Abstract
IL-17A is a cytokine critical for tissue repair, but in excess, it prolongs inflammation and impairs healing. In type 2 diabetic (T2D) wounds, keratinocyte functions, including migration and inflammation, are disrupted, though mechanisms remain unclear. Here, we demonstrate that IL-17A regulates keratinocyte dysfunction via induction of the histone demethylase Jumonji domain-containing protein 3 (JMJD3) through a TRAF6/NFκB pathway. JMJD3 removes repressive histone 3 lysine 27 (H3K27me3) marks at anti-migratory (Itga3, Timp1) and inflammatory (Ccl20, Cxcl1, Cxcl3, Cxcl5) gene promoters, increasing transcription. Human and murine diabetic wounds exhibit elevated IL-17A signaling, JMJD3, and expression of associated anti-migratory and inflammatory genes compared to controls. Importantly, keratinocyte-specific deletion of IL-17A signaling or JMJD3 in diabetic mice improves wound healing and decreases expression of JMJD3 target genes. These findings reveal an IL-17A/JMJD3-mediated mechanism driving keratinocyte dysfunction in T2D wounds and highlight the therapeutic potential of targeting this axis to enhance wound repair.
Similar content being viewed by others
Introduction
Wound healing progresses through discrete yet overlapping stages, where impaired progression through these phases results in pathologic healing. Keratinocytes are non-immune resident cells in the skin that play key roles (proliferation, migration, inflammation, differentiation) primarily during the re-epithelialization and remodeling phases of wound repair. In normal wound repair, keratinocytes migrate as a sheet towards the wound site and proliferate during the re-epithelialization phase1,2. During remodeling, they produce matrix metalloproteinases (MMPs) and tissue inhibitors of MMPs (TIMPs) to restructure the extracellular matrix and terminally differentiate to form a protective outer layer of the epidermis3. Directional/collective cell migration, rather than random/individual cell migration, is crucial for proper wound closure and is achieved partly via integrin-mediated adhesion and regulation of matrix topography4. Although previously thought to be passive cell types that solely act as a physical barrier, keratinocytes are now recognized as highly active cells that respond to external stimuli throughout all stages of wound healing5. In response to the cytokine milieu in the wound environment, keratinocytes produce inflammatory cytokines and chemokines that promote inflammation and recruit immune cells5,6; however, an alteration in the cytokine milieu can result in impaired keratinocyte functions7,8,9. Our lab and others have found that keratinocytes display impairments in migration, proliferation, and other functions during the re-epithelialization phase in nonhealing type 2 diabetic (T2D) wounds7,8,10; however, the precise molecular mechanisms that regulate keratinocyte function in normal or pathologic repair remain unclear.
T2D wounds are often characterized by a late prolonged inflammatory phase due, in part, to pathologic production of inflammatory cytokines by both immune and non-immune cells8,11,12,13,14,15. Importantly, serum IL-17A levels have been shown to be increased in diabetic patients, providing a potential mediator for the chronic low-grade inflammation seen in non-healing diabetic wounds16,17,18. Our lab and others have identified that IL-17A is increased in diabetic wound tissue as well as in other tissues/organs (blood, pancreas, kidneys, etc.)12,17,18,19,20. This is highly relevant since increased IL-17A has been shown to impair inflammatory cyto/chemokine production, proliferation, and differentiation functions in epithelial cells in other chronic inflammatory disease processes (i.e., psoriasis, Crohn’s disease, chronic obstructive pulmonary disease)21,22,23. Despite this, the effects of excessive IL-17A on keratinocyte function have not been examined in tissue repair. Further, the molecular mechanisms by which IL-17A signaling in keratinocytes controls their function are unknown. In this regard, epigenetic modifications have been shown to regulate changes in gene expression and, hence, cell function in the setting of tissue repair24,25,26. Epigenetic modifications have been well studied in immune cells during tissue repair, but considerably less is known about epigenetic modifications in non-immune cells during the wound healing process. Although not well-studied in keratinocytes, NFκB-mediated cytokine expression has been recently found to be regulated upstream in keratinocytes by chromatin-modifying enzymes (CMEs)27. Additionally, certain transcriptional factors have been shown to regulate keratinocyte migration-related genes during wound re-epithelialization possibly via recruitment of CMEs28. Despite these reports, studies on epigenetic regulation of wound keratinocyte function in the setting of normal or dysregulated wound repair are limited.
Here, we demonstrate that increased IL-17A signaling in the diabetic wound environment alters keratinocyte functions, including migration and inflammation, that are necessary for tissue repair. Using in vitro and in vivo assays, we identified that increased IL-17A signaling in keratinocytes leads to decreased wound closure rates via an increase in the expression of anti-migratory (i.e., Itga3, Timp1) and inflammatory (i.e., Ccl20, Cxcl1, Cxcl3, and Cxcl5) genes. Using multiple keratinocyte-specific murine models (Il17rafl/fl K14cre+, Traf6fl/fl K14cre+, Jmjd3fl/fl K14cre+), we identified that IL-17A signaling upregulates the CME, JMJD3, in keratinocytes, and that JMJD3 is regulated upstream in keratinocytes via a TRAF6/NFκB pathway. Chromatin immunoprecipitation (ChIP) analysis of JMJD3-mediated demethylation of the repressive H3K27me3 mark on Itga3, Timp1, Ccl20, Cxcl1, Cxcl3, and Cxcl5 gene promoters in keratinocytes stimulated with IL-17A revealed a decrease in H3K27me3 contributing to increased gene expression. Importantly, treatment with a pharmacological inhibitor specific for JMJD3 decreased the expression of Itga3, Timp1, Ccl20, Cxcl1, Cxcl3, and Cxcl5 genes and proteins in IL-17A-stimulated keratinocytes, resulting in improved keratinocyte migration rates. As a translational corollary, single-cell RNA-Sequencing (scRNA-Seq) in keratinocytes from T2D and non-T2D human wounds revealed increased JMJD3 as well as ITGA3, TIMP1, CCL20, CXCL1, CXCL3, and CXCL5 in keratinocytes from T2D patients compared with non-T2D controls. Importantly, keratinocyte-specific diabetic murine models deficient in IL-17A signaling or JMJD3 displayed increased wound repair and decreased Itga3, Timp1, Ccl20, Cxcl1, Cxcl3, and Cxcl5 gene expression in keratinocytes in vivo. Our findings reveal that expression of anti-migratory and inflammatory genes is regulated by an IL-17A/TRAF6/JMJD3-mediated mechanism, and manipulation of this pathway in keratinocytes may improve wound repair in the setting of diabetes.
Results
IL-17A signaling in keratinocytes controls wound repair via regulation of migratory and inflammatory genes
In order for wound repair to progress in an orchestrated fashion, chemokine and cytokine signals must occur at precise times in the wound microenvironment. In this regard, IL-17A and its downstream signaling are essential for wound repair, where it is increased early in wounds and serves to initiate inflammation and promote pathogen clearance19,29. It is well-established that delayed or exaggerated chemokine/cytokine-mediated signaling can result in pathologic healing25,26,30. For example, excessive IL-17A in wound tissue has been shown to promote prolonged inflammation in the late phases of wound repair and delay re-epithelialization and remodeling20. However, the precise cell-specific mechanisms for this IL-17A-mediated impaired re-epithelialization/migration and altered inflammation are unknown. In order to first examine IL-17A signaling in keratinocytes in wound repair, we isolated and cultured basal keratinocytes from C57BL/6 mice as we have previously described31, stimulated them with or without recombinant mouse IL-17A (rIL-17A) (40 ng/mL), and performed scratch wound assays over a 48-h period to compare migration rates. Since IL-17A increases proliferation in keratinocytes21, we pre-treated them with a mitosis inhibitor, Mitomycin C (10 μg/mL), for 2 h to eliminate the contribution of cell proliferation to gap closure and to specifically assess changes in migration32. We found that IL-17A-stimulated keratinocytes displayed a significant increase in percentage of initial gap area, indicating decreased gap closure rates at 48 h compared to unstimulated controls (Fig. 1a). To assess translational relevance, we repeated the scratch assay using N/TERT human keratinocytes, which closely mimic primary human keratinocyte epithelialization in vitro33. We confirmed that IL-17A-stimulated human keratinocytes displayed decreased gap closure rates at 8 and 12 h compared to unstimulated controls (Fig. 1b).
a, b Scratch assays of primary murine (n = 3 biological replicates, p = 0.0003 (48 h)) and N/TERT keratinocytes (n = 3 biological replicates, p = 0.0209 (8 h), p = 0.0324 (12 h)) with (red) or without (blue) IL-17A, n = 3 independent experiments. c qPCR analysis of migration-related gene expression in unstimulated (blue) and IL-17A-stimulated (red) keratinocytes. n = 3 biological replicates, Fgf2: p = 0.0242, Mmp2: p = 0.0274, Itga3: p = 0.0454, Timp1: p = 0.0374. n = 3 independent experiments. d RNA-seq analysis of inflammatory chemokine expression in unstimulated (blue) and IL-17A-stimulated (red) keratinocytes. n = 3 biological replicates. e qPCR analysis of inflammatory gene expression in Il17rafl/fl K14cre+ (red) and Il17rafl/fl K14cre- (yellow) keratinocytes. n = 3 biological replicates, Ccl20: p < 0.0001 (cre- medium vs. stimulation), p = 0.8426 (cre+ medium vs. stimulation), p < 0.0001 (cre- stimulation vs. cre+ stimulation), Cxcl1: p < 0.0001 (cre- medium vs. stimulation), p = 0.9949 (cre+ medium vs. stimulation), p < 0.0001 (cre- stimulation vs. cre+ stimulation), Cxcl3: p < 0.0001 (cre- medium vs. stimulation), p = 0.9835 (cre+ medium vs. stimulation), p < 0.0001 (cre- stimulation vs. cre+ stimulation), Cxcl5: p < 0.0001 (cre- medium vs. stimulation), p = 0.9983 (cre+ medium vs. stimulation), p < 0.0001 (cre- stimulation vs. cre+ stimulation). n = 3 independent experiments. Data were analyzed for variances, and 2-tailed Student’s t tests for (c) and 1-way ANOVA tests for (a, b), (e) were performed. Data are presented as mean ± SEM.
Several genes/proteins play key roles in keratinocyte migration including fibroblast growth factor 2 that accelerates migration by increasing epithelial-to-mesenchymal transition in keratinocytes34,35 and integrin alpha 3 that inhibits migration by promoting cell adhesion36,37,38,39. During ECM remodeling, wound edge keratinocytes control their migration path by modulating expression of MMPs and TIMPs40, where TIMP-1 is a strong inhibitor of most MMPs including MMP-2, which is linked with keratinocyte migration3,41,42. In normal wound repair, a balance of MMPs/TIMPs is maintained and promotes keratinocyte migration3; however, in a dysregulated setting, an abnormal increase in TIMP-1 expression decreases keratinocyte migratory capacity43. To examine the mechanisms underlying delayed wound closure in IL-17A-stimulated keratinocytes, we analyzed gene(s) involved in migration. Upon stimulating cultured murine keratinocytes with rIL-17A (40 ng/mL), we found that expression of pro-migratory genes, fibroblast growth factor 2 (Fgf2) and MMP-2 (Mmp2) were decreased3,34,35, while expression of anti-migratory genes, integrin alpha 3 (Itga3) and TIMP-1 (Timp1) were increased compared to their unstimulated controls (Fig. 1c). In order to identify other keratinocyte genes potentially regulated by IL-17A, we performed bulk RNA-Seq of IL-17A-stimulated murine keratinocytes and found an increase in expression of inflammatory genes Ccl20, Cxcl1, Cxcl3, Cxcl5 (Fig. 1d). These chemokines are regulated by IL-17A in keratinocytes in other chronic inflammatory skin diseases21,44 and recruit inflammatory cell types such as TH17 cells, neutrophils, and monocytes21,45, driving pathologic inflammation. Next, to further verify that IL-17A signaling regulates these inflammatory genes in keratinocytes, we generated a keratinocyte-specific murine model with a deletion of IL-17 receptor A (Il17rafl/fl K14cre+) on a C57BL/6 background. Keratinocytes isolated from Il17rafl/fl K14cre+ and their littermate controls (Il17rafl/fl K14cre-) were isolated, cultured, and stimulated with rIL-17A for 5 h, and gene expression was examined. Keratinocytes from Il17rafl/fl K14cre+ exhibited decreased Ccl20, Cxcl1, Cxcl3, Cxcl5 compared with littermate controls (Fig. 1e). Lastly, to assess whether these chemokines contribute to impaired migration in response to IL-17A, we performed scratch assays using neutralizing antibodies (1 μg/mL) against CCL-20, CXCL-1, CXCL-3, and CXCL-5, or an isotype control, in both murine (Supplementary Fig. 1A, B) and human keratinocytes (Supplementary Fig. 1C, D). In IL-17A-stimulated keratinocytes, both isotype control and neutralizing antibody treatments resulted in decreased migration, with no rescue observed upon chemokine blockade. These findings suggest that these individual chemokines play a minimal role in mediating IL-17A-induced migration defects. Collectively, these findings indicate that IL-17A signaling in keratinocytes impairs wound closure, likely through the regulation of both migratory and inflammatory genes.
IL-17A signaling upregulates expression of the histone demethylase, JMJD3, via a TRAF6/NFκB pathway
Epigenetic enzymes have been shown by our group and others to regulate immune cell functions in wound tissue; however, their role in non-immune cells associated with tissue repair is less studied. In order to examine epigenetic alterations in response to IL-17A in keratinocytes, we stimulated murine keratinocytes with rIL-17A (40 ng/mL) and performed a 96-well epigenetic super array8,12,25,46. We found that the histone demethylase, JMJD3, displayed significant changes in expression in response to IL-17A compared to other epigenetic enzymes examined (Supplementary Fig. 2). Next, to confirm our super array results, we examined Jmjd3 expression via qPCR and western blot in IL-17A-stimulated keratinocytes isolated from C57BL/6 mice and found that IL-17A significantly increases expression of JMJD3 mRNA and protein (Fig. 2a, b). Prior studies from our group have found that Jmjd3 expression is abnormally high in diabetic wound macrophages and controls inflammatory gene expression via an H3K27 mechanism8,46,47; however, the role of JMJD3 has not been examined in wound keratinocytes or in response to IL-17A. To further validate that JMJD3 is regulated via an upstream IL-17A signaling pathway, we used our Il17rafl/fl K14cre+ mice and their littermate controls (Il17rafl/fl K14cre-), isolated keratinocytes, stimulated them with rIL-17A (40 ng/mL), and examined Jmjd3 expression. We identified that Il17rafl/fl K14cre+ keratinocytes were unable to upregulate Jmjd3 following stimulation with IL-17A (Fig. 2c), suggesting that the IL-17A signaling pathway is involved in regulating Jmjd3 expression.
a Jmjd3 qPCR analysis of unstimulated (white) and IL-17A-stimulated (blue) keratinocytes. n = 3 biological replicates, p = 0.0021. n = 3 independent experiments. b Western blot of JMJD3 expression in unstimulated (white) and IL-17A-stimulated (blue) keratinocytes. n = 3 technical replicates, p = 0.0224. n = 3 independent experiments. Representative densitometry plot shown. c Jmjd3 qPCR analysis of Il17rafl/fl K14cre+ (red) and Il17rafl/fl K14cre- (yellow) keratinocytes. n = 3 biological replicates, p = 0.0008 (cre- medium vs. stimulation), p = 0.9131 (cre+ medium vs. stimulation), p = 0.0005 (cre- stimulation vs. cre+ stimulation), n = 3 independent experiments. d Jmjd3 qPCR analysis of Traf6fl/fl K14cre+ (red) and Traf6fl/fl K14cre- (yellow) keratinocytes. n = 3 biological replicates, p = 0.0036 (cre- medium vs. stimulation), p = 0.9007 (cre+ medium vs. stimulation), p = 0.0095 (cre- stimulation vs. cre+ stimulation), n = 3 independent experiments. e Jmjd3 qPCR analysis of keratinocytes treated with DMSO only (white), with IL-17A alone (blue), or with IL-17A and IKK inhibitor VII (2 µM) (red). n = 3 biological replicates, p = 0.0156 (DMSO vs. IL-17A), p = 0.0198 (IL-17A vs. IL-17A and inhibitor), n = 3 independent experiments. f Schematic of proposed IL-17A/TRAF6/NFκB-mediated regulation of Jmjd3. Data were analyzed for variances, and 2-tailed Student’s t tests for (a, b) and 1-way ANOVA tests for (c–e) were performed. Data are presented as the mean ± SEM.
The primary IL-17A receptor downstream signal pathway involves Act1/TRAF6 complex formation48,49,50,51. TRAF6 signaling has been shown to be responsible for activation of NFκB-mediated pathways involved in chronic inflammation in psoriasis21; thus, we first examined TRAF6 and its’ downstream signaling following IL-17A receptor stimulation. In order to examine the role of TRAF6 in keratinocytes, we generated a keratinocyte-specific Traf6-deficient murine model (Traf6fl/fl K14cre+) (on a C57BL/6 background), and isolated keratinocytes from Traf6fl/fl K14cre+ mice and their littermate controls (Traf6fl/fl K14cre-). Keratinocytes were then stimulated with rIL-17A (40 ng/mL) and examined for Jmjd3 expression. We found that keratinocytes deficient in TRAF6 were unable to upregulate Jmjd3 following stimulation with IL-17A (Fig. 2d). Next, to determine if NFκB alters Jmjd3 expression, we treated IL-17A-stimulated keratinocytes with a NFκB inhibitor (IKK inhibitor VII, 2 μM) and observed that inhibition of NFκB also resulted in decreased Jmjd3 expression (Fig. 2e). This suggests that both TRAF6 and NFκB are important for Jmjd3 expression. Taken together, these results indicate that IL-17A induces Jmjd3 in keratinocytes via a TRAF6/NFκB pathway (Fig. 2f).
Jmjd3 directly regulates anti-migratory and inflammatory genes in wound keratinocytes in response to IL-17A and direct inhibition of Jmjd3 in keratinocytes improves migration/wound closure rate
JMJD3 is a histone demethylase that has specificity for the lysine 27 site on histone 3 site (H3K27), resulting in the opening of chromatin that renders promoter sites accessible for transcription factor binding, thus effectively increasing gene expression47. To examine if IL-17A increases JMJD3-mediated H3K27 demethylation on specific gene promoters, we isolated keratinocytes, stimulated them with rIL-17A (40 ng/mL), and analyzed by chromatin immunoprecipitation (ChIP) for H3K27me3 methylation at gene promoters of anti-migratory (Itga3, Timp1) and inflammatory (Ccl20, Cxcl1, Cxcl3, and Cxcl5) genes. We found decreased H3K27me3 on the anti-migratory and inflammatory gene promoters in the IL-17A-stimulated keratinocytes compared to unstimulated controls (Fig. 3a). Since there are other chromatin modifying enzymes (CMEs) that methylate the H3K27 site, we stimulated primary murine keratinocytes with rIL-17A (40 ng/mL) and treated them with a pharmacological inhibitor, GSK-J4 (1 μM), that specifically blocks JMJD3 demethylase activity52. Treatment of IL-17A-stimulated keratinocytes with GSK-J4 resulted in both decreased gene (Fig. 3b) and protein (Fig. 3c, d) expression (Itga3, Timp1, Ccl20, Cxcl1, Cxcl3, Cxcl5) compared to keratinocytes stimulated with IL-17A alone. To address any potential off-target effects of GSK-J4, we treated keratinocytes with siRNA targeting Jmjd3 and found decreased anti-migratory and inflammatory gene expression compared to non-targeting controls (Fig. 3e). Next, we generated a keratinocyte-specific Jmjd3-deficient murine model (Jmjd3fl/fl K14cre+) and isolated keratinocytes from Jmjd3fl/fl K14cre+ and their littermate controls (Jmjd3fl/fl K14cre-), stimulated them with rIL-17A (40 ng/mL), and examined changes in anti-migratory and inflammatory gene expression. In response to IL-17A stimulation, Jmjd3fl/fl K14cre+ keratinocytes had decreased gene expression (Itga3, Timp1, Ccl20, Cxcl1, Cxcl3, Cxcl5) compared to their Jmjd3fl/fl K14cre- controls (Fig. 3f). To specifically interrogate the role of JMJD3 on keratinocyte function, we performed a scratch assay of both murine and human keratinocytes treated with or without GSK-J4 over a 48-h or 12-h period. With GSK-J4 treatment, murine keratinocytes displayed improved wound closure rates at 48 h (Fig. 3g), while human keratinocytes displayed improved wound closure rates at 12 h compared to their untreated controls (Fig. 3h), suggesting that inhibiting JMJD3 demethylase activity improves keratinocyte migration. Additionally, we repeated the scratch assay using an alternative pharmacological inhibitor called GSK-J1 to confirm the specificity of JMJD3-mediated migration defects. We demonstrated that GSK-J1 treatment resulted in improved migration rates in both murine and human keratinocytes (Fig. 3i, j). To further assess changes in migration rates using our genetic model and control for off-target effects of pharmacological inhibitors, we performed a scratch wound assay of keratinocytes isolated from Jmjd3fl/fl K14cre+ mice and littermate controls (Jmjd3fl/fl K14cre-) over a 48-h period. At 48 h, Jmjd3fl/fl K14cre+ keratinocytes displayed increased wound closure rates compared to controls (Fig. 3k). To exclude the role of JMJD3 in keratinocyte proliferation, we conducted proliferation assays using keratinocytes isolated from Jmjd3fl/fl K14cre+ mice and littermate controls (Jmjd3fl/fl K14cre-). Proliferation-inhibited controls confirmed no change in proliferation (Supplementary Fig. 3A). Both Jmjd3fl/fl K14cre- and Jmjd3fl/fl K14cre+ keratinocytes resulted in around a 2-fold increase in proliferation at 48 h (Supplementary Fig. 3B), and the difference between the two groups was not significant, suggesting that the role of JMJD3 on keratinocyte proliferation is minimal. Together, these data indicate that JMJD3 impairs wound closure by upregulating the expression of anti-migratory and inflammatory genes in wound keratinocytes.
a ChIP analysis of H3K27me3 or IgG at indicated promoters in keratinocytes with (white) or without (blue) IL-17A stimulation. Itga3: n = 5 (unstimulated), n = 4 (IL-17A-stimulated) technical replicates, p = 0.0034, Timp1: n = 3 technical replicates, p = 0.0046, Ccl20: n = 3 technical replicates, p = 0.0059, Cxcl1: n = 3 technical replicates, p = 0.0020, Cxcl3: n = 3 technical replicates, p = 0.0258, Cxcl5: n = 3 technical replicates, p = 0.0174. n = 3 independent experiments. b qPCR analysis of keratinocytes treated with DMSO only (white), with IL-17A alone (blue), or with IL-17A and GSK-J4 (1 µM) (red). Itga3: n = 4 biological replicates, p < 0.0001 (DMSO vs. IL-17A), p = 0.0017 (IL-17A vs. IL-17A and inhibitor), Timp1: n = 6 biological replicates, p < 0.0312 (DMSO vs. IL-17A), p = 0.0029 (IL-17A vs. IL-17A and inhibitor), Ccl20: n = 3 biological replicates, p = 0.0008 (DMSO vs. IL-17A), p = 0.0428 (IL-17A vs. IL-17A and inhibitor), Cxcl1: n = 3 biological replicates, p = 0.0003 (DMSO vs. IL-17A), p = 0.0294 (IL-17A vs. IL-17A and inhibitor), Cxcl3: n = 3 biological replicates, p < 0.0001 (DMSO vs. IL-17A), p = 0.0003 (IL-17A vs. IL-17A and inhibitor), Cxcl5: n = 3 biological replicates, p < 0.0001 (DMSO vs. IL-17A), p = 0.0007 (IL-17A vs. IL-17A and inhibitor), n = 3 independent experiments. c Western blot of ITGA-3 expression in keratinocytes treated with DMSO only (white), with IL-17A alone (blue), or with IL-17A and GSK-J4 (red). Representative densitometry plot is shown. n = 3 independent experiments. d Protein quantification of lysates from keratinocytes treated with DMSO only (white), with IL-17A alone (blue), or with IL-17A and GSK-J4 (red). TIMP-1: n = 6 biological replicates, p = 0.0012 (DMSO vs. IL-17A), p = 0.0068 (IL-17A vs. IL-17A and inhibitor), CCL-20: n = 3 biological replicates, p < 0.0001 (DMSO vs. IL-17A), p = 0.0199 (IL-17A vs. IL-17A and inhibitor), CXCL-1: n = 3 biological replicates, p < 0.0001 (DMSO vs. IL-17A), p = 0.0158 (IL-17A vs. IL-17A and inhibitor), CXCL-3: n = 3 biological replicates, p < 0.0001 (DMSO vs. IL-17A), p = 0.0167 (IL-17A vs. IL-17A and inhibitor), CXCL-5: n = 3 biological replicates, p < 0.0001 (DMSO vs. IL-17A), p = 0.0005 (IL-17A vs. IL-17A and inhibitor), n = 3 independent experiments. e qPCR analysis of keratinocytes treated with a non-targeting control (siNTC) (white) or siJmjd3 (gray). Jmjd3: n = 6 (siNTC), n = 4 biological replicates (siJmjd3), p = 0.0192, Itga3: n = 6 (siNTC), n = 4 biological replicates (siJmjd3), p = 0.0019, Timp1: n = 3 (siNTC), n = 4 biological replicates (siJmjd3), p = 0.0205, Ccl20: n = 6 (siNTC), n = 4 biological replicates (siJmjd3), p = 0.0015, Cxcl1: n = 6 (siNTC), n = 4 biological replicates (siJmjd3), p = 0.0019, Cxcl3: n = 6 (siNTC), n = 4 biological replicates (siJmjd3), p = 0.0011, Cxcl5: n = 6 (siNTC), n = 4 biological replicates (siJmjd3), p = 0.0011, n = 3 independent experiments. f qPCR analysis of Jmjd3fl/fl K14cre+ (red) and Jmjd3fl/fl K14cre- (yellow) keratinocytes. n = 3 biological replicates, Itga3: p = 0.0184, Timp1: p = 0.0044, Ccl20: p = 0.0160, Cxcl1: p = 0.0371, Cxcl3: p = 0.0042. n = 3 independent experiments. g, h Scratch assays of primary murine (n = 3 biological replicates, p < 0.0001 (48 h)) and N/TERT (n = 3 biological replicates, p = 0.0340 (12 h)) keratinocytes treated with IL-17A alone (blue) or IL-17A and GSK-J4 (red). n = 3 independent experiments. i, j Scratch assays of primary murine (n = 3 biological replicates, p = 0.0016 (48 h)) and N/TERT (n = 6 biological replicates (IL-17A alone), n = 4 biological replicates (IL-17A and GSK-J1), p = 0.0223 (8 h), p < 0.0001 (12 h)) keratinocytes treated with IL-17A alone (blue) or IL-17A and GSK-J1 (red). n = 3 independent experiments. k Scratch assay of Jmjd3fl/fl K14cre+ (red) and Jmjd3fl/fl K14cre- (blue) keratinocytes. n = 3 biological replicates, p = 0.0198 (48 h). n = 3 independent experiments. Data were analyzed for variances, and 2-tailed Student’s t tests for (a), (e), (f) and 1-way ANOVA tests for (b), (d), (g–k) were performed. Data are presented as the mean ± SEM.
Human and murine diabetic wounds display increased IL-17A signaling, JMJD3, and expression of anti-migratory / inflammatory genes in wound keratinocytes
Recent studies have identified that IL-17A is increased in diabetes12,17,18,19,20,53, hence, we analyzed our human bulk RNA-Seq dataset isolated from T2D and non-T2D patients for IL-17A signaling47,54,55. We identified that there is increased IL-17A signaling in human T2D wounds compared to non-T2D controls (Fig. 4a). Since these samples are not cell-specific and IL-17A signaling could be increased in many wound cell types, we sought to specifically examine diabetic wound keratinocytes for IL-17A signaling. Using a well-established murine model of T2D, db/db mice, which lack a functional leptin receptor56, we wounded mice and isolated keratinocytes on day 5 as previously described31 and performed bulk RNA-Seq. Analysis of bulk RNA-Seq of diabetic wound keratinocytes demonstrated increased expression of IL-17A-induced genes in the diabetic wound keratinocytes compared to littermate controls (Fig. 4b). To obtain kinetic and spatial data of IL-17A expression in diabetic wounds, we performed immunofluorescence (IF) staining for IL-17A, H3K27me3, and Keratin 14 in control and diabetic wounds collected at multiple timepoints (days 1, 3, 7) post-wounding (Supplementary Fig. 4A, B). We utilized a second model of diabetes, diet-induced obesity (DIO) mice, that were generated by feeding mice with a high fat diet (HFD) for 12–16 weeks to simulate a “prediabetic” state that is physiologically similar to humans57. Mice were subjected to 6 mm punch biopsy wounds and collected at days 1, 3, and 7 post-wounding for histology, as we have previously described7. We found that DIO wounds displayed elevated IL-17A at days 1, 3, 7 while control wounds only displayed elevated IL-17A at day 1. We observed that IL-17A was highly localized near the wound edge, close to the epidermis.
a Human bulk RNA-seq heatmap of IL-17A pathway genes in T2D (N = 4) vs. non-T2D (N = 3) wounds. b Bulk RNA-seq heatmap of IL-17A-induced genes in keratinocytes from db/db (n = 3) vs. db/+ wounds (n = 3). c–e Spatial scRNA-seq of T2D (N = 3) and control (N = 1) wounds: UMAP clustering (c), lineage annotation (d), and IL-17A expression (e). f–g Proximity analysis of keratinocytes near IL-17A+ cells in T2D (pink) vs. non-T2D (teal) wounds, with differential expression of CCL20, CXCL1, CXCL3, CXCL5 in keratinocytes near (light blue) vs. far (dark blue) from IL-17A+ cells in T2D wounds (g). n = 21734 (diabetic far), n = 1629 (diabetic near) cells. h, i Jmjd3 mRNA (n = 3 technical replicates, p < 0.0001, n = 3 independent experiments) (h) and JMJD3 protein (n = 3 technical replicates, p = 0.0127, n = 3 independent experiments) (i) in keratinocytes from control (yellow) vs DIO (blue) wounds. j, k scRNA-seq of T2D (N = 10) and control (N = 10) wounds: UMAP clustering (j) and JMJD3 expression in keratinocyte subsets (k). l qPCR analysis of migration-related and inflammatory gene expression in keratinocytes from control (yellow) vs. DIO (blue) wounds. n = 3 technical replicates, Fgf2: p = 0.0059, Mmp2: p = 0.0008, Itga3: p = 0.0430, Timp1: p < 0.0001, Ccl20: p < 0.0001, Cxcl1: p < 0.0001, Cxcl3: p < 0.0001, Cxcl5: p = 0.0002. n = 3 independent experiments. m ChIP for H3K27me3 at indicated promoters in keratinocytes from control (yellow) vs. DIO (blue) wounds. Itga3: n = 3 technical replicates, p = 0.0027, Timp1: n = 3 technical replicates, p = 0.0104, Ccl20: n = 6 technical replicates, p = 0.0347, Cxcl1: n = 3 technical replicates, p = 0.0072, Cxcl3: n = 3 technical replicates, p = 0.0005, Cxcl5: n = 3 technical replicates, p < 0.0001. n = 3 independent experiments. n ChIP for JMJD3 at indicated promoters in keratinocytes from control (yellow) vs. DIO (blue) wounds. n = 3 technical replicates. Itga3: p = 0.0170, Timp1: p = 0.0350, Ccl20: p = 0.0021, Cxcl1: p = 0.0037, Cxcl3: p = 0.0003, Cxcl5: p = 0.0005. n = 3 independent experiments. o ChIP for JMJD3 at indicated promoters in keratinocytes from db/+ (yellow) vs. db/db (blue) wounds. n = 3 technical replicates. Itga3: p = 0.0407, Timp1: p < 0.0001, Ccl20: p = 0.0026, Cxcl1: p = 0.0004, Cxcl3: p = 0.0143, Cxcl5: p = 0.0060. n = 3 independent experiments. p, q scRNA-seq of T2D vs control wounds showing keratinocyte subsets (p) and expression of indicated genes (q). Data were analyzed for variances, and 2-tailed Student’s t tests for (h, i), (l–o) were performed. Data are presented as the mean ± SEM.
To identify potential sources of IL-17A in diabetic wounds, we analyzed our spatial RNA-seq dataset of wounds from T2D and non-T2D patients (Fig. 4c). Cell clusters were annotated using lineage-specific markers to visualize the organization of distinct cell types (Fig. 4d). We observed an increased number of IL-17A-expressing cells adjacent to keratinocytes in diabetic wounds (Fig. 4e). Proximity-based interaction analysis further revealed a higher proportion of keratinocytes interacting with IL-17A-expressing cells in the diabetic setting (Fig. 4f). In non-diabetic wounds, IL-17A-expressing cells were mainly restricted to the epidermis, whereas in diabetic wounds, they were more abundant and dispersed throughout the wound bed. To characterize the cellular sources of IL-17A, we subclustered IL-17A-expressing cells based on lineage markers and identified myeloid cells, CD8+ T cells, neutrophils, NK cells, and CD4+ T cells as the predominant producers (Supplementary Figs. 5A–C). Additionally, we found that keratinocytes located near IL-17A-expressing cells expressed higher levels of CCL20, CXCL1, CXCL3, and CXCL5 (Fig. 4g), suggesting a localized inflammatory response driven by IL-17A in diabetic wounds. Specifically, given that diabetic wound keratinocytes near IL-17A+ cells exhibit increased expression of myeloid chemoattractants (CXCL1, CXCL3, and CXCL5) (Fig. 4g), we used our spatial RNA-seq to classify immune cell types adjacent to keratinocytes in diabetic and non-diabetic wounds. Proximity analysis revealed a higher frequency of keratinocyte interactions with myeloid cells and neutrophils in diabetic wounds (Supplementary Figs. 5D, E), suggesting that keratinocyte-derived chemokines contribute to increased immune cell recruitment.
Since IL-17A is increased in diabetic wounds, and IL-17A signaling is increased in diabetic keratinocytes, we examined whether Jmjd3 is elevated in diabetic murine wound keratinocytes. Mice were subjected to 6 mm punch biopsy wounds and wound keratinocytes were collected from control and DIO mice as we have previously described7 on day 5, a timepoint when IL-17A production is significantly increased in mice12. We observed increased expression of JMJD3 mRNA and protein in murine diabetic wound keratinocytes compared to their nondiabetic controls (Fig. 4h, i), suggesting a link between elevated IL-17A levels in the diabetic wound environment and increased Jmjd3 expression in keratinocytes. To assess the spatial and temporal dynamics of JMJD3 activity in relation to IL-17A expression during wound healing, we examined our IF staining of H3K27me3 and Keratin 14 in control and DIO wounds at days 1, 3, and 7 post-wounding. Due to JMJD3’s nuclear localization, low abundance, and the lack of a reliable antibody, direct detection via immunohistochemistry or IF remains challenging. Instead, we used H3K27me3 as an indirect readout of JMJD3 activity. We observed increased colocalization of H3K27me3 with keratinocytes in control wounds at days 3 and 7, while DIO wounds showed decreased H3K27me3 signal in keratinocytes at day 7 (Supplementary Fig. 4B). These findings suggest enhanced H3K27 demethylation—and thus increased JMJD3 activity—in diabetic keratinocytes at later stages. Notably, this increase in JMJD3 activity coincides with increased IL-17A protein expression near keratinocytes (Supplementary Fig. 4A), supporting a mechanistic link between IL-17A signaling and JMJD3 activation.
In order to determine the translational relevance and keratinocyte-specificity of JMJD3 expression, we analyzed our scRNA-Seq dataset on wounds from T2D patients and non-T2D controls7,8,47,55. Cluster analysis was performed using uniform manifold approximation and projection (UMAP) as previously described58; 10 cell clusters were identified in wounds following injury. We identified 3 clusters of keratinocytes, including keratinized, differentiated, and basal keratinocytes (Fig. 4j). We identified that JMJD3 expression is increased in human T2D wound keratinocytes compared to non-T2D controls (Fig. 4k).
Next, to confirm that downstream JMJD3 target genes are altered in diabetic wound keratinocytes, we collected wound keratinocytes from control and DIO mice and examined for anti-migratory and inflammatory genes. We found that expression of pro-migratory genes fibroblast growth factor 2 (Fgf2) and MMP-2 (Mmp2) was decreased, while expression of anti-migratory genes integrin alpha 3 (Itga3) and TIMP-1 (Timp1) was increased in diabetic wound keratinocytes (Fig. 4l). Additionally, we found that expression of inflammatory genes (Ccl20, Cxcl1, Cxcl3, and Cxcl5) was increased in murine diabetic wound keratinocytes compared to controls (Fig. 4l). We further confirmed increased expression of JMJD3-mediated anti-migratory (Integrin α3, TIMP-1) and inflammatory (CCL-20, CXCL-1, CXCL-3, CXCL-5) target genes in DIO wounds at the protein level via western blot or ELISA (Supplementary Fig. 6A–F).
To confirm that JMJD3 directly mediates H3K27me3 demethylase activity on anti-migratory and inflammatory gene promoters in diabetic wound keratinocytes, we performed ChIP studies analyzing K27 trimethylation and JMJD3 occupancy in wound keratinocytes isolated from DIO and db/db mice. K27me3 deposition on anti-migratory (Itga3, Timp1) and inflammatory (Ccl20, Cxcl1, Cxcl3, Cxcl5) gene promoters was significantly decreased in DIO wound keratinocytes compared to controls (Fig. 4m). Additionally, we observed a significant increase in JMJD3 binding to these promoters in both DIO and db/db keratinocytes, further providing direct evidence for JMJD3-mediated K27me3 demethylation in diabetic wound keratinocytes (Fig. 4n, o). As a translational corollary to our murine data, using our scRNA-Seq dataset, we identified that expression of anti-migratory genes ITGA3 and TIMP1 and inflammatory genes CCL20, CXCL1, CXCL3, and CXCL5 was all increased in human T2D basal keratinocytes compared to their non-T2D controls (Fig. 4p, q). Altogether, these data suggest that increased IL-17A signaling in human and murine diabetic wounds drives the upregulation of JMJD3 and regulates downstream anti-migratory and inflammatory genes in diabetic wound keratinocytes, providing a potential mechanism for impaired wound repair in diabetic wounds.
Disruption of the IL-17A/JMJD3 cascade in diabetic wound keratinocytes improves wound repair via decreased expression of anti-migratory / inflammatory genes
Since JMJD3 appears to be playing a pathologic role in diabetic keratinocytes, we first examined if pharmacological modulation of JMJD3 in vivo would result in improved healing in diabetes. We first utilized the genetic T2D murine model, db/db mice, created 6 mm punch biopsy wounds, and administered daily injections of GSK-J4 (1 mg/kg) or vehicle only (DMSO) as we have previously described58. We found that there is increased wound healing in db/db mice treated with local wound GSK-J4 injections at later timepoints, days 8 and 9, compared to their vehicle only controls (Supplementary Fig. 7). Representative images on day 9 are shown.
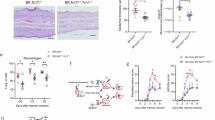
To cell-specifically examine the effects of manipulating the IL-17A/JMJD3 axis in keratinocytes on wound healing, we utilized our mice with keratinocyte-specific IL-17A signaling deficiency (Il17rafl/fl K14cre+). We first determined whether these effects were specific to the diabetic context by analyzing wound healing in non-diabetic Il17rafl/fl K14cre+. Mice were subjected to 6 mm punch biopsy wound, and following injury, wound closure was monitored daily using NIH ImageJ software, and the epithelial gap was measured by histology, as we have previously described7,8,12,25,46,54. Il17rafl/fl K14cre+ mice did not show a significant improvement in wound closure compared to their littermate controls (Il17rafl/fl K14cre-) (Supplementary Fig. 8). Next, we examined if a reduction of IL-17A signaling in diabetic keratinocytes would result in decreased JMJD3 and improved healing in diabetes using the same model (Il17rafl/fl K14cre+) and inducing diabetes. Il17rafl/fl K14cre+ mice were fed a high fat diet (HFD) for 12 – 16 weeks to create a diet-induced obesity (DIO) model57. Oral glucose tolerance test (OGTT) studies confirmed glucose intolerance in these mice. DIO Il17rafl/fl K14cre+ mice demonstrated improved wound closure rates compared to their littermate controls (DIO Il17rafl/fl K14cre-) (Fig. 5a). Representative images from day 7 are shown. These findings indicate that excessive IL-17A signaling in diabetes, rather than IL-17A signaling under homeostatic conditions, drives impaired healing. To further evaluate the contribution of wound contraction in this acute (non-splinted) healing model, we performed histomorphometric analyses of wounds from DIO Il17rafl/fl K14cre+ mice. To assess the involvement of myofibroblasts, the key mediators of contraction, we stained wound sections for alpha smooth muscle actin (αSMA). DIO Il17rafl/fl K14cre+ mice exhibited minimal improvement in re-epithelialization (Supplementary Fig. 9A) but significantly greater wound contraction (Supplementary Fig. 9B, C) and earlier αSMA+ myofibroblast transition (Supplementary Fig. 9D, 9E) compared to controls (DIO Il17rafl/fl K14cre-).
a Non-splinted wound closure in DIO Il17rafl/fl K14cre+ (red) (n = 4 mice) and littermate controls (blue) (n = 6 mice). Day 3: p = 0.0497, day 4: p = 0.0336, day 5: p = 0.0358, day 6: p = 0.0276, day 7: p = 0.0056. n = 2 independent experiments. b Splinted wound closure in DIO Il17rafl/fl K14cre+ (red) (n = 6 mice) and littermate controls (blue) (n = 9 mice). Day 1: p = 0.0155, day 2: p = 0.0048, day 3: p = 0.0042, day 4: p = 0.0118, day 7: p = 0.0422, day 9: p = 0.0295. c, d Day 7 wound diameter (c) and trichrome staining for collagen deposition (d). DIO Il17rafl/fl K14cre+ (red) (n = 4 mice) and littermate controls (blue) (n = 4 mice). Epithelial gap: p = 0.0008, collagen: p = 0.0211. e Non-splinted wound closure in DIO Jmjd3fl/fl K14cre+ (red) (n = 5 mice) and littermate controls (blue) (n = 5 mice). Day 2: p = 0.0336, day 4: p = 0.0267, day 6: p = 0.0419. n = 2 independent experiments.f Splinted wound closure in DIO Jmjd3fl/fl K14cre+ (red) (n = 10 mice) and littermate controls (blue) (n = 8 mice). Day 1: p = 0.0225, day 3: p = 0.0431, day 4: p = 0.0272, day 5: p = 0.0202, day 6: p = 0.0019, day 7: p = 0.0031, day 8: p = 0.0023, day 9: p = 0.0020, day 10: p = 0.0007. g, h Day 6 wound diameter (g) and trichrome staining for collagen deposition (h). DIO Jmjd3/fl K14cre+ (red) (n = 4 mice) and littermate controls (blue) (n = 4 mice). Epithelial gap: p = 0.0031, collagen: p = 0.0257. i qPCR of anti-migratory and inflammatory genes in wound keratinocytes from Jmjd3fl/fl K14cre+ (red) (n = 3 technical replicates) vs. littermate controls (blue) (n = 3 technical replicates). Itga3: p = 0.0004, Timp1: p = 0.0010, Ccl20: p < 0.0001, Cxcl1: p < 0.0001, Cxcl3: p < 0.0001, Cxcl5: p = 0.0233. n = 2 independent experiments. Data were analyzed for variances, and 2-tailed Student’s t tests for (a–i) were performed. Data are presented as the mean ± SEM.
To specifically assess re-epithelialization independent of contraction, we performed a well-established model of chronic (splinted) wound healing of DIO Il17rafl/fl K14cre+ mice for 10 days as we have previously described7. The chronic (splinted) wound model also displayed significantly improved wound healing in the IL-17A signaling deficient mice on days 1, 2, 3, 4, 7, 9 (Fig. 5b). Representative images from days 3, 7, and 9 are shown, and decreased epithelial gap by histology from day 7 are shown (Fig. 5c). Histologically, DIO Il17rafl/fl K14cre+ wounds harvested on day 7 post injury also exhibited improved wound healing and increased wound collagen deposition compared to DIO Il17rafl/fl K14cre- mice (Fig. 5d). Further, we performed single cell RNA-sequencing in wounds from keratinocyte-specific IL-17RA-deficient mice (Il17rafl/fl K14cre+) and wildtype (WT) controls to examine alterations in keratinocyte function and other cell types due to deletion of IL-17A signaling in keratinocytes. We first confirmed that Jmjd3 expression was decreased in Il17rafl/fl K14cre+ keratinocytes compared to their controls (Supplementary Figs. 10A, B). To further validate that JMJD3 is regulated via an IL-17A pathway in our model, we examined expression of our JMJD3-mediated target genes in wound keratinocytes. Expression of inflammatory target genes Cxcl3, Cxcl1, and Ccl20 was decreased in Il17rafl/fl K14cre+ keratinocytes compared to their controls (Supplementary Fig. 10C). Expression of JMJD3-mediated anti-migratory gene Itga3 was decreased while expression of pro-migratory Mmp9 that is regulated by Timp1 was increased in Il17rafl/fl K14cre+ keratinocytes compared to their controls, suggesting improved migratory gene profile in the keratinocytes lacking IL-17A signaling (Supplementary Fig. 10C).
Next, we examined whether loss of JMJD3 in diabetic wound keratinocytes improves diabetic wound repair by altering migratory function. We used our murine model with a keratinocyte-specific deletion of JMJD3 (DIO Jmjd3fl/fl K14cre+) and their littermate controls (DIO Jmjd3fl/fl K14cre-), placed them on a HFD for 12 – 16 weeks, confirmed glucose intolerance, and created 6 mm punch biopsy wounds. Following injury, wound closure was monitored daily, and the epithelial gap was measured by histology. DIO Jmjd3fl/fl K14cre+ mice demonstrated significantly improved wound healing compared to their littermate controls on days 2, 4, 6 (Fig. 5e). Representative images from days 2 and 6 are shown. We performed the chronic (splinted) wound healing model with these mice and observed an identical increase in wound closure rates on days 1, 3, 4, 5, 6, 7, 8, 9, and 10 (Fig. 5f). Representative images from days 4, 7, and 10 are shown, and decreased epithelial gap by histology from day 6 are shown (Fig. 5g). Histologically, DIO Jmjd3fl/fl K14cre+ wounds harvested on day 6 post injury exhibited improved wound healing and increased wound collagen deposition compared to DIO Jmjd3fl/fl K14cre- (Fig. 5h). Additionally, we harvested wound keratinocytes from these mice on day 6 and examined anti-migratory and inflammatory gene expression. We found that DIO Jmjd3fl/fl K14cre+ keratinocytes displayed decreased Itga3, Timp1, Ccl20, Cxcl1, Cxcl3, and Cxcl5 (Fig. 5i), suggesting that Jmjd3-deficiency downregulates anti-migratory and inflammatory gene expression in wound keratinocytes and improves diabetic wound repair.
Since keratinocyte crosstalk with other wound-resident cells is essential for coordinated tissue repair, we examined how deletion of IL-17A signaling and JMJD3 in keratinocytes affects other cell populations involved in wound healing. We characterized T cells, specifically TH17 cells, that are recruited in response to CCL-20 as well as neutrophils that are recruited in response to CXCL-1, CXCL-3, and CXCL-5. Through our scRNA-Seq of Il17rafl/fl K14cre+ and control wounds, we first confirmed that deletion of IL-17A signaling in keratinocytes leads to decreased overall T cell counts (Supplementary Fig. 10D). Next, we performed flow cytometric analysis of wounds from our DIO Il17rafl/fl K14cre+ and DIO Jmjd3fl/fl K14cre+ mice to analyze TH17 cell and neutrophil numbers. We observed a decreased percentage of TH17 cells within the CD4+ T cell population in DIO Il17rafl/fl K14cre+ and DIO Jmjd3fl/fl K14cre+ wounds and a decreased percentage of Ly6G+ neutrophils within the CD45+ immune cell population (Supplementary Fig. 11A–D). We further examined T cell phenotypes by sorting CD4+ T cells (CD11b-, CD45R-, Ter119-, CD8a-, CD49b-, CD19-, CD11c-, TCRgd-, CD24-, CD4+) from Il17rafl/fl K14cre+ and Jmjd3fl/fl K14cre+ wounds and analyzing expression of inflammatory (Tnf, Il17a) and reparative (Il10) genes. T cells from both Il17rafl/fl K14cre+ and Jmjd3fl/fl K14cre+ displayed decreased Tnf and increased Il10, potentially suggesting increased reparative T regulatory cell phenotypes (Supplementary Fig. 11E, F). Additionally, Jmjd3fl/fl K14cre+ T cells displayed decreased Il17a (Supplementary Fig. 11F), corresponding with the decreased number of TH17 cells (Supplementary Fig. 11D). Next, to examine macrophage phenotypes in these mice, we sorted macrophages (CD3-, CD19-, Ly6G-, CD11b+) from Il17rafl/fl K14cre+ and Jmjd3fl/fl K14cre+ wounds and analyzed expression of inflammatory (Il1b, Il6, Tnf, Nos2) genes (Supplementary Fig. 11G, H). We found that Il17rafl/fl K14cre+ macrophages expressed decreased inflammatory Il1b, Tnf, Nos2 expression compared to their littermate (Il17rafl/fl K14cre-) controls (Supplementary Fig. 11G). Jmjd3fl/fl K14cre+ macrophages expressed decreased inflammatory Il1b, Il6, Tnf, Nos2 expression compared to their littermate (Jmjd3fl/fl K14cre-) controls (Supplementary Fig. 11H). Lastly, given the importance of keratinocyte-fibroblast crosstalk in regulating fibroblast functions such as collagen deposition and differentiation into reparative myofibroblasts, we assessed fibroblast function in these mice. Myofibroblasts are essential for wound contraction, and previous work from our group and others has shown that their transition is impaired in diabetic wounds59. We sorted wound fibroblasts (CD45-, CD31-, EpCAM-, TER119-, Tie2-) from Il17rafl/fl K14cre+ and Jmjd3fl/fl K14cre+ mice and analyzed expression of collagen production genes (Col1a1, Cald1) and myofibroblast genes (Myl9, Acta). We found increased Col1a1 and Cald1 in Il17rafl/fl K14cre+ fibroblasts and increased Myl9 and Acta in Jmjd3fl/fl K14cre+ fibroblasts (Supplementary Fig. 11I, J). Additionally, through our single cell RNA-sequencing analysis of wounds from Il17rafl/fl K14cre+, we identified that myofibroblast gene Acta expression was increased in Il17rafl/fl K14cre+ fibroblasts compared to their controls (Supplementary Figs. 12A, B), suggesting increased reparative myofibroblast transition. Collectively, these findings demonstrate that keratinocyte-specific targeting of JMJD3 in diabetic wounds may be a viable therapeutic strategy to improve keratinocyte migration and decrease inflammation, thereby limiting excessive recruitment of pathologic cell types and promoting a reparative landscape in diabetic wounds.
Discussion
Recent work has identified that increased inflammatory signals in the diabetic wound environment contribute to the reprogramming of critical immune and reparative cell types12,60,61. Prior work done by our lab highlights that the functions of diabetic wound cells, including keratinocytes, are significantly altered leading to delayed wound re-epithelialization7,8. Additionally, our lab has identified that IL-17A is linked to increased TH17 cell differentiation in wound tissues12. While IL-17A is essential for normal keratinocyte function, the downstream IL-17A signaling pathway in wound keratinocytes and the impact of excessive IL-17A levels on diabetic wound keratinocytes have not been studied. In this study, we found that high levels of IL-17A impair keratinocyte migration and induce an impaired keratinocyte phenotype by altering the expression of anti-migratory (Itga3, Timp1) and inflammatory (Ccl20, Cxcl1, Cxcl3, Cxcl5) genes, as summarized in our working model (Fig. 6). Upregulation of Itga3 and Timp1 may impair keratinocyte migration by enhancing adhesion36,37,38,39 and limiting matrix remodeling42, while increased inflammatory chemokine expression promotes immune cell recruitment21,45, contributing to a pro-inflammatory wound environment and delayed healing. We further identified that JMJD3, a histone demethylase, is upregulated in response to IL-17A via a TRAF6/NFĸB pathway downstream of the IL-17A receptor and results in a decrease in H3K27 trimethylation on the promoter(s) of anti-migratory and inflammatory genes (i.e., Itga3, Timp1, Ccl20, Cxcl1, Cxcl3, and Cxcl5). Importantly, treatment of IL-17A-stimulated keratinocytes with a JMJD3-specific inhibitor reduced both anti-migratory and inflammatory gene expression and protein levels. Our human bulk RNA-Seq and spatial scRNA-Seq analyses revealed increased IL-17A signaling and IL-17A-expressing cells in diabetic wounds, corresponding with our human single-cell RNA-Seq data demonstrating elevated expression of JMJD3 and associated anti-migratory (Itga3, Timp1) and inflammatory (Ccl20, Cxcl1, Cxcl3, Cxcl5) genes in diabetic wound keratinocytes. Finally, using our diabetic murine models with keratinocyte-specific deficiencies in IL-17A signaling or JMJD3 (DIO Il17rafl/fl K14cre+ and DIO Jmjd3fl/fl K14cre+), we found that loss of IL-17A signaling or JMJD3 in keratinocytes improved diabetic wound repair. Taken together, these findings suggest that the manipulation of the IL-17A/TRAF6/JMJD3 pathway in keratinocytes could be therapeutically manipulated to improve diabetic wound repair.
(Top) In normal wounds, keratinocytes receive balanced environmental cues that support regulated JMJD3 activity, allowing appropriate H3K27me3 demethylation and expression of pro-migratory and inflammatory genes, promoting keratinocyte migration and timely recruitment of reparative immune cells. (Bottom) In diabetic wounds, elevated IL-17A engages the IL-17 receptor and signals through the TRAF6/NFκB pathway to drive increased JMJD3 expression, resulting in excessive H3K27me3 demethylation and increased expression of Itga3, Timp1, Ccl20, Cxcl1, Cxcl3, and Cxcl5. This cascade contributes to decreased keratinocyte migration, increased inflammatory immune cell recruitment, and impaired diabetic wound healing.
Although it has been previously identified that JMJD3 is relevant in keratinocytes following injury27, the exact upstream regulation and the downstream genes that JMJD3 regulates were previously not explored. In this regard, our single cell RNA-Seq data from human wounds also supports JMJD3 as an upregulated enzyme in keratinocytes following injury, as it was highly expressed in keratinocytes compared to other cell types. Our findings also indicate that primary keratinocytes isolated from our murine model with a keratinocyte-specific Jmjd3-deficiency exhibited improved migration rates. This contrasts with an existing scratch migration study, which shows worse gap closure in HaCaT keratinocyte cell lines where JMJD3 was inhibited via siRNA27. However, our studies utilized primary keratinocytes, which are known to behave differently from commercial cell lines, and we employed genetic deletion rather than siRNA treatment, which could result in a compensatory increase in the expression of other genes involved in wound healing, potentially explaining these observed differences. Additionally, these mice displayed increased wound repair rates in vivo in the diabetic setting, suggesting that a fine balance of Jmjd3 expression may be needed for proper keratinocyte function in tissue repair processes.
Our study demonstrates that the increased expression of JMJD3 in diabetic wound keratinocytes is induced via an IL-17A-mediated mechanism. While the role of IL-17A in the pathogenesis of other chronic inflammatory skin diseases (i.e., psoriasis, lupus) is well characterized, its involvement in diabetic wound healing has not been previously explored. IL-17A-mediated induction of TRAF6 has been shown to result in downstream activation of canonical NFĸB pathways in keratinocytes48. A previous study showed that NFĸB can bind to the Jmjd3 promoter, regulating its expression in macrophages under chronic inflammation62. Further, through the use of keratinocyte-specific murine models and pharmacological approaches, we provide evidence linking the IL-17A/TRAF6/NFĸB axis to epigenetic CMEs (i.e., JMJD3) for the first time. Thus, our studies find that the NFĸB/JMJD3 axis is conserved in wound keratinocytes and identify IL-17A as an upstream regulator of this pathway in diabetic wounds. Although other sources of NFĸB-mediated JMJD3 regulation in diabetic keratinocytes (i.e., PRRs, TLRs, TNF-α receptor, and IL-1β receptor)63 may exist, our data suggest that IL-17A is a significant driver, particularly given its established role in regulating keratinocyte migration and inflammatory responses21,44,64.
We identified that the IL-17A/JMJD3 axis increases keratinocyte production of inflammatory chemokines, including Cxcl1, Cxcl3, and Cxcl5, which are known, amongst other functions, to promote neutrophil infiltration65. This is highly relevant as diabetic wounds are characterized by an increased neutrophil population66, although precise mechanisms remain unknown. In diabetic wounds, these neutrophils persist into the late stages of wound healing, contributing to prolonged inflammation. While IL-17A is known to directly promote neutrophil-induced inflammation67, the additional production of neutrophil chemoattractants by keratinocytes may further elevate neutrophil levels, particularly during the late phases of re-epithelialization. Our flow cytometric analysis revealed decreased neutrophils in diabetic wounds from mice with keratinocyte-specific deletion of IL-17A signaling or JMJD3, indicating that the IL-17A/JMJD3 axis may drive the excessive neutrophil recruitment observed in diabetic wounds. Additionally, we observed that IL-17A/JMJD3 upregulates Ccl20 in keratinocytes, which recruits CCR6+ TH17 cells21. Consistently, keratinocyte-specific deletion of IL-17A signaling or JMJD3 resulted in decreased TH17 cells, suggesting a potential positive feedback loop that further increases IL-17A levels in diabetic wounds. While this study focuses on the role of IL-17A in keratinocyte function, we acknowledge that IL-17A also directly regulates other cell types involved in wound healing. Notably, fibroblast function is significantly altered in diabetic wounds, leading to dysfunctional epithelialization59. Diabetic wound fibroblasts are known to exhibit impaired transition into reparative myofibroblasts59,68. Under normal conditions, IL-17A enhances fibroblast survival and proliferation and promotes a profibrotic phenotype69; however, the impact of elevated IL-17A levels, as observed in diabetic wounds, on fibroblast dysfunction has not been explored. Therefore, additional studies investigating how abnormal IL-17A levels in tissues affect other cells within the wound environment would enhance our knowledge of the complex interplay in wound tissue environments.
While our study provides strong evidence for a keratinocyte-intrinsic IL-17A/JMJD3 axis contributing to impaired wound healing in diabetes, there are limitations that warrant further exploration. The use of mouse models, though well-established for studying wound healing mechanisms, may not fully recapitulate the complexity of human diabetic wounds. To help bridge this gap, we performed spatial RNA-sequencing on human diabetic and non-diabetic wound tissue, which allowed us to identify IL-17A-expressing immune populations and their spatial proximity to keratinocytes. Interestingly, we identified myeloid cells, CD8⁺ T cells, neutrophils, and NK cells as prominent sources of IL-17A in diabetic wounds, highlighting less conventional producers beyond the well-established IL-17A-producing cell populations (i.e., RORγT⁺ TH17 and γδ T cells). Thus, this human dataset offers initial insights into a broader range of cellular sources contributing to elevated IL-17A levels. Nonetheless, further validation of this pathway and its upstream regulators in larger human cohorts will be critical to confirm its translational relevance.
Moreover, while our study identifies IL-17A-producing immune cells in diabetic wounds, we did not directly investigate the upstream factors driving increased IL-17A expression. Chronic inflammation, metabolic dysfunction, and microbial persistence in the diabetic wound environment have been implicated as potential contributors that may skew immune cell phenotypes toward IL-17A production20,60,70. For example, dysregulated IL-17A expression has been shown to exacerbate inflammatory responses and alter the wound microbiome21, while the diabetic microbiome itself may be more pathogenic, further promoting IL-17A-driven inflammation20. These factors likely contribute to a sustained pro-inflammatory milieu in diabetic wounds, and future studies will be necessary to define how these upstream cues influence IL-17A regulation in this context.
To conclude, our study highlights the role of the IL-17A/JMJD3 axis in diabetic keratinocytes, contributing to the dysregulated wound healing observed in diabetic tissue and identifies several promising therapeutic targets. While epigenetic regulation in non-immune cells remains underexplored, emerging evidence suggests that targeting these cells could offer therapeutic approaches7,71. While global inhibition of IL-17A likely disrupts essential functions in keratinocytes and other cell types, future therapies should involve cell-specific targeting to minimize off-target effects. In this regard, selective targeting of JMJD3 or its binding partners in keratinocytes may offer promise for restoring healing in diabetic patients.
Methods
Study approval
All experiments using human samples were approved by the IRB at the University of Michigan (IRB#: HUM00098915) and were conducted in accordance with the principles in the Declaration of Helsinki. All mice used were on a C57BL/6 background. We used 2 diabetic models, diet-induced obesity (DIO) and db/db, which were used in acute and/or chronic wound models. DIO mice were generated by feeding mice with a high fat diet (HFD) for 12 – 16 weeks to simulate a “prediabetic” state that is physiologically similar to humans; mice were used at 20 weeks of age or older. Female C57BL/6 J mice do not develop glucose intolerance/insulin resistance metabolic syndrome or “prediabetes” on a high-fat diet; therefore, they cannot be used in this T2D model. For db/db mice, both male and female mice were used in experiments starting at 7 weeks of age. Mice were housed at the University of Michigan Biomedical Sciences and Research Building in the Unit for Laboratory and Animal Medicine (ULAM) - a pathogen free animal facility. Mouse experiments were conducted with approval from our Institutional Animal Care and Use Committee (IACUC; Protocol no. PRO00009811) and all regulatory and safety standards were strictly adhered to.
Mice
All mice were housed at the University of Michigan animal facilities. C57BL/6 J, db/db, db/+, K14cre+ mice were purchased from The Jackson Laboratory at 6 - 7 weeks of age and maintained in breeding pairs in the Unit for Laboratory Animal Medicine (ULAM) facilities. Il17rafl/fl mice [RRID:IMSR_JAX:030849] on a C57BL/6 J background were a gift from B. Moore (University of Michigan). Traf6fl/fl mice [RRID:IMSR_JAX:030849] on a C57BL/6 J background were a gift from S. Soleimanpour (University of Michigan)72. Jmjd3fl/fl mice were created as previously reported by our laboratory73. Il17rafl/fl, Traf6fl/fl, Jmjd3fl/fl mice were then bred with K14cre+ [RRID:IMSR_JAX:018964] mice from the Jackson Laboratory to obtain mice expressing keratinocytes deficient in IL-17RA, TRAF6, and JMJD3, respectively. Floxed-cre mice were genotyped regularly after birth with custom primers. All mice were housed on a 14-h light/10-h dark cycle with free access to food, water, and bedding (Andersons Lab Bedding Bed-o’Cobs combination) at ambient temperatures of 65-75 °F (18-23 °C) and 40-60% humidity.
Wound-healing assessment
Mice were anesthetized with isoflurane using a vaporizer with the station flowmeter set at 2.0 liters per minute (LPM). Mice were depleted of hair on their dorsum via Veet (Reckitt Benckiser), and two 6 mm punch biopsy wounds were created per mouse54. For the chronic wound-healing model, a splinted full-thickness wound was used to minimize wound contracture54. Briefly, a 6 mm full-thickness excisional wound was created on the shaved dorsum of the anesthetized mice. A 10 mm doughnut-shaped silicone splint was then centered on the wound and fixed to the skin using interrupted 6-0 nylon sutures (Ethicon). In GSK-J4 rescue experiments, 1 mg/kg GSK-J4 (Tocris, Catalog 4594) or 100 μL DMSO control injection was performed s.c. at 2 points along the wound edge, as described previously by our group58. An iPad camera (8-megapixel) was used to take digital photographs daily. The wound surface area was calculated using ImageJ software (NIH) and was expressed as a percentage of the original wound size/time. The wounds were collected at the indicated time points and immediately snap-frozen and placed at –80 °C for RNA isolation, prepared for keratinocyte isolation or placed in formalin for histology. For wound keratinocyte isolation experiments, 6 mm wound biopsies were obtained, encompassing a 2 mm margin around the wound.
Some wounds were harvested and formalin fixed overnight, followed by embedding the tissue in paraffin. Murine tissue slides were stained with H&E74. The wound diameter was obtained by calculating the distance between the leading wound epithelial edges compared with the maximum diameter of the wound. Images were taken using a Zeiss Axioskop 2 microscope.
Primary keratinocyte isolation
Primary keratinocytes were isolated from the tails or wounds of mice31. First, skin from tails and wounds was incubated in 1% trypsin for 1.5 h and 2 h, respectively, at 37 °C. Following incubation, the epidermis from tails and wounds was peeled off, and epidermal cells (containing primarily keratinocytes) were agitated off into high-calcium (HiCa) medium (8% nonmixed FBS in EpiLife, with 60 μM Ca2+ (Gibco)). Next, primary keratinocytes from the tail were cultured in low-calcium (LoCa) medium (8% mixed FBS in EpiLife, with 60 μM Ca2+ (Gibco), at 0.05 mM final Ca2+ concentration) for 1 - 2 days at 37 °C. Following isolation of primary keratinocytes (tail) and wounds, cells were harvested for RNA or fixed in 1% paraformaldehyde for ChIP.
N/TERT human keratinocyte culture
N/TERT human keratinocyte cell line was a gift from J. Gudjonsson (University of Michigan) that was used with permission from James G. Rheinwald (Brigham and Women’s Hospital, Boston, Massachusetts, USA)75. N/TERT human keratinocytes were cultured in Keratinocyte Serum-Free Medium (Gibco) supplemented with 30 μg/mL bovine pituitary extract and 0.2 ng/mL human epidermal growth factor. Cells were maintained below 60% confluency and used for scratch wound assays at passages less than 10 to ensure consistent behavior and viability.
Scratch wound assay
N/TERT human keratinocytes or cultured primary keratinocytes isolated from tails of mice were subjected to scratch wound assays32,76. Briefly, cells were grown to 90 – 95% confluency at 37 °C. Since IL-17A increases proliferation in keratinocytes21, cells were pre-treated with a mitosis inhibitor, Mitomycin C (10 μg/mL), for 2 h to eliminate the contribution of cell proliferation to gap closure and to specifically assess changes in migration32. Mitomycin C medium was removed, and keratinocytes were washed once with HBSS (without Ca2+, Mg2+). P20 pipette tips were used to scratch the wells horizontally and vertically. A new tip was used for each scratch. Wells were washed with 1XHBSS and appropriate medium (containing peptide stimuli, inhibitors, etc.) was added. For neutralizing antibody experiments in murine keratinocytes, IgG isotype control (R&D, Cat# AB-108-C, Lot# ES4524111) or a combination of anti-mouse anti-CCL-20 (R&D, Cat# AF760-SP, Lot# CZQ1024101), anti-CXCL-1 (R&D, Cat# AF-453-SP, Lot# VF062403), anti-CXCL-3 (R&D, Cat# AF5568-SP, Lot# CDEO022502B), and anti-CXCL-5 (R&D, Cat# AF433-SP, Lot# BCH032502A) at 1 μg/mL was used. For neutralizing antibody experiments in human keratinocytes, IgG isotype control (R&D, Cat# AB-108-C, Lot# ES4524111) or a combination of anti-human anti-CCL-20 (R&D, Cat# AF360-SP, Lot# ATU0824011), anti-CXCL-1 (R&D, Cat# AF275-SP, Lot# BEP082503A), and anti-CXCL-5 (R&D, Cat# AF254-SP, Lot# ARO0424101) at 1 μg/mL was used. 0 h, 8 h, 12 h, 24 h, or 48 h images of the same sections of each well were taken using an inverted microscope (EVOS Cell Imaging System). Wound gap area was measured using ImageJ (NIH) software and comparisons were made using images taken at the same location of each well. Percentage of initial gap area was calculated using the following formula: \(\%{\rm{Initial}} \; {\rm{Wound}} \; {\rm{Area}}=\frac{{\rm{Area}} \; {\rm{at}}8,12,24,{\rm{or}}48{\rm{hr}}}{{\rm{Area}} \; {\rm{at}}0{\rm{hr}}}\times 100\%\)
Keratinocyte proliferation assay
Primary keratinocytes isolated from our murine models with the keratinocyte-specific JMJD3 deficiency (Jmjd3fl/fl K14cre+) and their littermate controls (Jmjd3fl/fl K14cre-) were seeded at identical densities (400,000 cells / mL), cultured for 24, 48 h, and alamarBlue (Invitrogen) reagent was added to get viability readouts at each timepoint after 4 h of incubation at 37 °C. Viability readouts or fluorescence intensities were compared to that of the initial timepoint to calculate proliferation rates or % initial viability at each timepoint. A subset of cells from each group was pre-treated with Mitomycin C (10 μg/mL) for 2 h and washed with HBSS (without Ca2+, Mg2+).
RNA extraction
Total RNA extraction was performed with TRIzol (Invitrogen) using manufacturer’s directions. RNA was extracted using chloroform, isopropanol and ethanol. Superscript III Reverse transcriptase (ThermoFisher Scientific) kits were used to synthesize cDNA from extracted RNA. cDNA primers for Jmjd3 (Mm01332680), Fgf2 (Mm01285715), Mmp2 (Mm00439498), Itga3 (Mm00442910), Timp1 (Mm01341361), Ccl20 (Mm01268754), Cxcl1 (Mm04207460), Cxcl3 (Mm01701838), Cxcl5 (Mm00436451), and 18 s (no. 4318839) were purchased from Applied Biosciences. RT-PCR was conducted with 2x Taqman Fast PCR mix and run on a 7500 Real-Time PCR system (Applied Biosciences), and data was then reviewed in a relative quantification analysis to the 18 s ribosomal RNA. All samples were assayed in triplicate. Data was then compiled in Excel (Microsoft) and presented using Prism software (GraphPad version 10.2.2).
PCR array analysis
Keratinocytes were isolated from tails of C57BL/6 mice, cultured, and stimulated with rIL-17A (40 ng/mL) for 1 h. RNA was isolated and DNase digested using the RNAeasy mini kit, and reverse transcription was performed using the RT2 first strand kit (QIAGEN). Gene expression was analyzed using PCR array PAMM-085Z (Qiagen), which includes PCR primers for 84 chromatin-modifying enzymes. Data were analyzed using the PCR Array Data Analysis Web Portal.
siRNA Knockdowns
Primary murine keratinocytes were transfected with 1 μM siRNA (Dharmacon Cat# E-063799-00-0005) targeted to Jmjd3 or a non-targeting control (NTC) siRNA for 72 h. Accell Delivery Medium (Dharmacon Cat# D-001910-01-05) was used as the transfection reagent.
Murine bulk RNA sequencing analysis
Keratinocytes were isolated from tails of C57BL/6 mice, cultured, and stimulated with rIL-17A (40 ng/mL) for 5 h. Keratinocytes were isolated from day 5 wounds from db/+ and db/db mice. RNA isolation was performed using a RNeasy Kit (Qiagen) with DNase digestion. Library construction and analysis of reads was performed as described previously70. Briefly, reads were trimmed using Trimmomatic and mapped using HiSAT277,78. Read counts were performed using the feature-counts option from the subRead package followed by the elimination of low reads, normalization and differential gene expression using edgeR79,80. Differential expression was performed on mapped reads using the taqwise dispersion algorithm in edgeR.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assay was performed as described previously using a kit purchased from Abcam (ab500)81. Briefly, following ex vivo studies, keratinocytes were cross linked in 1% formaldehyde for 10 min at RT and pellets stored at -80 °C until analyzed. Following manufacturer’s instructions, cells were lysed for 10 min on ice in an SDS-containing lysis buffer (Abcam), supplemented with a protease inhibitor cocktail (Abcam), sonicated to generate 100-300 base pair fragments. Five percent of the total chromatin volume was put aside for the input control. The rest of the chromatin was subsequently incubated with antibodies against JMJD3 (Cat# ab169197, Lot# GR3263105-3, Abcam) at 3 µg/sample, trimethylated H3K27 (Active Motif, Cat# 39155, Lot# 16021022) at 1 µg/sample, or rabbit polyclonal IgG (Diagenode, Cat# C15410206, Lot# RIG002C) at 1 or 3 µg/sample overnight at 4 °C. This was followed by addition of Protein A sepharose beads (Abcam) for 1 h at 4 °C. The pellet was washed, and a DNA purifying slurry (Abcam) was added. The eluates were reverse cross linked for 10 min at 98 °C, followed by a proteinase K digestion for 30 min at 55 °C and 10 min at 98 °C. Precipitated DNA was analyzed by quantitative real time PCR on a Taqman 7500 sequence detection system. The following primers were used to amplify DNA in samples: Itga3: (forward) 5’ GGGCTGCAAAGATCTCAAAC 3’ and (reverse) 5’ GCCACCACTATGTAAACCCA 3’, Timp1: (forward) 5’ CCAGGGAATACTCCTGCATC 3’ and (reverse) 5’ CTCAGGCAGGACACAATCTT 3’, Ccl20: (forward) 5’ TCACCACCATCTTTGGACTG 3’ and (reverse) 5’ ACAACATGACAGAGGTGTCC 3’, Cxcl1: (forward) 5’ AGGGACCTAACCTTGACAGT 3’ and (reverse) 5’ CTACAGTGATTTGCGGGGAT 3’, Cxcl3: (forward) 5’ GCGAAAAGTACTGGGGTACA 3’ and (reverse) 5’ ACTCCTGATTGTGACTGCTG 3’, Cxcl5: (forward) 5’ AAACTCCTTCCTTTGGCTCC 3’ and (reverse) 5’ TGAGGATGCCCTTCTTTTCC 3’
Enzyme linked immunosorbent assay (ELISA)
Keratinocytes were isolated, cultured, scratched, and stimulated in culture for 24 or 48 h in low-calcium (LoCa) medium. After stimulation, cell free supernatant was collected and analyzed by the University of Michigan Immune Monitoring Shared Resource Core for CCL-20, CXCL-1, CXCL-5 or specific enzyme immunoassay kits for CXCL-3 (Boster Bio) and TIMP-1 (R&D Systems) according to the manufacturer’s instructions.
Western blot
Cell suspensions were lysed in radioimmunoprecipitation assay (RIPA) buffer (Sigma) and standardized for protein concentrations using a Bradford protein assay (BioRad) to generate a standard curve. Equal amounts of protein were then loaded onto to 4 to 12% sodium dodecyl sulfate gel electrophoresis under reducing conditions. Protein bands were then transferred to nitrocellulose membranes and probed with primary antibodies anti-mouse ITGA3 (Cat# ab190731, Lot# 1043494-4, Abcam, 1:1000), anti-mouse beta actin (Cat# A5316-100UL, Lot# 0000127607, Sigma Aldrich, 1:1000) at 4 °C for 48 hr. For JMJD3 western blots, nuclear extraction was performed using a kit (Active Motif) and lysates were probed with primary antibodies anti-JMJD3 (Cat# ab169197, Lot# GR3263105-3, Abcam, 1 µg/mL) or anti-H3 (Cat# 3638S, Lot# 11, Cell Signaling, 1:1000). All primary antibodies were diluted 1:1000 in 1% BSA in 0.1% Tween Tris-buffered saline (TBS-T) solution. Nitrocellulose membranes were then washed and incubated with horseradish peroxidase−labeled secondary antibody (Cell Signaling) for 1 hr at room temperature and visualized with chemiluminescence (Thermo Fisher Scientific). Blot images were analyzed using NIH ImageJ software to obtain sample densitometry readings normalized to beta actin.
Histology and immunofluorescence
Tissues were harvested from mice and fixed in 10% formalin for 24 h then stored in 70% ethanol. Specimens were embedded in paraffin and sectioned onto microscope slides. After deparaffinization, sections underwent Trichrome staining or were processed for immunofluorescence. For histomorphometric analysis of wounds, re-epithelialization rates were calculated as the distance between the two leading epithelial tongues expressed as a percentage of the initial wound diameter. Wound contraction rates were determined by measuring the distance between the two edges of intact dermis or solid blue collagen bands and expressing this as a percentage of the initial wound diameter. For immunofluorescent staining, slides underwent antigen retrieval in citric acid buffer (pH 6.0). Samples were then permeabilized, blocked, and incubated with primary antibodies, rabbit polyclonal IgG (Diagenode, Cat# C15410206, Lot# RIG002C), anti-cytokeratin14 (Cat# LL002, Abcam, 5 μg/mL), anti-IL-17A (Cat# ab91649, Lot# 1017634-1, Abcam, 5 μg/mL), anti-H3K27me3 (Cat# 39155, Lot# 16021022, Active motif, 2 μg/mL), or anti-αSMA (Cat# A5228-100UL, Lot# 0000348067, Sigma-Aldrich, 2 μg/mL) in blocking solution overnight at 4 C in a humidity chamber. The following day, slides were washed in PBS, then incubated with fluorophore-conjugated secondary antibody (Vector Labs, 1:400) in PBS for 2 h at room temperature or with DAPI. Slides were washed with PBS, mounted, allowed to dry overnight, and then imaged. Images were taken with Nikon Ti2 Widefield Fluorescence Microscope at 20X magnification to capture the wound edge or 4X magnification to capture the whole wound. To quantify αSMA staining, brightness/contrast adjustments and background subtraction were first applied to all images using identical settings. Granulation tissue regions of interest (ROIs) were analyzed in ImageJ. Mean fluorescence intensity (MFI) of αSMA within the ROI was quantified and normalized to DAPI signal.
Flow cytometry
For T cell analysis, single-cell suspensions from murine wound tissue were prepared and filtered through a 70 μm cell strainer. Cells were stained with BD Horizon™ Fixable Viability Stain 510 (BD Biosciences, Cat# 564406, RRID: AB_2869572) to exclude dead cells. Fc receptors were blocked using anti-CD16/32 (BioXCell, Cat# CUS-HB-197) for 10 minutes on ice. For lineage exclusion, cells were stained with FITC-conjugated anti-CD8 (Invitrogen, Cat# 11-0081-81, RRID: AB_464915), anti-CD19 (BioLegend, Cat# 115506, RRID: AB_313641), and anti-Gr-1 (BioLegend, Cat# 108406, RRID: AB_313371). For T cell phenotyping, surface markers included CD3 APC (Invitrogen, Cat# 17-0031-82, RRID: AB_469315), and CD4 BV605 (BioLegend, Cat# 100547, RRID: AB_11125962). Following surface staining, cells were fixed in 2% formaldehyde and permeabilized using BD Perm/Wash buffer (BD Biosciences, Cat# 00-8333-56). Intracellular staining was performed with Foxp3 PE (BioLegend, Cat# 126404, RRID: AB_1089117) for regulatory T cells and RORγt PerCP-Cy5.5 (BD Pharmingen, Cat# 562683, RRID: AB_2737720) for TH17 cells. For myeloid cell analysis, single-cell suspensions from murine wound tissue were similarly prepared and stained with BD Horizon™ Fixable Viability Stain 510 (BD Biosciences, Cat# 564406, RRID: AB_2869572) for viability discrimination. To exclude lymphoid and erythroid lineages, cells were stained with biotin-conjugated antibodies: anti-CD3 (BioLegend, Cat# 100304, RRID: AB_312669), anti-CD19 (BioLegend, Cat# 115504, RRID: AB_313639), and anti-TER-119 (BioLegend, Cat# 116204, RRID: AB_313705). Streptavidin-Pacific Orange (BioLegend, Cat# 405233) was used to label these biotinylated antibodies. Lineage-positive and dead cells were excluded from analysis. Myeloid populations were then identified using: CD45 PE-Cy7 (BioLegend, Cat# 103114, RRID: AB_312979), F4/80 PE (BioLegend, Cat# 111704, RRID: AB_2936728), CD11b APC (BioLegend, Cat# 101212, RRID: AB_312795), Ly6C PerCP (BioLegend, Cat# 128028, RRID: AB_10897805), and Ly6G FITC (BioLegend, Cat# 127606, RRID: AB_1236494). All flow cytometric analyses were performed using FlowJo version 10.10.0 software.
Magnetic-activated cell sorting (MACS) of murine wound monocyte, CD4+ T Cell, and fibroblast isolates
MACs sorting of wound cell isolates was performed. Wound tissue from mice was digested into single-cell suspensions as described previously. To enrich for monocytes, magnetic-activated cell sorting (MACS) was performed using a two-step process. First, cells were subjected to negative selection to deplete lymphocytes and neutrophils. Briefly, single-cell suspensions were incubated with biotin-conjugated antibodies against lineage markers: anti-mouse CD3 (BioLegend, Cat#100304, RRID: AB_312669), CD19 (BioLegend, Cat#115504, RRID: AB_313639), and Ly6G (BioLegend, Cat#127604, RRID: AB_1186108). Following incubation, cells were labeled with streptavidin-conjugated magnetic particles using the EasySep™ Mouse Streptavidin RapidSpheres™ Isolation Kit (Stemcell Technologies, Cat#19860) according to the manufacturer’s instructions. The lineage-negative fraction was then subjected to positive selection for CD11b+ myeloid cells using the EasySep™ Mouse CD11b Positive Selection Kit II (Stemcell Technologies, Cat#18970). The resulting CD11b+ cell population was enriched for monocytes and used for downstream applications. For CD4+ T cell isolation, CD4⁺ T cells were isolated from single-cell suspensions of murine wound or lymphoid tissues using the EasySep™ Mouse CD4⁺ T Cell Isolation Kit II (Stemcell Technologies, Cat#18952 A). The procedure was performed according to the manufacturer’s instructions to positively select for CD4⁺ T cells. Isolated cells were used for downstream applications such as gene expression analysis. Lastly, to enrich for wound fibroblasts, single-cell suspensions from digested murine wound tissue were subjected to negative selection to remove hematopoietic, endothelial, and epithelial cell populations. Cells were incubated with biotin-conjugated antibodies targeting non-fibroblast lineages: anti-mouse CD45 (BioLegend, Cat#103104, RRID: AB_312969), CD31 (BioLegend, Cat#102404, AB_312899), EpCAM (BioLegend, Cat#118204, AB_1134178), TER119 (BioLegend, Cat#116204, RRID: AB_313705), and Tie2/CD202b (BioLegend, Cat#124006, RRID: AB_2203221). Labeled cells were then incubated with streptavidin-conjugated magnetic particles and depleted using the EasySep™ Mouse Streptavidin RapidSpheres™ Isolation Kit (Stemcell Technologies, Cat#19860), following the manufacturer’s protocol. The resulting lineage-negative cell population was enriched for fibroblasts and used for downstream analysis.
Human wound isolation
All experiments using human samples were approved by the IRB at the University of Michigan and were conducted in accordance with the principles in the Declaration of Helsinki. Written informed consent was obtained from all subjects involved in human studies. Human subject data is de-identified and these patients are not followed after the sample is collected. Per our IRB, at initial sample collection, we obtain information on age, sex, and diabetes status. For the bulk RNA sequencing cohort, biopsies from human diabetic wounds (N = 4) versus normal skin samples (N = 38) were collected. For a separate scRNA-Seq cohort, biopsies from human diabetic wounds versus normal skin samples were collected. Within the diabetic patient samples for scRNA-Seq, the average age was 60 years, with all patients having diabetes, hypertension, hyperlipidemia, and coronary artery disease. Within the nondiabetic patient samples, the average age was 70 years, with 50% of patients having hypertension, hyperlipidemia, and coronary artery disease. Wounds were obtained from the lateral edge of wound specimens using an 8 mm punch biopsy tool and processed for RNA extraction and sequencing as described for the murine wounds, and the material was fully consumed during analysis. RNA with RNA integrity number scores of greater than 8 were used, and all values were done with comparison to 28S/18S ratios and other housekeeping genes.
Human Bulk RNA-Seq and scRNA-Seq Analyses
Single-cell suspensions for scRNA-Seq were generated as follows. Wound tissue from diabetic and non-diabetic patients was collected by punch biopsy and immediately processed. Samples were incubated overnight at 4 °C in 0.4% Dispase (Life Technologies, Thermo Fisher Scientific) prepared in HBSS (Gibco, Thermo Fisher Scientific). The epidermal and dermal layers were then separated. The epidermis was enzymatically digested with 0.25% Trypsin-EDTA (Gibco, Thermo Fisher Scientific) containing 10 U/mL DNase I (Thermo Fisher Scientific) for 1 h at 37 °C, followed by quenching with FBS (Atlanta Biologicals) and filtration through a 70 µm strainer. Dermal tissue was finely minced and digested in 0.2% Collagenase II (Life Technologies, Thermo Fisher Scientific) and 0.2% Collagenase V (MilliporeSigma) for 1.5 h at 37 °C, then passed through a 70 µm strainer. Epidermal and dermal cells were combined in a 1:1 ratio and submitted to the University of Michigan Advanced Genomics Core for scRNA-Seq library generation on the 10x Genomics Chromium platform. Libraries were sequenced on the Illumina NovaSeq 6000 to produce 151 bp paired-end reads. Read trimming and quality control followed standard pipelines82,83. Processed reads were aligned to the human reference genome (GRCh37) using STAR84. Gene-level counts were obtained with HTSeq85 and normalized using DESeq286, which was also used for differential expression testing with negative binomial models. To expand control sample representation, additional skin biopsy data from our prior study87 were incorporated. Bulk RNA-seq and scRNA-seq data have been deposited in the Gene Expression Omnibus under accession numbers GSE154556 and GSE154557, respectively. For scRNA-Seq processing, Cell Ranger (10x Genomics) was used for alignment, quality filtering, and quantification. Subsequent normalization, data integration, and clustering were carried out in Seurat88. Cell identities were assigned by comparing cluster marker genes with known lineage-specific signatures. Spatial scRNA-Seq data from diabetic (N = 3) and non-diabetic (N = 1) wounds were analyzed to assess keratinocyte proximity to IL-17A-expressing cells using Giotto (v4.2.0). A Delaunay triangulation–based spatial network was constructed with a maximum intercellular distance of 50 µm. Cells with detectable IL-17A transcripts (raw counts > 0) were defined as IL-17A+. Keratinocytes were designated “Near” if they had at least one direct spatial edge connecting to an IL-17A+ cell and “Far” if no such connections were present. Log-normalized expression values for CXCL1, CCL20, CXCL3, CXCL5, ITGA3, and TIMP1 were compared across proximity (“Near” vs. “Far”) and diabetic status groups.
Statistics
GraphPad Prism software (RRID:SCR_002798) version 10.2.2 was used to analyze the data. Data were analyzed for normal distribution and then statistical significance between multiple groups was determined using a one-way analysis of variance test followed by Newman-Keuls post hoc test. For all single group comparisons, if the data passed the normality test, we used a two-tailed Student’s t test. Otherwise, data were analyzed using the Mann–Whitney U-test. All data are representative of at least two independent experiments as detailed in the figure legends. A P value of less than or equal to 0.05 was significant. Values smaller than 0.0001 are reported as P < 0.0001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided with this paper. All sequencing data generated in this study have been deposited in the Gene Omnibus (GEO) or Zenodo repositories. The human bulk RNA-sequencing data have been deposited under accession code GSE154556, and the human scRNA-sequencing data under accession code GSE154557. The human spatial scRNA-sequencing data of diabetic and non-diabetic wounds are available at [https://doi.org/10.5281/zenodo.17508360]. The mouse bulk RNA-sequencing data of IL-17A-stimulated keratinocytes are available at [https://doi.org/10.5281/zenodo.17504145], and the mouse bulk RNA-sequencing data of db/+ and db/db wounds at [https://doi.org/10.5281/zenodo.17508179]. The mouse scRNA-sequencing data of wounds from keratinocyte-specific IL-17A signaling-deficient mice are available at [https://doi.org/10.5281/zenodo.17508202]. Source data are provided with this paper.
References
Dekoninck, S. & Blanpain, C. Stem cell dynamics, migration and plasticity during wound healing. Nat. Cell Biol. 21, 18–24 (2019).
Wang, Y. & Graves, D. T. Keratinocyte function in normal and diabetic wounds and modulation by FOXO1. J. Diab. Res. 28, 3714704 (2020).
Caley, M. P., Martins, V. L. & O’Toole, E. A. Metalloproteinases and wound healing. Adv. Wound Care (N. Rochelle) 4, 225–234 (2015).
Petrie, R. J., Doyle, A. D. & Yamada, K. M. Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 10, 538–549 (2009).
Piipponen, M., Li, D. & Landén, N. X. The immune functions of keratinocytes in skin wound healing. Int. J. Mol. Sci. 21, 8790 (2020).
Jiang, Y. et al. Cytokinocytes: the diverse contribution of keratinocytes to immune responses in skin. JCI Insight 5, e142067 (2020).
Wolf, S. J. et al. IFN-κ is critical for normal wound repair and is decreased in diabetic wounds. JCI Insight 7, e152765 (2022).
Wolf, S. J. et al. Diabetic wound keratinocytes induce macrophage jmjd3-mediated Nlrp3 Expression via IL-1R Signaling. Diabetes 73, 1462–1472 (2024).
Kelly, G. et al. Dysregulated cytokine expression in lesional and nonlesional skin in hidradenitis suppurativa. Br. J. Dermatol. 173, 1431–1439 (2015).
Li, L. et al. High glucose suppresses keratinocyte migration through the inhibition of p38 MAPK/Autophagy pathway. Front. Physiol. 10, 24 (2019).
Moura, J. et al. Impaired T-cell differentiation in diabetic foot ulceration. Cell Mol. Immunol. 14, 758–769 (2017).
Audu, C. O. et al. Histone demethylase JARID1C/KDM5C regulates Th17 cells by increasing IL-6 expression in diabetic plasmacytoid dendritic cells. JCI Insight 9, e172959 (2024).
Wang, Q. et al. Dysregulation of circulating CD4+CXCR5+ T cells in type 2 diabetes mellitus. APMIS 123, 146–151 (2015).
Cai, B. et al. Micro-inflammation characterized by disturbed Treg/Teff balance with increasing sIL-2R in patients with type 2 diabetes. Exp. Clin. Endocrinol. Diab. 121, 214–219 (2013).
Dworacka, M. et al. Circulating CD3+56+ cell subset in pre-diabetes. Exp. Clin. Endocrinol. Diab. 122, 65–70 (2014).
Hu, W. et al. Skin γδ T cells and their function in wound healing. Front. Immunol. 13, 875076 (2022).
Lavoz, C. et al. Could IL-17A be a novel therapeutic target in diabetic nephropathy?. J. Clin. Med. 9, 272 (2020).
Ma, J. et al. Interleukin 17A promotes diabetic kidney injury. Sci. Rep. 9, 2264 (2019).
Mu, X. et al. IL-17 in wound repair: bridging acute and chronic responses. Cell Commun. Signal. 22, 288 (2024).
Hadian, Y., Bagood, M. D., Dahle, S. E., Sood, A. & Isseroff, R. R. Interleukin-17: Potential Target for Chronic Wounds. Mediators Inflamm. 2019, 1297675 (2019).
Furue, M., Furue, K., Tsuji, G. & Nakahara, T. Interleukin-17A and Keratinocytes in Psoriasis. Int. J. Mol. Sci. 21, 1275 (2020).
Majumder, S. & McGeachy, M. J. IL-17 in the Pathogenesis of Disease: Good Intentions Gone Awry. Annu. Rev. Immunol. 26, 537–556 (2021).
Gurczynski, S. J. & Moore, B. B. IL-17 in the lung: the good, the bad, and the ugly. Am. J. Physiol. Lung Cell Mol. Physiol. 314, L6–L16 (2018).
Jaenisch, R. & Bird, A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254 (2003).
Kimball, A. S. et al. The histone methyltransferase Setdb2 modulates macrophage phenotype and uric acid production in diabetic wound repair. Immunity 51, 258–271 (2019).
Kimball, A. S. et al. The Histone Methyltransferase MLL1 Directs Macrophage-Mediated Inflammation in Wound Healing and Is Altered in a Murine Model of Obesity and Type 2 Diabetes. Diabetes 66, 2459–2471 (2017).
Na, J. et al. Histone H3K27 Demethylase JMJD3 in Cooperation with NF-κB Regulates Keratinocyte Wound Healing. J. Invest. Dermatol. 136, 847–858 (2016).
Kashgari, G. et al. GRHL3 activates FSCN1 to relax cell-cell adhesions between migrating keratinocytes during wound reepithelialization. JCI Insight 6, e142577 (2021).
Konieczny, P. et al. Interleukin-17 governs hypoxic adaptation of injured epithelium. Science 377, eabg9302 (2022).
Wikramanayake, T. C., Borda, L. J., Miteva, M. & Paus, R. Seborrheic dermatitis-Looking beyond Malassezia. Exp. Dermatol. 28, 991–1001 (2019).
Lichti, U. et al. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat. Protoc. 3, 799–810 (2008).
Vang Mouritzen, M. & Jenssen, H. Optimized scratch assay for in vitro testing of cell migration with an automated optical camera. J. Vis. Exp. 138, 57691 (2018).
Smits, J. P. H. et al. Immortalized N/TERT keratinocytes as an alternative cell source in 3D human epidermal models. Sci. Rep. 7, 11838 (2017).
Koike, Y., Yozaki, M., Utani, A. & Murota, H. Fibroblast growth factor 2 accelerates the epithelial-mesenchymal transition in keratinocytes during wound healing process. Sci. Rep. 10, 18545 (2020).
Meyer, M. et al. FGF receptors 1 and 2 are key regulators of keratinocyte migration in vitro and in wounded skin. J. Cell Sci. 125, 5690–5701 (2012).
Margadant, C. et al. Integrin alpha3beta1 inhibits directional migration and wound re-epithelialization in the skin. J. Cell Sci. 122, 278–288 (2009).
Kim, J. P., Zhang, K., Kramer, R. H., Schall, T. J. & Woodley, D. T. Integrin receptors and RGD sequences in human keratinocyte migration: unique anti-migratory function of α3β1 epiligrin receptor. J. Invest. Dermatol. 98, 764–770 (1992).
Hodivala-Dilke, K. M., DiPersio, C. M., Kreidberg, J. A. & Hynes, R. O. Novel roles for α3β1 integrin as a regulator of cytoskeletal assembly and as a transdominant inhibitor of integrin receptor function in mouse keratinocytes. J. Cell Biol. 142, 1357–1369 (1998).
DeHart, G. W., Healy, K. E. & Jones, J. C. The role of α3β1 integrin in determining the supramolecular organization of laminin-5 in the extracellular matrix of keratinocytes. Exp. Cell Res. 283, 67–79 (2003).
Rousselle, P., Montmasson, M. & Garnier, C. Extracellular matrix contribution to skin wound re-epithelialization. Matrix Biol. 75-76, 12–26 (2019).
Arpino, V., Brock, M. & Gill, S. E. The role of TIMPs in regulation of extracellular matrix proteolysis. Matrix Biol. 44-46, 247–254 (2015).
Rohani, M. G. & Parks, W. C. Matrix remodeling by MMPs during wound repair. Matrix Biol. 44-46, 113–121 (2015).
Huang, S. M. et al. High glucose environment induces M1 macrophage polarization that impairs keratinocyte migration via TNF-α: An important mechanism to delay the diabetic wound healing. J. Dermatol. Sci. 96, 159–167 (2019).
Chiricozzi, A. et al. Integrative responses to IL-17 and TNF-α in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J. Invest. Dermatol. 131, 677–687 (2011).
Korbecki, J., Maruszewska, A., Bosiacki, M., Chlubek, D. & Baranowska-Bosiacka, I. The Potential Importance of CXCL1 in the Physiological State and in Noncancer Diseases of the Cardiovascular System, Respiratory System and Skin. Int. J. Mol. Sci. 24, 205 (2022).
Gallagher, K. A. et al. Epigenetic changes in bone marrow progenitor cells influence the inflammatory phenotype and alter wound healing in type 2 diabetes. Diabetes 64, 1420–1430 (2015).
Audu, C. O. et al. Macrophage-specific inhibition of the histone demethylase JMJD3 decreases STING and pathologic inflammation in diabetic wound repair. Cell. Mol. Immunol. 19, 1251–1262 (2022).
Swaidani, S. et al. TRAF regulation of IL-17 cytokine signaling. Front. Immunol. 10, 1293 (2019).
Qian, Y. et al. IL-17 signaling in host defense and inflammatory diseases. Cell. Mol. Immunol. 7, 328–333 (2010).
Chang, S. H. et al. Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. J. Biol. Chem. 281, 35603–35607 (2006).
Xu, S. & Cao, X. Interleukin-17 and its expanding biological functions. Cell. Mol. Immunol. 7, 164–174 (2010).
Dalpatraj, N., Naik, A. & Thakur, N. GSK-J4: An H3K27 histone demethylase inhibitor, as a potential anti-cancer agent. Int. J. Cancer 153, 1130–1138 (2023).
Qiu, A. W., Cao, X., Zhang, W. W. & Liu, Q. H. IL-17A is involved in diabetic inflammatory pathogenesis by its receptor IL-17RA. Exp. Biol. Med. (Maywood) 246, 57–65 (2021).
Davis, F. M. et al. Epigenetic regulation of the PGE2 pathway modulates macrophage phenotype in normal and pathologic wound repair. JCI Insight 5, 138443 (2020).
Melvin, W. J. et al. The histone methyltransferase mixed-lineage-leukemia-1 drives T cell phenotype via notch signaling in diabetic tissue repair. JCI Insight 9, e179012 (2024).
Tartaglia, L. A. et al. Identification and expression cloning of a leptin receptor, OB-R. Cell 83, 1263–1271 (1995).
Parekh, P. I. et al. Reversal of diet-induced obesity and diabetes in C57BL/6J mice. Metabolism 47, 1089–1096 (1998).
Davis, F. M. et al. Palmitate-TLR4 signaling regulates the histone demethylase, JMJD3, in macrophages and impairs diabetic wound healing. Eur. J. Immunol. 50, 1929–1940 (2020).
Liu, Y. et al. Fibroblasts: Immunomodulatory factors in refractory diabetic wound healing. Front. Immunol. 13, 918223 (2022).
Ling, C. & Rönn, T. Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab. 29, 1028–1044 (2019).
Nilsson, E. et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes 63, 2962–2976 (2014).
De Santa, F. et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130, 1083–1094 (2007).
Liu, T. et al. NF-κB signaling in inflammation. Sig. Transduct. Target. Ther. 2, 17023 (2017).
Zhang, X., Yin, M. & Zhang, L. J. Keratin 6, 16 and 17-Critical Barrier Alarmin Molecules in Skin Wounds and Psoriasis. Cells 8, 807 (2019).
Rajarathnam, K., Schnoor, M., Richardson, R. M. & Rajagopal, S. How do chemokines navigate neutrophils to the target site: Dissecting the structural mechanisms and signaling pathways. Cell Signal 54, 69–80 (2019).
Ma, X. X. et al. The role of neutrophils in diabetic ulcers and targeting therapeutic strategies. Int. Immunopharmacol. 124, 110861 (2023).
Takagi, N. et al. IL-17A promotes neutrophilic inflammation and disturbs acute wound healing in skin. Exp. Dermatol. 26, 137–144 (2017).
Wan, R. et al. Diabetic wound healing: The impact of diabetes on myofibroblast activity and its potential therapeutic treatments. Wound Repair Regen. 29, 573–581 (2021).
Wang, J. & Ding, X. IL-17 signaling in skin repair: safeguarding metabolic adaptation of wound epithelial cells. Signal Transduct. Target. Ther. 7, 359 (2022).
Kimball, A. S. et al. Ly6CHi Blood Monocyte/macrophage Drive Chronic Inflammation And Impair Wound Healing in Diabetes Mellitus. Arterioscler. Thromb. Vasc. Biol. 38, 1102–1114 (2018).
Zhang, T. et al. EZH2-dependent epigenetic modulation of histone H3 lysine-27 contributes to psoriasis by promoting keratinocyte proliferation. Cell Death Dis. 11, 826 (2020).
Kobayashi, T. et al. TRAF6 is a critical factor for dendritic cell maturation and development. Immunity 19, 353–363 (2003).
Kroetz, D. N. et al. Type I interferon induced epigenetic regulation of macrophages suppresses innate and adaptive immunity in acute respiratory viral infection. PLoS Pathog. 11, e1005338 (2015).
Boniakowski, A. M. et al. SIRT3 regulates macrophage-mediated inflammation in diabetic wound repair. J. Invest. Dermatol. 139, 2528–2537 (2019).
Dickson, M. A. et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell Biol. 20, 1436–1447 (2000).
Martinotti, S. & Ranzato, E. Scratch Wound Healing Assay. In: Turksen, K. (eds) Epidermal Cells. Methods Mol. Biol. 2109, 225–229 (2019).
Bolger, A. M. et al. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Kim, D. et al. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Liao, Y. et al. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Robinson, M. D. et al. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Ishii, M. et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 114, 3244–3254 (2009).
Li, B. et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-Seq provides insights into disease mechanisms. J. Invest. Dermatol. 134, 1828–1838 (2014).
Tsoi, L. C. et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 16, 24 (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-Seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15, 550 (2014).
Tsoi, L. C. et al. Progression of acute-to-chronic atopic dermatitis is associated with quantitative rather than qualitative changes in cytokine responses. J. Allergy Clin. Immunol. 145, 1406–1415 (2020).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Acknowledgements
The authors wish to thank Robin Kunkel for her artistic expertise. This work was supported in part by National Institute of Health grants R01-HL137919 (KAG), R01-DK124290 (KAG), R01-HL15627401 (KAG), R01-DK127531 (KAG), T32-AI007413 (BBM), and F31-DK136199 (JYM).
Author information
Authors and Affiliations
Contributions
J.Y.M., S.J.W., A.D.J., J.S., C.O.A., W.J.M., L.C.T., J.E.G., B.B.M., and K.A.G. designed research; J.Y.M., S.J.W., C.O.A., W.J.M., A.D.J., K.D.M., T.M.B., J.S., L.D.H., E.C.B., S.R., G.S.J., A.L.E., M.N., H.Z., Q.L., R.B., L.C.T., J.E.G., B.B.M., and K.A.G. performed research; L.C.T., F.M.D., A.T.O., J.E.G., B.B.M., B.L., and K.A.G. contributed new reagents/analytic tools; J.Y.M., S.J.W., C.O.A., W.J.M., A.D.J., L.C.T., H.Z., Q.L., R.B., J.E.G., B.B.M., and K.A.G. analyzed data; and J.Y.M. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Irena Pastar, Weifeng He and Sabine Werner for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Moon, J.Y., Wolf, S.J., Joshi, A.D. et al. IL-17A is increased in diabetic wounds and impairs keratinocyte function via histone demethylase JMJD3. Nat Commun 17, 780 (2026). https://doi.org/10.1038/s41467-025-67456-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-67456-3