Abstract
In image-based drug discovery, accurately capturing cellular phenotypic responses to chemical perturbations is crucial for understanding drug mechanisms and predicting efficacy. However, existing approaches often depend on complex, multi-step pipelines that are computationally intensive and prone to error. PhenoProfiler addresses these challenges with an efficient, end-to-end deep learning framework that directly transforms high-content, multi-channel cellular images into low-dimensional quantitative representations. Evaluated on nearly 400,000 high-content images and 8.42 million single-cell images, PhenoProfiler consistently outperforms state-of-the-art methods by up to 20% in both accuracy and robustness. Its tailored phenotype correction strategy further emphasizes treatment-induced variations, improving the detection of biologically meaningful and reproducible signals. PhenoProfiler also effectively clusters treatments with shared molecular pathways and biological annotations, facilitating mechanistic interpretation and target discovery. Collectively, PhenoProfiler establishes a scalable, interpretable, and generalizable framework for high-throughput phenotypic profiling, paving the way for next-generation AI-driven drug screening, precision therapeutics, and systems-level understanding of cellular responses.
Similar content being viewed by others
Introduction
In image-based drug discovery1,2, particularly with techniques like Cell Painting, learning robust image representations is essential for extracting meaningful insights from complex, high-throughput image datasets. Cell Painting involves using multiple fluorescent dyes to label various organelles and cellular components, producing multi-channel images that capture phenotypic changes in response to different drugs and perturbations3. These high-dimensional images are rich in information, making automated methods for learning image representations crucial. These representations enable the development of predictive models for drug discovery, allowing for better identification of therapeutic compounds, understanding drug mechanisms, and predicting off-target effects2,3,4. Many laboratories and companies have generated extensive Cell Painting datasets5,6,7,8, facilitating the identification of phenotypic changes in response to drug treatments and supporting various downstream applications. For example, analyzing changes in cell morphology can provide insights into drug targets and mechanisms of action9,10, while comparisons of cell morphology before and after treatment can help evaluate drug efficacy and identify promising candidates with significant impacts7,11. Additionally, these image representations can be integrated with other data types, such as gene expression profiles6,12, allowing for comprehensive multimodal analyses that further advance drug discovery and biomedical research8,13.
The high-dimensional nature of Cell Painting images often introduces redundancy and noise, necessitating extensive preprocessing steps such as normalization, segmentation, and artifact removal7,14. Furthermore, the large-scale nature of these datasets demands substantial computational resources for scalable processing, and the extracted morphological features may lack biological interpretability, making it challenging to directly utilize these images for meaningful analysis. General-purpose models pre-trained on ImageNet15, such as ResNet5016 and ViT17, offer general solutions for image processing and representation learning. ResNet50 provides hierarchical feature representations through residual connections, while ViT captures long-range dependencies in image data. ImageNet pre-training further enhances their generalizability for computational image analysis across diverse datasets. To address the specific challenges posed by Cell Painting images, tailored methods including CellProfiler18, DeepProfiler9, SPACe19 and OpenPhenom20 have been developed to extract informative and compact representations of cell morphology. CellProfiler is a versatile, open-source tool for high-throughput image analysis, enabling biologists to extract physical cellular features (e.g., size, shape, intensity, texture) through modular pipelines. SPACe enhances computational efficiency using less but refined feature set and GPU-accelerated texture analysis. DeepProfiler adopts a deep learning approach, utilizing the EfficientNet21 model architecture to generate morphological profiles from Cell Painting sub-images. OpenPhenom leverages Vision Transformer (ViT)-based masked autoencoders to analyze large-scale microscopy images. By transforming complex image data into concise and interpretable representations, these methods enable phenotypic profiling, providing valuable insights into drug effects and cellular perturbations.
Despite these advancements, existing methods for morphological representation learning face several critical limitations, particularly when applied to high-dimensional Cell Painting images. First, these methods often process whole multi-channel images by decomposing them into multiple sub-images, resulting in a complex and resource-intensive workflow. These approaches typically involve segmenting the whole images to identify individual cell locations, extracting sub-images for each cell, applying models to extract features from these sub-images, and finally integrating those features to generate a comprehensive representation of the original whole multi-channel image. This multi-step process not only increases computational and acquisition costs but also introduces additional sources of error, such as inaccuracies in segmentation and feature integration. Second, these methods rely on drug treatment conditions as classification labels9,22, which provide limited information for capturing the diversity and complexity of cellular responses. This reliance often results in less biologically meaningful phenotypic representations, as those condition labels may fail to capture subtle morphological changes. Moreover, these labels lack universality and are often specific to certain plates or experimental setups. Consequently, existing models trained on these limited labels struggle to generalize effectively across diverse experimental conditions, reducing their scalability and applicability. These limitations highlight the need for more streamlined, efficient, and robust method to obtain biologically meaningful representations.
In this paper, we introduce PhenoProfiler, an innovative tool for learning phenotypic representations of cell morphology from high-throughput images. Unlike existing methods, PhenoProfiler functions as an end-to-end framework that directly encodes high-content multi-channel images into low-dimensional feature representations, without the need for extensive preprocessing such as segmentation and sub-image extraction. PhenoProfiler consists of three main modules: a gradient encoder, a transformer encoder, and a multi-objective learning module that integrates classification, regression, and contrastive learning. These modules establish a unified and robust feature space for representing cellular morphology. Additionally, PhenoProfiler incorporates a tailored phenotype correction strategy, designed to emphasize relative changes in cell phenotypes under different treatment conditions, enhancing its ability to capture meaningful biological signals. Extensive benchmarking on nearly 400,000 multi-channel images demonstrates PhenoProfiler’s state-of-the-art performance in extracting accurate and interpretable phenotypic representations. By addressing the complexity, high costs, and limited generalization capabilities of existing workflows, PhenoProfiler represents a significant advancement in phenotypic profiling and offers a powerful tool for accelerating image-based drug discovery. PhenoProfiler is freely accessible at https://phenoprofiler.org.
Results
Overview of the PhenoProfiler model
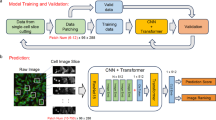
PhenoProfiler learns morphological representations and extracts phenotypic changes of treatment effects from high-throughput images. Different with existing methods (Fig. 1a), PhenoProfiler is designed as an end-to-end model with three key modules (Fig. 1b): a gradient encoder using difference convolution23,24, which enhances cell edge information and improves the clarity and contrast of cell morphology, and thus enhances the model’s adaptive perception of cells. A transformer encoder with multi-head self-attention mechanisms22 further captures long-range dependencies and intricate relationships within the data. A multi-objective learning module, composed of two multi-layer perceptron layers, is then utilized to enhance accuracy and generalization. This module integrates classification, regression, and contrastive learning. The classification learning maps image representations to categorize treatment conditions based on their corresponding labels. The regression learning leverages the rich and continuous supervisory information provided by regression objective to capture detailed morphological representations across different treatments, thereby significantly improving model performance. The contrastive learning improves robustness and generalization by maximizing similarity among representations of similar treatment conditions and minimizing it among dissimilar ones. After well training, PhenoProfiler identifies a unified and robust feature space for representing cell morphology. During the inference phase (Fig. 1c), PhenoProfiler utilizes a phenotype correction strategy to emphasize relative phenotypic changes under different treatment conditions, thereby revealing associated biological matches and treatment-associated representations. The detailed rationale for the design of PhenoProfiler is provided in Supplementary Note 1.
a Flowchart comparison of end-to-end PhenoProfiler with existing non-end-to-end methods. b PhenoProfiler includes a gradient encoder to enhance edge gradients, improving clarity and contrast in cell morphology. A transformer encoder then captures high-dimensional dependencies and intricate relationships, enriching image representations. A designed multi-objective learning module is utilized for accurate morphological representation learning. c For model inference, PhenoProfiler uses phenotype correction strategy (PCs) with hyperparameter α to identify morphological changes between treated and control conditions. Created in BioRender. Song, Q. (2025) https://BioRender.com/v0nqs11.
Superior performance of PhenoProfiler in biological matching tasks
For comprehensive and robust evaluation of the PhenoProfiler, we compare it with established methods, including DeepProfiler9, OpenPhenom20, ResNet5016, and ViT17 (Benchmarking Methods) using the leave-perturbation-out strategy. Two evaluation metrics (Evaluation Metrics) are used: Folds of Enrichment (FoE) and Mean Average Precision (MAP).
First, we use over 230,000 images from three datasets (BBBC02225, CDRP-BIO-BBBC03626, and TAORF-BBBC03727), covering 231 plates and 4285 treatments, including both compound and gene overexpression perturbations. The experimental results are illustrated in Fig. 2, demonstrating that PhenoProfiler surpasses all competing methods across three benchmarking datasets in both FoE and MAP metrics. As shown in Fig. 2a, PhenoProfiler achieves FoE improvements of 23.8%, 2.1%, and 12.9% over the second-best method (DeepProfiler) on the BBBC022, CDRP-BIO-BBBC036, and TAORF-BBBC037 datasets, respectively. For MAP evaluation, PhenoProfiler outperforms the second-best method by significant margins of 3.3%, 7.3%, and 7.1% across these datasets. Additionally, as depicted in Fig. 2b, we conducted comprehensive recall analysis at multiple thresholds (Recall@1, @3, @5, @10), where recall rates measure the proportion of biologically relevant treatments retrieved within top-ranked predictions (Evaluation Metrics). Using Recall@10 as a representative benchmark, PhenoProfiler exhibits performance enhancements of 17.4%, 4.3%, and 10.5% compared to DeepProfiler on the BBBC022, CDRP-BIO-BBBC036, and TAORF-BBBC037 datasets, respectively.
a Comparison of end-to-end feature representation performance across different methods in biological matching tasks using three benchmark datasets (BBBC022, CDRP-BIO-BBBC036, and TAORF-BBBC037) under leave-perturbations-out setting, evaluated with two evaluation metrics (MAP, FoE), and four comparison methods (DeepProfiler, ResNet50, ViT, OpenPhenom). b Performance comparison of different methods at different recall rates (recall@1, recall@3, recall@5, and recall@10). c Ablation experiments of PhenoProfiler, showing performance changes after sequential removal of each module. Specifically, “-MSE”, “-Con”, and “-CLS” represent the removal of regression, contrastive, and classification learning in the multi-objective module, while “-Gradient” represents the exclusion of difference operations. d Performance curve of PhenoProfiler under solely classification learning, showing variations in MAP and FoE as the classification loss decreases. e Sensitivity analysis of multi-objective learning of PhenoProfiler, exploring the impact of regression and contrastive learning (\({\lambda }_{2}\) and \({\lambda }_{3}\)) while maintaining fixed classification learning. f Hyperparameter analysis of θ₁ and θ₂ in the gradient encoder in parallel branches. MAP: Mean Average Precision; FoE: Folds of Enrichment; MSE: Mean Squared Error; Con: Contrastive Learning; CLS: Classification Learning. Source data are provided as a Source Data file.
To further illustrate the contributions of each module within PhenoProfiler, we have performed extensive ablation experiments using BBBC022 dataset in end-to-end pipeline (Fig. 2c). First, we remove the regression learning component within the multi-objective learning module (i.e., “-MSE” option), retaining only classification and contrastive learning. The results show that removing the regression learning results in a notable performance drop, with FoE and MAP decreasing by 12.0% and 12.7%. Next, we test various combinations of loss functions (e.g., “-Con”, “-CLS”, “-MSE-Con”, “-Con-CLS”, and “-CLS-MSE”). For example, the removal of both regression and classification learning leads to more performance decrease, with FoE and MAP reduced by 28.0% and 20.6%, respectively. Moreover, compared to the average performance of models without gradient encoding (i.e., “-MSE-Con-Gradient”, “-Con-CLS-Gradient”, and “-CLS-MSE-Gradient”), this modification results in a reduction of 25.2% in FoE and 11.5% in MAP, highlighting the effectiveness of gradient-based feature encoding. Additionally, the measurement metrics do not consistently improve as classification loss decreases. As illustrated in Fig. 2d, while both MAP and FoE initially increase with decreasing classification loss, they eventually decline. This observation highlights the importance of PhenoProfiler’s multi-objective learning design. The optimal weights for multi-objective learning were systematically determined through sensitivity analysis on the BBBC022 datasets. Figure 2e–f presents the tuned parameters of PhenoProfiler achieved optimal performance. Detailed evaluation of leave-sites-out are shown in Supplementary Notes 2 and 3.
PhenoProfiler demonstrates robust generalization and applicability
To evaluate the generalization of PhenoProfiler, we conduct experiments on the benchmarking datasets using the leave-plates-out and leave-dataset-out evaluation strategies. For the leave-plates-out strategy, A subset of plates are used as the test set while the remaining plates are used for training. For the leave-dataset-out strategy, one dataset is used for training and the other two serve as test sets. For example, “BBBC022 → BBBC036” indicates training on the BBBC022 dataset and validating on BBBC036 (CDRP-BIO-BBBC036). Regarding the leave-plates-out scenario (Fig. 3a), PhenoProfiler consistently surpasses other methods in both FoE and MAP. Specifically, PhenoProfiler has higher FoE than the second-best method by 57.0%, 10.7%, and 13.7%. For the MAP metric, PhenoProfiler outperforms the second-best method by 16.9%, 17.3%, and 6.0%. Figure 3b illustrates the performance comparison in the leave-dataset-out scenario, highlighting the consistent superior performance of PhenoProfiler. For example, in the BBBC022 → BBBC036 scenario, PhenoProfiler outperforms the next best method with higher FoE and MAP metrics by 13.0% and 17.1%, respectively. Similarly, in the BBBC022 → BBBC037 scenario, PhenoProfiler surpasses the next best method with higher FoE and MAP metrics by 6.9% and 5.4%, respectively. Collectively, PhenoProfiler demonstrates better generalization than existing methods, enhancing more accurate downstream tasks in drug discovery.
a Performance evaluation of different models using leave-plates-out validation. b Performance evaluation of different models using leave-dataset-out validation. c Performance evaluation of different models using out-of-distribution validation. The test plates include BR00115125-BR00115134 (cpg0001) and SQ00014812-SQ00014816 (cpg0004). FoE: Folds of Enrichment; MAP: Mean Average Precision; BBBC036: CDRP-BIO-BBBC036; BBBC037: TAORF-BBBC037. Source data are provided as a Source Data file.
To further validate PhenoProfiler’s generalization capability, we performed an out-of-distribution (OOD) evaluation using 10 diverse plates (BR00115125-BR00115134) from cpg0001 dataset, comprising 76,800 images covering 83 unique treatments and 47 annotated mechanisms of action (MoA). As shown in the upper panel of Fig. 3c, we directly applied PhenoProfiler models pretrained on BBBC022, BBBC036, and BBBC037 to these OOD plates. Remarkably, PhenoProfiler outperformed the second-best model (DeepProfiler) by an average of 45.8% in FoE and 27.3% in MAP across all test plates (upper panel of Fig. 3c), demonstrating robust generalization and OOD robustness. Moreover, since these evaluations were exclusively from the U2OS cell line, we further evaluated PhenoProfiler on five A549 cell line plates (SQ00014812, SQ00014813, …, SQ00014816 from cpg0004 dataset, lower panel of Fig. 3c). Remarkably, PhenoProfiler outperforms the second-best model (OpenPhenom) by 21.4% (FoE) and 20.3% (MAP) on average (lower panel of Fig. 3c), demonstrating robust generalization across different cell lines.
PhenoProfiler effectively removes batch effects for robust phenotypic representations
Variations stemming from technical and instrumental factors9,28 can introduce batch effects between plates subjected to the same treatment, which obscure true biological phenotypic signals and compromise downstream analyses. To evaluate PhenoProfiler’s ability to mitigate batch effects, we employed the Inverse Median Absolute Deviation (IMAD) metric, which quantifies the dispersion of image representations. Higher IMAD values indicate reduced dispersion, reflecting successful batch effect correction (details provided in the Evaluation Metrics section).
Figure 4 illustrates the well-level representation features, with plate IDs distinguished by different colors to highlight batch effects. For DeepProfiler, its representation features extracted from the BBBC022 dataset exhibit clear separation between plate IDs (IMAD = 0.326), indicating the presence of significant plate-specific biases. This separation shows strong technical variations among the extracted representation features, which can obscure true biological signals and hinder downstream analyses. Upon applying extra batch correction step to DeepProfiler’s representation features, the UMAP projections displayed improvements in feature cohesiveness (IMAD = 0.458). Notably, the representation features learned by PhenoProfiler exhibit a distinctly more integrated distribution (IMAD = 0.603). This superior performance suggests that PhenoProfiler learns harmonized representation features across different plates, effectively addressing batch effects without additional corrections. This pattern is consistently observed across all three datasets, further validating PhenoProfiler’s reliability in generating phenotypic representations that are robust to technical confounders. The inherent capacity of PhenoProfiler to learn harmonized representations directly from raw data not only reduces the need for computationally intensive post-processing but also ensures that biological signals are preserved.
UMAP visualizations of well-level features predicted by DeepProfiler and PhenoProfiler across three benchmark datasets. Features are colored by plate IDs and treatment conditions (control vs. treatment). IMAD quantifies the cohesiveness of the UMAP patterns, with higher values indicating better batch effects removal. UMAP: Uniform Manifold Approximation and Projection; BC: Batch Correction; IMAD: Inverse Median Absolute Deviation.
Phenotype correction strategy of PhenoProfiler improves biological matches
To effectively capture relative changes under treatments, PhenoProfiler is tailor designed with a phenotype correction strategy (PCs, Fig. 5a, details in Materials and Methods section) to refine learned phenotypic presentations, which distinguishes PhenoProfiler from existing methods. As illustrated in Fig. 5a, PhenoProfiler with PCs corrects image representations by leveraging controlled and treated wells within one plate and emphasizing relative changes in cell phenotypes under treatments.
a The conceptual motivation for the design of PCs. Phenotypic differences between treated- and controlled- wells capture the treatment response. b Ablation experiments demonstrating the impact of PCs across three datasets, showing a consistent increase in the FoE with the inclusion of PCs. c Sensitivity analysis of the hyperparameter α in the PCs. d UMAP visualizations of feature representations generated by PhenoProfiler, with and without PCs, evaluating the harmonization of well-level features using IMAD. PCs: Phenotype correction strategy; IMAD: Inverse Median Absolute Deviation; FoE: Folds of Enrichment; MAP: Mean Average Precision; UMAP: Uniform Manifold Approximation and Projection. Source data are provided as a Source Data file. Created in BioRender. Song, Q. (2025) https://BioRender.com/y4tl93e.
As shown in Fig. 5b, we assess the impact of the PCs through ablation experiments on three benchmark datasets. The results demonstrate that PCs consistently improves the FoE metric with minimal impact on the MAP metric (Supplementary Fig. 3). We also analyze the hyperparameter α in PCs, which determines the relative weight of controlled and treated wells. As illustrated in Fig. 5c, the outer circle of the radar chart corresponds to the value of α, with α = 0 representing PhenoProfiler without PCs. As α increases from 0 to 1, the relative weight of controlled wells increases, resulting in more pronounced differential effects. Meanwhile, the FoE generally trends upward while the MAP metric remains relatively stable. When α is greater than or equal to 0.7, the FoE begin to reach its maximum. These results demonstrate that PhenoProfiler with PCs can effectively capture the relative changes of cell phenotypes under treatments.
Furthermore, we analyze the aggregation of features before and after incorporating PCs across the three benchmark datasets. UMAP with well-level image representations are quantitatively measured for their dispersion before and after adding PCS using the IMAD metric (Fig. 5d). After implementing PCs, the representation features from different plates become significantly more clustered. Additionally, the IMAD metrics increases notably, with substantial improvements of 51.5%, 69.7%, and 11.6% across the three benchmark datasets, respectively.
PhenoProfiler efficiently captures representations of treatment effects
To visually illustrate the treatment effects, we obtain phenotypic representations using PhenoProfiler under various treatment conditions. Figure 6 displays a UMAP projection of PhenoProfiler representations across three benchmark datasets, providing a clear demonstration of how PhenoProfiler captures and organizes biological patterns in both non-end-to-end (Fig. 6a) and end-to-end (Fig. 6b) scenarios. For the BBBC022 and CDRP-BIO-BBBC036 datasets, which involve compound treatments, distinct clusters emerge based on their mechanisms of action (MoA). These clusters represent the functional similarities between compounds with shared MoAs, effectively demonstrating how PhenoProfiler translates phenotypic profiles into meaningful groupings that align with known biological functions. Similarly, the TAORF-BBBC037 dataset, which involves gene overexpression perturbations, reveals clear clusters of treatments corresponding to their genetic pathways, such as MAPK and PI3K/AKT. This clustering reaffirms established biological relationships and validates PhenoProfiler’s capability to accurately discern pathway-specific features. Notably, these groupings remain consistent across different wells, showcasing PhenoProfiler’s robust performance in feature extraction and its reliability in accurately representing treatment effects across experimental variations. However, some clusters are less distinctly discerned in the UMAP projections, suggesting that incorporating additional features, such as compound structures or gene expression data, could enhance the understanding of MoAs.
a UMAP projections of treatment profiles using well-level features provided by PhenoProfiler in non-end-to-end scenario. b UMAP projections of treatment profiles using well-level features provided by PhenoProfiler in end-to-end scenario. Well-level profiles, control wells, and treatment-level profiles are included. Text annotations highlight clusters where all or most points share same biological annotations for treatment-level profiles. UMAP: Uniform Manifold Approximation and Projection. c Heatmap visualization of PhenoProfiler-derived features demonstrates significant feature shifts in drug-treated groups compared to DMSO controls. d Biological interpretability of key drug-responsive phenotypic features captured by PhenoProfiler. The directionality of feature changes captured by PhenoProfiler (with red indicating increase and blue representing decrease) shows strong concordance with established pharmacological mechanisms.
To further assess PhenoProfiler’s ability to identify clinically actionable phenotypic patterns while maintaining biological interpretability, we performed an in-depth evaluation using the cpg0004-LINCS dataset, which includes treatments with well-characterized MoAs. As shown in Fig. 6c, all drug-treated groups exhibited significant feature shifts compared to DMSO controls. Notably, quadruplicate wells of the same drug class demonstrated high consistency, while distinct separation was observed between different drug categories. These results confirm PhenoProfiler’s dual capability: sensitive detection of drug-induced phenotypic perturbations and precise discrimination between pharmacological mechanisms. To elucidate the model’s biological interpretability, we focused on key phenotypic features responsive to pharmacological perturbations. From five major feature categories (granularity, correlation, spatial distribution, intensity, and area shape), we identified 15 most drug-sensitive biomarkers (three per category). Quantitative analysis in Fig. 6d demonstrates that PhenoProfiler-captured feature dynamics (red for upregulation, blue for downregulation) show remarkable concordance with established pharmacological mechanisms. For example, for the aclidinium, PhenoProfiler detected significantly increased cellular compactness and decreased eccentricity, aligning with its known mechanism of vesicle trafficking inhibition through mAChR blockade29,30. More remarkably, PhenoProfiler also captured a decrease in Golgi fluorescence intensity (Cells_Intensity_MeanIntensity_AGP) and an increase in RNA distribution heterogeneity (Cells_Intensity_MADIntensity_RNA), which highly coincides with the secretory dysfunction caused by aclidinium inhibiting muscarinic receptors and leading to signaling blockade29,30. Additionally, we specifically analyzed a typical case of the PKC activator TPA (12-O-Tetradecanoylphorbol-13-acetate). PhenoProfiler accurately captured the cell area expansion induced by this drug, which perfectly matches the theory of PKC-mediated cytoskeletal reorganization31,32.
Discussion
In this study, we present PhenoProfiler, an advanced tool for phenotypic representations of cell morphology in drug discovery. PhenoProfiler operates as a fully end-to-end framework, transforming multi-channel images into low-dimensional, biologically meaningful representations. By integrating a multi-objective learning module that incorporates classification, regression, and contrastive learning, PhenoProfiler effectively captures a unified and robust feature space. The extensive benchmarking conducted in this study, involving over 400,000 images from seven publicly available datasets, demonstrates that PhenoProfiler significantly outperforms existing state-of-the-art methods in both end-to-end, leave-plates-out, leave-dataset-out, and out-of-distribution validation scenarios. Moreover, PhenoProfiler achieves greater computational efficiency compared to existing non-end-to-end approaches (Supplementary Note 4). Furthermore, in non-end-to-end settings, we utilize over 8.42 million single-cell images for comparative analysis. PhenoProfiler’s superior performance on these large-scale datasets across all scenarios highlights its robustness, generalizability, and scalability, positioning it as a highly effective tool for advancing image-based drug discovery.
A key challenge in phenotypic representation learning lies in the heterogeneity of cellular responses to treatments. To address this, PhenoProfiler incorporates a multi-objective loss function that effectively guides the model’s learning process. First, the classification loss serves as the foundation, capturing phenotypic distinctions across treatments and enabling macro-level differentiation while aligning the learned representations with biologically meaningful semantics. Second, the regression component, supervised by pre-extracted morphological features, directs the model’s attention to cells exhibiting significant phenotypic perturbations, filtering out noise from unaffected cells and mitigating the effects of intracellular heterogeneity. Third, contrastive learning improves robustness to batch effects and morphological variability by encouraging similar representations for similar treatments and distinguishing between different treatments. Together, these objectives enable PhenoProfiler to learn rich hierarchical features, from local morphological details (via regression) to global treatment-specific patterns (via classification and contrastive learning), without relying on segmentation or sub-image extraction. Finally, the Phenotype Correction strategy (PCs) enhances the learned features by contrasting control and treated wells, amplifying biologically meaningful signals related to spatial distributions. In practical applications, PhenoProfiler supports flexible strategies for obtaining the pre-extracted morphological features used in regression supervision. These can be derived either from CellProfiler-extracted features, commonly available in Cell Painting assays, or from morphology profiles generated by a non-end-to-end EfficientNet model (Supplementary Note 5, Supplementary Figs. 4, 5).
While PhenoProfiler sets a new benchmark in phenotypic representation learning, there are several areas for future explorations and enhancement. First, the design of the multi-objective learning module can be further refined by exploring the interconnections and synergies among classification, regression, and contrastive objectives. Understanding the dependencies between these objectives33,34 could lead to more cohesive learning strategies. Additionally, while PhenoProfiler currently employs a stepwise training approach to address conflicts between objectives35,36, future work could focus on developing joint training and optimization techniques to balance these objectives more effectively. Second, recent advancements in large biomedical language models37,38,39,40 offer opportunities to integrate extensive domain knowledge into computational frameworks. Integrating these models’ embeddings into PhenoProfiler could enhance its generalizability, robustness, and effectiveness. Last, future efforts should prioritize integrating assays with complementary data modalities, such as genetic profiles12,41 and chemical structures42,43. Combining these multi-modal data would enable more comprehensive representations, offering a holistic view of cell states and phenotypic responses to various treatments.
The ability of PhenoProfiler to consistently capture and organize complex biological information across diverse datasets and treatment types underscores its versatility and utility in high-throughput drug screening and discovery. By addressing critical challenges in phenotypic profiling, such as scalability, robustness, and interpretability, PhenoProfiler enhances our understanding of treatment effects at the phenotypic level. Additionally, its potential for integrative multi-modal analysis, combining phenotypic data with complementary modalities such as genetic profiles, transcriptomics, and chemical structures, opens new opportunities to explore drug mechanisms and novel drug targets.
Methods
The PhenoProfiler model
PhenoProfiler is an end-to-end model, with input as multi-channel images and output as phenotypic representations under treatments. The overall framework includes three key modules: a gradient encoder, a transformer encoder, and a multi-objective learning module.
Gradient encoder
To enhance the model’s ability to understand cell morphology, we design a gradient encoder based on difference convolution (\({DC}\))23,24. This \({DC}\) function enhances gradient information around cell edges, improving the perception of morphological structures44,45. Formally, let \({p}_{0}\) represent the central position of a local receptive field \(R({p}_{0})\), with \({p}_{{{\rm{i}}}}\in R\left({p}_{0}\right)\). The pixel value at position \({p}_{i}\) in the input image is denoted as \({{{\boldsymbol{x}}}}_{{p}_{{{\rm{i}}}}}\), and the \({DC}\) function is defined as:
where \({{{\boldsymbol{w}}}}_{{p}_{i}}\) is a learnable parameter, and \(\theta \in [{\mathrm{0,1}}]\) is a hyperparameter that controls the balance between semantic and gradient information. When \(\theta=0\), the \({DC}\) function reduces to traditional convolution (\({TC}\)), i.e., \({TC}\left({p}_{0}\right)={\sum }_{{p}_{i}\in R\left({p}_{0}\right)}{{{\boldsymbol{w}}}}_{{p}_{i}}\cdot {{{\boldsymbol{x}}}}_{{p}_{i}}\).
The gradient encoder consists of two components: gradient enhancement and residual feature extraction. The gradient enhancement includes two parallel branches of difference convolution: the Deep Gradient branch, denoted as \({DG}={DC}(\bullet {,\theta }_{1})\), and the Shallow Gradient branch, denoted as \({SG}={DC}(\bullet {,\theta }_{2})\). We conducted a systematic experimental analysis of the hyperparameters \({\theta }_{1}\) and \({\theta }_{2}\) in the gradient encoder module (see Parameter Tuning section, Supplementary Note 3). Through comprehensive evaluation of different weight combinations, we ultimately determined the optimal configuration of \({\theta }_{1}=0.7\) and \({\theta }_{{{\boldsymbol{2}}}}=0.3\) for the two-branch architecture. The multi-channel input images, represented as \({{{\boldsymbol{x}}}}_{{in}}\), are processed in parallel through the two branches. The outputs of these branches \({{{\boldsymbol{H}}}}_{1}\) are then concatenated and passed through a Multi-Layer Perceptron (MLP) layer as follows:
Here \({{{\boldsymbol{H}}}}_{1}\) is the enhanced latent features. Batch normalization (\({BN}\)) ensures stable and efficient training by normalizing the intermediate latent features.
The subsequent residual feature extraction component employs the ResNet5016 model, pre-trained on ImageNet15. By incorporating residual connections, the ResNet50 model effectively addresses the vanishing gradient problem encountered in training deep neural networks, enabling the network to extract deeper features. This step outputs a fixed-length feature vector \({{{\boldsymbol{H}}}}_{2}{{\mathbb{\in }}{\mathbb{R}}}^{B\times 2048}\), where \(B\) denotes the batch size.
Transformer encoder
The latent features \({{{\boldsymbol{H}}}}_{2}\) is passed through the transformer encoder17,46 with multi-head self-attention mechanism, effectively capturing global dependencies among features. Then, layer normalization and residual connections are applied to ensure stable gradient propagation. Subsequently, a Feed-Forward Neural Network (\({FFN}\)) is used to further extract high-level features, resulting in the output feature vector \({{{\boldsymbol{H}}}}_{4}{{\mathbb{\in }}{\mathbb{R}}}^{B\times 2048}\). This process is formulated as below:
Here, \({MultiHeadAttention}\) computes attention weights across multiple attention heads to capture diverse feature dependencies. The \({FFN}\) consists of two fully connected layers with non-linear ReLU activations, parametrized by weights \({{{\boldsymbol{W}}}}_{1}\), \({{{\boldsymbol{W}}}}_{2}\), and biases \({{{\boldsymbol{b}}}}_{1}\), \({{{\boldsymbol{b}}}}_{2}\). This transformer encoder effectively captures global and local information simultaneously, ensuring the generation of rich and informative feature representations.
Multi-objective learning
To facilitate efficient multi-objective learning, we use a feature projection that maps latent features to a lower-dimensional space, facilitating efficient multi-objective learning. Specifically, the latent feature \({{{\boldsymbol{H}}}}_{6}{{\mathbb{\in }}{\mathbb{R}}}^{B\times 2048}\) is first linearly transformed to a lower-dimensional space. A GELU activation function is then applied to introduce non-linear characteristics, followed by further processing through a fully connected layer with dropout to prevent overfitting. This feature projection process provides final output \(\hat{{{\boldsymbol{Z}}}}{{\mathbb{\in }}{\mathbb{R}}}^{B\times 672}\) as follows:
Then a classification head is implemented as a simple linear layer maps output representations \(\hat{{{\boldsymbol{Z}}}}\) to \(\hat{{{\boldsymbol{y}}}}\), representing the predicted treatment labels.
Classification learning
To characterize the phenotypic responses of cells under various treatments and enable the model to learn discriminative features among different treatments, we employ a cross-entropy loss function to quantify the discrepancy between the model’s predictions and the ground truth:
Here, \(N\) represents the number of treatment categories, and \({{{\boldsymbol{y}}}}_{i}\) is the one-hot encoding of the treatment label in ground truth. If the ground truth is treatment category \(i\), then \({{{\boldsymbol{y}}}}_{i}=1\); otherwise, \({{{\boldsymbol{y}}}}_{i}=0\).
Regression learning
Here, we have innovatively designed a regression learning component to learn cell morphological representations. Unlike discrete classification labels, this approach leverages richer and more nuanced feature information, enabling the model to focus on cells exhibiting the most pronounced phenotypic perturbations while effectively disregarding interference from unaffected cell populations. To obtain regression labels, we use the median of the morphology profiles of cpg0019 as the regression morphology labels and train the model using Mean Squared Error (MSE) loss:
where \({{{\boldsymbol{Z}}}}_{i}\) is the morphology profiles of the \(i\)-th image, \({\hat{{{\boldsymbol{Z}}}}}_{i}\) is predicted morphology features, and \(M\) is the number of images.
Contrastive learning
Contrastive learning enhances the model ‘s ability to distinguish features by maximizing the similarity between similar images and minimizing the similarity between dissimilar ones. Specifically, contrastive learning does not rely on ground truth labels but focuses on learning feature representations based on the relative relationships between images. This approach not only reduces the negative impact of noisy labels but also improves the model’s robustness and generalization when handling unseen data. The contrastive loss function is formulated as follows:
where \(B\) is the batch size, \({\hat{{{\boldsymbol{Z}}}}}_{i}\) denotes the predicted morphology features of the \(i\)-th image, and \({{{\boldsymbol{Z}}}}_{i}\) is the morphology profiles, and \(\tau\) is the temperature parameter. This objective trains the model to produce discriminative feature vectors by maximizing the similarity between matching image-embedding pairs while minimizing the similarity between non-matching pairs.
Multi-objective loss
By integrating classification, regression, and contrastive learning, PhenoProfiler provides a unified and robust feature space, comprehensively learns the image representations of multi-channel cell images. This multi-object learning architecture enhances the overall generalization performance of the model. To achieve an effective balance in multi-object learning, we assign a weight parameter to each object and adjust these weight parameters to balance the losses. The final total loss function can be expressed as:
where \({\lambda }_{1},{\lambda }_{2},{\lambda }_{3}\) are the weight parameters for the classification, regression, and contrastive learning, respectively. Based on extensive ablation experiments (see Fig. 2e), we set these weights to 0.1, 100, and 1, respectively. This multi-loss balancing strategy enables PhenoProfiler to find the optimal trade-off among different objects, thereby enhancing the overall performance and robustness of the model. Additionally, this strategy allows us to flexibly adjust the weights of each learning according to the specific requirements of the application scenario, achieving optimal feature representation and prediction performance.
Parameter tuning
PhenoProfiler is an end-to-end multi-channel image encoder designed to convert multi-channel images into corresponding morphology representations. During training, conflicts among objects in multi-objective joint training caused the model to struggle to converge to an optimal state35,36. To address this, we adopted a stepwise training strategy. Initially, we trained the regression learning using MSE loss to optimize the model. After approximately 100 epochs, we proceeded with joint optimization based on the multi-object learning architecture. The hyperparameter settings were as follows: a batch size of 300, a maximum of 200 training epochs, and 12 workers. The learning rate followed a staged decay strategy: 2e-3 for the first 10 epochs, 1e-3 for the next 50 epochs, 5e-4 for the subsequent 60 epochs, and 1e-4 for the final 80 epochs. The training environment was Ubuntu 22.04, utilizing four NVIDIA A100 GPUs (40GB version). Details on the tuning of loss function coefficients and the gradient encoder hyperparameters are provided in Supplementary Note 3 and Supplementary Fig. 2.
Model inference
In model inference, we first clarify the four levels of data involved in this task. The dataset comprises four levels of features: plate-level, treatment-level, well-level, and site-level. Specifically, a dataset contains P plates, each plate includes T treatments, each treatment corresponds to W wells, and each well contains S sites, with each site corresponds to a multi-channel image. Hence, the output of PhenoProfiler is a site-level feature \(\hat{{{\boldsymbol{Z}}}}\). The values of P, T, W, and S vary across datasets. Following the validation procedure used in DeepProfiler, we applied mean aggregation at both stages to get the next level aggregated features. After obtaining well-level features, we employed Sphering transform as a batch correction method to minimize confounders. Through these aggregation processes, we obtained the final treatment-level features for evaluation.
Phenotype Correction Strategy (PCs) directly optimizes the output of the PhenoProfiler in a plate. It aims to leverage the differences between treated wells and controlled wells within a plate to refine image representation. The implementation process of PCs is detailed in Fig. 1c. First, we calculate the mean of all control wells in the current plate, denoted as \({{{\boldsymbol{W}}}}^{i}\), where \(i\) represents the \(i\)-th plate. Assuming that a plate contains C controlled wells, the calculation is as follows:
Then, define \(\alpha\) as a hyperparameter that balances the weight between controlled wells and treated wells, \({\hat{{{\boldsymbol{E}}}}}_{{ij}}\) represent the predicted embedding vector for the \(j\)-th well in the \(i\)-th plate. \(\alpha*{{{\boldsymbol{W}}}}^{i}\) is subtracted from all predicted embeddings \({\hat{{{\boldsymbol{E}}}}}_{{ij}}\) in the current plate to obtain the refined embeddings \({{{\boldsymbol{E}}}}_{{ij}}\). The process is as follows:
PCs serves as a correction and optimization operation applied to the extracted features, making it virtually cost-free and plug-and-play.
Web server implementation
The PhenoProfiler web server integrates a JavaScript/TypeScript frontend (Next.js framework) with server-side rendering and Tailwind CSS styling, enhanced with React hooks for dynamic interactivity. The Python Django backend supports scalable data processing, coupled with an SQLite database for lightweight storage. A RESTful API coordinates client-server communication, enabling seamless file uploads and standardized JSON responses. Deployment utilizes the Caddy server with automated HTTPS/SSL and reverse proxy configurations, ensuring secure and efficient operations. The platform is publicly accessible at https://phenoprofiler.org.
Benchmarking methods
To intuitively evaluate PhenoProfiler’s performance within an end-to-end pipeline, we compared it against four benchmarking methods: DeepProfiler9, OpenPhenom20, ResNet5016, and ViT-Base17. DeepProfiler is a deep learning-based phenotyping tool that processes single-cell imaging data using an EfficientNet model architecture to generate single-cell morphological features. To adapt it for end-to-end image processing tasks, we trained the EfficientNet model on our end-to-end image dataset. OpenPhenom is a self-supervised learning method based on masked autoencoders. Since its training includes multi-cell images, we directly used the official pre-trained weights (available at https://huggingface.co/recursionpharma/OpenPhenom) for inference, resizing all input images to 256×256 resolution without further training or fine-tuning. ResNet50 and ViT-Base are widely recognized as general-purpose benchmark models in computer vision, serving as standard references for performance comparison. We made uniform adjustments to apply these methods to high-content multi-channel image processing. Specifically, to handle five-channel Cell Painting images, we included a convolutional layer at the front of each model to adjust the number of input image channels from five to three and reduce the dimensionality. The networks were initialized using the corresponding pre-trained weights provided by the “timm” library. For example, the ViT-Base model utilized the weights for vit_base_patch32_224. Notably, we adopted a fully trainable parameter mode for all models, without freezing any task weights, and maintained consistent training methods and hyperparameters throughout the training process.
In the non-end-to-end scenarios, we included three additional comparison methods: CellProfiler18, SPACe19, and EfficientNet21. CellProfiler is an open-source software tool designed for measuring and analyzing cell images, providing a robust platform for extracting quantitative data from biological images. It enables researchers to identify and quantify phenotypic changes effectively. SPACe is an open-source single-cell image analysis platform. Compared to CellProfiler, it achieves faster processing speeds while maintaining high accuracy by optimizing feature extraction and employing CellPose-powered cell segmentation. Following SPACe’s pipeline, we merged the five feature files extracted in its fourth step to obtain 423-dimensional features, encompassing intensity, morphology, and texture measurements, which were then used for downstream performance evaluation. It is worth noting that this tool has a relatively high entry barrier and learning curve, as it requires handling multiple prerequisites. For instance, users need to create customized guideline files tailored to their dataset characteristics and modify corresponding data-loading code accordingly. As for EfficientNet, adhering to the design framework of DeepProfiler, we utilized models pre-trained on the large-scale ImageNet dataset without additional training or fine-tuning on Cell Painting images. By incorporating these models, we aimed to benchmark our pipeline’s performance against established standards in the field, ensuring a comprehensive evaluation of its capabilities.
Benchmarking datasets
The PhenoProfiler model leverages seven distinct datasets: BBBC02225, CDRP-BIO-BBBC03626, TAORF-BBBC03727, LUAD-BBBC04347, LINCS48, cpg00017, and cpg000412. Among these, BBBC022, CDRP-BIO-BBBC036, LINCS, cpg0001, and cpg0004 focus on phenotypic responses to compound treatments, whereas TAORF-BBBC037 and LUAD-BBBC043 address phenotypic responses to gene overexpression. Together, these datasets form a comprehensive dataset of nearly 400,000 multi-channel images. These images encompass two treatment types (compounds and gene overexpression), two control types (empty and DMSO), and two cell lines (A549 and U2OS), collected from 246 plates. Detailed dataset information is summarized in Supplementary Data 1. To facilitate data storage and transmission, we apply image compression and illumination correction to convert the images from TIFF to PNG format, achieving approximately six times (via 2× bit-depth reduction from 16-bit to 8-bit and ~3× PNG lossless encoding) the compression without significant quality loss9. To further validate the cell morphology representation capability of PhenoProfiler in a non-end-to-end pipeline, we introduce the cpg0019 dataset9. This dataset comprises 8.4 million single-cell images, which were cropped from multi-channel images across these first five datasets. It includes 450 treatments, meticulously selected to represent a diverse array of phenotypic responses.
Given the distinct input requirements of the end-to-end and non-end-to-end pipelines, we trained and validated them on different datasets. For the end-to-end pipeline, we directly used the BBBC022, CDRP-BIO-BBBC036, and TAORF-BBBC037 datasets. For the non-end-to-end pipeline, cells from the first five datasets were segmented into single-cell sub-images and supplemented with a subset from the cpg0019 dataset. Because the other two datasets lacked ground truth, both pipelines were ultimately evaluated on BBBC022, CDRP-BIO-BBBC036, and TAORF-BBBC037, with the main distinction being whether the input images were cropped. To ensure robust evaluation, we applied two complementary validation strategies: leave-perturbations-out (Fig. 2) and leave-sites-out (Supplementary Fig. 2). In the leave-perturbations-out setting, the training and test sets contained entirely distinct perturbations, ensuring evaluation on unseen experimental conditions. The leave-sites-out strategy assessed performance at the site level, the smallest input unit, by systematically holding out a subset of sites from each well, providing an additional layer of fine-grained validation. Finally, the cpg0001 and cpg0004 datasets were used to test out-of-distribution generalization.
Data preprocessing involves three main components: (1) Image data preprocessing: We stack images from different channels in the order of [‘DNA’, ‘ER’, ‘RNA’, ‘AGP’, ‘Mito’] to obtain multi-channel images. The images are resized to a uniform size of (5, 448, 448) pixels and the pixel values are scaled to the range of 0 to 1. (2) Classification label preprocessing: We read the CSV file and initialize the label encoder. Labels are encoded based on the column names (Treatment or pert_name) in the CSV file. (3) Morphology profiles preprocessing for the regression and contrastive learning. For the supervisory labels in regression and contrastive learning, we select the median values of the morphology profiles provided in the cpg0019 dataset.
Evaluation metrics
We evaluate the model’s capacity to represent cell morphology using a reference collection of treatments to identify biological matches in treatment experiments. Following strategies outlined in previous studies9,49,50,51, we implement a biological matching task where users can search for treatments associated with the same MoA or genetic pathway, applicable to both compound and gene overexpression perturbations. Initially, we aggregate features from various methods at the treatment level and assess the relationships among these treatments within the feature space, guided by established biological connections. This approach allows us to identify treatments that are proximally situated, suggesting potential similarities in their biological effects or mechanisms of action. This evaluation not only demonstrates the model’s effectiveness in representing cell morphology but also offers a valuable framework for advancing biological research. To quantify the similarity between query treatments, we utilize cosine similarity and generate a ranked treatment list based on relevance, presented in descending order. A positive result is achieved if at least one biological annotation in the sorted list matches the query; otherwise, the result is regarded as negative.
For evaluating the quality of results for a given query, we employ two primary metrics: (1) Folds of Enrichment (FoE) and (2) Mean Average Precision (MAP).
(1) Folds of Enrichment (FoE): This metric assesses the overrepresentation of predicted features in the reference set, indicating the model’s ability to identify relevant biological treatments. We calculate the odds ratio using a one-sided Fisher’s exact test for each query treatment, which employs a 2 × 2 contingency table. The first row contains the counts of treatments with the same MoAs or pathways (positive matches) versus those with different MoAs or pathways (negative matches) above a pre-defined threshold. The second row contains the corresponding counts for treatments below the threshold. The odds ratio is computed as the sum of the first row divided by the sum of the second row, estimating the likelihood of observing treatments sharing the same MoA or pathway among the top connections. We average the odds ratios across all query treatments, with the threshold set at the top 1% of connections, thus anticipating significant enrichment for positive matches49.
(2) Mean Average Precision (MAP): For each query treatment, we calculate the average precision, which is the area under the precision-recall curve, following standard practices in information retrieval. The evaluation process starts with the result most similar to the query and continues until all relevant pairs (those with the same mechanism of action or pathway) are identified. MAP effectively captures both the precision and recall of the model’s predictions, offering insights into its reliability and robustness. The specific calculation process is as follows:
Here, \({TP}\) stands for true positives, \({FP}\) for false positives, and \({FN}\) for false negatives. MAP refers to the average precision across multiple queries. For each query, we calculate the area under the precision-recall curve and then average these values across all queries. Given that the number of MoAs or pathways varies, precision and recall are interpolated for each query to cover the maximum number of recall points. The interpolated precision at each recall point is:
The average precision of a query treatment is the mean of the interpolated precision values \({P}_{{inter}}\) at all recall points. The reported MAP is the average of the average precision values across all queries. Together, these metrics provide a rigorous assessment of PhenoProfiler’s performance in capturing biological relevance in treatment experiments and underscore its utility in phenotypic drug discovery.
We also used recall rates at various levels (recall@1, recall@3, recall@5, and recall@10) to evaluate the model’s performance. Recall@K is an important metric in information retrieval and recommendation systems, indicating the proportion of correct results within the top K returned results. For instance, recall@5 represents the proportion of biologically related treatments retrieved within the top five positions of a predicted treatments ranked list. This is crucial for user experience, as users typically only look at the first few results, and the relevance of these results directly impacts user satisfaction.
In addition to the main metrics mentioned above, we introduced the Inverse Median Absolute Deviation (IMAD) metric to quantitatively evaluate the aggregation degree of features. The calculation steps for the IMAD metric are as follows: First, Principal Component Analysis (PCA) is performed to reduce the dimensionality of the data, retaining 95% of the variance. Then, we use Uniform Manifold Approximation and Projection (UMAP) for further dimensionality reduction and embedding. Next, we combine the UMAP 1 and UMAP 2 columns into a coordinate array and calculate the pairwise distances between all points. Subsequently, we compute the Median Absolute Deviation (MAD) of these distances. Finally, we take the reciprocal of the MAD to obtain the IMAD. A higher IMAD indicates a tighter aggregation of the data.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All experiments in this study utilized publicly available datasets, which can be accessible from public S3 buckets. To download the data, you need to install the AWS CLI that matches your device by following the instructions at AWS CLI Installation Guide. Use the following command with the cp or sync command, along with the --recursive and --no-sign-request flags for data retrieval. For the BBBC022 dataset can be downloaded with the following command: “aws s3 cp s3://cytodata/datasets/Bioactives-BBBC022-Gustafsdottir/./ --recursive --no-sign-request”. For the other datasets, use the following commands: CDRP-BIO-BBBC036: “aws s3 cp s3://cytodata/datasets/CDRPBIO-BBBC036-Bray/./ --recursive --no-sign-request”; TAORF-BBBC037: “aws s3 cp s3://cytodata/datasets/TA-ORF-BBBC037-Rohban/./ --recursive --no-sign-request”; cpg0001-cellpainting-protocol: “aws s3 sync s3://cellpainting-gallery/cpg0001-cellpainting-protocol/source_4/images/2020_08_11_Stain3_Yokogawa/./2020_08_11_Stain3_Yokogawa --no-sign-request”; cpg0004-lincs: “aws s3 cp --recursive “s3://cellpainting-gallery/cpg0004-lincs/broad/workspace/profiles/2016_04_01_a549_48hr_batch1/SQ00014815/“ “./SQ00014815” --no-sign-request”. For the non-end-to-end dataset cpg0019: “aws s3 cp s3://cellpainting-gallery/cpg0019-moshkov-deepprofiler/./ --recursive --no-sign-request”. Source data are provided with this paper.
Code availability
All source codes and trained models in our experiments have been made publicly available under the MIT License at Github and Zenodo (https://github.com/QSong-github/PhenoProfiler)54.
References
Vincent, F. et al. Phenotypic drug discovery: recent successes, lessons learned and new directions. Nat. Rev. Drug Discov. 21, 899–914 (2022).
Chandrasekaran, S. N., Ceulemans, H., Boyd, J. D. & Carpenter, A. E. Image-based profiling for drug discovery: due for a machine-learning upgrade?. Nat. Rev. Drug Discov. 20, 145–159 (2021).
Seal, S. et al. Cell Painting: a decade of discovery and innovation in cellular imaging. Nat. Methods 22, 254–268 (2025).
Cross-Zamirski, J. O. et al. Label-free prediction of cell painting from brightfield images. Sci. Rep. 12, 10001 (2022).
Chandrasekaran, S. N. et al. Three million images and morphological profiles of cells treated with matched chemical and genetic perturbations. Nat. Methods 21, 1114–1121 (2024).
Haghighi, M., Caicedo, J. C., Cimini, B. A., Carpenter, A. E. & Singh, S. High-dimensional gene expression and morphology profiles of cells across 28,000 genetic and chemical perturbations. Nat. Methods 19, 1550–1557 (2022).
Bray, M.-A. et al. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat. Protoc. 11, 1757–1774 (2016).
Chandrasekaran, S. N. et al. JUMP Cell Painting dataset: morphological impact of 136,000 chemical and genetic perturbations. BioRxiv (2023) 2023.03. 23.534023.
Moshkov, N. et al. Learning representations for image-based profiling of perturbations. Nat. Commun. 15, 1594 (2024).
Tian, G., Harrison, P. J., Sreenivasan, A. P., Carreras-Puigvert, J. & Spjuth, O. Combining molecular and cell painting image data for mechanism of action prediction. Artif. Intell. Life Sci. 3, 100060 (2023).
Mirabelli, C. et al. Morphological cell profiling of SARS-CoV-2 infection identifies drug repurposing candidates for COVID-19. Proc. Natl. Acad. Sci. USA 118, e2105815118 (2021).
Way, G. P. et al. Morphology and gene expression profiling provide complementary information for mapping cell state. Cell Syst 13, 911–923.e9 (2022).
Wang, S. et al. PhenoScreen: a dual-space contrastive learning framework-based phenotypic screening method by linking chemical perturbations to cellular morphology. bioRxiv (2024) 2024.10. 23.619752.
Caicedo, J. C. et al. Data-analysis strategies for image-based cell profiling. Nat. Methods 14, 849–863 (2017).
Deng, J. et al. ImageNet: a large-scale hierarchical image database. in Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition 248–255.
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. in Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition 770–778.
Dosovitskiy, A. et al. An image is worth 16x16 words: transformers for image recognition at scale. In International Conference on Learning Representations (2021).
McQuin, C. et al. CellProfiler 3.0: Next-generation image processing for biology. PLOS Biol. 16, e2005970 (2018).
Stossi, F. et al. SPACe: an open-source, single-cell analysis of Cell Painting data. Nat. Commun. 15, 10170 (2024).
Kraus, O. et al. Masked autoencoders for microscopy are scalable learners of cellular biology. In Proc. 2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition 11757–11768.
Tan, M. & Le, Q. EfficientNet: rethinking model scaling for convolutional neural networks. in Proceedings of the 36th International Conference on Machine Learning 6105–6114 (PMLR, 2019).
Bao, Y., Sivanandan, S. & Karaletsos, T. Channel vision transformers: an image is worth 1 × 16 × 16 words. In International Conference on Learning Representations (2024).
Yu, Z. et al. Searching central difference convolutional networks for face anti-spoofing. In Proc. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition 5294–5304 (IEEE, 2020).
Yu, Z. et al. Dual-cross central difference network for face anti-spoofing. In Proc. International Joint Conference on Artificial Intelligence 1281–1287 (2021).
Gustafsdottir, S. M. et al. Multiplex cytological profiling assay to measure diverse cellular states. PLoS One 8, e80999 (2013).
Bray, M.-A. et al. A dataset of images and morphological profiles of 30,000 small-molecule treatments using the Cell Painting assay. GigaScience 6, giw014 (2017).
Rohban, M. H. et al. Systematic morphological profiling of human gene and allele function via Cell Painting. eLife 6, e24060 (2017).
Arevalo, J. et al. Evaluating batch correction methods for image-based cell profiling. Nat. Commun. 15, 6516 (2024).
Grando, S. A. Cholinergic control of epidermal cohesion. Exp. Dermatol. 15, 265–282 (2006).
Alagha, K., Bourdin, A., Tummino, C. & Chanez, P. An update on the efficacy and safety of aclidinium bromide in patients with COPD. Ther. Adv. Respir. Dis. 5, 19–28 (2011).
Kitajima, Y., Inoue, S. & Nagao, S. Biphasic effects of 12-O-tetradecanoylphorbol-13-acetate on the cell morphology of low calcium-grown human epidermal carcinoma cells: involvement of translocation and down regulation of protein kinase C[J]. Cancer research 48, 964–970 (1988).
El-Yazbi, A. F., Abd-Elrahman, K. S. & Moreno-Dominguez, A. PKC-mediated cerebral vasoconstriction: Role of myosin light chain phosphorylation versus actin cytoskeleton reorganization. Biochem. Pharmacol. 95, 263–278 (2015).
Chen, S., Zhang, Y. & Yang, Q. Multi-task learning in natural language processing: an overview. ACM Comput. Surv. 56, 1–32 (2024).
Allenspach, S., Hiss, J. A. & Schneider, G. Neural multi-task learning in drug design. Nat. Mach. Intell. 6, 124–137 (2024).
Gong, Z. et al. CoBa: convergence balancer for multitask finetuning of large language models. In Proc. 2024 Conference on Empirical Methods in Natural Language Processing 8063–8077 (2024).
Tiomoko, M., Tiomoko, H. & Couillet, R. Deciphering and optimizing multi-task learning: a random matrix approach. in Proceedings of the 9th International Conference on Learning Representations (2021).
Wang, B. et al. Pre-trained language models in biomedical domain: a systematic survey. ACM Comput. Surv. 56, 1–52 (2024).
Ma, T. et al. Y-Mol: A Multiscale Biomedical Knowledge-Guided Large Language Model for Drug Development. arXiv preprint arXiv:2410.11550, 2024.
Cui, H. et al. scGPT: toward building a foundation model for single-cell multi-omics using generative AI. Nat. Methods 21, 1470–1480 (2024).
Hao, M. et al. Large-scale foundation model on single-cell transcriptomics. Nat. Methods 21, 1481–1491 (2024).
Iyer, N. S. et al. Cell morphological representations of genes enhance prediction of drug targets. bioRxiv, 2024: 2024.06. 08.598076.
Sanchez-Fernandez, A., Rumetshofer, E., Hochreiter, S. & Klambauer, G. CLOOME: contrastive learning unlocks bioimaging databases for queries with chemical structures. Nat. Commun. 14, 7339 (2023).
Zheng, S. et al. SYSU-cross-modal graph contrastive learning with cellular images. Adv. Sci. 11, 2404845 (2024).
Li, B. et al. Gene expression prediction from histology images via hypergraph neural networks. Brief. Bioinform. 25, bbae500 (2024).
Li, B. et al. Exponential distance transform maps for cell localization. Eng. Appl. Artif. Intell. 132, 107948 (2024).
Chen, H. et al. Pre-trained image processing transformer. in Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition 12294–12305.
Caicedo, J. C. et al. Cell Painting predicts impact of lung cancer variants. Mol. Biol. Cell 33, ar49 (2022).
Way, G. P. et al. Predicting cell health phenotypes using image-based morphology profiling. Mol. Biol. Cell 32, 995–1005 (2021).
Rohban, M. H., Abbasi, H. S., Singh, S. & Carpenter, A. E. Capturing single-cell heterogeneity via data fusion improves image-based profiling. Nat. Commun. 10, 2082 (2019).
Wolf, T. et al. Transformers: state-of-the-art natural language processing. in Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations 38–45.
Ljosa, V. et al. Comparison of methods for image-based profiling of cellular morphological responses to small-molecule treatment. J. Biomol. Screen. 18, 1321–1329 (2013).
Hancock, D. Y. et al. Practice and Experience in Advanced Research Computing 2021: Evolution Across All Dimensions (Association for Computing Machinery, Boston, MA, USA, 2021).
Boerner, T. J., Deems, S., Furlani, T. R., Knuth, S. L., Towns, J. Practice and Experience in Advanced Research Computing 2023: Computing for the Common Good 173–176 (Association for Computing Machinery, Portland, OR, USA, 2023).
Li, B. et al. PhenoProfiler: advancing phenotypic learning for image-based drug discovery. QSong-github/PhenoProfiler https://doi.org/10.5281/zenodo.17407526 (2025).
Acknowledgements
B.Z. was supported by the University of Macau [Grant Number MYRG-GRG2024-00205-FST]; and the Science and Technology Development Fund, Macau SAR [Grant Number 0028/2023/RIA1]. This work partially used Jetstream252 through allocation CIS230237 from the Advanced Cyberinfrastructure Coordination Ecosystem: Services & Support (ACCESS)53 program, which is supported by National Science Foundation grants #2138259, #2138286, #2138307, #2137603, and #2138296. The authors thank Srinivas Niranj Chandrasekaran, Postdoctoral Associate at the Imaging Platform, Broad Institute of MIT and Harvard, for his guidance, discussions, and support throughout this project.
Author information
Authors and Affiliations
Contributions
Q.S., and B.Z. supervised the overall project. Q.S. and B.L. drafted the paper and led the revision process. B.L., C.Z., and S.W. were responsible for data collection, model implementation and optimization. M.Z. developed and maintained the web server. W.H., Q.W., M.L., and Y.Z. contributed to project discussions and assisted in refining the paper and visualizations. All authors reviewed and approved the final version of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Lassi Paavolaine, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, B., Zhang, B., Zhang, C. et al. PhenoProfiler: advancing phenotypic learning for image-based drug discovery. Nat Commun 17, 793 (2026). https://doi.org/10.1038/s41467-025-67479-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-67479-w