Abstract
Strand exchange is a key step in homologous recombination, enabling template-based repair of DNA double-strand breaks. Eukaryotic RAD51 forms an ATP-dependent helical presynaptic filament on single-stranded DNA (ssDNA), which then searches for homologous double-stranded DNA (dsDNA), and catalyzes the strand exchange to form a D-loop in an ATP hydrolysis-independent manner. The molecular mechanism by which RAD51 facilitates dsDNA unwinding and pairing remains unclear. Here, we present cryo-EM structures of RAD51 mini-filaments bound to homologous dsDNA, capturing five intermediates from dsDNA recruitment to D-loop formation. These structures, together with molecular dynamics simulations, suggest a stepwise mechanism: the conserved N-terminal domain (NTD) recruits and bends the dsDNA, weakening base pairing near the exchange site. Subsequent engagement with positively-charged regions, including the loop L2 and loop Arg303–Arg306, further bends the homologous dsDNA, thereby not only positioning it closer to the strand exchange site but also inducing local base-pair opening. Additionally, the loop L2 (Met278 and Phe279) inserts between strands, and the secondary DNA binding sites (S2 sites) capture the displaced strand to prevent strand reannealing. Together, our findings provide detailed insight into a spatially coordinated mechanism of strand exchange by RAD51.
Similar content being viewed by others
Introduction
Homologous recombination (HR) is central to the template-based repair of DNA double-strand breaks, maintaining genome integrity1,2,3,4. A central reaction in HR is the strand exchange, in which the damaged ssDNA pairs with the complementary strand from the homologous dsDNA5,6. To catalyze the strand exchange reaction, recombinases, such as prokaryotic RecA and eukaryotic RAD51, must assemble on ssDNA to form an ATP-dependent helical nucleoprotein filament–termed the presynaptic filament7,8,9,10. This presynaptic filament recruits the homologous dsDNA to enable homologous pairing, giving rise to a postsynaptic filament in which the complementary strand of dsDNA is paired with the invading ssDNA to form a heteroduplex dsDNA. This pairing process results in a characteristic D-loop complex, where the complementary strand of the dsDNA pairs with the ssDNA, and the displaced strand is sequestered by site II11,12. RAD51 harbors two distinct DNA-binding sites that coordinate homologous pairing and strand exchange: site I (S1), a high-affinity DNA-binding site composed of loops L1 and L2, and site II (S2), a lower-affinity DNA-binding site enriched in basic residues (Supplementary Fig. 1)13. Located along the inner groove of the filament, L1 (e.g., Arg235 in human RAD51) facilitates homologous recognition, while L2 promotes strand exchange. Adjacent to S1, S2 captures the displaced donor strand and is essential for stabilizing the strand exchange intermediates12,14,15,16,17,18,19,20,21,22,23. Although ATP is present, its hydrolysis is not required during the exchange reaction. The cryo-EM study of prokaryotic RecA has shown that its 64-residue C-terminal domain (CTD) recruits the dsDNA and directs it to the loop L2, which destabilizes base pairing and opens dsDNA12,24. While eukaryotic RAD51 and RecA share a conserved ATPase core for ssDNA and ATP interactions, RAD51 lacks the RecA CTD, and, instead, possesses an evolutionarily conserved 66-residue NTD (Supplementary Fig. 1)25. Although the RAD51 NTD is structurally unrelated to the RecA CTD, biochemical studies suggest it is involved in dsDNA binding25. However, RAD51 NTD is spatially distant from the loop L2, so how RAD51 destabilizes the homologous dsDNA to allow pairing of the complementary strand remains poorly understood. Helical reconstruction cryo-EM has been powerful in revealing atomic-resolution RAD51 presynaptic and postsynaptic filament structures8. However, dsDNA recruitment and D-loop formation only occur at specific protomers, disrupting the helical pattern. Consequently, the symmetry averaging in helical reconstruction obscures the D-loop structure.
In this work, we show a RAD51 mini-filament of approximately nine protomers, enabling cryo-EM structure determination using single-particle analysis without imposing symmetry.
Results
RAD51-SEAD mini-filaments resemble RAD51 filaments
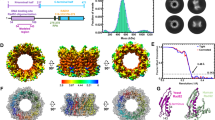
During cryo-EM sample preparation, wild-type RAD51 readily assembles into extended nucleofilaments, hindering single-particle analysis because their preferred lay-down orientation limits the acquisition of sufficient 2D projections necessary for the following 3D reconstruction. A previously characterized S208E/A209D double mutant (hereafter RAD51-SEAD), which disrupts BRC4 interaction but retains filament formation ability8,26,27,28, offers a solution to this challenge. Within the short timeframe of cryo-EM sample preparation, mouse RAD51-SEAD preferentially assembles into mini-filaments consisting of ~6–10 protomers on 27 nt ssDNA or 27 bp dsDNA in the presence of 5 mM CaCl₂ and 1 mM AMP-PNP, a non-hydrolyzable ATP analog (Fig. 1a). With prolonged incubation, it forms extended filaments similar to wild-type RAD51 (Supplementary Fig. 2)26,27,28. Importantly, RAD51-SEAD retains wild-type activity in presynaptic filament assembly and DNA strand exchange in vitro (Fig. 1b).
a Representative cryo-EM micrograph depicting both mini-filaments and extended RAD51 filaments formation using the SEAD mutant. The mini-filaments are highlighted in the red circles. The experiment was repeated at least three times with similar results. b DNA strand exchange activity of the mouse RAD51-SEAD mutant is comparable to that of the wild-type (WT) protein. (i) Schematic representation of the strand exchange reaction. An asterisk (*) indicates the Cy5 fluorescent dye. (ii) DNA strand exchange activity of RAD51-SEAD. The results were graphed. Data represent the mean ± standard error of the mean (s.e.m.) from three independent experiments (n = 3). Source data are provided as a Source Data file. c Cryo-EM density map (left) and atomic model (right) of the RAD51 presynaptic mini-filament. The N-terminal domain (NTD) is colored orange and the bound ssDNA is shown in red. d Cryo-EM density map (left) and atomic model (right) of the RAD51 postsynaptic mini-filament. The NTD, ssDNA, and the paired complementary strand are colored orange, red, and blue, respectively.
Single-particle cryo-EM reconstructions revealed structures of presynaptic and postsynaptic RAD51-SEAD mini-filaments at resolutions of 2.91 Å and 3.04 Å, respectively (Fig. 1c, d; Supplementary Fig. 3 and Supplementary Fig. 4). Each mini-filament comprises nine RAD51-SEAD protomers, each engaging DNA in nucleotide triplet clusters, consistent with previously reported human RAD51 filament structures solved by helical reconstruction7,8. Structural comparisons between our mouse RAD51-SEAD mini-filaments and reported RAD51 filaments show near-identical conformations, with root-mean-square deviations of 0.93 Å and 1.04 Å for presynaptic and postsynaptic filament, respectively, confirming that the mini-filaments structurally recapitulate wild-type RAD51 nucleofilament for our structural studies.
Partially-mismatched ssDNA/dsDNA trap RAD51 mini-filaments at different conformational states
The strand exchange reaction occurs when the RAD51 presynaptic filament recruits homologous dsDNA. Previous RecA studies utilized partially mismatched homologous dsDNA—engineered to contain a bubble of mismatched regions aligned with ssDNA—to open up dsDNA locally, thereby capturing RecA D-loop structures12. By systematically testing over two dozen dsDNA substrates containing various mismatched bubbles, we successfully captured five distinct structural intermediates using single-particle analysis and determined the corresponding structures (Fig. 2). The DNA recruitment complex in this study refers to the state of the RAD51 presynaptic filament upon engaging the homologous dsDNA. In this complex, the dsDNA is bound to the filament but has not yet undergone the conformational changes necessary for D-loop formation. A 4-bp G-C mismatched substrate yields a RAD51 mini-filament density map at 4.53 Å resolution (Supplementary Fig. 5). The density clearly showed that an unbent dsDNA interacts with two adjoining NTDs, presenting the early recruitment state of homologous dsDNA (Fig. 2a). Increasing the mismatch to 8-bp yielded another density map at 3.23 Å (Fig. 2b and Supplementary Fig. 6), where dsDNA displayed pronounced curvature, interacting with four adjoining NTDs (N + 1, N + 2, N + 3, and N + 4). These two structures represent the recruitment of homologous dsDNA via nonspecific interactions of NTDs. Positioning an 8-bp mismatch near the 5’-end allowed us to solve a 3.91 Å resolution map featuring a D-loop formation, with clear heteroduplex pairing involving 5’−1-⅓−3’ base-pair triplets (Fig. 2c and Supplementary Fig. 7). For clarity, we describe heteroduplex formation in the D-loop complex in terms of base-pair triplets, with each triplet comprising three base pairs accommodated by a single RAD51 protomer. A complete triplet represents three fully paired bases, whereas fractional terms (e.g., ‘1/3’ or ‘2/3’) indicate partial triplet formation. Our simulations (see section below) allow us to place the first three intermediates in a temporal sequence of RAD51 D-loop formation. Relocating the mismatch toward the 3’-end enabled us to resolve RAD51 D-loop structures at 3.2 Å resolution (Fig. 2d and Supplementary Fig. 8), with clear heteroduplex pairing involving 5’−2-⅔−3’ base-pair triplets. Extending to 12-bp mismatch yields additional RAD51 D-loop structures at 3.42 Å resolution, with heteroduplex pairing involving 5’-⅓–3–⅔−3’ triplets (Fig. 2e and Supplementary Fig. 9). The simulation (see section below) suggested a transitional intermediate between the last two intermediates for D-loop propagation.
a–e Cryo-EM reconstructions of RAD51 D-loop complexes assembled with donor dsDNA substrates containing 4-bp, 8-bp, another 8-bp, 8-bp, and 12-bp mismatched bubbles, respectively. For each panel, a schematic of the ssDNA and the corresponding dsDNA mismatched bubble is shown on the left, and the corresponding cryo-EM density map is shown on the right. RAD51 protomers are labeled from N + 1 to N + 9 along the filament axis. In the density maps, the N-terminal domain (NTD), displaced strand, and complementary strand are colored orange, green, and blue, respectively. The global resolution of each reconstruction is indicated in the respective panel.
Structural basis of D-loop formation by RAD51
Using a 48 bp dsDNA substrate containing an 8-bp mismatched bubble (Fig. 3a), the cryo-EM map at 3.91 Å revealed a RAD51 mini-filament comprising seven protomers (N + 1 to N + 7) and partial dsDNA (Fig. 2c). The dsDNA structure features a 3’-tilted 6-bp duplex, an 11-bp D-loop, and a 5’-tilted 12-bp duplex. To form this 11 bp D-loop, the nucleotides 5’-G25–A28 of the complementary strand form a heteroduplex with ssDNA containing 5’−1-⅓−3’ base-pair triplets at site IN+4 and site IN+5 (Fig. 3b and Supplementary Fig. 10a, b). Additionally, the 8 bp mismatched bubble is accompanied by unwinding of the adjacent flanking regions—2 bp at the 3′ end and 1 bp at the 5′ end relative to the complementary strand (Fig. 3a, b).
a–e D-loop complex containing a ⅓–1 base-pair triplet heteroduplex, (f–i) D-loop complex with a 2 and ⅔ base-pair triplet, (j–m) D-loop complex with a ⅓–3–⅔ base-pair triplet. (a, f, j) Nucleotide sequences of the 27-nt ssDNA and the corresponding 48-bp donor dsDNA. The invading ssDNA is shown in red, and the homologous regions (8 bp or 12 bp) on the dsDNA are highlighted. The green bases represent the displaced strand. b, g, k Atomic models of RAD51–DNA complexes illustrating progressive heteroduplex formation at different triplet stages. Nucleotide sequences are labeled to show specific base-pairing events. c, h, l Ribbon diagrams of the RAD51 D-loop complexes highlighting the heteroduplex core. The ssDNA, displaced strand, and complementary strand are colored red, green, and blue, respectively. Key RAD51 residues involved in DNA interaction are shown as stick-and-ball: S2 site residues in gray, L2 loop residues in yellow, N-terminal domain (NTD) residues in orange, and Arg235 in cyan. Corresponding RAD51 protomers are labeled as superscripts. d, i, m Close-up views of protein–DNA interactions near the strand separation site. – (d) Interactions of Met278N+6,Phe279N+6, and Arg306N+7 with the just-separated displaced strand. – (i) Interactions of Arg235N+6, M278N+7, F279N+7, and Arg306N+8 with the just-separated displaced and complementary strands. – (m) Interactions involving Arg235N+6, M278N+7, F279N+7, M278N+8, F279N+8, and Arg306N+8 with the just-separated displaced and complementary strands.
The dsDNA is bound by Lys64 and Lys70 of NTDN+1, NTDN+2, and NTDN+4, curving toward site IN+5 (containing loop L1N+5 and L2N+6) for the strand exchange (Fig. 3c). The dsDNA bound results in a 6.4° rotation of NTDN+1 relative to unbound NTD (such as NTDN+3) (Supplementary Fig. 11). Notably, hydrophobic Met278N+6, Phe279N+6, Met278N+7, and Phe279N+7 from loop L2 occupy positions between newly-separated strands, presumably preventing dsDNA reannealing (Fig. 3c)17,25. The positively-charged Arg306N+7 interacts with the phosphate backbone of T26 and T27 from the displaced strand (Fig. 3d and Supplementary Fig. 10c). Other residues in the S2 site, including Lys304N+7, Arg303N+7, Arg130N+7, and Lys284N+6, further stabilize the displaced strand, highlighting the functional significance of S2 (Fig. 3c)12. In contrast to the canonical role of Arg235 in base-pair triplet partition, Arg235N+6 is rotated outward to engage the newly-separated complementary strand (Fig. 3e and Supplementary Fig. 10d). Collectively, these structural observations reveal the precise spatial coordination and residue-specific interactions orchestrating RAD51-mediated dsDNA opening, displaced strand stabilization, and homologous pairing.
Some of the flanking bases, in addition to the heteroduplex, in Fig. 3c (h and l) on the duplex DNA, are not base-paired. The simulations (see section below) show that RAD51 induces significant bending in the homologous dsDNA. This bending introduces local mechanical strain that may destabilize the base pairing in the duplex DNA.
Structural basis of RAD51 D-loop formation and pairing propagation
By positioning an 8-bp mismatched bubble aligning with the 3’-end of the ssDNA region (Fig. 3f), we captured another cryo-EM structure of D-loop intermediate at 3.2 Å resolution (Fig. 2d). The resulting RAD51 mini-filament, comprising nine protomers (N + 1 to N + 9), resolves heteroduplex formation at the site IN+3, site IN+4, and site IN+5(Fig. 3h). The density map of homologous dsDNA reveals a 3’-tilted 9-bp duplex region followed by an 11 bp bubble (Fig. 3g). The 3’-titled duplex interacts with the positively-charged Lys64 and Lys70 of NTDN+3 likely via its negatively-charged phosphate backbone (Fig. 3h). Notably, eight nucleotides (5’-G23–G30) of the complementary strand form a 2 and ⅔ triplet heteroduplex with ssDNA (Fig. 3g and Supplementary Fig. 10e, f). The displaced strand is stabilized by positively-charged residues in S2 sites, while hydrophobic residues, including Phe279N+5, Met278N+5, Phe279N+6, and Met278N+6, occupy space between the heteroduplex and displaced strand to prevent the dsDNA reannealing (Fig. 3h). Slightly different interactions near the just-separated displaced strand include a π-stacking interaction between the benzyl group of Phe279N+7 and the nucleotide T17, as well as electrostatic contacts between Arg306N+8 and phosphate backbones of nucleotides T17 and C16 from the displaced strand (Fig. 3i and Supplementary Fig. 10g).
Extending the mismatch to 12 bp yielded another D-loop intermediate (Fig. 3j), where the complementary strand forms a ⅓–3–⅔ triplet heteroduplex with ssDNA, spanning five protomers (Fig. 3k and Supplementary Fig. 10h, i). The corresponding cryo-EM structure was determined at a resolution of 3.42 Å. The density map of homologous dsDNA reveals a 3’-tilted 9 bp duplex region followed by a 15 bp bubble and a 5’-tilted 6-bp duplex (Fig. 3k). Within the 9-protomer mini-filament, 12 nucleotides (5’-G19–G30) form heteroduplexes at sites IN+3 through the site IN+7 (Fig. 3l). The 3’-tilted duplex likely interacts with the positively-charged Lys70N+3 and Lys73N+3, whereas the 5’-tilted duplex interacts with the Lys70N. As observed previously, positively-charged residues at the S2 site stabilize the displaced strand, while hydrophobic Phe279N+5, Met278N+5, Phe279N+6, Met278N+6, Phe279N+7, Met278N+7from loop L2 occupy space between the heteroduplex and the displaced strand (Fig. 3l). Near the just-separated displaced strand, a π-stacking interaction between the benzyl group of Phe279N+7 and the nucleotide T19 and charge-charge interaction between Arg306N+8 and the nucleotide of A18 from the displaced strand was also observed (Fig. 3m and Supplementary Fig. 10j). Phe279N+8 and Met278N+8 from the loop L2 are near the base pairing between T17 and A32, contributing to hydrophobic packing (Fig. 3m).
Comparative analysis of different D-loop intermediates revealed that Arg235 in the loop L1 (Supplementary Fig. 12), known for its role in partitioning heteroduplex DNA into triplet units, adopts variable conformations away from the exchange site, contacting the newly separated complementary strand. This suggests an additional function for Arg235 in guiding the just-separated complementary strand toward the strand exchange, consistent with diminished exchange activity observed in the R235A mutant29. Additionally, the contact between the 3’-tilted duplex and the NTD differs slightly depending on the degree of heteroduplex pairing.
Overall, our cryo-EM structures support a conserved mechanism in which the displaced strand is sequestered by a positively-charged channel created by the S2 site, while the hydrophobic residues from the loop L2 wedge into the space between the heteroduplex and the displaced strand, to prevent reannealing. Conformational variability in Arg235 and in the 3’-titled duplex suggests local rearrangement to modulate pairing propagation30.
Previous functional studies showed that a minimal 8-nt tract of microhomology is required for stable D-loop formation20,31. This requirement likely reflects the structural organization of the RAD51 filament and its interaction with DNA. For productive strand pairing and stable D-loop initiation, the complementary strand must spatially align with the invading ssDNA, while its two terminal ends are anchored by the NTDs of protomers N + 1 and N + 4 within the RAD51 filament. Our cryo-EM analysis of the RAD51 D-loop intermediates indicates that an 8-nt homologous segment is the shortest length that enables this precise spatial alignment and pairing. When homology is reduced to 7 bp, as observed in the cryo-EM reconstruction with a 7 bp tract (Supplementary Fig. 13), the contacts with the NTDs remain intact, but the complementary strand fails to form a stable heteroduplex with the invading ssDNA. These findings demonstrate that 8-nt microhomology motifs represent the essential minimal unit required for homologous pairing and stable D-loop initiation, a critical step in homologous recombination.
Molecular dynamics simulation for base pair break
To assess whether homologous dsDNA recruitment by NTDN+1 and NTDN+2 is sufficient to initiate dsDNA bending required for D-loop formation, we performed molecular dynamics (MD) simulations using a straight B-form dsDNA model docked onto the dsDNA density shown in our 4 nt mismatched complex (Fig. 2a). In addition, the RAD51-SEAD double mutant was mutated back to the wild-type for simulation work. The simulation revealed that the B-form dsDNA bound to NTDN+1 and NTDN+2 can be attracted toward the positively charged channel near the ssDNA-binding site and further interacts with positively-charged NTDN+3, resulting in a subtle bending of the dsDNA with approximately 170° (Fig. 4a–d). Although the nucleotides remain base-paired at this stage, the dsDNA bending introduces local restraint that likely destabilizes the base pairing (Fig. 4d)24. As the simulation progresses, this 3′-tilted duplex transiently interacts with the positively charged loop 2 N+7 and loop Arg303–Arg306N+7. These contacts further bend dsDNA to approximately 140°, resulting in the first base-pair break with the proximity of the loop L2 (Figs. 4b and 4e and Supplementary Movie 1). After the base pair disruption, the 3’-titled dsDNA bypass NTDN+4 due to spatial constraints and engages with NTDN+5, resulting in an angular deflection of about 120° (Fig. 4f), closely mirroring the dsDNA conformation and its NTD interactions observed shown in Fig. 2c (Supplementary Fig. 14).
a The hydrogen bond distance variation between two specific nucleotide base pairs during the initiation of D-loop formation. The blue line indicates the distance variation for the base-pair 1 (A–T base pair hydrogen bond distance between the N1 atom of adenine and the N3 atom of thymine); the purple line indicated the distance variation for the base-pair 2 (A–T base pair hydrogen bond distance between the N1 atom of adenine and the N3 atom of thymine). Source data are provided as a Source Data file. b Snapshots of RAD51–dsDNA interaction at four key time points (1 ps, 10 ns, 19 ns, and 47 ns). The base-pair opening occurs around 19 ns. RAD51 is rendered as a surface colored by electrostatic potential (blue, positive; red, negative). dsDNA is shown in a cartoon representation. Regions of base-pair opening are highlighted with blue circles, and the positively charged Arg303–Arg306N+7 region is outlined with a green dashed box. c–f Top panels: Cartoon schematic representations showing nucleotide base-pair interactions at key simulation time points (1 ps, 10 ns, 19 ns, and 47 ns), depicting the progressive disruption of base pairs during dsDNA bending. Bottom panels: Structural snapshots illustrating the corresponding dsDNA bending conformations at 1 ps, 10 ns, 19 ns, and 47 ns, respectively.
To examine whether this unwinding mechanism continues during D-loop propagation, we used the 8 bp mismatched model (Fig. 2d) as the starting structure, modifying the 3′-tilted duplex to consist entirely of G:C base pairs to enhance the base-pairing stability and RAD51-SEAD double mutant was mutated back to the wild-type. In the starting model, a heteroduplex is already formed at sites IN+4 and IN+5, while the 3′-tilted duplex interacts with NTDN+4. G31 from the complementary strand remains base-paired with the displaced strand and is approximately 6 Å from site IN+6 (Fig. 2d). MD simulations revealed that the 3′-tilted duplex detaches from NTDN+4, allowing 9.7° upward translocation (Fig. 5 and Supplementary Movie 2). Engagement with the positively charged region of loop L2N+7, loop Arg303–Arg306N+7, and loop Arg303–Arg306N+8 permits the complementary strand to approach site IN+6, resulting in the breakage of the G31 base-pair (Fig. 5b, c–f). These observations indicate a stepwise mechanism of strand exchange propagation to the next RAD51 protomer, mediated by the detachment from the bound NTD and the engagement of the loop Arg303–Arg306, bringing dsDNA close to the strand exchange site and locally breaking the desired base-pair (Fig. 5b). Notably, the breakage event occurs in proximity to the loop L2N+7 and S2 siteN+7, allowing the L2N+7 insertion into the newly-separated dsDNA and enabling the S2 sites to capture the displaced strand, consistent with previous mutagenesis studies32.
a The hydrogen bond distance variation between two specific nucleotide base pairs during the propagation of D-loop formation. The blue line indicates the distance variation for the first pair (A–T base pair hydrogen bond distance between the N1 atom of adenine and the N3 atom of thymine); the purple line indicated the distance variation for the second pair (A–T base pair hydrogen bond distance between the N1 atom of adenine and the N3 atom of thymine). Source data are provided as a Source Data file. b Snapshots of RAD51–dsDNA interaction at four key time points (1 ps, 145 ns, 151 ns, and 180 ns). The base-pair opening occurs around 151 ns. RAD51 is rendered as a surface colored by electrostatic potential (blue, positive; red, negative). The dsDNA is shown in cartoon representation. Regions of base-pair opening are highlighted with blue circles, and the positively charged Arg303–Arg306 region is outlined with a green dashed box. c–f Top panels: Cartoon schematic representations showing nucleotide base-pair interactions at key simulation time points (1 ps, 145 ns, 151 ns, and 180 ns), depicting the progressive disruption of base pairs during dsDNA bending. Bottom panels: Structural snapshots illustrating the corresponding dsDNA bending conformations at 1 ps, 145 ns, 151 ns, and 180 ns, respectively.
Functional analysis of structure-based mutants
Structure-based mutagenesis analyses support the functional roles of the NTD, loop L2, and the Arg303–Arg306 segment in RAD51-mediated strand exchange. We found that the Lys58Ala/Lys64Ala double mutation in the NTD severely impairs ssDNA-binding affinity, consistent with a previous report, which makes it difficult to evaluate the specific contribution of these residues to strand exchange. In contrast, Pro83 is positioned at the linker between NTD and ATPase core. By substituting a rigid proline residue with flexible glycine at position 83, we were able to subtly perturb NTD movement without significantly compromising its ssDNA-binding affinity. On this basis, we selected the P83G mutant for functional analysis of NTD. Lys284, located within loop L2, contributes to its positively-charged surface. Although these residues, P83, K284, and R306, are spatially distant from the ssDNA-binding groove, the mutations exhibit a modest reduction in ssDNA-binding efficiency, requiring slightly higher protein concentration to reach saturation relative to the wild-type (Fig. 6a). This suggests their additional role in presynaptic filament formation or stability. Nevertheless, under saturating DNA-binding conditions, P83G, K284A, and R306A mutations exhibited persistently reduced strand exchange activity (Fig. 6b), reinforcing the notion that NTD, the loop L2, and Arg303–Arg306 region contribute to destabilizing base pairing in the donor duplex during strand exchange. In parallel, we conducted a comparative analysis of strand exchange activity between the wild-type and mutant proteins using both a fully homologous dsDNA substrate and a partially separated dsDNA substrate containing a 12-bp mismatched bubble. If these residues are involved in strand separation, the mutants would be defective in strand exchange with fully homologous dsDNA, but their activity could be rescued when tested with the partially separated substrate. As expected, all three mutants showed a ~ 50% increase in activity with the partially separated substrate compared to the fully homologous substrate (Supplementary Fig. 15), whereas wild-type RAD51 exhibited only a marginal change. These findings highlight the specific contributions of the NTD, loop L2, and the Arg303–Arg306 segment to strand separation, consistent with our proposed model. Lastly, our proposed mechanism is consistent with previous functional studies, supporting the conclusion that the NTD and loop L2 contribute to dsDNA recruitment and facilitate the transition from intact dsDNA to heteroduplex formation23,32.
a Quantification of ssDNA-binding activity of RAD51 variants P83G, K284A, and R306A. The percentage of bound ssDNA was calculated by the decrease in the percentage of free ssDNA. The results were plotted. Data represent the mean ± standard error of the mean (s.e.m.), n = 3 independent experiments. Source data are provided as a Source Data file. b D-loop formation assay of RAD51 variants. (i) Schematic illustration of the D-loop assay. The asterisk (*) marks the Cy5-labeled oligonucleotide. (ii) Quantification of D-loop formation by each RAD51 variant. Data represent the mean ± s.e.m. from four independent experiments (n = 4). Under DNA-saturating conditions, RAD51 variants exhibit intrinsically reduced D-loop formation activity compared to wild-type RAD51. Source data are provided as a Source Data file.
Discussions
In the present work, we resolved five distinct cryo-EM structures of RAD51–DNA mini-filaments that may represent sequential intermediates along the strand exchange pathway. These structures reveal mechanistic details of how RAD51 presynaptic filament recruits homologous dsDNA and promotes base-pair opening and strand exchange. Specifically, the conserved N-terminal domain (NTD) interacts with the duplex DNA, while a positively charged groove comprising Arg130, Lys284, Arg303, Lys304, Arg306, and Lys313 (designated as S2 site) engages the displaced strand (Supplementary Fig. 1)25. Concurrently, Met278 and Phe279 in the loop L2 insert between the complementary and displaced strands, potentially preventing dsDNA re-annealing.
Our molecular dynamics simulations not only provide a coherent timeline of our cryo-EM structures during the strand exchange event but also structural snapshots of D-loop initiation and propagation. Protein-induced DNA bending and local kinking is a common mechanism for destabilizing base pairs, observed in various biological systems including CRISPR-Cas12a and DNA glycosylases33,34,35,36,37. During D-loop initiation, the 5′-tilted duplex engages with NTDN+1 to NTDN+3, while the 3′-tilted duplex is attracted to the positively charged loop 2 N+7 and loop Arg303–Arg306N+7, inducing a bend of approximately 140° in the homologous dsDNA (Fig. 7a). This bending weakens the nucleotide base pairing and brings it near the exchange site located along the central axis of the RAD51 presynaptic filament. Further analysis of the dsDNA conformation shows that bending extends the major groove, reflecting a common feature of dsDNA unwinding induced by enzymes38,39,40. In our proposed mechanism, RAD51 induces dsDNA bending, suggesting its ability to bind to bent dsDNA, consistent with previous findings41. The bending toward the central axis to destabilize the local base-pair was also proposed by the simulation work of RecA-mediated strand exchange24. The following heteroduplex formation results in 3′-tilted duplex that bypasses NTDN+4 and instead interacts with NTDN+5 as shown in Fig. 2c. Similar loop Arg303–Arg306-mediated translocation and NTD disengagement are also observed during D-loop propagation, suggesting a recurring mechanism for strand exchange progression (Fig. 7b).
a Structural Illustration of D-loop Formation: This panel depicts the dynamic transition from a straight B-form dsDNA molecule to a bent conformation that displays the initial base-pair opening, as derived from our MD simulations. The initial straight model (from Fig. 2a) is overlaid in magenta for structural comparison. b Structural Illustration of D-loop Propagation: This image utilizes snapshots of cryo-EM D-loop intermediates to demonstrate the extension of homologous pairing. It contrasts the initial 4 bp heteroduplex structure (Fig. 2c, left) with the expanded 12-bp pairing (Fig. 2e, right). The overlay highlights the overall movement and propagation of the heteroduplex along the filament. c Filament Core and Functional Elements: This provides a simplified cartoon representation of the RAD51 presynaptic filament, highlighting the key functional elements involved in strand exchange: the NTD (blue), the L2 loop (orange), and the Arg303-Arg306 segment (green) of the structural core. d Proposed Stepwise Mechanism of Formation and Propagation: This comprehensive schematic integrates our cryo-EM structures and MD simulations into a unified model for RAD51-mediated D-loop activity. The process proceeds in the 3’ to 5’ direction (relative to the invading ssDNA, red strand): 1. dsDNA Recruitment: The RAD51 filament recruits homologous dsDNA. The N + 1 and N + 2 NTDs initiate engagement and minor unwinding (supported by Fig. 2a). 2. dsDNA Unwinding: Subsequent engagement by additional NTDs and positively charged regions (L2, Arg303-Arg306) induces pronounced dsDNA bending. This bending promotes local base-pair opening at the exchange site (supported by Fig. 2b and MD simulation, Fig. 4). 3. Strand Sequestration: Hydrophobic L2 residues (Met278 and Phe279) insert between the separated strands to prevent re-annealing. Concurrently, the displaced strand (green) is sequestered by the positive S2 sites. 4. Propagation: Once the initial D-loop forms (Fig. 2c), the cycle repeats in the 3’ to 5’ direction. This involves the sequential detachment of the 3’-tilted duplex from one NTD and its re-engagement with the next set of positively charged regions on adjacent protomers. This action drives further base-pair disruption and heteroduplex extension (derived from Fig. 2d, e, and MD simulation, Fig. 5).
Our findings bear similarities—but also key differences—to the mechanism proposed for RecA12. Previous RecA cryo-EM structures suggested that its CTD binding to dsDNA causes a steric clash with loop L2, resulting in L2 insertion and strand opening, followed by sequestration of the displaced strand by the S2 sites for opening propagation12. In contrast, our data suggest that in RAD51, the NTD first disengages from the 3’-tilted duplex, enabling repositioning of the dsDNA toward the positively charged surfaces of the loop L2 and the loop Arg303–Arg306 (Fig. 7c, d). This transition induces the bending of the dsDNA, promoting local base-pair opening and enabling insertion of the loop L2, and displaced strand sequestering by S2 which in turn delivers the complementary strand to the strand exchange site for homologous pairing32. Our cryo-EM analyses, combined with computational simulations, offer a stepwise visualization of the D-loop formation process, as shown in Fig. 7d. The dsDNA recruitment complexes are reflected in our cryo-EM results shown in Fig. 2a, b, where the filament engages the homologous dsDNA and induces bending, supported by our MD simulations. The structure in Fig. 2c represents an intermediate state of D-loop formation. Finally, the cryo-EM structures in Fig. 2d, e reflect subsequent stages of D-loop formation, with extended regions of homologous pairing, again supported by MD simulations, demonstrating how the heteroduplex propagates along the filament.
The structural comparison of the RAD51 D-loop with published RecA D-loop structures reveals both a fundamental conservation in the architecture and geometry of the invading strand and D-loop. As illustrated in Supplementary Fig. 16, the overall D-loop geometry and the conformation of the invading ssDNA strands are remarkably similar between RAD51 and RecA. In both complexes, the L2 loop actively inserts into the dsDNA to separate the strands, and the site II DNA-binding site sequesters the displaced strand. Despite differences in domain usage for dsDNA interactions, the D-loop geometry near the 5′-tilted duplex is strikingly conserved, even facilitating the formation of an additional base pair. However, the key distinction arises near the 3′-tilted duplex. In RAD51, the 3′-tilted duplex undergoes an additional ~60° outward twist compared to RecA. Consequently, the 3′-tilted duplex interacts with the NTD of the RAD51 N + 3 protomer, whereas in RecA it interacts with the CTD of the N + 4 protomer.
Lastly, our work reveals that RAD51-mediated strand exchange proceeds in a 3’ to 5’ direction relative to the invading single-stranded DNA (ssDNA). The high-resolution postsynaptic complex structure allowed us to unambiguously determine the polarity of the invading strand (Supplementary Fig. 17). This observation reveals that the RAD51 mini-filament structures are oriented with their NTDs pointing toward the 5’ end of the invading ssDNA, which is consistent with previously published models for human RAD518,23. Consequently, our numbering of the protomers (e.g., from N + 1 to N + 9) proceeds along the filament axis from the 3’ to the 5’ end of the invading ssDNA. With this polarity established, our D-loop structures and subsequent molecular dynamics simulations demonstrate that heteroduplex formation propagates from protomer NTD N + 4 to NTD N + 5 (Fig. 5), corresponding to movement from the 3′ end to the 5′ end of the invading ssDNA, thus providing direct structural evidence for a 3’ to 5’ propagation mechanism. This conclusion is in full agreement with several well-established biochemical studies that have also demonstrated a 3’ to 5’ polarity for strand exchange relative to the invading ssDNA (equivalently, a 5’ to 3’ polarity relative to the complementary strand)42,43,44. The recently published cryo-EM structure of the human RAD51 D-loop by Joudeh et al. proposed an opposite 5′ to 3′ polarity, based on the asymmetric binding of the L2 loops and the interaction of the RAD51 N-terminal domain with the donor DNA arms23, indicating that further experimental reconciliation is needed.
In summary, our cryo-EM structures and molecular simulation suggest a model in which RAD51 orchestrates a spatially and temporally coordinated series of events—duplex bending, base-pair destabilization, strand separation, and the sequestration of the displaced strand— to initiate and propagate homologous pairing during eukaryotic recombination (Fig. 7d).
Methods
RAD51 expression and purification
The plasmid harboring wild-type mouse RAD51 cDNA in vector pET51b (Novagen) was subject to site-directed mutagenesis to construct the different mRAD51 mutants28,45. The resulting plasmids were sequenced to ensure no unwanted mutation. Mouse RAD51-SEAD double mutant protein and other single mutant proteins such as P83G, K284A, and R306A were expressed in the E. coli RecA-deficient BLR strain harboring pRARE plasmid to supply tRNA for rare codons. Expression and purification of mutant proteins involves the following steps. Cells were grown in Luria broth at 37 °C until the A600 reached 0.8, followed by treatment with 1 mM IPTG for 3 h at 37 °C to induce protein expression. Cells were harvested and lysed in buffer K containing 20 mM K2HPO4 at pH 7.5, 0.5 mM EDTA, 300 mM KCl, 10% glycerol, 0.01% Igepal, and 1 mM 2-mercaptoethanol. A protease inhibitor cocktail—PMSF, benzamidine, aprotinin, chymostatin, leupeptin, and pepstatin A—was included to prevent protein degradation. The mRAD51 suspension was subjected to a Streptactin affinity step, specifically washed with buffer K and eluted with 4 mM D-desthiobiotin in buffer K containing 50 mM KCl, this was then followed by chromatographic fractionation using Source Q anion exchange chromatography column. The Source Q column (1 mL) was equilibrated with Buffer A (25 mM Tris-HCl, pH 7.5, 50 mM KCl, 10% glycerol, 0.5 mM EDTA, 0.01% Igepal, 1 mM beta-mercaptoehtanol). Protein was loaded at 0.8 mL/min and washed at 1 mL/min. Elution was achieved using an increasing concentration of Buffer B (Buffer A +1 M KCl) from 0% to 70% at 0.8 mL/min. Finally, the mRAD51-containing fractions were pooled, concentrated, and aliquoted for storage at –80 °C.
Electrophoretic mobility shift assay (EMSA)
All oligonucleotide sequences used in this study are listed in the Supplementary Table 1. To analyze ssDNA-binding activity, 2.4 μM nucleotides of 5′-Cy5-labeled 90-mer single-stranded DNA (oligo1) were incubated with mouse RAD51 variants at the indicated concentrations (0.1, 0.2, and 0.4 μM for wild-type; 0.2, 0.4, 0.8, and 1.2 μM for P83G, K284A, and R306A mutants, respectively) in a total volume of 10 μL of reaction buffer C (35 mM Tris-HCl, pH 7.5, 1 mM DTT, 2.5 mM MgCl₂, 5 mM CaCl₂, 0.1 mg/mL BSA, 45 mM KCl, and 1 mM ATP). The reaction mixtures were incubated at 37 °C for 10 minutes and resolved on 4% native polyacrylamide gels in TAE buffer (40 mM Tris, 20 mM acetate, 2 mM EDTA, pH 7.5) at 4 °C. Fluorescence signals were detected using an Amersham™ Typhoon™ Biomolecular Imager equipped with a Cy5 670BP30 (655–685 nm) filter, and band intensities were quantified using ImageQuant TL software (Cytiva).
DNA strand exchange assay
We conducted DNA strand exchange assays to validate the functional roles of specific residues in our cryo-EM studies.
Wild-type and SEAD mutant
To assemble presynaptic filaments, 3 μM nucleotides of 80-mer ssDNA (oligo2) were pre-incubated with either wild-type mouse RAD51 or the SEAD mutant at concentrations of 0.25, 0.33, and 0.5 μM in reaction buffer C. Strand exchange was initiated by adding 1.5 µM base pairs of homologous 40-bp dsDNA, which was formed by annealing a 5′-Cy5-labeled oligo3 with its complementary oligo4. The reaction, with a final volume of 10 µL, was incubated at 37 °C for 30 minutes. The reaction was then terminated with SDS and proteinase K, and the products were separated by electrophoresis on a 10% polyacrylamide gel. Fluorescence imaging and quantification were performed to analyze the results using the Amersham™ Typhoon™ Biomolecular Imager and ImageQuant TL software (Cytiva).
P83G, K284A, and R306A mutants
To validate the functional roles of specific residues identified in our cryo-EM structures, we performed a series of strand exchange assays using site-directed RAD51 mutants. The RAD51-P83G mutant was designed to probe the role of NTD flexibility, while the RAD51-K284A and RAD51-R306A mutants were constructed to assess the functional contributions of the L2 loop and Site II, respectively. These mutants were tested for their ability to catalyze strand exchange on a fully homologous dsDNA substrate and a dsDNA substrate containing a 12-bp mismatch. Presynaptic filaments were assembled by pre-incubating 3.6 μM nucleotides of 80-mer ssDNA (oligo2) with either wild-type mRAD51 or the three mutants at concentrations of 0.45 and 0.9 μM in reaction buffer C. Strand exchange was initiated by adding 1.5 µM base pairs of the homologous 40-bp dsDNA or the 12-bp mismatched dsDNA, which was formed by annealing a 5′-Cy5-labeled oligo3 with its complementary oligo5. The reaction, with a final volume of 10 µL, was incubated at 37 °C for 15 min. The results were analyzed by gel electrophoresis to quantify strand-exchanged product formation and compared to the activity of wild-type RAD51, allowing us to directly correlate our structural observations with their functional significance.
D-loop formation assay
For D-loop assays, 2.4 μM nucleotides of 5′-Cy5-labeled 90-mer ssDNA (oligo1) were pre-incubated with various concentrations of mouse RAD51 variants (0.4, 0.8, 1.2, and 2.4 μM) in reaction buffer C supplemented with 3 mM ATP for 5 minutes at 37 °C. The reaction was initiated by adding 37 μM base pairs of pBluescript replicative form I (supercoiled) dsDNA to a final volume of 10 μL. After 2 h of incubation at 37 °C, the reaction was stopped by adding 1 μL of 0.5% SDS containing proteinase K (8 μg), followed by incubation at 37 °C for 15 min. Samples were analyzed on 0.9% agarose gels in TBE buffer at 4 °C. Fluorescent signals were detected with an Amersham™ Typhoon™ Biomolecular Imager, and band intensities were quantified using ImageQuant TL software (Cytiva).
Cryo-EM sample preparation and data acquisition
The primary challenge in obtaining a high-resolution RAD51-DNA D-loop structure by single-particle cryo-EM is the propensity of wild-type RAD51 to form long, extended nucleoprotein filaments, which are unsuitable for this method. To overcome this, our goal was to generate short, discrete mini-filaments. While some studies have achieved this through chemical modification of DNA substrates (e.g., biotinylation)23, we were concerned that such modifications can potentially constrain DNA dynamics and alter the strand exchange reaction. Through extensive trials, we determined that the most effective and reproducible strategy for forming mini-filaments was to utilize the mouse RAD51-SEAD mutant. This mutant, carrying S208E and A209D substitutions, improves the yield of mini-filaments suitable for cryo-EM without requiring any chemical modification of the DNA. We previously established that this mutant retains comparable strand exchange activity to wild-type RAD5128 and confirmed this in our current study (Fig. 1b). Furthermore, the mini-filaments formed by this mutant closely resemble the structure of wild-type RAD51 filaments previously solved by helical reconstruction8, providing a reliable platform for our structural analysis.
In the present work, we strategically incorporated a GC-rich content within the heteroduplex-forming region, alongside partially repetitive sequences. This approach was partly inspired by the successful cryo-EM D-loop structure of the RecA recombinase12. Our primary rationale for this design was to enhance the thermodynamic stability of the RAD51 D-loop intermediates, a critical factor for their successful capture at high resolution via cryo-EM. Preliminary experiments using AT-rich sequences around the anticipated strand exchange sites yielded lower-resolution structures, likely reflecting the inherently reduced stability and increased dynamism of such complexes. The heightened stability conferred by the GC-rich regions proved indispensable for obtaining the well-resolved structural snapshots presented in this study. While acknowledging that sequence composition can influence DNA mechanics and protein-DNA interactions, we anticipate that the fundamental mechanisms elucidated—such as dsDNA bending by the NTD, active strand separation by loop L2 insertion, and displaced strand capture by the S2 site—are mediated by conserved RAD51 domains and are broadly applicable across various DNA sequences. Future investigations employing a more diverse set of DNA sequences, potentially coupled with single-molecule techniques, will be valuable for exploring any subtle sequence-dependent variations in RAD51-mediated strand exchange dynamics.
DNA oligonucleotides were synthesized and PAGE-purified by Integrated DNA Technologies (IDT). The D-loop DNA sequences used for cryo-EM studies were designed to facilitate the capture of stable, high-resolution intermediates of the strand exchange reaction. Our approach was guided by the successful design reported for the cryo-EM structure of the RecA recombinase12. We retained the flanking sequences from that design while modifying the overall length of the substrates and introducing specific mismatched regions. This strategy allowed us to trap distinct stages of the reaction for structural analysis. RAD51 D-loop complexes were prepared by first assembling presynaptic mini-filaments. Mouse RAD51 protein (6 µM) was incubated with single-stranded DNA (ssDNA) at a 1:2 protein-to-DNA ratio in assembly buffer (35 mM Tris-HCl at pH 7.5, 108 mM KCl, and 1 mM DTT) supplemented with 1 mM AMP-PNP, 5 mM CaCl2, and 2.5 mM MgCl2 at 22 °C. Once these presynaptic mini-filaments were formed, the desired homologous double-stranded DNA (dsDNA) was added to the solution at a 1:2 protein-to-dsDNA molar ratio. The mixture was then incubated for an additional 4 min to allow D-loop formation to occur. This method ensures that the D-loop is formed de novo within the pre-existing RAD51 filament, allowing us to capture distinct, stepwise intermediates in the strand exchange reaction.
Mini-filaments were applied on a pre-glow-discharged graphene-oxide-coated Quantifoil holey carbon grid and vitrified using a Vitrobot Mark IV system (Thermo Fisher Scientific) by blotting and plunge-freezing in liquid ethane. Cryo-EM data were collected on a Titan Krios G3 (Thermo Fisher Scientific) microscope operated at 300 kV and equipped with a Quantum K3 Summit direct electron detector (Gatan) and a BioQuantum energy filter set to an 18 eV slit width. Data acquisition was performed in a super-resolution mode at a nominal magnification of 105,000×, corresponding to a pixel size of 0.83 Å. Movies of 50 frames with a total exposure dose of 50 electrons/Å2 were collected with a defocus range of −1 to −2 μm.
Cryo-EM data processing and 3D reconstruction
Our cryo-EM data workflow of the RAD51 mini-filament is summarized in Supplementary Figs. 3–9). In brief, micrographs were subjected to motion correction and CTF estimation using MotionCor2 and Gctf, respectively, within the cryoSPARC v4.4.1 framework. Initial particle sets were manually picked to generate high-quality 2D templates for automated particle picking. 2D classification and selection were carried out to remove ice contaminants and poorly defined particles. Ab-initio 3D reconstruction and heterogeneous refinement were performed, resulting in distinct classes. These classes were further subjected to 3D classification. One class, showing the highest resolution features, was selected for homogeneous refinement, followed by Non-uniform (NU) refinement to improve the resolution. The overall resolution was assessed using the gold-standard Fourier shell correlation (GSFSC) criterion with a cutoff of 0.143. Local resolution was estimated using the utility in cryoSPRAC46, and the model-to-map FSC was calculated using phenix.mtriage47. The final reconstruction was visualized and analyzed using ChimeraX48.
Model building and refinement
An atomic model of the RAD51 mini filament was generated by Coot and refinement in Phenix real-space refinement using the 2.91 Å resolution cryo-EM map as a guide. The model was manually adjusted and built into the cryo-EM density map using Coot49. Real-space refinement was performed using Phenix50 to optimize the model against the experimental map and impose appropriate stereochemical restraints. Due to diffuse density at the termini of the mini-filament, particularly in the N-terminal regions of terminal protomers, precise atomic modeling was not feasible. In these regions, rigid-body docking was employed to maintain overall architectural integrity. To ensure consistent protomer numbering across structures, rigid-body modeling of the N-terminal domain (NTD) was used to generate protomer N + 1 in the 8-bp D-loop complex shown in Fig. 2d. The displaced strand DNA, which exhibited similarly diffused density, was positioned based on residual map features and guided by alignment to the ssDNA structure from the RecA S2 complex12. This provided a template for estimating the path of the displaced strand in low resolution density map. Selected secondary structure elements were identified and analyzed within the refined model. These included single-stranded DNA (ssDNA), α-helices spanning residues 197–215 and 238–260, and β-sheets spanning residues 156–161 and 262–269 (Supplementary Fig. 3e). The quality of the model fit to the cryo-EM density was assessed by visual inspection in ChimeraX48. Cryo-EM data collection, refinement, and validation statistics are listed in Table S2 and S3. Residues classified as Ramachandran outliers are typically located in flexible, poorly resolved regions—such as the protomer N-termini—where the density is diffused.
While our cryo-EM analysis yielded high-resolution structures of the overall D-loop intermediates, it is important to acknowledge the limitations in resolving highly dynamic regions. Structures of mismatch bubbles in Fig. 2a, b, which are inherently flexible and transient, often exhibit blurred electron density in cryo-EM maps due to their conformational heterogeneity. Therefore, even at high overall resolution, the local resolution in regions like the mismatch bubble can be significantly lower. The corresponding DNA backbone is modeled by cryoREAD, a DNA/RNA de novo atomic structure modeling based on cryo-EM map51. This common limitation in structural biology means that while our models accurately capture the overall D-loop geometry and key protein-DNA interactions, the precise atomic details of the unpaired bases within the bubble remain unresolved.
Molecular dynamics simulations
All-atom molecular dynamics (MD) simulations were performed in explicit water using the Amber 20 package52. Initial coordinates of the dsDNA-bound RAD51 presynaptic filament were derived from our cryo-EM structures (shown in Fig. 2b, e). Missing residues were rebuilt by Coot49. Simulations were performed using Amber ff14SB force field53 and a set of GAFF54 parameters was adopted for the description of ANP. The partial charge of standard amino acid and nucleotide were based on the Amber 20 libraries and AM1-BCC method for ANP. Periodic boundary conditions were imposed with box lengths of 154.7 × 141.8 × 203.6 Å3 for D-loop propagation and 143.7 × 117.5 × 205.1 Å3 for D-loop initiation. In our system, long-range electrostatic interactions are carried out using the smoothed particle mesh Ewald algorithm with a real-space cut-off length of 10 Å. The SHAKE algorithm was implemented to constrain the covalent bond. All MD simulations were carried out in the NPT ensemble with the time step of 1 fs. A Langevin thermostat was used to maintain the system temperature with collision frequency of 1 ps−1 to the target temperature 300 K. The system first underwent an annealing process from 0 to 300 K under a constant pressure of 1.0 bar over 7 ns, and maintained at this equilibrated point for the following simulation. After equilibrated steps, the final equilibrium system density was ~1.0 ± 0.01 g/cm3. Finally, we further performed 48 ns MD simulations (initiation system) and 180 ns MD simulations (propagation system) for trajectory analysis. MD movie were rebuilt by ChimeraX v1.348 and Power Director 2025.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The minimum dataset necessary to interpret, verify, and extend the research in this article is publicly available through the following depositions and repositories: Cryo-EM Maps and Atomic Models: The cryo-EM map and atomic model coordinates generated in this study have been deposited in the Protein Data Bank (PDB) and the Electron Microscopy Data Bank (EMDB). The accession codes are 9UI4 and EMD-64183 (presynaptic filament), 9UI5 and EMD-64184 (post-synaptic filament), 9UI7 and EMD-64186 (4-base-pair D-loop Complex), 9UI8 and EMD-64187 (8-base-pair D-loop Complex), 9UI9 and EMD-64188 (12-base-pair D-loop Complex), and EMD-65955 (7-base-pair D-loop Complex). The PDB model corresponding to EMD-65955 is not applicable to this study because the 7 bp mismatched dsDNA does not form a stable D-loop and therefore cannot be modeled. All additional coordinates, including molecular dynamics simulation trajectory data and low-resolution cryo-EM maps related to our proposed mechanism, are publicly available in the Figshare (https://figshare.com/projects/Mechanism_of_strand_exchange_of_eukaryotic_homologous_recombination_from_RAD51_D-loop_structures/245306). Source data are provided with this paper.
References
Kowalczykowski, S. C. An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb. Perspect. Biol. 7, a016410 (2015).
Wright, W. D., Shah, S. S. & Heyer, W.-D. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 293, 10524–10535 (2018).
Prakash, R., Zhang, Y., Feng, W. & Jasin, M. Homologous Recombination and Human Health: The Roles of BRCA1, BRCA2, and Associated Proteins. Cold Spring Harb. Perspect. Biol. 7, a016600 (2015).
Matos-Rodrigues, G., Guirouilh-Barbat, J., Martini, E. & Lopez, B. S. Homologous recombination, cancer and the ‘RAD51 paradox’. NAR Cancer 3, zcab016 (2021).
Sung, P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science 265, 1241–1243 (1994).
Lin, Z., Kong, H., Nei, M. & Ma, H. Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc. Natl. Acad. Sci. USA 103, 10328–10333 (2006).
Short, J. M. et al. High-resolution structure of the presynaptic RAD51 filament on single-stranded DNA by electron cryo-microscopy. Nucleic Acids Res 44, 9017–9030 (2016).
Xu, J. et al. Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange. Nat. Struct. Mol. Biol. 24, 40–46 (2017).
Galletto, R., Amitani, I., Baskin, R. J. & Kowalczykowski, S. C. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature 443, 875–878 (2006).
Candelli, A. et al. Visualization and quantification of nascent RAD51 filament formation at single-monomer resolution. Proc. Natl. Acad. Sci. USA 111, 15090–15095 (2014).
Cloud, V., Chan, Y. L., Grubb, J., Budke, B. & Bishop, D. K. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337, 1222–1225 (2012).
Yang, H., Zhou, C., Dhar, A. & Pavletich, N. P. Mechanism of strand exchange from RecA–DNA synaptic and D-loop structures. Nature 586, 801–806 (2020).
Petiot, V., White, C. I. & Da Ines, O. DNA-binding site II is required for RAD51 recombinogenic activity in Arabidopsis thaliana. Life Sci. Alliance 7, e202402701 (2024).
Story, R. M., Weber, I. T. & Steitz, T. A. The structure of the E. coli recA protein monomer and polymer. Nature 355, 318–325 (1992).
Ogawa, T., Yu, X., Shinohara, A. & Egelman, E. H. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science 259, 1896–1899 (1993).
Yu, X., Jacobs, S. A., West, S. C., Ogawa, T. & Egelman, E. H. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc. Natl. Acad. Sci. 98, 8419–8424 (2001).
Matsuo, Y., Sakane, I., Takizawa, Y., Takahashi, M. & Kurumizaka, H. Roles of the human Rad51 L1 and L2 loops in DNA binding. FEBS J. 273, 3148–3159 (2006).
Ragunathan, K., Joo, C. & Ha, T. Real-Time Observation of Strand Exchange Reaction with High Spatiotemporal Resolution. Structure 19, 1064–1073 (2011).
Renkawitz, J., Lademann, C. A. & Jentsch, S. Mechanisms and principles of homology search during recombination. Nat. Rev. Mol. Cell Biol. 15, 369–383 (2014).
Qi, Z. et al. DNA Sequence Alignment by Microhomology Sampling during Homologous Recombination. Cell 160, 856–869 (2015).
Greene, E. C. DNA Sequence Alignment during Homologous Recombination. J. Biol. Chem. 291, 11572–11580 (2016).
Piazza, A. et al. Dynamic Processing of Displacement Loops during Recombinational DNA Repair. Mol. Cell 73, 1255–1266 (2019).
Joudeh, L., Appleby, R. E., Maman, J. D. & Pellegrini, L. Structural mechanism of strand exchange by the RAD51 filament. eLife 14, RP107114 (2025).
Yang, D., Boyer, B., Prévost, C., Danilowicz, C. & Prentiss, M. Integrating multi-scale data on homologous recombination into a new recognition mechanism based on simulations of the RecA-ssDNA/dsDNA structure. Nucleic Acids Res. 43, 10251–10263 (2015).
Aihara, H., Ito, Y., Kurumizaka, H., Yokoyama, S. & Shibata, T. The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR. J. Mol. Biol. 290, 495–504 (1999).
Yu, D. S. et al. Dynamic control of Rad51 recombinase by self-association and interaction with BRCA2. Mol. Cell 12, 1029–1041 (2003).
Esashi, F., Galkin, V. E., Yu, X., Egelman, E. H. & West, S. C. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat. Struct. Mol. Biol. 14, 468–474 (2007).
Su, G. C. et al. Role of the RAD51-SWI5-SFR1 Ensemble in homologous recombination. Nucleic Acids Res. 44, 6242–6251 (2016).
Prasad, T. K., Yeykal, C. C. & Greene, E. C. Visualizing the Assembly of Human Rad51 Filaments on Double-stranded DNA. J. Mol. Biol. 363, 713–728 (2006).
Reymer, A., Frykholm, K., Morimatsu, K., Takahashi, M. & Nordén, B. Structure of human Rad51 protein filament from molecular modeling and site-specific linear dichroism spectroscopy. Proc. Natl. Acad. Sci. 106, 13248–13253 (2009).
Lee, J. Y. et al. Base triplet stepping by the Rad51/RecA family of recombinases. Science 349, 977–981 (2015).
Ito, K. et al. Real-time tracking reveals catalytic roles for the two DNA binding sites of Rad51. Nat. Commun. 11, 2950–2950 (2020).
Privalov, P. L., Dragan, A. I. & Crane-Robinson, C. The cost of DNA bending. Trends Biochem. Sci. 34, 464–470 (2009).
Harteis, S. & Schneider, S. Making the bend: DNA tertiary structure and protein-DNA interactions. Int J. Mol. Sci. 15, 12335–12363 (2014).
Miyazono, K. I., Wang, D., Ito, T. & Tanokura, M. Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI. Nucleic Acids Res 48, 5106–5118 (2020).
Basu, A., Bobrovnikov, D. G. & Ha, T. DNA mechanics and its biological impact. J. Mol. Biol. 433, 166861 (2021).
Soczek, K. M., Cofsky, J. C., Tuck, O. T., Shi, H. & Doudna, J. A. CRISPR-Cas12a bends DNA to destabilize base pairs during target interrogation. Nucleic Acids Res 53, gkae1192 (2025).
Gao, F. et al. Structural basis of σ(54) displacement and promoter escape in bacterial transcription. Proc. Natl. Acad. Sci. USA 121, e2309670120 (2024).
Glyde, R. et al. Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation. Mol. Cell 70, 1111–1120 (2018).
Chen, X. et al. Cutting antiparallel DNA strands in a single active site. Nat. Struct. Mol. Biol. 27, 119–126 (2020).
Danilowicz, C. et al. The differential extension in dsDNA bound to Rad51 filaments may play important roles in homology recognition and strand exchange. Nucleic Acids Res 42, 526–533 (2014).
Sung, P. & Robberson, D. L. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell 82, 453–461 (1995).
Sung, P. & Stratton, S. A. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem. 271, 27983–27986 (1996).
Murayama, Y., Kurokawa, Y., Mayanagi, K. & Iwasaki, H. Formation and branch migration of Holliday junctions mediated by eukaryotic recombinases. Nature 451, 1018–1021 (2008).
Tsai, S. P. et al. Rad51 presynaptic filament stabilization function of the mouse Swi5-Sfr1 heterodimeric complex. Nucleic Acids Res 40, 6558–6569 (2012).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D. Struct. Biol. 75, 861–877 (2019).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr 66, 486–501 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D. Struct. Biol. 74, 531–544 (2018).
Wang, X., Terashi, G. & Kihara, D. CryoREAD: de novo structure modeling for nucleic acids in cryo-EM maps using deep learning. Nat. Methods 20, 1739–1747 (2023).
Amber 2024 (University of California, San Francisco., 2024).
Maier, J. A. et al. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Wang, J., Wang, W., Kollman, P. A. & Case, D. A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 25, 247–260 (2006).
Acknowledgements
This work was supported by Academia Sinica (AS-TP-112-L01 to M.C.H) and National Taiwan University (P.C.), National Science and Technology Council (NSTC 113-2326-B-002-010 and NSTC 111 − 2311-B-002-006-MY3 to P.C. and NSTC 113-2811-B-001−085 to S.C.L.). The cryo-EM experiments were performed at the Academia Sinica Cryo-EM Center (ASCEM), and the cryo-EM data were processed at the Academia Sinica Grid-computing Center (ASGC). ASGC and ASCEM are supported by Academia Sinica (Grant numbers AS-CFII-114A-11 and AS-CFII-108-110, respectively).
Author information
Authors and Affiliations
Contributions
C.W.L., M.Y., H.Y.Y., and P.C. contributed to protein preparation. H.Y.Y. and P.C. contributed to biochemical analysis, and related experimental designs. M.Y. and S.C.L. performed cryo-EM related experiments. C.H.Y. performs MD simulation work. P.C., C.H.Y., S.C.L,. and M.C.H. wrote the paper. S.C.L. and M.C.H. designed the experiments. C.H.Y., S.C.L., and M.C.H. analyzed data. M.C.H. supervised the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Wataru Kagawa, Akira Shinohara and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Luo, SC., Yang, CH., Yeh, HY. et al. RAD51 D-loop structures reveal the mechanism of eukaryotic RAD51-mediated strand exchange. Nat Commun 17, 243 (2026). https://doi.org/10.1038/s41467-025-66925-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66925-z