Abstract
The mechanisms of the chronic inflammatory response in smoking-induced chronic obstructive pulmonary disease (COPD) remain unclear, with limited understanding of T cell responses to smoke exposure in lung tissue and circulation. We characterized the alterations in T cells associated with smoke exposure using single-cell RNA sequencing, TCR sequencing, and flow cytometry-based validation. Our experiments show significant recruitment of T cells within the airways, exhibiting a multifaceted immune profile, augmented TCR clonal diversity, and a prolonged CDR3 length. Notably, smoke-induced γδ T cells, particularly clonally expanded γ6δ4 T cells with high Il17a expression, accumulate in smoke-induced airway inflammation. Despite arising in inflammation, these cells function in a protective manner: γδT17 deficiency worsens smoke-induced lung injury, whereas adoptive transfer of γδT17 cells restores tissue protection in smoke-exposed lungs over time. Here, we show γδT17 cells as a potential target for addressing smoke-induced immune dysfunctions, providing alternative avenues for early prevention and treatment of COPD.
Similar content being viewed by others
Introduction
Chronic obstructive pulmonary disease (COPD) is the third leading cause of death worldwide and a global public health concern1,2. COPD is a highly heterogeneous lung disease characterized by chronic respiratory symptoms arising from airway (bronchitis, bronchiolitis) and/or alveoli (emphysema) abnormalities3. Tobacco smoking causes a decline in lung function and disrupts the immune balance in lung tissue, which can ultimately result in structural lung damage and the progression of COPD.
T cells are a vital component of the immune system and play a crucial role in maintaining immune homeostasis in the lungs affected by COPD. Numerous studies have shown that the number and percentage of T cell are increased in COPD, as are T cell-associated cytokines. Previous studies have found that the proportion of CD8+ T cells in lung tissue was higher in COPD patients4, and that CD8+ T cells in the peripheral circulation were negatively correlated with the frequency of acute exacerbations of COPD5. The presence of IL-17A-secreting T cells has also been confirmed in lung biopsies6,7, and their roles in neutrophil-induced inflammation by inducing the release of CXCL1 and CXCL88. The imbalance between anti-inflammatory and pro-inflammatory T regulatory subsets is suggested to be significant in COPD progression9. However, systematic studies on T cell subpopulation distribution, homeostatic maintenance, and functional roles in COPD are limited, leaving the underlying pathogenic mechanisms unclear. In addition, it remains unclarified whether these resident T cells are derived from selective clonal expansion or nonspecifically recruited.
γδ T cells, a distinct subset of lymphocytes, are widely distributed in mucosal tissues and possess a broader antigen recognition repertoire compared to αβ T cells, enabling them to mount robust responses to various stimuli. In lung tissues, γδ T cells are postulated to play crucial roles in modulating inflammation, maintaining immune homeostasis, and facilitating tissue repair. One study indicates an increased γδ T cell numbers is observed in the alveoli in lung biopsies10. Notably, γδ T cells were significantly increased in smokers with preserved lung function compared to nonsmokers11. A positive correlation has also been identified between the relative numbers of γδ T cell in induced sputum and FEV1% pred in COPD patients12. Lack of γδ T cells increased inflammatory cells in response to smoke exposure in mice13. These studies suggest that the number of γδ T cells is increased in patients with COPD, the phenotype and functional role of which are unclear, and that these variations appear to be closely linked to disease progression and prognosis.
In this work, we aim to investigate the phenotypes of T cells in COPD and their relationship with disease progression. Using a mouse model of COPD induced by tobacco smoke exposure, we utilize in vivo antibody labeling and flow cytometry, and isolate T cells from various lung sites and spleen for single-cell RNA (scRNA) and T cell receptor (TCR) sequencing to construct a T cell atlas. Furthermore, we employ in vitro expansion cultures of γδ T17 cells and adoptive transfers to explore the homeostatic maintenance and functional role of T cells in COPD pathogenesis. To summarize, this study describes the T cell landscape associated with smoking and identifies a hallmark of smoking-related pathology: the expansion of clonal γδ T17 cell populations in mice.
Results
T cells recruited to airways after tobacco smoke exposure
To study the phenotypic and functional mechanisms of T cells in chronic airway inflammation in COPD, we used tobacco smoke exposure to construct a mouse model of COPD and evaluated pathology as well as lung function to determine whether the model was generated successfully. C57BL/6J wild-type (WT) mice were randomly divided into two groups: an air control group and a tobacco smoke exposure group, which were exposed to tobacco smoke for 2 or 6 months, and body weights were monitored (Supplementary Fig. 1a, b). After 6 months, significant lung tissue destruction was observed (Supplementary Fig. 1c, d), and a significant lung function decline was observed in the smoke-exposed group (Supplementary Fig. 1e–j), thereby confirming the successful establishment of a robust COPD mouse model in this study.
T cells are crucial for immune regulation and homeostasis in lung tissues, particularly in COPD4,13. By intravenous (i.v.) and intranasal (i.n.) administration of CD90.2 and CD45 antibodies, we distinguished T cells in the airways, parenchyma, and vasculature using a previously described method14,15. Our findings revealed that a large proportion of T cells in the respiratory tract were localized in the vasculature (Fig. 1a). Moreover, the vast majority of cells in the airways were not labeled by i.v. antibody administration, indicating their anatomic compartmentalization (Fig. 1a). After 2 and 6 months of smoke exposure, we observed significant changes in T cell aggregation and migration patterns. The number of airway T cells increased, particularly after 2 months, suggesting enhanced immune activation during early inflammation (Fig. 1b). Conversely, the number of T cells in the vasculature decreased, indicating a recruitment of T cells to the airways (Fig. 1b). These findings were consistently reproduced in an independent replication study (Supplementary Fig. 2). Notably, the percentage of CD4+ and CD8+ T cells in the airways increased post-exposure, while CD4-CD8- T cells decreased (Fig. 1c). These observations suggest that specific T cell populations may play a significant role in COPD progression, with the increased presence of CD4-CD8- T cells potentially involved in disease development.
Mice were sacrificed at 0 months (n = 5), 2 months (n = 4), and 6months (n = 4) after tobacco smoke exposure. T cells from the airway, the area between the airway and parenchyma (air-par), parenchyma, vasculature, and spleen were analyzed by fluorescence-activated cell sorting (FACS). a To localize T cells in the respiratory tract after tobacco smoke exposure, 0.25 mg of fluorochrome-conjugated CD45 and 0.5 mg of fluorochrome-conjugated CD90.2 antibodies were administered intranasally (i.n.) and intravenously (i.v.), respectively, as described in the Methods. The frequency and/or numbers of T cells (b) or T cell subsets (c) are shown. Data are mean \(\pm \,\)SEM. One-way ANOVA was used to compare differences among multiple groups when data followed a normal distribution, whereas the non-parametric Kruskal–Wallis test was applied for data that did not meet this assumption. NS not significant, Air Airways, Air-par the airway-parenchyma interface, Par parenchyma, Spl Spleen, CON air control group, 2 M, 2-month tobacco smoke exposure group; 6 M, 6-month tobacco smoke exposure group.
Shared and specific changes in the smoke-induced T cell immune system
To investigate the subpopulation composition, sources, and functional roles of T cells following tobacco smoke exposure, we conducted a comprehensive analysis of the T cell immune system in C57BL/6J mice at early (2 months, 2 M) and later (6 months, 6 M) stages of COPD, alongside an air control (CON) group. Utilizing single-cell RNA sequencing (scRNA-seq) and T cell receptor sequencing (TCR-seq), we generated detailed profiles from 40,849 CD3+ T cells sorted from the spleen (an immune organ) and lungs (the respiratory organ) through single-cell targeted transcriptome analysis (TTA). Our systematic investigation identified three lung cell populations: airways, the airway-parenchyma interface (air-par), and parenchyma, with three parallel replicates for each group (CON, 2 M, 6 M) (Fig. 2a).
a Overview illustrating single-cell analysis of T cell immune system. Clustering of 40,849 T cells isolated from airway (9187 cells), air-par (6935 cells), parenchyma (11,404 cells), and spleen (13,323 cells) from air control group, the 2-month tobacco smoke exposure group, and the 6-month tobacco smoke exposure group of C57BL/6J male mice: b annotated T cell clusters, c cell distribution, and d percentage and e fold changes in cluster proportions associated with smoke exposure. CON: air control group, pooled from 3 biological replicates, each comprising cells from 20 mice (total n = 60 mice). 2 M: 2-month tobacco smoke exposure group, pooled from 3 biological replicates, each comprising cells from 4 mice (total n = 12 mice). 6 M: 6-month tobacco smoke exposure group, pooled from 3 biological replicates, each comprising cells from 4 mice (total n = 12 mice).
We identified 39 T cell clusters based on gene expression profiles (Supplementary Fig. 3a). Using 41 marker genes, these clusters were annotated into specific T cell subsets, including naïve CD4⁺ and CD8⁺ T cells (high Cd4/Cd8, Ccr7, Sell, Tcf7, Il17r; low Cd44, Cd69, Pdcd1), double-positive (DP) T cells (Trac, Cd4, Cd8a, Cd8b, Cd69), Th1 cells (high Cd4, Cxcr3; low Cxcr6, Rorc, Il17a, Il17f), Th17 cells (high Cd4, Cxcr6, Rorc, Il17a, Il17f; low Cxcr3), double-negative (DN) T cells (Trac, Cd44; low Cd4, Cd8a, Cd8b), γδ T cells (high Trdc; low Trac), Tregs (Cd4, Gata3, Foxp3, Il10, Il2ra), NKT cells (Klra1, Klrc1, Klrk1), and cytotoxic T cells (Cd8a, Cd8b, Cxcr3, Eomes, Tbx21, Ifng; low Cxcr6, Rorc, Il17a, Il17f) (Fig. 2b–d, Supplementary Fig. 3b). Comparative analyses highlighted significant smoking-induced changes in cell proportions and gene expression. Notably, there was an accumulation of γδ T cells in all lung tissues, while their percentage decreased in the airways with tobacco smoke exposure. These changes mirrored the trends observed for CD4-CD8- T cells via flow cytometry (Fig. 1c), indicating that tobacco smoke exposure affected the distribution characteristics of γδ T cells.
Altered T cell clonal expansion patterns in the airways after tobacco smoke exposure
Preliminary data from flow cytometry and scRNA sequencing indicate that tobacco smoke exposure significantly alters T cell subset proportions in various lung tissue areas. To explore whether these changes are related to clonal proliferation and antigen-specific recruitment in the COPD mouse model, we conducted single-cell TTA combined with TCR sequencing. We analyzed 17,524 paired TCR sequences from T cells in the airways (3472 cells), air-par (2872 cells), parenchyma (4744 cells), and spleen (6436 cells). The increase in Gini coefficients revealed that clonal diversity of T cells in airways increased with prolonged smoke exposure (Fig. 3a). Principal component analysis of TCR regions showed that, in contrast to the clonal aggregation of γδ T cells in control mice, smoke exposure resulted in an increased in clonal diversity among airway T cells at both 2 and 6 months (Fig. 3b), suggesting an antigen encounter following smoke exposure. Moreover, analysis of the lengths of CDR3 indicated that T cells enhanced their specific recognition of antigen by increasing the length and diversity of the CDR3 region in the exposure group (Fig. 3c). And all T cell subpopulations, except γδ T cells, showed increased TCR clonal diversity (Fig. 3d). Interestingly, the Gini coefficients for γδ T cells remained stable, indicating that a few clones dominated this population (Supplementary Fig. 4). These findings suggest that airway T cells are likely stimulated by various antigens after tobacco smoke exposure, leading to increased diversity and specificity in TCR clones, while γδ T cells appear to maintain a more stable clonal profile.
Gini coefficients show the pattern of TCR clone expansion in T cells from a different tissue sites and d different T cell subpopulations in the airways after 2 and 6 months of tobacco smoke exposure. Box plots indicate median (middle line), 25th, 75th percentile (box), and the minimum and maximum values (whiskers) as well as outliers (single points). b The PCA dot plots of T cell clones in the airways show the composition (indicated by dot color) and percentage (indicated by dot size) of TCR clones in the CON, 2 M and 6 M, with the PC1 value of each clone being the mean on the X-axis of the UMAP, and the PC2 being the mean on the Y-axis. c The CDR3 lengths corresponding to the prominent Vαγ, Jαγ, Vβδ, and Jβδ regions are shown for T cells on airways in the CON, 2 M and 6 M. The arrangement of the segment types follows a specific order, with the Vαγ and Jαγ regions on the left and the Vβδ and Jβδ regions on the right. CON: air control group, pooled from 3 biological replicates, each comprising cells from 20 mice (total n = 60 mice). 2 M: 2-month tobacco smoke exposure group, pooled from 3 biological replicates, each comprising cells from 4 mice (total n = 12 mice). 6 M: 6-month tobacco smoke exposure group, pooled from 3 biological replicates, each comprising cells from 4 mice (total n = 12 mice).
γδ T cells expressing Il17a expand with tobacco smoke exposure in multiple tissue sites
We next focused on the significant remodeling of the γδ T cell compartment following tobacco smoke exposure. Notably, γδ T cells in the spleen exhibited distinct characteristics compared to those in lung tissue (Fig. 4a), suggesting inconsistent changes across different organs. The smoke-induced γδ T cells displayed elevated levels of Pdcd1, Il17a, and Cd44, while showing decreased expression of Tigit, Havcr3, and Lag3 (Fig. 3b, c). This pattern indicates that tobacco smoke exposure drives γδ T cell differentiation towards the γδT17 phenotype rather than towards the functionally exhausted or terminally differentiated phenotypes. We further analyzed the TCR repertoires of γδ T cells and identified a dominant TCR clone (Top1) that accounted for a significant proportion of the γδ T cell population (Fig. 4d, e). In the air control group, about 50% of γδ T cells in the airways did not express Il17a. However, with continued smoke exposure, the percentage of γδ T cell clones expressing Il17a in the airways increased significantly (Fig. 4e). Similar clonal characteristics were observed in the air-par and parenchyma regions, where γδ T cells expressing Il17a constituted approximately 90% of the total population (Supplementary Fig. 5a). Although γδ T cells were less prevalent in the spleen, they exhibited greater clonal diversity relative to their numbers (Supplementary Fig. 5a). These findings highlight the dynamic role of γδ T cells in response to tobacco smoke exposure, particularly their potential involvement in inflammatory processes through Il17a expression across multiple tissue sites.
Cell clustering form single-cell targeted transcriptome analysis (TTA) data was performed on 2024 γδ T cells from total T cells, with 422 cells from airways, 793 cells from air-par, 763 cells from parenchyma, and 46 cells from spleen. a UMAP showing the scRNA-seq clusters of 4299 γδ T cells. b Differential representation of γδ T cell clusters. c Scaled gene expression. d Projection of the top 1 frequent TCR clones (red dots) onto γδ T cell clusters. e Parallel plots showing a correlation in the airways between the frequency of V gene usage and its pairing with Il17a expression in relation to the progression of tobacco smoke exposure. Clones (represented by lines) shared are colored: Top 1 (red), top 2 (blue), top 3 (yellow), and all other clones with low frequency (gray). The number of clones/cells for CON, 2 M and 6 M are also shown. f Bar graphs illustrating the frequency of TCR clones in γδ T cells in the airways over time of tobacco smoke exposure, with the number of cells at each time point (CON, 2 M, and 6 M) shown at the top of the bars. g Bar graphs showing the proportion of γ6δ4 T cells expressing Il17a in the airways in response to the duration of tobacco smoke exposure. The top of the bars indicates the number of γ6δ4 T cells at the three time points (CON, 2 M, and 6 M). CON: air control group, pooled from 3 biological replicates, each comprising cells from 20 mice (total n = 60 mice). 2 M: 2-month tobacco smoke exposure group, pooled from 3 biological replicates, each comprising cells from 4 mice (total n = 12 mice). 6 M: 6-month tobacco smoke exposure group, pooled from 3 biological replicates, each comprising cells from 4 mice (total n = 12 mice).
Increased frequency and Il17a expression of γ6δ4 T cells with tobacco smoke exposure
Our findings revealed that the proportion of γ6δ4 T cells, identified as the dominant clone (Top1 clone) in the airways, gradually increased with prolonged tobacco smoke exposure (Fig. 4f). This result indicates that tobacco smoke exposure may directly or indirectly stimulate the clonal expansion of γ6δ4 T cells. Notably, these cells were prevalent across all three lung tissue sites, while they were underrepresented in the spleen (Fig. 4e, f and Supplementary Fig. 5a, b). As exposure to tobacco smoke continued, γ6δ4 T cells in lung tissue also began to express Il17a (Fig. 4f). This trend was similarly observed in the air-par and parenchyma, with γ6δ4 T cells showing increased Il17a expression (Supplementary Fig. 5b). Particularly in the air-par, the proportion of γ6δ4 T cells expressing Il17a reached notable levels (2 M: 85.5%; 6 M: 91.2%; Supplementary Fig. 5b), which indicates that tobacco smoke exposure can enhance Il17a secretion in these cells. This result aligns with recent findings16 that γ6δ4 T cells are primarily derived from lung tissue and are classified as Rorc+ cells. In conclusion, smoke-induced γδ T cells were present in large numbers in lung tissues and gradually activated Il17a expressing, showing a phenotype similar to γδT17. Our results suggest that γδ T cells are aberrantly activated into γδT17 cells by smoke exposure, either directly or indirectly.
Differentiation of γδ T cells to γδT17 cells with tobacco smoke exposure
To investigate the developmental trajectory of γδ T cells, we analyzed 5295 γδ T cells extracted from single-cell whole transcriptome analysis (WTA). Cells lacking Trdc expression were excluded, and the remaining cells were re-clustered. RNA velocity was visualized on a UMAP plot by using scVelo method (Fig. 5a and Supplementary Fig. 6a). In the air control group, most γδ T cells in the airways and lung parenchyma were at the early transition stage (purple, Fig. 5b). However, after 6 months of smoke exposure, the majority of γδ T cells across all three anatomical sites had progressed to the later transition stage (yellow, Fig. 5b). Gene expression patterns of γδ T cells in the WTA were highly consistent with those in the TTA (Fig. 4b, c and Supplementary Fig. 6a, c). Partition-based graph abstractions (PAGA) highlighted pseudotime dynamics, indicating a clear transition from the control group to 2 months and then to 6 months of exposure (Fig. 5c). Notably, γδ T cells without Il17a expression showed a unidirectional flow towards those expressing Il17a across all four tissue sites (Fig. 5d), suggesting an irreversible differentiation process (Fig. 5d). In conclusion, our results illustrate the temporal dynamics of γδ T cells under tobacco smoke exposure, indicating that these cells begin to differentiate into γδT17 cells. However, the specific functional roles of these γδT17 cells within the context of tobacco smoke-induced COPD warrant further investigation.
Cell clustering from single-cell whole transcriptome analysis (WTA) data was performed on 5295 γδ T cells isolated from total T cells, including 1120 cells from the airways, 1920 cells from the air-par, 2039 cells from the parenchyma, and 216 cells from the spleen from air control group, 2-month tobacco smoke exposure group, and 6-month tobacco smoke exposure group of C57BL/6J male mice. a RNA velocity vectors projected onto a UMAP embedding using the scVelo package. b Gene-shared latent time based on transcriptional dynamics, representing the cell’s internal clock. The endpoints of the color scale correspond to the origin and end states of the velocity vectors, with dark color representing the start and yellow representing the endpoint of the velocity latent time. c, d Topology-preserving PAGA graph superimposed on the UMAP embedding to visualize potential cell type transitions. Nodes represent cell groups, and edge weights quantify the connectivity between groups. CON: air control group, pooled from 3 biological replicates, each comprising cells from 20 mice (total n = 60 mice). 2 M: 2-month tobacco smoke exposure group, pooled from 3 biological replicates, each comprising cells from 4 mice (total n = 12 mice). 6 M: 6-month tobacco smoke exposure group, pooled from 3 biological replicates, each comprising cells from 4 mice (total n = 12 mice).
Functional role of γδ T17 cells in ameliorating airflow limitation and the emphysema-like phenotype
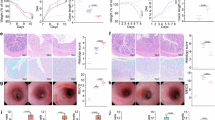
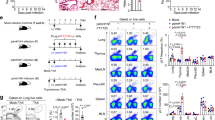
To explore the functional role of γδ T cells under tobacco smoke exposure, we used an in vitro expansion method17 to generate γδT17 cells (Supplementary Fig. 7). C57BL/6J WT and Tcrd-/- mice were divided into an air control (CON) group and a 6-months tobacco smoke exposure group. Twenty-four hours before exposure, Tcrd-/- mice were further split into a γδT17 cell transfer (T17) group and a PBS control group (Fig. 6a). The transferred γδ T17 cells successfully engrafted in the Tcrd-/- mice (Supplementary Fig. 8). Body weights were monitored (Fig. 6b). After 6 months of tobacco smoke exposure, lung tissue destruction was evident in all groups compared to the air control group (Fig. 6c). Notably, the transfer of γδT17 cells into Tcrd-/- mice significantly reduced mean alveolar intervals (MLI) compared to both the WT smoke-exposed group and the Tcrd-/- smoke-exposed group (Fig. 6d). This suggests that γδT17 cells can ameliorate the emphysema-like phenotype in Tcrd-/- mice, potentially delaying disease progression.
a, b C57BL/6J wild-type (WT) mice and Tcrd-/- mice were randomly divided into two groups: the air control (CON) group and the tobacco smoke-exposed group. Mice were placed in a fume box and exposed to tobacco smoke for 6 months, respectively. Twenty-four hours prior to tobacco smoke exposure, Tcrd-/- mice were randomly divided into two groups: the transferred γδT17 cell (T17) group and the PBS control group, with body weights continuously recorded during the exposure (WT CON group, n = 8 mice/time point; WT smoke-exposed group, n = 6 mice/time point; Tcrd-/- CON group, n = 6 mice/time point; Tcrd-/- smoke-exposed group, n = 6 mice/time point; Tcrd-/--T17 CON groups, n = 7 mice/time point; Tcrd-/--T17 smoke-exposed groups, n = 7 mice/time point). c Representative images of hematoxylin and eosin-stained paraffin sections (40×, scale bar = 50 µm) of the WT, Tcrd-/-, and Tcrd-/--T17 groups show that after 6 months of tobacco smoke exposure (6 M), the smoke-exposed group exhibited marked airway inflammation and fused alveolar damage compared to the CON group. The experiment was repeated independently at least three times with similar results. d The mean alveolar intervals (MLI) were significantly increased in the smoke-exposed group (n = 3 mice/group). Pulmonary function tests showed significant differences in e total lung capacity (TLC), f forced vital capacity (FVC), g vital capacity (VC), h inspiratory capacity (IC), i chord compliance between 0 and 10 cmH2O (Cchord), and j the ratio of forced expiratory volume (FEV) in the first 50 ms to FVC (FEV50/FVC) among the WT, Tcrd-/- and Tcrd-/--T17 groups under both air control and 6-month tobacco smoke exposure conditions (n = 6 mice/group). Data are mean \(\pm \,\)SEM. Two-sided unpaired Student’s t-test was used to analyze differences in the mean values between groups, while the Mann–Whitney test was applied to compare the central tendencies (mean or median) between the two groups. All P values presented are two-sided. NS not significant. Smoke, smoke-exposed.
Lung function assessments revealed that total lung capacity (TLC), forced vital capacity (FVC), vital capacity (VC), inspiratory capacity (IC), and chord compliance between 0 and 10 cm H2O (Cchord) significantly increased in all smoke-exposed groups compared to controls (Fig. 6e–i). However, the Tcrd-/--T17 smoke-exposed group exhibited significantly lower TLC, FVC, VC, IC, and Cchord compared to the Tcrd-/- smoke-exposed group (Fig. 6e–i). Cchord did not significantly differ between the Tcrd-/--T17 smoke-exposed and Tcrd-/--T17 CON groups (Fig. 6i), suggesting a protective role of γδT17 cells against the emphysema-like phenotype. Additionally, the Tcrd-/--T17 smoke-exposed group did not show a significant decrease in the ratio of forced expiratory volume (FEV) in the first 50 ms to FVC (FEV50/FVC) compared to the Tcrd-/--T17 CON group (Fig. 6j), providing additional evidence that γδT17 cells help mitigate airflow limitation associated with tobacco smoke exposure. Consistent with these findings, adoptive transfer of γδT17 cells into WT recipients attenuated airway inflammation and emphysema-like pathology after six months of tobacco smoke exposure (Supplementary Fig. 9). Together, these findings indicate that γδT17 cells play a protective role by promoting tissue repair during smoke-induced lung injury.
Discussion
COPD is a major cause of morbidity, mortality, and healthcare utilization worldwide, primarily caused by exposure to inhaled noxious particles, notably tobacco smoke18. It is associated with chronic inflammation induced by tobacco smoke exposure, predominantly affecting the lung parenchyma and peripheral airways, resulting in largely irreversible and progressive airflow limitation19. In this study, we have extensively and thoroughly described the immune characterization of T cells induced by tobacco smoke exposure. We revealed the percentage in T cell subsets and the characteristics of clonal expansion in different sites of lung tissues in the progression of COPD in mice. Our findings highlight the expansion of clonal γ6δ4 T cells as a significant cellular marker of tobacco smoke exposure.
In the absence of external stimuli and infections, the airways usually contain fewer T cell types and numbers, predominantly tissue-resident subsets. T cells are recruited and retained in lung tissues to fulfill their functional roles in clearing pathogens20,21; however, overactivation of T cells can lead to immune-mediated lung injury22,23. Our results show that both the proportion and number of T cells in the airways increased significantly following tobacco smoke exposure, with a richer variety of subpopulations. Notably, the higher number of T cells is associated with the early stages of inflammatory responses in the lungs. Taken together, abnormally activated and aggregated T cells may either drive the initiation of airway inflammation or represent a consequence of inflammatory response and tissue damage. Additionally, our findings showed the presence of a population of T cells in an intermediate state between the airways and the parenchyma that became more prominent with prologed tobacco smoke exposure. Chronic inflammation in COPD affects both the airways and parenchyma, altering tissue structure and immune microenvironment, which leads to T cell aggregation in this intermediate state. Therefore, we hypothesized that intermediate state T cells are result of lung tissue injury, warranting further study.
Our study was designed to test the hypothesis that tobacco smoke introduces exogenous stimuli—such as physicochemical agents or microbial products—that activate innate and structural airway cells, resulting in the release of inflammatory cytokines and the creation of a pro-inflammatory microenvironment. Simultaneously, smoke-induced epithelial damage may lead to regulated cell death (e.g., apoptosis or necrosis), exposing self-antigens that further contribute to immune activation and airway remodeling. Thus, both exogenous and self-antigens may contribute to the recruitment and clonal expansion of antigen-specific T cells. The observed increase in TCR diversity, including longer and more diverse CDR3 regions, is consistent with antigen-driven T cell activation, though it does not pinpoint specific antigenic triggers. We honestly acknowledge the limitations of our current data. Due to the extensive diversity of TCRs and the complexity of antigen-epitope-TCR interactions (where a single antigen can contain multiple epitopes, each potentially recognized by a variety of TCRs), our current results cannot directly clarify the nature of antigen specificity in CD4⁺ and CD8⁺ T cells within COPD airways. Future work may focus on elucidating the nature of these antigens using emerging technologies such as peptide–MHC tetramer screens or antigen discovery platforms.
Our study indicates that TCR clonal diversity increased in the airways after smoke exposure, with an extended CDR3 length, but γδ T cells show different characteristics. γδT17 cells play important roles in the early phase of host defense against pathogens and in inflammatory diseases24. Although we observed a reduction in total γδ T cell proportion in the airways of the tobacco smoke-exposed group, our single-cell data revealed clonal expansion of γ6δ4 T cells. Previous studies suggest that γδ T cell gene rearrangement γδ TCR repertoire is relatively limited, with both innate25 and adaptive26 immune cell characteristics. However, our experimental data provide evidence that γ6δ4 T cells occur clonal expansion after tobacco smoke exposure, potentially conferring a protective effect. Additionally, γδ T cells exposed to tobacco smoke exhibited high expression of Il17a and Pdcd1, while showing low expression of Sell and Cd44, which is similar to findings in the Vγ6+ γδT17 lineage. The γδ T cells residing in lung tissue are mainly γ6 T cells and γ4 T cells16,27,28, with γδT17 cells developing exclusively before birth and subsequently persisting in adult mice as self-renewing, long-lived cells29. These results suggest that smoke-induced γδ T cell expansion may occur through proliferating locally rather than recruitment from the circulatory system. Compared to previous studies, we provide a comprehensive description of the distribution and immunological characteristics of γδ T cells in lung tissue after tobacco smoke exposure, which serves as an important reference for further research. In summary, COPD progression due to tobacco smoke exposure is linked to the expansion and phenotypic changes of lung tissue-resident γδ T17 cells.
The airflow limitation that defines COPD is caused by increased resistance of the small conducting airways and increased compliance of the lung as a result of emphysematous destruction30. This condition is usually progressive and associated with an enhanced chronic inflammatory response in the airways and the lung to noxious particles or gases31. Our study found that γδT17 cells improved airflow limitation and the emphysema-like phenotype in Tcrd-/- mice after 6 months of tobacco smoke exposure. This may relate to interactions between γδT17 cells and other immune or structural cells in lung tissues, though their functions and mechanisms require further investigation. Although the long-term tissue residency dynamics and quantitative persistence of transferred γδT17 cells require further elucidation, we suggest that these cells play a significant role in the COPD model. Additionally, the impact of chronic smoke exposure on abundance, TCR repertoire diversity, and function of γδT17 cells warrants further exploration.
γδT cells play a critical role in restoring mononuclear phagocyte numbers, protecting the lung from inappropriate inflammation32. Their dysregulation by local microbiota can stimulate IL-17 production, promoting neutrophil expansion and tumor cell proliferation33. Moreover, neutralization of IL-17 or G-CSF, or the absence of γδ T cells, has been shown to prevent neutrophil accumulation and downregulate the T cell-suppressive phenotype of neutrophils34. However, whether smoke-induced γδ T cells serve a similar function remains unclear. IL-17A mediates inflammatory cell recruitment, such as neutrophils, and has been studied in COPD pathogenesis. While experimental models implicate IL-17A in lung damage, findings vary due to differences in experimental protocols, including smoke exposure duration and composition35. IL-17A produced by γδ T cells may have a dual role in acute infectious diseases, both promoting and reducing inflammation36,37 and tissue repair38,39. A recent study found that IL-17A/F supplied by RORγt+ γδ T cells directly interacts with epithelial IL-17RC, inducing migration and re-epithelialization40.
Our experimental observations reveal a distinct, context-dependent role when IL-17A is derived from γδ T cells. We firstly noted a reduced percentage of γδ T cells within the airways in COPD models, alongside clonal expansion of γ6δ4 T cells—a subset of γδ T cells with high Il17a expression. Functional studies further demonstrated that depletion of these γδ T cells exacerbated disease susceptibility, whereas reconstitution of γδT17 cells (IL-17A-producing γδ T cells) restored their protective capacity, rendering the system more resistant to COPD progression. These findings collectively indicate that IL-17A-secreting γδ T cells exert a protective role in this context. γδ T cells, as innate-like effector cells, possess functional versatility beyond mere cytokine secretion; their protective role may integrate not only IL-17A-mediated effects but also direct cellular interactions, antigen recognition, or secretion of other mediators. Elucidating these mechanistic nuances will require further investigation to dissect the multifaceted roles of γδ T cells and the context-specific functions of IL-17A in COPD.
In conclusion, our findings identify γδT17 cells as a hallmark of immune responses to smoking and highlight the critical role of these cells in advancing the understanding of the immunoinflammatory pathogenesis of COPD. These insights may offer potential diagnostic and therapeutic targets for addressing smoke-induced immune system dysfunctions.
Methods
Mice
WT mice (C57BL/6J background) were purchased from GemPharmatech Co., Ltd. B6.129P2-Tcrdtm1Mom/J (Tcrd-/-) and C57BL/6-Il17atm1Bcgen/J (IL17-GFP) mice were provided by Zhinan Yin from the Institute of Biomedical Translational Research, Jinan University. All animal experiments described here complied with the guidelines of the Committee on the Use and Care of Animals of Guangzhou Medical University (Guangzhou, China) and were approved by the Animal Subjects Committee of Guangzhou Medical University. All mice were bred in the animal facility at Guangzhou Medical University Laboratory Animal Centre, where they were maintained under specific pathogen-free conditions on a 12 light/12 dark cycle at 18–24 °C and 40–60% humidity. Mice were euthanized using carbon dioxide (CO₂) inhalation or anesthesia overdose, in accordance with institutional animal care and ethical guidelines.
Mice used for flow cytometry analysis were sacrificed at 0 months (n = 5), 2 months (n = 4), and 6 months (n = 4) following tobacco smoke exposure. For single-cell RNA sequencing (scRNA-seq) and single-cell TCR sequencing (scTCR-seq), mice were sacrificed at 0 months (air control, CON), 2 months (2 M), and 6 months (6 M) after exposure. The CON group was generated by pooling three biological replicates, each comprising cells from 20 mice (total n = 60). The 2 M and 6 M groups were each generated by pooling three biological replicates, with each replicate consisting of cells from 4 mice (total n = 12 per group). For adoptive transfer experiments, C57BL/6J WT and Tcrd-/- mice were randomly assigned to either the air control (CON) group or the 6-month tobacco smoke exposure group and exposed in a fume chamber. Twenty-four hours prior to smoke exposure, Tcrd-/-mice were further divided into two groups: one receiving in vitro–expanded γδT17 cells (T17 group) and the other receiving PBS as a control. Body weight was continuously monitored throughout the exposure period (WT CON group, n = 8 mice/time point; WT smoke-exposed group, n = 6 mice/time point; Tcrd-/- CON group, n = 6 mice/time point; Tcrd-/- smoke-exposed group, n = 6 mice/time point; Tcrd-/--T17 CON groups, n = 7 mice/time point; Tcrd-/--T17 smoke-exposed groups, n = 7 mice/time point). For lung histology, 3 mice per group were used, and for pulmonary function tests (PFT), 6 mice per group were included.
Tobacco smoke exposure
Commercial cigarettes (Hongmei brand; tar: 13 mg/cigarette, nicotine: 0.8 mg/cigarette) were combusted using a whole-body passive smoking system (Liuwei Hardware & Electromechanical Co., Ltd., Guangzhou, China). Each exposure session involved 12 cigarettes burned within a 60 × 57 × 100 cm chamber over 10 min, reaching a total particulate matter concentration of 500 mg/m³. Mice were exposed four times daily for 1 h per session, with 15-min fresh-air intervals between sessions to prevent CO₂ accumulation. This corresponds to a daily exposure of 48 cigarettes over 4 h. Exposures were conducted 6 days per week for 2 or 6 months. Concurrently, Age-matched controls were housed in a clean-air chamber under identical conditions.
Pulmonary function measurement
Mice were anesthetized with 2% avertin (1.5 mL/kg) via intraperitoneal injection, then tracheostomized and placed in a DSI PFT system (Wilmington, NC, USA) to assess their pulmonary function according to the manufacturer’s protocol. The quasistatic PV maneuver was used to measure TLC, FVC, VC, IC, and Cchord. The FVC and FEV in the first 50 ms (FEV50) were recorded with the Fast Flow Volume maneuver. Each maneuver was repeated at least three times to ensure reproducibility.
Mouse lung histology
The left lung without lavage was fixed in 4% phosphate-buffered paraformaldehyde (pH 7.4), embedded in paraffin, and sliced into 4-mm-thick sections. The paraffin sections were then stained with hematoxylin and eosin solution for morphological examination. Alveolar enlargements were determined by the MLI, defined as the ratio of the total length of alveoli to the number of alveoli per field.
Simultaneous intranasal and intravascular antibody labeling
Mice were lightly anesthetized with isoflurane and treated i.n. with 0.25 mg of fluorochrome-conjugated CD45 antibody in 100 µL PBS. After 2 min, mice were injected i.v. with 0.5 mg of fluorochrome-conjugated CD90.2 antibody for an additional 3 min prior to euthanasia as previously described14,15. Detailed information about the antibodies used is shown in Supplementary Table 1. Mice were perfused, and cells were collected from the airway, lungs, and spleen.
Cell preparation
Cells were collected from the airways, lungs, and spleen. Bronchoalveolar lavage fluid (BALF) was obtained via tracheal cannulation, while lung tissue was enzymatically digested. Splenocytes were isolated by mechanical dissociation, with cell suspensions filtered and red blood cells lysed when necessary.
For scRNA-seq and TCR-seq assays, the cell suspension was stained with sample tags (BD Mouse Single-Cell Sample Multiplexing Kit, 626545, BD Biosciences) and CD3-APC antibody. The designation of sample tags can be found in Supplementary Table 2. Fixable Viability Dye eFluor 520 (65-0867-14, eBioscience) was used to exclude dead cells. Detailed information is shown in Supplementary Table 1. Live CD3+ T cells were sorted from the airway (CD45+CD90.2- cells in BALF), the airway-parenchyma interface (air-par, CD45+CD90.2- cells in lungs), the parenchyma (CD45-CD90.2- cells in lungs), and the spleen into cold buffer (BD Rhapsody Cartridge Reagent Kit, 633731, BD, Biosciences) using a BD FACSAria III cell sorter. WTA and TTA combined with TCR-seq of FCAS enriched live T cells (airway, air-par, parenchyma, spleen) were performed using the BD Rhapsody Single-Cell Analysis System (BD, Biosciences). For flow cytometry, the filtered cell suspension was stained with antibodies as described in the Flow cytometry part of the Methods. For detailed operations, see Supplemental Methods in the Supplementary Information.
Flow cytometry
Single-cell suspensions were stained in 96-well round-bottom plates at 5 \(\times\)105 lymphocytes per well. For surface staining, cells were stained with the indicated antibodies (1:100 dilution) at room temperature (20–25 °C) in the dark for 30 min and then labeled with Fixable Viability Dye eFluor 520 (65-0867-14, eBioscience) at room temperature (20–25 °C) in the dark for another 15 min. Then cells were washed with fluorescence-activated cell sorting (FACS) buffer twice and resuspended in FACS buffer (DPBS + 2% heat-inactivated fetal bovine serum) for flow cytometer detection. Information about antibodies and products used for flow cytometry is shown in the Antibodies part of the Methods and Supplementary Table 1. Flow cytometry data were acquired on a BD LSRFortessa X-20 or FACSAria III and analyzed with FlowJo software (version 10.8.1). Gating strategies are detailed in Supplementary Fig. 10.
Antibodies
The following antibodies were used: APC anti-mouse CD3ε antibody clone 145-2C11, 100312, Biolegend, 1:100. PE anti-mouse CD4 antibody clone GK1.5, 100408, Biolegend, 1:100. eFluor 450 anti-mouse CD8a monoclonal antibody clone 53-6.7, 47-0081-82, eBioscience, 1:100. BV650 Anti-mouse CD45R/B220 clone RA3-6B2, 563893, BD Biosciences, 1:100. APC-Cy7 anti-mouse CD4 clone GK1.5, 552051, BD Biosciences, 1:100. APC-eFluor 780 anti-mouse CD90.2 (Thy-1.2) antibody clone 53-2.1, 47-0902-82, eBioscience, 0.5 mg per mouse. Brilliant Violet 510 anti-mouse CD45 antibody clone 30-F11, 103138, Biolegend, 0.25 mg per mouse. InVivoMAb anti-mouse TCRγ/δ clone UC7, BE0070, Bioxcell, 10 μg per milliliter. InVivoPlus anti-mouse IFNγ clone XMG1.2, BP0055, Bioxcell, 10 μg per milliliter. All antibodies used are commercial antibodies reported by the manufacturer to be validated for use.
scRNA-seq and scTCR-seq data preprocessing
Detailed protocols for BD Rhapsody sequencing library preparation, sequencing, data preprocessing, and subsequent analysis using the “Seurat 4.0.1” R package are provided in the Supplementary Methods. Unless otherwise specified, all analysis of scRNA-seq and scTCR-seq data throughout this study were performed using R (version 4.4.0). SCT normalizations were performed on both TTA and WTA data and the PCA was conducted to obtain the top 50 PCs for subsequent UMAP ploting. Single cell was then clustered with a resolution of 1.6 for TTA and 0.8 for WTA. Parameters that not declared were all done with default setting. TTA RNA data matrix was further normalized with the iSMNN 1.20 method. For TTA data, γδ T cells were defined as cells with VδTCR chains. Cell clustering was performed on TTA γδ T cells with a resolution 1 and resulted in 12 cell subclusters. For WTA data, γδ T cells were extracted based on Trdc expression. An independent cell clustering process was conducted on these γδ T cells with a resolution of 0.5 and yielded 7 cell subclusters. Cells from different sample tags and samples were well-mixed, as Supplementary Fig. 11 showed, which indicated our normalization effectively corrected for the batch effects deriving from different samples.
Gene marker and differential expression analysis
DotPlot was used to show marker gene expression in each cell cluster defined in UMAP. Average gene expression and the percentage of cells expressing each gene under specific conditions were showed as cell color and cell size, respectively. FeaturePlot was used to visualize the expression landscape for the interested genes within the dataset. For TTA data, the genes with a value larger than 5 were taken as 5 in the RNA data matrix for visualization purposes. For WTA data, the gene read count was transformed by log2, and the maximum value after conversion was set to 4 and the minimum value was set to 0 for further visualization. For both TTA and WTA datasets, the percentage of expressed genes was calculated according to the number of cells with a given gene read count >0 to the total number of cells.
TCR analysis
TCR information were loaded from files names “VDJ_perCell” files in the BD pipeline results and integrated into the Seurat object for TTA data analysis. Clonotypes were defined as single-cell TCRαβ/TCRγδ pairs that exhibited the same V, D, J, and amino acid sequences in two CDR3 regions. Cells with incomplete TCR information were filtered out. And cells with TCRα chain information but with TCRδ chain information, or cells with TCRγ chain information accompanied with TCRβ chain were also discarded. Meanwhile, principal component analysis of Vαγ, Jαγ, Vβδ, and Jβδ region elements was performed using paired single-cell TCRαβ and TCRγδ sequencing data. The UMAP coordinate was firstly collected for each cell. The PC1 value for each clone was calculated as the mean value on x-axis in UMAP, while PC2 was derived from the y-axis. The “geom_point” function from “ggplot2 3.5.1” was used to plot the data and dot color and size were assigned according to the genotype of Vαγ, Jαγ, Vβδ, and Jβδ regions and the number of cells with the same clone type. The length of the CDR3 region was calculated with “nchar” function in R program.
The Gini Simpson index and clonality
In this study, the Gini Simpson Index was calculated with “vegan 2.6.4” R package, while Clonality was calculated with “entropy 1.3.1” R package. We calculated the Gini Simpson Index to assess the diversity of TCR clones for a given group of cells, and clonality was used to measure T cell expansion. Higher Gini Simpson Index indicates a richer diversity of corresponding TCRs. Higher TCR clonality indicated that corresponding TCRs were more clonally expanded. If there was a singleton TCR, we defined the TCR clonality as 0; while a single TCR species with multiple copies was assigned a TCR clonality of 1. These approaches provided a quantitative measure of TCR repertoire dynamics within the studied populations.
RNA velocyto
The sequencing mapped BAM format file generated by the BD Rhapsody data processing pipeline was pre-processing using samtools 1.15.1 to replace the “MA:Z:” to “UB:Z:” before running the velocyto. The produced loom format file was then analyzed with scVelo v0.2.5 module in Python (version 3.9.11). RNA Velocity41 was calculated and Pseudotime was estimated based on it. The (PAGA)42 was used to deduce the trajectory of cell development for the given group of cells.
γδT17 cell expansion culture and adoptive transfers
Spleen cells from IL17-GFP mice were cultured as previously described17. For detailed operations, see Supplemental Methods in the Supplementary Information. On day 9, γδT17 cells were harvested and purified via FACS. The collected cells were washed twice and resuspended in PBS, and 2 × 106 cells were intravenously injected into Tcrd-/- mice. A solvent control group (PBS injection only) was also established. After confirming no procedural errors within 24 h, the mice were transferred to a smoke exposure box for exposure.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 9.5.0 software. Two-sided unpaired Student’s t-test was used to analyze differences in the mean values between groups. The Mann–Whitney test was used to compare the central tendencies of the two groups (mean or median). One-way ANOVA was used to compare differences among multiple groups when data followed a normal distribution, whereas the non-parametric Kruskal–Wallis test was applied for data that did not meet this assumption. In some cases, P values were transformed into Q values were calculated using Storey FDR procedure. In case of multiple comparisons, P values were adjusted using the Bonferroni corrections. Significant values (P < 0.05) are indicated for all figures where relevant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data generated or analysed in this study are provided in the article and its Supplementary Information/Source Data file. RNA and TCR sequencing data generated in this study are deposited in the NGDC under accession number CRA023351. Source data are provided with this paper.
References
Safiri, S. et al. Burden of chronic obstructive pulmonary disease and its attributable risk factors in 204 countries and territories, 1990-2019: results from the Global Burden of Disease Study 2019. BMJ 378, e069679 (2022).
Adeloye, D. et al. Global, regional, and national prevalence of, and risk factors for, chronic obstructive pulmonary disease (COPD) in 2019: a systematic review and modelling analysis. Lancet Respir. Med. 10, 447–458 (2022).
Celli, B. et al. Definition and nomenclature of chronic obstructive pulmonary disease: time for its revision. Am. J. Respir. Crit. Care Med. 206, 1317–1325 (2022).
Saetta, M. et al. CD8+ T-lymphocytes in peripheral airways of smokers with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 157, 822–826 (1998).
Ryu, M. H. et al. Blood gene expression and immune cell subtypes associated with chronic obstructive pulmonary disease exacerbations. Am. J. Respir. Crit. Care Med. 208, 247–255 (2023).
Di Stefano, A. et al. T helper type 17-related cytokine expression is increased in the bronchial mucosa of stable chronic obstructive pulmonary disease patients. Clin. Exp. Immunol. 157, 316–324 (2009).
Doe, C. et al. Expression of the T helper 17-associated cytokines IL-17A and IL-17F in asthma and COPD. Chest 138, 1140–1147 (2010).
Laan, M., Lötvall, J., Chung, K. F. & Lindén, A. IL-17-induced cytokine release in human bronchial epithelial cells in vitro: role of mitogen-activated protein (MAP) kinases. Br. J. Pharm. 133, 200–206 (2001).
Hou, J. et al. Imbalance between subpopulations of regulatory T cells in COPD. Thorax 68, 1131–1139 (2013).
Majo, J., Ghezzo, H. & Cosio, M. G. Lymphocyte population and apoptosis in the lungs of smokers and their relation to emphysema. Eur. Respir. J. 17, 946–953 (2001).
Pons, J. et al. Blunted gamma delta T-lymphocyte response in chronic obstructive pulmonary disease. Eur. Respir. J. 25, 441–446 (2005).
Urboniene, D., Babusyte, A., Lötvall, J., Sakalauskas, R. & Sitkauskiene, B. Distribution of γδ and other T-lymphocyte subsets in patients with chronic obstructive pulmonary disease and asthma. Respir. Med. 107, 413–423 (2013).
Shan, M. et al. Cigarette smoke induction of osteopontin (SPP1) mediates T(H)17 inflammation in human and experimental emphysema. Sci. Transl. Med. 4, 117ra119 (2012).
Zhao, J. et al. Airway memory CD4(+) T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity 44, 1379–1391 (2016).
Wang Y. et al. Simultaneous intranasal/intravascular antibody labeling of CD4(+) T cells in mouse lungs. Bio. Protoc. 7, e2099 (2017).
du Halgouet, A. et al. Multimodal profiling reveals site-specific adaptation and tissue residency hallmarks of γδ T cells across organs in mice. Nat. Immunol. 25, 343–356 (2024).
McKenzie, D. R. et al. IL-17-producing γδ T cells switch migratory patterns between resting and activated states. Nat. Commun. 8, 15632 (2017).
Christenson, S. A., Smith, B. M., Bafadhel, M. & Putcha, N. Chronic obstructive pulmonary disease. Lancet 399, 2227–2242 (2022).
Barnes, P. J. Inflammatory mechanisms in patients with chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 138, 16–27 (2016).
Teijaro, J. R. et al. Cutting edge: tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 187, 5510–5514 (2011).
Hogan, R. J. et al. Protection from respiratory virus infections can be mediated by antigen-specific CD4(+) T cells that persist in the lungs. J. Exp. Med. 193, 981–986 (2001).
Xu, L. et al. Cutting edge: pulmonary immunopathology mediated by antigen-specific expression of TNF-alpha by antiviral CD8+ T cells. J. Immunol. 173, 721–725 (2004).
Zhao, M. Q. et al. Alveolar epithelial cell chemokine expression triggered by antigen-specific cytolytic CD8(+) T cell recognition. J. Clin. Investig. 106, R49–58 (2000).
Akitsu, A. & Iwakura, Y. Interleukin-17-producing γδ T (γδ17) cells in inflammatory diseases. Immunology 155, 418–426 (2018).
Davies, D. et al. PD-1 defines a distinct, functional, tissue-adapted state in Vδ1(+) T cells with implications for cancer immunotherapy. Nat. Cancer 5, 420–432 (2024).
Holtmeier, W. & Kabelitz, D. Gammadelta T cells link innate and adaptive immune responses. Chem. Immunol. Allergy 86, 151–183 (2005).
Cheng, M. & Hu, S. Lung-resident γδ T cells and their roles in lung diseases. Immunology 151, 375–384 (2017).
Simonian, P. L. et al. IL-17A-expressing T cells are essential for bacterial clearance in a murine model of hypersensitivity pneumonitis. J. Immunol. 182, 6540–6549 (2009).
Haas, J. D. et al. Development of interleukin-17-producing γδ T cells is restricted to a functional embryonic wave. Immunity 37, 48–59 (2012).
Hogg, J. C. Pathophysiology of airflow limitation in chronic obstructive pulmonary disease. Lancet 364, 709–721 (2004).
Baraldo, S., Turato, G. & Saetta, M. Pathophysiology of the small airways in chronic obstructive pulmonary disease. Respiration 84, 89–97 (2012).
Kirby, A. C., Newton, D. J., Carding, S. R. & Kaye, P. M. Pulmonary dendritic cells and alveolar macrophages are regulated by gammadelta T cells during the resolution of S. pneumoniae-induced inflammation. J. Pathol. 212, 29–37 (2007).
Jin, C. et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell 176, 998–1013.e1016 (2019).
Coffelt, S. B. et al. IL-17-producing γδ T cells and neutrophils conspire to promote breast cancer metastasis. Nature 522, 345–348 (2015).
Ritzmann F., Lunding L. P., Bals R., Wegmann M., Beisswenger C. IL-17 cytokines and chronic lung diseases. Cells 11, 2132 (2022).
Li, C. et al. IL-17 response mediates acute lung injury induced by the 2009 pandemic influenza A (H1N1) virus. Cell Res. 22, 528–538 (2012).
Huang, H., Saravia, J., You, D., Shaw, A. J. & Cormier, S. A. Impaired gamma delta T cell-derived IL-17A and inflammasome activation during early respiratory syncytial virus infection in infants. Immunol. cell Biol. 93, 126–135 (2015).
Xi-zhi, J. G. et al. Lung γδ T cells mediate protective responses during neonatal influenza infection that are associated with type 2 immunity. Immunity 49, 531–544. e536 (2018).
Bonneville, M., O’Brien, R. L. & Born, W. K. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 10, 467–478 (2010).
Konieczny, P. et al. Interleukin-17 governs hypoxic adaptation of injured epithelium. Science 377, eabg9302 (2022).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Wolf, F. A. et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20, 59 (2019).
Acknowledgements
This work was supported by the Foundation of Guangzhou National Laboratory (SRPG22-016 and SRPG22-018), Major Project of Guangzhou National Laboratory (GZNL2023A02001, GZNL2024A02005), the Postdoctoral Fellowship Program of CPSF under Grant Number GZC20251431, National Natural Science Foundation of China (82270043, U22A20265 and 82200045), the Zhongnanshan Medical Foundation of Guangdong Province (ZNSA-2020003), the grant of State Key Laboratory of Respiratory Disease (SKLRD-Z-202315), and Research Capability Strengthening and Fundamental Improvement Project of Guangzhou Medical University (2024SRP080). We would like to thank Zhinan Yin from the Institute of Biomedical Translational Research, Jinan University, for providing B6.129P2-Tcrdtm1Mom/J (Tcrd-/-) and C57BL/6-Il17atm1Bcgen/J (IL17-GFP) mice.
Author information
Authors and Affiliations
Contributions
X.M. performed the experiments, analyzed data, contributed to single-cell data interpretation and wrote the paper. J.W. performed the experiments and contributed to data analysis. Y.W., J. Zhang analyzed scRNA-seq and scTCR-seq data. L.C., X.W., X.L., G.B., J. Zhong, M.H., S.H., P.H., J.C., Y.L., Q. Wen, R.S., E.Y., F.W., X.W., Z.Y., Q. Wan, X.Z., Y. Zuo and L.Z. contributed to mouse handling and performed the experiments. Z.W. and Y. Zhou supervised X.M., provided scientific inputs, supported funding, and contributed to data interpretation. P.R. conceptualized and coordinated the study, acquired funding, conducted experiments, analyzed the data, and wrote the paper. All authors have read and approved the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Alen Faiz, Rabih Halwani, and Yohannes Tesfaigzi for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mei, X., Wang, J., Wang, Y. et al. Comprehensive profiling of smoke-induced T cells in mice implicates clonal γδT17 cells as a hallmark of COPD. Nat Commun 17, 441 (2026). https://doi.org/10.1038/s41467-025-67120-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-67120-w