Abstract
Sialic acid-containing glycans (sialoglycans) on pathological cells interact with Siglecs, glyco-immune checkpoint receptors expressed on myeloid cells, suppressing the cytotoxic functions of these immune cells. Using targeted glycomic analyses and gene editing, we show that HIV infection reprograms the glycosylation machinery of infected cells to increase the expression of the sialoglycan ligands for Siglec-3, −7, and −9. These ligands engage Siglecs on myeloid cells, impairing their ability to target HIV-infected cells. Selective disruption of these interactions using 10-1074-SiaD, an HIV-specific antibody conjugated to sialidase, an enzyme that removes sialic acids, significantly enhances monocyte- and neutrophil-mediated killing of HIV-infected cells in autologous assays. Treatment with 10-1074-SiaD in female humanized mice infected with HIV reduces viral load and decreases inflammation. These findings reveal an immune evasion mechanism exploited by HIV to evade myeloid cell immune surveillance and highlight the potential of targeting sialoglycan-Siglec interactions to improve immune clearance of HIV-infected cells.
Similar content being viewed by others
Introduction
HIV‑infected cells employ multiple strategies to evade immune surveillance1,2,3,4,5. Understanding these mechanisms is critical, as many HIV eradication approaches rely on the immune system’s capacity to recognize and eliminate infected cells6,7,8,9. Therefore, elucidating how HIV enhances resistance to immune detection can improve current cure strategies and inform other immunotherapies that boost immune‑mediated clearance of infected cells.
Beyond classical protein–protein immune checkpoint interactions, a complementary mode of immune regulation has recently emerged: glyco‑immune checkpoints, wherein immune receptors recognize glycans on target cells to transmit inhibitory signals10,11,12. Classical checkpoints such as PD‑1/PD‑L1 act through protein ligands on pathological cells to restrain T‑cell activity; by contrast, glyco‑immune checkpoints recognize glycans to modulate immune responses13. Among these, Siglecs (sialic acid‑binding immunoglobulin‑like lectins) are prominently expressed on innate immune cells, especially myeloid cells (monocytes, macrophages, neutrophils) and natural killer (NK) cells14,15. Inhibitory Siglecs (e.g., Siglec‑2, ‑3, ‑5, ‑6, ‑7, ‑8, ‑9, ‑10, ‑11, ‑12) signal through intracellular immunoreceptor tyrosine‑based inhibitory motifs (ITIMs) or ITIM‑like motifs that, upon engagement, recruit Src‑homology‑2 domain‑containing phosphatases SHP‑1/2, thereby dampening activation pathways such as PI3K/AKT, MAPK, AP‑1, and NF‑κB11,14,15,16,17. By contrast, activating Siglec (e.g., Siglec‑14, ‑15, ‑16) pair with immunoreceptor tyrosine‑based activation motif (ITAM)‑containing adapters such as DAP10/12 to trigger downstream activation, whereas Siglec‑1 lacks canonical signaling motifs18,19.
When pathological cells express elevated levels of sialic acid‑containing ligands (sialoglycans), these ligands engage inhibitory Siglec receptors on immune cells and trigger their signaling cascades. As a result, immune cell activity is suppressed, enabling pathological cells to evade innate immune surveillance18,19,20,21. This immune evasion mechanism is a common strategy used by cancer cells to escape immune surveillance20,21,22,23,24,25,26. Beyond cancer, recent studies have shown that some viral infections, such as hepatitis B virus infection, may also exploit this mechanism to evade immune detection27. Recognition of aberrant glycosylation patterns in cancer and other pathological cells has spurred interest in strategies that disrupt these glycan‑mediated interactions to enhance immune‑mediated clearance10,11,20,21,22,23,24,25,26,27,28,29,30,31.
Here, we asked whether HIV leverages this glycan‑based strategy in infected CD4⁺ T cells. To focus on the most relevant Siglec receptors, we first examined a publicly available single‑cell RNA‑sequencing (RNA-Seq) dataset from the peripheral blood of people living with HIV (PLWH) and HIV‑negative donors. These analyses showed that the inhibitory Siglec‑3, ‑7, ‑9, and ‑10 are the most abundantly expressed Siglec family members across circulating effector immune cell populations. Guided by these data, we concentrated on Siglec‑3/7/9/10 for functional interrogation. We show that HIV infection reprograms the host glycosylation machinery of infected CD4⁺ T cells, increasing cell‑surface sialoglycan ligands for Siglec‑3, Siglec‑7, and Siglec‑9, while not increasing ligands for Siglec‑10. This ligand remodeling suppresses myeloid and NK effector functions against infected targets. Motivated by the therapeutic implications and the need for selectivity, we disrupted the sialic acid–Siglec axis using a sialidase enzyme (which removes sialic acid residues from glycans) conjugated to an HIV‑specific antibody. This conjugate selectively desialylates HIV‑infected cells and enhances effector‑mediated killing of infected cells in vitro, and reduced viremia in vivo in a humanized mouse model of HIV infection.
Results
Siglec‑3, ‑7, ‑9, and ‑10 are the predominant inhibitory Siglecs across circulating effector immune cell populations
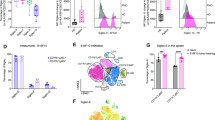
To identify the most relevant Siglec receptors on immune cells, we analyzed a publicly available single‑cell RNA‑seq dataset from the peripheral blood of people without HIV (PLWoH), PLWH who are not on antiretroviral therapy (ART), and PLWH on ART32. We restricted our analysis to immune populations involved in immune surveillance and cytotoxicity, such as CD8⁺ T cells, NK cells, monocytes, conventional dendritic cells (DCs), and plasmacytoid DCs (pDCs), and focused on inhibitory Siglecs bearing ITIM or ITIM‑like motifs: Siglec‑3, ‑5, ‑6, ‑7, ‑8, ‑9, ‑10, and ‑12. We excluded activating Siglecs (‑14, ‑15, and ‑16), which signal through ITAM‑coupled adapters such as DAP10/12; non‑signaling or tissue‑restricted family members (e.g., Siglec‑1, which lacks inhibitory motifs; Siglec‑4 and ‑11, which are central nervous system‑biased); and Siglec‑2, which is predominantly B‑cell‑restricted and thus outside our effector‑focused scope (Fig. 1a).
a UMAP projection of effector immune cells in single-cell analysis of PBMCs from people living without HIV (PLWoH) and people living with HIV (PLWH). Each dot represents a single cell, and the color denotes a distinct cell type. b Bubble heatmap showing the differential expression of inhibitory Siglecs across multiple effector immune cells in PLWoH, PLWH without ART (PLWH ART−) and PLWH on ART (PLWH ART+). The size of the bubble represents the percentage of cells expressing the Siglec, and color denotes the average expression. c–f Top panel shows the UMAP visualization of Siglec-3 (c), Siglec−7 (d), Siglec−9 (e), and Siglec-10 (f) expression across innate immune effector cells in PLWoH and PLWH. The bottom panel shows the pie chart distribution of the percentage of immune cells expressing each Siglec. Data in panels (a–f) are from the GEO dataset# GSE157829 consisting of single-cell RNA-seq data from PBMCs of four PLWoH and six PLWH. Data were analyzed using the Bioturing Browser 4.
Across PLWoH and PLWH (with and without ART), Siglec‑3, ‑7, ‑9, and ‑10 emerged as the most abundantly and consistently expressed inhibitory Siglec, based on both (i) the proportion of cells with detectable expression and (ii) the mean expression level per cell (Fig. 1b). The expression of these Siglecs was highest in monocytes, followed by NK cells and DCs (Fig. 1c–f). These findings suggest that, if HIV-infected cells exploit Siglec-sialic acid interactions as an immune-evasion mechanism, the receptors most likely to be involved are these four Siglecs, given their predominant expression across effector immune cells and their established role in the literature as potent inhibitory receptors10,14,25,26,27,33.
HIV infection induces the expression of sialoglycan ligands for Siglec‑3, ‑7, and ‑9, but not Siglec‑10, on the surface of primary CD4⁺ T cells
We next investigated whether HIV infection of primary CD4⁺ T cells (the principal cellular target of HIV) induces the expression of sialoglycan ligands for Siglec‑3, ‑7, ‑9, and ‑10. CD4⁺ T cells from HIV‑negative donors were activated with αCD3/αCD28 beads and infected with HIVTYBE in the presence or absence of benzyl‑2‑acetamido‑2‑deoxy‑α‑D‑galactopyranoside (BAGN), an inhibitor of O‑glycosylation. While previous studies have reported that BAGN increases HIV infection in primary CD4⁺ T cells34, in our experiments, BAGN treatment did not result in a significant change in infection levels, as measured by intracellular p24 staining (Fig. 2a). This discrepancy is likely due to differences in T‑cell activation protocols, viral strain, and timing/culture conditions. Therefore, in our system, BAGN can be reliably used as a functional inhibitor of O‑glycosylation without significantly altering HIV infection levels.
a Primary CD4+ T cells isolated from HIV-negative donors were activated with anti-CD3/CD28 Dynabeads and concurrently infected with a primary HIV isolate (HIVTYBE) in the presence or absence of the glycosylation inhibitor BAGN (2.5 mM). Intracellular p24 expression was assessed by flow cytometry 72 h post-infection. Statistical analysis was performed using paired one-way ANOVA, corrected by the two-stage step-up procedure of Benjamini, Krieger, and Yekutieli. N = 3 biological replicates. Error bars represent the mean ± standard deviation. b–e HIV-uninfected as well as HIVTYBE-infected primary CD4+ T cells cultured in the presence or absence of BAGN were stained with increasing concentrations of recombinant Siglec-3 Fc (b), Siglec-7 Fc (c), Siglec-9 Fc (d), and Siglec-10 Fc (e) 72 h post-infection. The left panels show changes in Siglec ligand expression, while the right panels depict the area under the curve (AUC) for recombinant Siglec Fc binding. Error bars represent the mean ± standard deviation. Statistical analysis was performed using paired one-way ANOVA, corrected by the two-stage step-up procedure of Benjamini, Krieger, and Yekutieli. Data in panels (b–e) were generated with n = 3 biological replicates. f Non-activated primary CD4+ T cells from HIV-negative donors were infected with HIVDH12 using CD44 microbeads and stained for intracellular p24 expression and CD69 surface expression as a marker of T cell activation. g Non-activated CD4+ T cells exposed to CD44 microbeads or exposed and infected with HIVDH12 were stained with 10 or 20 µg/ml of recombinant Siglec-7 Fc 48 h post-infection. Statistical analyses were performed using paired one-way ANOVA, corrected by the two-stage step-up procedure of Benjamini, Krieger, and Yekutieli. N = 4 biological replicates.
To quantify cell‑surface Siglec ligands, we stained with recombinant Siglec–Fc chimeras followed by a BV421‑conjugated secondary antibody and analyzed by flow cytometry. HIVTYBE infection significantly increased ligands for Siglec‑3 on activated CD4⁺ T cells (Fig. 2b). Similar induction was observed for Siglec‑7 (Fig. 2c) and Siglec‑9 ligands (Fig. 2d), whereas Siglec‑10 ligands were unchanged (Fig. 2e). As expected, BAGN reduced Siglec‑ligand display, consistent with inhibition of O‑glycan elongation and reduced terminal sialylation.
To isolate effects of viral infection from T‑cell activation and to test strain generalizability, we used CD44 microbeads to achieve high‑level infection of non‑activated primary CD4⁺ T cells with the HIVDH12 strain35. This approach generated a high frequency of p24⁺ cells without increasing T‑cell activation, as measured by CD69 expression (Fig. 2f). Consistent with findings in activated cells, HIVDH12 infection of non‑activated CD4⁺ T cells increased Siglec‑7 ligand expression compared with CD44‑bead controls (Fig. 2g). Beyond these four Siglecs, we also assessed global cell‑surface glycome changes using a microarray of 96 lectins (glycan‑binding proteins with defined specificities; Supplementary Table 1). As shown in Supplementary Fig. 1, lectin-microarray profiling of primary CD4⁺ T cells revealed broader glycome alterations caused by the HIV infection, including shifts in fucosylation and in mannose/galactose/GalNAc signals. Together, these data indicate that HIV infection remodels the surface glycome of infected CD4⁺ T cells, most notably increasing sialoglycan ligands for Siglec‑3/‑7/‑9, and that this induction is independent of the activation state of the target T cells, potentially facilitating immune evasion through Siglec‑mediated suppression.
HIV infection alters the glycosylation machinery of CD4⁺ T cells
We next investigated whether HIV infection alters the glycosylation machinery of CD4⁺ T cells, potentially explaining the induction of sialoglycan ligands for Siglecs on infected cells. To minimize confounding by global T‑cell activation, we infected non‑activated CD4⁺ T cells from three HIV‑negative donors with the HIVDH12 strain in the presence of CD44 microbeads, which facilitate infection without activation. After 16 h, cells were washed to remove unbound virus, cultured for an additional 24 h, and subjected to bulk RNA‑seq (Fig. 3a). For transcriptomic analyses, we focused on 560 genes involved in cellular glycosylation, enzymes and lectins mediating the addition, removal, or binding of glycans (Supplementary Data 1). RNA‑seq showed significant alterations in glycosylation‑related gene expression following infection (Fig. 3b, c). Notable genes implicated in sialic‑acid biosynthesis and glycan modification that were upregulated are GALNT6, NANP, B4GALNT1, ST8SIA4, and ST6GALNAC6; pathways essential for the biosynthesis of sialoglycan ligands recognized by multiple Siglecs. GALNT6 initiates O‑linked glycosylation by transferring GalNAc to serine/threonine residues; NANP regulates sialic‑acid precursor availability; B4GALNT1 extends glycan structures that can be terminally sialylated; and ST8SIA4 and ST6GALNAC6 catalyze α‑2,8‑ and α‑2,6‑sialylation, respectively (Fig. 3d)36,37,38,39. These changes likely enhance specific glycosylation pathways, contributing to increased Siglec‑ligand expression on HIV‑infected CD4⁺ T cells.
a Schematic representation of the workflow used to identify changes in the glycosylation machinery post-HIV infection. Non-activated primary CD4+ T cells from three HIV-negative donors were incubated with CD44 microbeads or a mixture of CD44 microbeads and HIVDH12 for 16 h, washed, and cultured for another 24 h before being processed for RNA sequencing. b, c Heatmaps showing glycosylation-related genes upregulated (b) or downregulated (c) in response to HIV infection. Red indicates higher gene expression, while blue indicates lower gene expression. Two-sided paired t tests with p-values corrected using the Benjamini-Hochberg procedure were applied to determine the false discovery rate (FDR) and control for multiple testing. d Schematic representation illustrating the role of key genes in sialic acid biosynthesis (NANP), core glycan biosynthesis (GALNT6), glycan elongation (B4GALNT1), and increased sialylation (sialyltransferases; ST6GALNAC6, ST8SIA4). Created in BioRender. Singh, S. (2025) https://BioRender.com/95tebnz. e Bar plot representing the relative mRNA expression of GALNT6 and NANP in HIV-infected CD4+ T cells compared to CD44 bead-treated control cells. Error bars represent the mean ± standard error of the mean. Statistical analysis was performed using two-sided paired t tests. N = 3 biological replicates. f Bubble heatmap illustrating the differential expression of key glycosylation-related genes as well as key sialyltransferases in primary CD4+ T cells from PLWoH and PLWH (top panel). UMAP visualization of CD4+ T cells in single cell analysis from PLWoH and PLWH representing the expression of ST6GAL1 and GALNT10 (bottom panel). Data are from GEO dataset# GSE157829, consisting of single-cell RNA-seq data from PBMCs of four PLWoH and six PLWH. Data were analyzed using the Bioturing Browser 4.
To validate the RNA‑seq findings, we performed qPCR for GALNT6 and NANP, key genes for O‑glycosylation initiation and sialic‑acid biosynthesis. Consistent with the RNA‑seq results, qPCR confirmed significant upregulation of both genes in HIV‑infected cells compared with CD44‑bead controls (Fig. 3e). To further validate these observations and assess their relevance in vivo, we queried a single‑cell RNA‑seq dataset32 to compare expression of these genes as well as 20 sialyltransferases (Supplementary Fig. 2) in CD4⁺ T cells from PLWH versus PLWoH. As shown in Fig. 3f, several of these genes as well as several sialyltransferases (such as ST3GAL1 and ST3GAL5) are upregulated in CD4⁺ T cells from PLWH, reinforcing the impact of HIV on glycosylation-pathway gene expression. The upregulation of several α2,3-sialyltransferase enzymes is consistent with an up-regulation of the ligands for Siglec-7 and Siglec-9 on both glycoproteins and glycolipids, as these receptors bind to α2,3-linked sialic acid40. Together, these findings indicate that HIV infection reprograms the glycosylation machinery of CD4⁺ T cells, including pathways critical for the biosynthesis of Siglec ligands, potentially facilitating immune evasion.
Disrupting sialic‑acid/Siglec interactions enhances the susceptibility of HIV‑infected CD4⁺ T cells to innate‑immune‑mediated cytotoxicity
To further validate the contribution of sialic‑acid/Siglec interactions to immune evasion by HIV‑infected cells, we first asked whether attenuating HIV‑induced glycosylation pathways in target cells would increase their vulnerability to innate effector cells. Guided by the transcriptomic results in Fig. 3, we used CRISPR-Cas9-mediated knockdown of a gene pool (GALNT6, B4GALNT1, and NANP) in primary CD4⁺ T cells and then assessed susceptibility to autologous monocyte‑mediated cytotoxicity (Fig. 4a). Knockdown reduced the expression of the targeted genes compared with a non‑targeting gRNA control (Fig. 4b). When co‑cultured with autologous monocytes, gene‑edited CD4⁺ T cells exhibited greater susceptibility to killing, as measured by reduced HIV p24 positivity relative to control cells (Fig. 4c).
a Schematic representation of the experimental workflow for generating CRISPR/Cas9-mediated knockout of GALNT6, NANP, and B4GALNT1 in primary CD4⁺ T cells infected with HIV. Some of the images in panel a were created in BioRender. Singh, S. (2025) https://BioRender.com/8d3lee4. b qRT-PCR analysis of GALNT6, NANP, and B4GALNT1 expression in gene-edited, HIV-infected primary CD4⁺ T cells. Statistical analysis was performed using two-sided ratio paired t tests. Each dot represents samples from an individual donor, run in triplicate and averaged. N = 4 donors. c NTC-targeted or gene-edited HIV-infected primary CD4⁺ T cells were co-cultured with autologous monocytes for 16 h (E:T ratio = 10:1). After overnight incubation, the co-cultures were stained with Aqua Live/Dead (BV510) and antibodies against CD3 and intracellular HIV p24. Representative contour plots (left panel) show a reduction in p24⁺ cells in gene-edited HIV-infected primary CD4⁺ T cells upon co-culture with autologous monocytes compared to NTC-targeted cells. The right panels depict normalized cytotoxicity, calculated based on either the percentage of p24⁺ cells or the reduction in p24 MFI. Two-sided Paired t tests. Each dot represents samples from an individual donor. N = 5 donors. d–f HIV-infected primary CD4⁺ T cells were co-cultured with autologous monocytes (E:T ratio = 10:1) in the presence of anti-Siglec-3 antibody or isotype control. After overnight incubation, the co-cultures were stained with Aqua Live/Dead (BV510) and antibodies against CD3 and intracellular HIV p24. d Representative flow cytometry plots. e Percentage of p24⁺ cells and cytotoxicity calculated based on % p24. f p24 MFI and cytotoxicity calculated based on p24 MFI. Statistical analysis was performed using paired one-way ANOVA, corrected by the two-stage step-up procedure of Benjamini, Krieger, and Yekutieli. Error bars represent mean ± standard deviation; n = 3 donors.
We next probed the role of Siglecs on effector cells. Because specific blocking antibodies to Siglec‑7 and ‑9 are not currently available, we targeted Siglec‑3 using a monoclonal anti–Siglec-3 (CD33) antibody (clone Hu9a, R&D Systems), which has been shown to bind the extracellular domain of human Siglec-3. We then asked whether Siglec‑3 blockade on monocytes enhances their ability to eliminate HIV‑infected targets. Indeed, blocking Siglec‑3 significantly increased monocyte cytotoxicity against autologous, HIV‑infected primary CD4⁺ T cells compared to isotype control (Fig. 4d–f). While anti-Siglec-3 antibodies can enhance phagocytosis via several non-exclusive routes, including an intrinsic mechanism driven by FcγR-dependent antibody‑dependent cellular phagocytosis (ADCP)41 as well as checkpoint blockade of Siglec-3’s ITIM signaling on myeloid cells27, considering all data together that: (1) HIV-mediated increases in specific Siglec ligands on CD4+ T cells (Fig. 2), (2) genetic perturbations of the Siglec-ligand axis that increase the susceptibility of infected cells to myeloid-cell-mediated cytotoxicity (Fig. 4a–c), and (3) the higher activity of anti-Siglec-3 versus isotype control in enhancing myeloid-mediated cytotoxicity (Fig. 4d–f), our results support a model in which sialic‑acid/Siglec interactions constrain innate immune clearance of HIV‑infected cells and that their disruption enhances killing by myeloid effectors.
Selective disruption of sialic‑acid–Siglec interactions on HIV‑infected cells enhances the cytotoxic activity of specific innate immune cells in vitro
Many cell types, not just HIV‑infected cells, express sialoglycans, and Siglec–sialic acid interactions function as negative immune checkpoints that help prevent autoimmunity17,31,42,43. We therefore sought an approach that selectively targets these interactions on HIV‑infected cells. To this end, we generated a conjugate of sialidase (SiaD; an enzyme that removes terminal sialic acids) and the HIV‑specific broadly neutralizing antibody (bNAb), 10‑1074. 10‑1074 is a well‑characterized bNAb that recognizes the V3‑glycan epitope on gp120 and exhibits potent neutralizing activity across diverse HIV‑1 isolates44,45,46. The conjugate, 10‑1074–SiaD, directs sialidase to the surface of HIV‑infected cells via antibody targeting, thereby removing sialoglycan ligands and disrupting Siglec engagement. This disruption alleviates glycan‑mediated immune suppression and enhances the cytotoxic capacity of Siglec‑expressing effector cells against HIV‑infected targets (Fig. 5a).
a Left panel: Increased expression of sialylated Siglec ligands on the surface of HIV-infected CD4+ T cells protects them from immune cell-mediated cytotoxicity. Right panel: Disruption of the sialoglycan-Siglec interaction using sialidase conjugated to 10-1074 (10-1074-SiaD) enhances the immune cell-mediated clearance of HIV-infected cells. b Schematic representation of the strategy used for the conjugation of sialidase (SiaD) to the HIV bNAb 10-1074, along with two SDS-PAGE gels showing the successful construction of the conjugate before (left) and after (right) concentration. c Workflow and equation used to evaluate antibody-mediated killing by multiple immune cell sub-populations. CEM.NKR CCR5+ Luc+ cells were infected with HIVIIIB for 72 h, followed by co-culture with whole PBMCs, NK cells, monocytes, monocyte-derived macrophages (MDMs), or neutrophils for 16 h in the presence of increasing concentrations of 10-1074 or 10-1074-SiaD. Cytotoxicity was assessed by measuring the decrease in relative luminescence units (RLU) compared to infected target cells alone. Created in BioRender. Singh, S. (2025) https://BioRender.com/an6q4co. d–f Representative histogram plots showing the expression of Siglec-3, Siglec−7, and Siglec−9 on monocytes, MDMs, neutrophils, NK cells, γδ T cells and CD8+ T cells respectively. g–j Antibody-mediated cytotoxicity of NK cells, monocytes, MDMs, and neutrophils, respectively, against HIVIIIB-infected CEM.NKR CCR5+ Luc+ target cells was measured in the presence of increasing concentrations of 10-1074 or 10-1074-SiaD by subtracting direct cytotoxicity from the total cytotoxicity (left panels), and AUCs were computed to assess the overall effect (right panels). Effector-to-target (E: T) ratios of 10:1, 10:1, 2.5:1, and 10:1 were used for NK cells, monocytes, MDMs, and neutrophils-based assays, respectively. Statistical analysis for AUC was performed using two-sided paired t tests. Data in panels (g–i) were generated from n = 3 donors while data in panel (j) were from n = 5 donors.
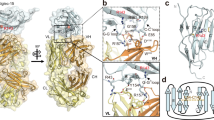
The 10‑1074–SiaD conjugate was synthesized using proximity‑induced antibody labeling (pClick), which enables site‑specific conjugation while preserving antigen and Fc‑receptor binding (Fig. 5b)26,47. Briefly, an antibody‑binding protein (FB) with low predicted immunogenicity was fused to the N‑terminus of sialidase via a flexible GGGS linker. Using genetic‑code expansion, the non‑canonical amino acid 4‑fluorophenyl carbamate lysine (FPheK) was introduced at position 25 of FB to enable site‑specific crosslinking. FB binds the CH2–CH3 junction of 10‑1074, positioning FPheK for proximity‑induced covalent coupling to a nearby lysine and yielding the 10‑1074–SiaD conjugate (Fig. 5b). Both 10-1074 and 10-1074-SiaD exhibited similar ability to bind to HIV gp120 (Supplementary Fig. 3a).
To test whether disrupting sialoglycan interactions enhances killing of infected cells, we used the cytotoxicity assay in Fig. 5c. CEM.NKR CCR5⁺ Luc⁺ cells infected with HIV served as targets. We first confirmed that HIV infection of this cell line induces sialoglycan expression, consistent with our primary CD4⁺ T cell data (Supplementary Fig. 3b). Infected targets were co‑cultured with effector cells in the presence or absence of 10‑1074 or 10‑1074–SiaD. Antibody‑mediated cytotoxicity was calculated as total killing minus direct killing (no antibody). 10‑1074–SiaD significantly increased the capacity of PBMCs to eliminate HIV‑infected cells compared with 10‑1074 alone, as shown by both total killing (Supplementary Fig. 4a) and antibody‑mediated killing (Supplementary Fig. 4b).
To identify which effector subsets are involved in Siglec‑mediated evasion, we profiled Siglec‑3, ‑7, and ‑9 protein expression across six immune populations: CD8⁺ T cells, γδ T cells, NK cells, monocytes, monocyte‑derived macrophages (MDMs), and neutrophils. In agreement with Fig. 1, peripheral CD8⁺ T cells and γδ T cells lacked these Siglecs (Fig. 5d–f). NK cells expressed moderate levels of Siglec‑3 and Siglec‑9 and high levels of Siglec‑7. Consistent with this profile, 10‑1074–SiaD modestly enhanced NK‑mediated killing relative to 10‑1074 alone, by both total killing (Supplementary Fig. 4c) and antibody‑mediated killing (Fig. 5g), aligning with prior evidence that Siglec engagement contributes to NK evasion29.
By contrast, myeloid cells, monocytes, MDMs, and neutrophils expressed the highest levels of Siglec‑3, ‑7, and ‑9 (Fig. 5d–f). Disrupting Siglec interactions with 10‑1074–SiaD markedly enhanced their cytotoxicity against HIV‑infected targets, evident in total killing (Supplementary Fig. 4d–f) and antibody‑mediated killing (Fig. 5h–j). Effects were most pronounced in monocyte cultures (~ 6‑fold increase) and neutrophil cultures ( ~ 19‑fold increase) relative to 10‑1074 alone. As shown in Supplementary Fig. 5, the 10‑1074–SiaD conjugate did not induce additional cell death in HIV‑infected cells culture alone, indicating that the enhancement of killing is immune‑mediated rather than due to direct cytotoxicity.
To further assess specificity, we perturbed O‑glycosylation globally with BAGN and profiled cell‑surface glycans using the 96‑lectin microarray. As shown in Supplementary Fig. 6a, BAGN treatment produced broad glycomic remodeling, including a decrease in sialic acid–associated signals and differential effects on additional features such as fucosylation and galactosylation. Functionally, BAGN increased the susceptibility of HIV‑infected target cells to immune‑mediated clearance. This increase was more pronounced with 10‑1074 than with 10‑1074–SiaD, consistent with a shared mechanism, reduction of cell‑surface sialylation, limiting further benefit when desialylation is already directed specifically to infected cells by the conjugate (Supplementary Fig. 6b, c). Together, these data indicate that HIV‑infected cells evade multiple myeloid effector populations by inducing specific sialoglycan ligands, and that selective desialylation of infected cells overcomes this glyco‑immune checkpoint, substantially boosting innate‑immune cytotoxicity.
10‑1074‑SiaD enhances the cytotoxicity of myeloid cells selectively against HIV‑infected cells
The 10‑1074‑SiaD conjugate used in the Fig. 5 cytotoxicity assays is designed to direct sialidase to the surface of HIV‑infected cells expressing viral antigens, thereby removing terminal sialic acids from these targets. This disrupts interactions between target‑cell sialoglycans and Siglecs on effector cells, increasing the susceptibility of infected cells to immune‑mediated clearance while sparing HIV‑uninfected cells. To test selectivity, we first asked whether 10‑1074‑SiaD removes more sialic acid from infected than from uninfected cells. We mixed HIV‑infected cells (Hut78/SF2) and HIV‑uninfected cells (Hut78) and treated them with 10‑1074 or 10‑1074‑SiaD. As shown in Supplementary Fig. 7, 10‑1074‑SiaD selectively removed a greater amount of sialic acid from HIV‑infected cells than from uninfected cells.
We next evaluated whether this selective desialylation translates into selective immune‑mediated killing. Uninfected CEM.NKR CCR5⁺ Luc⁺ cells (stained with Incucyte Rapid Red) and HIVIIIB–infected CEM.NKR CCR5⁺ Luc⁺ cells (stained with Incucyte Rapid Green) were co‑cultured to permit live‑cell visualization of each population. The mixed targets were then incubated with primary monocytes from four HIV‑negative donors in the presence of isotype control, 10‑1074, or 10‑1074‑SiaD. Live imaging (Fig. 6a, b) and cumulative data across donors (Fig. 6c) show that 10‑1074‑SiaD enhances monocyte‑mediated elimination of HIV‑infected cells while minimally affecting uninfected cells. These findings indicate that bNAb–sialidase conjugates can selectively potentiate myeloid cytotoxicity against HIV‑infected targets, offering a functional advantage over the bNAb alone.
a Primary monocytes from healthy donors were co-cultured with uninfected CEM.NKR CCR5+ Luc+ (stained with Incucyte Rapid red dye) and HIVIIIB-infected CEM.NKR CCR5+ Luc+ (stained with Incucyte Rapid green dye) target cells in the presence of isotype control, 10-1074, or 10-1074-SiaD, and imaged for 16 h using the Incucyte SX1. Representative images from each condition across four independent experiments are shown. b Kinetic measurement of monocyte-mediated cytotoxicity against HIV-infected cells (closed circles) and uninfected cells (open circles) was performed at 1-hour intervals over a 16-hour period. Each panel represents data obtained in triplicate from one donor (E:T ratio = 5:1; n = 4 donors). Percent cytotoxicity was calculated as the change in the green or red cell count under antibody-treated conditions compared to the target cells-only control. Error bars represent the mean ± standard error of the mean. c Kinetic data representing cytotoxicity from four donors (left panel). The AUC was calculated to demonstrate cytotoxic potential over time (right panel). Error bars represent the mean ± standard error of the mean. Statistical analysis was performed using paired one-way ANOVA, corrected by the two-stage step-up procedure of Benjamini, Krieger, and Yekutieli.
Disrupting Siglec interactions enhances monocyte and neutrophil targeting of autologous HIV‑infected primary CD4⁺ T cells and increases monocyte activation and cytotoxic capacity
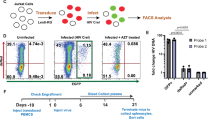
Our data thus far indicate that disrupting Siglec interactions significantly enhances the cytotoxicity of innate immune cells (especially monocyte‑ and neutrophil‑mediated cytotoxicity) against HIV‑infected cells. Because the assays in Figs. 5 and 6 used an HIV‑infected cell line as the target, we next tested whether these effects hold in autologous co‑cultures using primary cells from the same donors. Monocytes or neutrophils were isolated from six to seven HIV‑negative donors and co‑cultured with HIVTYBE‑infected autologous CD4⁺ T cells in the presence of isotype control, 10‑1074, or 10‑1074‑SiaD (Fig. 7a and Supplementary Fig. 8). As shown in Fig. 7b–e, disrupting Siglec interactions with 10‑1074‑SiaD significantly increased the ability of autologous monocytes and neutrophils to reduce intracellular HIV p24 levels.
Primary CD4+ T cells were infected with HIVTYBE for 72 h, followed by co-culture with autologous monocytes in the presence of isotype controls, 10-1074, or 10-1074-SiaD for 16 h (E:T ratio = 10:1). After overnight incubation, the co-cultures were stained with Zombie Aqua and antibodies against CD3 and intracellular HIV p24. a Representative contour plots showing the decline in p24+ HIV-infected CD4+ T cells when co-cultured with autologous effector cells in the presence of 10-1074 or 10-1074-SiaD. b, c The left panels show (b) the percentage of p24+ HIV-infected CD4+ T cells and (c) p24 median fluorescence intensity (MFI). The right panels depict normalized cytotoxicity based on either the percentage of p24+ cells (b) or reduction in p24 MFI (c). Data in panels b and c are generated from n = 7 donors. d, e HIVTYBE-infected primary CD4+ T cells were used as targets for autologous neutrophils, and cytotoxicity was evaluated by intracellular p24 staining 16 h after co-culture (E:T ratio = 5:1). The left panels show the percentage of p24+ cells (d) and p24 MFI (e), while the right panels depict cytotoxicity based on either percentage p24+ cells (d) or p24 MFI (e). Statistical analysis was performed using paired one-way ANOVA, corrected by the two-stage step-up procedure of Benjamini, Krieger, and Yekutieli. Data in panels d and e are generated from n = 6 donors. f Transcriptomic analysis was performed on monocytes co-cultured with autologous HIV-infected CD4+ T cells for 16 h in the presence of isotype control, 10-1074, or 10-1074-SiaD. Differential gene expression analysis was conducted using DESeq2. Heatmaps represent genes differentially regulated in MAPK/AP-1/NF-κB signaling (top left), PI3K-mTOR and metabolic reprogramming (bottom left), phagocytosis and effector functions (top right), and inflammatory mediators and innate immune sensing (bottom right). Red indicates higher gene expression, while blue indicates reduced gene expression. Two-sided paired t tests with p-values corrected using the Benjamini-Hochberg procedure were applied to determine the false discovery rate (FDR) and control for multiple testing. n = 3 donors.
To define immune programs associated with this enhanced killing, we performed bulk RNA‑seq on monocytes co‑cultured with autologous CD4⁺ T cells in the presence of isotype control, 10‑1074, or 10‑1074‑SiaD. Relative to both isotype‑ and 10‑1074‑treated cultures, 10‑1074‑SiaD significantly modulated genes and pathways consistent with enhanced monocyte activation and cytotoxicity (FDR < 0.05) (Fig. 7f). As expected from relieving Siglec‑SHP inhibitory signaling, we observed robust upregulation of NF‑κB components (NFKB1, NFKB2, NFKBIA, NFKBIZ, NFKBIL1), MAPK‑pathway kinases (e.g., MAP3K8, MAP3K12, MAP3K13, MAP3K15, MAP2K1, MAPK7, MAPKAPK2), and AP‑1–associated transcription factors (FOSL2, EGR1), consistent with increased pro‑inflammatory transcriptional output. In parallel, key negative regulators normally engaged by Siglec–SHP pathways, PTPN6 (SHP‑1), PTPN11 (SHP‑2), TOLLIP, and PPP6C, were downregulated, removing multiple “brakes” on NF‑κB/MAPK signaling (Fig. 7f, top left). We also observed activation of the PI3K–AKT–mTOR axis, with upregulation of PIK3CB and AKT3, and glycolytic enzymes PFKFB3/4, alongside reduced TSC2, collectively supporting a pro‑inflammatory, metabolically reprogrammed myeloid state (Fig. 7f, bottom left).
Concordantly, surface receptors and mediators that facilitate phagocytosis and effector functions were increased (Fig. 7f, top right): MARCO, CD300E, SLAMF9, FCAMR, and CCR7 expression increased in 10‑1074‑SiaD cultures, while inhibitory checkpoints SIRPA and SLAMF8 decreased; reduced NRROS supports a heightened oxidative burst. Finally, 10‑1074‑SiaD reconfigured inflammatory mediators and innate‑sensing pathways (Fig. 7f, bottom right): pro‑inflammatory chemokines CCL19, CCL20, CCL3, CCL5, CCL23, CCL28 and cytokines IL36A, IL36G, IL17C, as well as NLRP3 (inflammasome sensor) and PTGS2 (COX‑2), were upregulated, whereas inhibitory chemokines CCL17, CCL22, CCL13, CCL24, CCL8 and the IL‑1 decoy receptor (IL1R2) were downregulated. Together, these data support a model in which Siglec blockade (via selective desialylation of targets) re‑programs monocytes, reducing inhibitory checkpoints and elevating pro‑inflammatory, metabolic, and phagocytic pathways, consistent with the observed gains in cytotoxic activity.
Targeting sialic acid/Siglec interactions enhances the anti‑HIV activity of 10‑1074 in a humanized mouse model
Our data thus far suggest that HIV infection induces the overexpression of sialoglycan ligands for several Siglecs, primarily expressed on multiple myeloid cells and NK cells. These sialoglycans bind to Siglecs on immune cells, triggering inhibitory signals that contribute to the ability of HIV-infected cells to evade immune surveillance. We also show that disrupting these interactions using a conjugate of 10-1074 and sialidase can selectively target HIV-infected cells, enhancing the cytotoxic capacity of these immune cells against HIV-infected cells with minimal impact on HIV-uninfected cells, as shown by in vitro assays using both cell lines and autologous primary cells.
Next, we tested this strategy in vivo using a humanized mouse model of HIV infection48. In this model, six‑week‑old immunodeficient female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were humanized by engrafting isolated memory CD4⁺ T cells from healthy donors (Supplementary Fig. 9a). Thirty-four days post-engraftment, mice were evaluated for human cell engraftment (Supplementary Fig. 9b), and then infected with the transmitted/founder virus HIVSUMA. We first confirmed Siglec ligand (sialic acid) availability on human CD4+ T cells from this mouse model. Human CD4⁺ T cells were isolated (high purity) from the pooled blood of four uninfected humanized mice and four HIV‑infected mice (Supplementary Fig. 10a–d) and subjected to lectin microarray profiling of membrane proteins. As shown in Supplementary Fig. 10f, sialic‑acid signals were detected and increased in HIV‑infected mice relative to uninfected controls, alongside other glycomic alterations.
We then humanized 15 six‑week‑old female immunodeficient NSG mice (Fig. 8a). Thirty‑four days post‑engraftment, mice were randomized into three groups based on peripheral human CD4⁺ T cell counts (Fig. 8b). The mean concentration of human CD4⁺ T cells in blood was 92.2 cells/µL (median 42.5 cells/µL), with comparable reconstitution across groups (Fig. 8b). Mice were then infected with HIVSUMA; one week later, baseline viral loads were similar among groups (Fig. 8c). The mice were then treated weekly for five weeks with: (1) PBS (control), (2) 10‑1074 plus autologous monocytes and NK cells (Supplementary Fig. 11), or (3) 10‑1074‑SiaD plus autologous monocytes and NK cells.
a Study design outlining the generation of 15 female humanized NSG mice engrafted with human memory CD4+ T cells, followed by HIVSUMA infection and weekly treatment with 10-1074 or 10-1074-SiaD (0.75 mg/dose). Monocytes and NK cells were injected weekly. Control mice were treated with PBS only (n = 5 per group). b Peripheral blood human CD4+ T cell counts before HIVSUMA infection, measured by flow cytometry. c Humanized NSG mice were infected intravenously with 1500 TCID50 of HIVSUMA. Establishment of infection was confirmed by measuring HIV viral load (VL) prior to treatment initiation. d, e Weekly plasma viral load, measured as HIV copies/mL (y-axis) over the experimental duration (x-axis) (d), and presented as AUC (e) to represent cumulative HIV infection over time across the three groups. f, g Mice were weighed weekly, starting before engraftment and continuing until the end of the study. Data are presented as percentage changes from the initial weight (y-axis) over the experimental duration (x-axis) (f). AUC (g) was calculated to represent the cumulative weight change over time. h, i Absolute CD4+ T cell counts in peripheral blood (y-axis) was calculated weekly post-engraftment (h) and presented as AUC (i) to represent cumulative CD4+ T cell levels over time. Error bars represent the mean ± standard error of the mean. Statistical analyses were performed using the Kruskal-Wallis test, corrected by the two-stage step-up procedure of Benjamini, Krieger, and Yekutieli. j Heatmap representing the protein levels of several human pro-inflammatory markers in plasma across the three treatment groups, before (day 34) and after HIV infection and treatment initiation (day 48). Red indicates higher marker abundance, while blue indicates lower abundance. P-values were computed using two-sided paired t tests. Data in panels (b–j) were generated with n = 15 mice (five mice per each of the three groups).
While 10‑1074 + effectors did not significantly reduce viral load compared with PBS, 10‑1074‑SiaD + effectors produced greater viral‑load reduction (Fig. 8d, e). Importantly, body weight was maintained in 10‑1074‑SiaD–treated mice versus PBS controls (Fig. 8f, g), suggesting minimal off‑target toxicity from sialidase conjugation. CD4⁺ T cell depletion was attenuated in both 10‑1074 and 10‑1074‑SiaD groups relative to controls (Fig. 8h, i). Finally, whereas 10‑1074 + effectors did not reduce inflammatory markers, 10‑1074‑SiaD + effectors significantly reduced HIV‑induced inflammation, including TNF‑α, IFN‑γ, IP‑10, and MIP‑1α (Fig. 8j). Together, these data support 10‑1074‑SiaD as a selective and effective immunotherapeutic approach that enhances innate immune control of HIV in vivo.
Discussion
This study identifies an immune‑evasion mechanism employed by HIV, wherein infection reprograms the glycosylation machinery of infected CD4⁺ T cells to upregulate sialoglycans that engage multiple, yet distinct, inhibitory Siglecs on immune cells, particularly myeloid cells. These interactions suppress the cytotoxic functions of monocytes, neutrophils, and other effectors, facilitating escape from immune surveillance. By targeting this glyco‑immune checkpoint with a sialidase‑conjugated bNAb, 10‑1074‑SiaD, we show enhanced clearance of HIV‑infected cells in vitro and in vivo, supporting a promising therapeutic strategy against this mode of immune evasion.
The cell-surface glycome plays pivotal roles in modulating various cellular processes49 and mediating cell-cell50,51 and cell-pathogen52 interactions. For instance, the cell-surface glycosylation of T cells has recently been implicated in influencing HIV susceptibility and selection during transmission53. While aberrant glycosylation is a well‑established mechanism of cancer immune evasion, whether HIV leverages a similar strategy has been unclear. Our findings reveal that, beyond influencing susceptibility and selection, HIV‑induced glycomic alterations on infected cells actively contribute to immune evasion by promoting binding to inhibitory Siglecs on immune cells. This underscores the multifaceted role of cell‑surface glycosylation in HIV pathogenesis and persistence. Whether this glyco‑immune evasion mechanism contributes to the survival of infected cells during suppressive ART warrants further study; notably, elevated Siglec expression has been associated with a faster time to viral rebound after treatment interruption54, highlighting the potential of Siglec pathways to modulate anti‑HIV immunity, particularly innate responses.
Beyond checkpoint engagement, sialylation exerts broad biophysical and signaling effects, modulating electrostatic cell–cell repulsion, receptor clustering, viral binding, adhesion, and immune activation, that can shape viral entry, cellular adhesion, and the magnitude and quality of immune responses during HIV infection43,55. Our data indicate that HIV alters not only sialylation but also other glycan features. Defining how HIV-mediated alterations in sialylation and other glycans influence receptor localization, membrane organization, antigen presentation, cell trafficking, and survival, processes central to viral persistence, will be critical. For instance, modulation of sialic-acid levels on T cells affects their activation, function, and persistence56. The heightened sialylation we observe on HIV-infected CD4⁺ T cells may impair intrinsic T cell function, in addition to its role in facilitating immune evasion via Siglec engagement. Together, these effects can contribute to HIV persistence and immune dysfunction. Moving forward, integrated site-specific glycoproteomic and structural glycomic analyses are needed to define the precise glycans and their protein/glycolipid carriers and to determine how HIV-induced remodeling repositions receptors and rewires membrane dynamics, insights essential for designing interventions that restore immune recognition without disrupting homeostatic glycan functions. In addition, although our work focuses on HIV, related glycomic changes have been reported in other chronic viral infections (e.g., increased sialylation in hepatitis B virus infection and altered glycosylation in SARS-CoV-227,57,58), underscoring the need to determine whether the specificity and functional consequences of glycan remodeling are conserved across pathogens or are virus- and tissue-context dependent.
Myeloid cells, including monocytes, macrophages, and neutrophils, play critical roles in controlling HIV infection through cytotoxic and effector functions. These cells recognize and eliminate infected targets via mechanisms such as ADCP, contributing substantially to control of viral replication, particularly within tissues59,60,61,62. However, the capacity of HIV‑infected cells to evade these responses by inducing inhibitory glycans that engage Siglecs may present a significant barrier to effective clearance. While our study emphasizes the impact of Siglec interactions on direct innate cytotoxicity, Siglec signaling on myeloid cells likely has broader consequences, such as dampening antigen‑presentation programs and indirectly suppressing downstream T cell responses63,64,65. Thus, HIV‑driven induction of Siglec ligands may impair multiple myeloid functions, including their role in shaping adaptive immunity. Dissecting these mechanisms will be important for improving eradication strategies that aim to enhance anti‑HIV immune functions, including antibody‑mediated effector mechanisms enabled by bNAbs.
In this study, disrupting Siglec–ligand interactions with 10‑1074‑SiaD significantly enhanced myeloid‑cell cytotoxicity against HIV‑infected cells while minimally affecting uninfected cells. Mechanistically, blockade of the Siglec–sialoglycan axis reprogrammed monocytic effector cells, promoting phagocytic and activation pathways against infected targets. Approaches that augment innate effector functions via glyco‑immune checkpoint interference have been tested safely and effectively in oncology25,26,66, supporting the translational plausibility of this strategy in HIV. Critical next steps include evaluating its impact on myeloid and NK cell responses during suppressive ART and after analytical treatment interruption. Importantly, this strategy depends on viral antigen display on infected cells, positioning it as complementary to “shock‑and‑kill” approaches that reactivate latent reservoirs and render targets visible to the immune system67. Combining multiple bNAbs, each conjugated to sialidase and directed to distinct Env epitopes, may broaden coverage and potency, enabling robust and selective clearance. Together, these considerations position glycan‑focused interventions as promising components of combination cure strategies.
Although our findings provide evidence that HIV exploits sialoglycan–Siglec interactions to blunt innate immunity, several limitations should be acknowledged. First, our work focused on Siglec-3, -7, -9, and -10 because they are the predominant inhibitory Siglecs on circulating effector cells; however, Siglec expression is tissue-specific, and other family members may dominate in distinct microenvironments. For example, Siglec-9 is absent on peripheral T cells yet enriched on tumor-infiltrating lymphocytes (TILs), where its blockade restores cytotoxicity22,28. Whether HIV-induced sialoglycans similarly modulate tissue-resident immune responses, through the same or different Siglecs, remains to be determined. Second, glycomic profiling of HIV infected cells from PLWH on ART is hindered by the scarcity of markers that unambiguously distinguish infected from uninfected cells. Nonetheless, our recent reports show that transcriptionally active HIV-infected cells in the blood of ART-treated individuals display aberrant glycosylation, including elevated sialic-acid levels68, supporting in vivo relevance and motivating deeper tissue studies. Third, although 10-1074-SiaD was well tolerated in our humanized-mouse experiments, longer dosing regimens, broader immune and metabolic profiling, and evaluation in models with intact immune systems will be required to fully assess safety and off-target effects. Finally, testing sialidase-conjugated bNAbs in ART-suppressed infection models will be critical for gauging their ability to purge latent reservoirs and for mapping Siglec interplay across the full spectrum of antiviral immunity. Despite these limitations, the present work expands our understanding of glyco-immune checkpoints in HIV pathogenesis and provides a foundation for glycan-targeted strategies aimed at durable viral control or cure.
Methods
Inclusion & ethics statement
Blood and cryopreserved PBMCs from 40 HIV-negative donors were collected from the Wistar Institute and Human Immunology Core, University of Pennsylvania, and Northwestern University, respectively. Research protocols were approved by The Wistar Institute committee on Human Research (IRB# 2110176-6a) and Northwestern University (STU00222402). Written informed consent was obtained from all human donors, and all data and specimens were coded to protect confidentiality. All human experimentation was conducted in accordance with the guidelines of the US Department of Health and Human Services and those of the authors’ institutions. The age and sex details of the donors are documented in Supplementary Table 2.
Isolation of primary CD4+ T cells and effector cells
Primary human CD4+ T cells, CD8+ T cells, γδ T cells, monocytes and NK cells were isolated from fresh or cryopreserved PBMCs from healthy donors by immunomagnetic negative selection using the EasySep Human CD4+ T Cell Isolation Kit (Catalog #17952), EasySep Human CD8+ T Cell Isolation Kit (Catalog #17953), EasySep Human Gamma/Delta T Cell Isolation Kit (Catalog #19255), EasySep Human Monocyte Isolation Kit (Catalog #19359) and the EasySep Human NK Cell Isolation Kit (Catalog #17955; STEMCELL Technologies), respectively, following manufacturer’s protocol. Autologous myeloid cells, including monocytes and neutrophils, were obtained from the same HIV-negative donors who provided CD4+ T cells for HIV infection. Since neutrophils cannot be reliably cryopreserved and must be used fresh, we scheduled two separate blood draws from each donor: (1) During the first visit, we isolated CD4+ T cells and infected them with HIV. (2) During the second visit (days later), we isolated fresh myeloid effector cells, including neutrophils, from the whole blood of the same donor, using the EasySep Direct Human Neutrophil Isolation Kit (STEMCELL Technologies, Catalog #19666) following the manufacturer’s protocol to bypass the need for Ficoll density gradient separation.
Analysis of publicly available single-cell data
Bioturing Browser 469 was used to analyze the following deposited study: GSE15782932, consisting of single-cell RNA sequencing data from PBMCs of four PLWoH and six PLWH. For all studies, clustering was performed using the Louvain method. UMAP and bubble heatmaps were generated on the Talk2data platform using the Vinci software.
Cell culture
CEM.NKR CCR5+ Luc+ (Catalog #ARP-5198, contributed by Dr. John Moore and Dr. Catherine Spenlehauer), Hut78 (Catalog #ARP-89) and Hut78/SF2 (Catalog #ARP-279, contributed by Dr. Jay Levy) cells obtained from NIH AIDS Reagent Program were cultured in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 µg/ml) (complete R10) at 37 °C. PBMCs, primary NK cells and monocytes were cultured in complete R10 media. Primary CD4+ T cells were cultured using the same medium supplemented with 30 U/ml recombinant human IL-2 (PeproTech, Catalog #200-02-50UG). Primary monocyte-derived macrophages (MDMs) were differentiated for 7-8 days, as described previously14,70. Briefly, freshly isolated monocytes were cultured in IMDM (Gibco) supplemented with 10% AB human serum (Sigma) and 50 ng/mL M-CSF (Peprotech). After 3-4 days of differentiation, the cells were supplemented with 50 ng/mL TGF-β (Peprotech) and 50 ng/mL human IL-10 (Peprotech), and further cultured till day 8, for flow cytometry staining and cytotoxicity assays.
Primary CD4+ T cell infection with HIVTYBE for siglec ligand staining
CD4+ T cells (2 × 105) were concurrently exposed to concentrated HIVTYBE (4 µg/ml of p24; Penn Center for AIDS Research Virus and Molecular Core, University of Pennsylvania, Philadelphia) and Dynabeads Human T-Activator CD3/CD28 (Thermo Scientific, Catalog #11132D) and incubated at 37 °C for 72 h in 48-well plates. Activation beads were added at a ratio of 5 beads/cell. O-glycan biosynthesis inhibitor Benzyl 2-acetamido-2-deoxy-α-D-galactopyranoside, BAGN (Sigma, Catalog #B4894) was dissolved in DMSO and used at a final concentration of 2.5 mM, at time of virus exposure to block O-glycosylation. After 72 h, cells were separated from the beads, washed with 1X PBS supplemented with 0.5% BSA and 0.1% sodium azide (FACS buffer) and processed for intracellular p24 staining and cell surface siglec ligand staining.
Primary CD4+ T cell infection with HIVDH12 without T cell activation
1 mL of CEMx174-grown HIVDH12 (312,500 TCID50) was mixed with 50 μl of HIV Infectivity Enhancement Reagent (Miltenyi Biotec, Catalog # 130-095-093). This reagent works by targeting CD44, a cell-surface adhesion molecule, which facilitates the attachment and fusion of HIV particles to primary CD4+ T cells, particularly in the absence of strong cellular activation. This method has been validated and described as a strategy to enhance HIV-1 infection efficiency in non-activated T cells without inducing broad activation profiles35. This mixture was incubated at 4 °C for 30 min. 250 μl of the virus-CD44 microbead mixture was then added to 300,000 unstimulated CD4+ T cells in 250 μl IL-2 supplemented complete R10 media for 16 h. After overnight incubation, cells were washed with 5 x volume of the R10 medium and incubated for an additional 24 h and 48 h at 37 °C for RNAseq and recombinant Siglec-7 Fc staining, respectively. Aliquots of virus-exposed cells were taken from culture and washed with FACS buffer. HIV infection and CD4+ T cell activation status was evaluated by staining for intracellular p24 using KC57-RD1, anti-p24 antibody (Beckman Colter) and anti-CD69 FITC (Biolegend, Clone FN50) markers, respectively.
Detection of cells surface siglec ligands by flow cytometry
1 × 105 uninfected or HIV-infected CD4+ T cells were resuspended in 100 µl FACS buffer. Indicated amounts of recombinant human Siglec-3, -7, -9 and -10 Fc proteins (R&D Systems) resuspended in PBS were added to the cells and incubated for 45 min at room temperature. Following incubation, the cells were washed twice with FACS buffer and incubated with BV421 Fc-specific rat anti-human IgG (BioLegend, Clone - M1310G05); at 1:20 dilution, for 20 min at room temperature. Cells were further washed twice, fixed for 15 min at room temperature (Biolegend Cytofix/Cytoperm), and acquired by flow cytometry on BD LSR18. Flow cytometry data were analyzed using FlowJo software.
Transcriptomic analysis of glycosylation related genes
RNA extraction was performed using the Qiagen RNeasy mini kit (Catalog # 74104) according to the manufacturer’s protocol. 6-12 ng of total RNA was used as input to prepare libraries using the Stranded Total RNAseq with Ribo-zero Plus kit (Illumina, San Digo,CA) as per manufacturer’s instructions. Library size was assessed using the 4200 Tapestation and the High-Sensitivity DNA assay (Agilent, Santa Clara, CA). Concentration was determined using the Qubit Fluorometer 2.0 (Thermofisher, Waltham, MA). Next Generation Sequencing with a paired-end 2 x 150bp run length was done on the NovaSeq X series platform (Illumina, San Diego, CA). A minimum of 30 M reads per sample were acquired for each sample. RNA-seq data were aligned to GRCh37 using the RSEM (RNA-Seq by Expectation-Maximization) V1.3.3 software in conjunction with bowtie271,72. Raw counts below 10 were filtered out, and differential gene expression analysis was performed using the DESeq273. DESeq2 normalized count values were used to determine the log2 fold change and adjusted p-values. P-values were adjusted using the Benjamini-Hochberg procedure to determine the False Discovery Rate (FDR) and control for multiple testing.
cDNA synthesis and quantitative real-time PCR (qPCR)
15 μL of RNA was used to prepare cDNA using the SuperScript VILO cDNA Synthesis Kit (Thermo, Catalog #11754050) according to the manufacturer’s protocol. Briefly, the reaction was carried out at 25 °C for 10 min, 42 °C for 60 min, 85 °C for 5 min, and 4 °C, hold. qPCR for GALNT6 and NANP was performed on QuantStudio 6 Pro (Thermo Fisher), and the following FAM-MGB probes were obtained from Thermo: GALNT6 (Hs00926629_m1), NANP (Hs00600979_m1). 18S (Thermo, Catalog #4352930E) was used as the endogenous control to calculate relative copy number.
CRISPR/Cas9 gene editing of primary CD4⁺ T cells
Cryopreserved PBMCs from five to six healthy donors were thawed and rested overnight, followed by isolation primary CD4+ T cells by StemCell negative selection kit. Primary CD4+ T cells were cultured in complete R10 supplemented with 25 mM HEPES, 1 mM sodium pyruvate and 30 U/mL rhIL-2. 2 × 105 CD4+ T cells were simultaneously activated using Immunocult Human CD2/CD3/CD28 T Cell activator (STEMCELL; Catalog # 10970) and infected with concentrated HIVTYBE at ~ 4 µg/ml of p24 for 72 h. 72 h post-infection (hpi), the cells were washed and prepared at a density of 0.5 × 106 cells per reaction, for nucleofection. Gene-edited CD4+ T cells were generated by CRISPR/Cas9 mediated knockout of glycosylation related genes – GALNT6, NANP and B4GALNT1; using specific synthetic guide RNA (sgRNA) targeting these genes (Horizon, GALNT6 - Catalog# SG-012366-01-0005 (TATGATCTCCTTGAGCAAGA); NANP – Catalog# SG-007285-01-0005 (CTGATTTAAGGACTTCACAT); B4GALNT1 – Catalog# SG-011279-01-0005 (CTGAACTTCCACACCCTGTA)). A non-targeting gRNA served as a control (catalog# U-009502-01-02). CRISPR ribonucleoprotein (crRNP) complexes were assembled and nucleofected as described previously74. Briefly, 1 µL of each sgRNA was combined with 1 µL tracrRNA and incubated at 37 °C for 30 min to allow duplex formation. 1.6 µL of Poly-L-glutamic acid sodium salt (100 mg/mL) was added to the duplex solution, followed by the slow addition of recombinant Cas9 protein (2 µL). The final crRNP complex was incubated at 37 °C for an additional 15 min to complete assembly. Assembled crRNPs (5.6 µL per reaction) were aliquoted into PCR strip tubes and stored on ice prior to transfection. To knockout the three genes simultaneously, equal volume of crRNP complexes targeting each gene were mixed. Electroporation was performed using the Lonza 4D-Nucleofector system with the P3 Primary Cell 4D-Nucleofector® X Kit S (Lonza, Catalog # V4XP-3032). 20 µL nucleofection buffer was prepared by combining 3.6 µL of Supplement1 with 16.4 µL of P3 Primary Cell Nucleofector Solution. 0.5 × 106 HIV-infected CD4+ T cells were resuspended in 20 µL nucleofection buffer and mixed with 3.5 µL of the crRNP complex prior to nucleofection using program EH-115. Immediately following electroporation, 100 µL of pre-warmed complete R10 supplemented with 25 mM HEPES, 1 mM sodium pyruvate and 30 U/mL rhIL-2 was added to each reaction for recovery at 37 °C for 30 min. Cells were then transferred to round-bottom 96-well plates containing 220 μL of medium per well. Immunocult human CD2/CD3/CD28 T cell activator was added to each well at a concentration of 12.5 μL/mL media to facilitate survival and continued activation of infected CD4+ T cells.
Knockout efficiency was assessed 72 h post-nucleofection by qPCR. Briefly, RNA was isolated using the Qiagen RNeasy Micro Kit (Catalog #74004). 12 μL of RNA was used to prepare cDNA using the SuperScript VILO cDNA Synthesis Kit (Thermo, Catalog #11754050) according to the manufacturer’s protocol. qPCR for GALNT6, NANP and B4GALNT1 was performed on QuantStudio 6 Pro (Thermo Fisher) and the following FAM-MGB probes were obtained from Thermo: GALNT6 (Hs00926629_m1), NANP (Hs00600979_m1), and B4GALNT1 (Hs00155195_m1). 18S (Thermo, Catalog #4352930E) was used as the endogenous control to calculate relative copy number.
Autologous cytotoxicity assays against gene-edited HIV-infected primary CD4+ T cells
2 × 104 gene-edited HIV-infected primary CD4+ T cells were harvested 72 hpi and cultured in the presence of autologous monocytes (freshly isolated from cryopreserved PBMCs) at an effector to target ratio (E:T ratio) of 10:1 for 16 h at 37 °C. Following incubation, the co-cultures were harvested and evaluated for intracellular p24 by staining with Zombie Aqua fixable viability dye (BioLegend), anti-CD3 BV421 (BioLegend, Clone UCHT1), and anti-p24 KC57-RD1 (Beckman Colter).
Autologous cytotoxicity assay against HIV-infected primary CD4+ T cells using the Siglec-3 blocking antibody
For Siglec-3 blocking experiments, monocytes were treated with anti-Siglec-3 antibody (50 µg/mL; clone: Hu9a; R&D Systems; catalog #MAB9635-100) or isotype-matched control antibody for 30 min at 37 °C in 96-well microplates before co-culture with autologous HIV-infected CD4+ T cells (2 × 104 cells/well) at an E:T ratio of 10:1. The co-cultures were incubated for 16 h at 37 °C. Following incubation, cells were harvested and evaluated for intracellular p24 by staining with Zombie NIR fixable viability dye (BioLegend), anti-CD3 BV421 (BioLegend), and anti-p24 KC57-RD1 (Beckman Colter).
Viability assay
Uninfected and HIV-infected CEM NKR CCR5+ Luc+ cells were cultured in the presence or absence of increasing concentrations of 10-1074 or 10-1074-SiaD for 16 h. After the incubation, cells were harvested and stained with Aqua live/dead viability stain (Invitrogen) and analyzed on FACS Symphony A5.
Selectivity of 10-1074-SiaD against HIV-infected CD4+ T cells
The selectivity of 10-1074-SiaD conjugated antibodies against HIV-infected cells was determined as described previously29. Briefly, equal number of uninfected CD4+ T cell line, Hut78 and HIV-infected CD4+ T cell line, Hut78/SF2 were cultured in the presence of 400 nM 10-1074 or 10-1074-SiaD antibodies for 1 h at 37 °C. Following incubation, cells were washed twice with PBS (with Ca2+/Mg2+) supplemented with 2% FBS and co-stained with FITC-labeled-SNA and fixable Aqua Live/Dead dye for 30 min at 4 °C. Cells were washed, fixed, permeabilized and stained with intracellular p24 antibody, KC57-RD1, to label the HIV-infected cells. Samples were analyzed by flow cytometry. Cell surface sialylation levels were revealed with SNA-FITC, and HIV-infected cells were determined by p24 expression.
Expression of Siglec-7 ligands on HIV-infected CEM.NKR CCR5+Luc+ cells
CEM.NKR CCR5+Luc+ cells were infected with 30 μL HIVIIIB (1 × 106 TCID50/mL) by spinoculation for 12 h in the presence or absence of 2.5 mM BAGN. Cells were harvested after 12 h and stained with 10 μg recombinant Siglec-7 Fc to determine the expression of Siglec-7 ligands.
Glycomic profiling by Lectin microarray
Membrane proteins isolated from activated, primary CD4+ T cells, uninfected or HIVTYBE-infected in the presence and absence of BAGN, were subjected to a 96-plex lectin microarray to determine cell-surface glycans as described previously. Briefly, 0.4 μg of protein is labeled with Cy3, washed, loaded onto the lectin chips and incubated overnight with the probing solution. After incubation, the chips are washed and scanned on Bio-Rex Scan 300.
Detection of cell surface Siglec-3, -7 and -9 receptors
Primary γδ T cells, CD8+ T cells, NK cells, monocytes, macrophages and neutrophils were stained with anti-Siglec-3 (Biolegend; Clone P67.6), Siglec-7 (Biolegend; Clone 6-434) and Siglec-9 (Biolegend; Clone K8) antibodies to evaluate their cell surface expression by flow cytometry.
Generation of 10-1074-SiaD conjugate
To construct the pET151-FB-E25TAG-GGGS-STSia plasmid, the FB-E25TAG gene was generated by PCR using primers CY060/JP001 and template pET-22b-T5-FB-E25TAG and inserted into the pET151-STSia vector amplified with primers CY061/JP002 (Supplementary Tables 3, 4). The pET151-FBE25-GGGS-STSia and pUltra-KcrRS were then co-transformed into E. coli BL21(DE3) strain. Cells were grown in LB media, supplemented with ampicillin (50 μg/mL) and spectinomycin (25 μg/mL) at 37 °C. When the OD reached to 0.6, 1 mM Isopropyl β- d-1-thiogalactopyranoside (IPTG) and 10 mM FPheK were added to the culture, and the culture was grown overnight at 30 °C. The cells were harvested by centrifugation at 5840 x g for 10 min. The cell pellet was collected, resuspended in PBS and lysed by sonication. The supernatant was filtered, and the Sia-FB protein was purified by running through a Ni-NTA column using the FPLC (buffer A: 50 mM Imidazole in PBS, pH 7.8; buffer B: 500 mM Imidazole in PBS, pH 7.8). The eluted Sia-FB protein was concentrated and buffer exchanged into PBS using a 30 K concentrator and characterized by ESI-MS analysis. Endotoxin was removed using PierceTM High-Capacity Endotoxin Removal Spin Column, 0.50 ml. 10-1074 antibody was then co-incubated with 10 equivalent amounts of Sia-FB at 37 °C for 48 h. After showing the success of the conjugation by SDS-PAGE analysis, the conjugate was concentrated using a 50 K cutoff concentrator and further purified by running through a size-exclusion column using the FPLC. The purified conjugate was collected from the 96-well plate and characterized by SDS-PAGE analysis. For molecular weight calibration, the pre-concentration gel was run with ExcelBand 3-Color Regular Range Protein Marker [PM2500] (SMOBIO; Cat# PM2500). The post-concentration gel was run with Prestained Protein Marker (APExBIO; Cat# F4005).
Binding of 10-1074 and 10-1074-SiaD to HIV gp120
High-binding 96-well ELISA plates (Costar) were coated overnight with 100 ng/well of purified gp120 (Acrobiosystems; Catalog #GP4-V14227) in PBS. After washing, the plates were blocked for 1 h with superblock buffer at RT, washed and then incubated with 100 nM of 10-1074 or 10-1074-SiaD (in quadruplets) for 1 h at room temperature. After washing, the plates were developed by incubation with goat HRP-conjugated anti-human IgG antibodies (Jackson ImmunoReseach) for 1 h and by addition of HRP chromogenic substrate (R&D Systems, Catalog #DY999), and absorbance was measured at 450 nM.
Immune cell mediated killing of HIV-infected CEM.NKR CCR5+ Luc+ cells
CEM.NKR CCR5+ Luc+ cells were infected with HIVIIIB (1 × 106 TCID50/ml) on RetroNectin precoated dishes as described previously29. After 72 h, the infected cells were washed, resuspended in complete R10 and plated at 2 × 104 cells/well in a V-bottom 96-well microplate. HIV-infected CEM.NKR CCR5+ Luc+ were treated with indicated concentrations of 10-1074 or 10-1074-SiaD conjugates for 2 h at 37 °C. Following incubation, human PBMCs were added to the wells at an effector-to-target (E:T) ratio of 100:1. Similarly, cytotoxicity assays against HIVIIIB-infected CEM.NKR CCR5+ Luc+ cells using NK cells, monocytes or freshly isolated neutrophils were performed at an E:T ratio of 10:1 in the presence or absence of 10-1074 or 10-1074-SiaD. Monocyte-derived macrophages differentiated for 8 days were co-cultured with the HIV-infected target cells at an E:T ratio of 2.5:1, to evaluate their cytotoxic potential in the presence or absence of 10-1074 and 10-1074-SiaD. The cell co-cultures were centrifuged at 200 g for 2 min and incubated for 16 h at 37 °C. Following incubation, 100 µl supernatant was removed from all wells and replaced with 100 µl Bright-Glo luciferase substrate reagent (Promega, Catalog # E2620). After 2 min, the well contents were mixed and transferred to a clear-bottom black 96-well microplate and luminescence (RLU) measurements were integrated over 1 s per well.
Incucyte imaging
Uninfected or HIVIIIB-infected CEM.NKR CCR5+ Luc+ cells were stained 800 nM of cytolight rapid red dye (Incucyte, Catalog #4706) or 400 nM of cytolight rapid green dye (Incucyte, Catalog #4705) according to the manufacturer’s instructions. Briefly, the cells were washed with PBS to remove serum and incubated with the indicated concentrations of dye in PBS at 37 °C for 20 min. Following this, the cells were washed with 5 x volume of 10% FBS containing RPMI 1640 media to quench and wash off unbound dye. The cells were resuspended in complete R10, and a mix of 20,000 uninfected (red) and HIV-infected (green) target cells were added to 96-well flat bottom plates (10,000 cells each/well) and co-cultured with 3.75 nM of isotype control, 10-1074 or 10-1074-SiaD antibodies for 2 h at 37 °C for opsonization. After 2 h, primary monocytes from 4 HIV-negative donors were added as the effector cells (E:T = 5:1), and the cells were imaged on Incucyte SX1 Live Cell Imager (Incucyte) at 1 h intervals for a time period of 16 h. % cytotoxicity was calculated based on the live red and green cell count at each time point using the Incucyte analysis software.
Autologous cytotoxicity assays
Primary CD4+ T cells from six to seven HIV-negative donors were infected with HIVTYBE as described earlier. Autologous monocytes and HIVTYBE-infected CD4+ T cells were cocultured at E:T ratio of 10:1 in the presence of 50 nM isotype control, 10-1074 or 10-1074-SiaD, for 16 h at 37 °C. Similarly, freshly isolated neutrophils were co-cultured with autologous HIV-infected target cells at an E:T ratio of 5:1 in the presence of 10 nM isotype control, 10-1074 or 10-1074-SiaD for 16 h at 37 °C. Since neutrophils cannot be reliably cryopreserved and must be used fresh, we scheduled two separate blood draws from each donor: (1) During the first visit, we isolated CD4+ T cells and infected them with HIV. (2) During the second visit (days later), we isolated fresh myeloid effector cells, including neutrophils, from the whole blood of the same donor, which bypasses the need for Ficoll density gradient separation. Following incubation, the co-cultures were harvested and evaluated for intracellular p24 by staining with Zombie Aqua fixable viability dye (BioLegend), anti-CD3 BV421 (BioLegend, Clone UCHT1), and anti-p24 KC57-RD1 (Beckman Colter).
Transcriptomic analysis from autologous monocyte-HIV-infected CD4+ T cell co-cultures
Monocytes co-cultured with autologous HIV-infected CD4+ T cells in the presence of 50 nM isotype control, 10-1074 and 10-1074-SiaD for 16 h were harvested, and RNA isolation was performed using the RNAeasy micro kit (Qiagen, Catalog #74004) with on-column DNA digestion. Libraries were constructed on the Stranded mRNA Library Prep Kit (Illumina) using 25 ng input RNA. The sequencing was performed on a NovaSeq X Plus 10B flowcell as 50 bp single-end, and a minimum of 30 M reads were captured for each sample. The RNA-seq data analysis was performed by Bencos Research Solutions Pvt. Ltd., Mumbai, India. For the analysis, the raw data obtained after performing the RNA sequencing were aligned to the Reference Human Genome assembly GRCh38 using the salmon (v 1.10.1) software in conjunction with the STAR aligner (v 2.7.10a)75. Raw read counts were used as the input to perform differential gene expression analysis using the R package DESeq2 (v 1.34.0)73. DESeq2 normalized read counts were used to calculate the log2 fold change and adjusted p-values. P-values were adjusted using the Benjamini-Hochberg procedure to determine the False Discovery Rate (FDR) and control for multiple testing.
Human memory CD4+ T cell isolation and engraftment into immunodeficient mice
All mice were maintained in appropriate environmental conditions, housed under controlled temperature (22 ± 2 °C) with 40–60% humidity, a 12 h light-12-hour dark cycle, with ad libitum access to standard rodent chow and water. All animal protocols were approved by Wistar Institutional Animal Care and Use Committee (IACUC protocol # 201468) and Northwestern University IACUC (protocol # IS00028851). Euthanasia was performed using carbon dioxide (CO2) delivered as bottled gas, which is listed among the acceptable agents for euthanasia of laboratory rodents in the American Veterinary Medical Association (AVMA) Guidelines for the Euthanasia of Animals: 2013 Edition. All experiments were performed in a Biosafety Level 2+ (BSL2+) facility. Memory CD4+ T cells were isolated from the PBMCs of a female HIV-negative donor using the EasySep human memory CD4+ T cell enrichment kit (StemCell Technologies, Catalog #19157), according to the manufacturer’s recommendation. Non-irradiated 6-week-old female NSG mice (The Wistar Institute, Philadelphia) were engrafted with 5 × 106 memory CD4+ T cells via intravenous injection, as previously described48. Mice were bled weekly to assess human immune cell reconstitution and plasma viremia. Approximately 70 μL of peripheral blood was collected into EDTA-coated tubes via submandibular bleeding and processed immediately. Blood was centrifuged at 2500 × g for 10 min to separate plasma, which was stored at − 80 °C to further viral RNA extraction. Cell pellets were immediately stained for flow cytometry analysis as described below. One week post-infection, mice were treated with 0.75 mg of either 10-1074 or 10-1074-SiaD conjugate, with 5 × 106 effector cells, consisting of NK cells and monocytes in a 1:1 ratio, administered intravenously, once weekly for five consecutive weeks. The control group received only PBS using the same route. Mouse body weight was measured three times a week until the end of the study.
Flow cytometric analysis of mouse samples
Samples were stained using a compensated panel of the following human-specific antibodies: anti-CD3 APC-Cy7 (Biolegend, Clone SK7), anti-CD4 Alexafluor647 (Biolegend, Clone OKT4), anti-CD8 BV421 (Biolegend, Clone SK1), anti-CD45 BUV395 (Invitrogen, Clone HI30), anti-CD56 PerCP-eFluor710 (Invitrogen, Clone TULY56), anti-CD14 FITC (Biolegend, clone M5E2), anti-CD16 BUV661 (Invitrogen, Clone CB16), anti-CD19 PE (Biolegend, Clone HIB19), anti-CD45RA BV750 (Biolegend, Clone HI100), anti-CD45RO PE-Cy7 (Biolegend, Clone UCHL1), Live/Dead BV510 (Zombie Aqua, Catalog #423102), and anti-mouse CD45 BUV805 (Invitrogen, Clone 30-F11) for 30 min at room temperature. After surface staining, red blood cells were lysed using ACK lysis buffer (Gibco, Catalog #A104920) and washed with PBS twice. Cells were then fixed, and fluorescent counting beads (Invitrogen; Catalog #C36950) were added to each sample to determine the absolute cell count. Flow cytometric analysis of human cells was conducted using Symphony A5 and FlowJo version 10 software.
In vivo HIV infection of humanized mice
Mice were infected via intravenous tail vein injection with HIV-1SUMA at a dose of 1500 TCID50 per mouse. The efficiency of infection was assessed post-infection weekly by measuring plasma HIV-1 RNA using qRT-PCR. All infected mice had detectable viremia, confirming 100% infection efficiency. Plasma samples from each time point was spiked in with a known quantity of RCAS virus to use as an internal control, and RNA was isolated using QIAmp Viral RNA mini kit (Qiagen, Catalog #52906). cDNA was generated using SuperScript VILO Master Mix (Invitrogen, Catalog #11755500) on a thermal cycle at 25 °C for 10 min, 42 °C for 60 min, 85 °C for 5 min, and 4 °C. HIV quantitation was done from cDNA using Real-Time PCR. The reaction was carried out using Universal Master Mix II with UNG (Applied Biosystems, catalog #4440038) and following HIV-1 primers and probe: F522-43 Kumar- 5’ GCCTCAATAAAGCTTGCCTTGA 3’, R626-43 Kumar- 5’ GGGCGCCACTGCTAGAGA 3’, Probe Kumar (FAM/ZEN/BHQ-1)- 5’ CCAGAGTCACACAACAGACGGGCACA 3’. The run was done on a Quant Studio 6 Real-Time PCR system using the following cycling parameters: 50 °C for 2 min, 95 °C for 10 min, followed by 60 cycles of 95 °C for 15 s and 59 °C for 1 min. Cycle threshold values were compared with the validated HIV RNA standard run on each plate to determine HIV RNA copies.
Glycomic profiling of HIV-infected memory CD4+ T cells isolated from humanized mice
Untouched human CD4+ T cells were isolated from the pooled blood of 4 uninfected and 4 HIVSUMA-infected humanized mice by EasySep Mouse/Human Chimera Isolation Kit (StemCell; Catalog # 19829 A). Membrane protein was isolated and subjected to a 96-plex lectin microarray to determine the changes in cell surface glycans post HIV infection, as described above.
Measurement of plasma inflammatory markers from mice
Plasma levels of human INF-γ, IP-10, TNF-α, MIP-1α, SDF-1a, IL-2, IL-6, IL-10, and IL-21 were determined using U-PLEX kits from Meso Scale Diagnostics (Biomarker Group 1 (hu) Assay; Catalog #K151AEM-2, Custom Immuno-Oncology Grp (hu) Assay; Catalog #K15067L-2, according to the manufacturer’s instruction.
Statistical analysis
Statistical analysis was performed on GraphPad Prism, v.10 and R. The analysis used for each figure has been described in the corresponding figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that data supporting the findings of this study are available within the paper and its Supplementary Information files. In addition, raw RNAseq data in Fig. 3 have been deposited in the Gene Expression Omnibus (GEO) under GEO accession #: GSE289893. Raw RNAseq data in Fig. 7 have been deposited in the ArrayExpress database at EMBL-EBI under accession number E-MTAB-15808. Single-cell analyses in Fig. 1 and Fig. 3 used publicly available data deposited in the GEO under GEO accession #: GSE15782932. Source data are provided in this paper.
References
Collins, K. L., Chen, B. K., Kalams, S. A., Walker, B. D. & Baltimore, D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391, 397–401 (1998).
Duette, G. et al. The HIV-1 proviral landscape reveals that Nef contributes to HIV-1 persistence in effector memory CD4+ T cells. J. Clin. Invest. 132, e154422 (2022).
Li, Y. L. et al. The structural basis for HIV-1 Vif antagonism of human APOBEC3G. Nature 615, 728–733 (2023).
Mangeat, B. et al. HIV-1 Vpu neutralizes the antiviral factor Tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog. 5, e1000574 (2009).
Schwartz, O., Marechal, V., Le Gall, S., Lemonnier, F. & Heard, J. M. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 2, 338–342 (1996).
Board, N. L., Moskovljevic, M., Wu, F., Siliciano, R. F. & Siliciano, J. D. Engaging innate immunity in HIV-1 cure strategies. Nat. Rev. Immunol. 22, 499–512 (2022).
Deeks, S. G. et al. Research priorities for an HIV cure: international AIDS society global scientific strategy 2021. Nat. Med 27, 2085–2098 (2021).
Siliciano, J. D. & Siliciano, R. F. HIV cure: The daunting scale of the problem. Science 383, 703–705 (2024).
Ward, A. R., Mota, T. M. & Jones, R. B. Immunological approaches to HIV cure. Semin Immunol. 51, 101412 (2021).
Hudak, J. E., Canham, S. M. & Bertozzi, C. R. Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat. Chem. Biol. 10, 69–75 (2014).
Jandus, C. et al. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J. Clin. Invest. 124, 1810–1820 (2014).
Laubli, H. et al. Engagement of myelomonocytic Siglecs by tumor-associated ligands modulates the innate immune response to cancer. Proc. Natl. Acad. Sci. USA 111, 14211–14216 (2014).
RodrIguez, E., Schetters, S. T. T. & van Kooyk, Y. The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. 18, 204–211 (2018).
Barkal, A. A. et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 572, 392–396 (2019).
Chen, G. Y., Tang, J., Zheng, P. & Liu, Y. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science 323, 1722–1725 (2009).
Kang, E. A. et al. Soluble Siglec-9 alleviates intestinal inflammation through inhibition of the NF-kappaB pathway. Int. Immunopharmacol. 86, 106695 (2020).
Laubli, H. & Varki, A. Sialic acid-binding immunoglobulin-like lectins (Siglecs) detect self-associated molecular patterns to regulate immune responses. Cell Mol. Life Sci. 77, 593–605 (2020).
Macauley, M. S., Crocker, P. R. & Paulson, J. C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 14, 653–666 (2014).
Duan, S. & Paulson, J. C. Siglecs as Immune Cell Checkpoints in Disease. Annu. Rev. Immunol. 38, 365–395 (2020).
Beatson, R. et al. The mucin MUC1 modulates the tumor immunological microenvironment through engagement of the lectin Siglec-9. Nat. Immunol. 17, 1273–1281 (2016).
Stanczak, M. A. et al. Self-associated molecular patterns mediate cancer immune evasion by engaging Siglecs on T cells. J. Clin. Invest. 128, 4912–4923 (2018).
Choi, H. et al. Development of siglec-9 blocking antibody to enhance anti-tumor immunity. Front. Oncol. 11, 778989 (2021).
Haas, Q. et al. Siglec-9 Regulates an effector memory CD8(+) T-cell subset that congregates in the melanoma tumor microenvironment. Cancer Immunol. Res. 7, 707–718 (2019).
Daly, J. et al. Targeting hypersialylation in multiple myeloma represents a novel approach to enhance NK cell-mediated tumor responses. Blood Adv. 6, 3352–3366 (2022).
Xiao, H., Woods, E. C., Vukojicic, P. & Bertozzi, C. R. Precision glycocalyx editing as a strategy for cancer immunotherapy. Proc. Natl. Acad. Sci. USA 113, 10304–10309 (2016).
Gray, M. A. et al. Targeted glycan degradation potentiates the anticancer immune response in vivo. Nat. Chem. Biol. 16, 1376–1384 (2020).
Tsai, T. Y. et al. SIGLEC-3 (CD33) serves as an immune checkpoint receptor for HBV infection. J. Clin. Invest. 131, e141965 (2021).
Ibarlucea-Benitez, I., Weitzenfeld, P., Smith, P. & Ravetch, J. V. Siglecs-7/9 function as inhibitory immune checkpoints in vivo and can be targeted to enhance therapeutic antitumor immunity. Proc. Natl. Acad. Sci. USA 118, e2107424118 (2021).
Adeniji, O. S. et al. Siglec-9 defines and restrains a natural killer subpopulation highly cytotoxic to HIV-infected cells. PLoS Pathog. 17, e1010034 (2021).
Zhao, D. et al. Decreased siglec-9 expression on natural killer cell subset associated with persistent HBV replication. Front. Immunol. 9, 1124 (2018).
Schwarz, F. et al. Siglec receptors impact mammalian lifespan by modulating oxidative stress. Elife 4, e06184 (2015).
Wang, S. et al. An atlas of immune cell exhaustion in HIV-infected individuals revealed by single-cell transcriptomics. Emerg. Microbes Infect. 9, 2333–2347 (2020).
Wen, R. M. et al. Sialylated glycoproteins suppress immune cell killing by binding to Siglec-7 and Siglec-9 in prostate cancer. J. Clin. Invest. 134, e180282 (2024).
Olvera, A. et al. Benzyl-2-acetamido-2-deoxy-alpha-d-galactopyranoside increases human immunodeficiency virus replication and viral outgrowth efficacy in vitro. Front Immunol. 8, 2010 (2017).
Terry, V. H., Johnston, I. C. & Spina, C. A. CD44 microbeads accelerate HIV-1 infection in T cells. Virology 388, 294–304 (2009).
Keppler, O. T. et al. UDP-GlcNAc 2-epimerase: a regulator of cell surface sialylation. Science 284, 1372–1376 (1999).
Lewis, A. L., Chen, X., Schnaar, R. L. & Varki, A. Sialic acids and other nonulosonic acids. in Essentials of Glycobiology. (2022).
Li, Y. & Chen, X. Sialic acid metabolism and sialyltransferases: natural functions and applications. Appl. Microbiol. Biotechnol. 94, 887–905 (2012).
Tian, E. & Ten Hagen, K. G. Recent insights into the biological roles of mucin-type O-glycosylation. Glycoconj. J. 26, 325–334 (2009).
Gonzalez-Gil, A. & Schnaar, R. L. Siglec Ligands. Cells 10, 1260 (2021).
Kellner, C. & Peipp, M. Fc-optimized antibodies quickly pull the trigger. Blood 124, 3180–3181 (2014).
Dharmadhikari, G. et al. Siglec-7 restores beta-cell function and survival and reduces inflammation in pancreatic islets from patients with diabetes. Sci. Rep. 7, 45319 (2017).
Varki, A. & Gagneux, P. Multifarious roles of sialic acids in immunity. Ann. N. Y. Acad. Sci. 1253, 16–36 (2012).
Caskey, M. et al. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 23, 185–191 (2017).
Mouquet, H. et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. USA 109, E3268–E3277 (2012).
Shingai, M. et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature 503, 277–280 (2013).
Yu, C. et al. Proximity-Induced Site-Specific Antibody Conjugation. Bioconjugate Chem. 29, 3522–3526 (2018).
McCann, C. D. et al. A participant-derived xenograft model of HIV enables long-term evaluation of autologous immunotherapies. J. Exp. Med. 218, e20201908 (2021).
Barrera, C., Espejo, R. & Reyes, V. E. Differential glycosylation of MHC class II molecules on gastric epithelial cells: implications in local immune responses. Hum. Immunol. 63, 384–393 (2002).
de Freitas Junior, J. C. et al. Inhibition of N-linked glycosylation by tunicamycin induces E-cadherin-mediated cell-cell adhesion and inhibits cell proliferation in undifferentiated human colon cancer cells. Cancer Chemother. Pharmacol. 68, 227–238 (2011).
Hu, P., Berkowitz, P., Madden, V. J. & Rubenstein, D. S. Stabilization of plakoglobin and enhanced keratinocyte cell-cell adhesion by intracellular O-glycosylation. J. Biol. Chem. 281, 12786–12791 (2006).
Everest-Dass, A. V. et al. Comparative structural analysis of the glycosylation of salivary and buccal cell proteins: innate protection against infection by Candida albicans. Glycobiology 22, 1465–1479 (2012).
Itell, H. L., Guenthoer, J., Humes, D., Baumgarten, N. E. & Overbaugh, J. Host cell glycosylation selects for infection with CCR5- versus CXCR4-tropic HIV-1. Nat. Microbiol. 9, 2985–2996 (2024).
Duran-Castells, C. et al. Plasma proteomic profiling identifies CD33 as a marker of HIV control in natural infection and after therapeutic vaccination. EBioMedicine 95, 104732 (2023).
Varki, A., Schnaar, R. L. & Schauer, R. Sialic acids and other nonulosonic acids. in Essentials of Glycobiology (2015).
Edgar, L. J. et al. Sialic acid ligands of CD28 suppress costimulation of T cells. ACS Cent. Sci. 7, 1508–1515 (2021).
Gong, Y., Qin, S., Dai, L. & Tian, Z. The glycosylation in SARS-CoV-2 and its receptor ACE2. Signal Transduct. Target Ther. 6, 396 (2021).
Sheng, Y. et al. Host cell glycocalyx remodeling reveals SARS-CoV-2 spike protein glycomic binding sites. Front. Mol. Biosci. 9, 799703 (2022).
Krause, C., Bergmann, E. & Schmidt, S. V. Epigenetic modulation of myeloid cell functions in HIV and SARS-CoV-2 infection. Mol. Biol. Rep. 51, 342 (2024).
Mehraj, V., Jenabian, M. A., Vyboh, K. & Routy, J. P. Immune suppression by myeloid cells in HIV infection: new targets for immunotherapy. Open AIDS J. 8, 66–78 (2014).
Snow, B. J. et al. Potent broadly neutralizing antibodies mediate efficient antibody-dependent phagocytosis of HIV-infected cells. PLoS Pathog. 20, e1012665 (2024).
Tay, M. Z., Wiehe, K. & Pollara, J. Antibody-dependent cellular phagocytosis in antiviral immune responses. Front. Immunol. 10, 332 (2019).
Egan, H. et al. Targeting stromal cell sialylation reverses T cell-mediated immunosuppression in the tumor microenvironment. Cell Rep. 42, 112475 (2023).
Mei, Y. et al. Siglec-9 acts as an immune-checkpoint molecule on macrophages in glioblastoma, restricting T-cell priming and immunotherapy response. Nat. Cancer 4, 1273–1291 (2023).
Wang, J. et al. Siglec receptors modulate dendritic cell activation and antigen presentation to T cells in cancer. Front. Cell Dev. Biol. 10, 828916 (2022).
Stanczak, M. A. et al. Targeting cancer glycosylation repolarizes tumor-associated macrophages allowing effective immune checkpoint blockade. Sci. Transl. Med. 14, eabj1270 (2022).
Nixon, C. C. et al. Systemic HIV and SIV latency reversal via non-canonical NF-kappaB signalling in vivo. Nature 578, 160–165 (2020).
Colomb, F. et al. Sialyl-Lewis(X) glycoantigen is enriched on cells with persistent HIV transcription during therapy. Cell Rep. 32, 107991 (2020).
Le, T. et al. BBrowser: Making single-cell data easily accessible. Preprint at https://doi.org/10.1101/2020.12.11.414136 (2020).
Martinez F. O. Analysis of gene expression and gene silencing in human macrophages.Curr. Protoc. Immunol. https://doi.org/10.1002/0471142735.im1428s96 (2012).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Hultquist, J. F. et al. CRISPR-Cas9 genome engineering of primary CD4(+) T cells for the interrogation of HIV-host factor interactions. Nat. Protoc. 14, 1–27 (2019).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Acknowledgements
This study is supported by NIH R01AI165079 to M.A.-M. and H.X. M.A.-M. is also supported by NIH grants (R01AG092241, R01AA029859, R01DK123733, and R01NS117458). M.A.-M. is also funded by the NIH-funded BEAT-HIV Martin Delaney Collaboratory to cure HIV-1 infection (1UM1Al126620). P.W.D. has research time supported by P20GM103427 and R15AI178516. The publication of this manuscript was supported by a dissemination grant from the Emerging and Re-Emerging Pathogens Program (EREPP) in the Division of Infectious Diseases at Northwestern University. We would like to thank Drs. Michel Nussenzweig, Costin Tomescu, and Luis J. Montaner for providing the wild-type 10-1074.
Author information
Authors and Affiliations
Contributions
M.A.-M. and H.X. conceived and designed the study. S.S., S.M.S., R.L., O.S.A., L.B.G., P.S. carried out the experiments. L.M.S. and J.F.H. supervised and helped perform the CRISPR-Cas9 experiments. H.T. produced the lectin microarray. A.D., P.W.D., R.B.J. interpreted data. S.S., S.M.S. and M.A.-M. wrote the manuscript, and all authors edited it.
Corresponding author
Ethics declarations
Competing interests
J.F.H. received research support, paid to Northwestern University, from Gilead Sciences and is a paid consultant for Merck and Ridgeback Biotherapeutics. All other authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Singh, S., Islam, S.M.S., Liu, R. et al. HIV-induced sialoglycans on infected CD4+ T cells promote immune evasion from myeloid cell-mediated killing. Nat Commun 17, 18 (2026). https://doi.org/10.1038/s41467-025-66540-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66540-y