Abstract
The T-cell receptor (TCR) initiates T-lymphocyte activation, but the mechanism of TCR activation remains uncertain. Here, we present cryogenic electron microscopy structures for the unliganded and human leukocyte antigen (HLA)-bound human TCR–CD3 complex in nanodiscs that provide a native-like lipid environment. Distinct from the open and extended conformation seen in detergent, the unliganded TCR–CD3 in nanodiscs adopts two related closed and compacted conformations that represent its physiologic resting state in vivo. By contrast, the HLA-bound complex adopts the open and extended conformation, and conformation-locking disulfide mutants show that ectodomain opening is necessary for maximal ligand-dependent T-cell activation. These structures also reveal conformation-dependent protein–lipid and glycan–glycan interactions within the TCR. Together, these results establish allosteric conformational change during TCR activation, reveal avenues for immunotherapeutic engineering, and highlight the importance of native-like lipid environments for membrane protein structure determination.
Similar content being viewed by others
Introduction
T cells are central to both adaptive and pathologic immune responses1. The T-cell receptor (TCR), in association with CD3 proteins, allows T cells to recognize and respond to antigenic peptides presented on human leukocyte antigen (HLA) complexes of other cells. While the constituents of the TCR–CD3 complex have been known for decades, questions remain regarding its activation mechanism2,3,4.
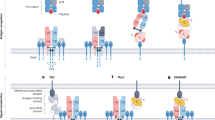
The TCR–CD3 is a hetero-octameric membrane protein complex5,6,7,8. The αβ-type TCR is comprised of two variable chains, TCRα and TCRβ, which directly interface with the HLA–antigen complex. Antigen specificity is generated through somatic recombination of the TCRα and TCRβ variable segment genes. The remainder of the core complex is comprised of six invariant CD3 family chains: the CD3ϵ-CD3γ and CD3ϵ-CD3δ heterodimers (CD3ϵγ and CD3ϵδ, respectively), and a CD3ζ homodimer. Herein, we differentiate the CD3ϵ protomers by the CD3 component with which each pairs (CD3ϵG pairs with CD3γ and CD3ϵD pairs with CD3δ) and the CD3ζ protomers as CD3ζ and CD3ζ’.
While the TCRα and TCRβ chains lack intracellular signaling domains, the CD3 proteins possess cytoplasmic immunoreceptor tyrosine activation motifs (ITAMs) that become phosphorylated upon TCR interaction with HLA–antigen complex and then bind and activate downstream signaling kinases. Dynamic interactions between ITAMs and the membrane regulate signaling9,10, but the mechanism by which ligand binding to the extracellular TCRα and TCRβ variable domains results in phosphorylation of cytoplasmic CD3 ITAMs remains a matter of debate. Specifically, it is unclear whether ligand binding and activation involves conformational change in the TCR–CD3 complex. Reconstitution of TCR–CD3 signaling in vivo with minimal constructs argues against transmembrane conformational change11. However, biochemical evidence in support of conformational change12,13,14,15 – specifically, for reversible structural extension of the TCR under mechanical force – suggests the opposite16, although the structural details of such conformational transitions remain to be determined.
Structures of the hetero-octameric αβ-type TCR–CD3 have previously been determined by cryogenic electron microscopy (cryo-EM)5,6,7,8. These studies revealed an antenna-like structure, with the TCRα and TCRβ variable domains projecting over a ring formed by the CD3 ectodomains and the TCRα and TCRβ constant domains, with numerous interactions between their respective transmembrane (TM) helices (Supplementary Fig. 1a). Addition of antigen–β2-microglobulin–HLA (herein, “HLA”) to this complex did not meaningfully alter the conformation of the TCR–CD37,8, arguing against models of intrinsic conformational change in ligand-dependent TCR–CD3 activation (Supplementary Fig. 1a, b).
However, the TCR–CD3 used in these studies were solubilized in detergent. Biochemical data have shown important roles for the lipid bilayer in TCR–CD3 signaling6,17,18, raising questions as to the physiologic meaning of the determined structures. Membrane protein structure determination is generally preferable in a native-like membrane environment that increases the likelihood that important protein–lipid interactions are captured in the obtained molecular models19,20.
Here, we report cryo-EM structures of a human TCR–CD3 both in detergent micelles and nanodiscs. The structures reveal conformations for the TCR–CD3 in a native-like lipid environment distinct from those previously reported in detergent, and cross-linking data support the notion that these conformations represent the resting state of an unstimulated TCR–CD3 on T cells. Determination of the structure of the same nanodisc-embedded TCR–CD3 bound to HLA reveals substantial conformational change upon physiologic ligand binding, and functional data show that exit from the resting state conformation is required for maximal TCR–CD3 signaling.
Results
The TCR–CD3 complex is in an open and extended conformation in detergent micelles
In this study, we investigated the structure of the 1G4 (LY-variant) TCR–CD3. We chose the 1G4 TCR, which binds to an HLA-A*02:01-restricted 9-amino acid epitope derived from the NY-ESO-1 cancer-testis antigen, because it is both a model immunoreceptor and being developed as an anti-cancer therapeutic21,22,23,24. To confirm that 1G4 has a similar structure to the other TCR–CD3 complexes analyzed in detergent, we purified this complex in glyco-diosgenin (GDN) and determined its structure by cryo-EM to a nominal resolution of 3.3 Å (Fig. 1a, g, Supplementary Figs. 1c–h, 3; Supplementary Tables 1–3). Herein, this model will be abbreviated as “GDN.” Overall, GDN was very similar to the structures of three other TCR–CD3 constructs, including those with and without HLA bound (Fig. 1b). This is unsurprising, as their sequences only differ meaningfully in the TCR variable domains (Fig. 1c).
a Nickel-nitrilotriacetic acid (NiNTA)-affinity purification scheme for TCR–CD3 in detergent (black lines) and its reconstitution into nanodiscs (red lines). b Backbone representations of GDN and prior TCR–CD3 structures (HLA not shown). c GDN model colored by sequence identity to the three TCR–CD3 complexes previously determined in detergent. Representative purification of nanodisc-embedded TCR–CD3, showing size-exclusion chromatogram (d; asterisk indicates peak), silver-stained SDS-PAGE (e; constituents identified by expected molecular weight, which does not distinguish CD3γ, δ, and ε), and negative-stain electron micrograph (f; inset, zoomed-in view). Colored asterisks in e indicate molecular weight values: magenta, 250 kDa; orange, 150 kDa; green, 100 kDa; white, 75 kDa; cyan, 50 kDa; yellow, 37 kDa; navy, 25 kDa. Data shown are representative of more than three experiments. g Cryo-EM maps (top, middle) and model surfaces (bottom) of TCR–CD3 in GDN and in nanodiscs. Top row shows B-factor-sharpened cryo-EM maps without local refinement, contoured to a low threshold to highlight micelle and nanodisc positioning. Red dashed box indicates approximate plasma membrane span (extracellular and intracellular faces indicated as e.c. and i.c., respectively). Middle row shows DeepEMhancer-sharpened maps after local refinement, accentuating the protein and glycan densities. Protein regions indicated at far right abbreviated as follows: var variable domains, ecto ectodomains, const constant domains, JM juxtamembrane linkers, TM transmembrane helices.
The TCR–CD3 is in a distinct closed and compacted conformation in nanodiscs
To determine the structure of the 1G4 TCR–CD3 in a native-like lipid environment, we reconstituted it into membrane scaffold protein-based nanodiscs loaded with mammalian polar lipids and cholesteryl hemisuccinate (Fig. 1a, d–f). Computational processing of cryo-EM data yielded two distinct maps determined to nominal resolutions of 3.3 to 3.1 Å (Fig. 1g, Supplementary Figs. 2, 3; Supplementary Tables 1–3). These maps, which we denote as nanodisc (ND)-I and ND-II, were clearly different from GDN (Fig. 1g) and the previously published TCR structures. Overall, reconstitution of the TCR–CD3 into nanodiscs shortens the TCR–CD3 by approximately 35 Å along the membrane-normal axis and induces interdomain contacts (Fig. 1g). Attempts to bias the particle search towards the published conformations with projections of the structure in detergent or that same structure docked into a lipid nanodisc model did not yield any maps approximating the previously published conformation (Supplementary Fig. 2c). Further, analysis of 2D classes from the nanodisc dataset does not show a subset of particles in the conformation seen in detergent (Supplementary Fig. 2i, j). Computational modeling of the conformation from detergents into the nanodisc used in these studies reveals no geometric constraints preventing the TCR–CD3 from adopting this conformation in a nanodisc, arguing against a nanodisc geometry-induced artifact (Supplementary Fig. 2k–m).
Despite equal or higher nominal overall resolutions than GDN, the ND-I and ND-II maps were not as high quality (compare local resolution estimations in Supplementary Fig. 2d with Supplementary Fig. 1h). This was likely due to a higher degree of conformational variability, but also due in part to map anisotropy (compare Supplementary Fig. 2f, h with Supplementary Fig. 1g). Nonetheless, with much of the density having estimated local resolution better than 4 Å, these maps were of sufficient quality to build models into the density.
Molecular models were built for ND-I and ND-II by docking the GDN structure into the maps and refining them by standard approaches (see Methods for details; Supplementary Fig. 3c–k; Supplementary Tables 2, 3). These models reveal two distinct but related conformations of the TCR–CD3 (Figs. 1g and 2). The ND structures show two large conformational changes relative to GDN: a closure of the TCR ectodomains against the CD3 ectodomains (Fig. 2a–c) and a compaction of the juxtamembrane (JM) linker peptides with associated tilting of the TM helices (Fig. 2d–h). Closure of the TCR ectodomains (Fig. 2a) results in part from rotation of the variable domains towards their respective constant domains about the interdomain hinge region (Fig. 2b, c). Relative to the extended JM linker conformations seen in GDN, the CD3 JM linkers are compacted, with closer approximation of the CD3 ectodomains to the amino-terminal ends of their TM helices (Fig. 2e–g). The conformational changes in the TCR ectodomains are accompanied by tilting of TM helices. While the in-plane arrangement of the TM helices is largely similar when viewed along the membrane normal (Fig. 2d), examination from orthogonal axes reveals differences in TM helix conformation. Compaction of the CD3 JM linkers requires opening of the angle between the TM helices of CD3ϵδ by 23° (from 31° to 54°) and CD3ϵγ by 28° (from 39° to 67°), when comparing ND-II with GDN (Fig. 2e, f). For ND-I, the TM helices of CD3ϵδ are bent, with the first helical turn displaying a wide splay (76°, Fig. 2g), but the overall angle between the modeled helices (46°) is intermediate to ND-II and GDN. The CD3ϵG TM helix was not modeled, as its register could not be confirmed, and thus the CD3ϵγ TM angle was not measured. Additionally, there is splaying of the TCRβ and distal CD3ζ TM helices away from the membrane normal (Fig. 2h). In GDN, the TCRα TM helix extends outward past the amino-terminal ends of the CD3 TM helices, where it is ensconced by the extended CD3 JM linkers. Compaction of the JM linkers in nanodiscs necessitates reorganization of the first turn of the TCRα TM helix, which is unstructured and displaced towards the lipid bilayer (Fig. 2h). Taken together, these cryo-EM maps reveal marked conformational differences between the TCR–CD3 in detergent micelles and a lipid bilayer.
a Comparison of the GDN and ND-I models (cartoon inside transparent model surfaces), highlighting ectodomain closure in nanodiscs. b Cartoon representations of TCRα in ND-I (solid color) and GDN (transparent gray), aligned on the TCRα constant domain, highlighting variable domain movement comprising closure (arrow). c Same as in b but for TCRβ. d Overlay of the GDN, ND-I, and ND-II TM helices viewed from the extracellular space showing similar overall arrangement. e, f Comparison of GDN and ND-II models, highlighting JM linker compaction (green dashed box) and coincident splay of the TM helices (cyan lines and angular measures). CD3ϵδ and CD3ϵγ are shown in (e) and (f), respectively. g The bent TM helices of ND-I CD3ϵδ. Splay measured at the level of the first helical turn (cyan) is greater than that of the helices overall (orange). h Comparison of GDN (transparent gray) and ND-II (solid, color-coded) TM helices aligned to TCRα residues 245-260 and viewed parallel to the membrane. Green and orange arrows highlight increased splay of the TCRβ TM helix and the CD3ζ TM helix carboxy-terminus, respectively. Blue arrow highlights unstructuring and deflection of the first helical turn at the amino-terminus of the TCRα TM helix in nanodiscs. i Cartoon representations with basic and aromatic residues highlighted as space-filling spheres. Blue dashed box highlights the cluster of basic and aromatic residues visualized at the carboxy-termini of the TM helices in GDN, but unstructured in ND-I/II. j Examples of densities (dashed ovals) at ND-II TM carboxy-termini that could not be confidently modeled, but likely correspond to continuation of the polypeptides indicated.
Additionally, the TM helices of ND-I and ND-II appear shorter than in GDN. These structures lack strong density for the cluster of basic and aromatic amino acids visualized at the carboxy-terminal ends of the GDN TM helices (Fig. 2i). In a membrane, these extensions would likely be interfacial or extend into the cytoplasm25. Although there is additional density beyond the modeled carboxy-termini for some of these shortened helices in ND-I and ND-II, it is not of sufficient quality to model with confidence (Fig. 2j). This suggests the TCR–CD3 TM helices are less rigid in the inner leaflet of a lipid bilayer than in a detergent micelle.
The TCR–CD3 interacts with the lipid bilayer
The TCR–CD3 is known to interface directly with membrane cholesterol26, and binding sites for two cholesterols have been visualized in previously published structures6. In GDN, ND-I, and ND-II, density corresponding to these cholesterols was again apparent (Fig. 3a). The two cholesterol molecules in ND-I and ND-II are oriented tail-to-tail in a groove between CD3ζ’, TCRα, TCRβ, and CD3γ, as in GDN (Fig. 3b). However, the compaction-associated changes in the TM helices cause sliding of the cholesterol molecules towards each other, with deflection of their branched aliphatic tails to prevent clashing (Fig. 3b, insets). We refer to the cholesterol molecules as CholA and CholB, corresponding to the TCR–CD3 TM helix with which each is approximated. The deflected tail of CholB is interposed between adjacent TM helices, accommodated by their increased splay in the compacted conformation (Fig. 3c, insets, dashed black ovals). The cholesterol-binding groove is more open to the membrane bilayer adjacent to CholB in nanodiscs, creating a possible path for cholesterol loading into the assembled TCR–CD3 and/or exchange with the membrane environment (Fig. 3c, bottom right, dashed red oval).
a Cholesterol densities in ND-I, ND-II, and GDN. Transparent green surface is the locally refined, B-factor-sharpened map, cropped to a zone of 2.2 Å around the cholesterols (black). b Comparison of GDN and ND-I cholesterol-binding sites. Insets show zoomed-in views with (left) and without (right) modeled protein highlighting the deflection of the branched aliphatic tails. c Model sections parallel to the membrane through the cholesterol-binding groove at the levels of CholA and CholB, as indicated. Locally refined, B-factor-sharpened maps are shown as black mesh, and cholesterols as green sticks. Red dashed circle highlights structured lipid density in the nanodisc adjacent to the opened cholesterol-binding groove. Insets show the cholesterol-binding groove from the same angle as the main panels, but with both cholesterols included in the slice, highlighting intercalation of the deflected CholB aliphatic chain between TM helices in nanodiscs (black dashed oval). d–g Additional densities attributable to lipids in the ND-II map. Proteins are shown as model surfaces, and cholesterol as sticks. Yellow surface is unmodeled density from the ND-II B-factor-sharpened map after local refinement (d) or non-uniform refinement (e–g). Color legend applies to (d–g).
Imaging of the TCR–CD3 in a lipid bilayer revealed additional sites of protein–lipid interaction. Unmodeled tubular densities consistent with phospholipid acyl chains are present at several sites adjacent to the TCR–CD3 TM helices (yellow densities in Fig. 3d–g). Structured lipid is seen parallel to the tail-to-tail cholesterol dimer, filling the aforementioned channel between the cholesterol-binding groove and the bulk membrane lipids (Fig. 3d). Probable sites of structured lipid include a shroud of density over the CD3ζζ’ TM helices (Fig. 3e) and in the cleft over the TCRαβ TM helices between CD3δ and CD3ϵG (Fig. 3f). Splay of the CD3ζ TM helix away from that of CD3ϵD enlarges a cavity, which is filled by several presumptive lipid acyl chains in the nanodisc (Fig. 3g).
The ND-I and ND-II maps show clear density for the proteolipid nanodisc enshrouding not only the TM helices, but also the membrane-proximal faces of the TCR–CD3 ectodomains (Fig. 4a, left). Lipid contacts with the CD3 ectodomains are apparent even in maps contoured at a higher threshold, consistent with high occupancy (Fig. 4a, right). While the CD3 ectodomain surfaces are poorly conserved in comparison to the TM helices, patches on the membrane-proximal surfaces of the CD3 ectodomains do show high sequence conservation (Fig. 4a, c). These highly conserved ectodomain surface patches (black triangles in Fig. 4b–e) interface with the extracellular membrane leaflet and are neutral to electropositive (Fig. 4e), facilitating phospholipid head group interactions. Of note, we observe a similar conserved electropositive patch at the interface of CD79αβ27, the analogous ITAM-bearing chains in the B-cell receptor, raising the prospect that this interface may also interact with membrane lipids and favor a different conformation in the lipid bilayer than that reported in detergent (Supplementary Fig. 4).
a ND-II model surface colored by sequence conservation (Consurf score) with yellow unmodeled map density shown at two contouring thresholds. Lipid density adjacent to the CD3 ectodomains persists at high threshold (red dashed box). b Top, ND-II model with arrows and eyes indicating viewing directions for surfaces below. Bottom, overviews orienting the zoomed-in views of CD3ϵγ (left) and CD3ϵδ (right) in (c) through (e). Red dashed lines encircle TM surfaces. c Zoomed-in views of the membrane-proximal CD3 heterodimer surfaces. Black triangles highlighting the conserved surface patch at the subunit interface. d Lipid density (yellow) over this conserved patch, visualized at high contouring threshold (as in a, right). e Model surfaces colored by electrostatic potential.
The unliganded TCR–CD3 is closed and compacted in vivo
Which conformation of the TCR–CD3 is the physiologic resting state on the cell membrane: the open/extended conformation seen in detergent micelles, or the closed/compacted conformations seen in the lipid bilayer of the nanodisc? Given that the lipid bilayer of a nanodisc more closely approximates the physicochemical milieu of the plasma membrane, we hypothesized that the TCR–CD3 is closed and compacted in the absence of activating ligand in vivo. To test this notion, we engineered a series of disulfide bonds that would only be geometrically favorable in ND-I and/or ND-II but not GDN (or vice versa), and we read out their presence or absence by one of the following approaches: (1) non-reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot of whole-cell lysates for interchain disulfides, or (2) mass spectrometry on affinity-purified protein for intrachain disulfides. Analysis of disulfide bonds in the Protein Databank have shown that they almost never have a Cys-Cys Cα-Cα distance greater than 7 Å, with most being between 5 and 6.5 Å28. Accordingly, we sought residue pairs in the TCR–CD3 ectodomains with Cα-Cα distances below 7 Å in one conformation and above 7 Å in the other as candidates for disulfide engineering.
The interface between CD3δ and TCRα contains several pairs of residues with geometry favorable for engineered disulfide-bond formation in ND-I but not GDN (Fig. 5a). As TCRα and TCRβ are linked by an interchain disulfide natively (Supplementary Fig. 5a), formation of a TCRα–CD3δ disulfide would result in an upward gel shift of the TCRαβ heterodimer in nonreducing SDS-PAGE. For all three tested engineered disulfide pairs meeting the 7 Å Cα-Cα distance constraint, a gel shift consistent with TCRα–CD3δ disulfide formation was observed (Fig. 5b, asterisks; full-length blots in Supplementary Fig. 5b). Confirming that this mass shift was due to disulfide formation, no gel shift was apparent under reducing conditions (Supplementary Fig. 5b). Moreover, the mass shift was due to the formation of the engineered disulfide bond and not an off-target bond, as adduct formation was only seen when both residues in a given target pair were mutated to cysteines (Fig. 5b). As an internal negative control, a permutation of cysteine mutants at this interface not meeting geometric criteria for disulfide-bond formation in ND-I (TCRα S78C – CD3δ N38C, Fig. 5a, right, orange labels) was produced. It did not form a disulfide bond in vivo (Fig. 5b, orange) despite S78C and N38C successfully pairing with cognate cysteine mutants that did meet the inter-Cα threshold. This control shows the structural precision and specificity of the engineered disulfide-bond approach. These experiments were performed using whole-cell lysates, but we obtained similar results with NiNTA-affinity-purified protein (Supplementary Fig. 5c), and we were able to reconstitute the two G79C-crosslinked mutants into nanodiscs (Supplementary Fig. 5d, e), suggesting proper folding and assembly of TCR–CD3.
a Comparison of Cα-Cα distances between apposing faces of CD3δ and TCRα in GDN and ND-I, highlighting plausible engineered disulfide positions in ND-I (distances in blue shades) that are not geometrically plausible in GDN (distances in warm shades). b Non-reducing SDS-PAGE of whole-cell lysates from HEK cells infected with the indicated TCR–CD3 virus genotypes Western blotted for EGFP-tagged TCRβ. Engineered disulfides yielding a TCRαβ–CD3δ covalent adduct resulted in an upward gel shift (asterisks). Color coding and distances as in (a). Full blots from two experimental replicates are shown in Supplementary Fig. 5b. Design of intrachain disulfide bonds in the closed conformation of TCRα (c) and TCRβ (d). Distances color-coded as above. Mass spectrometric detection of intrachain disulfide bonds in TCRα (e) and TCRβ (f). EThcD fragment spectra of disulfide-linked dipeptides shows gas-phase reduced linear peptides (red, peptide A; blue, peptide B; reduced Cys, bold) and their fragment ions (magenta, peptide A; cyan, peptide B). Charge-reduced precursor ions are in gray.
The region around the TCRα and TCRβ interdomain hinges is also notably different in the closed/compacted conformation compared to the open/extended one, with the variable domains coming into closer apposition with their respective constant domains, allowing the engineering of conformation-specific disulfide pairs in this region (Fig. 5c, d). We probed formation of these intrachain disulfides by mass spectrometry. Proteolytic digestion of the TCR–CD3 with trypsin and chymotrypsin yielded distinct peptides, several of which were linked by disulfide bonds. Electron-transfer/higher-energy collision dissociation (EThcD) fragmentation was used to fragment the disulfide-bonded peptides providing in vacuo reduction to the linear peptide constituents along with peptide backbone fragmentation, thereby allowing unambiguous identification of the engineered disulfides29. All native TCR–CD3 disulfides spanning two proteolytic fragments (Supplementary Fig. 5a) were observed in the wild-type 1G4 TCR–CD3 by this approach (Supplementary Table 4). For both ND conformation-compatible engineered disulfides tested (Fig. 5c, d), we observed formation of the engineered disulfide (Fig. 5e, f and Supplementary Table 4). In contrast, an engineered disulfide that would only be present in the open/extended conformation (Supplementary Fig. 5f) did not form in vivo (Supplementary Table 4). Taken together, these data support the conclusion that TCR–CD3 in the unliganded, resting state adopts the closed/compacted conformation in vivo.
The unliganded TCR–CD3 is dynamic in a lipid bilayer
To allow direct comparison of the two distinct, but related, closed/compacted structures (ND-I and ND-II), they were aligned by their TCRα TM helices. TCRα was chosen because it is relatively perpendicular to the membrane plane and thus might be less affected by differences in TM helix splay. Indeed, this alignment strategy brought the nanodisc density of their maps into alignment as well (Supplementary Fig. 6a), giving both models the same orientation relative to the membrane bilayer. In contrast to the strong overall alignment of the TCR–CD3 TM helices by this approach (Supplementary Fig. 6b, c), the ectodomains of ND-I and ND-II have marked conformational differences, as seen in the aligned cryo-EM densities (Supplementary Fig. 6a, b) and the resulting protein models (Fig. 6a). Comparison of the structures reveals the motions allowing exchange between these conformations (Fig. 6b, c and Supplementary Movie 1). ND-I is the most closed of the three structures, and transition to ND-II requires several motions: the depression of the CD3ϵD (Fig. 6b) and CD3γ ectodomains (Fig. 6c) away from the TCR variable domains and towards the bilayer outer leaflet; rotation of the TCRα variable domain about the interdomain hinge (Fig. 6b); and retraction of the TCRβ variable domain (Fig. 6c). This is accompanied by sliding of CD3ζ along its helical axis towards the ectodomains (Fig. 6b). While the TCRα and CD3ζ‘ TM helices remain relatively fixed in this frame of reference, the TCRβ and CD3γ TM helices shift away from the cholesterol-binding groove (Fig. 6d and Supplementary Fig. 6c). To accommodate this movement, there is rotation of CholA and elevation of CholB (Fig. 6d), showing cholesterol to be an integral and dynamic component of the TCR–CD3 structure.
a Comparison of ND-I (transparent cyan) and ND-II (red) model surfaces, aligned on the TCRα TM helix. b, c Cartoon representation of select domains in ND-I (cyan) and ND-II (red), highlighting domain movements between them (gradient arrows). d TCRβ and CD3γ TM helices move away from the ND-I cholesterol-binding site in the transition to ND-II. Arrows indicate accompanying cholesterol movements. e Comparison of the TCRα and CD3δ lateral glycosylation sites (red and cyan dashed lines, respectively) in the maps, with stacking of the glycan array on ectodomain closure (purple dashed line). Insets highlight N-linked glycan cores modeled into these densities in GDN and ND-I. f Comparison of the CD3δ N38 residue in GDN, ND-II and ND-I. Gray surface is the locally refined, DeepEMhancer-sharpened map. g Modeling HLA (gray) binding to the TCR–CD3 by docking the TCR–HLA crystal structure 2BNQ59. Cross section through the modeled HLA–TCR interface in ND-I with the ND-I model surfaces (h) or locally refined, DeepEMhancer-sharpened map (i) shown.
The GDN map displays densities for many of the known asparagine (N)-linked glycans in the TCR–CD3 (Supplementary Fig. 6d). Fewer densities could be modeled as N-linked glycans in the ND-I and ND-II maps (Supplementary Fig. 6d). However, all three maps show an array of densities lateral to the CD3δ and TCRα ectodomains, corresponding to sites of known N-linked glycosylation. In ND-II, TCRα (linkage unclear) and CD3δ N74-linked glycans are visible, emerging radially from the TCR–CD3, as in GDN (Fig. 6e). By comparison, ND-I reveals clear reorganization of the glycans (Fig. 6e), with stacking of adjacent glycans brought into close proximity by tight ectodomain closure. This arraying of N-linked glycans is reminiscent of that seen on human immunodeficiency virus-1 Env30, where interacting adjacent glycans form a structured arcade (Supplementary Fig. 6e). Two densities in the array are attributable to CD3δ N74- and TCRα N82-linked glycans, with a third density emerging between them without definitive attribution (purple question mark in Fig. 6e, right inset). In GDN, as in other structures of TCR–CD3 in detergent, the CD3δ N38-linked glycan is oriented inward towards the cleft between the TCR and CD3 ectodomains (Fig. 6f, top). In ND-II, CD3δ N38 is unstructured (Fig. 6f, middle), but is grossly positioned at the entrance to the cleft, and the linked glycan could conceivably orient either into or away from the cleft. In ND-I, CD3δ N38 is structured (Fig. 6f, bottom), but its glycan cannot be confidently modeled. However, density protrudes from N38 into the open cleft (Fig. 6f, asterisk) where additional unmodeled density is seen, consistent with the glycan remaining within the cleft in the ND-I conformation.
The different degrees of ectodomain closure between ND-I and ND-II could affect TCR variable domain accessibility to HLA. To assess this possibility, we modeled the binding geometry of HLA onto GDN, ND-I, and ND-II by alignment of the TCR–HLA crystal structure to the TCR variable domains in our models (Fig. 6g). There appears to be adequate clearance for the TCR variable domains to interact with HLA in GDN and ND-II. For ND-I, the modeled fit is tighter, with the CD3ϵD ectodomain predicted to be in close contact with HLA. However, on assessment of space-filling model cross-sections, no overt steric clashes are evident (Fig. 6h). Because a portion of a potentially clashing surface loop in the CD3ϵD ectodomain is not modeled in ND-I, the TCR-aligned HLA model was docked into the ND-I cryo-EM density (Fig. 6i), but no unmodeled density appears to clash with HLA. The CD3ϵD ectodomain from GDN is completely modeled and aligns closely with that from ND-I (Supplementary Fig. 6f, inset). We modeled the GDN CD3ϵD ectodomain into ND-I and again assessed the HLA interaction. While there is a small overlap in their space-filling models (Supplementary Fig. 6f), no large clashes are observed. Thus, while it remains unclear what quantitative effect ectodomain cleft closure might have on HLA affinity, particularly in the physiologic context of a glycosylated HLA molecule, it is at least plausible that both ND-I and ND-II may be receptive to HLA binding.
While our modeling data suggest that HLA can bind to TCR–CD3 in the closed/compacted resting state conformations, TCR–CD3 opening and/or extension may be necessary for coreceptor interaction with HLA bound to the TCR–CD3. Modeling the HLA class II–CD4 complex31 onto ND-I and ND-II reveals an unfavorable geometry for CD4 interaction with HLA because of the proximity of the TCR variable regions to the membrane (Supplementary Fig. 6g). In contrast, CD4–HLA can be accommodated in the open/extended GDN conformation (Supplementary Fig. 6g). The full ectodomain structure of the CD8 coreceptor has not been described, precluding a similar analysis applicable to TCRs recognizing HLA class I-restricted epitopes, like the one used in this study.
HLA binding is sufficient to trigger TCR–CD3 conformational change
Whether or not TCR–CD3 undergoes conformational change upon binding to HLA remains an open question. To determine the structure of the HLA-liganded TCR–CD3 in a lipid bilayer, we reconstituted TCR–CD3 in nanodiscs as above, incubated the complex with an excess of single-chain trimeric NY-ESO-1(C9V)–β2-microglobulin–HLA, and determined the structure by single-particle cryo-EM (Fig. 7a and Supplementary Fig. 7; Supplementary Tables 1–3). Despite picking particles using a template model of the closed/compacted TCR–CD3 complex bound to HLA, cryo-EM reconstruction showed the HLA-bound TCR–CD3 to be open and extended (Fig. 7a). Indeed, the HLA-bound conformation was nearly identical to that seen in GDN (Fig. 7b). Comparison of the TCR variable domains in the HLA-bound nanodisc and GDN structures showed them to be nearly identical, despite the absence of ligand in the GDN structure, and both were similar to the crystal structure of the 1G4 TCRαβ ectodomain heterodimer bound to HLA (Fig. 7b). There was subtle tilting of HLA relative to the TCR when comparing the HLA-bound nanodisc structure to the crystal structure, but the binding interface was otherwise similar (Fig. 7c). By contrast, ligand binding induced several structural changes in the TCR variable domains relative to ND-I and ND-II. Specifically, in ND-I and ND-II, the amino-terminal strands and several surface loops in the variable domains were unstructured, but these were ordered in the HLA-bound structure (Supplementary Table 3). Qualitatively, the HLA-bound map showed better resolved protein density throughout (compare Fig. 7a with Fig. 1g), consistent with a general rigidification of the TCR structure upon HLA binding, as has been observed previously for the TCR ectodomains by NMR15 and hydrogen-deuterium exchange32. This rigidification was apparent in the density corresponding to the TCR variable domains, particularly for TCRβ (Fig. 7d, e).
a Locally refined, DeepEMhancer-sharpened maps (outer) and model surfaces (inner) of HLA-bound TCR–CD3 in nanodiscs. b Overlay of HLA-bound and GDN models with zoomed-in insets. The HLA-bound model is colored by chain as in (a), and GDN is transparent gray. Lower inset highlights TM regions. Upper inset compares backbone traces of the TCR variable domains from HLA-bound (cyan), GDN (green), and 2BNQ59 (red), aligned on the TCRα variable domain. c Cartoon representation of the TCR–HLA interface from the cryo-EM structure in nanodiscs (colored by chain) and the crystal structure (2BNQ59, colored red), aligned on the variable domains. Comparison of the cryo-EM density for the TCR variable domains in ND-I, ND-II, and the HLA-bound form zoomed-in on TCRα (d) and TCRβ (e) to highlight progressive structural rigidification. Top row, cartoon representation of model; bottom row, cryo-EM map. Legend in e also applies to (d). f Proposed model of TCR–CD3 conformational dynamics. The unliganded TCR–CD3 explores resting subconformations ND-I and ND-II. HLA binding induces a conformational transition from ND-II to an open/extended conformation. Magenta arrows indicate movements required to transition to the next conformation in the series. g Changes in the TM helices between resting (ND-I shown) and HLA-bound conformations. h Increasingly zoomed-in views of the TCRα variable and constant domains in the HLA-bound conformation highlighting the S104-V182 Cα-Cα distance. i T-cell activation as quantified by flow cytometry for ZsGreen expression in J8Zb2m-α-β- reporter T cells after anti-CD3ϵ enrichment and exposure to NY-ESO-1 peptide-pulsed COS-7-A2 cells (n = 3 replicates). T cells were untransduced or transduced with a virus expressing TCRαβ 1G4 with the TCRα genotype indicated prior to enrichment, and COS-7-A2 cells were pulsed with the indicated concentrations of NY-ESO-1 peptide. PMA/ionomycin response is shown as a TCR-independent positive control. Data shown are mean +/- standard deviation using biologic triplicates, and P values were calculated by unpaired, two-tailed t-tests without adjustment for multiple comparisons (P < 0.05 highlighted in bold). See Supplementary Table 7 for source data.
Comparison of the resting and HLA-bound TCR–CD3 in lipid bilayers demonstrates that HLA binding is sufficient to induce substantial conformational changes in TCR–CD3. Our data support a model in which the unliganded resting TCR–CD3 explores a thermally accessible spectrum of closed/compacted conformations from the most closed ND-I to the more open ND-II, with HLA binding triggering a change to the open/extended conformation (Fig. 7f). We favor this transition occurring via the ND-II conformation, which is more open than ND-I and thus closer to the HLA-bound state along this trajectory. Morphing the closed/compacted conformation into the open/extended one (Supplementary Movie 2) highlights the straightening of the TM helices towards the membrane normal (that is, decreased splay, Fig. 7g). These changes in TM conformation add to the plausibility of the model, as they provide a means to transmit conformational changes in the ectodomains to the cytoplasmic ITAMs.
While some published data support allosteric activation models for the TCR, in which HLA binding is sufficient to induce TCR activation33, other data support models in which ligand-induced conformational change is necessary but not sufficient, requiring subsequent receptor clustering12 or mechanical force application34. However, all of these models propose that TCR–CD3 conformational change is a necessary step in the activation process, and thus we hypothesized that constraining conformational change would impair ligand-induced T-cell activation. Transition from the closed/compacted to the open/extended conformation requires further ectodomain opening from the ND-II state (Fig. 7f). We hypothesized that the intra-TCRα disulfide bond-forming double mutation S104C–V182C (characterized above, Fig. 5c, e) would prevent ectodomain opening to the HLA-bound conformation, as the Cα-Cα distance between these residues in that conformation is outside the limits of a disulfide bond (Fig. 7h). To investigate the impact of restrained TCR–CD3 opening on ligand-dependent T-cell activation, we introduced either wild-type or TCRα S104C–V182C mutant TCR constructs into TCRαβ-negative Jurkat T cells bearing an NFAT-responsive fluorescent reporter. TCR–CD3 assembly and surface localization (as assessed by anti-CD3ϵ antibody binding) as well as HLA-binding competence were confirmed by flow cytometry. These parameters were found to be comparable for cells transduced with the mutant and wild-type 1G4 (Supplementary Fig. 8a). In the absence of stimulation, there was no detectable reporter fluorescence in these cells; however, upon exposure to HLA–antigen tetramers, cells expressing wild-type TCR–CD3 became activated, with reporter fluorescence peaking at 16 h (Supplementary Fig. 8b). Reporter fluorescence was significantly decreased in the TCRα S104C–V182C mutant, with peak activation attenuated by 26% relative to wild type on exposure to 8 μg/mL of tetramers (P < 0.05, Supplementary Fig. 8b; source data in Supplementary Table 5). Similarly, using peptide pulsed COS7-A2 cells, we observed a 42% reduction in activated T cells transduced with the TCRα S104C–V182C mutant compared to wild-type TCR (Fig. 7i, P = 0.00009; Supplementary Fig. 8c; source data in Supplementary Tables 6 and 7). This finding confirms that the conformation-constraining double mutant also restrains T-cell activation when antigen is presented in a cellular context. In contrast, both the TCRα S104C–V182C mutant and wild-type controls were equally responsive to phorbol 12-myristate 13-acetate and ionomycin (PMA/IMY), which bypass TCR–CD3 signaling to activate T cells, showing this phenotype is specific to ligand-induced TCR–CD3 activation. Similar results were observed for the TCRβ T130C/H172C mutant (characterized above, Fig. 5d, f), which would also be hypothesized to prevent TCR–CD3 opening. For this mutant, peak response to HLA–antigen tetramers was reduced by approximately 50%, despite surface expression of TCR–CD3 complex and PMA/IMY responsiveness greater than those of wild type (Supplementary Fig. 8e–g; source data in Supplementary Tables 8 and 9). Efforts were undertaken to constrain TCR–CD3 extension with two additional disulfide mutants in TCRβ; however, the surface expression of these mutants was significantly lower than that of wild type, precluding unambiguous analysis of their ligand-dependent activation (Supplementary Fig. 8e, f; source data in Supplementary Table 8). Taken together, these data indicate that TCR–CD3 conformational change is associated with optimal receptor signaling following binding to peptide-HLA.
Discussion
Here, we report structures of a human TCR–CD3 both in detergent micelles and in a lipid bilayer. We find that TCR–CD3 assumes a spectrum of more closed and compacted conformations when embedded in a lipid bilayer relative to that seen in detergent micelles, and that the closed/compacted conformation is indeed the native unliganded state of the TCR–CD3. Addition of HLA, the physiologic ligand for the TCR, induced a change in conformation from the resting state to an open/extended conformation like that previously seen in detergent, and exit from the resting conformation by ectodomain opening is necessary for maximal ligand-dependent activation of the TCR–CD3. While this latter finding does not rule out roles for TCR–CD3-extrinsic phenomena (e.g., higher-order clustering, steric exclusion of inhibitory phosphatases) in the activation of the TCR–CD3 or regulation thereof, it is consistent with prior biochemical studies suggesting specific conformational changes during TCR activation13,35,36 (including CD3ζ dimer apposition37) and suggests that intrinsic conformational change in the TCR–CD3 is necessary for early TCR–CD3 activation33. These findings challenge previous models that have excluded this requirement11,38. We note that our results do not rule out a role for mechanical force application or catch-bond formation in the maximal activation of the TCR–CD3 but show that the earliest stage of TCR activation (i.e., conformational change out of the resting state) can be mediated by ligand binding alone.
Complementing the engineered disulfide crosslinking data presented here, independent support for our resting-state model is provided by a recently published in situ photo-crosslinking study of the mouse 2B4 TCR39. Although Natarajan et al. conclude that their crosslinking data support an open/extended resting-state conformation for the TCR–CD3, a number of their reported crosslinks could not be explained by this conformation and are instead consistent with ND-I and ND-II. For example, Natarajan et al. report an unexpected photo-crosslink between mouse TCRβ CC’ loop residues S170 and G171 (human 1G4 S186 and G187) and CD3δ. In our models, compaction of the TCR–CD3 in conformation ND-I brings CD3δ residues 26-30 in closer proximity to TCRβ residues 186 and 187 than in the GDN conformation (red arrows, Supplementary Fig. 9a), making TCRβ CC’–CD3δ crosslinking plausible. Similarly, the reported crosslinks between the TCRβ FG-loop and CD3γ would also not be predicted by the conformation of the TCR–CD3 in detergent (Supplementary Fig. 9b, right). However, closure and compaction in ND-I bring the FG-loop in proximity to the CD3γ amino terminus (Supplementary Fig. 9b, left, dashed circle), explaining this finding. Lastly, they report crosslinks between the helix 4–F-strand region of TCRβ, and CD3ϵ and CD3δ, which they wrote could not be explained by the structure of the TCR–CD3 in detergent. The mouse TCRβ R207 (human R223) crosslink to CD3ϵ is indeed incompatible with the GDN conformation (Supplementary Fig. 9c, right), but this residue is brought into proximity of CD3ϵ in the ND-I conformation (Supplementary Fig. 9c, left). In contrast to their assessment, the mouse TCRβ F202 (human F218) crosslink to CD3δ is compatible with both the open/extended and ND-II closed/compacted conformations that we report (Supplementary Fig. 9d). Natarajan et al. also directly assessed crosslinks based on our reported TCRα and CD3δ interface (Fig. 5a, b) and their crosslink-based, open/extended computational model. One mutant carrying a crosslink designed from each of these structural hypotheses was successfully expressed on the cell surface. The reporter based on our cryo-EM models – mouse CD3δ E55 (human D77) – did indeed form a crosslink with TCRα (see Natarajan et al. Supplementary Fig. 16c), whereas the reporter based on their open/extended model did not form a crosslink. Taken together, the photo-crosslinking data reported by Natarajan et al.39 are consistent with our closed/compacted resting conformation for the TCR–CD3 complex in vivo.
Natarajan et al. went on to analyze the effect of activating ligands on TCR–CD3 conformation using their photo-crosslinking assay and concluded that there was no ligand-dependent conformational change. However, we contend that the low spatial resolution, inherent non-specificity, and mutation site choice in their analysis would not exclude ligand-induced conformational change. Most of the p-azido phenylalanine substitution sites used in their study are at the core interfaces between the CD3 ectodomains and/or the TCR constant domains. Photo-crosslinking at these sites would not be expected to show ligand-induced changes in subunit-level crosslinking patterns based on our models. They also show that the aforementioned mouse CD3δ E55 (human D77) to TCRα crosslink persists after activating ligand exposure. However, this is actually what we would expect given the proximity of CD3δ D77 to TCRα in both our resting state and ligand-bound models. For both of our open/extended conformations (GDN and ND-HLA), the CD3δ D77 Cβ remains 8-9 Å from TCRα (Supplementary Fig. 9e), comparable to the 9-10 Å Cβ-to-any-heavy-atom radius previously used by the Krogsgaard group to predict plausible crosslinking40. Modeling the UV-activated nitrene intermediate of CD3δ D77 p-azido phenylalanine into ND-HLA as the most likely rotamer of tyrosine places the reactive nitrene less than 4 Å from the TCRα backbone (Supplementary Fig. 9e). In sum, we argue that the experimental approach used by Natarajan et al. cannot exclude the conformational changes described in our models.
We employed an engineered disulfide TCR mutant to constrain conformational change and found that it significantly reduced peak T-cell activation in response to HLA. There are several possible reasons for the partial amelioration of signaling instead of complete extinction. The disulfide may not form reliably in 100% of cell surface-localized receptors, resulting in a mixed population. Even if formed, the disulfide may not completely prevent conformational change, perhaps allowing formation of intermediate conformations, if not the open/extended conformation, per se. Lastly, ligand-induced clustering may otherwise change the membrane milieu in ways that tip the scales to allow for partial (suboptimal) signaling even without conformational change. None of these hypothetical caveats are mutually exclusive. Nonetheless, the ability of this mutant to reduce peak signaling with surface expression and ligand binding grossly comparable to wild type is evidence of the importance of conformational change in the process of ligand-induced receptor activation.
Broadly, these findings reinforce the importance of structural interrogation of membrane proteins in lipid environments that closely resemble their physiologic environment. Comparison of TCR–CD3s in detergent micelles in the absence and presence of activating ligand had revealed no change in TCR–CD3 structure7,8, consistent with models excluding TCR–CD3 conformational change in the activation process. Although these studies were technically well performed, our results now reveal that they did not identify the true resting state of the TCR–CD3 in lipid bilayers, preventing observation of the conformational changes during ligand-dependent activation.
Additionally, the structures in detergent did not capture TCR–CD3–membrane interactions. Not only are membrane lipids closely interwoven with the TM helices, but conserved ectodomain surface patches mediate lipid–protein interactions, likely partially counteracting the thermodynamic costs of JM linker compaction and spring loading the TCR with potential energy for conformational change during activation. In this model, ligand binding provides the activation energy to release the membrane-binding surfaces of the CD3 ectodomains and allow JM extension. Detergent extraction overcomes this thermodynamic barrier by removing the lipid bilayer entirely. During reconstitution into nanodiscs, we first extracted the TCR–CD3 in detergent, opening and extending the receptor, which suggests that the addition of a lipid bilayer is sufficient to reset the TCR–CD3 to its resting conformation without the need for extrinsic chaperones. Lastly, we note that a similar conserved surface patch is present on the CD79 heterodimer in the B-cell receptor27, raising the possibility that other hetero-multimeric ITAM-containing immunoreceptors exploit protein surface interactions with the lipid bilayer to constrain conformation.
As with the TCR–CD3 structures in detergent, the CD3 ITAMs and other cytosolic components are not visible in our structures. The ITAMs are membrane-associated in the unliganded state9,10, but are likely to be radially disordered along the intracellular membrane face. However, the TM helix movements seen between our closed/compacted and the open/extended conformations suggest a means for communication of extracellular conformational changes in the TCR–CD3 complex to the ITAMs. Within the cytoplasmic domains of CD3 proteins, the basic residues proximal to the CD3 TM helices are critical for ITAM association with the membrane9,10. Upon transition from the resting to the HLA-bound conformation, the decreased splay in the CD3 TM helices could alter basic patch interaction with the membrane and favor ITAM release.
The basal conformational heterogeneity of the TCR–CD3 suggests additional means of TCR–CD3 regulation. The TCRα and CD3δ glycan ordering observed in the ND-I conformation is notably different from the glycan arrangement in the more open ND-II conformation or the TCR–CD3 in detergent. While molecular modeling of the branched glycan sugars was not possible with sufficient confidence, the stacked arrangement of glycan core densities raises the possibility of extensive hydrogen bonding between the glycan branches distally and perhaps also with TCR–CD3 surface residues. While glycan–glycan and glycan–protein interactions are well described in trans, there are fewer examples of structured glycan–glycan interactions within a given protein or stable complex. The human immunodeficiency virus-1 Env protein is an example of the latter30,41 and the arrangement of the radial glycan array in ND-I is reminiscent of that seen in crystallographic studies of Env. However, the stacking of glycans observed in the TCR–CD3 stands out in being protein conformation-dependent. Breaking an extensive glycan hydrogen-bonding network to transition out of ND-I would be energetically costly, and thus modification of TCR–CD3 glycosylation could serve as a means of posttranslational regulation of the TCR–CD3 activation threshold by increasing or decreasing the free energy barrier to ND-I exit.
Previous studies have explored the role of cholesterol in the regulation of TCR–CD3 activation6,18,26. The substantive movements of the two cholesterol molecules seen between TCR–CD3 conformations raise the possibility that changes in the sterol composition of the T-cell membrane (either global or local) could alter TCR–CD3 activity even prior to ligand binding by shaping the basal TCR–CD3 conformational landscape. Moreover, our structures reveal channels for cholesterol loading or exchange with the lipid bilayer not previously observed, making it plausible that already assembled TCR–CD3 could respond to changes in membrane sterol content. Non-sterol lipid species are also known regulators of T-cell signaling17, including phosphoinositol lipids42,43 and oxidized lipids in the tumor microenvironment44. This work delineates multiple sites of structured interaction between the TCR–CD3 and membrane lipids beyond the sterol-binding site, which may allow specific lipids to modulate TCR–CD3 conformation and activity.
The extensive contacts between TCR variable domain residues and those of the TCR constant domains or CD3 ectodomains in the closed/compacted conformations raises questions about how variation at these sites might confer specific baseline sensitivities to given TCR alleles. Whether use of certain V or J alleles changes the occupancy of different segments of the TCR–CD3 conformational landscape will need to be determined and has broad implications for vaccine design and immunotherapy engineering. Likewise, whether individually rare germline-coding variants in the TCR constant domain loci or CD3 proteins affect an individual’s basal T-cell reactivity is unclear outside of known deleterious loss-of-function alleles45. Such information might predict individual propensity for autoimmunity or responsiveness to immune-oncology agents and will provide insights for engineering TCRs with favorable activity profiles46.
Methods
Plasmids and sequences
A modified version of the BacMam expression system47 was used for the expression of the TCR–CD3 complex in HEK cells with furin cleavage and P2A ribosomal skipping sites separating each protein sequence (see below, Protein expression and purification and Engineered disulfide crosslinking). The modified viral vector also expressed the VSV-g sequence (kind gift from Titia de Lange, Rockefeller University) under a baculoviral promoter to enhance mammalian cell tropism. A self-inactivating retroviral plasmid encoding the 1G4(LY) TCRαβ sequence (wild type for this study) was a kind gift from S. A. Rosenberg, NIH. CD3ζ was cloned from Jurkat cell cDNA using standard restriction enzyme-based approaches. Similarly, CD3ϵ, CD3γ, CD3δ, EGFP, mCherry, and 8xHis tags were cloned from synthetic double-stranded DNA blocks (IDT DNA). Mutant TCR–CD3 sequences were produced by overlap-extension PCR or synthesized de novo (Genscript). Construct sequences can be found in and following Supplementary Table 10.
E. coli strains
Bacteria were grown under standard conditions: 37 °C, ambient CO2; LB broth shaking 200 rpm for liquid cultures, LB agar for plates. DH5α (Thermo Fisher Scientific, 18265017) were used for cloning and amplification of plasmid DNA, and BL21(DE3) (Thermo Fisher Scientific, EC0114) were used for MSP expression. DH10Bac cells (Thermo Fisher Scientific, 10361012) were grown as per manufacturer instructions for the recombination of plasmid DNA into bacmids.
Eukaryotic cell lines
Insect and human cells lines were maintained under standard sterile conditions using aseptic technique with the following parameters: SF-9 cells (ATCC, CRL-1711) were maintained at 5 × 105-1 × 106 cells/mL in Sf-900 II SFM medium (Thermo Fisher Scientific) supplemented with antibiotic-antimycotic solution (Thermo Fisher Scientific) at 27 °C, ambient CO2. HEK293GP cells (Takara Bio, 631458) and derivatives were maintained at 10-90% confluency in RPMI medium 1640 (RPMI, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS, Sigma Aldrich), 10 mM HEPES, pH 7.5, and penicillin/streptomycin at 37 °C, 5% CO2. HEK293F FreeStyle cells (Thermo Fisher Scientific, R79007) were maintained at 5 × 105 to 2 × 106 cells/mL in FreeStyle 293 expression medium (Thermo Fisher Scientific) supplemented with antibiotic-antimycotic solution at 37 °C, 8% CO2. Modified Jurkat T cells (see below, Generation of an NFAT-reporter cell line) and COS-7-A2 cells were maintained at 2 × 105 to 2 × 106 cells/mL in RPMI medium supplemented with 10% FBS and penicillin-streptomycin solution at 37 °C, 5% CO2. HEK cells are of female human origin and Jurkat cells are of male human origin. COS-7-A2 cells was gifted by Dr. Steven Rosenberg (NIH) and derived from the male African green monkey cell line COS-7 with stable transduction of human HLA-A*02:01. Cell line identify was validated by fluorescence-activated cell sorting (FACS).
Baculovirus production
Bacmid recombineering was performed as per manufacturer specifications with the FastBac system in DH10Bac cells (Thermo Fisher Scientific). Bacmid was purified by phenol/chloroform/isoamyl alcohol extraction, and approximately 25 μg bacmid DNA was transfected into SF-9 cells adherent in 6-well plates using Cellfectin II reagent (Thermo Fisher Scientific) in Grace’s Insect Medium (Thermo Fisher Scientific) for 6 h at 27 °C before transfer to Sf-900 II SFM medium supplemented with antibiotic-antimycotic solution. After 5 days, the cell supernatant (P1 virus) was removed and used to inoculate a 40-mL culture of SF-9 cells at 1106 cells/mL. After >50% of the cells had died ( ~ day 5), supernatant (P2 virus) was harvested by centrifugation of the culture at 3500×g for 20 min, decanted, supplemented with 2% FBS, and stored in the dark at 4 °C. For large-scale expression, P3 virus was produced by inoculating 40-mL cultures of SF-9 cells at 3 × 106 cells/mL with 2 mL P2 virus and cultured, harvested, and stored as for P2 virus.
Protein expression, purification, and nanodisc reconstitution
See overview schematic in Fig. 1a. For structural studies, ~250-mL cultures of HEK293F FreeStyle cells at 2 × 106 cells/mL shaking in 1-L baffled polycarbonate flasks were inoculated with 15 mL each of P3 virus, and sodium butyrate (Alfa Aesar) was added to a final concentration of 10 mM to enhance expression. For preparation of the TCR–CD3 in detergent for cryo-EM, TCR–CD3 was expressed using two baculoviruses: (1) TCRα-[furin/P2A]-TCRβ::mCherry, and (2) CD3ϵ-[furin/P2A]-CD3ζ-[furin/P2A]-CD3δ-[furin/P2A]-CD3γ::8HIS. Cells were incubated at 30 °C, 5% CO2 for 3 days to allow expression and harvested by centrifugation at 4000×g for 10 min at 4 °C. Cell pellets were resuspended in 25 mM HEPES (Sigma Aldrich), pH 7.5, 1 mM phenylmethanesulfonylfluoride (Sigma Aldrich) and lysed with a Dounce homogenizer. Lysate was clarified by centrifugation at 4000×g for 10 min at 4 °C, and the supernatant was decanted and ultracentrifuged at 100,000×g for 1 h at 4 °C to recover the membrane fraction. Membrane pellets were resuspended in 25 mM HEPES, pH 7.5, 150 mM NaCl (Thermo Fisher Scientific), herein HBS, and n-dodecyl-β-D-maltoside (DDM, Anatrace) and cholesteryl hemisuccinate (CHS, Anatrace) were added to final concentrations of 1% and 0.2%, respectively. The membrane pellets were solubilized on a rotator for 2 h at 4 °C. Debris was removed by centrifugation at 4000×g for 30 min at 4 °C. The clarified supernatant was loaded onto NiNTA-agarose resin (Qiagen). Resin was washed with 50 column volumes of HBS with 0.02% glyco-diosgenin (GDN) and 30 mM imidazole (Thermo Fisher Scientific). The TCR–CD3 was eluted from the NiNTA-agarose resin with 360 mM imidazole and 0.02% GDN in HBS. The mCherry fusion to TCRβ was removed by incubation with human rhinovirus protease 3 C overnight at 4 °C. Protein was concentrated with Amicon centrifugal filters (Millipore) and further purified by size-exclusion chromatography in HBS with 0.02% GDN over a Superose6 column (GE Healthcare).
For expression of the TCR–CD3 for affinity purification and nanodisc reconstitution, we found that co-expression of three viral constructs gave acceptable yields of material for cryo-EM analysis: (1) TCRα-[furin/P2A]-TCRβ::mCherry-[furin/P2A]-CD3δ-[furin/P2A]-CD3γ-[furin/P2A]-CD3ϵ-[furin/P2A]-CD3ζ::EGFP, (2) CD3δ-[furin/P2A]-CD3γ::8HIS, and (3) CD3ϵ-[furin/P2A]-CD3ζ::EGFP. This three-virus expression scheme was later simplified to two viruses for studies with HLA-bound (below), in detergent (above), or TCR–CD3 crosslinking experiments, as tandem affinity purification was not found to be necessary for intact complex recovery. Cells were cultured, infected, harvested, and lysed as above. Lysate was clarified, membranes isolated and extracted, and extracts loaded onto NiNTA-agarose resin as above. Resin was washed with 50 column volumes of HBS with 0.1% DDM, 0.02% CHS, and 30 mM imidazole. The resin was resuspended in HBS with 0.1% DDM, 0.02% CHS for on-bead nanodisc reconstitution. For the TCR–CD3 complex in nanodiscs subsequently incubated with HLA–β2M–NY-ESO-1, a simplified two-virus expression scheme was used: (1) TCRα-[furin/P2A]-TCRβ::EGFP, and (2) CD3ϵ-[furin/P2A]-CD3ζ-[furin/P2A]-CD3δ-[furin/P2A]-CD3γ::8HIS. Protein complexes were otherwise purified as above.
T-cell subsets differ in plasma membrane lipid composition, and individual T cells have spatially heterogenous membrane compositions17. To create a lipid environment with a diversity of lipid constituents, including the negatively charged phosphatidylserine and phosphatidylinositol lipids important for ITAM–membrane interactions9,10, porcine brain polar lipids (Avanti Polar Lipids #141101 C) were used to reconstitute the lipid bilayer. Avanti reports the composition of this lipid mixture as follows: 12.6% phosphatidylcholines, 33.1% phosphatidylethanolamines, 4.1% phosphatidylinositides, 18.5% phosphatidylserines, 0.8% phosphatidic acid, 30.9% unknown. The porcine brain polar lipids dissolved in 10% DDM were added to a final concentration of 33 mg/mL (approximately 43 μM using the molecular weight of 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine as a standard phospholipid) and membrane-scaffold protein (MSP) 1D1ΔH5 was added to a final concentration of 20 μM. MSP 1D1ΔH5 was expressed in E. coli BL21(DE3), NiNTA-affinity purified, and affinity tags removed with TEV protease as previously described48 after subcloning from pET28a-MSP1D1ΔH5 (a gift from Gerhard Wagner, Addgene plasmid # 71714)49. Nanodisc reconstitution components were allowed to incubate for 15 min on ice before addition of SM-2 BioBeads (Bio-Rad) to approximately ¼ of the reaction volume. The mixture was incubated overnight on a rotator at 4 °C. The reaction mixture was again returned to a disposable plastic column and washed with 20 column volumes of HBS, 50 column volumes of HBS with NaCl increased to 300 mM and 30 mM imidazole, washed with 20 column volumes of HBS, then eluted in HBS with 360 mM imidazole. Protein was concentrated with Amicon centrifugal filters (Millipore) and further purified by size-exclusion chromatography in HBS over a Superose6 column (GE Healthcare). Peak fractions were collected and their quality ensured by SDS-PAGE followed by silver staining (Invitrogen), Coomassie blue staining, or Acquastain (Bulldog Bio) as per manufacturer specifications, and by negative-stain electron microscopy as described previously50. Briefly, samples were adsorbed for 30 s to glow-discharged, carbon-coated copper grids (Electron Microscopy Sciences), washed three times in deionized water, stained in freshly prepared 0.7% uranyl formate, and air dried before imaging on a CM-10 transmission electron microscope (Philips) equipped with XR-16 detector (Advanced Microscopy Techniques).
Cryo-EM specimen preparation and data collection
For the TCR–CD3 in GDN, Superose6 peak fractions were concentrated with Amicon centrifugal concentrators to 5 mg/mL as determined with a NanoDrop spectrophotometer (Thermo Fisher Scientific) assuming 1 AU = 1 mg/mL. Aliquots of 4 μL were applied to glow-discharged 400 mesh R1.2/1.3 Cu grids with graphene oxide (Quantifoil) using a Vitrobot Mark VI (Thermo Fisher Scientific) set at 4 °C and 100% humidity. After 20 s, grids were blotted for 0.5 s with a blot force of −2 and plunged into liquid nitrogen-cooled ethane.
For the TCR–CD3 in nanodiscs, Superose6 peak fractions were concentrated with Amicon centrifugal concentrators to 330 μg/mL as determined with a NanoDrop spectrophotometer assuming 1 AU = 1 mg/mL. Aliquots of 4 μL were applied to glow-discharged 400 mesh R1.2/1.3 Cu grids using a Vitrobot Mark VI set at 4 °C and 100% humidity. After 5 s, grids were blotted for 4 s with a blot force of −2 and plunged into liquid nitrogen-cooled ethane. For the TCR–CD3 in nanodiscs with HLA–β2M–NY-ESO-1, the Superose6 peak fractions were concentrated with Amicon centrifugal concentrators to 600 μg/mL. Lyophilized HLA single-chain trimer (herein, “HLA”; fusion of NY-ESO-1 C9V [SLLMWITQV], β2M I21-M119, and HLA-A*02:01 G25-T305; Kactus Biologics) was dissolved in deionized water per manufacturer instructions and then re-purified by size-exclusion chromatography over a Superose6 column into HBS to remove PBS and trehalose and concentrated to 1 mg/mL. The TCR–CD3 and HLA single-chain trimer were mixed at a 2:1 volume ratio (1:5 approximate molar ratio). Aliquots of 4 μL were applied to glow-discharged graphene oxide-coated 400 mesh R1.2/1.3 Cu grids (Quantifoil) using a Vitrobot Mark VI (Thermo Fisher Scientific) set at 4 °C and 100% humidity. After a wait time of 20 s, grids were blotted for 0.5 s with a blot force of −2 and plunged into liquid nitrogen-cooled ethane.
Cryo-EM imaging was performed in the Cryo-EM Resource Center at the Rockefeller University using SerialEM v4.1.0beta51. Data collection parameters are summarized in Supplementary Table 1. In brief, for all samples, data were collected on a 300-kV Titan Krios electron microscope at a nominal magnification of 64,000×, corresponding to a calibrated pixel size of 1.08 Å on the specimen level. Images were collected with a K3 direct electron detector (Gatan) in super-resolution counting mode. The superfast mode in SerialEM was used, in which 3 × 3 holes are exposed using beam tilt and image shift before moving the stage to the next position52.
For the TCR–CD3 in detergent sample, images were collected using a defocus range of −0.8 to −2.5 μm. Exposures of 2.0 s were dose-fractionated into 50 frames (40 ms per frame) with a dose rate of 30 electrons per pixel per s (approximately 1.03 electrons per Å2 per frame), resulting in a total dose of 51 electrons per Å2. For the TCR–CD3 in nanodisc sample, images were collected using a defocus range of −0.8 to −2.0 μm. Exposures of 2.2 s were dose-fractionated into 55 frames (40 ms per frame) with a dose rate of 30 electrons per pixel per s (approximately 1.03 electrons per Å2 per frame), resulting in a total dose of 57 electrons per Å2. For the HLA-bound TCR–CD3 in nanodiscs sample, images were collected using a defocus range of −0.8 to −2.0 μm. Exposures of 2.0 s were dose-fractionated into 50 frames (40 ms per frame) with a dose rate of 30 electrons per pixel per s (approximately 1.03 electrons per Å2 per frame), resulting in a total dose of 51 electrons per Å2.
Cryo-EM image processing for the TCR–CD3 in detergent
The 15,726 collected movie stacks were gain-normalized, motion-corrected, dose-weighted, and binned over 2 × 2 pixels in MotionCor2 v1.1.053 and imported into cryoSPARC v4.2 for all further processing54 (see data processing overview and motion-corrected micrograph examples in Supplementary Fig. 1). A subset of the larger dataset, comprising 4,278 micrographs, was used to generate an initial model. The contrast transfer function (CTF) parameters were determined with CTFFIND v455 implemented in cryoSPARC. Micrographs were curated by average defocus value, CTF fit resolution and CTF cross correlation, yielding 3,292 motion-corrected micrographs with CTF estimations for downstream processing. Particles were picked with the cryoSPARC blob-picker using a size range of 80 to 180 Å. These initial particles were filtered by power level and normalized cross-correlation score. Particles were subjected to serial rounds of 2D classification, wherein particles with the best alignment, as determined quantitatively by resolution estimate or qualitatively by appearance of fine structure, were advanced to the next round of 2D classification and selection. The best 82k particles were used for ab initio modeling, specifying three models. One model was clearly recognizable as TCR–CD3 in a detergent micelle; the other two models were amorphous blobs. All three models were used to seed 3D heterogeneous refinement using the same 82k particles, and the resulting TCR–CD3 model was used as an initial model in downstream processing.
We returned to the full dataset of 15,726 motion-corrected micrographs. CTF parameters were estimated using Patch CTF in cryoSPARC. Micrographs were curated by average defocus value, defocus range, CTF fit resolution, and relative ice thickness, yielding 15,153 micrographs for particle picking. Particles were picked with 20-Å low pass-filtered templates from the model of the TCR–CD3 in detergent determined above and filtered by power level and normalized cross-correlation score. Particles were subjected to serial 2D classification as above and 3D classification by multireference refinement (using the heterogeneous refinement job in cryoSPARC). Serial rounds of 3D classification were seeded with the best available model along with decoy noise models to remove poorly aligning particles. Decoy classes were created by aborting ab initio modeling jobs after the first round of modeling before any structure was visible. The best aligning particles emerging from 3D classification were then used to train a Topaz (v 0.2.5a) particle picking model56 in cryoSPARC and the resulting particle stacks subjected to iterative rounds of 2D classification, 3D variability analysis, 3D classification, Topaz retraining, and particle pooling until map quality reached a plateau.
Picking from the 15,153-micrograph set with the best Topaz model yielded 2 M particles. These particles were subjected to serial 2D classification, and the best 951k particles were entered into three rounds of serial 3D classification seeded with the best prior model and three noise decoys. This yielded 311k particles which were subjected to an additional round of 2D classification, from which 248k particles were selected. These particles, along with those from the best prior Topaz-picked model (242k), were re-extracted in 300×300-pixel boxes and pooled to ensure that all high-quality particles were captured for final reconstruction. One round of 2D classification was performed on the extracted particles and duplicates removed, yielding 388k unique particles. Of these, the 310k best-aligning particles were selected to advance into a final cascade of five rounds of serial 3D classification seeded with the best prior model and 3 noise decoys. The final 252k particle set emerging from these classifications was subjected to non-uniform refinement, yielding a map with nominal resolution of 3.3 Å. For this and all nominal resolutions listed herein, resolutions are reported from gold-standard Fourier shell correlation (FSC) curves with a cut-off criterion of 0.143. Local refinement with a mask including the protein components but excluding the micelle density further improved map quality and interpretability.
Cryo-EM image processing for the TCR–CD3 in nanodiscs
The 17,766 collected movie stacks were gain-normalized, motion-corrected, dose-weighted, binned over 2 × 2 pixels, and CTF parameters estimated as above for GDN (see data processing streams and motion-corrected micrograph examples in Supplementary Fig. 2). Micrographs were curated by CTF fit resolution and cross correlation, yielding 14,852 motion-corrected micrographs with CTF estimations for downstream processing.
Three parallel processing streams were pursued based on different approaches to particle picking (Supplementary Fig. 2). Particles were picked in cryoSPARC using (1) an unbiased blob-picking approach using a particle size range of 60 to 120 Å, (2) a template-based approach with a model of the TCR–CD3 in detergent (PBD 6JXR5) docked into the MSP 1D1DH5 nanodisc using the CHARMM-GUI web server [https://www.charmm-gui.org/]57 nanodisc builder58, and (3) a template-based approach using the structure of the TCR–CD3 in detergent (PBD 6JXR5). For each processing stream, 2D image classification was performed on 4×4 binned particle images to eliminate classes of noise or poorly aligning particles and ab initio modeling was performed. At this stage, it was clear that the particles picked using 6JXR templates were of poor quality and did not yield reconstructions substantially different from those obtained with the other two processing streams (see below) and this avenue was thus aborted. For the other two streams, particle stacks were subjected to iterative rounds of 2D classification, ab initio modeling, 3D variability analysis, and 3D classification until map quality plateaued. The best aligning particles from these two streams were pooled and iterative rounds of 2D classification, 3D classification, Topaz training, and particle pooling was performed until map quality plateaued.
Having now exhaustively sampled the micrographs for well-aligning particles, the best aligning 2.2 M unique particles were extracted into 256×256-pixel boxes without binning. After one round of 2D classification and selection, 1.8 M particles remained. These particles were subjected to two rounds of serial 3D heterogenous refinement seeded with the best two available maps along with two noise maps, yielding 630k and 809k particles in the two best classes. After re-extraction into 300×300-pixel boxes without binning, these two classes, which we now denote ND-I and ND-II, contained 620k and 797k particles and refined to nominal resolutions of 3.2 Å and 3.0 Å, respectively. Local refinement with a mask including the protein components but excluding the nanodisc density further improved map quality and interpretability.
Cryo-EM image processing for the TCR–CD3 in nanodiscs incubated with HLA
The 13,644 collected movie stacks gain-normalized, motion-corrected, dose-weighted, binned over 2 × 2 pixels, and CTF parameters estimated as above for GDN, ND-I, and ND-II (see data processing stream and motion-corrected micrograph examples in Supplementary Fig. 7). Micrographs were curated by average defocus, astigmatism, CTF fit resolution, and CTF fit cross correlation, yielding 10,610 motion-corrected micrographs with CTF estimations for downstream processing. Particles were initially picked by a template-based approach using a model of the TCR–CD3 in nanodiscs ND-II conformation with HLA from PDB 2BNQ59 docked by alignment of the TCR variable domains. Templates were 20-Å low pass-filtered prior to particle picking. These initial particles were filtered by power level and normalized cross-correlation score and subjected to iterative 2D classification, ab initio modeling, 3D classification (by heterogeneous refinement and by 3D classification without alignment), and Topaz training.
To ensure exhaustive sampling of the available data, particle picks from the best three Topaz models were pooled and duplicate particles removed, yielding 3.3 M unique particles. These particles were extracted in 400×400-pixel boxes (binned 2×2) to ensure adequate accommodation for delocalized signal around the elongated TCR–CD3–HLA particles. One round of 3D heterogenous refinement was performed, seeded with the best two maps (one showing strong HLA density and one showing weaker HLA density) from the prior 3D classifications alongside 2 decoy maps. The 895k particles aligning to the HLA-strong model were subjected to an additional cycle of 3D classification without alignment, seeded by principal component analysis and specifying six classes. The six output volumes were used as the starting classes for one round of heterogeneous refinement with the 895k-particle set. One class comprising 192k particles refined to higher resolution than the others. The same analysis (3D classification without alignment followed by 3D heterogenous refinement) was performed for the HLA-weak set, yielding a single high-resolution class with 215k particles. These two sets of particles were re-extracted into a 400 × 400-pixel box without binning, and combined with the prior best particle stack, comprising 297k particles, also extracted now in a 400 × 400-pixel box. Duplicate particles were removed, yielding 587k unique particles. These particles were subjected to four rounds of serial 3D heterogeneous refinement seeded with the HLA-strong and HLA-weak models, with the particles classifying in the HLA-strong class advancing to the next round. This yielded 155k particles in the final set. Non-uniform refinement of this particle set yielded a map with a nominal resolution of 3.6 Å. Local refinement was performed with a custom mask excluding the bulk lipid of the nanodisc, yielding a map with a nominal resolution of 3.5 Å. This map was used for refinement in Phenix. Additional local refinements on subsets of the structure (e.g., TCR variable domains–HLA) were performed and used to guide manual model building but were not used for final refinement or calculation of refinement statistics.
Cryo-EM map assessment and post-processing
Gold-standard FSC curves were generated in cryoSPARC during refinement and nominal resolutions are reported with a cut-off criterion of 0.143. FSC curves for final maps are shown in Supplementary Figs. 1, 2, 7. Map anisotropy was assessed with 3DFSC60 implemented in cryoSPARC, and local resolution analysis was also performed in cryoSPARC (Supplementary Figs. 1, 2, 7). Although all models were refined against unsharpened maps (see Model building and refinement, below), sharpened maps were used for visual interpretation, manual model building, and figure generation. Map sharpening (Supplementary Fig. 3) was performed either by global B-factor approaches (better visualization of lipid densities) or DeepEMhancer v0.1461 (better visualization of protein densities).
Model building and refinement
The model of TCR–CD3 in GDN micelles was built starting with rigid-body docking of prior models in ChimeraX v1.6.1-1.1062. Specifically, PDB 6JXR was used as a starting model for the CD3 proteins and the TCR constant regions and transmembrane helices, PDB 7FJD was used to dock the cholesterols, and PDB 6BNU59 was used as a starting model for the 1G4 TCR variable regions. This starting model readily docked into the map, anchored by the clear alignment of immunoglobulin folds to the map features and continuous density for all chains through the ectodomains, juxtamembrane linkers, and transmembrane helices (see example densities in Supplementary Fig. 3). As with prior TCR–CD3 structures, the cytoplasmic unstructured regions were not resolved in the map. The model was improved by iterative cycles of refinement with phenix.real_space_refine in PHENIX v1.20.163, manual adjustment in Coot v0.9.8.764, and brief map-constrained relaxation in ISOLDE v1.965. Residues without clear side-chain density beyond Cβ were manually modeled as stubs. Cores for the N-linked glycans were built in Coot using tools included in the package66 (modeled glycans indicated graphically in Supplementary Fig. 6d). For all models, refinement data are shown in Supplementary Table 2, and model completeness is shown in Supplementary Table 3. All cryo-EM map and molecular model figures were generated in ChimeraX.
For the ND-I and ND-II models, the TCR–CD3-in-GDN model built above was used as a starting model and domains were docked into the map in ChimeraX. Again, the immunoglobulin folds in the TCR and CD3 ectodomains were readily anchored into the map features, with bulky hydrophobic residues and disulfide bonds periodically confirming register (map density overviews and sections shown in Supplementary Fig. 3). Regions without clear density in the maps were removed and residues without clear side-chain density beyond Cβ were manually modeled as stubs. For the CD3ϵ, CD3γ, and CD3δ proteins, the register for the juxtamembrane linker and transmembrane helices were anchored by the placement of the C-X-X-C lariats at the beginning of the juxtamembrane region (Supplementary Fig. 3f). Bulky hydrophobic side chains provided further confirmation of register within the transmembrane helices. In ND-I, the register at the amino-terminus of the CD3ϵG transmembrane helix could not be unambiguously determined, so this helix was left unmodeled. For CD3ζ, the GDN model of these chains was docked in place and manually refit, using the interchain disulfide bond and bulky hydrophobic residues to confirm register (Supplementary Fig. 3g). For TCRα and TCRβ, the linker peptides between their constant domains and the transmembrane helices were poorly structured and left unmodeled. Their transmembrane helices were docked from the GDN structure relative to the surrounding helices and bulky side chains were used to confirm register (Supplementary Fig. 3h). Model building and refinement was performed in Phenix, Coot, ChimeraX, and ISOLDE as described above for the TCR–CD3-in-GDN structure.
For the TCR–CD3–HLA structure, the GDN structure readily docked into the cryo-EM map. The HLA–β2M–NY-ESO-1 structure from PDB 2BNQ59 was docked into the map. The structure was refined in Phenix as described above. Example density with constructed model is shown in Supplementary Fig. 7j.
Sequence conservation analysis
TCR and CD79-BCR sequences were analyzed using the Consurf67 web server [https://consurf.tau.ac.il/] and colored according to relative sequence conservation level in ChimeraX. For computation of sequence identity among GDN and the published TCR–CD3 structures in detergent, sequences were aligned with the Clustal Omega web server [https://www.ebi.ac.uk/jdispatcher/msa/clustalo]68.
Engineered disulfide-bond analysis
Cysteine mutant pairs were designed with the Cα-Cα parameters described in the main text. For Western blot analysis, HEK293F cells were plated at 1.5 × 106 cells/mL in 6-well plates, 2 mL per well. Cells were infected with baculovirus expressing the indicated TCR–CD3 mutants at a volume 1/30 the culture volume and the indicated CD3 mutants at 1/15 the culture volume. All TCRα mutants were paired with the same wild-type TCRβ sequence. 10 mM sodium butyrate was added at the same time as viral infection to enhance expression. Cultures were incubated for 2 days at 37 °C, 8% CO2. Cells were harvested by centrifugation at 20,800×g, and resuspended in Laemmli sample buffer with or without 100 mM dithiothreitol (Gold Bio). Samples were heated for 5 min at 95 °C prior to separation by SDS-PAGE. Gels were electroblotted to immobilon-P PVDF membranes (Millipore), rinsed in 200 mM NaCl, 20 mM Tris (Research Products International), pH 8 (TBS), supplemented with 0.05% Tween-20 (TBST, Sigma Aldrich), and blocked in neat Intercept Blocking Buffer (Li-Cor) for 1 h at room temperature. Blots were incubated with mouse anti-GFP antibody (1:3,000, Roche/Sigma Aldrich, Cat. # 11814460001) in TBST with 10% block overnight at 4 °C. Blots were washed with TBST at 25 °C, incubated with Alexa 700-conjugated goat anti-mouse IgG (1:10,000, Invitrogen, Cat. # A-21036) in TBST with 10% block at 25 °C for 1 h, and again washed sequentially with TBST and TBS. Blots were imaged on an Amersham Typhoon infrared fluorescence scanner.
To confirm the results seen in whole-cell lysates, mutant TCR–CD3 was also detergent extracted from membrane pellets and NiNTA-affinity purified, as was done for cryo-EM analyses, and NiNTA elutions in DDM/CHS were processed for Western blot analysis. To further confirm that these crosslinks were formed in properly assembled TCR–CD3 complexes, select disulfide-bond mutants were also reconstituted into nanodiscs and purified by size-exclusion chromatography, by the same protocol used for producing TCR–CD3 in nanodiscs for cryo-EM analysis. For these samples, protein constituents were visualized in gels using Acquastain as per manufacturer specifications.
Mass spectrometry
Protein was produced as for structural studies but using the indicated mutant TCR–CD3 constructs with TCRβ fused to EGFP and CD3ζ untagged. The CD3 proteins, including 8xHis-tagged CD3γ were expressed from a single virus. Instead of reconstitution into nanodiscs, the TCR–CD3 complex was eluted from NiNTA-agarose resin in HBS containing DDM/CHS, free cysteines were blocked with 20 mM iodoacetamide (Sigma Aldrich) in the dark for 1 h at room temperature. The reaction was quenched, and detergent was removed by acetone precipitation. Protein complexes were deglycosylated with PNGase F and sequentially digested using sequencing-grade trypsin for 3 h, followed by a second digestion step using chymotrypsin for another 3 h. Enzymatic activity was quenched by acidification using trifluoroacetic acid and the resulting peptides were purified using in-house constructed C18 micropurification tips prior to LC-MSMS analysis69. Peptides were separated across a 70-min linear gradient, in which the fraction of solvent B (80% acetonitrile, 0.1% formic acid) over solvent A (1% acetonitrile, 0.1% formic acid) was increased from 2% to 35% using an Easy nLC 1200 Ultra-High Performance Liquid Chromatography system (Thermo Fisher Scientific) equipped with a 100 μm-by-120 μm pulled-emitter column packed with 3-μm C18 material (Nikkyo Technos). Analysis was carried out using a Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific) operating in positive-ion DDA, FT-mode. Ions were fragmented using EThcD. Disulfide bonds were identified using the XlinkX software through Proteome Discoverer v. 2.5 (Thermo Fisher Scientific).
Generation of an NFAT-reporter cell line
The J8Zb2m-α-β- cell line for TCR–CD3 display was generated as follows: The Jurkat-derived cell line J.RT3-T3.5 (ATCC, TIB-153) lacking endogenous TCRβ chain was modified to express an NFAT reporter and CD8 via transduction with retroviral supernatant from HEK293GP-GLV9 producer cells transfected with the 8xNFAT-ZsG-hCD8 plasmid (a gift from Maki Nakayama, Addgene plasmid # 153417)70,71. 48 h post transduction, transductants were stimulated with PMA/Ionomycin overnight and sorted on CD8+ZsGreen+ cells using a Sony SH800. Sorted cells were allowed to recover in vitro for 5 days and subsequently nucleofected using a Nucleofector IIb device (Program X-001) with β2M sgRNA (5’-CGCGAGCACAGCUAAGGCCA-3’; Synthego) pre-complexed with TrueCut Cas9 V2 (Invitrogen, A26498) according to the manufacturer’s recommendation. Nucleofected cells were allowed to recover in RPMI + 2 mM L-glutamine + 20% FCS without antibiotics for 2 days and thereafter HLA-ABC- live cells were sorted using a Sony SH800.
To then render the resulting cell line deficient for the TCRα chain, TRAC gene sgRNA (5’-TCTCTCAGCTGGTACACGGC-3’; IDT) and Alt-R S.p. Cas9 Nuclease V3 (IDT 1081058) were introduced using a Lonza 4D-Nucleofector. TRAC knock-out was confirmed by ICE CRISPR Analysis Tool v3 (Synthego) using Sanger sequencing data flanking the edited cut site. Primer pair: Forward 5’-AGCTTGTGCCTGTCCCTGAG-3’, Reverse 5’-GGGCTGGGGAAGAAGGTGT-3’. The sorted cell line, termed J8Zb2m-α-β-, was expanded and maintained in vitro in RPMI + 2 mM L-glutamine + 10% FCS + 100 μg/mL penicillin/streptomycin.
Viral transduction of T-cell lines
SFG-1G4 wild-type and mutant plasmids were synthesized by Genscript. For transfection of the plasmids mentioned above, 293GP cells (Takara Bio, 631458) were co-transfected with 4.5 μg SFG plasmid and 2.25 μg RD114 feline envelop plasmid (provided by S. A. Rosenburg, NIH) using Lipofectamine™ 3000 Transfection Reagent (Thermo Fisher Scientific, L3000075). Viral supernatant was harvested on day 2 post transfection and always used freshly.
T-cell line transduction was mediated by retronectin (Takara, T100B). In brief, retronectin was coated on a non-tissue-culture-treated plate at 10 μg/mL in PBS overnight. Coated plates were washed by PBS and blocked by 2% FBS in PBS for 30 min at room temperature. The blocking solution was aspirated, viral supernatant was added to the coated plates, and the plates were centrifuged at 2500×g for 2 h at 32 °C. Viral supernatant was aspirated after centrifugation. Cells were loaded on plates and spun at 300 g for 30 min at 32 °C. On day 2 post transduction, transduction rates were measured by flow cytometry (see below) after staining with phycoerythrin-labeled anti-human TCR Vβ13.1 antibody (3% v/v, Biolegend, Cat. # 362410). For the TCRα mutant and matching wild-type controls, cells were stained by iTAg tetramer/APC – HLA-A*02:01 NY-ESO-1 (SLLMWITQC) (5% v/v, MBL, Cat. # TB-M011-2; prior to HLA-tetramer activation assay) or APC anti-human CD3 antibody (5% v/v, Biolegend, 317318; prior to NY-ESO-1 peptide-pulsed COS-7-A2 cell activation assay) and enriched using the EasySep™ Human APC Positive Selection Kit II (Stemcell Technologies, 17661). TCRβ mutants showed high transduction efficiency and were not immuno-enriched (Supplementary Fig. 8e). Data shown in Supplementary Fig. 8c, f are mean +/- standard deviation using biologic triplicates. Raw data are provided in Supplementary Tables 6 and 8.
Flow cytometry
For cell-surface staining, cells were stained with antibodies for 45 min in PBS + 0.5% FBS at 4 °C. Cells were washed twice with PBS + 0.5% FBS and flow cytometry data were acquired on an LSRFortessa X-20 using FACSDIVA v9.2 (BD) and analyzed in FlowJo v10.6.2 (BD). Antibodies used in our surface staining are Brilliant Violet 785™ anti-human CD3 antibody (5% v/v, Biolegend, 317330), and PE anti-human TCR Vβ13.1 antibody (3% v/v, Biolegend, 362410). To visualize HLA binding, iTAg tetramer/APC-HLA-A*02:01 NY-ESO-1 (SLLMWITQC) (5% v/v, MBL, TB-M011-2) was used. Cells were counterstained with the viability reagent LIVE/DEAD Fixable Aqua Dead Cell Stain Kit as per manufacturer’s specifications (0.7% v/v, Thermo Fisher Scientific, L34966). Singlet, live cells were identified for further analysis by a standard gating hierarchy (Supplementary Fig. 8d).
NY-ESO-1–HLA tetramer activation assay
J8Zb2m-α-β- cells with or without transduction were loaded onto 96-well plates (Corning, 353072) at 2e5 cells per 100 μL complete medium in triplicates. Plates were left at room temperature for 20 min and transferred to Incucyte SX1 system (Sartorius) for baseline images at 37 °C and 5% CO2. The plate was then removed from Incucyte and loaded with 100 μL of iTAg tetramer/APC-HLA-A*02:01 NY-ESO-1 at 8 μg/mL, 4 μg/mL or 0 μg/mL as designated. Cell Stimulation Cocktail (PMA and ionomycin, Thermo Fisher Scientific, 00-4970-93) was added at 1:1000 as a positive control. Again, the plate was left at room temperature for 20 min and transferred to Incucyte system for imaging at 37 °C and 5% CO2. Instrumental setting: 20x objective lens, 9 images per well, 2-h scan interval, using Sartorius Incucyte v2023A. Analysis was done using Incucyte Basic Analysis Software v2023A. Data shown in Supplementary Fig. 8b are mean +/- standard deviation using biologic triplicates. Raw data are provided in Supplementary Table 5. P values were determined by unpaired, two-tailed t-tests using Prism 10 software (GraphPad), and the predetermined significance threshold was P < 0.05.
NY-ESO-1 peptide-pulsed COS-7-A2 activation assay
COS-7-A2 cells were treated with NY-ESO-1 (157-165) peptide (customized synthesis, Genscript, purity 96.4%) at designated concentrations at 37 °C and 5% CO2 for 1 h and washed twice with PBS. J8Zb2m-α-β- cells were mixed with COS-7-A2 cells at 1:2 ratio or treated with Cell Stimulation Cocktail (PMA and ionomycin, Thermo Fisher Scientific, 00-4970-93) at 1:1000 as positive control. Cells were incubated at 37 °C and 5% CO2 for 18 h and NFAT activity (ZsGreen positive rate) was read out by flow cytometry. ZsGreen positivity was gated against the untransduced controls. Data shown in Fig. 7i are mean +/- standard deviation using biologic triplicates. Raw data are provided in Supplementary Table 7. P values were determined by unpaired, two-tailed t-tests without adjustment for multiple comparisons using Prism v10 software (GraphPad) or Excel v16.102 (Microsoft), and the predetermined significance threshold was P < 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The source data underlying Fig. 7i and Supplementary Fig. 8 can be found in Supplementary Tables 5–9 and are provided as a Source Data file. Construct sequences used in this work are provided in the Supplementary Information. For cryo-EM structures, the atomic coordinates have been deposited in the Protein Data Bank (PDB) under accession codes 9BBC (TCR–CD3 in GDN detergent); 8TW4 (TCR–CD3 in nanodisc, conformation ND-I); 8TW6 (TCR–CD3 in nanodisc, conformation ND-II); 9C3E (TCR–CD3 in nanodisc bound to HLA). Primary unsharpened, as well as B-factor-sharpened and DeepEMhancer-sharpened cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-44417 (TCR–CD3 in GDN detergent); EMD-41658 (TCR–CD3 in nanodisc, conformation ND-II); EMD-41660 (TCR–CD3 in nanodisc, conformation ND-II); EMD-45166 (TCR–CD3 in nanodisc bound to HLA). The following previously published cryo-EM and x-ray crystallographic protein structures were referenced in this work: 6JXR; 7FJD; 7FJE; 7FJF; 8ES7; 8ES8; 7PHR; 2BNQ; 2BNU; 5FYJ; 3T0E; 7XQ8. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD070648 Source data are provided with this paper.
References
Janeway, C. A. et al. Immunobiology (Garland Science, 2001).
Reinherz, E. L. The structure of a T-cell mechanosensor. Nature 573, 502–504 (2019).
Courtney, A. H., Lo, W.-L. & Weiss, A. TCR signaling: mechanisms of initiation and propagation. Trends Biochem. Sci. 43, 108–123 (2018).
Mariuzza, R. A., Agnihotri, P. & Orban, J. The structural basis of T-cell receptor (TCR) activation: an enduring enigma. J. Biol. Chem. 295, 914–925 (2020).
Dong, D. et al. Structural basis of assembly of the human T cell receptor–CD3 complex. Nature 573, 546–552 (2019).
Chen, Y. et al. Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility. Mol. Cell 82, 1278–1287.e5 (2022).
Sušac, L. et al. Structure of a fully assembled tumor-specific T cell receptor ligated by pMHC. Cell 185, 3201–3213.e19 (2022).
Saotome, K. et al. Structural analysis of cancer-relevant TCR-CD3 and peptide-MHC complexes by cryoEM. Nat. Commun. 14, 2401 (2023).
Xu, C. et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3ɛ cytoplasmic tyrosine-based motif. Cell 135, 702–713 (2008).
Zhang, H., Cordoba, S.-P., Dushek, O. & Anton van der Merwe, P. Basic residues in the T-cell receptor ζ cytoplasmic domain mediate membrane association and modulate signaling. Proc. Natl. Acad. Sci. USA 108, 19323–19328 (2011).
James, J. R. & Vale, R. D. Biophysical mechanism of T cell receptor triggering in a reconstituted system. Nature 487, 64–69 (2012).
Minguet, S., Swamy, M., Alarcón, B., Luescher, I. F. & Schamel, W. W. A. Full activation of the T cell receptor requires both clustering and conformational changes at CD3. Immunity 26, 43–54 (2007).
Gil, D., Schamel, W. W. A., Montoya, M., Sánchez-Madrid, F. & Alarcón, B. Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell 109, 901–912 (2002).
Natarajan, K. et al. An allosteric site in the T-cell receptor Cβ domain plays a critical signalling role. Nat. Commun. 8, 15260 (2017).
Rangarajan, S. et al. Peptide-MHC (pMHC) binding to a human antiviral T cell receptor induces long-range allosteric communication between pMHC- and CD3-binding sites. J. Biol. Chem. 293, 15991–16005 (2018).
Das, D. K. et al. Force-dependent transition in the T-cell receptor β-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc. Natl. Acad. Sci. USA 112, 1517–1522 (2015).
Wu, W., Shi, X. & Xu, C. Regulation of T cell signalling by membrane lipids. Nat. Rev. Immunol. 16, 690–701 (2016).
Wang, F., Beck-García, K., Zorzin, C., Schamel, W. W. A. & Davis, M. M. Inhibition of T cell receptor signaling by cholesterol sulfate, a naturally occurring derivative of membrane cholesterol. Nat. Immunol. 17, 844–850 (2016).
Notti, R. Q. & Walz, T. Native-like environments afford novel mechanistic insights into membrane proteins. Trends Biochem. Sci. 47, 561–569 (2022).
Hiotis, G., Notti, R. Q., Bao, H. & Walz, T. Nanodiscs remain indispensable for Cryo-EM studies of membrane proteins. Curr. Opin. Struct. Biol. 92, 103042 (2025).
Robbins, P. F. et al. Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. J. Immunol. 180, 6116–6131 (2008).
Robbins, P. F. et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1–reactive T-cell receptor: long-term follow-up and correlates with response. Clin. Cancer Res. 21, 1019–1027 (2015).
D’Angelo, S. P. et al. Letetresgene autoleucel in advanced/metastatic myxoid/round cell liposarcoma. J. Clin. Oncol. 43, 1777–1788 (2025).
Hatipoglu, E., Furness, A. J. S. & Jones, R. L. Letetresgene autoleucel: a milestone in treatment of myxoid/round cell liposarcoma. J. Clin. Oncol. 43, 1755–1757 (2025).
Ulmschneider, M. B. & Sansom, M. S. P. Amino acid distributions in integral membrane protein structures. Biochim. Biophys. Acta Biomembr. 1512, 1–14 (2001).
Swamy, M. et al. A cholesterol-based allostery model of T cell receptor phosphorylation. Immunity 44, 1091–1101 (2016).
Su, Q. et al. Cryo-EM structure of the human IgM B cell receptor. Science 377, 875–880 (2022).
Gao, X., Dong, X., Li, X., Liu, Z. & Liu, H. Prediction of disulfide bond engineering sites using a machine learning method. Sci. Rep. 10, 10330 (2020).
Liu, F., van Breukelen, B. & Heck, A. J. R. Facilitating protein disulfide mapping by a combination of pepsin digestion, electron transfer higher energy dissociation (EThcD), and a dedicated search algorithm slinks*. Mol. Cell. Proteom. 13, 2776–2786 (2014).
Stewart-Jones, G. B. E. et al. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 165, 813–826 (2016).
Yin, Y., Wang, X. X. & Mariuzza, R. A. Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4. Proc. Natl. Acad. Sci. USA 109, 5405–5410 (2012).
Hawse, W. F. et al. Cutting edge: evidence for a dynamically driven T cell signaling mechanism. J. Immunol. 188, 5819–5823 (2012).
Lanz, A.-L. et al. Allosteric activation of T cell antigen receptor signaling by quaternary structure relaxation. Cell Rep. 36, 109375 (2021).
Kim, S. T. et al. The αβ T cell receptor is an anisotropic mechanosensor. J. Biol. Chem. 284, 31028–31037 (2009).
Gil, D., Schrum, A. G., Alarcón, B. & Palmer, E. T cell receptor engagement by peptide–MHC ligands induces a conformational change in the CD3 complex of thymocytes. J. Exp. Med. 201, 517–522 (2005).
Brazin, K. N. et al. The T cell antigen receptor α transmembrane domain coordinates triggering through regulation of bilayer immersion and CD3 subunit associations. Immunity 49, 829–841.e6 (2018).
Lee, M. S. et al. A mechanical switch couples T cell receptor triggering to the cytoplasmic juxtamembrane regions of CD3ζζ. Immunity 43, 227–239 (2015).
Fernandes, R. A. et al. T cell receptors are structures capable of initiating signaling in the absence of large conformational rearrangements. J. Biol. Chem. 287, 13324–13335 (2012).
Natarajan, A. et al. In situ cell-surface conformation of the TCR-CD3 signaling complex. EMBO Rep. 25, 5719–5742 (2024).
Wang, W. et al. Quantitative analysis of T cell receptor complex interaction sites using genetically encoded photo-cross-linkers. ACS Chem. Biol. 9, 2165–2172 (2014).
Lee, J. H., de Val, N., Lyumkis, D. & Ward, A. B. Model building and refinement of a natively glycosylated HIV-1 Env protein by high-resolution cryoelectron microscopy. Structure 23, 1943–1951 (2015).
Chouaki Benmansour, N. et al. Phosphoinositides regulate the TCR/CD3 complex membrane dynamics and activation. Sci. Rep. 8, 4966 (2018).
Costello, P. S., Gallagher, M. & Cantrell, D. A. Sustained and dynamic inositol lipid metabolism inside and outside the immunological synapse. Nat. Immunol. 3, 1082–1089 (2002).
Xu, S. et al. Uptake of oxidized lipids by the scavenger receptor CD36 promotes lipid peroxidation and dysfunction in CD8+ T cells in tumors. Immunity 54, 1561–1577.e7 (2021).
Kent, A., Longino, N. V., Christians, A. & Davila, E. Naturally occurring genetic alterations in proximal TCR signaling and implications for cancer immunotherapy. Front. Immunol. 12, 658611 (2021).
Klebanoff, C. A., Chandran, S. S., Baker, B. M., Quezada, S. A. & Ribas, A. T cell receptor therapeutics: immunological targeting of the intracellular cancer proteome. Nat. Rev. Drug Discov. 22, 996–1017 (2023).
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014).
Zhang, Y. et al. Visualization of the mechanosensitive ion channel MscS under membrane tension. Nature 590, 509–514 (2021).
Hagn, F., Etzkorn, M., Raschle, T. & Wagner, G. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J. Am. Chem. Soc. 135, 1919–1925 (2013).
Ohi, M., Li, Y., Cheng, Y. & Walz, T. Negative staining and image classification—powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34 (2004).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Cheng, A. et al. High resolution single particle cryo-electron microscopy using beam-image shift. J. Struct. Biol. 204, 270–275 (2018).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Qi, Y., Lee, J., Klauda, J. B. & Im, W. CHARMM-GUI nanodisc builder for modeling and simulation of various nanodisc systems. J. Comput. Chem. 40, 893–899 (2019).
Chen, J.-L. et al. Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J. Exp. Med. 201, 1243–1255 (2005).
Aiyer, S., Zhang, C., Baldwin, P. R. & Lyumkis, D. Evaluating local and directional resolution of cryo-EM density maps. Methods Mol. Biol. 2215, 161–187 (2021).
Sanchez-Garcia, R. et al. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 4, 1–8 (2021).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. Publ. Protein Soc. 27, 14–25 (2018).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. Sect. Struct. Biol. 74, 531–544 (2018).
Casañal, A., Lohkamp, B. & Emsley, P. Current developments in Coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. Publ. Protein Soc. 29, 1069–1078 (2020).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. Sect. Struct. Biol. 74, 519–530 (2018).
Emsley, P. & Crispin, M. Structural analysis of glycoproteins: building N-linked glycans with Coot. Acta Crystallogr. Sect. Struct. Biol. 74, 256–263 (2018).
Ashkenazy, H. et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016).
Sievers, F. & Higgins, D. G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27, 135–145 (2018).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003).
Mann, S. E. et al. Multiplex T cell stimulation assay utilizing a T cell activation reporter-based detection system. Front. Immunol. 11, e3883 (2020).
Ghani, K. et al. Efficient human hematopoietic cell transduction using RD114- and GALV-pseudotyped retroviral vectors produced in suspension and serum-free media. Hum. Gene Ther. 20, 966–974 (2009).
Acknowledgements
The authors thank the members of the Walz, T. de Lange (RU), and R. K. Hite (MSKCC) labs and W. D. Tap (MSKCC) for insightful discussions. We thank M. Ebrahim, J. Sotiris, and H. Ng at the Evelyn Gruss Lipper Cryo-EM Resource Center of The Rockefeller University (RRID:SCR_021146) for assistance with cryo-EM data collection. Mass spectrometry data were generated by the Proteomics Resource Center at The Rockefeller University (RRID:SCR_017797). The authors acknowledge the following funding sources: NIH T32CA9207 (R.Q.N.), NIH KL2TR001865 (R.Q.N.), NIH UL1TR001866 (R.Q.N.), The Black Family Metastasis Center at Rockefeller University (R.Q.N.), Shapiro-Silverberg Fund for the Advancement of Translational Research (R.Q.N.), The Mark Foundation for Cancer Research (R.Q.N. and T.W.), NIH R37 CA259177 (C.A.K.), NIH R01 CA269733 (C.A.K.), NIH R01 CA286507 (C.A.K.), NIH P30 CA008748 (C.A.K.), NIH P50 CA217694 (C.A.K.), Cycle for Survival (C.A.K.), The Shteinbuk and Mead Family (C.A.K.), Metropoulos Family Foundation (C.A.K.), The Parker Institute for Cancer Immunotherapy (C.A.K.), The Sarcoma Center at MSKCC (C.A.K.), Sohn Conferences Foundation (H.M.), Leona M. and Harry B. Helmsley Charitable Trust (H.M.).
Author information
Authors and Affiliations
Contributions
Conceptualization: R.Q.N. and T.W.; Methodology: R.Q.N., F.Y., S.H., M.W.B., Z.M., H.M., C.A.K., and T.W.; Investigation: R.Q.N., R.Y., S.H., M.W.B., Z.M., and P.D.; Visualization: R.Q.N., F.Y., S.H., and M.W.B.; Funding acquisition: R.Q.N., H.M., C.A.K., and T.W.; Project administration: R.Q.N., H.M., C.A.K., and T.W.; Supervision: R.Q.N., H.M., C.A.K., and T.W.; Writing—original draft: R.Q.N.; Writing—review & editing: R.Q.N., F.Y., S.H., M.W.B., H.M., C.A.K., and T.W.
Corresponding author
Ethics declarations
Competing interests
C.A.K. is an inventor on patents related to TCR discovery and public neoantigen-specific TCRs and is recipient of licensing revenue shared according to MSKCC institutional policies. C.A.K. has consulted for or is on the scientific advisory boards for Achilles Therapeutics, Affini-T Therapeutics, Aleta BioTherapeutics, Bellicum Pharmaceuticals, Bristol Myers Squibb, Catamaran Bio, Cell Design Labs, Decheng Capital, G1 Therapeutics, Klus Pharma, Obsidian Therapeutics, PACT Pharma, Roche/Genentech, Royalty Pharma, and T-knife. C.A.K. is a scientific co-founder and equity holder in Affini-T Therapeutics. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Michelle Krogsgaard, who co-reviewed with Aswin Natarajan, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Notti, R.Q., Yi, F., Heissel, S. et al. The resting and ligand-bound states of the membrane-embedded human T-cell receptor–CD3 complex. Nat Commun 16, 10996 (2025). https://doi.org/10.1038/s41467-025-66939-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66939-7
This article is cited by
-
Mechanotransduction through T cell receptors: consensus, controversies and future outlooks
Experimental & Molecular Medicine (2026)