Abstract
Excessive fibrosis, rather than endogenous tissue regeneration, often results from peripheral tissue injury repair, presenting a significant clinical challenge due to its poor reversibility and limited treatment options. A key factor contributing to this outcome is the aberrant commitment of mesenchymal stromal cells (MSCs) to a fibrotic lineage. However, the upstream signals driving this pathological differentiation remain unclear. In this study, we identify nociceptive sensory neuron rather than local cell-derived nerve growth factor (NGF) as a critical signaling factor guiding the formation of the fibrotic MSCs neurogenic niche, thereby driving MSC-mediated fibrosis. Using nociceptive sensory denervation and NGF knockout genetic models, we demonstrate that disrupting sensory nerve innervation significantly reduces MSC-mediated fibrosis. In vitro, NGF promotes MSC fibrotic commitment through the TrkA and hypoxia-inducible factor-1α (Hif1α) signaling pathways. Therapeutically, local administration of TPX-0005 (also known as repotrectinib), a clinically used TrkA inhibitor, partially reverses excessive fibrosis and improves long-term healing outcomes. Our findings reveal a previously unrecognized neurogenic axis, highlighting the functional specificity of nociceptive sensory neuron-derived NGF in peripheral tissue repair and emphasizing the regulation of MSC neurogenic niche as a promising strategy for fibrosis prevention and treatment.
Similar content being viewed by others
Introduction
Fibrosis, characterized by excessive scarring and tissue dysfunction, remains challenging to manage due to limited understanding of its molecular drivers1,2,3. Emerging evidence suggests that sensory nerves play a role beyond transmitting pain and external stimuli; they actively regulate tissue homeostasis, repair, and regeneration4,5,6,7,8,9,10,11,12. However, their involvement in fibrosis, particularly in the early processes that prime mesenchymal stromal cells (MSCs) for a fibrotic fate, remains poorly understood.
MSCs are pivotal in tissue maintenance and repair, and its niche, a microenvironment composed of MSCs themselves, spatially associated cellular structures, extracellular matrix, and cytokines that regulate MSCs, determines its differentiation direction13,14,15,16. In ideal condition, MSCs are normally recruited to injury sites to promote origin tissue regeneration and repair, but most of the time, excessive commitment to the fibrotic lineage leads to myofibroblast formation, causing pathological fibrosis and impairing tissue function17,18,19,20. Deciphering the mechanisms that regulate this fibrotic niche is critical for developing effective anti-fibrotic therapies. Tendon repair, for instance, is often hindered by peritendinous adhesion (PA), a hallmark fibrotic complication21,22,23. Current treatments, including surgery, frequently induce cyclical trauma and re-adhesion23,24,25, highlighting the urgent need for innovative, mechanism-based interventions.
While MSCs recruitment and fibrotic differentiation are established contributors to fibrosis pathogenesis, the upstream regulators and nerve-related influences remain elusive. Nociceptive sensory nerves, which secrete factors such as NGF, CGRP, SP, and NPY, are known to mediate various physiological and pathological processes, including wound healing and inflammation26,27,28,29,30,31,32,33,34. Among these, NGF, a key axon guidance molecule, has been implicated in tissue-nerve interactions in peripheral systems21,34,35. However, it is still unclear whether nociceptive sensory neuron-derived NGF contributes to fibrosis or healing in ways that are different from local cell-derived NGF. We hypothesized that nociceptive sensory neuron-derived NGF serves as a critical instructive signal driving fibrotic MSCs neurogenic niche formation, thereby promoting fibrosis.
In this study, our findings highlight the central role of sensory nerve-derived NGF in MSC niche commitment. Nociceptive sensory neuron-derived NGF is functionally specific, its mediation of MSC neurogenic niche formation drives MSC aberrant fibrotic commitment via TrkA/Hif1α pathways to form excessive fibrosis in peripheral tissue repair. However, TrkA inhibition with TPX-0005 significantly attenuates fibrosis and improves healing, providing a strategy for targeting this axis.
Results
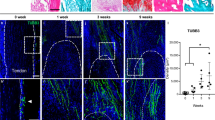
Nociceptive sensory nerve innervation after peripheral trauma with fibrogenesis in the fibrotic niche
Human peritendinous fibrotic tissue samples were collected at various time points following injury and categorized into the following groups: control, 2–3 days, 8–12 days, and 3 months and beyond (Fig. 1A). Functional assessments of total active mobility (TAM) at different time points revealed that tendon injury leads to a progressive decline in joint mobility (Fig. 1B). Histological examination confirmed that this decline in function is attributed to the progressive accumulation of collagen in the fibrotic regions (Fig. 1C). Furthermore, Prrx1 is a well-established marker of MSCs in the musculoskeletal system, and previous studies have demonstrated that the Prrx1-CreERT2 tool can be used to trace the fate of MSCs during musculoskeletal injury repair36,37,38. Immunofluorescence analysis revealed that the expression of nociceptive sensory nerve fibers gradually increased over time, peaking at 8-12 days (Fig. 1D, A). This upregulation coincided with the activation of αSMA⁺ myofibroblasts and Prrx1⁺ MSCs (Supplementary Fig. 1B), suggesting a potential interaction between sensory nerves and fibrotic remodeling. Cross-species validation was conducted using a mouse tendon injury model, which reproduced the pathophysiological changes observed in humans: specifically, fibrosis and adhesions that reduced the range of motion (ROM) and increased collagen deposition (Supplementary Fig. 1C-F). Temporal analysis of PGP9.5⁺ nociceptive sensory nerve fiber infiltration showed that nerve fiber density peaked 14 days post-injury in the mouse model (Supplementary Fig. 1G), providing experimental evidence for subsequent time-dependent analyses. Further analysis through tissue clearing revealed region-specific distribution of nociceptive sensory nerves: newly formed sensory nerves predominantly localized to the peritendinous region at the injury site, with minimal penetration into the tendon substance itself (Fig. 1E, F). These findings suggest that nociceptive sensory nerves may primarily mediate peritendinous fibrosis following tendon injury. Co-localization experiments of CGRP and αSMA demonstrated synchronous interactions between nerves and myofibroblasts, with this interaction gradually decreasing by day 28 post-injury (Supplementary Fig. 1H), a trend consistent with human case models. Next, we constructed Prrx1-CreERT2::Rosa26-LSL-tdTomato (Prrx1Tomato) transgenic mice to trace the spatial distribution of MSCs. Results showed that nociceptive sensory nerves converged with MSC-enriched regions, forming a functional fibrotic niche (Fig. 1G). In addition, the distribution of Prrx1⁺ MSCs was highly specific: they were predominantly localized in the tenosynovium, peritendinous fibrotic areas, and to a lesser extent, in tendon tissues, with minimal labeling in muscle tissues. Low-magnification analysis showed that Prrx1⁺ MSCs were almost exclusively enriched in the AD region, including most fibrotic tissue and epitenon cells, with a small subset labeling tenocytes, and co-localization of Prrx1⁺ MSCs with myofibroblasts revealed that the majority of myofibroblasts originated from Prrx1⁺ MSCs (Fig. 1H). Continuous time-point analyses of the same anatomical locations further confirmed the synergistic activation of Prrx1⁺ MSCs and myofibroblasts. Post-injury, the biomechanical function of mouse tendons declined, manifesting as reduced maximum load and stiffness, which mirrored the progressive decline in human TAM scores (Supplementary Fig. 1J, K).
A Fibrosis tissues of control individuals, injury after 2–3 days, 8–12 days, and over 3 months groups after injury. B Quantitative analysis of TAM of human peritendinous tissue (n = 4). C H&E and Masson staining of human peritendinous tissues (n = 4). Scale bars: 100 μm. D Representative double-immunofluorescent (IF) staining images of PGP9.5 (green) and CGRP (red) in human peritendinous tissue (n = 4). Scale bars: Low power image is 300 μm, and the high power image is 100 μm. E Tissue clearing for reconstruction of CGRP in WT mouse hind limb at 14 days (n = 3). Scale bars: Low power image is 3000μm, and the high power image is 250 μm. F Immunofluorescent staining showing CGRP distribution in WT mice at 14 days (n = 3). Scale bars: Low power image is 1000 μm, and the high power image is 200 μm. G Representative double-immunofluorescent 3D staining images of CGRP (green) and Tomato (red) of peritendinous tissue from Prrx1Tomato mice at 14 days after modeling. Scale bars: 50 μm. H Immunofluorescent co-staining of Prrx1-Td and αSMA at fibrotic sites (n = 6). Scale bars: Low power image is 1000 μm, and the high power image is 200 μm. Yellow dashed line shows space between tendon and surrounding tissues. Black or white dashed line shows space occupied by fibrosis tissues. T represents tendon; AD represents fibrosis tissue; M represents muscle (mean ± SD; B–D: ANOVA with multiple comparisons; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; A(hand), G(mouse) created in Adobe. Author, Yanhao Li. (2025). Contact the author for publication if needed; A(tendon and fibrosis tissue), G(MSCs) created in BioRender. Author, li, y. (2025)) https://BioRender.com/0hlz1hf).
In summary, these findings suggest that nociceptive sensory nerves contribute to the formation of fibrosis following tendon injury, and this involvement is synchronized with the activation of MSCs and myofibroblasts. The interaction between nerves and local cells is evolutionarily conserved.
Ablation of nociceptive sensory innervation in the fibrotic niche attenuates fibrosis
The Advillin-CreERT2::TrkA (AdvTrkA) transgenic mouse model was used to achieve genetic ablation of nociceptive sensory neurons (Fig. 2A). Tamoxifen (TMX) pretreatment activates the Cre recombinase, thereby preventing the infiltration of newly formed nociceptive sensory neurons into the fibrotic areas, without inducing endogenous fibrotic changes in the tendons themselves (Supplementary Fig. 2A). Co-staining experiments of NeuN and TrkA in the dorsal root ganglion (DRG) further confirmed the efficiency of TrkA knockout (Supplementary Fig. 2B), while a significant reduction in the density of CGRP⁺ nociceptive sensory nerve innervation was observed (Fig. 2B, Supplementary Fig. 2C).
A AdvTrkA mice experimental procedure. B IF of CGRP (red) in AdvTrkA mice injected with Veh or TMX at 14 days after Modeling. (n = 6). Scale bars: low power image is 100 μm, and the high power image is 50 μm. C H&E and Masson staining of peritendinous tissue from AdvTrkA mice (n = 6). Scale bars: 300 μm. D Analysis of ROM of AdvTrkA mice (n = 6). E IHC of Col1 and Col3 expression of peritendinous tissue from AdvTrkA mice (n = 6). Scale bars: 100 μm. F Protein expressions of Col 1 and Col 3 in fibrosis tissue from AdvTrkA mice (n = 3). G IF of Prrx1 (red) from AdvTrkA mice (n = 6). Scale bars: 50 μm. H Double IF of αSMA (green) and Col1 (red) of peritendinous tissue from AdvTrkA mice (n = 6). Scale bars: low power image is 100 μm, and the high power image is 50 μm. I AdvDTA mice experimental procedure. J Analysis of ROM of AdvDTA mice (n = 6). K IHC of Col1 and Col3 expression of peritendinous tissue from AdvDTA mice. Scale bars: 100 μm. L Protein expressions of Col 1 and Col 3 in fibrosis tissue of AdvDTA mice. M IF of αSMA (green) of peritendinous tissue from AdvDTA mice. (n = 6). Scale bars: 50 μm. Yellow dashed line shows space between tendon and surrounding tissues. Black or white dashed line shows space occupied by fibrosis tissues. T represents tendon; AD represents fibrosis tissue; M represents muscle (mean ± SD; B–H/J–M: unpaired t-test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; A/I created in Adobe. Author, Yanhao Li. (2025). Contact the author for publication if needed).
Nociceptive sensory denervation alleviated the progression of fibrosis at days 14 and 28 post-injury, as evidenced by widened tendon gaps, reduced collagen deposition, and restored range of motion (ROM) (Fig. 2C, D; Supplementary Fig. 2D). Additionally, the gene and protein expression levels of Col1/Col3 were significantly downregulated (Fig. 2E, F; Supplementary Fig. 2E-G). However, it is noteworthy that despite nociceptive sensory neuron ablation, tendon biomechanical properties remained impaired, as indicated by a decrease in maximum load and stiffness (Supplementary Fig. 2H, I), suggesting that nociceptive sensory neurons not only participate in fibrosis formation but also play a crucial role in tendon repair processes. Interestingly, on day 14 post-injury, Prrx1⁺ MSCs accumulated in the denervated tissue, while the number of αSMA⁺ myofibroblasts was reduced (Fig. 2G, H; Supplementary Fig. 2J), indicating that nociceptive sensory neurons may mediate the fate determination of MSCs into myofibroblasts. To further validate the regulatory role of nociceptive sensory neurons in fibrosis, we constructed Advillin-CreERT2::DTA (AdvDTA) transgenic mice for orthogonal verification. The results successfully recapitulated the fibrosis inhibition phenotype observed in the AdvTrkA mice (Fig. 2I-M; Supplementary Fig. 2K-O). The hot plate test showed that the paw withdrawal latency of mice in the TMX-treated group was more than twice as long as that in the Veh-treated group (Supplementary Fig. 2P), confirming the successful loss of targeted nociceptive sensory function in the model.
Grip strength testing revealed no significant difference between the TMX-treated group and the Veh control group (Supplementary Fig. 2Q), ruling out off-target effects on motor neurons. Serum glucose levels measured at euthanasia were within the normal range (Supplementary Fig. 2R).
In conclusion, the above analyses confirm that nociceptive sensory neurons may significantly influence the fibrosis process by regulating the fate transition of MSCs and myofibroblasts.
Nociceptive sensory neuron-derived NGF possess functional specificity to promote fibrosis
Fibrosis mediated by nociceptive sensory nerves occurs via local neuroendocrine signaling. Retrograde neuronal tracing with Cholera Toxin B (CTB) revealed that sensory neurons innervating the tendon are mainly localized to the L4–L6 dorsal root ganglia (DRG) (Fig. 3A, B; 3A). Given the complexity of NGF sources, we investigated whether NGF specifically derived from nociceptive sensory neurons exerts a unique function. We found a spatial segregation between the synthesis and secretion of NGF at both the gene and protein levels. Injury upregulated NGF transcription in sensory neurons of the DRG. Nociceptive sensory denervation (in AdvTrkA mice) eliminated the infiltration of peripheral nociceptive sensory nerves but not DRG neurons, thus leaving DRG NGF mRNA levels unchanged (Fig. 3C, D). Peripheral sensory nerve terminals secrete NGF into the local niche; despite no changes in mRNA levels in either the DRG or local tissues, nociceptive sensory denervation (AdvTrkA mice) eliminated the infiltration of peripheral nociceptive sensory nerves and reduced the pool of NGF protein derived from peripheral nociceptive sensory neurons (Fig. 3E, F, G). Subsequently, intrathecal injection of AAV-GFP-siNGF achieved DRG-specific NGF knockdown (Fig. 3H; Supplementary Fig. 3B). This manipulation not only reduced NGF expression in neurons (Supplementary Fig. 3C) but also attenuated fibrosis (Fig. 3I, D), decreased collagen deposition (Supplementary Fig. 3E, F), and diminished the accumulation of αSMA⁺ myofibroblasts (Supplementary Fig. 3G). These findings suggested that NGF derived from nociceptive sensory neurons plays a specific pro-fibrotic role. We therefore generated Advillin-CreERT2::NGF (AdvNGF) transgenic mice. After ablating NGF derived from nociceptive sensory neurons, we observed that the innervation pattern remained normal (Fig. 3J, K; Supplementary Fig. 4A), accompanied by improved range of motion (ROM) (Fig. 3L) and suppressed myofibroblast formation (Fig. 3M; Supplementary Fig. 4B).
A Schematic diagram of cholera toxin B (CTB) retrograde neuronal tracing. B Representative NeuN/CTB staining of DRG neurons (n = 6). C Relative mRNA expression of NGF, NPY, SP, and CGRP in WT mice at normal, 7, 14, and 28 days (n = 6). D Relative mRNA expression of NGF at the fibrosis site and DRG at 14 days (n = 6). E NGF protein concentration at fibrotic sites in WT and AdvTrkA mice (pg/mg protein, n = 6). F Double IF of NGF (green) and NeuN (red) after modeling on 14 day in DRG from normal, AdvTrkA mice injected with Veh or TMX (n = 6). Scale bars: low power image is 100 μm, and the high power image is 50 μm. G IF of NGF (green) after modeling on 14 day in fibrosis site from normal, AdvTrkA mice injected with Veh or TMX (n = 6). Scale bars: 50 μm. H Experimental workflow of intrathecal AAV-GFP-siNGF injection. I H&E and Masson staining of WT mice with intrathecal injection of AAV-GFP-siNGF or NC at 14 days. Scale bars: 300 μm. J AdvNGF mice experimental procedure. K CGRP (red) staining in AdvNGF mice injected with Veh or TMX at 14 days after Modeling (n = 6). Scale bars: low power image is 100 μm, and the high power image is 50 μm. L Analysis of ROM of AdvNGF mice (n = 6). M Double IF of αSMA (green) and Col1 (red) of AdvNGF mice. (n = 6). Scale bars: low power image is 100 μm, and the high power image is 50 μm.Yellow dashed line shows space between tendon and surrounding tissues. Black or white dashed line shows space occupied by fibrosis tissues. T represents tendon; AD represents fibrosis tissue; M represents muscle(mean ± SD; F, G: ANOVA with multiple comparisons, I/K–M: unpaired t-test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; A/H(mouse, injector and intrathecal space)/J created in Adobe. Author, Yanhao Li. (2025). Contact the author for publication if needed; H(tendon and fibrosis tissue) created in BioRender. Author, li, y. (2025)) https://BioRender.com/0hlz1hf).
We ruled out the possibility that NGF knockout exerts confounding effects via motor nerves. Immunofluorescence staining was performed on motor nerves, and their distribution was analyzed under low magnification. Results showed that Choline Acetyltransferase (ChAT)-positive motor nerves were almost exclusively localized to muscle tissue, with minimal infiltration into the peritendinous fibrotic niche (Supplementary Fig. 4C). Furthermore, NGF staining of motor neurons in the anterior horn of the spinal cord revealed negligible NGF expression in these neurons (Supplementary Fig. 4D). Therefore, even if potential confounding effects from motor nerves exist, such effects are not mediated by NGF.
Collectively, these complementary experiments confirm that NGF derived from nociceptive sensory neurons serves as a key mechanistic axis governing the formation of the fibrotic niche.
Nociceptive sensory neurogenic NGF mediates the formation of MSCs neurogenic niche driving excessive fibrosis
Prrx1 is a well-validated marker for mesenchymal stromal cells (MSCs) in the musculoskeletal system. The previously reported Prrx1-CreERT2 has been used to trace the fate of MSCs during musculoskeletal injury repair and is critical for osteogenesis, adipogenesis, and dermogenesis36,39. However, its contribution to fibrogenesis after injury remains unclear. Therefore, we first performed unbiased 10× single-cell RNA sequencing (scRNA-seq) on fibrotic and tendon tissues from WT mice at sequential time points (Fig. 4A). Subpopulations were annotated with reference to the recognized single-cell classification of tendon pathology39. In addition to conventional immune cells, endothelial cells, and pericytes, we categorized key stromal cells (widely expressing Dcn, Col1a1, Col3a1, and Prrx1) into epitenon cells (Slc1a3), bridging cells, tenocytes (Scx, Tnmd, Mkx, Fmod), IAF cells, and MTJ cells (Fig. 4B; Supplementary Fig. 5B). From the perspective of cell proportions, stromal cells gradually resembled the sham group as time progressed (Fig. 4C). Notably, epitenon cells and some bridging cells exhibited higher differentiation potential and displayed two distinct differentiation directions: fibrogenesis and tenogenesis (Supplementary Fig. 6A–C), which is consistent with previous literature reports39. We found that Prrx1 actually serves as a reliable marker for mesenchymal stromal cells in tendon injury model. This is because it labels most of stromal cells with higher stemness, as verified by trilineage differentiation assays, clonal proliferation assays, and FACS detection of CD105, CD73, and CD90 (Supplementary Fig. 7A). We quantified the proportions of Prrx1⁺Col3a1⁺ MSCs and Prrx1⁺Scx⁺ MSCs. Both cell populations showed increased proportions over time after injury, indicating that Prrx1⁺ MSCs contribute to both fibrogenesis and tendon repair. However, the proportion of Prrx1⁺Col3a1⁺ cells reached nearly 90% at 28 days post-injury, while that of Prrx1⁺Scx⁺ cells was less than 30%. This suggests that Prrx1⁺ MSCs are primarily involved in mediating fibrosis after tendon injury (Fig. 4D, E). To ensure Prrx1+MSC spatial localization, we traced the distribution of Prrx1⁺Col3a1⁺ cells using 14-day paraffin sections from Prrx1Tomato mice. Results showed that these cells were mainly distributed in fibrotic tissues (Fig. 4F2, F3) with a small number localized on the epitenon surface (Fig. 4F1).
A Single-cell sequencing workflow for fibrotic tissues from WT mice. B UMAP plot showing major cell clusters. C Cell type composition dynamics across time points. D, E Proportion of Prrx1+ fibroblast subsets classified by Col3a1 and Scx expression. F Immunofluorescent co-staining of Prrx1-Td and Col3a1 at fibrotic sites (n = 6). Scale bars: Low power image is 1000 μm, and the high power image is 250 μm. G Single-cell sequencing workflow for fibrotic tissues from AdvNGF mice and control mice. H UMAP plot showing major cell clusters. I GO analysis. J KEGG analysis. K Pseudotime analysis of Prrx1+ MSCs. L–Q Violin plots show the expression of related genes (Col3a1, Cxcl14, Pi16, Col1a1, Scx, Tnmd) in WT and AdvNGF mice. Black or white dashed line shows space occupied by fibrosis tissues. T represents tendon; AD represents fibrosis tissue; M represents muscle(mean ± SD; I-J: ANOVA with multiple comparisons; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; A(mouse)/G(mouse) created in Adobe. Author, Yanhao Li. (2025). Contact the author for publication if needed; A(tendon and fibrosis tissue) created in BioRender. Author, li, y. (2025) https://BioRender.com/0hlz1hf; A(Chromium 10x)/G(Chromium 10x) created in BioRender. Author, li, y. (2025) https://BioRender.com/zu1reum).
Collectively, these findings confirm that Prrx1 can serve as a reliable marker for mesenchymal stromal cells after tendon injury, enabling cell tracing related to fibrosis and repair following tissue damage.
Next, we also performed scRNA-seq analysis on 14-day tissues from AdvNGF mice and control mice (Fig. 4G), and similar cell subpopulations could be distinguished (Fig. 4H; Supplementary Fig. 7D, E). Differential gene analysis, Gene Ontology (GO) analysis, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis indicated that NGF depletion significantly altered pathways related to tissue migration, mesenchymal cell differentiation, stem cell differentiation, and extracellular matrix adhesion (Fig. 4I, J). Pseudotime analysis revealed that NGF depletion blocked the early stages of Prrx1⁺MSC differentiation, leading to cell accumulation at these stages. However, some cells eventually progressed to later differentiation stages, suggesting that the NGF signaling pathway influences the differentiation process—with some cells entering a transient quiescent state while others continue along the differentiation trajectory (Fig. 4K). This implies that NGF derived from nociceptive sensory neurons can drive the differentiation of Prrx1⁺ MSCs into myofibroblasts. After knocking out NGF derived from nociceptive sensory neurons, the expression of fibrosis-related genes in Prrx1⁺ MSCs was significantly downregulated (Col3a1, Cxcl14, Pi16) (Fig. 4L–N), whereas the expression of tenogenesis-related genes showed no significant changes (Col1a1, Scx, Tnmd) (Fig. 4O–Q). This is consistent with the conclusion that Prrx1⁺ MSCs are primarily involved in mediating fibrosis after tendon injury, and also indicates that blocking NGF does not exert strong side effects on tendon repair. In addition, we analyzed the cellular characteristics of cells labeled by two previously reported mesenchymal stromal cell markers: Prrx1 and Nestin. Although both cell populations could differentiate into myofibroblasts, endothelial cells, and pericytes, Prrx1⁺ MSCs exhibited lower relative stemness and higher profibrotic potential compared to Nestin⁺ MSCs (Supplementary Fig. 7F, G). GO analysis further confirmed that Prrx1⁺ MSCs are more prone to fibroblastic differentiation and lack angiogenic capacity (Supplementary Fig. 7H, I). Nociceptive sensory neurons-derived NGF does not affect cell-cell communication among local cells, but significantly influences the activation of Hif1α and its downstream target Twist1 in Prrx1⁺ MSCs(Supplementary Fig. 7J, K).
These results demonstrate that NGF derived from nociceptive sensory neurons plays a critical role in guiding the differentiation of Prrx1⁺ MSCs toward the fibroblastic lineage, further reinforcing its importance in the development of fibrosis.
Nociceptive sensory neuron-derived NGF mediates MSCs excessive fibrotic commitment
Experimental validation of the aberrant commitment of MSCs toward fibrosis that sensory neuron-derived NGF driven was performed using Prrx1Tomato mice with intrathecal AAV-GFP-siNGF (Fig. 5A). Successful NGF inhibition in DRG neurons was confirmed by immunofluorescence (Supplementary Fig. 8A). At days 14 and 28 post-injury, NGF blockade attenuated fibrosis progression, evidenced by expanded tendon gaps, reduced collagen deposition, improved ROM, and suppressed Col1/Col3 expression (Supplementary Fig. 8B-D). Co-staining revealed diminished αSMA+Tomato+ cells (Fig. 5B,C), confirming NGF-driven MSC-myofibroblast differentiation.
A Prrx1Tomato mice experimental procedure and phenotypic diagram. B, C Double IF of αSMA (green) and Tomato (red) of peritendinous tissue from Prrx1Tomato mice intrathecal injected with AAV-GFP-siNGF or NC at 14 and 28 days after modeling (n = 6). Scale bars: 50 μm. D Transwell_co-culture strategy and immunofluorescent staining of Tomato (red) cells and α-SMA (green) cells from four groups in vitro. Scale bars: 20 μm. E, F Relative mRNA expression and protein expressions of Col1 and Col3 in cell experiment groups intrathecal injected with AAV-GFP-siNGF or NC (n = 3). G Double IF of CD31 (green) and Tomato (red) in Prrx1Tomato mice and NesTomato mice at 14 days after modeling (n = 6). Scale bars: low power image is 100 μm, and the high power image is 40 μm. H Double IF of αSMA (green) and Tomato (red) in Prrx1Tomato mice, αSMA (green) and Engrailed-1 (red) in WT mice at 14 days after modeling (n = 6). Scale bars: 50 μm. I Double IF of Engrailed-1 (green) and Tomato (red) in Prrx1Tomato mice at 14 days after modeling (n = 6). Scale bars: 50 μm. Yellow dashed line shows space between tendon and surrounding tissues. Black or white dashed line shows space occupied by fibrosis tissues. T represents tendon; AD represents fibrosis tissue; M represents muscle(mean ± SD; C/E–H: unpaired t-test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; A(mouse, injector)/D(mouse) created in Adobe. Author, Yanhao Li. (2025). Contact the author for publication if needed; A(tendon, fibrosis tissue, MSCs and myofibroblasts)/D(MSCs and neurons) created in BioRender. Author, li, y. (2025)) https://BioRender.com/0hlz1hf).
Mechanistic co-culture studies demonstrated direct NGF-MSC crosstalk. BMSCs from AAV-GFP-siNGF-treated mice exhibited impaired differentiation compared to controls (Fig.5D), with parallel reductions in Col1/Col3 expression (Fig. 5E,F). Orthogonal validation in Nestin-CreERT2::Rosa26-LSL-tdTomato(NesTomato) mice reinforced NGF’s role in MSC lineage specification (Supplementary Fig. 8E-H).
Single-cell analysis uncovered MSC heterogeneity. Prrx1+ MSCs preferentially adopted fibroblastic fates under sensory neuron-derived NGF signaling, whereas Nestin+ MSCs retained broader differentiation potential (Fig. 5G). Lineage tracing identified multiple αSMA+ progenitor sources that approximately 60% originating from Prrx1+ MSCs versus 22% from Engrailed-1+ precursors (Fig. 5H,I), highlighting cellular diversity within fibrotic niches.
These findings establish sensory neuron-derived NGF as an important regulator of MSC fibrotic commitment while mapping progenitor contributions to fibrosis. The mechanistic delineation of this axis provides actionable targets for post-traumatic fibrosis intervention.
Depletion of TrkA receptor in MSCs attenuates fibrosis formation
Our study highlights the critical role of sensory neuron-derived NGF in driving MSCs differentiation into fibrotic lineages via TrkA receptors. Single-cell analysis identified TrkA as a key receptor specifically expressed in PT subpopulations. Genetic ablation of TrkA in Prrx1-CreERT2::TrkA (Prrx1TrkA) mice (Fig. 6A; Supplementary Fig. 9A) significantly attenuated fibrosis at days 14 and 28, evidenced by expanded tendon gaps, reduced fibrotic area, and improved ROM (Fig. 6B; Supplementary Fig. 9B). Sensory innervation patterns remained intact despite TrkA knockout, with nonsignificant CGRP+ fiber increases suggesting compensatory adaptation (Fig. 6C,D).
A Prrx1TrkA mice experimental procedure. B Analysis of ROM of Prrx1TrkA mice injected with Veh or TMX at 14 and 28 days after modeling (n = 6). C, D Double IF of CGRP (red) in Prrx1TrkA mice (n = 6). Scale bars: low power image is 100 μm, and the high power image is 50 μm. E Double IF of αSMA (green) and Col1 (red) in Prrx1TrkA mice (n = 6). Scale bars: low power image is 100 μm, and the high power image is 50 μm. F IHC of Col1 and Col3 expression of peritendinous tissue from Prrx1TrkA mice injected with Veh or TMX at 14 and 28 days after modeling (n = 6). Scale bars: 100 μm. G RNA-Sequencing experimental procedure of Prrx1TrkA mice. H GO analysis of Prrx1TrkA mice. (I) KEGG analysis of Prrx1TrkA mice. J Heatmap of key genes expression in GO and KEGG analysis of Prrx1TrkA mice. (K) Heatmap of EMT related gene set expression of Prrx1TrkA mice. L IF of Engrailed-1 (green) in WT, AdvTrkA, AdvNGF, Prrx1TrkA mice injected with TMX at 14 days after modeling. (n = 6). Scale bars: 50 μm.Yellow dashed line shows space between tendon and surrounding tissues. Black or white dashed line shows space occupied by fibrosis tissues. T represents tendon; AD represents fibrosis tissue; M represents muscle; S represents skin. (mean ± SD; B/D–F: unpaired t-test, H/I/L: ANOVA with multiple comparisons; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; A (mouse)/G(mouse) created in Adobe. Author, Yanhao Li. (2025). Contact the author for publication if needed; G (tendon, fibrosis tissue and MSCs) created in BioRender. Author, li, y. (2025) https://BioRender.com/0hlz1hf; G (Mechanical and enzymatic digestion, FACS) created in BioRender. Author, li, y. (2025) https://BioRender.com/1qycjy7).
TrkA deletion reduced Col1+αSMA+ cells and suppressed Col1/Col3 expression (Fig. 6E,F; Supplementary Fig. 9C). Orthogonal validation using Nestin-CreERT2::TrkA (NesTrkA) mice confirmed conserved TrkA functionality across MSC subtypes (Supplementary Fig. 9D-G).
RNA sequencing of TrkA-null MSCs revealed pathway enrichment in wound healing, stem cell differentiation, and extracellular matrix regulation (ECM) (Fig. 6G-I), with downregulation of Hif1α, Twist1, Col3a1, and Acta2 (Fig. 6J). Mesenchymal markers (Twist1, Fn1, Vim) and invasion-related genes (Loxl1, Loxl3, Acta2) were selectively suppressed(Fig. 6K), indicating that the NGF-TrkA axis promotes MSCs differentiation into fibrotic lineages through a quasi-EMT mechanism, which enhances MSCs stemness, rendering them more prone to myofibroblast differentiation. Specificity analysis across AdvTrkA, AdvNGF, and Prrx1TrkA models demonstrated unchanged Engrailed-1+ precursor populations (Fig. 6L), confirming selective targeting of MSC to myofibroblast transition.
These findings show NGF-TrkA signaling as the master regulatory axis driving MSC fibrotic commitment through stemness modulation, providing mechanistically informed targets for antifibrotic therapy development.
Nociceptive sensory neuron-derived NGF mediates multi-model fibrosis through TrkA-Hif1a signaling
NGF is typically thought to be secreted by local cells to exert its axon-guiding function on peripheral nerves, but we have previously demonstrated that sensory neuron-derived NGF functions independently. Based on RNA sequencing (Fig. 6G,K), we hypothesized that TrkA activation induces the Hif1α-Twist1 signaling pathway to drive MSC differentiation into myofibroblast. Genetic manipulation of Hif1α through Prrx1-CreERT2::Hif1α (Prrx1Hif1α) mice(Hif1α knockout) and Prrx1-CreERT2::VHL (Prrx1VHL)(Hif1α over expression) mice(Supplementary Fig. 10A-C) confirmed its pivotal role in fibrosis. Hif1α deletion attenuated fibrosis at days14 and 28, evidenced by expanded tendon gaps, reduced fibrotic area, improved ROM, and diminished Col1+αSMA+ cells (Fig. 7A-F; Supplementary Fig. 10D). Local NGF administration failed to rescue this phenotype, establishing Hif1α as the essential intracellular mediator of NGF signaling. Pharmacological inhibition of TrkA by TPX-0005 recapitulated anti-fibrotic effects, while Hif1α over expression reversed TrkA blockade (Fig. 7C-F; Supplementary Fig. 10E-G). Orthogonal validation in Nestin-Cre::Hif1α (NesHif1α) mice corroborated these findings (Supplementary Fig. 10H-N). In vitro differentiation assays demonstrated necessity of Hif1α for NGF-induced MSC differentiation (Fig. 7G). Molecular profiling revealed coordinated Hif1α-Twist1 downregulation in Prrx1Hif1α, AdvNGF, and Prrx1TrkA MSCs (Fig. 7H-L), defining Twist1 as the downstream transcriptional regulator of the NGF-TrkA-Hif1α axis. ChIP-qPCR results showed that NGF-mediated activation of Hif1α does not directly drive the transcription of Col1/Col3; instead, it induces Col1/Col3 transcription through activating Twist1 (Supplementary Fig. 11A).
A, B experimental procedure and phenotypic diagram of Prrx1Hif1a mice and Prrx1VHL mice. C H&E and Masson staining and double IF of αSMA (green) and Col1 (red) of peritendinous tissue from WT mice injected with Veh or TPX0005, Prrx1Hif1α mice injected with Veh or NGF, Prrx1VHL mice injected with TPX0005 at 14 days after modeling (n = 6), and investigation analysis of ROM (n = 6). Scale bars of of H&E and Masson staining: 300 μm. Scale bars of double-immunofluorescent staining: 100 μm. D, E Analysis of fibrosis percentage and ROM of WT mice, Prrx1Hif1α mice and Prrx1VHL mice (n = 6). F Analysis of Col1+αSMA+ cells of WT mice, Prrx1Hif1α mice and Prrx1VHL mice (n = 6). G IF of Prrx1 (red) cells and α-SMA (green) cells from four groups in vitro. Scale bars: 20 μm. H The diagram of sorting Prrx1+MSCs from fibrosis tissues of four types of mice. I, J Protein expressions of Hif1α in fibrosis tissue of Prrx1TrkA mice and AdvNGF mice (n = 3). K, L Protein expressions of Twist1 in fibrosis tissue of WT mice, Prrx1Hif1a mice, Prrx1TrkA mice and AdvNGF mice (n = 3). M AdvNGF mice, Prrx1TrkA mice and Prrx1Hif1a mice experimental procedure and phenotypic diagram in a skin wound fibrotic healing model. N, O Representative macroscopic illustration and macroscopic quantification of individual wound areas at D0-14 in AdvNGF mice, Prrx1TrkA mice and Prrx1Hif1a mice (n = 6). P H&E stained sections of wound areas on D14 in AdvNGF mice, Prrx1TrkA mice and Prrx1Hif1a mice used for morphometric analysis of percentage of (Q) wound closure (length of newly formed epithelium (NFE)/length of NFE + length of gap between edges of wound epithelium (black dotted line) × 100), (R) area of HPE, (S) wound contraction (distance between wound border HFs (blue dotted line)), and (T) re-epithelialisation (length of NFE) (n = 6). Yellow dashed line shows space between tendon and surrounding tissues. Black or white dashed line shows space occupied by fibrosis tissues. T represents tendon; AD represents fibrosis tissue; M represents muscle(mean ± SD; D–F: ANOVA with multiple comparisons, J/L/O/Q–T: unpaired t-test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; A(mouse)/B(mouse)/H(mouse)/M created in Adobe. Author, Yanhao Li. (2025). Contact the author for publication if needed; A(tendon, fibrosis tissue, MSCs and myofibroblasts)/B(tendon, fibrosis tissue, MSCs and myofibroblasts)/G(MSCs)/H(tendon, fibrosis tissue and MSCs) created in BioRender. Author, li, y. (2025)) https://BioRender.com/0hlz1hf).
The skin wound model confirmed the functional conservation of sensory neuron-derived NGF. AdvNGF, Prrx1TrkA, and Prrx1Hif1a mice exhibited delayed fibrotic healing versus controls, despite preserved sensory nerve infiltration (Fig. 7M-O; Supplementary Fig. 12A). NGF/TrkA/Hif1α blockade reduced fibrotic wound closure (%), hyperproliferative epidermis (HPE), and neo-epithelium length (NFE), while increasing hair follicle spacing (Fig. 7N-T). Histology revealed diminished granulation tissue organization (Supplementary Fig. 12B), collagen deposition (Supplementary Fig. 12C,D), and αSMA+ myofibroblasts (Supplementary Fig. 12E). Lineage tracing in Prrx1Tomato mice with intrathecal AAV-GFP-siNGF recapitulated these phenotypes (Supplementary Fig. 13A-J), demonstrating reduced Tomato+αSMA+ cells in granulation tissue (Supplementary Fig. 13K).
These results delineate a linear signaling hierarchy (NGF/TrkA/Hif1α/Twist1) governing MSC fibrotic differentiation and define sensory neuron-derived NGF as an conserved regulator of MSC niche reprogramming during peripheral repair, providing therapeutic relevance across fibrotic pathologies.
Inhibition of TrkA signaling with TPX-0005 partially reverses excessive fibrosis and improve long-term healing outcomes
The moderate fibrosis is important for early wound closure, so reversing the fibrotic MSCs neurogenic niche can reduce excessive fibrosis while minimizing its impact on the healing process itself. Directly blocking nerves or downstream conserved transcription factor significantly impacts tendon healing, whereas intervening with TrkA avoids this issue (Supplementary Fig. 14A-C), so defining TrkA blockade (TPX-0005) as the optimal therapeutic window (Fig. 8A)40,41.
A Experimental procedure of WT mice injected with Veh or TPX-0005. B Representative macroscopic illustration of wound healing at D0-D14 in WT mice. C H&E analysis of D HPE area, E re-epithelialization, and F contraction (n = 6). Boxed regions in granulation tissue (delineated by dotted lines) are shown at higher magnification in insets. (n = 6). Scale bars: low power image is 500 μm, and the high power image is 100 μm. G H&E and Masson staining of peritendinous tissue from WT mice and investigation analysis of ROM (n = 6). Scale bars: 300 μm. H Histological fibrosis percentage of peritendinous tissue from WT mice (n = 6). I Quantitative analysis of ROM (n = 6). J, K IF of Prrx1 (red) and α-SMA(green) cells in WT mice (n = 6). Scale bars: 100 μm. L Relative mRNA expression of Col1 and Col3 in peritendinous tissue of WT mice (n = 6). M Quantitative analysis of maximum load and stiffness of repaired tendons in normal WT mice (n = 6). N Double IF of αSMA (green) and Col1 (red) in fibrosis tissue of Prrx1TrkA mice and NesTrkA mice (n = 6). Scale bars: low power image is 100 μm, and the high power image is 50 μm. O Investigation analysis of ROM of Prrx1TrkA mice and NesTrkA mice (n = 6). Black or white dashed line shows space occupied by fibrosis tissues. T represents tendon; AD represents fibrosis tissue; M represents muscle(mean ± SD; 8D–F/H/I/K–O: unpaired t-test; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; A(mouse) created in Adobe. Author, Yanhao Li. (2025). Contact the author for publication if needed; A(tendon) created in BioRender. Author, li, y. (2025)) https://BioRender.com/0hlz1hf).
TPX-0005 administration in skin wound models accelerated healing post-day7 (Fig. 8B) without compromising final closure (Fig. 8C; Supplementary Fig. 14D). Treated skin showed enhanced epithelial regeneration (increased HPE/NFE, reduced HF spacing) (Fig. 8C-F) and reduced collagen deposition (Supplementary Fig. 14E, F), indicating fibrotic-to-regenerative niche conversion. Tendon injury models mirrored this phenotype. TPX-0005 reduced fibrosis (Fig. 8G-I; Supplementary Fig. 14F,G), detained Prrx1+ MSCs, and suppressed αSMA+ myofibroblast accumulation (Fig. 8J,K). qPCR confirmed Col1/Col3 downregulation (Fig. 8L) and biomechanical improvement, especially in long term stage (Fig. 8M; Supplementary Fig. 14H).
Specificity validation in TrkA CKO mice (Prrx1TrkA and NesTrkA) demonstrated TrkA-dependent action of TPX-0005, with no additive anti-fibrotic effects (Fig. 8N,O). Safety profiling revealed no ROS dysregulation or apoptotic changes at therapeutic doses (Supplementary Fig. 14I-L).
These findings underscore niche-targeted intervention as a superior strategy: modulating fibrotic commitment without compromising healing integrity. TPX-0005 exemplifies translational potential for fibrosis management through precise TrkA-Hif1α axis inhibition (Fig. 9A).
A Nociceptive sensory nerves infiltrate the peripheral trauma area, mediating the biased repair response due to the formation of fibrotic mesenchymal stromal cells (MSCs) neurogenic niche. Mechanistically, nociceptive sensory neuron-derived nerve growth factor (NGF) activates the TrkA receptors in MSCs and triggers the Hif1α-Twist1 pathway, driving the differentiation of MSCs into myofibroblasts to mediate fibrosis. And TPX-0005 exerts therapeutic effects by inhibiting TrkA to reverse fibrosis (A(hand, DRG, nerve, microenvirenment) created in Adobe. Author, Yanhao Li. (2025). Contact the author for publication if needed; A(tendon, fibrosis tissue and other cells) created in BioRender. Author, li, y. (2025))ve sensory nerves infiltrate the peripheral trauma area, mediating the biased repair response due to the formation of fibrotic mesenchymal stromal cells (MSCs) neurogenic niche. Nociceptive sensory neuron-derived nerve growth factor activates the TrkA receptors in MSCs and triggers the Hif1α-Twist1 pathway, driving the differentiation of MSCs into myofibroblasts to mediate fibrosis. TPX-0005 exerts therapeutic effects by inhibiting TrkA to reverse fibrosis.https://BioRender.com/0hlz1hf).
Discussion
Myofibroblast generation and activation are central events in fibrogenesis following peripheral trauma42,43,44,45. Mounting evidence supports that MSCs serve as an important cellular reservoir for myofibroblast formation18,20. Upon injury, MSCs are abundantly recruited to the lesion site during the early fibrotic phase, where their fate is shaped by niche cues46,47. While our previous work revealed that macrophage-derived TGF-β guides MSCs recruitment, the signals that determine MSCs lineage commitment remain poorly understood21. In this study, we demonstrate that nociceptive sensory nerve innervation profoundly influences MSCs niche change for differentiation and fibrosis progression. Disrupting nociceptive sensory nerve domination significantly reduced peritendinous fibrosis and skin defect healing, while specifically inhibiting MSCs conversion into myofibroblasts. Mechanistically, we identified NGF secreted by nociceptive sensory nerves as a pivotal mediator for the formation of fibrotic MSCs neurogenic niche. Ablating NGF secretion from nociceptive sensory neurons mimicked the anti-fibrotic effects observed in nociceptive sensory-denervated models, highlighting that nociceptive sensory neuron-derived NGF is indispensable for directing MSCs toward a fibrotic fate in the peripheral trauma microenvironment. As for the heterogeneity of myofibroblast sources, in addition to the MSCs and Engrailed-1+ precursors we have already mentioned, tissue-resident fibroblasts are also considered a major origin. They can be activated under various pathological stimuli and differentiate into myofibroblasts48. Epithelial-mesenchymal transition can convert epithelial cells into myofibroblasts, and endothelial-mesenchymal transition can have a similar effect on endothelial cells48. Moreover, pericytes and fibrocytes, which originate from the bone marrow, can also contribute to the myofibroblast population49.
NGF has traditionally been viewed as a peripherally produced axon guidance molecule essential for sensory nerve patterning, maturation, and plasticity26,34,35. However, neurons can also synthesize NGF50,51,52, and the physiological significance of neuron-derived NGF remains largely unknown. As Lee et al. focus on local cell-derived NGF in trauma-induced HO53. Their work demonstrates that local NGF acts as an axon-guiding signal to operate in an upstream axon-recruiting mode that local NGF first establishes neural innervation, and the nerves then dictate progenitor fate toward osteogenesis. Our study expands this understanding by uncovering nociceptive sensory neuron-derived NGF as a distinct regulatory axis in tendon pathological fibrosis. This neuron-derived NGF exerts a function independent of local cell-secreted NGF to function in a downstream cell-fate-directing mode that sensory neurons themselves secrete NGF, which directly targets MSCs to regulate fibrosis. This alignment positions our findings within a broader musculoskeletal NGF framework, highlighting the sensory nerve-MSC axis as a context adaptive mechanism in pathological remodeling, providing a robust basis for targeted therapies against fibrosis, HO, and related disorders. Our data reveal that NGF secreted locally by nociceptive sensory nerves creates a niche enabling MSCs fibrotic differentiation. In contrast, NGF from other local cellular sources may be insufficient in either spatial concentration or localization to drive this lineage commitment. Using Prrx1Tomato mice as a model for in vivo MSCs fate mapping54,55,56, we showed that approximately 60% of αSMA+ myofibroblasts originate from MSCs.
Our work focused on TrkA as the primary receptor mediating NGF’s effects on MSCs. However, NGF also binds the p75 receptor, which has been implicated in fibrosis by promoting fibroblast migration and myofibroblast formation in liver and lung57,58,59. The interplay between TrkA and p75 signaling in post-peripheral trauma fibrosis remains an intriguing avenue for further research. Additionally, our results highlight the role of Hif1α as a key downstream effector. Hif1α is known to modulate collagen-modifying enzymes and ECM alignment, influencing fibroblast behavior, ECM remodeling, and resistance to anoikis60,61. Thus, the NGF-TrkA-Hif1α signaling axis emerges as a central pathway enabling MSCs to adopt a fibrotic lineage.
Advillin is exclusively expressed in peripheral nociceptive sensory neurons and not in tissues related to vision, audition, or metabolism. The tissue specificity of the Advillin (Avil) promoter has been clearly validated in multiple studies: CreERT2 driven by this promoter is primarily expressed in peripheral sensory neurons, mostly nociceptive sensory neurons within DRG and trigeminal ganglia, which are responsible for pain and temperature perception—with negligible expression in the central nervous system62. DRG staining experiments by Hu et al. further confirmed the strict tissue specificity of this promoter63. AdvDTA mice have been applied to investigate pathophysiological processes associated with peripheral sensory nerves. For instance, Pei et al. constructed AdvDTA mice to explore the regulatory role of sensory nerves in incisor homeostasis; they did not report severe metabolic alterations or central sensory dysfunction following Avil ablation64. Therefore, genetic ablation achieved via AdvDTA does not induce sensory loss in the visual or auditory systems.
In scRNA-seq, while the overall cellular composition of the injured tissue gradually shifts toward the sham group pattern at later timepoints, reflecting the initiation of tissue repair, the Prrx1⁺Col3a1⁺ cells persists as the dominant pro-fibrotic subset, suggesting its important role in maintaining pathological fibrosis. This aligns with clinical observations that peritendinous adhesions often stabilize rather than resolve spontaneously in the late stages of healing.
Sensory nerves are classically understood as afferent pathways transmitting microenvironmental cues to the central nervous system for systemic metabolic regulation4,31. We cannot rule out that central neural circuits may also influence fibrosis remodeling in the late stage of fibrosis formation. In fact, our previous studies have shown that sensory nerves can perceive prostaglandin E2 (PGE2) signals to transmit bone mineral density signals to the central nervous system (CNS)4 and can also convey tendon injury signals to the CNS65. However, whether other signals trigger communication between tendons and the CNS, and how these signals guide the formation of tendon fibrosis via the CNS, are questions that require further investigation in our subsequent research. Moreover, NGF is one of several nerve-derived factors, while we show NGF mediates the formation of MSC neurogenic niche, other neuropeptides may have opposing effects. For example, CGRP can inhibit cardiac fibrosis66,67, suggesting that sensory nerves could exert dual or context-dependent roles in mediating MSC neurogenic niche. We demonstrated that Prrx1 can serve as a reliable marker for MSCs in tendon fibrosis models. The MSCs labeled by Prrx1 are primarily involved in promoting the fibrotic process, as evidenced by a high proportion of Col3a1-expressing cells and a low proportion of Scx-expressing cells. However, Prrx1 cannot effectively distinguish stromal cell subpopulations, which limits its application in studies investigating subpopulation crosstalk. For instance, the epitenon acts as a key coordinator of the early tendon healing response68. A novel epitenon cell marker Slc1a3, identified by Anne E. C. Nichols et al., labels epitenon cells with higher specificity, explicitly analyzing the dual contribution of epitenon cell to fibrosis and healing after tendon injury39.
Methods
Human subjects
The research was approved by Wuxi Ninth People’s Hospital internal review board (KT201803) and Shanghai Sixth People’s Hospital internal review board (2023-YS-260). We strictly controlled the baseline characteristics of all patients. All patients underwent corresponding surgeries due to isolated mechanical trauma (traffic accidents, knife wounds) and had no underlying chronic diseases (such as diabetes mellitus or autoimmune diseases) or pre-injury infections. Samples in the control group were obtained from patients who required amputation surgery due to mechanical trauma. Human peritendinous tissues at the other three time points were all derived from 12 patients. These specimens were divided into three groups: 2–3 days post-tendon injury, 8–12 days post-tendon injury, and more than 3 months post-tendon injury. For the acquisition of control group samples, we first performed emergency amputation surgery. Subsequently, from the amputated tissues, we selected regions that remained relatively intact, were not subjected to mechanical trauma, and had no obvious other diseases to obtain the samples, and the normal status of collagen tissue was confirmed histologically (Fig. 1C). For samples at the other three time points, tissues were collected during elective surgery or reoperation, and only peritendinous tissues obtained during adhesiolysis were harvested, with no damage to tendon tissues or surrounding tissues. Additionally, a list of sources for all samples (with patient names removed) is provided, including information on age, gender, diagnosis, and treatment (Supplementary Tables 1–4). After surgical removal, the tissue samples were divided into three parts: the part intended for scRNA-seq was immediately transferred to serum-free DMEM on ice and subjected to standardized scRNA-seq processing right away; the samples used for pathological studies were quickly placed into 4% paraformaldehyde (PFA) for fixation over 24 h, followed by subsequent processing for frozen sectioning or paraffin sectioning; the last part was stored in liquid nitrogen for protein-level research.
TAM was assessed to quantify functional recovery of the injured tendon, following modified protocols from American Society for Surgery of the Hand (ASSH) guidelines, with all measurements performed by two blinded independent orthopedic surgeons to minimize bias. For human participants (traumatic tendon injury groups: 8–12D, >3 M; the control group measures normal volunteers), TAM of the affected/corresponding finger was measured via a standard goniometer, calculated as the sum of active flexion (metacarpophalangeal + proximal interphalangeal + distal interphalangeal joints) minus active extension deficit (difference between full passive and active extension).
Mice and in vivo treatment
All mouse experiments were performed in accordance with the guidelines published by the NIH and the Shanghai Sixth People’s Hospital (Shanghai, China) internal review board. All mice were maintained in the animal facility of the Shanghai Sixth People’s Hospital. C57BL/6 J (wild-type) mice (referred to as WT in the test) were purchased from the Jackson Laboratory.
All genetically modified mice were constructed using the Cre/loxP system. Advillin-CreERT2, TrkA loxp, Nestin-CreERT2, Rosa-LSL-tdTomato, Hif1α loxp, Prrx1-CreERT2, VHL loxp, NGF loxp, Rosa26-DTA were purchased from the Jackson Laboratory and GemPharmatech Co., Ltd. Homozygous. Then, Advillin-CreERT2::TrkA mice, Nestin-CreERT2:: Rosa-LSL-tdTomato mice, Nestin-CreERT2::TrkA mice, Nestin-Cre::Hif1α mice, Prrx1-CreERT2::TrkA mice, Prrx1-CreERT2::Hif1α mice, Prrx1-CreERT2::VHL mice, Advillin-CreERT2::NGF mice, Advillin-CreERT2::DTA mice, Prrx1-CreERT2::Rosa-LSL-tdTomato mice were generated by crossing them. The PA model (n = 6 per group) was established in eight-week-old genetically modified male mice treated with Veh or TMX (100 mg/kg, i.p., for 5 consecutive days before modeling). The mice were euthanized after 14 days and 28 days for further analysis. All mice were served with mouse housing conditions (temperature: 22 ± 2 °C, humidity: 50 ± 10%, 12 h light/dark cycle) and food type (standard rodent chow: Jiangsu Xietong Pharmaceutical Bioengineering, Catalog No. XTI01SL-002).
For flexor tendon injury mode, eight-week-old male mice were anesthetized by inhalation of isoflurane. After anesthetization, the skin on the right hind paw was disinfected with 75% ethanol, the FDL tendon was exposed through a posterior midline incision and transversally transected, and the tendon was then repaired with a modified Kessler pattern to create the tendon trauma model. The wound was closed with 5-0 sutures. All mice were treated identically and allowed to move freely in the cage (Supplementary Fig. 15A)69,70,71,72,73.
For skin wound model, mice were anesthetized with 5% isoflurane and maintained on 3%. The dorsal skin was shaved, cleaned, and disinfected, and a 5 mm full-thickness wound was created. Post-operative analgesia was provided via buprenorphine in drinking water for 5 days. Sterile dressings were applied and changed on days 3, 7, 10, and 14.
For retrograde tracing of DRG, CTB (Sigma-Aldrich, C9903) was dissolved in sterile PBS to prepare a 1% (w/v) working solution, freshly prepared for each experiment to maintain tracer activity. For tracer injection, mice with established FDL tendon injury model were anesthetized with 1.5% isoflurane at 14D. A 3 mm longitudinal incision was made at the posterior midline of the right hind paw (2 mm proximal to the metatarsophalangeal joint) to expose the tendon repair site. Using a 10 μL Hamilton microsyringe, 2 μL of the 1% CTB solution was injected into the repair site via a multi-point, shallow-penetration approach: 3 injection points were distributed at the 12 o’clock, 4 o’clock, and 8 o’clock positions of the tendon cross-section, with each injection penetrating 0.5 mm into the tendon tissue (to avoid damaging underlying bone or neurovascular bundles) and delivering 0.6–0.7 μL of tracer. The syringe needle was retained in place for 30 seconds post-injection to prevent tracer reflux, after which the incision was closed with 5-0 absorbable sutures. Mice were housed under standard conditions for 7 days to allow retrograde transport of CTB to the DRG; thereafter, mice were euthanized, and all DRG segments were harvested for subsequent immunofluorescence analysis to identify CTB-positive neurons.
For treatment experiments, TPX-0005 was dissolved in DMSO at a concentration of 20 ng/ml. Eight-week-old male WT mice (n = 6 per group) were locally treated with TPX-0005 every 7 days, with 10 μl each time.
For intrathecal injection of mice, the fur on the back of each mouse was shaved between the bilateral pelvic girdles, and then the skin was disinfected with 75% ethanol. The L5-L6 intervertebral space is generally located on the cross point of the midline and the connection line of the bilateral anterior superior iliac spines. The needle was gently inserted with a slight tilt of 70–80° at the cross point, and the syringe was kept in the central sagittal plane. Then, the needle was slipped into the intervertebral space, which was considered successful when the tail slightly shook. NC or AAV-siNGF was then intrathecally injected (8 μl per mouse, 4.41 × 10⁸ TU/mL) 3 weeks before modeling.
The drugs and compounds used in this study were as follows: AAV-GFP-siNGF (OBIO Technology (Shanghai) Co., Ltd., custom service); TPX-0005 (MACKLIN, T872765); tamoxifen (Sigma, T5648); nerve growth factor(NGF)(Sigma, SRP4304); capsaicin(Sigma, 211275).
Phenotype assessment
For ROM assessment, we amputated the hind limb to the knee and removed the skin from the ankle (J = 6 per group). First, the tibia was fixed with clips. Then, the FDL was pulled with a weight of 19 grams, and the angle change of the metatarsophalangeal joint was measured74.
The hot plate test was conducted to assess nociceptive sensitivity. Mice were placed on a heated plate maintained at 50 °C (IITC Inc., Model 39). The latency to paw withdrawal (in seconds) was recorded as the time from when the mouse was placed on the plate until it lifted or licked its hind paw, with a 30-second cutoff to prevent tissue damage. Each mouse was tested in triplicate with a 5-minute interval between trials, and the mean value was calculated for analysis.
Forelimb grip strength was measured using a digital grip strength meter (Columbus Instruments). Mice were gently pulled by the tail until they released their grip from the metal grid, and the maximum force (in grams) was recorded. Each mouse performed 5 consecutive trials with 1-minute rest periods between attempts, and the highest value from valid trials (excluding those where the mouse clasped the tail or struggled excessively) was used for statistical analysis.
Biomechanical testing
We used the dynamic mechanical analyzer Q800 (TA instrument) to collect the maximum load and stiffness of the repaired tendon on day 14 and day 28 after repair. The proximal end was stretched at a rate of 30 mm/min until it broke, and the force displacement curve was obtained by digital calculation(Supplementary Fig. 16A). Ultimate tensile strength (UTS) was calculated as the maximum force at failure divided by the cross-sectional area of the tendon (measured using a digital caliper), expressed in megapascals (MPa).
Quantitative real-time polymerase reaction chain (qPCR)
Total RNA was purified from tissue or cells in culture using TRIzol (Invitrogen, 15596026) and then reverse transcribed into cDNA according to the manufacturer’s protocol. qPCR was performed using SYBR qPCR Master Mix (Vazyme, Q111) on a CFX Connect instrument (Bio-Rad) with analysis of dissociation curves for every experiment. The sequences of the primers used for each gene were from PrimerBank.
Specimen collection
Mice were euthanized by carbon dioxide inhalation followed by body perfusion with 4% paraformaldehyde through the femoral artery for 5 minutes. The feet were then retrieved, immersed in 4% PFA for 24 h and decalcified in 0.5 M ethylenediaminetetraacetic acid (VWR, 0105) (pH 7.5) for one week. Finally, the specimens were dehydrated with 30% sucrose (Sigma‒Aldrich, S9378) at 4 °C before embedding in optimal cutting temperature compound (OCT) at -80 °C. For qPCR, WB and ELISA, the tissue we collected is peritendon tissue, which is dissected away from the peritendon, not the whole tendon.
Histochemistry and immunofluorescence analysis
Frozen sagittal sections were cut into 8 μm (for routine staining) and 20-μm (for nerve staining) sections. The sections were stained with HE, Masson and IHC staining. Histologic adhesion scoring systems and the percentage of fibrosis area system were used to assess fibrosis.
IHC staining and immunofluorescence staining of human or mouse frozen sections were performed. Briefly, the sections were hydrated with PBS before blocking endogenous peroxidase and nonspecific sites with 5% blocking serum (Solarbio, SL050); then, the sections were incubated with primary antibodies at 4 °C overnight. Subsequently, the secondary antibodies were incubated with the sections at room temperature for 1 h, and for IHC, the sections were mounted with DAB coloration, and then label the nuclues with Hematoxylin; for immunofluorescence staining, the sections were mounted with Antifade Mounting Medium with DAPI (Beyotime, P0131). Images were captured with a Zeiss 780 confocal microscope. The quantification of positive cells was performed using ImageJ software. Specifically, all αSMA+ cells were referred to as αSMA+ myofibroblasts, as we eliminated the αSMA+ vessels in the statistical analysis.
The adhesion grading scale and healing grading scale were assessed as before.(Supplementary Fig. 17A; Supplementary Fig. 18A)
The primary antibodies used in this section were as follows: α-smooth muscle actin (αSMA, Cell Signaling, 19245 and 48938, 1:200); calcitonin gene-related peptide (CGRP, Cell Signaling, 14959, 1:200); nerve growth factor (NGF, Abcam, 52918, 1:150); neuron nucleus (NeuN, Cell Signaling, 94403 s, 1:100); Nestin (Aves Labs, NES, 1:200); TrkA (BIOSS, bs-0193R, 1:200); and Hif1α (NOVUS, H1alpha67, 1:300); Prrx1(NOVUS, NBP1-06067, 1:200); Collagen 1(Abcam, 138492, 1:200 for IF, 1:500 for WB); Collagen 3(Abcam, 184993, 1:200 for IF, 1:500 for WB); PGP9.5(Cell Signaling, 60702, 1:200); VHL(Abcam, 140989, 1:500); Engrailed-1(Sigma, AB5732, 1:200).
In vitro study
Primary MSCs were collected from the bilateral tibias. The mouse was euthanized by carbon dioxide, and then the bilateral legs were removed and placed in 75% ethanol for 30 minutes. Subsequently, the medullary cavity was rinsed with 1 ml of sterile PBS, and then the cells were collected by centrifugation and transferred to a 10 cm dish with complete medium (alpha-MEM, 10% FCS, 1% penicillin‒streptomycin). After 24 h, the dishes were washed with PBS to eliminate nonadherent cells. Then, flow cytometry was applied to sort Nestin+ cells. The sorted cells were passaged every 2 days. Cells at passages 3 to 6 were used for the experiment. In in vitro experiments, MSCs were induced using recombinant mouse beta-NGF. The induction was performed with a NGF dose of 10 ng/mL, and 10 μL of the NGF-containing medium was added per well. The medium was refreshed every 2 days, and the induction period lasted for 1 week.
DRG cells were isolated from MSCs collected from the L4-L6 levels of the spine levels in the same mice via digestion and trituration with collagenase I (Roche, 10103578001) at 37 °C. After isolation, the cells were resuspended and seeded into the upper chamber of a 0.4 μm Transwell system for the Transwell coculture experiment.
The drugs and compounds used in this study were as follows: AAV-GFP-siNGF (OBIO) and recombinant mouse beta-NGF (Novoprotein, 120AA).
Analysis of NGF concentrations and serum glucose
The concentration of NGF in the fibrosis tissues around the repaired tendon was determined with Mouse NGF-β ELISA Kit (BOSTER, EK0470).
Prior to euthanasia, mice were fasted for 4 h. A small drop of blood was collected from the tail vein using a sterile lancet, and the glucose concentration (in mmol/L) was immediately measured with a handheld glucose meter. Three measurements were performed within 5 minutes, and the average value was recorded to ensure accuracy.
Protein expression analysis
The protein expression analysis was performed by western blot targeting Hif1α (Cell Signaling, 36169S, 1:500), Twist1(Abcam, 50887, 1:400), actin (Abcam, ab8226, 1:500), Collagen 1(Abcam, 138492, 1:500) and Collagen 3(Abcam, 184993, 1:500). The results were visualized by automatic chemiluminescence imaging analysis system and analyzed by ImageJ software for normalization to that of actin.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed using the Simple ChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology, 9003 s). Sheared chromatin was then immunoprecipitated with Hif1a (Abcam, 308433), Twist1 (Active Motif, 91323) and IgG (Cell Signaling Technology, 2729) antibodies. SYBR green master mix (Applied Biosystems) was used to measure amplification of DNA using QuantStudio 3 Fast Real-Time PCR system (Applied Biosystems). After normalization to the Input DNA, the amount of output DNA of each target protein was calculated by subtracting that of the IgG control.
Flow cytometry analysis
The sample was minced and digested into single cells using collagenase type I. Subsequently, incubate with fluorescently labeled αSMA (Abcam, 208844, 1:500) and Col1 (Abcam, 310848, 1:500), Prrx1(NOVUS, NBP1-06067, 1:200) flow cytometry antibodies at room temperature for 1 hour, then wash with PBS, and proceed to cell sorting.
ROS detection
Cells were treated and resuspended with 1 mL 10 μM Dichlorodihydrofluorescein diacetate (DCFH-DA) (Beyotime, S0035S) diluted with the serum-free medium. Cells were incubated for 30 min at 37 °C in a cell incubator. The cells were washed with a serum-free cell culture medium three times and detected by flow cytometry. FlowJo was used to analyze the fluorescence intensity.
Cell cycle detection
The cells were cultured in 6 well plate and treated with TPX-0005 for 24 h. After collecting the cells, they were washed with cold PBS and fixed in 70% ethanol for 30 min at 4 °C. After being washed with PBS, the fixed cells were stained with the PI cocktail (50 g/ml PI and 50 g/ml RNase) for 30 min at room temperature in the dark. Cells were collected and detected by flow cytometry, and data were analyzed by FlowJo.
Mitochondrial membrane potential (MMP)
The MMP was assessed using the JC-10 fluorescent probe (YEASEN, 40707ES) in line with the manufacturer’s protocol. After preparing the working buffer, cells were collected and incubated with JC-10 working buffer (500 μL) at 37 °C for 30 min away from light. Cells were washed with PBS and determined by flow cytometry. The fluorescence intensity of FL1 and FL2 channels was used to measure MMP.
Apoptosis detection by Annexin V-FITC/PI
The BMSCs were treated with TPX-0005 for 24 h. Cells were collected, resuspended and washed twice with PBS. After centrifugation, cells then were resuspended in 195 μL Annexin V-FITC binding buffer. After adding Annexin V-FITC (5 μL) and PI (10 μL), cells were incubated at room temperature for 15 min away from light. Cells were collected and detected by CytoFLEX LX, and data were analyzed by FlowJo.
Single-cell RNA sequencing analysis
For the sham group (normal tendon), tissues were collected from the region spanning the heel to the area above the digital fork; for injured groups (3D, 7D, 14D, 28D post-injury), the entire scar region was carefully separated from surrounding tissues, and the scar tissue plus ~1 mm of tendon stumps on both sides of the repair site were collected.
Single-cell RNA-seq libraries were generated using the SeekOne® MM system and single-cell 3’ library preparation kit, following the manufacturer’s instruction program. SEEKGENE Biotechnology Co., Ltd. performed scRNA-Seq data analysis with the SeekSoul Online Platform. Used the Seurat package for cell normalization and regression. Used the fastMNN function in the R package scran to apply the mutual nearestneighbor method to correct for batch effects among specimens. Finally, got the unsupervised cell cluster result by using the graph-based cluster method (resolution = 0.8). For quality control (QC) standards, low-quality cells with fewer than 200 detected genes or mitochondrial gene content >5% were excluded (Supplementary Fig. 5A). After QC, the final valid cell numbers for each group were: 14,125 cells (sham group), 17,387 cells (3D group), 20,686 cells (7D group), 19,401 cells (14D group), and 20,633 cells (28D group). Sequencing was performed using the 10× Genomics Chromium Single Cell 3’ v3.1 platform, with an average read depth of ~50,000 reads per cell and a median gene count per cell of 1,800–2,200—parameters that align with standard scRNA-seq requirements for musculoskeletal tissue studies. The annotation of all cells was performed with reference to the public gene database of SeekGene and published articles39,71. aSMA transcript data in the re-analyzed scRNA-seq results is only used as a preliminary indicator of myofibroblast activation, and all conclusions about myofibroblasts are supported by protein-level, functional, and lineage tracing evidence. Based on the gene set which collected from Molecular signatures database and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database, QuSAGE (2.16.1) was used to characterize the relative activation of the given gene set. We used GO enrichment analysis to performed functional enrichment analysis. Each enriched ontology hierarchy (false discovery rate (FDR) < 0.05) conform to the highest significance value and the highest specificity. Single-cell trajectory analysis was performed by utilizing Monocle2/3. The fibroblast clusters were taken from Seurat data then the differentially expressed genes based on the fold-change expression were selected for Monocle to order the cells by applying DDR-Tree. We got branch fate-determined gene analysis by utilizing branch expression analysis modeling. Using CytoTracy to analyze the cell differentiation status of single-cell RNA sequencing data. Firstly, extract gene expression information from the data, and then calculate the number of unique genes expressed in each cell to evaluate the diversity of gene expression. By measuring the diversity of gene expression, the degree of cell differentiation can be inferred, where cells with higher diversity are in a more primitive differentiation state, while cells with lower diversity are more mature. Regulon analysis was performed using the SCENIC package (v1.3.1). Briefly, gene regulatory networks were inferred by (1) identifying candidate TF-target gene pairs based on TF motif enrichment in gene promoters; (2) building co-expression modules for each TF; (3) pruning modules to retain only motif-supported targets to form regulons; and (4) quantifying regulon activity in each cell using the AUCell algorithm. Only regulons with AUCell scores > 0.5 in at least 10% of cells were retained for subsequent analysis. By calculating the Regulon Specificity Score (RSS) for each cell population to analyze the corresponding relationship between Regulons and cell populations.
Animal stratification and statistical analysis
Animals were randomly assigned to experimental treatment groups and outcome groups prior to surgery, with each data point representing one mouse. Images were analyzed using Fiji software. Regions of interest (ROIs) were selected based on target signals, which allowed us to exclude αSMA⁺ or Prrx1-Td⁺ signals from the tendon (T) and muscle (M) regions. Additionally, we subtracted αSMA⁺ signals derived from pericytes with obvious vascular morphology within the AD region. These signals were distinguished by their characteristic linear, tube-like distribution consistent with vascular structures. The remaining αSMA⁺ or Prrx1-Td⁺ signals were subjected to mean fluorescence intensity (MFI) quantification, and the resulting data were analyzed using independent samples t-tests to verify statistical significance. MFI values were calculated by Fiji Measure function, with background signal subtracted to ensure accuracy75. We would like to clarify that the “AD” described in the Figure is a collective term for this pathological region, not a single component (pure fibrotic tissue or expanded epitenon alone). After tendon injury, disordered cell proliferation and inflammation drive the expansion of the tendon epitenon/peritenon. This expansion cross-links with excessive collagen deposition mediated fibrosis, ultimately forming the overall adhesive structure. All statistical analyses were performed using GraphPad Prism 9. For all quantitative results, the Shapiro-Wilk test was used to assess data normality (p < 0.05). Differences between the experimental group and the control group at each time point were determined by unpaired t-test, one-way analysis of variance (ANOVA) with multiple comparisons, or least significant difference (LSD) test for comparisons between two groups. Data are presented as mean ± standard deviation (SD), and statistical significance is indicated on the figures using the following conventions: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Mouse single-cell RNA-sequencing data have been deposited at SRA: For scRNA-seq of AdvNGF: PRJNA1224531, PRJNA1224599; For scRNA-seq of Veh: PRJNA1224718, PRJNA1225136; For scRNA-seq of WT: PRJNA976191; For RNA-seq of Prrx1TrkA and control: PRJNA1201315, and they are publicly available after the date of publication. All other relevant data supporting the key findings of this study are available within the article, its Supplementary Information and Source Data file. Source data are provided with this paper.
References
Vazir, A., Fox, K., Westaby, J., Evans, M. J. & Westaby, S. Can we remove scar and fibrosis from adult human myocardium? Eur. Heart J. 40, 960–966 (2019).
Zhang, J. et al. A pulsatile release platform based on photo-induced imine-crosslinking hydrogel promotes scarless wound healing. Nat. Commun. 12, 1670 (2021).
Zhang, Q. et al. Correction to “down-regulating scar formation by microneedles directly via a mechanical communication pathway”. ACS Nano 17, 11070–11071 (2023).
Chen, H. et al. Prostaglandin E2 mediates sensory nerve regulation of bone homeostasis. Nat. Commun. 10, 181 (2019).
Huang, S. et al. Lgr6 marks epidermal stem cells with a nerve-dependent role in wound re-epithelialization. Cell Stem Cell 28, 1582–1596.e1586 (2021).
Pinho-Ribeiro, F. A., Verri, W. A. & Chiu, I. M. Nociceptor sensory neuron-immune interactions in pain and inflammation. Trends Immunol. 38, 5–19 (2017).
Snyder, D. J. & Bartoshuk, L. M. Oral sensory nerve damage: causes and consequences. Rev. Endocr. Metab. Disord. 17, 149–158 (2016).
Yoneda, T., Hiasa, M., Okui, T. & Hata, K. Sensory nerves: a driver of the vicious cycle in bone metastasis? J. Bone Oncol. 30, 100387 (2021).
Lucas, D. et al. Chemotherapy-induced bone marrow nerve injury impairs hematopoietic regeneration. Nat. Med. 19, 695–703 (2013).
Fukuda, T. et al. Sema3A regulates bone-mass accrual through sensory innervations. Nature 497, 490–493 (2013).
Okano, M. et al. Interleukin-33-activated neuropeptide CGRP-producing memory Th2 cells cooperate with somatosensory neurons to induce conjunctival itch. Immunity 55, 2352–2368.e2357 (2022).
Huang, W.-K. et al. Generation of hypothalamic arcuate organoids from human induced pluripotent stem cells. Cell Stem Cell 28, 1657–1670.e1610 (2021).
Kramann, R. et al. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell 16, 51–66 (2015).
Hasani-Sadrabadi, M. M. et al. An engineered cell-laden adhesive hydrogel promotes craniofacial bone tissue regeneration in rats. Sci. Transl. Med. 12, eaay6853 (2020).
Soliman, H. et al. Multipotent stromal cells: one name, multiple identities. Cell Stem Cell 28, 1690–1707 (2021).
Wang, Y., Fang, J., Liu, B., Shao, C. & Shi, Y. Reciprocal regulation of mesenchymal stem cells and immune responses. Cell Stem Cell 29, 1515–1530 (2022).
Wang, Y., Chen, X., Cao, W. & Shi, Y. Plasticity of mesenchymal stem cells in immunomodulation: pathological and therapeutic implications. Nat. Immunol. 15, 1009–1016 (2014).
El Agha, E. et al. Mesenchymal stem cells in fibrotic disease. Cell Stem Cell 21, 166–177 (2017).
Hoang, D. M. et al. Stem cell-based therapy for human diseases. Signal Transduct. Target Ther. 7, 272 (2022).
Li, Y. et al. Neutralization of excessive levels of active TGF-β1 reduces MSC recruitment and differentiation to mitigate peritendinous adhesion. Bone Res. 11, 24 (2023).
Zhang, Q., Yang, Y., Yildirimer, L., Xu, T. & Zhao, X. Advanced technology-driven therapeutic interventions for prevention of tendon adhesion: design, intrinsic and extrinsic factor considerations. Acta Biomater. 124, 15–32 (2021).
Freedman, B. R. et al. Enhanced tendon healing by a tough hydrogel with an adhesive side and high drug-loading capacity. Nat. Biomed. Eng. 6, 1167–1179 (2022).
Cai, C. et al. Self-healing hydrogel embodied with macrophage-regulation and responsive-gene-silencing properties for synergistic prevention of peritendinous adhesion. Adv. Mater. 34, e2106564 (2022).
Altay, M. A., Erturk, C. & Isikan, U. E. De Quervain’s disease treatment using partial resection of the extensor retinaculum: a short-term results survey. Orthop. Traumatol. Surg. Res 97, 489–493 (2011).
Murray, E., Challoumas, D., Putti, A. & Millar, N. Effectiveness of sodium hyaluronate and ADCON-T/N for the prevention of adhesions in hand flexor tendon surgery: a systematic review and meta-analysis. J. Hand Surg. Am. 47, 896.e891–896.e820 (2022).
O’Keeffe, G. W., Gutierrez, H., Pandolfi, P. P., Riccardi, C. & Davies, A. M. NGF-promoted axon growth and target innervation requires GITRL-GITR signaling. Nat. Neurosci. 11, 135–142 (2008).
Zhang, Y. et al. Implant-derived magnesium induces local neuronal production of CGRP to improve bone-fracture healing in rats. Nat. Med. 22, 1160–1169 (2016).
Liu, X., Yan, D. & Guo, S.-W. Sensory nerve-derived neuropeptides accelerate the development and fibrogenesis of endometriosis. Hum. Reprod. 34, 452–468 (2019).
Li, Z. et al. Fracture repair requires TrkA signaling by skeletal sensory nerves. J. Clin. Investig. 129, 5137–5150 (2019).
Appelt, J. et al. The neuropeptide calcitonin gene-related peptide alpha is essential for bone healing. EBioMedicine 59, 102970 (2020).
Hu, B. et al. Sensory nerves regulate mesenchymal stromal cell lineage commitment by tuning sympathetic tones. J. Clin. Investig. 130, 3483–3498 (2020).
Ye Li, N. et al. Biodegradable magnesium combined with distraction osteogenesis synergistically stimulates bone tissue regeneration via CGRP-FAK-VEGF signaling axis. Biomaterials 275, 120984 (2021).
Wu, Q. et al. Nerve growth factor (NGF) with hypoxia response elements loaded by adeno-associated virus (AAV) combined with neural stem cells improve the spinal cord injury recovery. Cell Death Discov. 7, 301 (2021).
Eu, W. Z. et al. The effect of nerve growth factor on supporting spatial memory depends upon hippocampal cholinergic innervation. Transl. Psychiatry 11, 162 (2021).
Scott-Solomon, E. & Kuruvilla, R. Prenylation of axonally translated Rac1 controls NGF-dependent axon growth. Dev. Cell 53, 691–705.e697 (2020).
Liu, H. et al. Prrx1 marks stem cells for bone, white adipose tissue and dermis in adult mice. Nat. Genet. 54, 1946–1958 (2022).
Lee, K.-W. et al. PRRX1 is a master transcription factor of stromal fibroblasts for myofibroblastic lineage progression. Nat. Commun. 13, 2793 (2022).
Qin, T. et al. A population of stem cells with strong regenerative potential discovered in deer antlers. Science 379, 840–847 (2023).
Nichols, A. E. C. et al. Epitenon-derived progenitors drive fibrosis and regeneration after flexor tendon injury in a spatially-dependent manner. Nat. Commun. 16, 5448 (2025).
Carr, R. & Frings, S. Neuropeptides in sensory signal processing. Cell Tissue Res. 375, 217–225 (2019).
Wang, Z., Song, K., Kim, B. S. & Manion, J. Sensory neuroimmune interactions at the barrier. Mucosal Immunol. 17, 1151–1160 (2024).
Nagpal, V. et al. Response to letter regarding article, “MiR-125b is critical for fibroblast-to-myofibroblast transition and cardiac fibrosis. Circulation 133, e714 (2016).
Lagares, D. et al. Corrigendum: ADAM10-mediated ephrin-B2 shedding promotes myofibroblast activation and organ fibrosis. Nat. Med. 23, 1499 (2017).
Cocco, E., Scaltriti, M. & Drilon, A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 15, 731–747 (2018).
Drilon, A. et al. Repotrectinib (TPX-0005) is a next-generation ROS1/TRK/ALK inhibitor that potently inhibits ROS1/TRK/ALK solvent- front mutations. Cancer Discov. 8, 1227–1236 (2018).
Tang, Y. et al. TGF-beta1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat. Med. 15, 757–765 (2009).
Maxson, S., Lopez, E. A., Yoo, D., Danilkovitch-Miagkova, A. & Leroux, M. A. Concise review: role of mesenchymal stem cells in wound repair. Stem Cells Transl. Med. 1, 142–149 (2012).
Darby, I. A., Zakuan, N., Billet, F. & Desmoulière, A. The myofibroblast, a key cell in normal and pathological tissue repair. Cell Mol. Life Sci. 73, 1145–1157 (2016).
Wynn, T. A. Cellular and molecular mechanisms of fibrosis. J. Pathol. 214, 199–210 (2008).
Delcroix, J.-D. et al. NGF signaling in sensory neurons: evidence that early endosomes carry NGF retrograde signals. Neuron 39, 69–84 (2003).
Tomlinson, R. E. et al. NGF-TrkA signaling in sensory nerves is required for skeletal adaptation to mechanical loads in mice. Proc. Natl. Acad. Sci. USA 114, E3632–E3641 (2017).
Peng, T. et al. Nerve growth factor (NGF) encourages the neuroinvasive potential of pancreatic cancer cells by activating the warburg effect and promoting tumor derived exosomal miRNA-21 expression. Oxid. Med. Cell Longev. 2022, 8445093 (2022).
Lee, S. et al. NGF-TrkA signaling dictates neural ingrowth and aberrant osteochondral differentiation after soft tissue trauma. Nat. Commun. 12, 4939 (2021).
Zhen, G. et al. Inhibition of TGF-β signaling in mesenchymal stem cells of subchondral bone attenuates osteoarthritis. Nat. Med. 19, 704–712 (2013).
Nobre, A. R. et al. Bone marrow NG2+/Nestin+ mesenchymal stem cells drive DTC dormancy via TGFβ2. Nat. Cancer 2, 327–339 (2021).
Velloso, F. J. et al. Modestly increasing systemic interleukin-6 perinatally disturbs secondary germinal zone neurogenesis and gliogenesis and produces sociability deficits. Brain Behav. Immun. 101, 23–36 (2022).
Micera, A. et al. Nerve growth factor displays stimulatory effects on human skin and lung fibroblasts, demonstrating a direct role for this factor in tissue repair. Proc. Natl. Acad. Sci. USA 98, 6162–6167 (2001).
Shen, W.-Y. et al. Up-regulation of proBDNF/p75NTR signaling in antibody-secreting cells drives systemic lupus erythematosus. Sci. Adv. 8, eabj2797 (2022).
Kendall, T. J. et al. p75 Neurotrophin receptor signaling regulates hepatic myofibroblast proliferation and apoptosis in recovery from rodent liver fibrosis. Hepatology 49, 901–910 (2009).
Gilkes, D. M., Bajpai, S., Chaturvedi, P., Wirtz, D. & Semenza, G. L. Withdrawal: Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J. Biol. Chem. 299, 105144 (2023).
Rohwer, N. et al. Hypoxia-inducible factor 1alpha mediates anoikis resistance via suppression of alpha5 integrin. Cancer Res. 68, 10113–10120 (2008).
Zurborg, S. et al. Generation and characterization of an Advillin-Cre driver mouse line. Mol. Pain. 7, 66 (2011).
Pei, F. et al. Sensory nerve niche regulates mesenchymal stem cell homeostasis via FGF/mTOR/autophagy axis. Nat. Commun. 14, 344 (2023).
Hasegawa, H., Abbott, S., Han, B.-X., Qi, Y. & Wang, F. Analyzing somatosensory axon projections with the sensory neuron-specific Advillin gene. J. Neurosci. 27, 14404–14414 (2007).
Zhang, X. et al. Myeloid cells and sensory nerves mediate peritendinous adhesion formation via prostaglandin E2. Adv. Sci. 11, e2405367 (2024).
Li, W. et al. CGRP derived from cardiac fibroblasts is an endogenous suppressor of cardiac fibrosis. Cardiovasc. Res. 116, 1335–1348 (2020).
Li, W.-Q. et al. Calcitonin gene-related peptide inhibits the cardiac fibroblasts senescence in cardiac fibrosis via up-regulating klotho expression. Eur. J. Pharm. 843, 96–103 (2019).
Garner, W. L., McDonald, J. A., Koo, M., Kuhn, C. & Weeks, P. M. Identification of the collagen-producing cells in healing flexor tendons. Plast. Reconstr. Surg. 83, 875–879 (1989).
Loiselle, A. E. et al. Remodeling of murine intrasynovial tendon adhesions following injury: MMP and neotendon gene expression. J. Orthop. Res. 27, 833–840 (2009).
Best, K. T., Lee, F. K., Knapp, E., Awad, H. A. & Loiselle, A. E. Deletion of NFKB1 enhances canonical NF-κB signaling and increases macrophage and myofibroblast content during tendon healing. Sci. Rep. 9, 10926 (2019).
Ackerman, J. E. et al. Defining the spatial-molecular map of fibrotic tendon healing and the drivers of Scleraxis-lineage cell fate and function. Cell Rep. 41, 111706 (2022).
Korcari, A., Muscat, S., McGinn, E., Buckley, M. R. & Loiselle, A. E. Depletion of Scleraxis-lineage cells during tendon healing transiently impairs multi-scale restoration of tendon structure during early healing. PLoS One 17, e0274227 (2022).
Muscat, S., Nichols, A. E. C., Gira, E. & Loiselle, A. E. CCR2 is expressed by tendon resident macrophage and T cells, while CCR2 deficiency impairs tendon healing via blunted involvement of tendon-resident and circulating monocytes/macrophages. FASEB J. 36, e22607 (2022).
Hasslund, S. et al. Adhesions in a murine flexor tendon graft model: autograft versus allograft reconstruction. J. Orthop. Res. 26, 824–833 (2008).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
Y.L., S.P., X.Z., and M.L. contributed equally to this study. This work was supported by the National Key Research and Development Program of China (2024YFC2418105), National Natural Science Foundation of China (Grant No. 82172408, 82425035 and 9236810338), Shanghai Jiao Tong University Medical College “Two-hundred Talent” Program (No.20191829), Program of Shanghai Academic/Technology Research Leader (No. 22XD1422600), Shanghai Municipal Health Commission (No. 2022YQ073), Shanghai “Medical Star” Young Medical Talent Training Funding Program (Outstanding Youth), and Shanghai Health Commis-sion Health Industry Research Project (Excellent Project), Shanghai Municipal Health Commission Research Project (No. 20234Z0008). Fundamental Research Funds for the Central Universities (YG2024ZD18). Shanghai Science and Technology Industry High-quality Development Program (25S11901600). Shanghai Key Technology Research and Development Program (25CL2900400). Shanghai Leading Talent Program of Eastern Talent Plan (82272531). We are also grateful to Professor Jun Qin (Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences) for his technical assistance.
Author information
Authors and Affiliations
Contributions
Study conception and design: Y.L., S.L., B.H.; Acquisition of data: Y.L., S.P., X.Z., M.L.; Analysis and interpretation of data: Y.L., Z.L., H.C.; Drafting of manuscript: Y.L., J.Q., J.Z., R.W., J.H., J.J.; Revision: Y.L.; Approval of manuscript: All authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Peer review
Peer review information
Nature Communications thanks Stavros Thomopoulos and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, Y., Pang, S., Zhang, X. et al. Nociceptive sensory neuron-derived NGF orchestrates a fibrotic mesenchymal stromal cell neurogenic niche to drive tendon pathological fibrosis. Nat Commun 17, 650 (2026). https://doi.org/10.1038/s41467-025-67396-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-67396-y