Abstract
Pooled CRISPR screening enables large-scale interrogation of gene functions but typically measures simple phenotypes such as fitness. High-content methods like Perturb-seq extend dimensionality to transcriptomics but are costly and limited in scope. Optical pooled screening (OPS) combines pooled CRISPR screening with imaging to yield scalable, information-rich readouts, yet existing implementations remain pathway-specific. Here we describe an OPS-compatible Cell Painting platform that enables hypothesis-free reverse genetic screening through multiplexed morphological profiling. We validate this technique using a well-defined morphological gene set, compare classical image analysis to self-supervised learning methods using a mechanism-of-action library, and perform discovery screening with a druggable genome library. By combining rich morphological data with deep learning, gene networks emerge without the need for target-specific biomarkers, leading to unbiased discovery of gene functions.
Similar content being viewed by others
Introduction
CRISPR-based genetic screens allow researchers to causally connect genes to their cellular phenotypes and functions. While such studies can be conducted in an arrayed format, pooled CRISPR screening methodologies, stemming from seminal work utilizing barcoded shRNA technology1,2, are generally more cost-effective and scalable3. Pooled CRISPR screens typically assay for low dimensional readouts, such as cell survival4,5,6,7,8 or sortable fluorescent biomarkers9,10,11,12. While these CRISPR screens can produce valuable insights, the limited dimensionality of readouts leads to a reliance on a well-defined phenotypic marker, which prevents hypothesis-free exploration. Perturb-seq was developed to combine high dimensional single cell RNAseq phenotypes with pooled CRISPR screening13,14,15,16,17; however, it is cost prohibitive at large scales. In addition, transcriptomic data do not fully capture cell state and morphological data can provide complementary information in mechanism-of-action (MOA) predictions18.
Recently, pooled optical screening methods have employed bespoke phenotypic assays designed for studying specific biological questions such as the NFκB pathway, antiviral response, cytoskeletal organization, and essential genes19,20,21,22. In contrast, a generic morphological assay would enable hypothesis-free biological exploration. Inspired by the Cell Painting assay23, which has been successfully applied towards clustering genes by similar functions24, virtual screening for small molecules25 and MOA prediction18,26, we sought to address the current incompatibility between pooled optical screening and Cell Painting, which includes the spectral collision between Cell Painting and 4-color in situ sequencing (ISS) and the RNA degradation caused by the Cell Painting workflow. By combining Cell Painting with pooled optical screening, we aim to build a platform that would provide datasets conducive for machine learning (ML)27, enable unbiased mapping of genes to functions, and help improve drug discovery.
We report here Cell Painting Pooled Optical Screening in Human cells (CellPaint-POSH). We modified the original Cell Painting technique in several ways to provide compatibility with ISS. We compared self-supervised ML representation learning with classical image analysis, and demonstrated that machine learnt features achieved higher predictive performance than expert-engineered morphological features28. We demonstrated that morphological phenotypes successfully cluster genes by known functions, and that this unbiased profiling method is capable of revealing gene-gene associations and networks without explicit pathway-specific reporters.
Results
Development and optimization of CellPaint-POSH
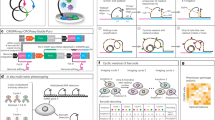
We made several modifications to optimize the Cell Painting assay23 for pooled optical screening (workflow described in Fig. 1a, detailed plasmid maps for lentiviral delivered Cas9 and single-guide RNA (sgRNA) shown in Supplementary Fig. 1a, b). First, MitoTracker used in Cell Painting is a stress-inducing live cell stain, and the sustained fluorescence of MitoTracker into the sequencing stages prohibits reliable in situ base calling. We thus developed an RNA-based label for mitochondria that we term “Mitoprobe.” Briefly, using the structure of the human mitochondrial ribosome from the Protein Data Bank (3J9M)29, we identified the most likely RNA sequences to bind to the human mitochondrial ribosome’s 12 s and 16 s ribosomal RNA (rRNA) (Fig. 1b). We then optimized on solvent-accessible surface area (SASA)30, probe length, GC content, absence of repetitive bases, and intra-sequence distance (Supplementary Fig. 1c), and conjugated the 5′ end of the resulting probe with Cy5. Simultaneous co-staining of fixed adenocarcinomic human alveolar basal epithelial (A549) cells with Mitoprobe and Tomm20 antibody gives concordant staining patterns (Fig. 1c and Supplementary Fig. 1d), demonstrating a POSH-compatible mitochondrial RNA probe that can be applied to fixed cells (Table 1), streamlining all cellular stains into a single step in the workflow. The RNA-FISH based mitochondrial label is washed out during ISS chemistry and heating steps, avoiding optical interference with 4-color sequencing. Second, the RNAse inhibitor Ribolock was implemented during all staining phases to prevent degradation of mitochondrial rRNA, sgRNA spacer-containing transcripts and other cellular RNAs. Third, we observed that subjecting cells to Cell Painting prior to reverse transcription (RT) led to degradation of RNAs and ISS signals. Moving RT prior to Cell Painting preserved RNAs and led to successful ISS (Fig. 1d).
a A general overview of the Pooled Optical Screening in Human cells platform (POSH). A pool of cells is generated that contain both constitutively-active Cas9, as well as lentivirally-delivered sgRNAs targeting a select number of genes. Cells are fixed and the sgRNA sequences are amplified via rolling circle amplification (RCA). The Cell Painting imaging assay is then conducted on the cells followed by in situ sequencing by synthesis to match image data with transfected sgRNA and gene knockout. b The protein structure of the human mitochondrial ribosome (transparent), 16 s, 12 s, and Val highlighted in teal, green, and orange, respectively. The exposed regions of the 12 s and 16 s rRNAs are highlighted in red. c 60× water-immersion confocal fluorescence images of A549s treated with Hoechst, mitoprobe, and anti-Tomm20 antibody, reveals close overlap between mitoprobe and Tomm20 without nuclear staining. Representative images from a single experiment with 6 wells per condition. d Representative images from 163,090 single cells analyzed showing the multiple Cell Painting tiles, as well as the several SBS tiles paired to the phenotypic data. rRNA ribosomal RNA, tRNA transfer RNA, Val valine.
The finalized 5-stain morphological assay panel includes Hoechst, concanavalin A (ConA), wheat germ agglutinin (WGA), phalloidin, and mitoprobe, which stain the nucleus, endoplasmic reticulum (ER), membranes, actin, and mitochondria, respectively. This leaves one vacant channel for flexible use, such as a hypothesis-specific biomarker. After imaging cells, ISS was conducted to identify the sgRNA within each cell (Fig. 1d, “Methods”).
We created a ML enabled data pipeline for high-throughput processing of phenotypic imaging and sequencing by synthesis (SBS) acquisitions in order to create a [morphological feature] * [genetic perturbation] matrix for downstream analysis (Fig. 2a). The process utilized Hoechst staining from all acquisitions to create registration tables, ensuring all morphological phenotyping and SBS acquisitions were spatially aligned and removing the need for manual coarse alignment of field of views during image acquisition, simplifying the lab process (Supplementary Fig. 2a, “Methods”). Our approach also includes an improved methodology for base-calling by training a 3-layer fully convolutional neural network (FCN) (Fig. 2a, “Methods”), which increases the deconvoluted cell recovery rate from 66.6% to 78.8% (Fig. 2b), and reduced the fraction of sgRNA miscalls that did not match the original library design at base-call threshold = 0.5 (Supplementary Fig. 2b, c, “Methods”). When compared against other state-of-the-art base-calling approaches (Starfish31 and BarDensr32), our approach shows better precision (percentage of barcodes sequenced that matched with the input sgRNA library) and cell recovery rate (percentage of segmented cells recovered with a valid barcode) (Supplementary Fig. 2d). The median amplicons detected per amplicon-presenting cell were ~2.0, ~4.0, and ~4.0 across three separate screens respectively (Fig. 2c). Samples were sequenced beyond the minimum number of nucleotides needed to call a specific sgRNA within the library (10 to 13 cycles depending on the size of the library). We targeted a minimum Hamming distance of 3 between any two sgRNAs, enabling sgRNA correction in the event of single-nucleotide mismatch. Across the 3 screens, Hamming correction recovered an additional 5.9%, 5.3%, and 34.8% of cells, respectively (Fig. 2d), making the ISS workflow more robust against occasional low-quality cycles or mistakes in base calling. We developed a simple metric for measuring the fidelity of POSH dots using the signal-to-total-ratio (STR, “Methods”), and found the quality of ISS to be consistently high across cycles of sequencing and across experiments in general (Supplementary Fig. 2e–h). A single-cell dataset is generated by cropping cell image tiles centered on each nucleus, masked by its corresponding cell mask and associated with a sgRNA identity based on the mapped barcode locations (Fig. 2a). Morphological features are determined using both classical feature extraction and self-supervised learning described below.
a A general overview of the computational pipeline for converting raw phenotyping and SBS images into usable tiles and feature matrices. SBS and readout images are registered using Hoechst staining, followed by amplicon base calling and alignment, in parallel with illumination processing, nucleus/cell segmentation, and tiling. Multiple analysis methods can then be implemented, such as deep learning-based methods or direct featurization, followed by embeddings and gene correlation calling. b % cells recovered with a valid sgRNA barcode from Feldman et al.19 (grey), our study using classical blob detector (orange), and our data using ML (green). c Number of valid amplicons per cell across the three screens. Most cells contain at least one valid amplicon. d Improvement of cell count based on Hamming correction of miscalled sgRNAs. FCN fully convolutional network, KD k-dimensional, POC proof of concept.
Proof-of-concept (POC) with morphological regulators
To demonstrate that high-content imaging using Cell Painting captures rich morphological information and can be used to classify gene functions, we set out to perform a small-scale CellPaint-POSH experiment against 124 genes with known morphological impact33. A sgRNA library was synthesized to target six key pathways: mitochondrial translation (25 genes), proteasome (27 genes), actin/kinesin (23 genes), unfolded protein response (10 genes), Golgi-ER retrograde processes (13 genes), and microtubule/dynein (19 genes), along with 7 miscellaneous genes and 400 nontargeting/intergenic control sgRNAs (Fig. 3a and Supplementary Data 1), at a representation of 10 sgRNAs per gene for robust analysis. A549 cells were processed with the CellPaint-POSH protocol with the aforementioned staining panel. We first assess the fitness effect to validate gene knockouts (KOs). As expected, DepMap-determined common essential genes34 like COPA, KIF11, and HSPA5, as well as several of the proteasome genes, had lower representation than other sgRNAs (Supplementary Fig. 3a); this provides validation that the KOs are effective and that sgRNAs are correctly sequenced and decoded.
a Summary of genes targeted in the screen in order to create distinct morphological effects. b Comparison of morphology network gene-gene edges to those from StringDB. Edges defined as strong by StringDB show similarly high edge strengths in our morphological network. Based on StringDB thresholds, n = 4736, 1961, and 563 gene-gene edges were binned as low-similarity, mid-similarity, and high-similarity, respectively. P value determined by two-sided, two-sample K–S test. White dot = median; box = 25%–75% percentile; whisker = 0%–100% percentile. Low-Mid p-value = 5.9e–28; Mid-High p-value = 1.6e–101; Low-High p-value = 5.7e–162. c AUC-ROC of CellPaint-POSH screen using StringDB network as ground truth, showing strong overlap (AUC = 0.83, TPR @ 5% FPR = 0.55). TPR true positive rate, FPR false positive rate. d UMAP projection of CellStats features showing genes clustered by their biological functions. Color and shading represents the anticipated biological grouping shown by (a), node size represents the median cosine similarity of sgRNA embeddings targeting the same gene. e Volcano plot to identify genes that show significant variation in nucleus size compared to intergenic control sgRNAs. Insets show example tile images of the nuc_sizehi and nuc_sizelo cells that led to the overall disruption in the distribution of nuclear sizes for their respective knockout populations. f Volcano plot of features comparing MRPL57 KOs to intergenic controls. g Histogram of the cytoplasmic 95th-percentile mitoprobe intensity feature in intergenic-sgRNA-presenting and MRPL57-sgRNA-presenting cells. h Example images of the cells with low cytoplasmic mitoprobe staining leading to the skewed distribution shown in (g). i Volcano plot of features comparing ACTB KO to intergenic controls. j Distribution of mean cytoplasmic phalloidin intensity of control and ACTB KO cells. k Example images of the cells with high cytoplasmic phalloidin staining leading to the skewed distribution shown in (j). l Volcano plot of features comparing GOSR2 KO to intergenic controls. m Distribution of cytoplasmic ConA intensity of intergenic and GOSR2 KO cells. n Example images of the cells with high cytoplasmic ConA intensity leading to the skewed distribution shown in (m). ESDB, StringDB edge strength. AUC-ROC, area under curve receiver operating characteristic. TPR true positive rate, FPR false positive rate, FDR false discovery rate, BH Benjamini–Hochberg. Representative images from 163,090 analyzed single cells were shown.
In total, 163,090 cells were analyzed using a classical image featurization engine similar to CellProfiler35 that extracted 1301 morphological features in four broad categories: (i) localized pixel intensity statistics, (ii) geometric features of cell segmentation masks, (iii) features characterizing textures that emerge from the different types of staining, and (iv) correlations of intensities across multiple channels (“Methods”). We term this classical featurization engine “CellStats.” Each unique sgRNA was assigned a “morphology signature” based on the mean features of all cells containing that sgRNA (“Methods”). Cosine similarity was determined between these averaged feature vectors to assign similarity scores among all sgRNAs. Similarities between sgRNAs targeting the same gene were specifically compared (Supplementary Fig. 3b), with a high similarity indicating effective sgRNA cutting and strength of a KO’s morphological phenotype. sgRNA signatures were then further aggregated to the gene level. To systematically assess the accuracy of the biological network emerging from our morphological analysis against prior literature, we compared it to StringDB36, which constructs a gene-gene interaction network in which interactions are weighted by a combination of literature scrubbing, gene-gene interaction databases, co-expression analysis, and organism transfer, and used this as a ground truth comparator. We first compared low, medium, and strong edges in our network to those formed by StringDB and found a significant increase in correlation between networks at higher StringDB cutoff score (Fig. 3b). A Spearman correlation of 0.517 (p-value < 1e-5) was calculated between morphology edges and StringDB edges for which a value was assigned. We additionally determined the area under the curve of the receiver operating characteristic (AUC ROC) using StringDB edge > 0.95 to define true positives, resulting in a 0.84 AUC, indicating strong overlap with the established network for the same genes (Fig. 3c). We visualize the aggregate gene-KO embeddings using Uniform Manifold Approximation and Projection (UMAP)37 grouped by known pathway function to further illustrate the core clusters, notably recreating functional clusters corresponding to the Golgi/ER, proteasome, mitochondrial translation (2 distinct clusters), and microtubule/dynein classes (Fig. 3d). Comparison with StringDB and visual inspection of gene clustering both demonstrate that CellPaint-POSH can reconstruct known gene-gene interaction networks.
We took two approaches to further investigate the morphological features driving the gene clustering. First, we identify genes affecting a given morphological feature, such as nuclei size, by computing each KO’s Kolmogorov–Smirnov (K–S) statistic for this feature (Fig. 3e). Genetic drivers of enlarged nuclei size included gene KOs that yielded strikingly large nuclei (e.g., KIF11), or multiple nuclei within the same cell (e.g., KIF23), which are consistent with nuclear size, mitotic arrest and spindle polarity phenotypes reported on the same genes in Funk et al20. We also observed a significantly smaller nuclear size in TLN1 KO cells, likely linked to the inability of the cells to form adhesion with their substrate or neighboring cells due to lack of talin-1 protein. Other strong modifiers of nucleus size tended to fall within the Microtubule/Dynein subgroup or encode for members of the actin-related protein (Arp) family, which form key chromatin-remodeling complexes38.
In the second analysis approach, which we term “differential morphology analysis,” we examine all CellStats features modulated by a particular genetic perturbation, relative to the control cells. Differential morphological features are similar in concept to differentially expressed gene analysis commonly used in comparing two sets of transcriptomics data. As an example, we selected one gene that represented a single point in one of the tightly-connected clusters within the network (MRPL57, Fig. 3d). We conducted K–S tests between MRPL57 cell features and intergenic-targeting sgRNA controls, and found that perinuclear mitochondrial intensity, variance, and texture were the most significantly different features of MRPL57 KO (Fig. 3f, g), which matches the mitochondrial dysfunction anticipated with the KO. Representative plots of MRPL57 KO cells driving these mitochondrial scores depict the low-mitochondria phenotype (Fig. 3h). We next examined cytoskeletal protein ACTB. Interestingly, phalloidin scoring indicated a significant increase in actin presence within the cells; this provides evidence for compensatory up-regulation of other actin isoforms upon loss of β-actin39 (Fig. 3i–k). Lastly, we examine Golgi SNAP Receptor Complex Member 2 (GOSR2), whose loss of function likely leads to progressive myoclonus epilepsy in patients40. Examining the differential morphological features upon GOSR2 KO (Fig. 3l), its unique morphological phenotype was driven largely by increased intensity, variance, and texture of Golgi/ER-specific ConA staining (Fig. 3m, n), matching its likely biological mechanism41, and providing a disease phenotype that could be adapted into a disease-modifier screen.
Mapping MOA with self-supervised vision transformer learning
Encouraged by our morphology-driven POC, and inspired by previous efforts that use Cell Painting information to cluster compounds by their MOAs18,26, we next attempted to assess the performance of CellPaint-POSH at phenotypic clustering of genes by their annotated MOA, irrespective of previous reported morphological phenotypes. We curated a list of 300 genes whose gene products are targeted by tool compounds with well annotated MOAs42,43,44 (Supplementary Data 2), using a more compact library design of top 4 sgRNAs per gene (“Methods”). We observed increased gene-matched sgRNA cosine similarity when compared with the 10-sgRNA-per-gene design used in the POC screen (Supplementary Fig. 4a). We built self-supervised vision transformer (DINO-ViT) models45 that can extract meaningful image representation with no labels, and compared between the following imaging representation techniques: (1) classical morphological featurization (CellStats); (2) self-supervised DINO-ViT model, trained on ImageNet46 containing ~1.2 million natural image data: e.g., animals, vehicles, and tools (ImageNet-DINO); (3) DINO-ViT embeddings trained on ~1.5 M single cell Cell Painting images from the 300-gene MOA experiment (CP-DINO 300, “Methods”). All three featurization methods are capable of morphologically classifying many genetic perturbations from intergenic control cells (Fig. 4a, b and Supplementary Fig. 4c, d), including ImageNet-DINO, which is not trained on cellular morphology data. This is consistent with previous reports of transfer learning47. Nevertheless, CP-DINO 300 trained on bioimaging data yielded a more informative embedding that has higher median prediction area under the receiver operating characteristic curve (AU-ROC) than the other two models (Fig. 4a–c and Supplementary Fig. 4b, f), and correctly classified more perturbations with significant phenotypic differences between intergenic controls (Fig. 4c, “Methods”). CP-DINO 300 also recovered more known biological associations from StringDB as measured by cosine similarity of the aggregate gene-KO embeddings (“Methods”) than the other two models (Fig. 4d).
a, b Comparison of feature embedding methodologies based on median AUC of binary classification of KO from intergenic controls for each genetic perturbation. a CP-DINO 300 vs. CellStats. b CP-DINO 300 vs. ImageNet-DINO. c Number of genetic perturbations classified at AUC > 0.6 by different representation models. d True positive StringDB edges predicted (at 5% false positive rate) performance using gene-gene similarity calculated from different representation models. e UMAP projection of DINO-ViT features showing genes (filtered by AUC > 0.53) clustered by their cellular component localization. Color and shading represents Leiden communities, node size represents the median similarity of aggregate sgRNA embeddings targeting the same gene. Cell tile images and top neighbors of select KOs (bolded) are shown with their respective cosine similarities. AUC area under curve.
Similar to CellStats, CP-DINO 300 representation of the Cell Painting assay allows reconstruction of gene networks, and the identification of pathway components in a hypothesis-free way using Leiden community detection (Fig. 4e)48. Specifically, we were able to reconstruct the genetic modifiers of glycoprotein biosynthesis, mitochondrial translation, actin/cytoskeletal organization, lipid metabolism, PI3K/Akt activation, mTORC1 signaling, and PRC2 complex, where morphological similarity associates genes from the same pathways/protein complexes. For example, genes with the highest similarities to SUZ12 included the other two PRC2 complex members EZH2 and EED49 (Fig. 4e); genes similar to mTORC1 activator RHEB included other pathway activators MTOR and PDPK150. Interestingly, the network was capable of clustering key components of the lipogenesis pathways without the use of an explicit lipid stain, including the core fatty acid synthesis enzymes (ACLY: ATP Citrate Lyase, ACACA: Acetyl-CoA Carboxylase Alpha, and FASN: Fatty Acid Synthase), and upstream AKT signaling regulators (PIK3C3: PI3K Regulatory Subunit 3, PIK3R4: PI3K Regulatory Subunit 4) that all contribute to lipogenesis51 (Fig. 4e). Representative images of gene KOs with the most distinguished phenotypes from several clusters are shown (Fig. 4e).
Deep learning-enabled discovery via druggable genome pooled optical screen
We next sought to scale CellPaint-POSH to the druggable genome to demonstrate the throughput needed for discovery screens, and to further assess the generalizability of Cell Painting across more diverse sets of genes and pathways. To this end, a library of 1640 genes was assembled based on the Tier 1 list of the Druggable Genome library52, spiked in with a subset of morphological genes and known mTORC1 pathway genes for controls (Supplementary Data 3). In addition to Cell Painting, anti-phospho-S6 (pS6) antibody with DyLight 755-conjugated secondary antibody was used in the 6th channel as an established biomarker53 used extensively in mTORC1 studies, including genome-wide screens10,54,55. In order to improve throughput and reduce labor-intensive manual work, we built and deployed fully automated liquid handling and microscopy for the ISS workflow (“Methods”, video demonstration in Supplementary Movie 1). We formally assessed sgRNA library abundances as determined by ISS with that measured by genomic DNA next-gen sequencing (NGS), by comparing the enrichment and depletion of sgRNAs targeting fitness genes. We again re-discovered known essential genes34, and the measured fitness effects were comparable between POSH and NGS (Supplementary Fig. 5a–c), indicating that there is little detection bias in POSH ISS.
We explored if the added diversity in the 1640-Gene druggable genome POSH dataset would lead to better self-supervised image representation. To that end we trained CP-DINO 1640, and found that it performs better than CP-DINO 300 at binary classification of genetic KOs vs. negative controls in the 1640 dataset (Fig. 5a, b), and that it captures more semantically meaningful structure in the data as demonstrated by its more accurate predictions of StringDB gene-gene associations than CP-DINO 300, ImageNet-DINO, or CellStats (Fig. 5c). Additionally, we compared a recently published DL model trained on single-cell segmented Cell Painting images56 with CP-DINO-1640 on the StringDB evaluation task on MOA-300 and Druggable Genome datasets (Supplementary Fig. 5d). Our model performed comparably despite being trained on a smaller dataset (~1.5 million vs. ~8 million cells). Lastly, in order to improve the binary classification (intergenic vs. gene KOs) accuracy despite label noise inherent in the single cell dataset (e.g., due to incomplete CRISPR editing), we explored using multiple instance learning57,58 by using the mean embedding over bags of 10 cells for the classification task instead of single cell embeddings. We found that bagged mean embeddings greatly improved the classification AUC compared to single cell embeddings across all gene KOs (Supplementary Fig. 5e).
a Comparison of feature embedding methodologies (CP-DINO 1640 vs. CP-DINO 300) based on median AUC of binary classification of KO from intergenic controls for each genetic perturbation. b Number of genetic perturbations classified at AUC > 0.6 by different representation models. c True positive StringDB edges predicted (at 5% false positive rate) from 1640 druggable genome dataset using gene-gene similarity calculated from different representation models. d True positive StringDB edges predicted (at 5% false positive rate) from a held-out 124 gene POC dataset using gene-gene similarity calculated from different representation models. e True positive StringDB edges predicted (at 5% false positive rate) from 300 gene MOA dataset using gene-gene similarity calculated from different representation models. f UMAP projection of DINO-ViT features showing genes (filtered by AUC > 0.54) clustered by their biological functions. Color and shading represents Leiden communities, node size represents the median similarity of aggregate sgRNA embeddings targeting the same gene. Cell tile images and top neighbors of select KOs (bolded) are shown.
We next evaluated all image representation models on held out data: the 124-gene morphology POC perturbation dataset. While all 3 self-supervised DINO-ViT models showed an ability to generalize out of experiment and outperform CellStats, CP-DINO 1640 in particular surpassed prediction accuracy from CP-DINO 300 and ImageNet-DINO at predicting StringDB gene-gene associations and physical interactions (Fig. 5d and Supplementary Fig. 5f). The superior performance of CP-DINO 1640 suggests a low likelihood of significant overfitting or trivial memorization, as the 1640-genes druggable genome library and 300-gene MOA library share similar numbers of overlapping genes with the 124 POC library (30 and 26 genes, respectively). When evaluated on the 300-MOA dataset, both CP-DINO 1640 and CP-DINO 300 performed similarly in StringDB prediction (Fig. 5e), likely because the latter already learned the full variance in the corresponding dataset.
To further test generalizability, we applied CP-DINO 1640 in a zero-shot setting to a large-scale, external dataset: the recently published genome-wide CRISPR Cell Painting dataset PERISCOPE59. We benchmarked our model against the exact analysis used in the PERISCOPE study, providing a direct measure of robustness to overfitting and ability to generalize across laboratories and protocols. On this dataset, CP-DINO 1640 detected 2,812 genetic perturbations (at 5% FDR, “Methods”) with significant phenotypic changes (Supplementary Fig. 5g)—2.5× more than the standard, non-deep-learning CellProfiler features to the same data with the same FDR threshold (Fig. 4a, Ramezani et al. 2025). In addition, CP-DINO 1640 embeddings captured more biologically meaningful structures, as evidenced by a higher overlap with known protein-protein interactions from StringDB and CORUM compared to other baselines (Supplementary Fig. 5h–j, “Methods”). These results demonstrate that our model’s improved sensitivity translates into greater power for uncovering biologically relevant relationships.
We constructed gene networks from the druggable genome dataset using CP-DINO 1640 model and Leiden community detection (Fig. 5f). Overall, many genetic pathways and known protein complexes emerged from network analysis of imaging features from the unbiased morphological profiling. As an example, the EP300 complex CREBBP and EP300 cluster together60,61,62,63. As another example, top related genes to TGFBR1 included other known TGF-β genes such as TGFBR2, TGFBR3, and SMAD3 (Fig. 5f)64. The TGFBR cluster also includes NCSTN and PSENEN, both of which encode for the gamma-secretase complex components, consistent with reports indicating that TGF-β receptors are substrates of γ-secretase65,66,67. Genes with the highest similarities to an essential gene POLD1 include COPS5, ACTR10, PLK4, CDK7, TOP2A, and TUT1. Interestingly, while they share similar KO phenotypes, these genes perform different biological functions (e.g., TUT1 encodes a nucleotidyl transferase that functions as both a terminal uridylyltransferase and a nuclear poly(A) polymerase, while PLK4 is involved in centriole duplication). We note that most of these are reported essential genes34 and confirmed by sgRNA abundances in our experiment (Supplementary Fig. 5b, c). Indeed, PLK4 is a known essential regulator of cell cycle68, and it is possible that nucleotidyl transferase TUT1 may also play an important role in cell division. Consistent with this hypothesis, CellStats analysis suggested that TUT1 KO induces large nuclei size and cell size (Fig. 3e and Supplementary Fig. 5k). In addition, Funk et al. recently reported both TUT1 and PLK4 KOs produce larger cell area, nucleus area, and nuclear DNA integrated intensity, consistent with the phenotype of cell cycle regulator genes such as CDC proteins20. Both Deep Learning (DL) and CellStats analysis thus nominate the potential role of TUT1 in cell cycle regulation.
Another major pathway that emerged from gene network analysis is mTORC1, where known mTORC1 complex proteins RPTOR and mTOR, form a tight cluster (Fig. 5f)69. Other genes that share the highest phenotypic similarity with mTORC1 include known regulators of the mTOR pathway, such as PDPK1, mLST8, and RHEB. In contrast, mTORC1 inhibitors TSC1, TSC2, DDIT4, and TBC1D7 cluster away from mTORC1 activators as expected (Fig. 5f).
Given the clustering of mTORC1 regulators, we asked if the inclusion of the pathway biomarker pS6 may improve the discovery of relevant genetic perturbations. We thus trained CP-DINO models with Cell Painting + pS6 channel information, and found that while both CP-DINO 1640 and the new CP-pS6-DINO 1640 have similar sensitivity in predicting genetic perturbations, pS6 notably improves the detection of many mTORC1 inhibitors (TSC1, TSC2, DDIT4, TBC1D7) (Fig. 6a). Indeed, while mTORC1 activators (defined as top genes morphologically similar to mTOR) are similarly predicted with or without pS6 (Fig. 6b), interestingly pS6 improves the cosine similarities of mTORC1 downregulators with respect to TSC2, such as TBC1D7 and DDIT470,71. In order to further validate morphologically discovered mTORC1 regulators against the ground truth biomarker, we systematically analyzed genes that influence pS6 intensity with two methods: (1) a binning approach, grouping cells with similar levels of pS6 intensity changes, simulating enriched bins of cells produced by FACS-based CRISPR screens (Supplementary Fig. 6a); (2) a distribution based approach, taking full advantage of the single cell resolution of our data (Fig. 6c, “Methods”). Both analyses resulted in comparable hits, effectively identifying many of the established regulators of the mTORC1 pathway10,54 (Fig. 6d and Supplementary Fig. 6b). Most top mTORC1 activators as defined by morphological similarity to mTOR, including mTOR, PDPK1, RPTOR, mLST8 mentioned above (Fig. 6b), are also top regulators of the pS6 biomarker (Fig. 6d).
a Comparison of feature embedding methodologies (CP-DINO 1640 with and without pS6 information) based on median AUC of binary classification of KO from WT for each genetic perturbation. b Top neighbor genes to MTOR and TSC2 (mTORC1 activator and inhibitor, respectively, bolded) predicted with and without pS6 are shown. Numbers indicate cosine similarity. c Pipeline for conducting single-cell based analysis via K–S 2-sided, 2-tailed statistical test with Benjamini–Hochberg correction, in contrast to methods used for flow-based screens such as MAUDE (Fig. S6a). d Results of single-cell based, one-feature analysis of cytoplasmic phospho-S6. Teal genes are well established mTOR upregulators, orange genes are well established mTOR downregulators, black genes are candidate pS6 modifiers. FDR false discovery rate. e Representative pS6 field with segmentation. f Example pS6 and mitoprobe staining images from control, known, and potentially novel regulators of mTORC1. g Representative images (from a single experiment, with 2 technical replicates per condition) of Tomm20 and pS6 staining from intergenic, AURKAIP1, HSD17B10, and CYP11B1 arrayed ribonucleoprotein (RNP) KO cells. h, i Violin plot of Tomm20 and pS6 median cellular intensity with a two-sample K–S test (two-sided) between the intergenic knockout and AURKAIP1, HSD17B10, and CYP11B1 arrayed RNP-KO. Solid lines indicate median, dotted lines indicate first and third quartiles. pS6 phosphorylated S6 kinase.
Interestingly, mTORC1 hits discovered by the pS6 assay can be further stratified into different subclusters by generic morphological analysis (Fig. 5f). For example, certain pS6 hits GFM1, HSD17B10, MRPS5, MRPL21, and AURKAIP1 form a morphological cluster that is separate from other pS6 hits and core mTORC1 regulators (e.g., mTOR, PDPK1, and RPTOR) (Fig. 5f), suggesting that the former group of genes may regulate mTOR biology through a distinct mechanism. Based on their biological functions72,73,74, we hypothesize that mitochondrial translation and metabolism may be involved in mTORC1 regulation. While the role of mTORC1 in regulating mitochondrial function has been reported75, the reverse causation is not fully understood. We performed differential feature analysis and visual inspection on these gene KOs, and showed that genes such as AURKAIP1 and HSD17B10 not only lower pS6 levels in the cell but also have a significant impact on mitochondria-related morphological features (Fig. 6e, f and Supplementary Fig. 6c, d). Consistent with our findings, HSD17B10 was recently identified as an mTORC1 regulator in a genome-wide screen against pS610. In addition, mitochondrial translation defects caused by the deletion of another mitochondrial elongation factor mtEF4 in C. elegans and mouse have been shown to regulate mTOR and cytoplasmic translation76,77 as a compensatory mechanism to cope with mitochondrial stress. We hypothesize that other mitochondrial genes like AURKAIP1 and HSD17B10 may have an effect on mTORC1 in our experiment. Notably, mutations in HSD17B10 have been linked to recurrent seizures and intellectual disability78,79, two symptoms also commonly observed in mTORopathies80.
We used transient Cas9 nucleofection to efficiently KO AURKAIP1 and HSD17B10 (Supplementary Fig. 6e, “Methods”). Consistent with the pooled screening results, we observed in this orthogonal array-based validation experiment that AURKAIP1 and HSD17B10 gene-KOs significantly decreased mitochondrial abundances and mTORC1 activity, as measured by Tomm20 and pS6 staining intensity, respectively. In comparison, a non-hit protein, CYP11B1, does not change TOMM20 levels, and has a much weaker effect on pS6 levels than the two hit proteins (pS6 Z-scores for CYP11B1, AURKAIP1, and HSD17B10 are −0.34, −2.37, and −2.39, respectively, Fig. 6g–i).
Discussion
Pooled CRISPR screens have emerged as a powerful technique to interrogate multiple genetic interventions in parallel; they are considerably more cost-effective and scalable than arrayed screens, and are able to greatly ameliorate batch effects. CellPaint-POSH combines the benefits of pooled CRISPR screening with the rich cell state information provided by Cell Painting. While biomarker based screens can enumerate pathway regulators along a unidimensional axis (e.g., pS6 intensity), morphology analysis can identify rich structures spanning multiple biologies. Indeed, across 3 experiments, morphological data clustered genes that affect the same cellular components (e.g., proteasome, Golgi/ER, mitochondria inner membrane), molecular functions (e.g., protein glycosylation, cytoskeletal organization, fatty acid synthetic process, chromatin modification) or biological pathways (e.g., EGF, mTOR, TGFb), and was able to do so without prior hypothesis or explicit biomarkers/assays. Morphological guilt-by-association analysis also helps generate hypotheses, for example, linking uridylyltransferase to cell cycle regulation. In the same example, we note that functionally distinct genes (e.g., PLK4 and TUT1) may have similar morphological phenotypes when deleted, suggesting shared functions for genes that are not known to be related.
We also expand the Cell Painting panel with an additional channel, and show that the resulting Cell Painting+1 design enables the use of a biomarker (e.g., pS6) that provides built-in validation of morphological discoveries from the same experiment. We demonstrate that the rich morphology readout can stratify genes that manifest similarly in biomarker studies (e.g., pS6) into sub-clusters representing molecular mechanisms (e.g., core mTORC1 vs. mitochondria translation), demonstrating the benefit of coupling high-dimensional data with ML analysis. We envision that our hypothesis-free approach can be a useful tool for disease modeling and drug target discovery, particularly when the disease biological pathway or biomarker is not completely understood, similar to recent applications of perturb-seq in disease research81.
Pooled optical screening does have some important technical limitations. First, pooled optical screening, like any other high-content image-based assay, can be sensitive to imaging artifacts such as poor focus and imaging channel bleedthrough, requiring careful image QC and staining panel design. Second, ISS signals can be low in certain cell-types with lower expression levels of sgRNA spacer-containing transcripts, requiring higher cell input to compensate for lower data recovery. Third, pooled optical screening requires instance (per cell) segmentation, which is difficult in cell-types that have overlapping structures (e.g., neurons with extensive and long dendrites, hepatocytes that grow in 3D). Fourth, our current efforts are centered on loss-of-function screening and are therefore limited in the set of gene function hypotheses that they can generate. Our screening platform is designed to be modular and could be extended to work with other CRISPR constructs to accommodate gain-of-function POSH, which holds promise for broadening our understanding of genetic functions. Fifth, it is challenging to directly infer gene functions from morphological phenotypes alone, and hence functional inference (including our CP-DINO evaluation) generally relies on known biology data sources—anchoring on curated gene-gene relationships (e.g., StringDB) or associations with genes of known function. Leveraging both DL analysis for better image representation and classical image analysis for interpretability may be a good combination strategy to partially mitigate this issue, as they did in providing an explanation as to why TUT1 may be related to cell cycle regulators, for example. In addition, incorporating orthogonal readouts such as multiplexed protein/RNA detection82,83 may expand phenotypic features and enhance interpretability, and will extend the utility of the approach to studying genes with weak morphological phenotypes.
During the final preparation of this manuscript, a similar method (PERISCOPE) emerged that combined Cell Painting and pooled optical screening to build an unbiased morphological atlas59. Our work is differentiated in a number of ways that enable the broad applicability and scaling of our approach. On the experimental side, we have addressed the spectral overlap challenges between the Cell Painting panel and the ISS barcodes by developing a structural biology-inspired RNA-FISH-probe for mitochondrial staining, avoiding the need for custom-conjugation antibodies. We also developed a closed-loop automation system for ISS, which was successfully utilized in our druggable-genome discovery screen (“Methods”); this capability is important for scaling throughput and supporting process and data robustness. Computationally, our use of ML enabled improvements to many critical steps during POSH analysis, including base calling, cell segmentation, and image representation. For base calling, our approach using a 3-layer convolutional network followed by global registration and transformation of coordinates can be executed without manual alignment of field-of-view images at acquisition time and without manual parameter tuning, yielding improved sensitivity and a simplified lab workflow. Moreover, the use of ML for cell segmentation and guide calling enhanced our ability to utilize more of the cells and hence increased the throughput of our platform. Finally, we note that while we have focused our presentation on our work in A549 cells, the fundamental components of our platform are not cell-type specific. We therefore anticipate that CellPaint-POSH can be readily adapted to most cell types that could be grown and transduced in vitro.
Morphological analysis is heavily influenced by the features extracted, and we rigorously compared classical image feature extraction with self-supervised DL methods. Classical image analysis uses expert-engineered features such as intensity, texture, shape, and size of cells and their organelles. While such an approach is simple and interpretable, it incorporates human bias and does not capture the full extent of biological variability in the dataset. Deep learning is an alternative approach that is less susceptible to human bias, but existing models are trained on natural image datasets and may not align well with the statistical patterns in bioimaging data. Recently, weakly supervised DL and self-supervised methods have been used to learn embedding directly from cellular images21,56,84,85,86. Our work leverages DINO and Vision Transformer (ViT) models45, which have been shown to achieve state-of-the-art performance in learning representations from natural images. We demonstrated that self-supervised models with ViT architecture trained on single cell feature embeddings provide better predictive accuracy and improved gene interaction prediction when compared with classical single cell featurization methods and ImageNet pretrained self-supervised models. Single cell CRISPR KO image datasets are inherently noisy due to biological and technical variability. To address this, we applied multiple instance learning, demonstrating its effectiveness in improving prediction accuracy. While multiple instance learning is one viable strategy, alternative strategies such as transfer learning, feature expansion, and additional regularization techniques could further reduce noise, enhance predictive performance, and improve generalizability.
The information richness of a high throughput and high dimensionality CellPaint-POSH dataset, combined with the power of deep learning featurization, allows a diverse range of analyses to be conducted based on a single experiment. For global gene network analysis, we borrowed heavily from standard single-cell RNAseq methods and found that guilt by association was one of the most effective ways of identifying known and potentially novel components of pathways. Our work demonstrates that this approach helps validate known biology and nominate new hypotheses via a simple and uniform analysis. As a future direction, other dimensionality reduction techniques that define and explore multi-dimensional latent spaces, similar to singular value decomposition or non-negative matrix factorization used in single-cell RNAseq analysis87,88, may further improve the computational resolution of mapping gene-function relationships and help reveal pleiotropic gene function (such as in the case of TUT1).
Importantly, we also showed that the single-cell feature representation models trained from one dataset (CP-DINO 1640) generalized well to unseen datasets (124-gene POC and 300-gene MOA datasets), allowing a pre-trained model to be easily deployable in interpreting future experiments without expensive and time-consuming de-novo training every time. We note that when comparing CP-DINO 1640 with CP-DINO 300, the former captures more semantic structure in the data, even though it is trained with a similarly sized dataset as the latter (both ~1.5 million cell tile images). We reason that this is due to the greater degree of phenotypic diversity present in the 1640 dataset due to the fivefold more genetic perturbations. When applied in a zero-shot manner to an external, genome-wide Cell Painting dataset (PERISCOPE), CP-DINO 1640 identified over 2.5 times more perturbations with significant phenotypic effects than standard CellProfiler features, while producing embeddings that were more sensitive and biologically informative. These results highlight the model’s robustness across laboratory environments and experimental protocols. We predict that increasing the in-house training data size and (even more importantly) phenotypic variance—e.g., scaling to genome-wide POSH experiments or even experiments done in different cell types—may further improve the DL model and make it more generalizable. In addition, publicly available non-POSH datasets such as the JUMP Cell Painting collection89 can be incorporated into the training data. Notably, the self-supervised ViT architecture requires no labels, and thus can be trained on bioimaging datasets without upfront annotations, circumventing a bottleneck and enabling model training on a broad range of internal and external data sets. Thus, over time, we can build richer models that provide an unbiased distillation of the phenotypic landscape of cell morphology. These models hold the potential for identifying new gene-phenotype relationships and provide further insights regarding diverse pathways and biologies.
Methods
No animal, human subjects, or other regulated materials were used during the execution of this project.
sgRNA design, synthesis, and ordering
Oligo libraries were designed that targeted (1) genes that lead to morphological phenotypes (124 genes, 10 sgRNAs per gene), (2) genes with established mechanisms of action (300 genes, at least 4 sgRNAs per gene), and (3) the druggable genome (1640 genes, 4 genes per sgRNAs). The gene list was then put through a sgRNA selection pipeline that pulls sgRNAs (~20 bps long) from Brunello90 and Toronto KnockOut version 3.091 sgRNA repositories. sgRNAs targeting the same region were removed by filtering on a Levenshtein distance of less than 3 and a max subsequence match length of greater than 15. sgRNAs with a BsmBI restriction enzyme cutting site used for Golden Gate cloning were discarded. The sgRNAs were then sorted by their on-target score per gene as reported if available90. A greedy approach was used to iteratively pick N sgRNAs per gene in ascending order of the number of sgRNA designs available per gene. In order to allow for correction of at least 1 base-pair error throughout the library, we also set a hamming distance cutoff requirement of 3 between all sgRNAs and selected the sgRNAs iteratively until we reached the target number of sgRNAs for all genes in our input design list. Non-targeting and intergenic sgRNAs were then placed in each library as controls. The last step of the pipeline appends the oligo sequence flanking the sgRNA that allows cloning into our backbone, while including the dialout primers needed for library-specific amplification. Libraries were duplicated or triplicated before submitting to a vendor for synthesis, based on the size of the initial sgRNA list. This helped to prevent jackpotting and propagation of errors while also keeping the relative ratio of controls to targeting sgRNAs the same. We then ordered the library from Twist Bioscience.
Library cloning
Library pools received from Twist Bioscience were spun down at 11,000 rcf for 2 min, then resuspended to 5–10 ng/mL in IDTE pH8.0 1xTE solution (11-05-01-13, IDT). We incubated the freshly resuspended library pool for 5 min at room temperature, then vortexed for 20–30 s, then spun down for 2 min at 11,000 rcf.
All of our oligo libraries were cloned into a customized lentiviral CROPseq-like vector (plasmid backbone) of which some features include puromycin resistance, TagBFP, padlock sites, and Esp3I/BsmBI sites for cloning (Supplementary Fig. 1b)17. We chose mU6 for sgRNA expression as demonstrated in prior publications13,92. Per library to be cloned, the backbone was pre-digested using 2 µg of plasmid backbone, 2 µL of FastDigest Esp3I (BsmBI) enzyme (ThermoFisher, FD0454), 0.5 µL 100 mMM DTT (Bioworld, 3483-12-3), 2 µL of 10X FastDigest buffer, and molecular biology (molbio) grade water (46-000-CI, Corning) added to a final volume of 50 µL. The solution was then incubated in a thermocycler at 37 °C for 2 h. Then 1 µL of fast alkaline phosphatase (ThermoFisher, EF0652) was added and the sample was incubated at 37 °C for 45 min then heat inactivated at 75 °C for 5 min. We then followed the E-Gel CloneWell II Agarose Gel protocol to gel-purify the digested backbone. Sample was eluted and DNA was quantified using a Nanodrop. Typical recovery of DNA was 50–70% of the starting solution. Digested vector was then stored at −20 °C.
A 200 µL dialout PCR solution was made in molbio grade water (46-000-CI, Corning) to a final concentration of 1× Q5 PCR (M0492S, NEB), oligo pool (2.5 ng final), 1× evagreen (Biotium, 31000T), and 0.25 µM dialout primers (see Table 1). qPCR was run on the sample according to the following protocol: 1 cycle of 98 °C for 10 s; 16 cycles of 56 °C for 30 s, 50–72 °C for 20–30 s, and 72 °C for 30 s; 1 cycle of 72 °C for 120 s. qPCR cycles were run until the last sample entered the exponential phase, up to a maximum of 16 cycles. The PCR product was cleaned using SPRIbeads (Beckman Coulter, B23318) at 0.65× ratio (ex: 32.5 µL SPRI beads and 50 µL of sample). DNA was eluted typically in 50 µL of the elution buffer after two 80% ethanol washes. We then quantified the DNA sample using a Qubit 4 Fluorometer (Invitrogen, Q33238) and the high-sensitivity 1× dsDNA kit (Invitrogen, Q33231).
Golden gate assembly was done using the NEB Golden Gate Assembly Kit (BsmBI-v2) E1602 protocol. In short, the NEB reaction was prepared in a PCR strip tube to a final amount of 200 ng of pre-digested CROP-seq backbone, 80 ng of the insert (sgRNA library), 2.5 µL T4 DNA Ligase Buffer, 2.5 µL of NEB Golden Gate Enzyme Mix (BsmBI-v2) Mix (NEB, E1602L), and molbio grade water added to a total of 50 µL reaction solution. The PCR strip tube with solution was then placed in a pre-warmed thermocycler with a temperature profile of 42 °C for 60 min; 60 °C for 5 min; hold at 4 °C.
The libraries were then cleaned and concentrated with SPRI using a 1× template:bead ratio (see SPRI cleaning above). Samples were then eluted by adding 15 µL of molbio grade water to the dried beads and keeping ~12 µL, being sure not to carry through any beads. Next the samples were transformed into electrocompetent cells (Biosearch Technologies, 60242-2). One 0.1 cm cuvette (Bio-Rad, 1652089) for each transformation and clean DNA in water were prepared on ice. 1 mL of recovery media (Biosearch Technologies, 80026-1) was transferred to each culture tube in advance. The electrocompetent cells were then thawed on ice. The entire volume of the clean plasmid library (~10–12 µL) was added to the entire volume of cells (~50 µL), then mixed gently with a pipette tip 10 times while avoiding bubbles. Entire contents of the DNA+Cell solution was then transferred to the cuvette. The cuvette was then gently tapped to drop cell solution between the metal plates while continuing to avoid bubbles. The solution was then electroporated with the following settings on a Bio-Rad GenePulser Xcell machine (Bio-Rad, 1652660): 10 µF, 600 Ohms, 1800 Volts. Immediately after the electrical pulse, 1000 µL of Recovery Media was added to the cuvette, then cells were resuspended by pipetting up and down, and the solution was transferred to the 15 mL culture tube (Falcon, 352057). They were recovered at 37 °C for 1 h in an incubator shaking at 300 rpm. Samples were then transferred to 100 mL of LB broth (Corning, 46-050-CM) + 0.1 mg/mL Carbenicillin (C2135, Teknova) in 500 mL culture flask (Corning, 431401) and incubated at 32.5 °C at 160 rpm overnight (16 h). Next, we plated dilutions to estimate total transformation efficiency as a quick validation. A 0.5 mL aliquot from the culture flask was pipetted into a 1.5 mL Eppendorf tube. We then made two 100 μL dilutions; the first with a 1:10 (v/v) dilution and the second with a 1:100 (v/v) dilution. Each dilution was then seeded into its respective prewarmed carbenicillin agar plates (Teknova, L1011) by adding the full volume of the tube to the plate and then distributing the solution by shaking with seeding beads (Cole-Parmer, UX-01850-33). Plates were then incubated overnight (~16 h) at 37 °C. The colonies were counted the next day and coverage was calculated by:
Calculated coverage was compared to expected coverage for any major deviations. The remaining was grown in 100 mL cultures in a 500 mL flask at 30 °C overnight at 300 rpm and harvested 16 h post-transformation. Midiprep was then performed on the samples using NucleoBond Xtra EF plasmid purification, with a final elution volume of 1500 µL. Samples were stored at −20 °C for long term storage. Plasmid library distributions were verified by deep sequencing as described in the Next Generation Sequencing Sample processing section below.
Lentivirus production
Lenti-X HEK293T cells (632180, Takara) were thawed and cultured in 10% FBS (Gemini, 100–800) in high glucose DMEM (Gibco, 10569-010) for at least 1 week before transfection. Cells were passaged via 12-min trypLE incubation at 37 °C every 3 to 4 days when they reached 70–80% confluence. On day 0 of transfection, cells were passaged into 6-well plates (Falcon, 353046) at 1 million cells per well and 2mLs per well of media. On Day 1, cells were transfected using an edited version of the Lipofectamine 3000 reagents and protocol (ThermoFisher, L3000075). In short, two solutions were first made. The first with 167 µL per well of OptiMEM (Gibco, 51985034) and 14.4 µL of L-3000 reagent. The second solution was made with the following values per well: 167 µL of OptiMEM, 1056 ng of psPAX2 (addgene, plasmid #12260), 704 ng of pMD2.G (addgene, plasmid #12259), 1408 ng of plasmid library, and 12 µL of P3000 reagent. For the second mix it was important to add all of the plasmids first before adding P3000, to prevent the plasmids from crashing out of solution. After mixing the two solutions independently by quickly vortexing, we then added the first solution to the second solution, mixed well and then let incubate for 10 min. We then added 333 µL of solution drop wise to each well. On day 2 cells were treated with a final concentration of 2 mM viralboost (Alstem, VB100) solution in media by adding dropwise to the wells. On day 3, the virus was harvested by collecting the supernatant and straining it through a 0.45 µm syringe filter (millipore sigma, SLHV004SL). The virus solution was aliquoted into cryovials and immediately frozen at −80 °C for later use. Virus was titered on A549s at a wide range of concentrations to determine appropriate volumes to achieve an MOI of between 0.10 and 0.30 for screens. Titer was measured via flow by percent BFP positive cells and then the following equation was used to scale the transduction for the screen:
A549-Cas9 line generation
CAG-Blast-Cas9 plasmid was purchased from Horizon Discovery (Horizon Discovery, CAS10141). A Cas9 lentivirus was created as described in the Lentivirus Production section. A549s in culture were then transduced with the Cas9 lentivirus. Using a selection titer strategy with blasticidin, we selected cells at an MOI of 1. The A549-Cas9 line was then selected with 10 µg/mL blasticidin (Gibco, A11139-03), expanded and banked in 10% DMSO (Sigma, D2650-100ML), 10% FBS (Gemini, 100–800), in low glucose DMEM (Gibco, 11885-084).
Cell culture and lentivirus transduction
For all three screens, A549-Cas9 line was thawed into T225 flasks (Thermo Scientific, 159934) and then expanded and selected for Cas9-positive cells with 10 µg/mL blasticidin (Gibco, A11139-03) in 10% FBS (Gemini, 100–800) in low-glucose DMEM (Gibco, 11885-084) for 6 days. They were passaged at 70–80% confluence every 3 or 4 days using TrypLE (Gibco, 12604039) for 12 min at 37 °C. On day 6 post-thaw, blast selection was discontinued and cells were infected with the lentivirus pool at a target MOI of 0.1 for the morphological phenotypes and mechanism of action studies (to reduce number of doublets) and a target MOI of 0.3 for the druggable genome screen (higher MOI for better guide coverage) for 24 h. On day 1 post transduction, cells were then passaged into multiple T225s for expansion and selected with 1 µg/mL puromycin (Gibco, A11138-03) for 3 days. For the POC and MOA studies, cells were passaged on day 4 post-transduction and puro selection was continued to day 7. Cells were then passaged and seeded into two 24-well glass-bottom, black plates (Cellvis, P24-1.5H-N) for the POC screen and one 6-well glass-bottom, black plate (Cellvis, P06-1.5H-N) for the MOA screen. At this point, they were taken off of puromycin selection and cultured to day 10 post-transduction, when cells were fixed for phenotyping and in-situ sequencing. For the druggable genome screen, cells were banked on day 7 post-transduction, then seeded into two 6-well glass-bottom, black plates (Cellvis, P06-1.5H-N) with no puromycin selection. Cells were cultured in 10% FBS in DMEM medium until day 10 post-transduction, when cells were fixed for phenotyping and in-situ sequencing. Transductions and selections were designed to ensure that cell coverage never fell below 300x cells per sgRNA. For the druggable genome screen, cell pellets with a target coverage of 1000× cells per guide coverage were collected for next-generation sequencing at day 1, 4, 7, and 10 post-transduction by spinning down passaged cells at 300 × g for 5 min, aspirating supernatant, and immediately storing at −80 °C (Supplementary Fig. 5a).
Next generation sequencing sample processing
For cell samples, NucleoSpin Blood kit (Macherey-Nagel, 740951.50) was used to isolate the genomic DNA. Plasmid library samples were used after midi prep. We ran indexing PCR on all our NGS samples by creating the following mix per sample: 1× Kapa Hifi readymix HS (Roche, 7958935001), 0.5 µM common primer (Table 1), 0.5 µM indexing primer (Table 1), 5% DMSO (Sigma, D2650-100ML), 1–10 µg of DNA sample, filled up to 100 µL with molbio grade water. The thermal profile for the PCR is the following: 1 cycle of 95 °C for 5 min; 24 cycles of 95 °C for 30 s, 56 °C for 30 s, 72 °C for 20 s; 1 cycle of 72 °C for 10 min; hold at 12 °C. Samples were split into at least 4 wells of a 96wp PCR plate to minimize jackpotting. DNA was cleaned and quantified using the protocol described in the PCR cleanup section above. Samples were sequenced via MiSeq with PE50 V2 kit with custom primer according to manufacturer’s recommendations.
Demultiplexing was done by running the BCL files through a BCL-to-fastq pipeline. Fastqs were then used to align sgRNAs to our sgRNA library using an exact match approach of up to 18 base pairs of the 20 total base pairs of the sgRNA. Log2 enrichment was calculated by first getting the cell fraction as:
Fold Change was then calculated as:
Log2 of Fold Change (FC) was then calculated. Lastly, the FoldChange was then normalized to the intergenic by doing the following:
Plots were made using normalized log2 FC.
MitoProbe
Coordinates for the human mitochondrial ribosome from the Protein DataBase (PDB) (3J9M)29 were used. We used Visual Molecular Dynamics (VMD) for initial inspection of the PDB. For all other calculations, we used standard Python libraries and MDTraj30.
Within 3J9M, for all RNA sequences between the lengths of 15 to 25 (inclusive), we computed the SASA for all atoms in the sequence using MDTraj. We then computed the mean, median, and standard deviation of the SASA for all RNA sequences. We then filtered the sequences to only keep sequences that had a fully continuously observed sequence in the PDB. This yielded roughly 4379 sequences, though this also contains overlapping sequences (i.e., length 15 sequences are in length 16 to 25 length sequences). We sorted these sequences by their mean and median atomic SASA values. We sorted the list by its SASA values and then filtered the sequences by their GC content, absence of repetitive bases, and intra-sequence distances. Out of the final 8 sequences, we picked the first 2 sequences (lengths 20 and 23 base pairs) with the highest SASA (Fig. 1) and a large physical separation within the PDB. These selected sequences occupy distinct positions on the mt-ribosome. Oligonucleotides of these sequences were ordered with all uracil (U) bases in the sequence replaced with thymine (T), coupled to 5′ dyes, and purified by HPLC (Table 1).
Cell-Paint POSH protocol
Reverse transcription, gap fill, ligation, and amplification
On day 10 post transduction, cells in the glass bottom plates were fixed by aspirating the medium and replacing it with 4% paraformaldehyde (Electron Microscopy Sciences, 15710-S) in a 1× RNase-free PBS solution made from 10× RNase-free PBS (ThermoFisher, AM9625) and molecular biology grade water (Corning, 46-000-CV) for 30 min at room temperature. The cells were then washed two times with RNase-free 1× PBS and carried through the CP-POSH protocol, which modifies and integrates optical pooled screening techniques19 with CellPainting23.
Plates were permeabilized in 70% ethanol (Sigma Aldrich E7023-500ML) in molbio grade water (Corning, 46-000-CV) for 30 min at room temperature. Ethanol was removed by a 1:1 serial dilution with 0.05% Tween in 1× PBS (PBST) six times to prevent the samples from drying out from ethanol evaporation. Ex: 1 mL of ethanol per well, remove 0.5 mL and add 0.5 mL PBST—repeat 6×. Samples were then washed fully twice with PBST.
A RT solution was then made in molbio grade water (Corning, 46-000-CV) over ice to a final concentration of 1× RevertAid RT Buffer (comes with enzyme), 250 µM dNTPs (ThermoFisher, R1122), 0.2 mg/mL BSA (New England Biosciences, B9000S (discontinued)), 1 µM RT Primer (see Table 1), 0.8 U/µL Ribolock (ThermoFisher, EO0384), and 4.8 U/µL Revert Aid Reverse Transcriptase (ThermoFisher, EP0452). PBST in all the wells was aspirated and replaced with the RT solution, then sealed with sealing film (Applied Biosystems, 4306311) and seal roller (Sigma-Aldrich, R1275-1EA), then incubated overnight (16 h) at 37 °C. Samples were then washed five times with PBST.
Samples were then fixed with a solution of 3.2% paraformaldehyde and 0.1% glutaraldehyde (Sigma-Aldrich, G7651-10ML) in 0.8× PBS for 30 min at room temperature. Samples were then washed three times with PBST.
A solution for gap fill, ligation and extension (GF) was made in molbio grade water over ice to a final concentration of 1× Ampligase Buffer (Biosearch Technologies, A1905B), 0.4 U/µL RNaseH (Enyzmatics, Y9220L), 0.2 mg/mL BSA, 0.1 µM padlock probe (see Table 1), 0.02 U/µL TaqIT (Qiagen Beverly Inc, P7620L), 50 nM dNTPs, and 0.5 U/µL Ampligase (Biosearch Technologies, A3210K). PBST was replaced with the GF solution and the plates were sealed and incubated at 37 °C for 5 min then 45 °C for 90 min. Plates were then washed twice with PBST at room temperature.
A rolling circle amplification (RCA) solution was made in molbio grade water over ice to a final concentration of 1× phi29 buffer (comes with enzyme), 250 µM dNTPs, 0.2 mg/mL BSA, 5% glycerol (Invitrogen, 15514-011), and 1 U/µL phi29 polymerase (ThermoFisher, EP0094). PBST was replaced with the RCA solution, plates were sealed, and incubated at 30 °C overnight (16 h). Samples were then washed twice with PBST. Please note that padlock can only prime to and amplify the sgRNA cassette transcribed with the pol II puromycin and tagBFP transcript, not the pol III transcribed sgRNA. As a result, only the pol II transcript is sequenced by ISS, and not the actual sgRNA, similar to the CROP-seq approach published from Feldman et al.19.
Staining and imaging
The clear lid on the Celvis 6 well plates were replaced with a black lid (GreinerBio-One, 656199) to protect the samples from light. All staining steps with fluorophores were protected from light as best as possible. All the staining steps with a fluorophore were done on the same day and imaging was started no more than 24 h after staining for each plate. Any plates not being stained at the time were kept sealed at 4 °C in PBST until ready to be stained. Staining for all plates was done within 4 days post RCA. If antibodies were used, we first blocked those plates for an hour at room temperature with antibody buffer made of 1% w/v Nuclease-free BSA (Sigma-Aldrich, 126609-100GM), 0.01% Sodium Azide (VWR, BDH7465-2), 0.1% Nuclease-free TritonX (Sigma Aldrich, 93443-500 ML), and 1× PBS (ThermoFisher, AM9625) in molbio grade water. Ribolock was added to the blocking buffer right before incubation at a final concentration of 1 U/µL to prevent RNA degradation, which would affect mitoprobe signal. Rabbit anti-human pS6 primary antibody (Cell Signaling Technologies, 2215S) solution was made by diluting pS6 primary Ab 1:250 in antibody buffer solution plus 1 U/µL Ribolock. After blocking incubation, blocking solution was exchanged for pS6 primary antibody solution; plates were then sealed and incubated in 4 °C overnight (~16 h). Plates were then washed 3 times with PBST (0.05% Tween in 1× PBS). They were then incubated for an hour at room temperature in the secondary antibody mix made of 1:1000 dilution of donkey anti-rabbit DyLight 755 (Invitrogen, SA5-10043) in the antibody buffer. Samples were then washed three times with PBST at room temperature. Next, our mitoprobe mix was prepared, consisting of a final concentration of 0.25 µM 12s-1 Cy5 probe (Table 1), 0.25 µM 16s-1 Cy5 probe (Table 1), 10% formamide (Fisher Scientific, AC327235000), 10 mM RVC (New England Biosciences, S1402S), and 2× SSC Nuclease-free buffer (ThermoFisher, AM9763) in molbio grade water. Cells were then incubated in Mitoprobe mix at 37 °C for 30 min protected from light. They were then washed twice at room temperature with PBST. The Cell Painting (CP) staining base was then made in molbio grade water to a final concentration of 1% w/v of Nuclease Free BSA (Sigma-Aldrich, 126609-100GM), 1× HBSS (Gibco, 14065-056), and 0.01% Sodium Azide (VWR, BDH7465-2). The CP base buffer was made in bulk (500 mLs) and kept at 4 °C for up to 4 months. A Cell Painting mix was made in CP base buffer to a final concentration of 0.033 µM Phalloidin Alexa 568 (ThermoFisher, A12380), 12.5 µg/mL ConA Alexa 488 (ThermoFisher, C11252), 1.5 µg/mL WGA Alexa 555 (ThermoFisher, W32464), 0.5 µg/mL Hoechst (ThermoFisher, 33342), and 1 U/µL Ribolock (ThermoFisher, EO0384). Cell Painting mix was then added to the cells and incubated in the dark at room temperature for 30 min. The cells were then washed 5 times with PBST. Cells were immediately imaged or kept in 4 °C and imaging was started within 24 h of staining.
Cells were imaged on a Nikon Ti2 Eclipse with 20 × 0.75 NA objective. The settings and equipment used for phenotyping of the 1640 druggable genome screen are listed in Supplementary Table 1. The other two screens were done with slightly different filter settings.
In situ sequencing by synthesis
MiSeq Reagent Kit v2 500-cycles (Illumina, MS-102-2003) were used for ISS by synthesis. Samples were primed for in situ SBS by hybridizing 1 µM anchor primer in 2× SSC for 15 min at room temperature, then washed twice with PR2 incorporation buffer. Incorporation mix for fluorescent labeling of nucleotides was added and incubated for 3 min at 60 °C. The plate was then washed 4 times with PR2 at room temperature, then incubated at 60 °C in PR2 for 6 min. Washing and incubation was repeated a total of 2 times. Then the wells were washed twice with PR2. PR2 was then replaced with an imaging buffer made of 200 ng/mL Hoechst (Thermofisher, 33342) in 2× SSC and the plate was imaged.
After imaging, cleavage was done to remove fluorescent tags by washing the plate once with PR2, followed by cleavage mix addition and incubation at 60 °C for 2 min. It was then washed 3 times with PR2 at room temperature, then incubated in PR2 at 60 °C for 2 min. It was then washed twice with PR2 at room temperature and incorporation was performed again to add the next base (see above). Incorporation, imaging, and cleavage were repeated until all the desired bases were imaged (about 13 cycles for all screens). Imaging was done using a Nikon Ti2 Eclipse with 10 × 0.45NA objective (see Supplementary Table 2 for more details).
Druggable genome screen improvements
Cell-Paint POSH protocol improvements
To improve the efficiency of our Cell-Paint POSH protocol, three modifications were made to improve Cell-Paint and POSH dot quality. First, cell permeabilization was done with 0.1% RNase Free Triton-X (Sigma Aldrich, 93443-500 ML) in 1× PBS solution made from 10× RNase Free PBS (ThermoFisher, AM9625) and molecular biology grade water (Corning, 46-000-CV) for 30 min at room temperature instead of 70% ethanol. This was done to avoid actin degradation by ethanol, which leads to low phalloidin signal. Two washes with PBST were then performed after permeabilization with Triton-X solution. Second, we added a pre-RT fixation step, as described by Labitigan et al.21. Briefly, after Triton-X permeabilization, we incubated cells in 1 µM RT primer diluted in PBST for 30 min at room temperature. We then replaced the RT primer solution with 3% paraformaldehyde, 0.1% glutaraldehyde in PBST and incubated for 30 min at room temperature. We then washed three times with PBST and continued on to RT as previously described. Third, after phenotypic imaging was completed, we actively degraded phalloidin signals with ethanol to avoid spectral overlap with the ISS. Briefly, prior to addition of sequencing primer, cells were incubated in 70% ethanol (Sigma Aldrich E7023-500ML) in molbio grade water (Corning, 46-000-CV) for 30 min at room temperature. Ethanol was removed by serial dilution with PBST by a factor of 1:2 six times to prevent the samples from drying out from ethanol evaporation. Samples were then washed fully twice with PBST.
In situ sequencing by synthesis with automation
The ISS process is repetitive, time intensive, and tedious, which made it a good candidate for automation (equipment listed in Supplementary Table 3). In short, the developed system cleaved and incorporated each base pair using a multiflo FX (Biotech, MFXP) for liquid handling, an inheco heater/shaker (inheco, 7100146-A Rev.:04) for heated incubation steps with shaking, a Nikon Ti2 eclipse, and a KX2 robot arm (PAA KX-2, KX2-500) to transfer the plate between all three. Briefly, cycle one begins with incorporation of the first nucleotide in Incorporation Mix (Illumina MiSeq), followed by incubation with shaking at 60 °C for 3 min, 4 washes with PR2 buffer (PR2, Illumina MiSeq), and 6 min incubation at 60 °C with shaking, followed by 4 washes was performed a total of 3 times. The plate is then imaged on the Nikon Ti2. Each subsequent cycle begins with the addition of Cleavage Reagent (Illumina MiSeq) and incubation for 2 min with shaking at 60 °C. This is followed by four iterations of four washes (PR2) and incubation for 2 min with shaking at 60 °C. The cleaved plate then undergoes the same incorporation process as is found in cycle 1. This is repeated for sufficient cycles to deconvolute POSH barcodes in the library (typically 13 cycles). A time lapse video demonstration is provided in Supplementary Movie 1.
RNP arrayed validation studies
A549-Cas9 line was thawed into a T225 flask and cultured for 6 days as described in the Cell Culture and Lentivirus Transduction section. On day 6 post thaw, ribonucleoprotein (RNP) editing was conducted using the Lonza Amaxa SF Cell Line 4D-Nucleofector X Kit S (Lonza, V4XC-2032) and protocol with some modifications. In short, 1 µL of 100 µM gRNAs in TE Buffer were combined with 0.5 µL of 10 µg/µL Cas9 (Aldevron, 9212-0.25 mg) and incubated for 20 min at room temperature, then 4 °C until ready to use. Cells were then passaged, spun down at 90 × g for 10 min and resuspended in SF buffer and 4 µM enhancer (IDT, 075916) at 250,000 cells per reaction. Then, cells were nucleofected with gRNA Human Gene KO kits from Synthego for HSD17B10, AURKAIP1, and CYP11B1 which included three gRNAs per gene. A single intergenic gRNA (AAAGGCCAGTAACATCAGAC) from the Druggable Genome Library from IDT was also used. Cells were passaged and expanded on Day 3 and Day 7 post RNP while accounting for varying growth rates from the knockouts. On day 7, cells were seeded into black, glass-bottom 24-well plates and an extra plate was seeded for Inference of CRISPR Edits (ICE). On day 10, cells were fixed and then immediately blocked and stained as described in the Staining and Imaging section above. The staining panel for this included Hoechst, WGA, Phalloidin, and Mitoprobe. WGA and Phalloidin were used for segmentation. pS6 was stained at a 1:1000 dilution for this study because there were no POSH steps that would hinder antibody signal. Mouse anti Human Tomm20 primary antibody (Abcam, AB56783) at a 1:2000 dilution was also added in place of ConA following the same protocol as pS6. Goat anti Mouse Alexa Fluor 488 was the secondary (Invitrogen, A11029) used for Tomm20. pS6 secondary antibody was used as described in the Staining and Imaging section. Cells were imaged with the following settings: hoechst at 15% laser power and 25 ms, Tomm20 (488 nm) at 15% laser power and 50 ms, WGA(555 nm) at 15% laser power and 50 ms, phalloidin(568 nm) at 15% laser power and 30 ms, mitoprobe(647 nm) at 80% laser power and 25 ms, and pS6(755 nm) at 100% laser power and 100 ms.
For ICE analysis, cell samples were passaged at day 10, counted, spun down at 300 × g for 5 min and then resuspended in 200 µL of QuickExtract DNA Extraction solution (Lucigen, QE09050). Total cells used ranged from 80,000 to 100,000 cells depending on the knockout. We then followed the QuickExtract protocol. Primers for PCR were designed by choosing regions 300 bps upstream and downstream of the target editing region, and identified using NCBI Primer Blast. Designs with similar melting temperatures and GC content of 45–60% were chosen, then filtered for specificity using UCSC BLAT. PCR was performed using the AmpliTaq Gold 360 Master Mix (Applied Biosystems, 43-988-86) protocol. Samples were sent out to Elim Biopharm for sequencing and results were analyzed with Synthego’s ICE CRISPR Analysis tool. The reported indel % was the average of the functioning primers.
Data capture and storage
The images acquired on the microscope are exported in raw tiff format and the corresponding metadata for each image as specified in Supplementary Table 4 are recorded.
Cell Painting image processing
The raw fluorescence field of view images acquired from the microscope have uneven illumination due to camera and vignetting artifacts. In order to create a flat-field illumination image, the raw images are scaled by an illumination correction function as estimated from the mean over field of view images followed by Gaussian smoothing93. The illumination corrected images are then min-max percentile (min percentile = 0.1, max percentile = 99.9) scaled based on per-channel intensity percentiles computed from plate-level intensity histograms and converted to 8-bit unsigned-integer type (min value = 0, max value = 255) for subsequent processing. The field of view images are segmented using CellPose94 to obtain single cell and nuclei boundaries. The field of view images are then cropped around the centroids of each of the segmented nuclei and masked by the corresponding cell segmentation mask to create tiles with a single cell in context.
in-situ sequencing by synthesis image processing
The SBS procedure described generates a dataset in which the full plate is imaged several times, with 4-color stationary dots showing variable fluorescent signatures corresponding to A/C/T/G that need to be converted to sequencing base calls.
Baseline
The baseline methodology for base calling and sequencing was obtained from Feldman et al., that first computes locally registered and aligned images across all the SBS cycles, followed by blob detection that requires manual fine tuning of parameters. The baseline methodology involves applying a Laplacian-of-Gaussian linear filter (kernel width s = 1 pixel) to subtract low-frequency background and enhance the point-like sequencing spots followed by calculating the per-pixel, per-channel standard deviation over sequencing cycles, averaging over color channels, and finding local maxima in the resulting image. The base intensity at each cycle was defined as the maximum value in a 3 × 3 pixel window centered on the read. A linear transformation to correct for optical cross-talk and intensity differences between color channels was then estimated from the data and applied. Finally, each base was called according to the channel with maximum corrected intensity, and a per-base quality score was defined as the ratio of intensity for the maximum channel to total intensity for all channels. For Bardensr baseline, we use the following parameters: estimate_density_singleshot (noise floor = 0.05), find_peaks (threshold = 0.72). For Starfish baseline, we use the following parameters: WhiteTopHat (masking radius = 15), BlobDetector (min_sigma = 1, max_sigma = 10, num_sigma = 30, threshold = 0.01, measurement_type = “mean”).
Proposed methodology
We propose a three-stage ISS methodology that involves first computing base call locations, transforming and projecting the base calls per cycle to the first SBS cycle and stitching of base calls by nearest neighbor matching.
(i) Base calling using convolutional neural network
In order to obtain the base call identities and locations per cycle, we trained a 3-layer fully convolutional network \([({Conv}2d(3x3),{BatchNorm}2d,{ReLU}){x}2,{Conv}2d(3x3),{Sigmoid}]\) that takes as input the 4-channel SBS image and predicts a 4-channel base identity probability map95. For training the base calling model, we collected a dedicated in-situ sequencing dataset on A549 imaged over 9 cycles, which is distinct from the datasets generated from the 3 screens. The complete datasets from the 3 screens were reserved exclusively for evaluation. By keeping the evaluation datasets separate from training data, we avoid Type-1 data circularity.
Concretely, to create our training dataset, we cropped the in-situ sequencing images across all the cycles in the training data into 275,754 image crops of size 4 × 256 × 256 pixels. To generate high-quality labels for training, we used a blob detector-based baseline method (Feldman et al.19) to obtain pseudo-ground truth, and filter the results to retain only barcodes that matched the sgRNA barcodes in the experiment library. This ensures that the false positive detections (Type I error) from the baseline method (Feldman et al.19) are removed from our ground truth. The ground truth 4-channel base location/identity images are created by setting a 3 × 3 pixel region around each baseline detected base call location to 1 and all other pixels to 0, and the model is trained to predict at each spatial coordinate the likelihood of each base. The convolutional network was trained with supervision on the proxy labeled dataset (275,754 images of size 4 (channels) × 256 × 256 pixels) on 8 Tesla K80 GPUs for 50 epochs by optimizing dice loss with a stochastic gradient descent optimizer as described below (\(y\) = ground truth base label mask, \(\overline{p}\) = predicted probability mask).
The predicted base call probabilities are converted to binary predictions by thresholding, labeled by connected components and converted to a table of base call locations and identities per image. We chose a threshold of 0.5 on the base call probabilities based on F1 score calibration (Supplementary Fig. 2c). We use complete datasets from the 124 gene morphology, 300 gene MOA, and 1640 gene druggable genome datasets as our evaluation datasets for the comparison of barcode and cell recovery efficiencies between our proposed and baseline methodologies in Fig. 2b–d. We use a single held-out field of view from the 300-gene MOA dataset for comparison of our proposed approach with additional state of the art baseline methods in Supplementary Fig. 2d.
(ii) Registration
In order to transform base calls detected in each SBS cycle imaged at different time points to a single acquisition’s coordinate space, we first convert the field of view level base call coordinates (\({x}_{k,f},\,{y}_{k,f}\)) to well coordinate space (\({x}_{k,w},\,{y}_{k,w}\)). We then compute an odometry transformation matrix (\({T}_{k,1}\)) to transform the well coordinates (\({x}_{k,w},\,{y}_{k,w}\)) in each SBS cycle \(k\) to well coordinates in the first SBS cycle as \((\underline{{x}_{k,w}},\,\underline{{y}_{k,w}})\) = \({T}_{k,1}\)(\({x}_{k,w},\,{y}_{k,w}\)). This process allows computational alignment of SBS per cycle post-image acquisition, without the need to do image alignment during. We compute the affine odometry transformation matrix \({T}_{k,1}\) using an FFT based phase cross correlation algorithm96 on a small center crop region of fiduciary markers (Hoechst) between the acquisitions (Supplementary Fig. 2a).
(iii) Transformation of base calls and barcode stitching
The base calls computed in each cycle are all transformed to the first SBS cycle using the odometry transformation matrix computed above. The transformed base calls are then chained together based on a KD Tree Nearest Neighbor matching algorithm across cycles to generate a sgRNA barcode readout in the coordinate space of the first SBS cycle.
Details for model architecture and training are included in the published code. During the preparation of this manuscript, a ML based method for in-situ barcode sequencing was published that uses pseudo-labels provided by fully reconstructed barcodes to enhance in-situ sequencing97.
Additional baseline
In addition to a convolutional neural network for base calling, our proposed algorithm also introduces a registration methodology that does not require the field of views to be in alignment across all the SBS cycles. The baseline methodology from Feldman et al., utilizes the base signal across all SBS cycles to determine a base call. So, as an additional baseline, we also compare the performance of blob detection (with a Laplacian-of-Gaussian filter) per SBS cycle followed by registration and coordinate transformation.
Evaluation and quality metrics
The performance and quality of detected base calls and barcodes were assessed using the following metrics
(i) Signal to total ratio per cycle
The fidelity of POSH dots throughout several sequencing cycles was measured using the STR equation below for all detected amplicons in each SBS cycle.
where \({I}_{g},\,{I}_{t},\,{I}_{a},\,{I}_{c}\) are the demultiplexed intensities of base fluorophores the detected base locations
(ii) Percent of cells with a valid barcode
We define a valid barcode as a stitched barcode that matches with a barcode in our perturbed sgRNA library. For each methodology, we compute the percentage of cells with a valid barcode as the ratio of # of cells with any valid barcode (including multiple valid barcodes) to the total number of segmented cells in the dataset.
Single cell sgRNA identity assignment
To assign sgRNA barcode identities to cropped single cell images obtained from Cell Painting, we transform the barcode locations, sgRNA sequence and their corresponding gene KO identities from the first cycle of SBS to the Cell Painting acquisition space using the same odometry transformation methodology described in the registration section above. The barcodes in Cell Painting acquisition space are then assigned to single cell contexts using the cell boundaries and then mapped to the corresponding cropped tiles to obtain a labeled perturbation dataset of single cell images.
Explicitly engineered image featurization (CellStats)
To localize the signal, we capitalize on the two segmented areas and derive features that are computed in the nucleus, the cytoplasm, and also the perinuclear regions by iteratively dilating the segmentation mask corresponding to the nucleus. First, we start by capturing characteristics of the pixel intensity empirical distributions corresponding to each localization mask. In particular, we extract percentiles from the empirical distributions at discrete steps, along with their means, variances, and standard deviations. Additionally, we compute pairwise Pearson and Spearman correlations among the different channels in our assays. Second, focusing on the localization masks of the nucleus and the cytoplasm, we capture characteristics of each cell’s geometry. Key attributes of cell geometry include the area of the nucleus and the cytoplasm and its convexity98. Lastly, we extract features that have been heavily utilized by the computer vision community to characterize the textures emerging in the nucleus and the cytoplasm from the selected types of staining. In particular, statistics derived from suitable wavelet transforms and region covariance descriptors are employed towards capturing such information. A total of 1301 features are extracted for each cell.
Self-supervised learning of single cell image representations
Self-supervised learning pertains to methodologies that are capable of learning latent space representations over input distributions without having an explicit label assigned to them. Recently, self-supervised learning methods [DINO, SimCLR, CLIP] have been shown to improve generalization and quality of learned representations as compared to supervised learning methods45,99,100. In this work, we use DINO45 a self-supervised learning technique that uses knowledge distillation between student (\({g}_{{\theta }_{s}}\)) and teacher (\({g}_{{\theta }_{t}}\)) networks parameterized by \({\theta }_{s}\) and \({\theta }_{{t}}\) respectively, to learn rich representations from images. Both networks are trained to output a probability vector over \(K\) dimensions computed as:
where \({\tau }_{s}\) is the temperature parameter that controls the sharpness of the output distribution. The student network is trained by minimizing the cross-entropy loss over outputs (as given by Eq below) with the teacher network using stochastic gradient descent.
where \(H(a,{b})\,=\,-{a}\log {b}\), \({x}_{g1\,}\) and \({x}_{g2}\) are augmented global views and \(V\) is a set of augmented local crop views of the input images. The teacher network is updated using an exponential moving average of past weights of the student network as given by:
Our motivation to choose DINO as our self-supervised framework stems from their superior performance in k-Nearest Neighbor benchmarks on ImageNet dataset as reported45. We also use a Vision Transformer architecture101 to parameterize both the student and the teacher network parameters.
Baseline pre-trained vision transformer
We utilize an ImageNet pre-trained DINO Vision Transformer model (ViT-small) with a patch size of 8 trained as our SSL baseline. The pretrained model weights were obtained from [https://github.com/facebookresearch/dino/tree/main]. In order to adapt the pre-trained model trained on 3-channel inputs to 5-channel Cell Painting fluorescence image inputs, we re-parameterized the first layer of the model as:
where \({{\theta }^{(i)\,}}_{l1-3{ch}}\) corresponds to the parameters of the patch embedding layer of the pretrained network that takes 3 channels as input to the network and \({{\theta }^{(i)}}_{l1-5{ch}}\) corresponds to the redistributed weights of the network that takes 5 channels as input to the network. In addition to ImageNet pre-trained ViT-small, we also compared our model to the vit-small model using pre-trained weights published by Doron et al., trained on a collection of Cell Painting datasets.
Training DINO ViT for fluorescence microscopic images
(i) Dataset preparation
The training dataset was created from the single cell masked and cropped tile images from the 300-gene and 1640-gene CRISPR KO screens for CP-dino-300 and CP-dino-1640, respectively. The dataset contained 1,585,396 cells for CP-dino-300 and 1,559,011 cells for CP-dino-1640. The tile images were then normalized by subtracting by global mean and standard-deviation per channel computed over a small subset of the training images.
(ii) Fluorescence image augmentation
In order to construct the distorted global and local views of images for DINO training, we modified the set of augmentations to be more relevant for fluorescence microscopic datasets. Some of the key changes are (a) we removed the scale augmentations for both global and local views as all our microscopic images were imaged at a fixed magnification and physical scale. (b) added random rotation augmentations to our images, (c) removed solarization, color jitter, and (d) added defocus, coarse dropout and dropout augmentations to generate our global and local views. We also restrict the local crop regions to be within the single cell region to avoid local crops of masked out background regions.
(iii) DINO-ViT training
We train our model using the standard DINO (v1) training procedure that uses global crops of the input image passed as input to the teacher network and global + local crops of the input image to the student network. We follow the same procedure in this work as the original DINO implementation with a slight modification of how we generate the local crops. We constrain local crops to a smaller region in order to retain at least some region of the cell in every local crop. We trained a ViT-small model (Table 2) with patch size = 8, number of global crops = 2, number of local crops = 8 on 4 nodes × 8 NVIDIA-V100 GPUs per node (32 GPUs) for 100 epochs over the 300-gene MOA or the 1640-gene druggable genome (with or without pS6) datasets. We use the output cls token embeddings of dimension 384 as our feature representation for subsequent analysis.
(iv) DINO-ViT embedding aggregation
We aggregate gene-KO embeddings is as follows: (1) we extract single-cell representations; (2) represent each sgRNA perturbation by the mean over all cells of that sgRNA perturbation; (3) represent each gene perturbation as the median of sgRNA representations corresponding to each gene KO; (4) compute pairwise cosine similarity between gene KO representations.
Feature normalization
In order to adjust for intensity variations and batch effects introduced by assay and imaging systems, we normalize each feature embedding by the feature embeddings of non-targeting controls in each well. The UMAP of normalized embeddings shows better mixing of embeddings across wells compared to UMAP of un-normalized embeddings (Supplementary Fig. 4g). We employ Robust Center-Scale Normalization, where for each well, we subtract all the embeddings by the median of non-targeting controls in the well and scale by the median absolute deviation of the non-targeting controls. During explicitly engineered feature analysis, this normalization may result in a 0 in the denominator of a small number of features. Thus, a filtering step is applied to remove any features that result in Inf or NAN values during normalization. This results in the POC, MOA, and DG screens having 1278, 1299, and 1298 total post-filter CellStats features, respectively (Supplementary Data 4).
Feature aggregation
For extrinsic evaluation of the usefulness of embeddings and understanding biological relationships from embeddings, we aggregate the single cell feature embeddings to a single vector representation per gene KO. We first compute a sgRNA-level representation by taking the mean over all the PCA-whitened cell embeddings corresponding to that sgRNA KO. We found that PCA-whitening improves the recovery of known StringDB edges compared to aggregating raw embeddings (Supplementary Fig. 4h). The gene level aggregate representations are obtained by taking a median over all the representations of sgRNAs targeting the same gene. To evaluate the robustness of metric to sample sizes used for aggregation, we also report the StringDB AU-ROC and TPR @ 5% FPR metrics on downsampled aggregate embeddings (Supplementary Fig. 4e).
Differential morphology analysis and gene enrichment analysis
In several cases, more focused analysis of a single knockout or single feature was necessary. In the case of gene-wise differential morphology analysis, the Z-scored cell populations containing the given knockout, as well as the non-targeting/intergenic control cells within the same pool, were subset from the rest of the pool. Between these two samples, a two-sample K–S test was applied for each feature with the Z score of the sub-population relative to controls. Due to the large number of features, as well as their redundancy, clear trends in significantly-changed features were annotated; more specific feature results were determined for all genes within the Druggable Genome Screen and created as a report file. A similar approach was applied in the case of feature-wise gene enrichment analysis, in which, for the given feature, each subset of cells receiving the same genetic knockout was compared to non-targeting/intergenic via a two-sample two-tailed K–S test.
Comparison to FACS binned 1-dimensional analysis
We sought to compare how POSH-based single-cell data on a single feature would compare to a simulated binned FACS screen using the same data. For the single cell resolution based analysis, K–S testing and mean Z were determined for mean cytoplasmic pS6 intensities. To simulate a FACS screen using a pseudo-binning approach, we used the well-normalized Z score matrix and determined the 0–15%, 15–30%, 70–85%, and 85–100% percentile bins, and assigned all cells within the screen to these respective bins. We then assumed perfect sequencing and assignment of the sgRNAs within cells that had been assigned to the bins. MAUDE analysis102 was then conducted to predict the likely distribution shift of cytoplasmic phospho-S6 within each knockout subset, leading to predicted mean Z and FDR for each gene.
Evaluation of single cell embeddings by linear classification model
We evaluate the intrinsic representational power of the different embedding methodologies by measuring their performance on downstream predictive tasks. We evaluate the embeddings by their performance on multiple binary classification tasks for discriminating between non-targeting controls and samples corresponding to a gene KO. For each gene KO, we train an Elastic Net Logistic Regression model with the task of binary classification between nontargeting and gene KO samples with a leave-one-well-out cross validation schema for all the wells in the experiment. The binary prediction AU-ROC is computed on the validation samples for each gene in every validation split and the median over AU-ROC is obtained as the scoring function for each embedding. We report the number of gene KOs with an AU-ROC > 0.6 and the mean AU-ROC for the subset of genes on prediction tasks for each embedding methodology as an objective metric for evaluation and comparison. We did not choose a higher AU-ROC threshold because: (1) not all perturbations will have a phenotype in this particular cell type or experimental condition, and hence we do not expect to get to accurate predictions for all guides; (2) guide penetrance is partial, so even for guides that do have a phenotype, we do not expect to be able to classify all cells; (3) this analysis is performed on single cells with inherent cell-to-cell noise which may confound perturbation classification. In Supplementary Fig. 5g, we reported all perturbations with a significant phenotypic alteration by performing a KS-test of targeting vs non-targeting against a bootstrapped null distribution of intergenic vs non-targeting classification scores (5% FDR).
Evaluation of biological meaningfulness of aggregate embeddings
Similarity of representations across sgRNAs targeting the same gene
To evaluate the meaningfulness of the aggregate embeddings for each embedding methodology, we computed a pairwise cosine similarity of embeddings [Eq below] for all sgRNAs targeting the same gene.
For each embedding method, we then compute the mean sgRNA similarity on a subset of genes that are predictive across all methodologies to rank the embeddings by their meaningfulness in capturing biologically meaningful structure.
Evaluating the ability of embeddings to capture known gene-gene relationships in StringDB
While it is difficult to determine ground truth in biological datasets, we sought to compare our unbiased screening approach to well-established protein-protein interaction networks in the field. To this end, we used StringDB (v11.5)36, a well-curated network of protein-protein interaction networks based on literature scrubbing, interaction databases, co-expression analysis, and organism transfer. The similarity networks of the genes we screened for were downloaded and used for comparison. For each embedding methodology, using the aggregate gene representations, we compute a pairwise cosine similarity matrix of embeddings. We use the cosine similarity as the strength of similarity/connectivity between two genes learned by the embedding model. We then define the stringDB ground truth connections by thresholding the StringDB combined score at different thresholds and compute the AU-ROC of the overlap of the embedding cosine similarity matrix with the StringDB network. For Spearman calculation, the same stringDB combined score was used. We only used StringDB edges in which a value was assigned (edge strength > 0). In Supplementary Fig. 5i, j, we report the correlation of profiles on CORUM and StringDB using the methodology from Ramezani et al.59 on the reported gene set (n = 1089 genes).
UMAP visualization of embeddings
We visualize the aggregate gene-KO embeddings (robust center scaling) using a UMAP projection with cosine similarity as the metric. The size of each gene-KO node in the UMAP is adjusted based on the similarity of sgRNAs targeting the gene. The gene-KO nodes were colored using Leiden community detection algorithm48 on the UMAP projection.
Statistics and reproducibility
Data for the pooled screening experiments (POC, MOA, and druggable genome screens; Figs. 3–6) were acquired with high coverage. The mean number of individual cells analyzed per gene knockout (KO) was ~1300 (POC), ~4000 (MOA), and ~1500 (druggable genome). In this design, each perturbation is represented by hundreds to thousands of single-cell measurements, providing a replicate depth equivalent to or greater than standard pooled CRISPR screens. No data were excluded from the pooled screening analyses. The pooled format inherently randomizes perturbations across the culture plates, mitigating spatial confounders (e.g., plate-position effects) common to arrayed screens. Furthermore, data collection was intrinsically blinded, as the perturbation identity at any given x-y coordinate was unknown to the experimenter until the ISS and decoding were complete.
For the orthogonal, arrayed validation experiment (Fig. 6h, i), data were generated from two technical replicate wells per KO. The total number of images analyzed per condition was 567 (intergenic), 568 (AURKAIP1), 568 (HSD17B10), and 568 (CYP11B1). No data were excluded from this analysis. In contrast to the pooled screens, this specific follow-up experiment was not randomized, and investigators were not blinded during the experiment or outcome assessment.
For comparison to PERISCOPE data used in Supplementary Fig. 5, data was downloaded from Cell Painting Gallery on 2024-05-22 from https://registry.opendata.aws/cellpainting-gallery under accession number cpg0021-periscope.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The single cell tile image data generated in this study are accessible at (s3://insitro-research-2023-cellpaint-posh/, Zenodo accession ID: https://doi.org/10.5281/zenodo.17470307). The NGS data generated in this study are accessible through NCBI Sequence Read Archive (SRA) under BioProject accession PRJNA1359537. The data are provided under a CC BY-NC-SA 4.0 license. Source data are provided with this paper.
Code availability
Models trained in this study are available at https://huggingface.co/insitro/cp-posh. Code is available at https://github.com/insitro/cp-posh (accession ID: https://doi.org/10.5281/zenodo.17470307). The models and code are provided under a CC BY-NC-SA 4.0 license.
References
Silva, J. M. et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science 319, 617–620 (2008).
Schlabach, M. R. et al. Cancer proliferation gene discovery through functional genomics. Science 319, 620–624 (2008).
Bock, C. High-content CRISPR screening. Nat. Rev. Methods Primers 2, 9 (2022).
Shalem, O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Hart, T. et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 (2015).
Wang, T., Wei, J. J., Sabatini, D. M. & Lander, E. S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014).
Ihry, R. J. et al. Genome-scale CRISPR screens identify human pluripotency-specific genes. Cell Rep. 27, 616–630.e6 (2019).
Gilbert, L. A. et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661 (2014).
Wells, M. F. et al. Genome-wide screens in accelerated human stem cell-derived neural progenitor cells identify Zika virus host factors and drivers of proliferation. Preprint at bioRxiv https://doi.org/10.1101/476440 (2018).
Condon, K. J. et al. Genome-wide CRISPR screens reveal multitiered mechanisms through which mTORC1 senses mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 118, e2022120118 (2021).
Wan, C. et al. Genome-scale CRISPR-Cas9 screen of Wnt/β-catenin signaling identifies therapeutic targets for colorectal cancer. Sci. Adv. 7, eabf2567 (2021).
Parnas, O. et al. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 162, 675–686 (2015).
Adamson, B. et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882.e21 (2016).
Dixit, A. et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866.e17 (2016).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Jaitin, D. A. et al. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-Seq. Cell 167, 1883–1896.e15 (2016).
Datlinger, P. et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017).
Way, G. P. et al. Morphology and gene expression profiling provide complementary information for mapping cell state. Cell Syst. 13, 911–923.e9 (2022).
Feldman, D. et al. Optical pooled screens in human cells. Cell 179, 787–799.e17 (2019).
Funk, L. et al. The phenotypic landscape of essential human genes. Cell 185, 4634–4653.e22 (2022).
Labitigan, R. L. D. et al. Mapping variation in the morphological landscape of human cells with optical pooled CRISPRi screening. eLife 13, RP94964 (2024).
Carlson, R. J., Leiken, M. D., Guna, A., Hacohen, N. & Blainey, P. C. A genome-wide optical pooled screen reveals regulators of cellular antiviral responses. Proc. Natl. Acad. Sci. USA 120, e2210623120 (2023).
Bray, M. A. et al. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat. Protoc. 11, 1757–1774 (2016).
Rohban, M. H. et al. Systematic morphological profiling of human gene and allele function via Cell Painting. Elife 6, e24060 (2017).
Rohban, M. H. et al. Virtual screening for small-molecule pathway regulators by image-profile matching. Cell Syst. 13, 724–736.e9 (2022).
Haghighi, M., Caicedo, J. C., Cimini, B. A., Carpenter, A. E. & Singh, S. High-dimensional gene expression and morphology profiles of cells across 28,000 genetic and chemical perturbations. Nat. Methods 19, 1550–1557 (2022).
Chandrasekaran, S. N., Ceulemans, H., Boyd, J. D. & Carpenter, A. E. Image-based profiling for drug discovery: due for a machine-learning upgrade?. Nat. Rev. Drug Discov. 20, 145–159 (2021).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Amunts, A., Brown, A., Toots, J., Scheres, S. H. W. & Ramakrishnan, V. Ribosome. The structure of the human mitochondrial ribosome. Science 348, 95–98 (2015).
McGibbon, R. T. et al. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 109, 1528–1532 (2015).
Axelrod, S. et al. starfish: scalable pipelines for image-based transcriptomics. J. Open Source Softw. 6, 2440 (2021).
Chen, S. et al. BARcode DEmixing through Non-negative Spatial Regression (BarDensr). PLoS Comput. Biol. 17, e1008256 (2021).
Fuchs, F. et al. Clustering phenotype populations by genome-wide RNAi and multiparametric imaging. Mol. Syst. Biol. 6, 1–13 (2010).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170, 564–576.e16 (2017).
Stirling, D. R. et al. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinforma. 22, 433 (2021).
Szklarczyk, D. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612 (2021).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2018).
Klages-Mundt, N. L., Kumar, A., Zhang, Y., Kapoor, P. & Shen, X. The nature of actin-family proteins in chromatin-modifying complexes. Front. Genet. 9, 398 (2018).
Bunnell, T. M., Burbach, B. J., Shimizu, Y. & Ervasti, J. M. β-Actin specifically controls cell growth, migration, and the G-actin pool. Mol. Biol. Cell 22, 4047–4058 (2011).
Corbett, M. A. et al. A mutation in the Golgi Qb-SNARE gene GOSR2 causes progressive myoclonus epilepsy with early ataxia. Am. J. Hum. Genet. 88, 657–663 (2011).
Hay, J. C., Chao, D. S., Kuo, C. S. & Scheller, R. H. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell 89, 149–158 (1997).
Ljosa, V. et al. Comparison of methods for image-based profiling of cellular morphological responses to small-molecule treatment. J. Biomol. Screen. 18, 1321–1329 (2013).
States, C. et al. Multiplex cytological profiling assay to measure diverse. PLoS ONE 8, 1–7 (2013).
Canham, S. M. et al. Systematic chemogenetic library assembly. Cell Chem. Biol. 27, 1124–1129 (2020).
Caron, M. et al. Emerging properties in self-supervised vision transformers. In Proc. IEEE/CVF International Conference on Computer Vision 9650–9660 (IEEE, Montreal, QC, Canada, 2021).
Deng, J. et al. ImageNet: a large-scale hierarchical image database. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 248–255 (IEEE, Miami, FL, USA, 2009).
Alzubaidi, L. et al. Novel transfer learning approach for medical imaging with limited labeled data. Cancers 13, 1590 (2021).
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
Margueron, R. & Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011).
Dibble, C. C. & Cantley, L. C. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 25, 545–555 (2015).
Mashima, T., Seimiya, H. & Tsuruo, T. De novo fatty-acid synthesis and related pathways as molecular targets for cancer therapy. Br. J. Cancer 100, 1369–1372 (2009).
Finan, C. et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 9, 1–16 (2017).
Liu, G. Y. & Sabatini, D. M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 (2020).
Gan, L. et al. The lysosomal GPCR-like protein GPR137B regulates Rag and mTORC1 localization and activity. Nat. Cell Biol. 21, 614–626 (2019).
Yan, G. et al. Genome-wide CRISPR screens identify ILF3 as a mediator of mTORC1-dependent amino acid sensing. Nat. Cell Biol. 25, 754–764 (2023).
Doron, M. et al. Unbiased single-cell morphology with self-supervised vision transformers. Preprint at bioRxiv https://doi.org/10.1101/2023.06.16.545359 (2023).
Kraus, O. Z., Ba, J. L. & Frey, B. J. Classifying and segmenting microscopy images with deep multiple instance learning. Bioinformatics 32, i52–i59 (2016).
Gadermayr, M. & Tschuchnig, M. Multiple instance learning for digital pathology: a review of the state-of-the-art, limitations & future potential. Comput. Med. Imaging Graph. 112, 102337 (2024).
Ramezani, M. et al. A genome-wide atlas of human cell morphology. Nat. Methods 22, 621–633 (2025).
Young, C. D. et al. Activating PIK3CA mutations induce an epidermal growth factor receptor (EGFR)/extracellular signal-regulated kinase (ERK) paracrine signaling axis in basal-like breast cancer. Mol. Cell. Proteom. 14, 1959–1976 (2015).
Ichise, T., Yoshida, N. & Ichise, H. CBP/p300 antagonises EGFR-Ras-Erk signalling and suppresses increased Ras-Erk signalling-induced tumour formation in mice. J. Pathol. 249, 39–51 (2019).
Gao, X. et al. MAP4K4 is a novel MAPK/ERK pathway regulator required for lung adenocarcinoma maintenance. Mol. Oncol. 11, 628–639 (2017).
Sun, W. et al. MEK kinase 2 and the adaptor protein Lad regulate extracellular signal-regulated kinase 5 activation by epidermal growth factor via Src. Mol. Cell. Biol. 23, 2298–2308 (2003).
Tzavlaki, K. & Moustakas, A. TGF-β signaling. Biomolecules 10, 487 (2020).
Zhang, X., Li, Y., Xu, H. & Zhang, Y.-W. The γ-secretase complex: from structure to function. Front. Cell. Neurosci. 8, 427 (2014).
Blair, C. R., Stone, J. B. & Wells, R. G. The type III TGF-β receptor betaglycan transmembrane-cytoplasmic domain fragment is stable after ectodomain cleavage and is a substrate of the intramembrane protease γ-secretase. Biochim. Biophys. Acta 1813, 332–339 (2011).
Hou, P. et al. The γ-secretase substrate proteome and its role in cell signaling regulation. Mol. Cell 83, 4106–4122.e10 (2023).
Zhang, X., Wei, C., Liang, H. & Han, L. Polo-like kinase 4’s critical role in cancer development and strategies for Plk4-targeted therapy. Front. Oncol. 11, 587554 (2021).
Sabatini, D. M. Twenty-five years of mTOR: uncovering the link from nutrients to growth. Proc. Natl. Acad. Sci. USA 114, 11818–11825 (2017).
Brugarolas, J. et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 18, 2893–2904 (2004).
Dibble, C. C. et al. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol. Cell 47, 535–546 (2012).
Gao, J. et al. Cloning and characterization of human and mouse mitochondrial elongation factor G, GFM and Gfm, and mapping of GFM to human chromosome 3q25.1-q26.2. Genomics 74, 109–114 (2001).
He, X. Y., Merz, G., Mehta, P., Schulz, H. & Yang, S. Y. Human brain short chain L-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme. Characterization of a novel 17beta-hydroxysteroid dehydrogenase. J. Biol. Chem. 274, 15014–15019 (1999).
Koc, E. C. et al. The large subunit of the mammalian mitochondrial ribosome: ANALYSIS OF THE COMPLEMENT OF RIBOSOMAL PROTEINS PRESENT *. J. Biol. Chem. 276, 43958–43969 (2001).
de la Cruz López, K. G., Toledo Guzmán, M. E., Sánchez, E. O. & García Carrancá, A. mTORC1 as a regulator of mitochondrial functions and a therapeutic target in cancer. Front. Oncol. 9, 1373 (2019).
Yang, F. et al. Mitochondrial EF4 links respiratory dysfunction and cytoplasmic translation in Caenorhabditis elegans. Biochim. Biophys. Acta 1837, 1674–1683 (2014).
Gao, Y. et al. Mammalian elongation factor 4 regulates mitochondrial translation essential for spermatogenesis. Nat. Struct. Mol. Biol. 23, 441–449 (2016).
Seaver, L. H. et al. A novel mutation in the HSD17B10 gene of a 10-year-old boy with refractory epilepsy, choreoathetosis and learning disability. PLoS ONE 6, e27348 (2011).
Falk, M. J. et al. A novel HSD17B10 mutation impairing the activities of the mitochondrial RNase P complex causes X-linked intractable epilepsy and neurodevelopmental regression. RNA Biol. 13, 477–485 (2016).
Jeong, A. & Wong, M. Targeting the mammalian target of rapamycin for epileptic encephalopathies and malformations of cortical development. J. Child Neurol. 33, 55–63 (2018).
Jin, X. et al. In vivo Perturb-Seq reveals neuronal and glial abnormalities associated with autism risk genes. Science 370, eaaz6063 (2020).
Saka, S. K. et al. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat. Biotechnol. 37, 1080–1090 (2019).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Kim, V. et al. Self-supervision advances morphological profiling by unlocking powerful image representations. Sci. Rep. 15, 4876 (2025).
Pfaendler, R., Hanimann, J., Lee, S. & Snijder, B. Self-supervised vision transformers accurately decode cellular state heterogeneity. Preprint at bioRxiv https://doi.org/10.1101/2023.01.16.524226 (2023).
Moshkov, N. et al. Learning representations for image-based profiling of perturbations. Nat. Commun. 15, 1594 (2024).
Alquicira-Hernandez, J., Sathe, A., Ji, H. P., Nguyen, Q. & Powell, J. E. scPred: accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome Biol. 20, 264 (2019).
Kotliar, D. et al. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-seq. Elife 8, e43803 (2019).
Chandrasekaran, S. N. et al. JUMP cell painting dataset: morphological impact of 136,000 chemical and genetic perturbations. Preprint at bioRxiv https://doi.org/10.1101/2023.03.23.534023 (2023).
Sanson, K. R. et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 9, 1–15 (2018).
Hart, T. et al. Evaluation and design of genome-wide CRISPR/SpCas9 knockout screens. G3 7, 2719–2727 (2017).
Replogle, J. M. et al. Maximizing CRISPRi efficacy and accessibility with dual-sgRNA libraries and optimal effectors. Elife 11, e81856 (2022).
Singh, S., Bray, M.-A., Jones, T. R. & Carpenter, A. E. Pipeline for illumination correction of images for high-throughput microscopy. J. Microsc. 256, 231–236 (2014).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Lecun, Y., Bottou, L., Bengio, Y. & Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 86, 2278–2324 (1998).
Guizar-Sicairos, M., Thurman, S. T. & Fienup, J. R. Efficient subpixel image registration algorithms. Opt. Lett. 33, 156–158 (2008).
Haghighi, M. et al. Pseudo-labeling enhanced by privileged information and its application to in situ sequencing images. In Proc. Thirty-Second International Joint Conference on Artificial Intelligence https://doi.org/10.24963/ijcai.2023/531 (International Joint Conferences on Artificial Intelligence Organization, California, 2023).
Burger, W. & Burge, M. J. Principles of Digital Image Processing: Core Algorithms (Springer Science & Business Media, 2010).
Chen, T., Kornblith, S., Norouzi, M. & Hinton, G. A simple framework for contrastive learning of visual representations. In Proc. 37th International Conference on Machine Learning, 1597–1607. Preprint at https://arxiv.org/abs/2002.05709 (PMLR, 2020).
Radford, A. et al. Learning transferable visual models from natural language supervision. In Proc. of the 38th International Conference on Machine Learning, 8748–8763. https://proceedings.mlr.press/v139/radford21a.html (PMLR, 2021).
Dosovitskiy, A. et al. An image is worth 16x16 words: transformers for image recognition at scale. In Proc. International Conference on Learning Representations (2020).
De Boer, C. G., Ray, J. P., Hacohen, N. & Regev, A. MAUDE: inferring expression changes in sorting-based CRISPR screens. Genome Biol. 21, 1–16 (2020).
Acknowledgements
The authors would like to thank Deirdre O’Sullivan and Tina Mahatdejkul-Meadows for assistance with early iterations of plasmid designs. We would also like to thank Matt Rasmussen and Christoph Klein for helpful discussions regarding data sharing, and the insitro Discovery Labs for discussions and experimental/computational contributions.
Author information
Authors and Affiliations
Contributions
S.S., E.L., D.F., M.R.S., and C.C. designed the research. B.L., E.L., A.E., D.F., and M.R.S. collected the data. S.S., P.S., J.W.B., E.S., and M.R.S. conducted software engineering and/or computational analysis. E.L., M.M.S., and F.R. developed Mitoprobe. B.L., J.E.M., Z.F.P., Albert.K., J.C., and M.R.S. developed Automated POSH sequencing. N.R. and D.F. designed the sgRNA libraries. D.K., E.S., Ajamete.K., M.R.S., and C.C. supervised the work. S.S., B.L., D.K., Ajamete.K., M.R.S., and C.C. prepared and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Musa Mhlanga and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sivanandan, S., Leitmann, B., Lubeck, E. et al. A pooled Cell Painting CRISPR screening platform enables de novo inference of gene function by self-supervised deep learning. Nat Commun 17, 77 (2026). https://doi.org/10.1038/s41467-025-66778-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66778-6