Abstract
Mini-Chromosome Maintenance 10 (MCM10) is essential for maintaining genome stability by facilitating DNA replication and repair across various organisms. While the role of MCM10 in DNA replication is well-established, its mechanism in DNA repair remains less understood. In this study, we demonstrate that loss of AtMCM10 function leads to increased DNA damage under genotoxic or salinity stress in Arabidopsis thaliana. Detailed analysis reveals that AtMCM10 works primarily downstream of ATM and is crucial for intermolecular homologous recombination (HR) mediated by synthesis-dependent strand annealing (SDSA) in response to DNA damage. Further cytological and biochemical analyses reveal that AtMCM10 possesses DNA annealing activity, colocalizes with the double-strand break (DSB) sites, and undergoes liquid-liquid phase separation (LLPS) upon DNA damage, facilitated by single strand DNA (ssDNA) in vitro. Altogether, our findings indicate that AtMCM10 acts as a single-strand DNA (ssDNA) annealing protein to promote SDSA-mediated intermolecular HR repair via LLPS in somatic cells upon DNA damage, providing new insights into the HR repair mechanisms.
Similar content being viewed by others
Introduction
Maintaining genome stability is fundamental for preserving genetic information, preventing tumorigenesis, and ensuring normal cellular functions1. Mini-Chromosome Maintenance 10 (MCM10) is a key DNA replication factor that plays an essential role in safeguarding genome stability2. MCM10 functions at multiple stages of DNA replication, including initiation, elongation, termination and fork stability, and is critical for replication-coupled nucleosome assembly in eukaryotes3,4,5,6,7,8. In Saccharomyces cerevisiae, MCM10 exhibits potent strand-annealing activity and prevents replication fork reversal, which is required for genome integrity9. Its N-terminus interacts with the 9-1-1 checkpoint clamp in response to DNA damage10. Interestingly, MCM10 forms a complex with the nuclease Dna2 and the DNA damage response kinase Ataxia-Telangiectasia Mutated (ATM) in Xenopus laevis11. Moreover, MCM10 interacts with homologous recombination (HR) factor BRCA2 to suppress PRIMPOL-mediated repriming and prevent lesion skipping under replication stress in human12. These studies suggest that MCM10 may play important roles not only in DNA replication but also in DNA repair to protect genome stability. However, elucidating the mechanism of MCM10 in DNA repair remains challenging in yeast and animals, as its essential function in DNA replication renders loss-of-function lethal13,14. In contrast, the absence of AtMCM10 does not affect Arabidopsis growth and development8, making it an excellent model for exploring MCM10 function in DNA repair.
Double-strand breaks (DSBs) are considered the most cytotoxic and mutagenic type of DNA damage, constituting a major threat to genome stability15,16. In somatic cells, ATM acts as a master kinase that senses DSBs and orchestrates the DNA damage response, thereby coordinating downstream repair pathways, cell cycle checkpoints, and apoptosis16,17,18,19. DSBs are primarily repaired through two major pathways: nonhomologous end joining (NHEJ), which ligates DNA ends in a template-independent manner, and homologous recombination (HR), a more accurate mechanism using homologous templates for repair19,20,21. HR repair is categorized into two subtypes: intramolecular and intermolecular HR. Intramolecular HR relies on repeat sequences flanking the break, often leading to rearrangements such as deletions or inversions. In contrast, intermolecular HR takes place between sister chromatids or homologous chromosomes and is mainly mediated by synthesis-dependent strand annealing (SDSA), which enables accurate repair without crossover formation22. In both cases, the search for homologous sequences and strand annealing are critical steps in HR repair. In the SDSA pathway, the homologous recombinase RAD51 is recruited to DSB sites, where it binds to 3’ ssDNA overhangs to initiate homology search and strand invasion23,24,25,26. In yeast, RAD52 is reported to promote the annealing of a second single-stranded DNA after invasion27. However, to date, the mechanism of annealing in SDSA pathway is not clear in mammals and plants27,28,29.
In this study, we found that Atmcm10 mutant exhibits hypersensitivity to certain DNA damage agents. Loss of AtMCM10 compromises SDSA-mediated intermolecular HR repair under DNA-damage stress. Cytological and biochemical analyses revealed that AtMCM10 undergoes liquid-liquid phase separation (LLPS) at DSB sites and facilitates rapid ssDNA strand annealing. Our findings identify AtMCM10 as a novel regulator in HR, highlight its LLPS and DNA annealing activity in the SDSA-mediated HR repair pathway in somatic cells, and provide new insights into the HR mechanism.
Results
AtMCM10 is required for DNA damage response
Our previous RNA-seq analysis revealed that several DNA damage response genes, including TSO2, BRCA1, RAD51, SMR7, and GMI, were upregulated in the Atmcm10 mutant (Supplementary Fig. 1a)8. These transcriptional changes were further validated by qRT-PCR (Supplementary Fig. 1b), suggesting that loss of AtMCM10 triggers the DNA damage response, likely due to increased DNA damage.
To investigate whether AtMCM10 involves in DNA damage response, we examined the sensitivity of Atmcm10 mutant to classic genotoxic treatments, including Zebularine (Zeb), mitomycin C (MMC), and hydroxyurea (HU), which induce different DNA damage response. Notably, Zeb and MMC induce DNA damage predominantly through DNA-protein crosslinks and DNA inter-strand crosslinks30,31,32, respectively, whereas HU primarily elicits replication stress33. Interestingly, Atmcm10 mutants did not affect Arabidopsis growth on 1/2 MS medium, consistent with previous findings8, and were not sensitive to HU (Supplementary Fig. 2a, b), suggesting that AtMCM10 does not make a major contribution to the cellular response to HU-induced replication stress.
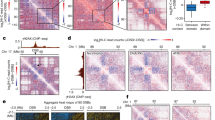
However, compared to wild type, Atmcm10 mutants were hypersensitive to Zeb and MMC, which can be rescued by complementation with functional AtMCM10 (Fig. 1a, b). Furthermore, we treated Atmcm10 mutants with high salinity stress, known to induce DNA damage via reactive oxygen species (ROS) accumulation32,34,35, we found Atmcm10 mutants exhibited severely limited root growth (Fig. 1c, d; Supplementary Fig. 3a, b), indicating their hypersensitivity to salinity stress. Comet assays assessing DNA integrity in Atmcm10 mutants showed significantly higher tail DNA content following Zeb treatment or 6-h recovery after high-salinity exposure compared to wild type (Fig. 1e, f), indicating increased DNA damage in the absence of AtMCM10. Given that increased DNA damage may lead to cell death, we assessed it in the root tip cells after treatments using propidium iodide (PI) that can indicate cell death. Consistently, Atmcm10 seedlings exhibited pronounced PI-positive signals upon Zeb or high salinity treatment, confirming elevated cell death in the mutant background (Fig. 1g, h). These findings demonstrate that AtMCM10 is required for maintaining genome stability under genotoxic and high salinity stress.
Representative phenotypes of WT, Atmcm10-1, Atmcm10-3 and com on genotoxins (a) or NaCl (c). Com represented the pro:AtMCM10:AtMCM10-NOS / Atmcm10-1 transgene line. Seeds were germinated on 1/2 MS medium with or without 5 mg/L zebularine (Zeb), 8 μM mitomycin (MMC) or 100 mM NaCl, and were grown for 7–10 d. Bars = 1 cm. b, d Statistical analyses of primary root lengths of seedlings presented in a–c. Data were presented as mean ± SD (n = 16 seedlings), and lowercase letters indicate significant differences by one-way ANOVA followed determined by Tukey’s HSD test (p ≤ 0.05). e Analysis of DNA damage level in response to Zeb or NaCl in WT, Atmcm10-1 and Atmcm10-3 by comet assay. Nuclei were extracted from 7-day-old seedlings treated with 5 mg/L Zeb or 150 mM NaCl for 24 h, or recovery 6 h on 1/2 MS medium after 150 mM NaCl treatment for 24 h. Bar = 100 μm. f Quantification of tail DNA percentages in (e). DNA content in comet tails was determined using OpenComet Score software. Data were presented as mean ± SEM (n = 100 nuclei). *** indicate significant differences by one-way ANOVA followed determined by Tukey’s HSD test (p ≤ 0.01). g Images of root meristem structure stained with propidium iodide (PI) in WT, Atmcm10-1 and Atmcm10-3 suffered with or without Zeb or NaCl. 5-day-old seedlings were transferred to 1/2 MS medium supplemented with or without 5 mg/L Zeb for 48 h or 50 mM NaCl for 24 h. Bar = 50 μm. h Quantification of dead cell area in (g). Data were presented as mean ± SEM (n = 15 seedlings). *** indicate significant differences by one-way ANOVA followed determined by Tukey’s HSD test (p ≤ 0.01).
Genetic epistasis analysis between AtMCM10 and ATM/ATR in DNA damage response
Upon DNA damage, the two core kinases in the DNA damage response, ATM and ATR, are activated and promote DNA repair. ATM is mainly triggered by DSBs, while ATR primarily responds to single-stranded DNA (ssDNA), particularly at stalled replication forks16,17,18,19. To further investigate the role of AtMCM10 in the DNA damage response, we analyzed its genetic interaction with ATM and ATR by generating the atm Atmcm10-1 and atr Atmcm10-1 double mutants, and assessed their sensitivity to various DNA damage treatments that reflect distinct DNA repair mechanisms.
Under normal conditions, the atr atm double mutant develops shorter roots compared to wild-type (WT) or either single mutant36, while the atr Atmcm10-1 mutant exhibited a root length similar to the atr atm double mutant (Fig. 2a, b), suggesting that AtMCM10 and ATR may have additive effects in regulating the DNA damage response. In contrast, the atm Atmcm10-1 mutant shows root length comparable to those of the atm and Atmcm10-1 single mutants (Fig. 2a, b). Notably, while the atr atm double mutant exhibits complete sterility36, the atr Atmcm10-1 mutant retains fertility, suggesting that AtMCM10 plays a limited role in meiotic cells.
a Representative phenotypes of WT, Atmcm10-1, atm, atm Atmcm10-1, atr, atr Atmcm10-1, and atm atr under genotoxic or salinity stress. Seeds were germinated on plates containing 1/2 MS medium or 1/2 MS supplemented with 5 mg/L Zeb, 2 mM HU or 100 mM NaCl, and were grown for 10 d. Bars = 1 cm. b Statistical analyses of primary root lengths of seedlings shown in (a). Data were presented as means ± SD (n = 16 seedlings), and lowercase letters indicate significant differences by one-way ANOVA followed determined by Tukey’s HSD test (p ≤ 0.05). c Representative phenotypes of WT, Atmcm10-1, atm, 35S:AtMCM10/atm, atr, 35S:AtMCM10/atr, and 35S:AtMCM10 under Zeb treatment. Seeds were germinated on plates containing 1/2 MS medium or 1/2 MS supplemented with 5 mg/L Zeb, and were grown for 10 d. Bars = 1 cm. d Statistical analyses of primary root lengths of seedlings shown in (c). Data were presented as means ± SD (n = 16 seedlings), and lowercase letters indicate significant differences by one-way ANOVA followed determined by Tukey’s HSD test (p ≤ 0.05).
When treated with Zeb, both atm and atr single mutants display hypersensitivity30. The atm Atmcm10-1 double mutant resembled the Atmcm10-1 single mutant, whereas atr Atmcm10-1 showed an additive hypersensitive phenotype, similar to atr atm (Fig. 2a, b). Overexpression of AtMCM10 significantly rescued the Zeb hypersensitivity phenotype in atm mutants but not atr mutants (Fig. 2c, d). These findings indicate that AtMCM10 primarily functions within the ATM-mediated pathway in response to the Zeb-induced DSB in somatic cells.
In contrast, under HU-induced replication stress, neither atm nor Atmcm10-1 mutants exhibited hypersensitivity, and the atm Atmcm10-1 double mutant responded similarly. By comparison, atr was highly sensitive to HU, and this hypersensitivity was not further increased by the additional loss of AtMCM10 or ATM (Fig. 2a, b), indicating that AtMCM10 is dispensable for the canonical ATR-dependent pathway in response to HU-induced replication stress.
Finally, under high salinity stress, Atmcm10-1 displayed pronounced hypersensitivity, while atm and atr single mutants showed no obvious phenotype (Fig. 2a, b). All double mutants, including atm Atmcm10-1, atr Atmcm10-1, and atr atm, phenocopied Atmcm10-1 (Fig. 2a, b), suggesting that AtMCM10 likely functions downstream of both ATM- and ATR-mediated pathways in response to salt-induced genotoxic stress.
Collectively, although genetic epistasis analysis of AtMCM10 with ATM/ATR yields varied results under different genotoxic conditions, all findings consistently support that AtMCM10 is required for the ATM/ATR mediated DNA damage responses. In particular, AtMCM10 predominantly acts through the ATM-mediated pathway in response to Zeb-induced DSB in somatic cells.
AtMCM10 is required for SDSA-mediated intermolecular HR repair
ATM orchestrates the DNA damage response and promotes the repair of DSBs primarily through HR, while also contributing to NHEJ pathways37,38,39. To examine how AtMCM10 functions in the ATM-mediated DNA damage response, we assessed its genetic interaction with core NHEJ component LIGASE IV (LIG4)40. Under normal conditions, the lig4 Atmcm10 double mutant exhibited shorter roots compared to WT or either single mutant (Supplementary Fig. 4a, b), suggesting that AtMCM10 and LIG4 are genetically additive in the DNA damage response pathway. Upon genotoxic stress, lig4 and the lig4 Atmcm10 double mutant both showed pronounced hypersensitivity to Zeocin, whereas Atmcm10 displayed no hypersensitivity (Supplementary Fig. 4a, b). Moreover, under Zeb treatment, the lig4 Atmcm10 double mutant phenocopied Atmcm10-1 (Supplementary Fig. 4a, b). These findings suggest that AtMCM10 likely functions independently of the NHEJ pathway.
Next, we explored whether AtMCM10 participates in HR pathway, which can be categorized into two subtypes: intramolecular HR and intermolecular HR. In Arabidopsis, HR efficiency can be quantitatively assessed using two established reporter lines: 1445/1406 (Intra-HR) and IC9C (Inter-HR, mediated mostly by SDSA). Both lines carry a non-functional β-glucuronidase (GUS) reporter gene split into two separate fragments (Fig. 3a, b)41,42. Successful HR-mediated repair restores GUS activity, which is detectable through histochemical staining (Fig. 3c).
a Schematic representation of the GUS-based recombination substrate in the 1445/1406 reporter lines. Intramolecular interactions between inverted repeats (U) lead to the restoration of a functional GUS gene through HR. b Schematic representation of the GUS-based recombination substrate and SDSA-mediated intramolecular HR in the IC9C reporter line. The spatial orientation of the two GUS fragments requires intermolecular interaction between homologous chromosomes or sister chromatids to restore a functional GUS gene through HR. c Representative GUS staining images of cotyledon in WT and Atmcm10-1 treated without or with Zeb or NaCl, assessed using the reporter lines 1445 and IC9C. Arrows indicate independent events. Bar = 1 mm. d Statistical analysis of recombination frequencies in WT and Atmcm10-1 seedlings following Zeb or NaCl treatment, assessed using the reporter lines 1445, 1406, and IC9C. Error bars represent mean ± SD of three biological replicates (n = 30 cotyledons per replicate). e The ratios of SDSA-mediated intermolecular HR versus intramolecular HR in WT and Atmcm10-1 after different treatments. p-values were calculated using a two-sided Fisher’s exact test. f DNA strand annealing assay for AtMCM10. AtMCM10 was incubated with two complementary oligonucleotides. The ssDNA was end-labeled with digoxin probe (DIG). The reactions were stopped at different time points and were used for DIG detection. GST was used as a negative control. g Schematic diagram of four truncated AtMCM10 proteins used for DNA strand annealing assay. h The results of DNA strand annealing assays for the truncated AtMCM10 proteins as shown in (g) at different incubation times (0, 0.5, 2 min). GST was used as a negative control. i DNA strand annealing activity analyses of AtMCM10 and the truncated proteins by fluorescence-based annealing assay. GST was used as a negative control.
We used these recombination substrate systems to assess the HR response to various DNA damage agents, including Zeb, Zeocin, HU, and high salinity. All of these agents led to an increase in both types of HR events (Fig. 3d, Supplementary Fig. 5a). Specifically, DNA damage induced by Zeocin and HU primarily activated intramolecular HR, as evidenced by a significant increase in HR events in the 1445 and 1406 systems (Supplementary Fig. 5a, b). In contrast, Zeb and high salinity-induced DNA damage were predominantly repaired via SDSA, as reflected by a dramatic increase in recombination events in the IC9C systems (Fig. 3c, d), consistent with previous studies30,34. Intriguingly, under normal growth conditions, Atmcm10 mutants exhibited HR frequencies similar to WT plants for both repair pathways. However, following Zeb or NaCl treatment, the frequency of SDSA-mediated intermolecular HR events was significantly reduced in Atmcm10 compared to WT (Fig. 3c–e). In contrast, intramolecular HR events showed a marked increase in the Atmcm10-1 mutant under the same stress conditions (Fig. 3c–e). These findings demonstrate that AtMCM10 plays a specific role in the SDSA-mediated HR repair pathway. The observed elevation in intramolecular HR frequency in Atmcm10-1 may represent a compensatory mechanism for the defective SDSA pathway (Fig. 3d).
AtMCM10 promotes SDSA-mediated intermolecular HR repair through its DNA annealing activity
In SDSA, strand annealing is essential for capturing the second ssDNA following RAD51-mediated strand invasion. Although RAD52 facilitates this process in yeast27, the mechanism of annealing in SDSA pathway is not clear in mammals and plants. Notably, ScMCM10 is reported to exhibit strong strand-annealing activity in vitro9, and AtMCM10 preferentially binds ssDNA via conserved zinc finger domain8. These findings prompted us to investigate whether AtMCM10 facilitates ssDNA annealing during SDSA repair in Arabidopsis. To test this hypothesis, we assessed the annealing activity of AtMCM10 by incubating the protein with two complementary single-stranded oligonucleotides for varying durations. The resulting single- and double-stranded oligonucleotides were subsequently analyzed by polyacrylamide gel electrophoresis (PAGE). As shown in Fig.3f, AtMCM10 efficiently annealed these two ssDNAs, whereas the control protein GST did not, indicating that AtMCM10 indeed possesses intrinsic strand annealing activity.
To identify the critical region of AtMCM10 responsible for annealing, we tested four truncations encompassing key functional regions (Fig. 3g)8. The AtMCM101-212 aa, containing the N-terminus and EOB fold, showed no annealing activity, whereas the AtMCM101-263 aa and AtMCM10264-396 aa exhibited significantly impaired activity (Fig. 3h). Notably, the AtMCM10213-396 aa truncation, comprising both the Zinc finger and C-terminus, retained annealing activity, though slightly reduced compared to the full-length AtMCM10 (Fig. 3h). These findings indicate that both the Zinc finger and C-terminal domains are essential for the annealing activity of AtMCM10. We further employed a fluorescence-based assay to better quantitatively assess the annealing activity of AtMCM10 and its truncated versions, using the fluorescent dye DAPI to monitor the formation of double-stranded DNA (Supplementary Fig. 6). Consistent with above results, the AtMCM10213-396 aa, which contains both the Zinc finger and C-terminal domains, exhibited a stronger annealing activity compared to both the CT-truncated (AtMCM101-212 aa and AtMCM101-263 aa) and C-terminal-only (AtMCM10264-396 aa) variants (Fig. 3i).
Taken together, these findings demonstrate that AtMCM10 plays a crucial role in SDSA-mediated intermolecular HR repair by facilitating DNA strand annealing.
AtMCM10 undergoes LLPS in vivo and in vitro upon DNA damage
A common regulatory feature of many HR-related factors is that they relocalize and form foci at DSB sites in response to DNA damage24,43,44. To further investigate the role of AtMCM10 in intermolecular HR repair, we analyzed the colocalization of AtMCM10 with RAD51, a key factor of HR process known to localize to DSB sites24,26,43,45. We co-expressed the mNeon-AtMCM10 and RAD51-mScarlet monomeric fluorescent proteins in Nicotiana benthamiana leaves. RAD51 foci were clearly observed following treatment with either 5 mg/L Zeb or 150 mM NaCl for 24 h (Fig. 4a), confirming the suitability of this system for monitoring DSBs responses. Notably, the subcellular localization of AtMCM10 changed upon DNA damage, resulting in foci that largely colocalized with RAD51 puncta (Fig. 4a), suggesting its recruitment to DSB sites in response to DNA damage. Subsequently, we investigated the subcellular localization dynamics of AtMCM10 in Arabidopsis. Transgenic line expressing proMCM10:MCM10:GFP was subjected to mock treatment, Zeb, or high salinity stress, followed by confocal microscopy analysis. In untreated controls, AtMCM10-GFP exhibited a diffuse and homogeneous nuclear distribution. Strikingly, within 24 h of Zeb or high salinity treatment, we observed the formation of distinct nuclear puncta (Fig. 4b, Supplementary Fig. 7a, b). These damage-induced puncta displayed sensitivity to 1,6-hexanediol (1,6-HD), a known liquid-liquid phase separation (LLPS) disruptor, while remaining unaffected by the control compound 2,5-hexanediol (2,5-HD) (Fig. 4c). This differential response strongly suggests that AtMCM10 undergoes LLPS in response to DNA damage.
a Subcellular co-localization of mNeon GREEN-AtMCM10 and RAD51-mScarlet upon DNA damage in N. benthamiana leaves. The mNeon GREEN-AtMCM10 and RAD51-mScarlet constructs were co-infiltrated into N. benthamiana leaves for transient expression. The subcellular co-localization of AtMCM10 and RAD51 was analyzed following treatment with 5 mg/L Zeb or 150 mM NaCl, or no treatment, for 24 h. Bars = 5 μm. b Dynamic changes of AtMCM10 upon Zeb or NaCl treatment in the root tip cells of Arabidopsis. The subcellular localization of AtMCM10 was examined in the root tip cells of proMCM10:MCM10:GFP transgenic line following treatment with 5 mg/L Zeb or 150 mM NaCl, or no treatment, for 24 h. Bar = 5 μm. c The disruption of AtMCM10 puncta treated by phase-separation disruptor 1,6-hexanediol (1,6 HD). 2,5-hexanediol (2, 5 HD) was used as the control reagent. Bar = 5 μm. d Phase-separation assay of AtMCM10 in vitro. The His-AtMCM10-GFP protein purified from the E. coli BL21 strain was added to the solution containing 10% PEG 3350, or PEG 3350 plus 1.6 HD. Bar = 5 μm. e Fusion of His-AtMCM10-GFP condensates in vitro. Left was time-course imaging. Right was circularity of fusing droplets over time, error bars represent mean ± SD, n = 4 fusing droplets. Bar = 2 μm. f, h FRAP analysis of AtMCM10 condensates in vitro (f) or vivo (h). FRAP analysis was performed using a Leica STELLARIS5 confocal microscope. Photobleaching was conducted at 90% laser power with an excitation wavelength of 488 nm. Bar = 2 μm. g, i Fluorescence intensity statistics were calculated in (f) or (h). Error bars represent mean ± SD, n = 3 droplets in (g), n = 6 droplets in (i). The fluorescent signals in (a–e, f, h) were observed using a Leica STELLARIS5 microscope.
To further investigate the phase separation properties of AtMCM10, we employed a multi-faceted computational and experimental approach. Analysis with MolPhase (https://molphase.sbs.ntu.edu), a machine-learning predictor that evaluates sequence-derived physicochemical motifs, indicated an exceptionally high LLPS propensity score of 0.996 (threshold = 0.5) for AtMCM10 (Supplementary Fig. 8)46. To corroborate these computational findings, we carried out in vitro reconstitution assays under polyethylene glycol (PEG)-induced molecular crowding conditions. Remarkably, AtMCM10 readily formed numerous spherical condensates under crowding conditions (Fig. 4d), which exhibited characteristic liquid-like behaviors including: (1) sensitivity to 1,6-HD treatment (Fig. 4d), (2) dynamic fusion events between droplets (Fig. 4e), and (3) rapid fluorescence recovery in fluorescence recovery after photobleaching (FRAP) assays both in vitro and in planta (Fig. 4f–i). These comprehensive analyses establish AtMCM10 as a bona fide phase-separating protein with dynamic liquid-like properties.
Taken together, these findings demonstrate that AtMCM10 undergoes LLPS in response to DNA damage, potentially facilitating its role in SDSA-mediated intermolecular HR repair.
Mechanisms driving AtMCM10 phase separation
We next explored the intrinsic determinants of AtMCM10 phase separation during DNA repair. Intrinsically disordered regions (IDRs) often mediate dynamic and multivalent interactions that frequently promote liquid-liquid phase separation47,48. Bioinformatic analysis of AtMCM10 with IUPRED3 (https://iupred.elte.hu) identified two prominent IDRs: an N-terminal IDR1 (1-85 aa) and a C-terminal IDR2 (287-396 aa) (Supplementary Fig. 9a). To assess their contribution to LLPS, we generated truncation variants of AtMCM10 (Supplementary Fig. 9b). In vitro phase-separation assays revealed that deletion of IDR1 had minimal impact on condensation, whereas deletion of IDR2 completely abolished liquid-like droplet formation (Fig. 5a). Consistently, IDR2-GFP, but not IDR1-GFP, formed liquid-like droplets in vitro (Fig. 5a). These findings indicate that IDR2 is the principal contributor to AtMCM10 LLPS in vitro.
a Phase separation assays of truncated AtMCM10s in vitro. AtMCM10 was truncated into IDR1, IDR2, ΔIDR1, ΔIDR2, and ΔIDR1/2 fragments. Bars = 5 μm. b The subcellular localizations of AtMCM10-ΔIDR1, ΔIDR2 and ΔIDR1/2 in Nicotiana benthamiana. The mNeon-MCM10-ΔIDR1, ΔIDR2 and ΔIDR1/2 were infiltrated into Nicotiana benthamiana leaves for transient expression. The transfected leaves were treated with 150 mM NaCl for 24 h. Bar = 5 (top) or 25 (bottom) μm. c Statistical analyses of AtMCM10 puncta upon NaCl treatment in (b). Data were presented as median with interquartile range, whiskers extending to 1.5× the interquartile range, and individual data points plotted as dots (n = 25 nuclei), and lowercase letters indicate significant differences by one-way ANOVA followed by Tukey’s HSD test (p ≤ 0.05). d Conditions for driving AtMCM10 phase separation in vitro. The ssDNA and dsDNA were labeled with Cy5.5 at 5’ end, and reactions were performed in annealing buffer. Bars = 5 μm. The fluorescent signals in (a, b, d) were observed using a Leica STELLARIS5 microscope.
To assess their LLPS behavior in planta, we examined puncta formation of the truncation variants in Nicotiana benthamiana leaves after 24 h of high salinity treatment. Corroborating the in vitro results, deletion of IDR2 or both IDRs abolished nuclear puncta formation after 24 h of NaCl treatment, suggesting that IDR2 is essential for AtMCM10 LLPS in vivo (Fig. 5b, c). Intriguingly, while IDR1 deletion minimally affected droplet formation in vitro, AtMCM10-ΔIDR1 did not form nuclear puncta in planta, indicating that IDR1 is necessary for LLPS in a physiological context (Fig. 5b, c). Notably, variants lacking IDR2 or both IDRs exhibited pronounced cytosolic as well as nuclear signals (Fig. 5b, c), suggesting that the nuclear localization signal (NLS) within IDR2 contributes to proper nuclear targeting of AtMCM10.
To further elucidate the molecular cues that trigger AtMCM10 LLPS during DNA repair, we established an in vitro reconstitution system mimicking key steps of HR-mediated repair. Under control conditions, AtMCM10 exhibited a homogeneous distribution in solution (Fig. 5d). Initial experiments using Cy5.5-labeled 48 bp dsDNA fragments (simulating blunt-ended DSBs) failed to induce LLPS, instead triggering protein aggregation (Fig. 5d), indicating that intact DSB termini cannot initiate proper AtMCM10 compartmentalization. Given that HR repair involves resection to generate 3’ ssDNA overhangs49, and that AtMCM10 preferentially binds ssDNA8, we next tested 48-nt Cy5.5-ssDNA. This substrate efficiently promoted discrete, spherical AtMCM10 condensates (Fig. 5d), demonstrating that resected ssDNA intermediates, rather than the initial DSB, may serve as the physiological trigger for AtMCM10 LLPS. Importantly, the AtMCM10-ΔIDR2 variant lost this responsive capacity, forming aggregates rather than droplets upon ssDNA addition (Fig. 5d), further confirming the essential role of IDR2 in proper phase separation.
Together, these findings reveal that IDRs and ssDNA cooperatively drive AtMCM10 LLPS, establishing a mechanistic link between its disordered regions and DNA damage response.
IDRs are critical for AtMCM10 function in DNA damage response
To further investigate the functional relevance of IDRs in DNA damage response, we expressed AtMCM10-ΔIDR1, AtMCM10-ΔIDR2, or AtMCM10-ΔIDR1/2 variants fused with GFP in the Atmcm10-1 background. Consistent with the observations in Nicotiana benthamiana leaves, AtMCM10-ΔIDR2 and AtMCM10-ΔIDR1/2 variants showed pronounced cytosolic and nuclear signals in Arabidopsis root tip cells and failed to undergo phase separation upon high-salinity treatment (Fig. 6a, b; Supplementary Fig. 10). Similarly, AtMCM10-ΔIDR1 also did not form nuclear puncta under stress (Fig. 6a, b; Supplementary Fig. 10), confirming that both IDR1 and IDR2 are indispensable for AtMCM10 LLPS in vivo in response to DNA damage.
a The subcellular localizations of truncated AtMCM10s upon NaCl treatment in the root tip cells of Arabidopsis. The pro35S:GFP:MCM10/Atmcm10-1, pro35S:GFP:MCM10-ΔIDR1/Atmcm10-1, pro35S:GFP:MCM10-ΔIDR2/Atmcm10-1, and pro35S:GFP:MCM10-ΔIDR1/2/Atmcm10-1 transgenic lines were treatment with or without 150 mM NaCl for 24 h. Bars = 5 μm. b Statistical analyses of AtMCM10 and variants puncta upon NaCl treatment in (a). Data were presented as median with interquartile range, whiskers extending to 1.5× the interquartile range, and individual data points plotted as dots (n = 25 nuclei). *** indicate significant differences by one-way ANOVA followed determined by Tukey’s HSD test (p ≤ 0.01). c Representative phenotypes of pro35S:GFP:MCM10/Atmcm10-1, pro35S:GFP:MCM10-ΔIDR1/Atmcm10-1 and pro35S:GFP:MCM10-ΔIDR2/Atmcm10-1 upon Zeb or NaCl treatment. Seeds were germinated on 1/2 MS medium with or without 5 mg/L Zeb or 100 mM NaCl, and were grown for 10 d. Bar = 1 cm. d Statistical analyses of primary root lengths of seedlings presented in (c). Data were presented as mean ± SD (n = 16 seedlings), and lowercase letters indicate significant differences by one-way ANOVA followed determined by Tukey’s HSD test (p ≤ 0.05). e A proposed working model of AtMCM10 in DNA damage response. During SDSA-mediated intermolecular HR repair, Zeb, MMC or high salinity stress induces DSBs that primarily activate ATM kinase, initiating a repair cascade through phosphorylation of downstream effectors. The process involves three coordinated mechanistic steps: (1) end resection generates ssDNA overhangs at DSB sites, (2) these ssDNA intermediates nucleate AtMCM10 LLPS, and (3) the resulting biomolecular condensates could create a specialized microenvironment that facilitates efficient strand annealing following RAD51-mediated invasion. Created in BioRender. Yang, Y. (2025) https://BioRender.com/ex5o3v5.
We next assessed the physiological consequences of IDRs disruption by analyzing the phenotypes of these transgenic lines. None of the IDR-deletion variants were able to rescue the hypersensitivity of Atmcm10-1 seedlings to Zeb or high-salinity stress (Fig. 6c, d), indicating that the N- and C-terminal IDRs are indispensable for AtMCM10 function during DNA repair.
Collectively, our findings establish IDRs as critical determinants that couple AtMCM10 phase separation and annealing activity with its role in SDSA-mediated intermolecular HR under genotoxic conditions.
Discussion
Collectively, our findings establish a comprehensive mechanistic model wherein ssDNA-triggered phase separation of AtMCM10 plays an essential role in the DNA damage response (Fig. 6e). We propose that during SDSA-mediated intermolecular HR repair, genotoxic or high salinity stress induces DSBs that primarily activate ATM kinase, initiating a repair cascade through phosphorylation of downstream effectors. The process involves three coordinated mechanistic steps: (1) end resection generates ssDNA overhangs at DSB sites, (2) these ssDNA intermediates nucleate AtMCM10 LLPS, and (3) the resulting biomolecular condensates could create a specialized microenvironment that facilitates efficient strand annealing following RAD51-mediated invasion. This model provides a structural framework for understanding phase separation in plant genome maintenance and suggests that AtMCM10’s LLPS property may enhance its ssDNA annealing activity to facilitate SDSA repair.
MCM10 is a highly conserved protein essential for genome stability, implicated in DNA repair through replication fork stabilization or interactions with repair-associated proteins9,10,11,12. Its direct role in repair remains unclear because knockout mutants are lethal in animals and yeast, obscuring replication-independent functions. In contrast, Arabidopsis MCM10 deletion mutants do not display overt replication defects, providing a tractable system to investigate the replication-independent functions of MCM10 in DNA damage repair. Unlike yeast and mammals, Atmcm10 were not hypersensitive to HU, which depletes dNTPs and stalls forks (Supplementary Fig. 2a). By contrast, Atmcm10 were hypersensitive to Zeb (Fig. 1a, b, e–h), a cytidine analog that traps DNA methyltransferases and generates replication-blocking protein-DNA crosslinks. This indicates that AtMCM10 is crucial for coping with replication-blocking lesions rather than nucleotide depletion-induced stalling. In addition, Atmcm10 exhibited sensitivity to other DNA-damaging agents, such as MMC and NaCl stress (Fig. 1a–d). Together, these results highlight a DNA repair-specific role for MCM10 in Arabidopsis, distinct from its canonical function in replication fork stabilization in yeast and animals, and establish this model as an advantageous system to elucidate its mechanistic contribution to DNA repair.
HR is a fundamental DNA damage repair pathway utilized by all organisms to maintain genome stability, with core mechanisms highly conserved across species18,19,22. ATM and ATR act as key upstream regulators that orchestrate HR by sensing DNA damage and coordinating downstream repair events. Our findings suggest that AtMCM10 primarily acts downstream of ATM in response to Zeb-induced DSBs, whereas under salt-induced genotoxic stress, its regulation may involve both ATM- and ATR-mediated signaling (Fig. 2a–d). This divergence likely reflects the multifaceted nature of salt stress, which provokes diverse cellular perturbations and activates multiple signaling cascades beyond canonical DNA damage signaling. Such context-dependent regulation implies that AtMCM10 integrates signals from distinct upstream kinases to safeguard genome integrity under various stress conditions.
HR is also critical during meiosis, playing an essential role in fertility and genetic diversity, with profound implications for genome evolution50. In Arabidopsis, the atr atm double mutant and rad51 single mutant are completely sterile51. Although meiotic recombination shares a common set of factors with somatic recombination, it operates through distinct mechanisms52,53. Here, we demonstrate that AtMCM10 functions primarily downstream of ATM in DNA damage response in somatic cells (Fig. 2a–d). The root length of atr Atmcm10-1 on 1/2 MS medium is similar to that of atr atm and is shorter than WT (Fig. 2a), supporting that AtMCM10 is involved in DNA repair in somatic cells. However, the atr Atmcm10-1 double mutant exhibits normal fertility, implying that AtMCM10 is not required for meiotic recombination, or that other proteins may compensate for its annealing function during meiosis.
ssDNA annealing is a pivotal step in HR, and this process is primarily mediated by RAD52 during SSA repair54. However, the role of RAD52 in ssDNA annealing during SDSA remains controversial. In yeast, RAD52 facilitates SDSA by recruiting RAD51 to ssDNA and promoting second ssDNA capture and anneal27; whereas in mammals and plants, RAD52 knockouts exhibit no discernible phenotypic defects, and BRCA2 has partially substituted for the mediator function of RAD52 in promoting RAD51 assembly24,55,56,57. These findings suggest that while SDSA is evolutionarily conserved, its regulatory mechanisms differ between yeast and higher eukaryotes. Our analyses reveal that AtMCM10 plays a critical role in SDSA pathway during DSBs repair (Fig. 3c–e), possesses intrinsic ssDNA annealing activity (Fig. 3f–i), and AtMCM10 variants lacking the C-terminal region required for annealing fail to complement the DNA repair defects of the mutant (Fig. 6c, d). These findings suggest that AtMCM10 promotes SDSA repair via its ssDNA annealing activity, raising the question of why this function appears largely restricted to SDSA. A likely explanation lies in the cell cycle context and lesion type: Zeb-induced replication-blocking lesions arise primarily during DNA replication, which are particularly suited for SDSA repair using sister chromatids as templates. Consistently, MCM10, as a replication-associated protein, undergoes cell cycle-dependent regulation and remains abundant and chromatin-bound during S/G2 phase in yeast and humans4,58. Given that SDSA relies on sister chromatids and homologous chromosomes as templates and occurs predominantly in S/G2 phase and in endoreduplicated nuclei, AtMCM10’s involvement in SDSA is likely coordinated with DNA replication. In line with this notion, a recent study in S. cerevisiae reported that rad52 mcm10 double mutants exhibited a more severe viability defect than either single mutant59, suggesting a conserved role of MCM10 in HR. The high conservation of MCM10 across eukaryotes raises important questions about the evolutionary trajectory of this function, warranting future comparative studies.
LLPS has emerged as a crucial mechanism regulating intracellular compartmentalization that has a range of intricate roles in cellular organization47. The membraneless organelles formed by LLPS provide more flexible and adaptive response48,60. Recently, multiple proteins involved in DNA repair have been found to undergo LLPS44,61. However, the molecular determinants governing phase separation during DNA damage repair remain largely unknown. The damaged ends of DSBs are processed into 3’ ssDNA overhangs, which are initially bound by RPA to prevent degradation and subsequently replaced by RAD51 to initiate HR repair22,49. Thus, ssDNA plays a critical role in recruiting repair-related proteins to damage sites. Notably, it is reported that AtMCM10 preferentially binds ssDNA over dsDNA8. Here, we further found that ssDNA can directly drive AtMCM10 LLPS in vitro (Fig. 5d), suggesting that AtMCM10 phase separation at DSBs may be triggered by the presence of 3’ ssDNA overhangs at DSB ends.
Beyond its role in LLPS, the disordered regions of AtMCM10 may also contribute to additional HR-related functions. The C-terminal IDR2, which contains a nuclear localization signal (NLS), is essential for annealing activity and nuclear targeting (Fig. 3h, I; 5a, b; 6a, b; and Supplementary Fig. 9), highlighting the functional interplay among these processes during SDSA repair. In contrast, the N-terminal IDR1, while dispensable for LLPS in vitro, is required for proper phase separation in vivo (Fig. 5a, b; 6a, b), highlighting its critical yet indirect contribution to proper phase separation under physiological conditions. Notably, in humans, the N-terminus of MCM10 mediates its interaction with BRCA2, thereby suppressing PRIMPOL-mediated repriming during DNA damage12. Arabidopsis encodes two highly conserved BRCA2-like proteins, BRCA2A and BRCA2B, which function redundantly in HR repair62. Given that both BRCA2s and AtMCM10 act downstream of the ATM-initiated SDSA pathway, and that BRCA2s functions in repair factor recruitment at DSB sites, it is plausible that BRCA2s mediates the recruitment of AtMCM10 and promotes its phase separation—a possibility that warrants further investigation.
Methods
Plant materials and growth conditions
All Arabidopsis thaliana plants used in this work were of the Columbia ecotype. The WT, Atmcm10-1, Atmcm10-3 mutant and proMCM10:MCM10:GFP transgenic line were described previously8. The T-DNA insertion mutants atm (SALK_040423), atr (SALK_032841), ku70 (SALK_123114C) and lig4 (SALK_044027C) were purchased from NASC (The Nottingham Arabidopsis Stock Centre). The HR reporter lines 1406, 1445, and IC9C have been previously studied34,41,63. Primers used for genotyping are listed in Supplementary Table 1. The sterilized Arabidopsis seeds were grown on plates containing 1/2 Murashige and Skoog (MS) medium with 0.8% (w/v) agar and 2% (w/v) sucrose for root observation and drug treatment, and seeds were stratified at 4 °C for 2 d then grown in a growth chamber at 22 °C with 16 h light and 8 h dark.
Treatment
For phenotype observation, the sterilized Arabidopsis seeds were grown on 1/2 MS containing 5 mg/L Zeb, 8 μM MMC, 2 mM HU, 40 μM Zeocin or 100 mM NaCl for 10 d. For PI staining, 5-day-old seedlings were treatment with 5 mg/L Zeb for 48 h or 50 mM NaCl for 24 h. For comet assay, 7-day-old seedlings were treatment with 5 mg/L Zeb or 150 mM NaCl for 24 h, or recovery 6 h after 150 mM NaCl treatment for 24 h. For LLPS observation, 5-day-old seedlings were treatment with 5 mg/L Zeb or 150 mM NaCl for 24 h.
PI staining and microscopy
For PI staining, 5-day-old seedlings were treatment with 5 mg/L Zeb for 48 h or 50 mM NaCl for 24 h, and then, roots were mounted in 100 μg/ml propidium iodide solution for 1 min and visualized using a Zeiss LSM 880 confocal laser-scanning microscope after dip washing in 1× PBS.
Comet assay
Comet assay was performed using Comet Assay Kit (AbbKine, KTA3040) with some modifications. 7-day-old seedlings were treatment with 5 mg/L Zeb or 150 mM NaCl for 24 h, or recovery 6 h after 150 mM NaCl treatment for 24 h. Nuclei of root tip cells were isolated by slicing tissues in cold 1× PBS buffer with 20 mM EDTA with a new razor blade under dim light. The slides containing the nuclei were treated with pre-chilled 1× Lysis buffer for 1 h, Alkaline solution (0.3 M NaOH, 5 mM EDTA, pH 13.5) for 30 min and subjected to electrophoresis (0.7 V/cm) in 1× TBE buffer (90 mM Tris-base, 2 mM EDTA, pH 8.4) for 10 min. The comets were visualized by staining with PI, captured with a Zeiss LSM 880 confocal laser-scanning microscope.
HR assays
GUS staining was described previously64. Briefly, 10-day-old seedlings treatment with or without genotoxins or NaCl were immersed in GUS staining solution (0.1% Triton X-100; 0.5 mM K4Fe(CN)6; 0.5 mM K3Fe(CN)6; 10 mM EDTA; 0.5 mg/ml X-Gluc in PBS) and incubated at 37 °C overnight. Seedlings were washed with 75% ethanol several times and photographed under a Stereo microscope.
Annealing activity
The Annealing activity assay was performed as described previously with small modifications29. Oligo A2 was labelled with DIG by Oligonucleotide 3’-End Labeling Kit (Roche 03353575910). The 10 µL reaction contained 10 ng protein, 0.5 nM DIG-labeled oligo A2 in an annealing buffer (30 mM Tris-Ac pH 7.5, 5 mM MgAc, 1 mM DTT). Incubated at 30 °C. Oligo A1 was added to a final concentration 1 nM which marked the start of the reaction. The sequences of Oligo A1 and Oligo A2 are listed in Supplementary Table 1. Reactions were stopped by adding 3.5 µL stop buffer (3.3% SDS, 3.3 µM unlabeled oligoA2, 7 mg/ml protein K) and incubating them for 15 min at 30 °C. The mixtures were resolved on 12% TBE-PAGE at 100 V and transferred to positively charged nylon membranes. DIG signal was detected by DIG detection starter kit (Roche 11585614910) according to the manufacturer's instructions.
DAPI fluorescence-based annealing assay was performed by mixing the indicated protein and 10 nM oligo A1 in 1× annealing buffer (30 mM Tris-acetate pH 7.5, 1 mM DTT) with 50 ng/ml DAPI at 30 °C. Background fluorescence was measured before starting a reaction by adding and mixing the complemented 10 nM oligo A2. Measurements were performed using a fluorescence spectrophotometer (HITACHI F-7000) under time scan mode with the following settings: excitation at 358 nm, emission at 461 nm, 5 nm slit widths each, scan speed at 0.2 s each time.
Subcellular localization
For subcellular localization observation in Arabidopsis, the 5-day-old transgenic seedlings with GFP tag were treatment with 1/2 MS liquid with or without 5 mg/L Zeb or 150 mM NaCl for 24 h. Root tip cells were viewed with a Leica STELLARIS5 confocal laser-scanning microscope, and the GFP signal was collected under 488 nm emission wavelength.
For subcellular co-localization in N. benthamiana, mNeon GREEN-AtMCM10 and RAD51-mScarlet constructs were co-infiltrated into leaves for transient expression. After 48 h, the same leaves were infiltrated with 5 mg/L Zeb, 150 mM NaCl, or left untreated for 24 h. A Leica STELLARIS5 confocal laser-scanning microscope was used to detect fluorescence signals at the emission wavelength of 488 and 561 nm.
In vitro protein expression and purification
All fusion proteins were expressed and purified from E. coli strain BL21. In brief, protein expression was induced by 0.4 mM IPTG at 16 °C overnight, and was purified by Ni-NTA agarose (MCLAB) using Lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM Imidazole, pH 8.0) and eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM Imidazole, pH 8.0).
Protein sequence prediction
The full-length amino acid sequence of AtMCM10 was analyzed using MolPhase (https://molphase.sbs.ntu.edu), a machine-learning engine that integrates prion-like motifs, intrinsically disordered regions (IDRs), charge distribution, and hydrophobicity to predict LLPS propensity46. ANCHOR for identifying IDR domains. PScore (http://pound.med.utoronto.ca/~JFKlab/Software/psp.htm) for predicting potential Pi-Pi interaction residues. Sequence charge and hydrophobicity parameters were predicted using CIDER.
In vitro phase-separation assay
His-MCM10-GFP and truncated variants purified from the E. coli BL21 strain were diluted to the desired concentrations. For molecular crowding-induced liquid–liquid phase separation, proteins were diluted to a final concentration of 10 μM in elution buffer supplemented with 10% (w/v) PEG3350 (Sigma, 25322-68-3). For annealing reaction-induced LLPS, proteins at a final concentration of 10 μM were mixed with annealing buffer (30 mM Tris-Ac pH 7.5, 5 mM MgAc, 1 mM DTT) containing either single-stranded or double-stranded DNA labeled with Cy5.5 at the 5’ end. The mixtures were then incubated at 30 °C for 5 min prior to imaging. Droplets were visualized using a STELLARIS5 confocal microscope, fluorescence signals were excited at 488 nm and collected within a 500-530 nm bandwidth.
FRAP
FRAP experiments were performed using a Leica STELLARIS5 confocal microscope. Photobleaching was performed with a 488-nm laser at 100% intensity (6 iterations for in vitro and 8 iterations for in vivo FRAP analysis). Fluorescence recovery was recorded at minimal time intervals (2.584 s). Time-lapse changes in fluorescence intensity were analyzed using ImageJ software, and the recovery curves were generated by normalizing the fluorescence intensity to the pre-bleach levels of the same region of interest.
Real-time quantitative RT-PCR
Total RNAs were extracted from 10-day-old seedlings using TRIzol (Invitrogen). Subsequently, 2 μg of RNAs were transcribed to cDNAs using the SuperScript VILO cDNA synthesis kit (Invitrogen). Real-time PCR was performed using the Step One Plus QPCR system with SYBR Premix Ex Taq mixture (Takara RR420A). The expression level was normalized against ACTIN2. Each experiment was repeated at least three times with three technical replicates each time.
Statistical analysis
Statistical analysis was performed in GraphPad Prism 8 using one-way ANOVA followed by Tukey’s multiple comparisons test. Different letters indicate statistically significant differences (p < 0.01).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The GO analysis in this study was performed using previously published RNA-seq datasets (accession code PRJNA839949; https://www.ncbi.nlm.nih.gov/sra/PRJNA839949)8. Due to file size limitations, raw microscopy data are not provided and will be made available upon request from the corresponding author. Source data are provided with this paper.
References
Wang, J. & Lindahl, T. Maintenance of genome stability. Genom. Proteom. Bioinform. 14, 119–121 (2016).
Ahmed, S. M. Q., Sasikumar, J., Laha, S. & Das, S. P. Multifaceted role of the DNA replication protein MCM10 in maintaining genome stability and its implication in human diseases. Cancer Metastasis Rev. 43, 1353–1371 (2024).
Wasserman, M. R., Schauer, G. D., O’Donnell, M. E. & Liu, S. Replication fork activation is enabled by a single-stranded DNA gate in CMG helicase. Cell 178, 600–611 e616 (2019).
Baxley, R. M. & Bielinsky, A. K. Mcm10: a dynamic scaffold at eukaryotic replication forks. Genes 8, https://doi.org/10.3390/genes8020073 (2017).
Chadha, G. S., Gambus, A., Gillespie, P. J. & Blow, J. J. Xenopus Mcm10 is a CDK-substrate required for replication fork stability. Cell Cycle 15, 2183–2195 (2016).
van Deursen, F., Sengupta, S., De Piccoli, G., Sanchez-Diaz, A. & Labib, K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 31, 2195–2206 (2012).
Campos, L. V. et al. RTEL1 and MCM10 overcome topological stress during vertebrate replication termination. Cell Rep. 42, 112109 (2023).
Zhao, X. et al. AtMCM10 promotes DNA replication-coupled nucleosome assembly in Arabidopsis. J. Integr. Plant Biol. 65, 203–222 (2023).
Mayle, R. et al. Mcm10 has potent strand-annealing activity and limits translocase-mediated fork regression. Proc. Natl. Acad. Sci. USA 116, 798–803 (2019).
Alver, R. C. et al. The N-terminus of Mcm10 is important for interaction with the 9-1-1 clamp and in resistance to DNA damage. Nucleic Acids Res. 42, 8389–8404 (2014).
Wawrousek, K. E. et al. Xenopus DNA2 is a helicase/nuclease that is found in complexes with replication proteins And-1/Ctf4 and Mcm10 and DSB response proteins Nbs1 and ATM. Cell Cycle 9, 1156–1166 (2010).
Kang, Z. et al. BRCA2 associates with MCM10 to suppress PRIMPOL-mediated repriming and single-stranded gap formation after DNA damage. Nat. Commun. 12, 5966 (2021).
Lim, H. J., Jeon, Y., Jeon, C. H., Kim, J. H. & Lee, H. Targeted disruption of Mcm10 causes defective embryonic cell proliferation and early embryo lethality. Biochim. Biophys. Acta. 1813, 1777–1783 (2011).
Wang, X. et al. S-CDK-regulated bipartite interaction of Mcm10 with MCM is essential for DNA replication. Front. Cell Dev. Biol. 12, 1420033 (2024).
Schubert, I. Boon and bane of DNA double-strand breaks. Int. J. Mol. Sci. 22, 5171 (2021).
Ciccia, A. & Elledge, S. J. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 (2010).
Hu, Z., Cools, T. & De Veylder, L. Mechanisms used by plants to cope with DNA damage. Annu. Rev. Plant Biol. 67, 439–462 (2016).
Herbst, J., Li, Q. Q. & De Veylder, L. Mechanistic insights into DNA damage recognition and checkpoint control in plants. Nat. Plants 10, 539–550 (2024).
Heyer, W. D., Ehmsen, K. T. & Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113–139 (2010).
Lieber, M. R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010).
Sonoda, E., Hochegger, H., Saberi, A., Taniguchi, Y. & Takeda, S. Differential usage of non-homologous end-joining and homologous recombination in double-strand break repair. DNA Repair 5, 1021–1029 (2006).
Li, J. et al. Pathways and assays for DNA double–strand break repair by homologous recombination. Acta Biochim. Biophys. Sin. 51, 879–889 (2019).
Kumar, R., Duhamel, M., Coutant, E., Ben-Nahia, E. & Mercier, R. Antagonism between BRCA2 and FIGL1 regulates homologous recombination. Nucleic Acids Res. 47, 5170–5180 (2019).
Seeliger, K., Dukowic-Schulze, S., Wurz-Wildersinn, R., Pacher, M. & Puchta, H. BRCA2 is a mediator of RAD51- and DMC1-facilitated homologous recombination in Arabidopsis thaliana. N. Phytol. 193, 364–375 (2011).
Grelon, M. et al. RAD54 is essential for RAD51-mediated repair of meiotic DSB in Arabidopsis. PLOS Genet. 17, e1008919 (2021).
Shioi, T. et al. Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site. Nature 628, 212–220 (2024).
Sugiyama, T., Kantake, N., Wu, Y. & Kowalczykowski, S. C. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. EMBO J. 25, 5539–5548 (2006).
Rossi, M. J., DiDomenico, S. F., Patel, M. & Mazin, A. V. RAD52: paradigm of synthetic lethality and new developments. Front. Genet. 12, https://doi.org/10.3389/fgene.2021.780293 (2021).
Sugiyama, T., New, J. H. & Kowalczykowski, S. C. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. USA 95, 6049–6054 (1998).
Liu, C.-H. et al. Repair of DNA damage induced by the cytidine analog zebularine requires ATR and ATM in Arabidopsis. Plant Cell 27, 1788–1800 (2015).
Dvorak Tomastikova, E. et al. The interplay of homology-directed repair pathways in the repair of zebularine-induced DNA-protein crosslinks in Arabidopsis. Plant J. 119, 1418–1432 (2024).
Roy, S., Choudhury, S. R., Sengupta, D. N. & Das, K. P. Involvement of AtPollambda in the repair of high salt- and DNA cross-linking agent-induced double strand breaks in Arabidopsis. Plant Physiol. 162, 1195–1210 (2013).
Spadafora, N. D. et al. Arabidopsis T-DNA insertional lines for CDC25 are hypersensitive to hydroxyurea but not to zeocin or salt stress. Ann. Bot. 107, 1183–1192 (2011).
Mahapatra, K. & Roy, S. SOG1 and BRCA1 interdependently regulate RAD54 expression for repairing salinity-induced DNA double-strand breaks in Arabidopsis. Plant Cell Physiol. 65, 708–728 (2024).
Boyko, A., Hudson, D., Bhomkar, P., Kathiria, P. & Kovalchuk, I. Increase of homologous recombination frequency in vascular tissue of Arabidopsis plants exposed to salt stress. Plant Cell Physiol. 47, 736–742 (2006).
Culligan, K. M., Robertson, C. E., Foreman, J., Doerner, P. & Britt, A. B. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48, 947–961 (2006).
Blackford, A. N. & Jackson, S. P. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell 66, 801–817 (2017).
Sekiguchi, J. et al. Genetic interactions between ATM and the nonhomologous end-joining factors in genomic stability and development. Proc. Natl. Acad. Sci. USA 98, 3243–3248 (2001).
Balmus, G. et al. ATM orchestrates the DNA-damage response to counter toxic non-homologous end-joining at broken replication forks. Nat. Commun. 10, 87 (2019).
Dvořák Tomaštíková, E. et al. The interplay of homology-directed repair pathways in the repair of zebularine-induced DNA–protein crosslinks in Arabidopsis. Plant J. 119, 1418–1432 (2024).
Gherbi, H. et al. Homologous recombination in planta is stimulated in the absence of Rad50. EMBO Rep. 2, 287–291 (2001).
Swoboda, P., Gal, S., Hohn, B. & Puchta, H. Intrachromosomal homologous recombination in whole plants. EMBO J. 13, 484–489 (1994).
Mishra, A. P. et al. BRCA2-DSS1 interaction is dispensable for RAD51 recruitment at replication-induced and meiotic DNA double strand breaks. Nat. Commun. 13, 1751 (2022).
Liu, Q. et al. The histone methyltransferase SUVR2 promotes DSB repair via chromatin remodeling and liquid-liquid phase separation. Mol. Plant 15, 1157–1175 (2022).
Krejci, L., Altmannova, V., Spirek, M. & Zhao, X. Homologous recombination and its regulation. Nucleic Acids Res. 40, 5795–5818 (2012).
Liang, Q. et al. MolPhase, an advanced prediction algorithm for protein phase separation. EMBO J. 43, 1898–1918 (2024).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Alberti, S., Gladfelter, A. & Mittag, T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019).
Symington, L. S. Mechanism and regulation of DNA end resection in eukaryotes. Crit. Rev. Biochem Mol. Biol. 51, 195–212 (2016).
Mercier, R., Mezard, C., Jenczewski, E., Macaisne, N. & Grelon, M. The molecular biology of meiosis in plants. Annu Rev. Plant Biol. 66, 297–327 (2015).
Li, W. et al. The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. USA 101, 10596–10601 (2004).
Cloud, V., Chan, Y. L., Grubb, J., Budke, B. & Bishop, D. K. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337, 1222–1225 (2012).
Chen, H. et al. RAD51 supports DMC1 by inhibiting the SMC5/6 complex during meiosis. Plant Cell 33, 2869–2882 (2021).
Liang, C. C. et al. Mechanism of single-stranded DNA annealing by RAD52-RPA complex. Nature 629, 697–703 (2024).
Hanamshet, K., Mazina, O. M. & Mazin, A. V. Reappearance from obscurity: mammalian Rad52 in homologous recombination. Genes 7, https://doi.org/10.3390/genes7090063 (2016).
Samach, A., Melamed-Bessudo, C., Avivi-Ragolski, N., Pietrokovski, S. & Levy, A. A. Identification of plant RAD52 homologs and characterization of the Arabidopsis thaliana RAD52-like genes. Plant Cell 23, 4266–4279 (2011).
Rijkers, T. et al. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell Biol. 18, 6423–6429 (1998).
Izumi, M., Yatagai, F. & Hanaoka, F. Localization of human Mcm10 is spatially and temporally regulated during the S phase. J. Biol. Chem. 279, 32569–32577 (2004).
Masnovo, C. et al. Stabilization of expandable DNA repeats by the replication factor Mcm10 promotes cell viability. Nat. Commun. 15, 10532 (2024).
Field, S., Jang, G. J., Dean, C., Strader, L. C. & Rhee, S. Y. Plants use molecular mechanisms mediated by biomolecular condensates to integrate environmental cues with development. Plant Cell 35, 3173–3186 (2023).
Oshidari, R. et al. DNA repair by Rad52 liquid droplets. Nat. Commun. 11, 695 (2020).
Siaud, N. et al. Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. EMBO J. 23, 1392–1401 (2004).
Gao, J. et al. NAP1 family histone chaperones are required for somatic homologous recombination in Arabidopsis. Plant Cell 24, 1437–1447 (2012).
Jefferson, R. A., Kavanagh, T. A. & Bevan, M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 (1987).
Acknowledgements
We thank Aiwu Dong (Fudan University) for providing reporter lines 1445/1406 and IC9C for the research. This work was supported by China Postdoctoral Science Foundation (2023M741146) to Xinjie Zhao, the Hunan Provincial Natural Science Foundation of China (2024JJ6312) to Xinjie Zhao, the Project of Education Department of Hunan Province of China (24B0074) to Xinjie Zhao, the National Natural Science Foundation of China (32500599) to Tianren Zhang, the Key Project of Developmental Biology and Breeding from Hunan Province (2000XKQ0203) to Dongping Li, the Natural Science Foundation of Changsha (kq2402157) to Dongping Li.
Author information
Authors and Affiliations
Contributions
X.Z. Z.G. D.L. and T.Z. conceived and designed the research. X.Z. performed most of the research. T.Z. worked on LLPS. D.J. performed DNA annealing. C.H. and S.X. constructed vectors. L.B. assisted with the generation of genetic constructs. X.H., L.H., L.T., Y.G. and Y.Z. supervised the study. X.Z, T.Z. and Y.Z. wrote the manuscript. All authors read and approved of its content.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Jonathan Dragwidge, Cecile Raynaud, Shunping Yan and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, X., Zhang, T., Jin, D. et al. AtMCM10 facilitates SDSA-mediated intermolecular homologous recombination repair via liquid-liquid phase separation in DNA damage response. Nat Commun 16, 11581 (2025). https://doi.org/10.1038/s41467-025-66705-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66705-9