of the de novo H3K36me3 deposition, we generated a negative control (S12N:SD2*) by substituting Arginine 1625 with Glycine in SETD2, a mutant previously shown to reduce H3K36me3 through de-stabilization of the SETD2 protein20 (Fig. 2a and Supplementary Fig. 2a). As expected, expression of the S12N:SD2, but not S12N:SD2*, led to a global increase (4-fold) in H3K36me3 (Supplementary Fig. 2b). Consistent with its reduced stability, expression levels of the mutant version of the S12N:SD2 fusion protein were significantly lower than those of the wild type fusion, resulting in H3K36me3 levels comparable to WT mECs. Consistently, we found strong H3K36me3 enrichment across the HoxA locus in S12N:SD2 mESCs – a region that is strongly enriched for H3K27me3 but devoid of endogenous H3K36me3 in wild-type mESCs (Fig. 2b). We further validated these results by performing ChIP followed by qPCR (ChIP-qPCR) at the Hoxa9 promoter (Supplementary Fig. 2c). Strikingly, the H3K36me3 pattern across the HoxA locus accurately resembled that of H3K27me3, and the same accuracy was observed for narrow peaks (Fig. 2b–e). Moreover, the enrichments of the S12N:SD2-driven H3K36me3 peaks have similar peak intensity to adjacent endogenous H3K36me3 peaks, suggesting the H3K36me3 levels deposited by S12N:SD2 are physiologically relevant (Fig. 2d, e).
a Schematic drawing of the domain architecture of the fusions S12N:SD2 and S12N:SD2* with indications of boundaries (amino acid numbers referring to the sequence in WT SUZ12 and SETD2). Asterisk indicates a point mutation introduced to generate catalytic inactive S12N:SD2*. b–e CUT&RUN tracks showing H3K27me3, H3K4me3, and H3K36me3 signals (RPKM) from the indicated cell lines within a representative genomic region that includes the HoxA cluster (b), PRC2 target genes Bmi1, Spag6, and Carlr (c), PRC2 target genes Fgf3 and Fgf15 and the endogenously H3K36me3 positive Fgf4 gene (d), and PRC2 target gene Ntng2 and the endogenously H3K36me3 positive Setx gene (e). The tracks are representative from one biological replicate, performed in biological duplicates (n = 2). CGI annotations (red) are shown below the track. f Mean H3K36me3 CUT&RUN signals (RPKM) in indicated cell lines within a 40 kb window centered on 7732 H3K27me3 positive promoter peaks identified in WT mESCs. g Plot of H3K36me3 CUT&RUN signals (RPM) enrichment within peak boundaries of all H3K27me3 peaks identified in WT mESCs (promoter and non-promoter; n = 22307) for the indicated cell lines quantified using BedTools Multicov. Smoothed lines represent generalized additive model (GAM) fits of mean H3K36me3 signal intensity (RPM) from two independent biological replicates (n = 2), as a function of H3K27me3 CUT&RUN signal intensity (RPM) in WT mESCs (n = 1); shaded ribbons show the 95% confidence interval of the fitted mean (not variability between biological replicates).
We then expanded the analysis to include all endogenous H3K27me3 peaks identified in WT mESCs (n = 22307). H3K36me3 was strongly enriched across all H3K27me3 peaks in S12N:SD2 mESCs, while undetectable in WT, Suz12 KO, and S12N:SD2* mESCs (Fig. 2f). Although average signal intensity analyses indicate an effective deposition of H3K36me3 by S12N:SD2, this approach does not distinguish between individual differences in H3K36me3 peak intensity across the H3K27me3 landscape. Thus, we quantified the H3K36me3 signal within the bounds of endogenous H3K27me3 peaks and directly compared it to the endogenous H3K27me3 signal pattern in WT mESCs. Strikingly, we found that the relative peak signal of ectopic H3K36me3 in S12N:SD2 mESCs strongly correlated with the relative WT H3K27me3 signal across all peaks (Fig. 2g and Supplementary Fig. 2d). Taken together, these data demonstrate that the S12N:SD2 chimeric complex can establish a H3K36me3 pattern that is very similar to the H3K27me3 pattern established by PRC2 genome wide.
H3K36me3 cannot substitute for H3K27me3 to restore gene repression and mESC pluripotency
To determine the transcriptional effect of establishing H3K36me3 at PRC2 target genes, we performed gene expression analysis by RNA sequencing (RNA-seq). Loss of H3K27 methylation in mESCs leads to deregulated gene expression, which can be rescued upon restoration of H3K27 methylation14. For this analysis, we first classified “direct” PRC2 target genes as genes having H3K27me3 high confidence peaks within +/− 3 kb of the transcriptional start site (TSS) showing differential expression upon loss of SUZ12. K-means cluster analysis (K = 4) revealed that loss of SUZ12 primarily led to increased expression of PRC2-targeted genes, consistent with previous reports (Fig. 3a)13. We also addressed the direct role of PRC2 in gene repression by performing RNA-seq during a time course of mAID-SUZ12 degradation. While protein degradation at early time points did not alter gene expression profiles, upregulation of PRC2 targets was observed 24 h onwards following auxin treatment, coinciding with reduced H3K27me3 levels upon SUZ12 loss. At 48 h, additional indirect effects became apparent, with both gene upregulation and downregulation detected (Supplementary Fig 3). Interestingly, while re-expression of WT SUZ12 restored gene expression to WT levels, we observed no general repression of direct PRC2 target genes by S12N:SD2 indicating that H3K36me3 cannot substitute for H3K27me3 in regulating gene expression (Fig. 3a).
a Cluster heatmap (k-means, 4) of RNA-seq data analyzed with Deseq2 for differential expression (FDR < 0.05). Heatmap shows differentially expressed PRC2 target genes identified by H3K27me3 positive promoters in WT mESCs (n = 4986) in the indicated cell lines based on three independent biological replicates. b MA plots showing mean changes in gene expression from Deseq2 analysis based on three independent biological replicates (n = 3). Indicated with red dots are significantly upregulated genes in Suz12 KO mESCs filtered with log2 expression foldchange > 1 and FDR < 0.05. The mean expression of the upregulated genes in Suz12 KO mESCs are traced in the indicated cell lines. c Bar plot showing the fraction of the 1326 upregulated PRC2 target genes in Suz12 KO mESCs that are fully or partially rescued in the indicated cell lines (see methods for filtering criteria). Statistical significance was calculated using Fisher’s exact test, two-tailed (*p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001). Source data are provided as a Source Data file. d Bar plot showing the percentage of beating clusters identified on day 10 after induction of embryoid body differentiation in the indicated cell lines. The plot is representative of at least three independent experiments. e Mean H3K4me3 CUT&RUN signals (RPKM) (n = 2) in indicated cell lines within a 40 Kb window centered on 7732 H3K27me3 positive promoter peaks identified in WT mESCs. f Custom annotation based DMR methylation (%, see methods) at CpG islands for indicated cell lines (n = 3). The horizontal lines mark the median; the boxes mark the interquartile range (IQR) and whiskers extend up to 1.5 times the IQR; individual data points beyond this range are plotted. Statistical significance was calculated for the means using an unpaired two-sample t test, two-tailed (*p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001). Source data are provided as a Source Data file.
Next, we sought to evaluate the repressive capacity of H3K36me3 more thoroughly by focusing specifically on the set of PRC2 target genes upregulated upon loss of H3K27me3. We quantified the number of significantly upregulated genes and found that 22% (1326/6026) of PRC2 target genes to be derepressed upon SUZ12 loss (Fig. 3a, b, and Supplementary Fig. 4a), consistent with previously published data13,14. We noticed that, in the wild type chromatin, the significantly upregulated gene group was decorated with considerably higher levels of H3K27me3 and H2AK119ub1 and a correspondingly lower H3K4me3 in WT mESCs relative to the unaffected gene group (Supplementary Fig. 4b–d). Further, H3K4me3 increased upon loss of H3K27me3 in the upregulated gene group, while the unaffected gene group showed increased H2AK119ub1 levels and decreased H3K4me3 (Supplementary Fig. 4e–g). Combined, this suggests that a higher repressive threshold may be required to restrict gene activation of the upregulated gene set, making the loss of H3K27me3 sufficient for derepressing these genes. As expected, restoring the H3K27me3 pattern in WT SUZ12-rescued mESCs reverted the chromatin environment to its original state for both gene groups (Supplementary Fig. 4h–j). To quantify the ability of ectopically expressed SUZ12 to fully restore repression, we applied stringent measures (see “Methods”). Using these criteria, the re-expressed WT SUZ12 reverted gene expression of 74.1% of the PRC2-targeted deregulated genes (983/1326; Fig. 3b, c). In contrast, S12N:SD2 chimera only led to minor effects on transcription, fully rescuing only 3.5% of the genes (46/1326; Fig. 3b, c).
Polycomb-specified H3K36me3 significantly reduces H3K4me3 levels
Consistent with the general inability of H3K36me3 to substitute for H3K27me3-mediated repression, the S12N:SD2 chimera, unlike WT SUZ12-rescued mESCs, failed to restore normal differentiation of mESC (Fig. 3d). However, the persistence of artificially deposited H3K36me3 may also contribute to impaired differentiation. Still, deposition of H3K36me3 was able to revert the increased expression of a significantly higher number of SUZ12 target genes than observed for S12N (0.8%) or S12N:SD2* (0.5%), suggesting H3K36me3 may act repressively albeit in a very specific chromatin context. Therefore, we sought to assess the direct effects of H3K36me3 deposition on the endogenous chromatin environment as well as to investigate whether any feature could explain the selective rescue. Unexpectedly, we found that deposition of H3K36me3 drastically reduced H3K4me3 levels at PRC2 target genes and this reduction was further exacerbated at the genes whose expression was repressed by H3K36me3 (Fig. 3e and Supplementary Fig. 5a). In contrast no global changes in H2AK119ub1 were observed upon H3K36me3 deposition (Supplementary Fig. 5b). We tested other elements of the chromatin environment including H3K36me3 deposition efficacy by S12N:SD2, H2AK119ub1 levels, peak width, genomic gene length, CGI annotation status, GC content and CpG density, initial gene expression level in WT mESCs, and fold-change upregulation upon SUZ12 depletion, but found no major difference between the two H3K36me3 gene-groups (Supplementary Fig. 5a,c–i).
To investigate the mechanism by which H3K36me3 leads to a reduction of H3K4me3 at TSSs, we examined whether H3K36me3 influences either the enzymatic activity of H3K4 methyltransferase complexes or their recruitment to chromatin. In vitro H3K4 methyltransferase assays were performed using a recombinant KMT2B (MLL2) complex with unmodified or H3K36me3 modified mononucleosomes as substrates. These assays revealed that H3K36me3 inhibits the catalytic kinetics of KMT2B, thereby reducing its ability to methylate H3K4 (Supplementary Fig. 6a, b). We next tested whether expression of the S12N:SD2 fusion proteins would affect the recruitment of the H3K4 methyltransferase complexes or H3K4me3 demethylases. No detectable changes were observed in the occupancy of RBBP5, a core subunit for all H3K4 methyltransferases, or of the KDM5B H3K4me3/me2 demethylase (Supplementary Fig. 6c, d). Together, these results suggest that the reduction of H3K4me3 at TSSs in response to H3K36me3 arises primarily from decrease in the catalytic efficiency of the H3K4 methyltransferases than altered complex recruitment.
Residual H3K4me3 obstructs H3K36me3 mediated repression by antagonizing de novo DNA methylation
Studies have linked H3K36me3-mediated gene repression to DNMT3B recruitment and de novo DNA methylation, a mechanism coupled to transcriptional elongation that prevents aberrant transcriptional initiation from cryptic start sites within gene bodies7,8. Notably, these regions are typically devoid of H3K4me3, which is largely confined to promoter regions. H3K4me3, in turn, has been shown to protect CpG island (CGI) promoters from de novo DNA methylation21. Based on this, we hypothesized that although our data indicate that H3K36me3 exerts dominance over H3K4me3, the inability of promoter-proximal H3K36me3 to mediate repression could be explained by the persistence of H3K4me3 at these sites, which blocks de novo DNA methylation otherwise recruited by H3K36me3.
To test this, we performed Reduced Representation Bisulfite Sequencing (RRBS) and focused our analyses to only include differentially methylated CpGs (DMCs) found within the bounds of WT H3K27me3 peaks. As expected, CGIs were refractory to DNA methylation, and DNA methylation levels increased with distance from CGIs in WT mESCs (Supplementary Fig. 7a). To explore the relationship between DNA methylation and gene expression, we called differentially methylated regions (DMRs) using a custom method based on CpG and gene annotations (See Supplementary Fig. 7b and “Methods”). Indeed, we found that promoter-proximal deposition of H3K36me3 by S12N:SD2 led to de novo DNA methylation specifically at CGIs confirming the previously observed link between H3K36me3 and DNA methylation (Fig. 3f and Supplementary Fig. 7a, c)7,8. Importantly, the observed accumulation of DNA methylation occurred at a frequency in the cell population likely insufficient for transcriptional repression, suggesting that the remaining H3K4me3 levels efficiently protected CGIs from this mode of repression. Given that we observed H3K4me3 levels that were mildly lower at genes rescued by H3K36me3, we tested if this correlated with increased de novo DNA methylation at these genes. Consistent with this, we detected significantly higher DNA methylation levels at CGIs in genes rescued by S12N:SD2 – the gene group with the lowest observed H3K4me3 levels (Supplementary Fig. 7d). Together, these results indicate that H3K36me3 has a repressive capacity mediated through de novo DNA methylation – a repressive mechanism that is generally incompatible with the chromatin context surrounding PRC2 target gene promoters.
To further investigate the role of H3K4me3 in preventing DNA methylation and H3K36me3-mediated repression, we combined our genome-wide H3K27me3 replacement system with an established RBBP5-FKBP12F36V knock-in mESC line by knocking out Suz12 in the cell line (R-Suz12 KO) and re-expressing of S12N:SD2 (RS + S12N:SUV2). RBBP5 is a core subunit of all H3K4 methylation complexes, and dTAG-13 treatment of these cells induces rapid RBBP5 degradation and complete H3K4me3 loss within two hours22. R-Suz12 KO mESCs expressing S12N:SD2 were treated with dTAG-13 for 24 h to allow H3K36me3 accumulation following H3K4me3 depletion (Supplementary Fig. 8b). As expected, H3K4me3 was fully depleted after 24 h, and S12N:SD2 expression was unaffected (Supplementary Fig. 8b).
To assess the effect of H3K4me3 depletion on H3K36me3-mediated transcriptional repression, we performed RNA-seq and observed a significant increase in the number of PRC2 target genes repressed by H3K36me3 (Supplementary Fig. 8c). Consistent a protective role of H3K4me3 against de novo DNA methylation, H3K4me3 depletion in the presence of increased H3K36me3 led to elevated DNA methylation at PRC2 target genes (Supplementary Fig. 8d, e). Together, these results show that H3K36me3 can repress Polycomb target genes in a context dependent manner, with efficiency in part dependent on the levels of H3K4 methylation.
Polycomb directed H3K9me3 gives continuous block-like accumulation, but different than endogenous H3K27me3
H3K9me3 is considered a strong repressive histone modification and a hallmark of constitutive heterochromatin and chromatin compaction. To further uncover whether histone PTMs are functionally specific or redundant in relation to transcriptional repression, we expressed S12N:SUV2 in Suz12 KO mESCs, along with a catalytic inactive control (S12N:SUV2*) in which two-point mutations of the active site (F259A and Y261A) were introduced and performed CUT&RUN for H3K9me3 (Fig. 4a and Supplementary Fig. 9a). As normalization of H3K9me3 high-throughput data is challenging due to the high enrichment of H3K9me3 at pericentric heterochromatin, we performed peak-normalization on H3K9me3 CUT&RUN samples relative to the enrichment at endogenous H3K9me3 peaks (see Methods, and Supplementary Fig. 10). We validated this method of normalization by performing ChIP-qPCR and found that the H3K9me3 signals at H3K9me3 endogenous peaks were comparable in all samples (Supplementary Fig. 10a).
a Schematic drawing of the domain architecture of S12N:SUV2 and S12N:SUV2* with indications of boundaries (amino acid numbers referring to the sequence in WT SUZ12 and SUV39H2). Asterisks indicate point mutations introduced to generate the methyltransferase dead S12N:SUV2*. b–e CUT&RUN track showing H3K27me3, H3K4me3, and H3K9me3 signals (RPKM and peak-normalization) from indicated cell lines within a representative genomic region that includes the PRC2 target genes Prdm12, Fibcd1, Lamc3, and Aif1l (b), PRC2 target gene Ntng2 with adjacent endogenous H3K9me3 peak (c), the PRC2 target gene Thbs2 with adjacent endogenous H3K9me3 peaks (d) and the Hox A cluster (e). Representative tracks from one biological replicate, performed in biological duplicates (n = 2). CGI annotations (red) are shown below the tracks. f Mean H3K9me3 CUT&RUN signals (peak-normalization) in indicated cell lines within a 40 Kb window centered on 7732 H3K27me3 positive promoter peaks identified in WT mESCs, scale on the right side. WT mESC H3K4me3 CUT&RUN signal (RPKM) is included for reference with the scale on the left. g Plot of H3K9me3 CUT&RUN signals (RPM) enrichment within peak boundaries of all H3K27me3 peaks identified in WT mESCs (promoter and non-promoter; n = 22307) for the indicated cell lines, quantified using BedTools Multicov. Smoothed lines represent generalized additive model (GAM) fits of mean H3K9me3 signal intensity (RPM) from two independent biological replicates (n = 2), as a function of H3K27me3 CUT&RUN signal intensity (RPM) in WT mESCs (n = 1); shaded ribbons show the 95% confidence interval of the fitted mean (not variability between biological replicates).
Like H3K36me3, expression of S12N:SUV2 resulted in the deposition of H3K9me3 at PRC2 target genes (Supplementary Fig. 9b), however the H3K9me3 profile differed from that of H3K27me3 in WT mESCs (Fig. 4b). First, we observed the H3K9me3 deposition by S12N:SUV2 frequently extended beyond the chromosomal intervals normally decorated by H3K27me3, which was further amplified with the proximity of an endogenous H3K9me3 peak (Fig. 4c–e and Supplementary Fig. 9c). Secondly, H3K9me3 did not show the same continuous block-like accumulation as endogenous H3K27me3 at these regions. (Fig. 4c). Instead, the S12N:SUV2-driven H3K9me3 peaks displayed dips that directly overlapped with CGI annotations and H3K4me3 enrichment, suggesting that H3K4me3 antagonizes H3K9me3 deposition. Consistent with this notion, we did not observe dips in H3K9me3 peaks at monovalent PRC2 target genes (Fig. 4d), and H3K9me3 displayed lower enrichment at the Hox clusters, which contain multiple H3K4me3 peaks (Fig. 4e). Still, we found a strong correlation in the relative peak intensity of S12N:SUV2-driven H3K9me3 and the H3K27me3 profile in WT mESCs, with the notable exception of Hox clusters that represent the strongest endogenous H3K27me3 peaks (Fig. 4f, g).
H3K9me3 can repress Polycomb target genes in a context-dependent manner, directly contingent on the H3K4me3 status
In contrast to the expression of S12N:SD2, expression of S12N:SUV2 in Suz12 knockout mESCs had a significant repressive effect on PRC2 target genes, reverting the gene expression of a substantial portion of genes upregulated upon SUZ12 loss (Fig. 5a, b). Moreover, this repressive effect could be ascribed to the deposition of H3K9me3 as no rescue was observed in mESCs expressing the catalytically dead S12N:SUV2* (Fig. 5a, b). Employing the same transcriptional analysis as for H3K36me3, we found that deposition of H3K9me3 resulted in complete rescue of 25.4% of PRC2 upregulated genes – approximately one third of the rescue achieved by SUZ12 re-expression and subsequent H3K27me3 deposition (Fig. 5c). Consequently, the expressing of S12N:SUV2 was insufficient to rescue mESC differentiation into beating clusters (Fig. 5d), even though we cannot discard that this effect could be responding to the lack of H3K9me3 removal.
a Cluster heatmap (k-means, 4) of RNA-seq data analyzed with Deseq2 for differential expression (FDR < 0.05) for the indicated cell lines. Heatmap shows z-score normalized counts of differentially expressed PRC2 target genes identified by H3K27me3 positive promoters in WT mESCs (n = 4986). Based on three independent biological replicates (n = 3). b MA plots showing mean changes in gene expression from Deseq2 analysis based on three independent biological replicates (n = 3). Indicated with red dots are significantly upregulated genes in Suz12 KO mESCs filtered with log2 expression fold change> 1 and FDR < 0.05. The mean expression of the upregulated genes in Suz12 KO mESCs are traced in the indicated cell lines. c Bar plot showing the fraction of the 1326 upregulated PRC2 target genes in Suz12 KO mESCs that are fully or partially rescued in the indicated cell lines (see methods for filtering criteria). Statistical significance was calculated using Fisher’s exact test, two-tailed (*p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001). Source data are provided as a Source Data file. d Bar plot showing the percentage of beating cluster identified on day 10 after induction of embryoid body differentiation in the indicated cell lines. The plot is representative of at least three independent experiments. e Mean H3K4me3 CUT&RUN signals (RPKM) (n = 2) in indicated cell lines within a 40 Kb window centered on 7732 H3K27me3 positive promoter peaks identified in WT mESCs. f Mean H3K4me3 CUT&RUN signals (RPKM) within 40 Kb window centered on PRC2 target promoters for the H3K9me3-rescued versus non-rescued genes in Suz12 KO mESCs expressing S12N:SUV2. g Mean H3K9me3 CUT&RUN signal (peak-normalized) within 40 Kb window centered on PRC2 target promoters for the H3K9me3-rescued versus non-rescued genes in Suz12 KO mESCs expressing S12N:SUV2 (n = 2). h Heatmaps of sequence depth normalized H3K4me3 CUT&RUN data (blue), SUZ12 ChIP-seq data (S12N fusion proteins) (red), and peak-normalized H3K9me3 CUT&RUN data (black) at PRC2 target promoters in RBBP5-FKBP12F36V + Suz12 KO mESCs expressing S12N:SUV2 treated with dTAG-13 or DMSO for 24hrs. Top: Average plots of the mean signal for the region displayed in heatmaps. i Bar plot of relative gene rescue (%) in RBBP5-FKBP12F36V + Suz12 KO mESCs expressing S12N:SUV2 treated with dTAG-13 for 24 h with DMSO treatment for 24 hrs as reference (n = 3).
Previous studies have shown that tethering the chromo-shadow domain (csd) of HP1α can induce H3K9me3-dependent gene silencing23,24. To test whether csdHP1α is more effective than SUV39H2-catalyzed H3K9me3 in repressing Polycomb target genes, we expressed S12N fused to csdHP1α in Suz12 KO mESCs. This resulted in the repression of a similar number of genes as observed with S12N fused to SUV39H2 (Supplementary Fig. 11), suggesting that the efficiency of repression by SUV39H2 or HP1α is determined by the mode of recruitment - whether mediated via Polycomb-directed H3K9me3 binding or direct artificial tethering to chromatin.
Given the established role of H3K9me3 in transcriptional repression, we also sought to understand why the deposition only partly rescued PRC2 target gene expression. We hypothesized that differences in the endogenous chromatin environment might determine whether H3K9me3 can induce repression. To test this, we compared the proximal chromatin environment of H3K9me3-rescued genes versus non-rescued genes. Similar to H3K36me3, we observed negligible differences between the two groups in H2AK119ub1 levels, H3K27me3 peak-width in WT cells, genomic gene length, basal gene expression level and fold-change upregulation upon SUZ12 depletion between the two groups (Supplementary Fig. 12a–d). In contrast to H3K36me3, however, deposition of H3K9me3 at PRC2 target promoters did not alter endogenous H3K4me3 levels (Fig. 5e). Instead, we found striking differences in pre-existing H3K4me3 levels. H3K9me3-rescued genes exhibited significantly lower endogenous H3K4me3 enrichment, which correlated with a markedly more efficient H3K9me3 accumulation (Fig. 5f, g and Supplementary Fig. 12f). Collectively, these findings suggest that pre-existing H3K4me3 acts as a barrier to H3K9me3 accumulation from reaching a required threshold for effective repression of PRC2 target genes.
Indeed, H3K4me3 has previously been shown to antagonize the binding of SUV39H1/2 in vitro25. Moreover, the H3K9-specific demethylases KDM4A and KDM4C both localize to H3K4me3-positive regions to promote active removal of H3K9me326. To investigate the mechanism limiting efficient H3K9me3 deposition in S12N:SUV2 mESCs, we utilized the RBBP5-FKBP12F36V Suz12 knockout (R-Suz12 KO) line described above and introduced S12N:SUV2 (RS + S12N:SUV2) (Supplementary Fig 13a). Cells were treated with dTAG-13 for 24 h to deplete H3K4me3 and allow sufficient time for subsequent H3K9me3 accumulation (Supplementary Fig. 13b).
To determine the effect of H3K4me3 depletion on H3K9me3 deposition, we performed ChIP-seq for SUZ12 and CUT&RUN for H3K4me3 and H3K9me3 in dTAG-13 treated and untreated cells. dTAG-13 treatment resulted in a complete loss of H3K4me3 peaks, which in turn markedly enhanced chromatin binding of the S12N:SUV2 chimera and promoted H3K9me3 accumulation at PRC2 target promoters (Fig. 5h and Supplementary Fig. 13c–e). Next, we tested whether the lack of H3K9me3 accumulation at H3K4me3 enriched sites could in part be explained by active demethylation through KDM4A/C. To this end, we treated S12N:SUV2 expressing mESCs with the KDM4-specific inhibitor QC6352 for 96 h27,28. However, we did not observe any additional H3K9me3 accumulation upon inhibitor treatment (Supplementary Fig 13 f). Together, these results indicate that pre-existing H3K4me3 directly antagonizes H3K9me3 deposition, not via rapid KDM4-mediated removal, but likely by preventing stable chromatin engagement of S12N:SUV2 through SUV39H2-associated components.
Finally, to corroborate this model, we performed RNA-seq in the H3K4me3-depleted cells. Consistent with our hypothesis, depletion of H3K4me3 and the consequent increase in H3K9me3 deposition resulted in an 82.4% increase in the number of PRC2 genes that were fully rescued. This effect could not be attributed to loss of H3K4me3 alone, as dTAG treatment of RBBP5-FKBP12F36V Suz12 KO mESCs resulted only in minor changes in overall mRNA levels (Fig. 5i and Supplementary Fig. 13g). Taken together, these results demonstrate that H3K9me3 can mediate repression of Polycomb targets in a context-dependent manner, critically determined by the pre-existing H3K4me3 state.
Discussion
Because histone modifications with comparable proposed effects on transcription are rarely found in the same genomic context, a comprehensive and direct comparison of their causal roles has not previously been assessed. In this study, we investigated whether different repressive histone modifications can substitute for one another genome-wide, taking advantage of the modular composition of PRC2. We focused on H3K9me3, a hallmark of constitutive heterochromatin, and H3K36me3, which normally marks gene bodies of active genes and prevents aberrant initiation from cryptic promoters.
We found that both the Polycomb-targeting chimeric complexes could deposit physiological levels of H3K9me3 and H3K36me3 at Polycomb target genes. However, unlike H3K27me3-mediated repression, the repression by H3K9me3 and H3K36me3, at most Polycomb target genes, was antagonized by H3K4me3, albeit for mechanistically distinct reasons and with different efficiencies. For H3K9me3, repression was not intrinsically impaired, but pre-existing H3K4me3 prevented stable accumulation of H3K9me3 at H3K4me3-positive nucleosomes, thereby blocking a repressive threshold. In contrast, H3K36me3 was unaffected by H3K4me3 and S12N:SD2-driven H3K36me3 deposition closely mirrored the H3K27me3 distribution genome-wide. Yet, repression still failed, apparently because residual H3K4me3 obstructed downstream DNA methylation. Thus, H3K9me3 and H3K36me3-mediated repression proved incompatible with the chromatin environment typical of Polycomb target genes, but through fundamentally different routes.
These findings further highlight the critical role of H3K4me3 in safeguarding CGI promoters, - not only from de novo DNA methylation, but also by blocking inappropriate invasion by H3K9me3. Since the absence of both DNA methylation and H3K9me3 has been shown to promote H3K27me3 recruitment29,30,31,32,33,34, we suggest that H3K4me3 indirectly ensures proper H3K27me3 recruitment at CGI promoters by excluding competing repressive pathways.
In summary, our work demonstrates that although H3K9me3, H3K27me3, and H3K36me3 are all classified as repressive histone modifications, their functions are not interchangeable. This supports the concept of a histone code35, but reframes it: rather than having single histone modifications dictating unique transcriptional outcomes, it is the crosstalk between histone modifications that ensures incompatible modifications are spatially separated. Such segregation results in groups of highly correlated modifications that are specialized for distinct genomic contexts. Our results, therefore, underscore the necessity for cells to maintain a repertoire of repressive histone modifications that are not redundant, but instead complementary, each tuned to operate within its compatible chromatin environment.
Methods
Cell culture
Mouse embryonic stem cells were cultured in serum/LIF medium (Glasgow Minimum Essential Media (GMEM) supplemented with 10% heat-inactivated Fetal Bovine Serum (HI-FBS), 1% Penicillin/Streptomycin, 2 mM L-glutamine, 1 mM Sodium Pyruvate (Gibco), 0.1 mM MEM Non-essential amino acids (Gibco), 100 µM ß-Mercaptoethanol (Gibco) and Leukemia Inhibitory Factor (LIF, produced in the lab)). mESCs were grown in gelatin-coated radiation sterilized dishes and kept in humidified incubators at 37 °C with 5 % CO2 and atmospheric O2 levels. Cells were routinely passed every 2-3 days by aspiration of medium, washed in PBS, dissociated using 0.25% trypsin-EDTA with gentle pipetting, followed by resuspension, centrifugation, and re-plating. Cells were re-plated at a density of 0.5-1 × 106 cells/10 cm dish. RBBP5- FKBP12F36V-2xHA cell lines were treated with dTAG-13 (Tocris) or DMSO for 24 h before being harvested for experiments. For KDM4 inhibitor experiments, Suz12 KO mESCs expressing S12N:SUV2 were treated with 50 nM or 100 nM of the KDM4C inhibitor QC635236 (MedChemExpress) for 96 h with media renewal every 24 h. An overview of cell lines can be found in Supplementary Table 1.
Embryoid body differentiation
1-1.5 × 106 cells were seeded into 10 cm low-attachment dishes in serum-containing medium without LIF (1:1 mix of DMEM-F12 (Gibco) and Neurobasal Medium (Gibco) supplemented with 1% Penicillin/Streptomycin, 2 mM L-glutamine, 1 mM Sodium Pyruvate (Gibco), 0.1 mM MEM Non-essential amino acids (Gibco) and 100 µM ß-Mercaptoethanol (Gibco)). The medium was changed on days 2, 4, 6, and 8. On days 4 and 8, a sub-fraction of embryoid bodies was collected for RNA or chromatin extraction. The remaining embryoid bodies were re-seeded onto new low-attachment plates (day 2, 4, and 6) or gelatin-coated plates (day 8). On day 10, colonies were analyzed for cardiomyocyte differentiation by quantification of spontaneous rhythmic contractions within colonies (beating clusters). Data were visualized with Prism v9 (Graphpad).
Generation of cell lines
Knockout mESCs
For knockout of Suz12, two gRNAs were designed to generate indels or large deletions in wild-type E14 mESCs using CRISPR-Cas9 nickase37. CRISPR sgRNAs were designed using the CRISPOR guide RNA selection tool38. Sequences are provided in Supplementary Table 2. For the construction of sgRNA-encoding plasmids, single-stranded oligonucleotides (IDT) containing the guide sequence of the sgRNAs were annealed, phosphorylated, and ligated into the BpiI site of pSpCas9(BB)-2A-GFP (PX461) and pSpCas9n(BB)-2A-Puro (PX462) vectors (Addgene #48140 and #62987)22. Plasmids were co-transfected into wild-type E14 mESCs followed by single-cell sorting based on GFP expression using a Sony MA900 Cell Sorter (Sony Biotechnology). The resulting clones were PCR-screened, verified by western blot, and genomic DNA was amplified by PCR and sequenced to verify out-of-frame indels or large deletions.
Knock-in mESCs
RBBP5-FKBP12F36V knock-in mESCs were generated using a STAR protocol39. In brief, the targeting construct was assembled from the PCR products into the pUC19 donor vector (Addgene plasmid #50005)40. The donor vector containing 1021 bp Rbbp5 left homology arm, 973 bp right homology arm, FKBP12F36V, 2×HA tags, P2A, and neomycin resistance gene were co-transfected with eSpCas9(1.1)-T2A-EGFP plasmid containing sgRNAs targeting the Rbbp5 stop codon into wild-type E14 mESCs using Lipofectamine 3000 transfection (Invitrogen) according to the manufacturer’s specifications. The transfected cells were selected with neomycin, single-cell sorted to obtain clonal cell lines and screened for correct bi-allelic integration. Sequences are provided in Supplementary Table 3.
For the insertion of an auxin-inducible degradation system into the endogenous Suz12 locus, an sgRNA targeting the stop codon region was cloned into the eSpCas9(1.1)-T2A-EGFP plasmid (a kind gift from Ian Chambers). The repair construct was assembled using the Gibson Assembly® Cloning Kit (NEB). Briefly, a fragment containing mAID-T2A-BFP was ordered as a gBlock (IDT), and the left and right homology arms—each including 25 bp overlaps with both the backbone plasmid and the mAID-T2A-BFP insert—were PCR-amplified from genomic DNA. All three fragments were assembled into a modified pUC19 backbone (a kind gift from Steve Pollard), which had been linearized with EcoRV, using the Gibson Assembly® Cloning Kit (3 h incubation at 50 °C). TCF2.2 cells were co-transfected with the sgRNA plasmid and the repair construct using Lipofectamine 3000 (Thermo Scientific), and GFP/BFP double-positive cells were single-cell sorted into 96-well plates 48 h later. Clones were genotyped by PCR 14 days post-sorting. Homozygous insertions were confirmed by Sanger sequencing and western blotting using a SUZ12 antibody. Verified homozygous clones were subsequently transfected with a pPB-OsTIR1-P2A-mCherry construct and selected with 100 μg/mL hygromycin B. After approximately two weeks of selection, stable pools were validated by western blot analysis. Sequences are provided in Supplementary Table 2.
Constitutive PiggyBac expressing mESCs
PiggyBac expression vectors of fusion proteins were constructed using Overhang Extension PCR. SnapGene in silico function was used to design primers with a 15 bp extension complementary to the ends of the partner PCR product. S12N was amplified from an in-house vector, SUV39H2 from GAL-SUV39H2-L (Addgene plasmid #89136)41, SETD2 catalytic domain from SETD2-GFP (Addgene plasmid #80653)42, and HP1 chromo shadow domain (amino acids 73 to 191) from an in house pDONHP1 alfa plasmid. Primers are provided in Supplementary Table 2. To generate the fusion proteins, PCR products were mixed for an overhang extension PCR reaction. Fused PCR products were cloned into pCR8 Gateway entry vector by TOPO cloning (Invitrogen) and transferred into PiggyBac Gateway expression vector by LR cloning (Invitrogen). Point mutations in S12N:SD2* (R1625G) and S12N:SUV2* (F259A and Y261A) were generated using Q5 site-directed mutagenesis kit (New England Biolabs) according to the manufacturer’s protocol. Expression vectors were introduced into Suz12 KO or R-Suz12 KO mESCs by Lipofectamine 3000 transfection (Invitrogen), and mESCs were kept under constant blasticidin selection to ensure continuous strong expression levels.
Western blot
Whole-cell extracts of mESCs were prepared in TOPEX+ buffer (50 mM Tris-HCl, pH 7.5, 300 mM NaCl, 0.5% Triton X-100, 1% SDS, 1 mM DTT, 33.33 U/ml Benzonase with Halt Protease Inhibitor cocktail (Roche)) and centrifuged at 20.000 g for 15 min at 4 °C. Protein concentrations were quantified by Bradford assay (Bio-Rad) according to the manufacturer’s specifications. The absorbances at OD595 were measured on a Nanodrop (Thermo Scientific), and protein concentrations were determined based on a Bradford standard curve. Lysates were diluted to equal concentrations in Laemmli sample buffer (LSB) (for a final concentration of 50 mM Tris, pH 6.8, 2% w/v SDS, 0.1% w/v bromphenol blue, 10% v/v glycerol, 100 mM DTT). Lysates were boiled for 5 min and separated by SDS-PAGE using acrylamide gels in running buffer (25 mM Tris base, pH 8.3, 0.25 M glycine, 0.1 % SDS) using the Bio-Rad gel system. Proteins separated by SDS-PAGE were transferred onto nitrocellulose membranes using the Trans-Blot Turbo Transfer System (Bio-Rad). Membranes were stained with Ponceau-red and scanned for initial loading control, followed by blocking in 5% skim milk (Sigma) in PBS-T (0.1% Tween-20 in PBS) for 30 min. Primary antibodies, listed in Supplementary Table 4, were diluted in 5% skim milk in PBS-T and incubated on the membranes for 1-2 hours, followed by three washes of 5 min in PSB-T. The membranes were then incubated for 45 min - 1 hr with IRDye fluorescent secondary antibodies diluted 1:15.000 in 5 % skim milk in PBS-T. After three washes of 5 min in PBS-T, membranes were imaged on Odyssey CLx Imaging System (LI-COR) and analyzed using the Image Studio Lite software (LI-COR). Uncropped scans of Western blots are provided in the Source Data file (for Fig. 1c) and in the Supplementary Information for the Western blots in the Supplementary Figs.
Intracellular FACS staining
Homogenous expression of piggyBac vectors was assessed by intracellular FACS staining using eBioscience Transcription Factor Staining Buffer Set (eBioscience). Cells were harvested, washed in PBS, and single-cell suspensions were prepared in a 96-well plate with 200 µl 1 x Fixation/Permeabilization working solution to a final concentration of 1 × 107 cell/ml. Samples were incubated for 1 h at 2–8 °C, protected from light, followed by 5 min centrifugation at 600 × g and aspiration of supernatants. 200 µl of 1x Permeabilization Buffer was then added to each well and centrifuged for 5 min at 600 × g before supernatants were removed. The step was repeated. Pellets were then resuspended and blocked for 15 min in 100 µl 1 x Permeabilization Buffer containing 2% goat serum at room temp. Without washing, 100 µl 1 x Permeabilization buffer containing 2% serum and SUZ12 primary antibody (final dilution 1:1000) was added and incubated at room temp for at least 30 min protected from light. Samples were then washed twice in 200 µl 1 x Permeabilization Buffer as described. Pellets were resuspended in 200 µl of 1 X Permeabilization Buffer spiked with 2% goat serum and Alexa flour 488 secondary antibody (Invitrogen; final dilution 1:2000) and incubated for at least 30 minutes at room temp protected from light. Samples were then washed twice in 1x Permeabilization Buffer before samples were resuspended in 100 µl 1x Permeabilization buffer and analyzed by flow cytometry.
Chromatin immunoprecipitation sample preparation
SUZ12 ChIP experiments were carried out according to a standardized protocol. Briefly, mESCs were prepared as a single-cell suspension in media containing 1% formaldehyde (Sigma-Aldrich) followed by a 10 min incubation before quenching with 0.125 M glycine. Cells were then washed 2 x in PBS and lysed in SDS buffer (100 mM NaCl, 50 mM Tris-HCl, pH 8.0, 5 mM EDTA, pH 8.0, 0.2% NaN3, 0.5 % SDS) and pelleted. Chromatin was prepared by resuspending pellets in 5x pellet-volume ice-cold IP buffer (100 mM NaCl, 67 mM Tris-HCl, pH 8.0, 5 mM EDTA, pH 8.0, 0.2% NaN3, 0.33 % SDS, 1.7 % Triton X-100) followed by 6 cycles of sonication for 30 sec using a Bioruptor (Diagenode) to shear DNA to ~ 200-500 bp. To ensure proper DNA fragmentation, small aliquots were de-crosslinked in elution buffer (1% SDS, 0.1 M NaHCO3) and incubated at 65 °C for 3 h before DNA was purified and analyzed by agarose gel electrophoresis. Chromatin was quantified using the BioRad protein assay, as described for western blot, and pre-cleared for 30 min at 4 °C using protein G-coupled sepharose beads (Cytiva).
SUZ12 ChIPs were carried out using 200 µg of chromatin diluted to 600 µl in ice-cold IP buffer containing 5% D. melanogaster spike-in chromatin – 1% input samples were taken from each ChIP. ChIP samples were then incubated on rotation with 4 µl SUZ12 antibody or IgG as a negative control at 4 °C overnight. Antibody-bound chromatin was then pulled down through a 4 h incubation at 4 °C with protein G-coupled Sepharose beads, before the chromatin was washed 3x with 150 mM wash buffer (150 mM NaCl, 20 mM Tris-HCl, pH 8.0, 2 mM EDTA, pH 8.0, 1 % Triton X-100, 0.1 % SDS) and 2x with 500 mM wash buffer (500 mM NaCl, 20 mM Tris-HCl, pH 8.0, 2 mM EDTA, pH 8.0, 1 % Triton X-100, 0.1 % SDS) separated by 1 min 1000 x g centrifugations. Precipitated chromatin and input samples were eluted from the beads by de-crosslinking in elution buffer (1 % SDS, 0.1 M NaHCO3) at 65 °C, shaking overnight. DNA was purified using the Qiagen PCR Purification kit (Qiagen) and quantified using the Qubit dsDNA HS assay kit (Thermo Fisher Scientific). Aliquots were taken out for qPCR analysis before libraries for ChIP-seq were prepared using NEBNext Ultra II DNA Library Prep kit (NEB). Library prep was performed according to the manufacturer’s protocol, using Ampure XP beads (Beckman) for size selection. Library DNA concentrations were quantified with the Qubit dsDNA HS assay kit, and size distributions were analyzed on a Tape Station 2200 (Agilent Technologies). Libraries were pooled based on molar concentrations, denatured, loaded onto an Illumina Nextseq 550 with SE75 high-output kits, and sequenced as paired-end 36/36 cycles.
ChIP-qPCR data was analyzed on a QuantStudio 6 Flex real-time qPCR machine (Applied Biosystems), enrichment calculated (accounting for primer efficacy) and visualized with Prism v9 (Graphpad).
For RBBP5 and KDM5B ChIP-seq experiments, six million mESCs were plated one day before the crosslinking. Attached mESCs were rinsed in PBS and crosslinked for 30 minutes at room temperature with 2 mM disuccinimidyl glutarate (DSG; cat. no. H58208.MC, Thermo Scientific) in PBS with 1 mM MgCl2. After removal of DSG solution and PBS rinses, the cells were further crosslinked with 1% formaldehyde for 10 minutes at room temperature. Formaldehyde was quenched with 125 mM glycine by rocking for an additional 5 min and the cells were then rinsed in PBS, harvested, pelleted and snap frozen. Crosslinked cell pellets were then resuspended in lysis buffer (50 mM HEPES pH 8, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100 supplemented with 1× Halt protease inhibitor cocktail and 0.2 mM PMSF), and the nuclei were pelleted and washed with ChIP rinse buffer (10 mM Tris pH 8, 1 mM EDTA, 0.5 mM EGTA, 200 mM NaCl with 1 × Halt protease inhibitor cocktail and 0.2 mM PMSF). The rinsed chromatin was further washed twice in sonication buffer (10 mM Tris pH 8, 1 mM EDTA, 0.2% LDS) and pelleted. The chromatin pellet was then resuspended in shearing buffer and sonicated in the Covaris E220 instrument for 20 minutes (peak power: 140; duty factor 5.0; cycles/burst: 200). Sheared chromatin was further centrifuged to remove residual debris and quantified using Pierce BCA assay. Chromatin preparations were then diluted 1:1 with 2X chromatin IP buffer (100 mM HEPES-KOH, pH 7.5, 600 mM NaCl, 1 mM EDTA, 2% Triton X-100, 0.2% DOC, 0.1% SDS with 2× Halt protease inhibitor cocktail) and precleared with protein G Sepharose beads for 1 h at 4 °C.
RBBP5 and KDM5B ChIPs were carried out with 500 µg chromatin in 1 mL 1X chromatin IP buffer. 1% input samples were taken from each ChIP sample. Samples were then incubated o/n with 4 µg RBBP5 (A300-109A, Bethyl Laboratories, Inc), 4 µg KDM5B (A301-813A, Bethyl Laboratories, Inc) or 4 µg rabbit control IgG (cat. no. 026102, Invitrogen). Antibody – target complexes were captured by incubating each IP with protein A Dynabeads (cat. no. 10002D, Invitrogen) for 3 h at 4 °C. IP medium was then removed and the beads were sequentially washed twice in low-salt wash buffer (50 mM HEPES pH 7.9, 1 mM EDTA, 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate), once in high-salt wash buffer (50 mM HEPES pH 7.9, 1 mM EDTA, 500 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate), once in LiCl wash buffer (20 mM Tris pH 7.5, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate) and twice in TE buffer. To elute DNA, the beads or input samples were resuspended in 200 µL fresh sodium bicarbonate-SDS buffer (1% SDS, 0.1 M NaHCO3) containing 1 µL RNase A/T1 (cat. no., 10775831, Thermo Scientific) and 1 µL proteinase K (cat.no., 10181030, Thermo Scientific). The beads were then incubated at 900 rpm in a thermoshaker (37 °C for 30 minutes; 65 °C for 15 h and 56 °C for 2 h). The eluted DNA was purified using ChIP DNA Clean & Concentrator™ (D5205, Zymo Research) and quantified using Qubit dsDNA HS assay kit (Thermo Fisher Scientific). The libraries were prepared using NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® (E7645S, NEB) according to the manufacturer’s protocol using SPRIselect beads (B23318, Beckman Colter) for cleanup without size selection. Library DNA concentrations were quantified with the Qubit dsDNA HS assay kit, and size distributions were analyzed on a Tape Station 2200 (Agilent Technologies). Libraries were pooled based on molar concentrations and sequenced using Illumina NextSeq 2000.
CUT&RUN sample preparation
Histone modifications were mapped with the standard CUT&RUN protocol43 using 400 K mESCs per sample. In brief, cells were prepared in biological duplicates as single-cell suspensions, counted, and washed 3x with Wash Buffer (20 nM HEPES pH 7.5, 150 mM NaCl, 0.5 mM, Roche Halt EDTA-free Protease Inhibitor cocktail), before cells were bound to BioMag Plus Concanavalin A beads (Bangs Laboratories) for 10 min. Cells were resuspended in 150 µL Antibody Buffer (20 nM HEPES pH 7.5, 150 mM NaCl, 0.5 mM, 0.05% digitonin, 2 mM EDTA, Roche Halt EDTA-free Protease Inhibitor cocktail) containing appropriate dilutions of indicated antibodies (See Supplementary Table 4) and incubated on a nutator overnight at 4 °C. Antibody-bound permeabilized cells were then washed twice in digitonin-containing Wash Buffer (20 nM HEPES pH 7.5, 150 mM NaCl, 0.5 mM, 0.05% digitonin, Roche Halt EDTA-free Protease Inhibitor cocktail), before incubated in a 1:1000 dilution of an in-house made Protein-A/G-MNase for 1-2 hrs on a nutator at 4 °C. After 2x wash with digitonin-containing Wash Buffer, samples were chilled to 0 °C in an ice bath before tethered pA/G-MNAse was activated with 1 mM CaCl2. Samples were incubated at 0 °C for 30 mins to allow for excision and release of pA/G-MNAse bound DNA while minimizing background cleavage. To stop the reaction 2XSTOP Buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA, 0.05% digitonin, 100 µg/mL RNAse A, 50 µg/mL glycogen) was added, and samples were incubated at 37 °C for 30 min release excised DNA fragments from insoluble nuclear chromatin. DNA was extracted with Phenol Chloroform Extraction using MaXtract phase-lock tubes (Qiagen) and quantified using Qubit dsDNA HS assay kit (Thermo Fisher Scientific). Libraries were prepared with NEBNext Ultra II DNA Library Prep kit (NEB) using Ampure XP beads (Beckman) for size selection and a protocol optimized for CUT&RUN to account for small DNA fragments44. Library DNA concentrations were quantified with the Qubit dsDNA HS assay kit, and size distributions were analyzed on a Tape Station 2200 (Agilent Technologies). Libraries were pooled based on molar concentrations, denatured, loaded onto an Illumina Nextseq 550 with SE75 high-output kits, and sequenced as paired-end 36/36 cycles.
RNA sequencing sample preparation
Total RNA was extracted from fresh cell pellets in biological triplicates using the RNeasy Plus Mini kit (Qiagen). RNA integrity was assessed on TapeStation 2200 (Agilent Technologies) using High Sensitivity RNA Screentape assay, and RNA was quantified with Qubit RNA HS assay kit. Libraries of mRNA were prepared using Illumina TruSeq RNA Sample Prep v.2 kit according to the manufacturer’s instructions using 0.5 µg input RNA. Libraries were quantified with the Qubit dsDNA HS assay kit, and size distributions were analyzed on a Tape Station 2200 (Agilent Technologies). Libraries were pooled based on molar concentrations, denatured, loaded onto an Illumina Nextseq 550 with SE75 high-output kits, and sequenced as paired-end 36/36 cycles.
Reduced representation bisulphite sequencing (RRBS) sample preparation
Mapping of DNA methylation was assessed in biological triplicates with RRBS. DNA was extracted from cells using the DNeasy Blood and Tissue kit (Qiagen), and concentrations were quantified with the Qubit dsDNA HS assay kit. For each sample, 2 μg genomic DNA was spiked with 2 ng unmethylated Lambda DNA (Promega) to allow assessment of the bisulfite-conversion reaction efficiency before DNA was digested for 5 hours at 37 °C using a methylation-insensitive restriction enzyme, MspI. Libraries were prepared using the NEXTFLEX Bisulfite-Seq kit (Bioo Scientific) with methylated adapters (Bioo Scientific) and 1 µg input DNA according to the manufacturer’s protocol. DNA was quantified again using the Qubit dsDNA HS assay kit before bisulfite conversion was performed with the EZ DNA Methylation-Lightning kit (Zymo Research). Bisulfite-converted DNA was PCR amplified and purified following the protocol of the NEXTFLEX Bisulfite-Seq kit (Bioo Scientific). RRBS libraries were quantified with the Qubit dsDNA HS assay kit, and size distributions were analyzed on TapeStation 2200 (Agilent Technologies). Libraries were first pooled based on molar concentrations and then combined with RNA-seq or CUT&RUN samples to compensate for the inherently low ATGC complexity of RBBS libraries. The final library pools were denatured, loaded onto an Illumina Nextseq 550 with SE75 high-output kits, and sequenced as paired-end 36/36 cycles.
In vitro H3K4 methylation assay
The H3K4 methyltransferase assay was performed as previous described45. Briefly, 0.2 µM of the KMT2B complex (Active Motif, cat. no 31499) was mixed with 2 µM unmodified mononucleosomes (Epicypher, cat. no 16-0009) or H3K36me3-modified mononucleosomes (Epicypher, cat. no 16-1320) in reaction buffer (25 mM Tris-Cl pH 8, 150 mM NaCl, 2 mM MgCl2, 0.02% Triton X-100, 1 mM DTT, 100 µM SAM). The individual reactions were incubated at 25 °C for 2’, 30’, 2 hrs, 6 hrs, 24 hrs. The reactions were quenched with Laemmli sample buffer; separated on 12% bis-tris precast gels and transferred to 0.2 µm nitrocellulose membranes. Membranes were blocked in 5% skimmed milk and probed o/n at 4 °C with primary antibodies against H3K4me0 (Active Motif, cat. no 91317, 1:10 000), H3K4me1 (Thermo Fisher, cat. no 710795, 1:3000), H3K4me2 (Thermo Fisher, cat. no MA5-33383, 1:2000), H3K4me3 (Cell Signaling Technology, cat. no 9751, 1:1000), H3K36me3 (Active Motif, cat. no 61021, 1:2000), WDR5 (Bethyl Laboratories, cat. no A302-429A, 1:1000), histone H4 (Cell Signaling Technology, cat. no 2935, 1:1000). Next, membranes were washed and incubated with IRDye 680RD or 800CW-labeled secondary antibodies (Licor) and imaged using Licor Odyssey M.
Bioinformatics analyses
Epigenome analysis
ChIP and CUT&RUN sequencing reads were trimmed and filtered for quality and adapter content using v0.4.5 of TrimGalore with a quality setting of 15 (https://www.bioinformatics. babraham.ac.uk/projects/trim_galore) and running version 1.15 of Cutadapt and version 0.11.5 of FastQC. Trimmed reads were aligned to the mouse assembly mm10 with v2.3.4.1 of bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). Mapped reads were deduplicated using MarkDuplicates in v2.16.0 of Picard Tools. To ascertain regions of enrichment, MACS2 (https://github.com/taoliu/MACS) was used with the --broad flag to capture H3K27me3 and using a p-value setting of 0.001. The BEDTools suite (http:// bedtools.readthedocs.io) was used to generate normalized read density profiles. First, blacklisted regions were removed to create a global peak atlas (http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/mm10mouse/mm10.blacklist.bed.gz). Then peaks within 500 bp were merged, and reads were counted with v1.6.1 of featureCounts (http://subread.sourceforge.net). No peaks were called for SUZ12, H3K4me3, H3K9me3, H3K36me3, or H2AK119ub1. H3K27me3 promoter peaks were identified as peaks overlapping regions +/− 3000 bp from transcriptional start sites (TSS) using R-packages TxDb.Mmusculus.UCSC.mm10.knownGene v3.10.0 and ChIPseeker v1.30.3.
Except for H3K9me3 CUT&RUN samples, data were normalized to sequencing depth (to ten million uniquely mapped reads), and normalized bigwig files were created using DeepTools v3.3.0 by running BamCoverage. For H3K9me3, 10,000 bp windows were generated from the mm10 genome using BedTools v2.27.1 by running Makewindows, and bins containing H3K27me3 peaks were removed by running Intersect with the setting -v. BedTools Multicov was used to count aligned reads from H3K9me3 bam files over the 10,000 bp windows, and size factors were estimated with the estimateSizeFactorsForMatrix function in DESeq2 v1.34.0. Size factors were used when running deepTools BamCoverage to create normalized bigwig files. For a list of scaling factors and validation of normalizations, refer to Supplementary Fig. 10.
For all samples, composite and tornado plots were created using v3.3.0 of DeepTools by running computeMatrix, plotHeatmap, and plotProfile on normalized bigwigs with signal sampled within 25 bp windows using H3K27me3 promoter peaks as reference point and flanking region defined by the surrounding 40 kb. Tracks were visualized with the IGV genome browser version 2.10.0 (http://software.broadinstitute.org/software/igv/). For quantifying peak intensities of histone modifications (H3K4me3, H3K9me3, H3K36me3, and H2AK119ub1) within wild-type H3K27me3 peaks, Bedtools Multicov was used to count aligned reads from BAM files within H3K27me3 peak boundaries. The resulting matrices were normalized to sequence depth (RPM) or by using the described scaling factors for H3K9me3. R-packages ggplot2 v3.3.5 and ggpubr v0.4.0 were used for data visualization.
Transcriptome analysis
RNA sequencing reads were 3’ trimmed for base quality with a setting of 15 and adapter sequences using TrimGalore v0.4.5 (https://www.bioinformatics.babraham.ac.uk/projects/ trim_galore), and read were then aligned to the mouse assembly mm10 using STAR v2.4 with default parameters. Data quality and transcript coverage were assessed using the Picard tool CollectRNASeqMetrics (http://broadinstitute.github.io/picard/). Read count tables were generated with HTSeq v0.9.1. The DESeq2 R-package was used with default parameters to evaluate normalization and expression dynamics, and potential outliers were assessed by sample grouping in principal component analysis (PCA). Differential expression was plotted as fold change from normalized filtered read counts, together with transcript level data represented as mean expression (log2), calculated as the average of the normalized count values, dividing by size factors, taken over all samples. Differentially expressed genes were identified from normalized filtered read counts as expression foldchange (FC) > 2 and false discovery rate-adjusted P (FDR) < 0.05 from pairwise contrasts and filtered based on H3K27me3 promoter peaks to identify direct PRC2 target genes. K-means clustering was performed using the superset of all differentially expressed PRC2 target genes, increasing k until the clusters became redundant. Significantly upregulated genes in Suz12 KO mESCs were defined as positive expression FC > 2 and FDR < 0.05, and the remaining genes were defined as unaffected. Fully rescued genes for a given cell line were identified from the upregulated gene set using the following criteria: (1) genes must be significantly downregulated (FC > 2, FDR < 0.05) in comparison to Suz12 KO mESCs expression, (2) genes may not be significantly upregulated in comparison to WT mESCs. Partially rescued genes were identified as significantly downregulated (FC > 2, FDR < 0.05) in comparison to Suz12 KO mESCs expression only. R-packages ggplot2 v3.3.5 and ggpubr v0.4.0 were used for data visualization.
DNA methylation analysis
RRBS sequencing reads were trimmed using v0.4.5 of TrimGalore with --rrbs setting enabled to remove 2 bp from the 3´end of Read 1 and 5´end of Read 2 in addition to adapter contamination. Raw reads were filtered for quality and adapter content using TrimGalore with a quality setting of 20 and running v1.15 of Cutadapt and v0.11.5 of FastQC. Trimmed reads were then aligned to the mm10 in silico bisulfite transformed genome with the bisulfite sequencing aligner Bismark v0.19.0 (https://www.bioinformatics.babraham.ac.uk/ projects/bismark/) using default Bowtie2 (v2.3.4.1) settings within Bismark. Duplicated reads were discarded, and the remaining alignments were used for cytosine methylation calling by the Bismark methylation extractor. Initial visualization with principal component analysis revealed no outliers or batch effects.
Differentially methylated CpGs (DMCs) were identified using the DSS R-package v2.42.0 using the dispersion shrinkage method followed by the Wald statistical test for beta-binomial distributions. Any CpGs with FDR < 0.05 and methylation percentage differences greater than 10% were considered significant DMCs. Custom annotation was built from the H3K27me3 peak-set using the R-package Annotatr v1.20.0, and DMCs were filtered for direct PRC2 target DMCs. Annotatr was then used to add annotations for CpG-type, gene, and transcriptional rescue groups to each DMC. Custom DMRs were called by grouping DMCs based on shared identifiers by combining the gene ID with CpG type, resulting in up to 4 DMRs per gene (CpG-island, CpG-shore, CpG-shelf, and CpG-opensea). Mean methylation (%) was then calculated for each DMR. R-packages ggplot2 v3.3.5 and ggpubr v0.4.0 were used for data visualization.
Statistical testing and reproducibility
Statistical analyses were performed using R. Depending on the data characteristics, a two-sided unpaired Student’s t test, Welch´s unequal variance t test, or Fisher’s exact test was used for the comparison of two groups. P-values are provided in the figures, and the nature of the statistical tests can be found in the associated figure legends. A combination of biological replicates, technical replicates and independent repetition was used to ensure reproducibility, which has been explicitly mentioned in the method sections and figure legends wherever applicable.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
ChIPseq/CUT&RUN data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE263059 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi].
RNA-seq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE263060 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi]. RRBS data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE263062 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi]. Source data are provided in this paper.
References
Lawrence, M., Daujat, S. & Schneider, R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 32, 42–56 (2016).
Zhao, Y. & Garcia, B. A. Comprehensive catalog of currently documented histone modifications. Cold Spring Harb. Perspect. Biol. 7, a025064 (2015).
Allshire, R. C. & Madhani, H. D. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 19, 229–244 (2018).
Li, J., Ahn, J. H. & Wang, G. G. Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell Mol. Life Sci. 76, 2899–2916 (2019).
Wagner, E. J. & Carpenter, P. B. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13, 115–126 (2012).
Bannister, A. J. et al. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 280, 17732–17736 (2005).
Baubec, T. et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520, 243–247 (2015).
Neri, F. et al. Intragenic DNA methylation prevents spurious transcription initiation. Nature 543, 72–77 (2017).
Laugesen, A., Hojfeldt, J. W. & Helin, K. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol. Cell 74, 8–18 (2019).
Guo, Y., Zhao, S. & Wang, G. G. Polycomb gene silencing mechanisms: PRC2 chromatin targeting, H3K27me3 ‘Readout’, and phase separation-based compaction. Trends Genet. 37, 547–565 (2021).
McKay, D. J. et al. Interrogating the function of metazoan histones using engineered gene clusters. Dev. Cell 32, 373–386 (2015).
Pengelly, A. R., Kalb, R., Finkl, K. & Muller, J. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev. 29, 1487–1492 (2015).
Sankar, A. et al. Histone editing elucidates the functional roles of H3K27 methylation and acetylation in mammals. Nat. Genet. 54, 754–760 (2022).
Hojfeldt, J. W. et al. Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat. Struct. Mol. Biol. 25, 225–232 (2018).
Ku, M. et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 4, e1000242 (2008).
Tanay, A., O’Donnell, A. H., Damelin, M. & Bestor, T. H. Hyperconserved CpG domains underlie Polycomb-binding sites. Proc. Natl. Acad. Sci. USA 104, 5521–5526 (2007).
Azuara, V. et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8, 532–538 (2006).
Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Hojfeldt, J. W. et al. Non-core subunits of the PRC2 complex are collectively required for its target-site specificity. Mol. Cell 76, 423–436 (2019).
Chen, K. et al. Methyltransferase SETD2-mediated methylation of STAT1 is critical for interferon antiviral activity. Cell 170, 492–506 (2017).
Hughes, A. L., Kelley, J. R. & Klose, R. J. Understanding the interplay between CpG island-associated gene promoters and H3K4 methylation. Biochim. Biophys. Acta Gene Regul. Mech. 1863, 194567 (2020).
Wang, H. et al. H3K4me3 regulates RNA polymerase II promoter-proximal pause-release. Nature 615, 339–348 (2023).
Braun, S. M. G. et al. Rapid and reversible epigenome editing by endogenous chromatin regulators. Nat. Commun. 8, 560 (2017).
Hathaway, N. A. et al. Dynamics and memory of heterochromatin in living cells. Cell 149, 1447–1460 (2012).
Binda, O. et al. Trimethylation of histone H3 lysine 4 impairs methylation of histone H3 lysine 9: regulation of lysine methyltransferases by physical interaction with their substrates. Epigenetics 5, 767–775 (2010).
Pedersen, M. T. et al. Continual removal of H3K9 promoter methylation by Jmjd2 demethylases is vital for ESC self-renewal and early development. EMBO J. 35, 1550–1564 (2016).
Chen, Y. K. et al. Design of KDM4 inhibitors with antiproliferative effects in cancer models. ACS Med. Chem. Lett. 8, 869–874 (2017).
Metzger, E. et al. KDM4 Inhibition targets breast cancer stem-like cells. Cancer Res. 77, 5900–5912 (2017).
Brinkman, A. B. et al. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 22, 1128–1138 (2012).
Cooper, S. et al. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 7, 1456–1470 (2014).
Fukuda, K. et al. Epigenetic plasticity safeguards heterochromatin configuration in mammals. Nucleic Acids Res. 51, 6190–6207 (2023).
Hagarman, J. A., Motley, M. P., Kristjansdottir, K. & Soloway, P. D. Coordinate regulation of DNA methylation and H3K27me3 in mouse embryonic stem cells. PLoS ONE 8, e53880 (2013).
Reddington, J. P. et al. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol. 14, R25 (2013).
Zhang, Y. et al. Targets and genomic constraints of ectopic Dnmt3b expression. Elife 7, https://doi.org/10.7554/elife.40757 (2018).
Jenuwein, T. & Allis, C. D. Translating the histone code. Science 293, 1074–1080 (2001).
Martin Gonzalez, J. et al. Embryonic stem cell culture conditions support distinct states associated with different developmental stages and potency. Stem Cell Rep. 7, 177–191 (2016).
Haeussler, M. et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Damhofer, H., Radzisheuskaya, A. & Helin, K. Generation of locus-specific degradable tag knock-ins in mouse and human cell lines. STAR Protoc. 2, 100575 (2021).
Norrander, J., Kempe, T. & Messing, J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26, 101–106 (1983).
Askew, E. B., Bai, S., Parris, A. B., Minges, J. T. & Wilson, E. M. Androgen receptor regulation by histone methyltransferase suppressor of variegation 3-9 homolog 2 and Melanoma antigen-A11. Mol. Cell Endocrinol. 443, 42–51 (2017).
Carvalho, S. et al. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. Elife 3, e02482 (2014).
Janssens, D.H. CUT&RUN. Targeted in situ genome-wide profiling with high efficiency for low cell numbers V.3 https://www.protocols.io/view/cut-amp-run-targeted-in-situ-genome-wide-profiling-14egnr4ql5dy/v3 (2019).
Liu, N. Library prep for CUT&RUN with NEBNext® Ultra™ II DNA library prep Kit for illumina® (E7645) V.1 https://www.protocols.io/view/library-prep-for-cut-amp-run-with-nebnext-ultra-ii-kxygxm7pkl8j/v1 (2019).
Li, Y. et al. Structural basis for product specificities of MLL family methyltransferases. Mol. Cell 82, 3810–3825 (2022).
Acknowledgements
We thank all members of the Helin laboratory for insightful discussions and continuous support. We are also grateful to Lydia Finley (MSKCC) and Emily Bernstein (Mount Sinai) for hosting L.H. in their laboratories during the final part of her work. This work was supported by a center grant from the Novo Nordisk Foundation to the NNF Center for Stem Cell Biology (no. NNF17CC0027852), a Tri-Institutional Stem Cell grant (no. 2019-035), the Memorial Sloan Kettering Cancer Center Support Grant (no. NIH P30 CA008748) and funds from The Institute of Cancer Research. L.H. was supported by R253-A14918-19-544 from the Danish Cancer Society.
Author information
Authors and Affiliations
Contributions
L.H. and K.H. conceived the strategy. L.H., A.M.M., A.A., R.L.A., T.T., and K.H. designed the experiments, which were performed by L.H., A.M.M., A.A., R.L.A., and T.T. L.H., A.M.M., A.A., R.K., and H.W. performed bioinformatics analysis. L.H. and K.H. wrote the manuscript, followed by review and editing by all authors. K.H. supervised the study and acquired funding.
Corresponding author
Ethics declarations
Competing interests
K.H. is a co-founder of Dania Therapeutics Aps and a scientific advisor for Hannibal Innovation. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Mazhar Adli, Andrey Krokhotin and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hedehus, L., Mas, A.M., Alpsoy, A. et al. Addressing the specific roles of histone modifications in transcriptional repression. Nat Commun 16, 11326 (2025). https://doi.org/10.1038/s41467-025-66426-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66426-z