Abstract
The mammalian airway epithelium contains specialized cells that detect and respond to environmental injury, yet the mechanisms that coordinate epithelial repair remain poorly defined. Pulmonary neuroendocrine cells act as neurosensory sentinels within this epithelium and can reprogram to support regeneration. Here we show that the calcium-binding protein S100B is essential for maintaining the stem cell-like properties of pulmonary neuroendocrine cells during airway repair. Following epithelial injury, S100B expression is induced in these cells and released to engage macrophages that express the enzyme arginase-1, which in turn promotes the expansion of pulmonary neuroendocrine cells and their transition into regenerative intermediate cells. Modulating S100B activity, either by blocking S100B with pentamidine or by enhancing signaling driven by the protein kinase p110-alpha in airway epithelial cells, alters the reparative capacity of pulmonary neuroendocrine cells. These findings identify an S100B-arginase-1 signaling axis that governs macrophage-epithelial communication and highlight S100B as a potential therapeutic target for lung repair.
Similar content being viewed by others
Introduction
Throughout life, multicellular organisms possess a remarkable capacity for cellular regeneration, ensuring the maintenance of integrity and function following injury. The lungs, being in direct contact with the external environment, are frequently exposed to a range of airborne pollutants, allergens, pathogens, and other toxins. Such exposure can trigger a variety of respiratory ailments, including infections, chronic obstructive pulmonary disease, asthma, and lung cancer1. The ability to swiftly detect and respond to injuries is crucial for both regeneration and survival. Neuroimmune interactions within the respiratory tract facilitate the rapid sensing and reaction to pathogens, and these interactions have been linked to a spectrum of disease states, such as viral and bacterial infections, acute lung injury, and lung fibrosis2,3. Given that the ongoing function of stem cells after injury is essential for reestablishing airway homeostasis, comprehending the role and molecular mechanisms of neuroimmune interactions that govern stem cell behavior is of great importance for the development of effective therapeutic strategies.
Over the years, various types of neurons have been identified in different respiratory regions, including the trigeminal, vagus, and dorsal root nerves of the spinal cord4, which interact with specialized epithelial cells like pulmonary neuroendocrine cell (PNEC), brush cells, and solitary chemosensory cells to form sensory recognition units. Among these epithelial cells, PNECs are indeed a unique and rare cell type, representing less than 1% of the lung epithelial population5. The clustered PNECs, called neuroepithelial bodies (NEBs), are predominantly located at airway bifurcations, where they play key roles in oxygen sensing6, mechanotransduction, pulmonary blood flow modulation, chemosensation7, and inflammation regulation8. They may also serve as progenitor cells niches, with in vitro and in vivo studies suggesting that calcitonin gene-related peptide (CGRP), their predominant peptide, may link stimuli to stem cell modulation9. However, the genetic ablation of CGRP did not impair homeostasis or repair airways10, challenging current understanding of PNEC-immune crosstalk. Intriguingly, the general neuronal and neuroendocrine marker, protein gene product 9.5 (PGP9.5, also known as UCHL1), labels expanded cell populations during injury repair11, suggesting that injury-responsive neuroendocrine-like cells or nerve fibers may contribute to regenerative processes.
More recently, it becomes increasingly clear that PNECs are not terminally differentiated as once thought, but can reenter cell cycle and transdifferentiate into other cell fates after injury12. Upon extensive airway epithelial injury, such as that caused by the club cell toxicant naphthalene (NAPH) or genetic ablation of club cells, PNECs rapidly proliferate and repair the surrounding epithelium11,13. Understanding the mechanisms by which PNEC contribute to lung repair is an active area of research. Previous studies provide evidence that tumor suppressors Rb1 and Trp53 act as an early signal to control PNEC renewal in response to injury14. By reactivation of the slithering signals or loss of attractive signals, PNEC is promoted to dispersal from their niche and migrate to the site of injury8,15. In the later stages of injury repair, the Notch pathway conducts as deprogramming and reprogramming signal, helping PNEC to acquire mature club and other cell fates to restore the epithelium13,14. The major challenge is uncovering the precise molecular mechanisms by which these signals are activated by injury and how they interact to control PNEC behavior. Beyond acting as individual progenitor cells, increased evidence also supports that NEB microenvironment appears to play a critical role in maintaining a reservoir for pollutant-resistant stem and progenitor cells at the airway bifurcations12,16 and the bronchioloalveolar junction17,18. The precise role of NEBs as either producers or recipients of stem cells remains unclear.
S100B, a member of S100 protein family, is a calcium sensor protein expressing in astrocytes and other non-neural cell types19. Intracellularly, S100B appears to regulate a variety of activities by interacting with different molecules in different cell types, intervening in cell proliferation, survival and differentiation20, but data currently available do not appear to converge toward a clearly defined function. In contrast, extracellular S100B has been extensively used as a biomarker for nervous system injury and tumor both in blood and in pathologic tissue21,22. S100B is overexpressed in the sera of lung cancer patients with brain metastasis and implicated in regulating enzyme activities, cell growth, and differentiation23,24,25. Moreover, pharmacological blockade of S100B activity with pentamidine, a drug disrupts S100B-p53 interactions, results in a significant tumor growth inhibition26,27. A complete picture of intracellular and extracellular regulatory effects of S100B in injury regeneration may shed light on developing new strategies for managing lung diseases and promoting lung health.
In this study, we have identified that S100B is selectively expressed in specific airway epithelial cells, namely club cells and ciliated cells, and is notably absent in PNECs in the lungs of adult mice under normal conditions. However, upon lung injury induced by NAPH, the expression of S100B in PNECs is activated, promoting their proliferation and reprogramming into club cells. The stem cell-like function of PNECs in the injury repair process is hindered by pentamidine (PTM) treatment, which suppresses the S100B protein’s activity. In contrast, augmenting P110α levels in airway epithelial cells, as exemplified by the generation of P110*/*; Gata5-Cre mice, enhances S100B expression and fosters club cell regeneration. Moreover, extracellular S100B, in addition to its roles within PNECs, engages with ARG1, attracting ARG1+ anti-inflammatory (M2) macrophages and accelerating epithelial cell proliferation during the repair phase. Our findings highlight the critical role of S100B in PNEC and their interactions with M2 macrophage, efficiently mobilizing PNECs to serve as stem cells for the regeneration of airway club cells.
Results
S100B is expressed in club and ciliated cells, but not in PNECs
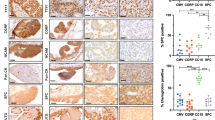
The distal conducting airway of the mammalian lung are composed of three major epithelial cell types namely club cells, ciliated cells and PNECs. During a critical window of embryonic development, E15.5 to E18.5 in mice, the progenitors undergo differentiation to form mature cell types. Analyses of protein and RNA revealed that S100B expression dramatically increased from E18.5 onwards, remaining elevated postnatally (Fig. 1A, B), suggesting a role in epithelial cell function rather than in driving progenitor differentiation. Single-cell RNA sequencing analysis (GSE202325 dataset) further confirmed S100B’s specific expression of in mouse lung epithelial cells (Fig. 1C and Supplementary Figs. 1 and 2). To identify S100B-expressing cell types in adult lung, multiple markers, including CC10 (club cell), β-tubulin (ciliated cell), PGP9.5 (PNEC), CGRP (PNEC), and SPC (alveolar type II cell), were applied. Notably, S100B was detected in club and ciliated cells but not in PNECs (Fig. 1D, Supplementary Fig. 3), implying it may not contribute significantly to PNEC physiology.
A Immunofluorescence (IF) staining of S100B (red) expression at E12.5, E15.5, E18.5, and PN1. Scale bar: 50 μm. B qRT-PCR quantification of S100b mRNA levels across developmental stages (n = 3 mice per stage, biological replicates). Data are presented as mean ± SEM and analyzed using two-sided Student’s t-tests assuming equal variance. C Single-cell expression profiles of S100b visualized by dot plot (left) and violin plot (right); outliers (mean±3) were excluded. D Double IF staining of S100B (red) with airway epithelial markers (CC10, β-TUBLIN), neuroendocrine markers (PGP9.5, CGRP), and alveolar type II marker (SPC) (green) in adult lungs. Scale bar: 50 μm. Arrows highlight S100B-negative PGP9.5+ or CGRP+ cells. All images are representative of ≥3 independent experiments. Source data are provided as a Source Data file.
S100B is required for the ability of PNECs to serve as a reservoir for club cell regeneration
To investigate the response of S100B+ cells to airway injury and subsequent repair, we intraperitoneally administered NAPH to 6-week-old C57BL6 mice to induce club cell ablation and monitored S100B+ cell dynamics at days 1, 2, 3, 5 and 7 post-injury. NAPH treatment triggered rapid club cells death and detachment from airway, followed by gradual reconstitution of the club cell population and epithelial architecture (Supplementary Fig. 4A, B). No significant differences were observed between male and female mice, likely reflecting the use of a relatively high NAPH dose (300 mg/kg) and the younger age of animals, which together may minimize sex-related variation in metabolic activation and repair (Supplementary Fig. 4C, D). Surprisingly, despite extensive club cell loss, S100B expression persisted at day 2 post-NAPH and peaked during new club cell differentiation at days 3–5, before declining as regeneration concluded by day 7 (Fig. 2A, B, Supplementary Figs. 5 and 6). Notably, the S100B+/PGP9.5+ cells expanded markedly by day 2, coinciding with injury-induced PNEC proliferation. Concurrently with CC10+ club cell repopulation at day 7, S100B+/PGP9.5+ cell numbers returned to baseline (Fig. 2C). Temporal profiling showed PGP9.5+ cell expansion during active repair at day 3 and 5 post-injury, whereas CGRP+ cells exhibited near-complete overlap with PGP9.5+ cells only at baseline and completion phases at day 0 and 7 (Supplementary Fig. 7), implying CGRP may specifically mark terminally differentiated PNECs. These dynamic S100B expression patterns within dispersed PGP9.5+ cells during club cell regeneration suggest that this population may contribute to airway repair, potentially through mechanisms distinct from the classical NEB-localized PNECs with established stem cell functions.
A Immunofluorescence co-staining of S100B (red) and PGP9.5 (green) in NAPH-injured lungs at days 0, 1, 2, 3, 5 and 7 post-injury. Scale bar: 50 μm. B qRT-PCR analysis of S100B, PGP9.5 and CC10 mRNA expression during the injury time course (days 0–7) (n = 3 mice). C The percentage of S100B+, PGP9.5+, and double-positive cells among total airway epithelial cells were quantified by manual cell counting. Airway cells were identified by DAPI staining (≥30 random fields per time point, n = 5 mice per group). D CC10 (green) and PGP9.5 (red) localization in injured lungs with/without PTM treatment at day 0, 3, 5 and 7. Scale bar: 50 μm. E, F qRT-PCR analysis of CC10 and PGP9.5 expression in PTM-treated vs. control lungs (n = 3 biological replicates). Data are presented as mean ± SEM and analyzed using two-sided t-test. G Quantification of CC10+, PGP9.5+, and transitional (CC10+/PGP9.5+) cell populations with/without PTM treatment (≥30 random fields per time point, n = 5 mice per group). Data are represented as mean ± SEM and analyzed using two-sided Student’s t-tests assuming equal variance. All images are representative of ≥3 independent experiments. Source data are provided as a Source Data file.
To elucidate the role of S100B in club cell regeneration, pentamidine (PTM)28 was administered daily to NAPH-treated mice to inhibit S100B function. Club cells and PNECs were identified using CC10 and PGP9.5 antibodies, respectively. While PTM treatment alone had no effect on club cells or airway structure (Supplementary Fig. 8), NAPH + PTM-treated lungs exhibited distinct regeneration patterns, with the airway surface largely populated by PGP9.5+ PNECs and relatively few club cells remaining by day 3. Emerging club cells appeared by day 5 alongside declining PNEC numbers, and while control lungs nearly fully reconstituted club cells by day 7, PTM-treated lungs showed significantly impaired regeneration (Fig. 2D and Supplementary Fig. 9). Consistently, this impairment was corroborated by reduced CC10 mRNA levels at days 5 and 7 in PTM-treated lungs (Fig. 2E). Additionally, PGP9.5 expression decreased on day 3 but increased at later timepoints with PTM treatment (Fig. 2F). The ratio of club cell and PNEC over the total number of airway cells were determined by manual counting on multiple samples (Fig. 2G). PTM-treated lungs showed a markedly reduced PNEC population on day 3 (60.50% vs 72.66% in control) and an increased population on day 5 (45.68% vs 32.95% in control). During recovery, control lungs exhibited more CC10+/PGP9.5+ transitional cells (13.49% vs 7.81%) and differentiated club cells on day 5 (28.07% vs 14.07%). Notably, PTM-treated lungs retained elevated PNECs (37.46%) and fewer club cells (25%) at day 7, while ciliated cells remained unaffected by PTM treatment throughout repair (Supplementary Fig. 10). These findings demonstrated that S100B inhibition disrupts PNEC proliferation and the PNEC-club cell transitional phase, ultimately compromising club cell regeneration.
S100B interacts with ARG1 and P110α during club cell regeneration
S100B is unique in that it is found both intracellularly and extracellularly, and it plays significant roles in numerous biological processes by interacting with various target proteins. These interactions can be direct or indirect and can lead to changes in cell behavior and signaling pathways. To identify proteins that interact with S100B in response to airway injury, co-immunoprecipitation (Co-IP) was performed on lung tissues 3 days post-NAPH injury (Fig. 3A). Proteins were extracted, and specific antibody was utilized to bind to S100B and any interacting proteins. The antibody-antigen complex was precipitated out of the protein mixture and subjected to mass spectrometry analysis (Supplementary Table 3). To further validate the role in the context of airway injury and interaction with S100B, the expression of potential interacting proteins in PTM-treated NAPH injury lungs was analyzed by qRT-PCR. The expression of Arginase 1 (Arg1) and the catalytic subunit of PI3K (phosphoinositide 3-kinase), p110α, were significantly decreased after PTM treatment, suggesting that S100B might regulate their expression or activity (Fig. 3B). To further strengthen the evidence of interaction, a reciprocal Co-IP using antibody against S100B to precipitate the S100B complex, followed by detection of ARG1 and P110α was performed (Fig. 3C). Western blot analysis showed a significant upregulation of ARG1 and P110α after NAPH injury, which was suppressed by PTM treatment (Fig. 3D). Consistently, immunohistochemical staining revealed a marked decrease in ARG1+ cells in the airways at days 3, 5, and 7 post-PTM treatment (Fig. 3E). Additionally, qRT-PCR confirmed reduced Arg1 expression upon S100B inhibition after injury (Fig. 3F). Together, the Co-IP and western blot data suggest a protein-protein network involving S100B, ARG1, and P110α, which may play a pivotal role in orchestrating cellular responses to airway injury.
A Coomassie blue staining of S100B protein complexes isolated from day 3 post-NAPH injury lungs. B qRT-PCR analysis of S100B-interacting genes in injured lungs with/without PTM treatment at day 3 (n = 3 mice, biological replicates). Data are represented as mean ± SEM and analyzed using two-sided Student’s t tests assuming equal variance. C Co-immunoprecipitation (co-IP) analysis of protein interactions between S100B, ARG1, and P110α at day 3 post-injury. D Western blot analysis of P110α and ARG1 protein levels during injury time course (days 0–7) with/without PTM treatment, grayscale intensity normalized to β-ACTIN. E Immunofluorescence localization of ARG1 (green) in injured lungs with/without PTM treatment at day 0, 3, 5 and 7. Scale bar: 50 μm. F qRT-PCR analysis of Arg1 mRNA levels across treatment groups and time points (n = 3 mice, biological replicates). Data are represented as mean ± SEM and analyzed using two-sided Student’s t tests assuming equal variance. All images are representative of ≥3 independent experiments. Source data are provided as a Source Data file.
S100B recruits ARG1+ macrophages to promote epithelial cell proliferation
To evaluate the role of macrophages in NAPH injury repair, we depleted them by administering clodronate liposomes (CL) 48 h prior to NAPH treatment. Macrophage depletion was confirmed using anti-F4/80 staining (Supplementary Fig. 11). Depletion led to a striking reduction in PNECs by day 3 (16.29% vs 64.94% in controls). During recovery, control lungs exhibited significantly higher numbers of CC10+/PGP9.5+ transitional cells (20.03% vs 0.72%) and differentiated club cells by day 5 (25.99% vs 2.17%). Conversely, CL-treated lungs maintained elevated PNEC levels (41.67% vs 20.21%) and generated fewer club cells by day 7 (7.33% vs 56.44%) (Fig. 4A, B). Given that ARG1 marks anti-inflammatory (M2) macrophage26, we isolated these cells from lung tissues for flow cytometry analysis. The gating strategy began with excluding debris and granulocyte based on FSC-A/SSC-A scatter, followed by doublet exclusion using FSC-W/FSC-A to isolate single cells. After selecting CD45⁺ leukocytes, M2 macrophages were identified through F4/80⁺/ARG1⁺ double-positive cells (Supplementary Fig. 12). The number of ARG1+ macrophage increased during repair, but this recruitment was markedly reduced by PTM treatment. In uninjured wild-type mice, ARG1+ macrophages constituted approximately 7.17% of total lung cells. Injury trigged an influx of inflammatory and apoptotic macrophages, decreasing the M2 subset to 4.29% by day 3. As repair progressed, this population expanded to 5.38% by day 5 and 8.68% by day 7. Strikingly, S100B inhibition with PTM severely impaired ARG1+ macrophage recruitment, with proportions falling to 1.74% (day 3), 2.51% (day 5) and 4.00% (day 7) (Fig. 4C). Flow cytometry analysis of bronchoalveolar lavage (BAL) fluid further confirmed that PTM treatment significantly diminished macrophages levels during regeneration (Fig. 4C).
A Immunolocalization of F4/80 (macrophages) (red), CC10 (green), and PGP9.5 (red) in NAPH-injured lungs at day 0, 3, 5 and 7, with or without clodronate liposomes (CL) treatment. Scale bar: 50 μm. B Quantification of CC10+, PGP9.5+, and CC10+/PGP9.5+ transitional cell populations with or without CL treatment (≥30 random fields per time point, n = 5 mice per group). Scale bar: 50 μm. Data are represented as mean ± SEM and analyzed using two-sided Student’s t-tests assuming equal variance. C Flow cytometric analysis of M2 macrophages (ARG1+/F4/80+) in lung tissue and macrophages (CD11b+/F4/80+) in bronchoalveolar lavage (BAL) at the indicated time points with or without PTM treatment. Red borders highlight the M2 or macrophage populations (percentage shown). D Quantification of MLE-15 cells after 12- and 20-day co-culture with ARG1+ or ARG1- macrophages. n = 5 independent biological replicates. Data are represented as mean ± SEM and analyzed using two-sided Student’s t tests assuming equal variance. E Colony formation assay of MLE-15 cells co-cultured with macrophage subsets. Scale bars: 400 μm (12-day) and 1000 μm (20-day). All images show representative results from ≥3 independent experiments. Source data are provided as a Source Data file.
To examine the impact of ARG1+ macrophages on lung epithelial cells, AGR1+ macrophages were collected from lungs at day 5 post-NAPH injury and co-cultured with the mouse lung epithelial cell line, MLE-15, within Matrigel (Supplementary Fig. 13). The proliferation and clonal expansion of MLE-15 were assessed on days 12 and 20. The results indicated that ARG1+ macrophages promoted epithelial cell proliferation (Fig. 4D), leading to the formation of larger monoclonal colonies (Fig. 4E). These findings suggest that PNECs attract ARG1+ macrophages via the S100B-ARG1 interaction, thereby facilitating their proliferation in response to NAPH-induced lung injury.
Activation of p110α enhances S100B-ARG1-regulated club cell regeneration
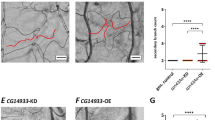
Above studies discovered that S100B interacted with P110α, to clarify the role of P110α in injury repair, we specifically activated P110α in airway epithelial cells by generating P110*/*; Gata5-cre mice (Fig. 5A). As previously reported, Gata5 expression in the lung is uniformly restricted to endoderm-derived epithelium29. Western blot analysis confirmed P110α accumulation in P110*/+; Gata5-cre lungs (Fig. 5B). Histological examination of H&E-stained lung sections from P110*/*; Gata5-cre mice at E12.5, E15.5, E18.5 and 3 months of age showed normal pulmonary architecture comparable to controls (Supplementary Fig. 14). To systematically evaluate potential effects of epithelial P110α activation on airway epithelium and mesenchyme composition, cell-specific markers for club, ciliated, PNEC, type II, type I and smooth muscle cells were examined at E15.5 and E18.5. Comparative analysis revealed no significant differences in marker expression patterns or distribution between P110*/*; Gata5-cre and control lungs (Supplementary Fig. 15A–C). Quantification confirmed normal club cell proportions in E18.5 P110*/*; Gata5-cre lungs (Supplementary Fig. 15D). Following NAPH exposure in adult mice, we analyzed lungs on post-injury days 3, 5 and 7 using CC10 (club cells), β-tubulin (ciliated cells), and PGP9.5 (PNECs) markers (Fig. 5C). While ciliated cells remained unaffected (Supplementary Fig. 16), P110*/*; Gata5-cre lungs showed increased ratios of PGP9.5+ cells and CC10+/PGP9.5+ cells on days 3 and 5. Subsequently, on day 7, CC10+ cells in P110*/*; Gata5-cre lungs reached 58.40%, whereas only 45.35% in control lungs. The markedly larger population of CC10+/PGP9.5+ cells on day 5 (19.61% vs 11.13% in controls) suggested that P110α activation enhances club cell regeneration by modulating PNEC proliferation and differentiation (Fig. 5D). This interpretation was further supported by increased numbers of anti-Ki67 labeled proliferating PGP9.5+ cells in P110*/*; Gata5-cre lungs at days 3 and 5 (Fig. 5E).
A Generation of P110*/*; Gata5-cre mice by crossing P110*/* with Gata5-cre mice. B Western blot analysis of P110α, phospho-AKT, and total AKT in P110*/* and P110*/*; Gata5-cre lungs. Quantification was performed on grayscale images normalized to β-ACTIN, with n = 3 independent biological replicates, each representing separate samples. Data are represented as mean ± SEM and analyzed using two-sided Student’s t-tests assuming equal variance. C Immunofluorescence co-staining of CC10 (green) and PGP9.5 (red) in P110*/* and P110*/*; Gata5-cre lungs at day 0, 3, 5, and 7 post-injury. Scale bar: 50 μm. D Quantification of CC10+, PGP9.5+, and CC10+/PGP9.5+ transitional cell populations across time points (≥30 random fields per time point, n = 5 mice per group). Data are represented as mean ± SEM and analyzed using two-sided Student’s t-tests assuming equal variance. E Proliferating PNECs identified by PGP9.5 (green) and Ki67 (red) co-staining at indicated time points. Scale bar: 50 μm. All images show representative results from ≥3 independent experiments. Source data are provided as a Source Data file.
We next examined S100B expression in P110*/*; Gata5-cre lungs to elucidate the impact of P110α during repair. qRT-PCR revealed significant upregulation of S100B mRNA by day 3 post-injury (Fig. 6A). Western blot showed elevated S100B protein levels both basally (day 0) and throughout repair (Fig. 6B). Notably, while S100B transcription declined by day 5 in P110*/*; Gata5-cre lungs, potentially reflecting transdifferentiatiation of PGP9.5+ progenitors into CC10+/PGP9.5+ intermediates and mature CC10+ club cells, S100B protein levels remained elevated at days 5 and 7. This divergence suggests that P110α may promote post-translational or post-translational stabilization of S100B, a mechanism that could support airway stem cell-mediated repair by maintain S100B protein availability during critical phases of epithelial regeneration. Consistent with the role of S100B in epithelial repair, PTM-treated P110*/*; Gata5-cre mice showed impaired club cell regeneration (Fig. 6C, D). Furthermore, we observed increased peribronchial ARG1+ macrophages in mutants at all time points during repair, correlating with upregulated Arg1 expression (Fig. 6E, F).
A qRT-PCR analysis of S100B mRNA levels in P110*/* and P110*/*; Gata5-cre lungs during injury time course (day 0–7) (n = 3 mice, biological replicates). Data are represented as mean ± SEM and analyzed using two-sided Student’s t tests assuming equal variance. B Western blot analysis of S100B protein expression across time points. Quantification was performed on grayscale images normalized to β-ACTIN, with n = 3 independent biological replicates, each representing separate samples. Data are represented as mean ± SEM and analyzed using two-sided Student’s t tests assuming equal variance. C Immunofluorescence co-staining of CC10 (green) and PGP9.5 (red) in P110*/*; Gata5-cre lungs with/without PTM treatment. Scale bar: 50 μm. D Quantification of CC10+, PGP9.5+, and CC10+/PGP9.5+ transitional cell populations across time points (≥30 random fields per time point, n = 5 mice per group). Data are represented as mean ± SEM and analyzed using two-sided Student’s t tests assuming equal variance. E ARG1+ (red) macrophage localization in P110*/* vs. P110*/*; Gata5-cre lungs (≥30 random fields per time point, n = 5 mice per group). Scale bar: 50 μm. F qRT-PCR analysis of Arg1 expression in P110*/* and P110*/*; Gata5-cre lungs on day 0, 3, 5 and 7 post-injury (n = 3 mice, biological replicates). Data are represented as mean ± SEM and analyzed using two-sided Student’s t tests assuming equal variance. All images show representative results from ≥3 independent experiments. Source data are provided as a Source Data file.
Discussion
PNECs are considered stem-like cells that play a crucial role in the regeneration of airway epithelial cells13,14. In response to NAPH-induced injury, PNECs undergo proliferation to generate additional cells (self-renew), and the daughter cells then reprogram into a PNEC/club cell transitional state, eventually leading to the formation of new club cells. Our current results define that this highly coordinated progression of PNECs is dependent on S100B activity. NAPH injury triggers the expression of S100B within PNECs, and the subsequent release of S100B protein attracts ARG1+ macrophages. These macrophages enhance the proliferation of PNECs, facilitating their differentiation into transitional cells and the subsequent production of new club cells. The modulation of S100B function through PTM treatment or the enhancement of P110α in airway epithelial cells significantly impacts the reparative capabilities of PNECs following injury (Fig. 7).
Following NAPH-induced injury, S100B is activated within PNECs. Secreted S100B recruits ARG1+ macrophages to the injury site, which promote PNEC proliferation through reciprocal signaling. This cellular crosstalk drives PNEC differentiation through a transitional cell state (CC10+/PGP9.5+), ultimately restoring the club cell population.
S100B is an important member of the multigenic S100 family and highly expressed in astrocytes within the brain, Schwann cells of the peripheral nervous system, and in non-neural tissues such as melanocytes, adipocytes and chondrocytes30,31. The primary objective of our study was to ascertain the localization of S100B in the mouse lung. The results reveal selective expression of S100B in specific airway epithelial cells, including club and ciliated cells, but not in PNECs. PNECs are integrated into the airway epithelium, forming junctions with adjacent club and ciliated cells and originating from common bronchial progenitors during early embryonic development. Cell lineage analysis indicates that S100B expression in the airway epithelium commences at E18.5, suggesting its involvement in establishing cell functions rather than progenitor cell differentiation. PNECs are specialized sensory cells that monitor airway conditions and transmit signals to other lung cells and the brain via synapses with sensory neurons14,32. In contrast, club cells secrete various substances, while ciliated cells contribute to mucociliary clearance through the coordinated beating of their cilia. The absent of S100B in mature PNECs suggests that alternative regulatory mechanisms may be involved in their sensory and paracrine signaling functions, which include monitoring airway status and regulating respiration. How S100B signaling contribute to the function of other highly specialized epithelial cell types in the airway are queries that had not been hitherto addressed.
PNECs typically remain quiescent under homeostatic conditions but exhibit remarkable plasticity following airway injury, whether induced by NAPH-mediated epithelial damage13 or genetic club cell ablation33. This injury-induced activation enables potential transdifferentiation into alternative lineages, highlighting their crucial role in epithelial repair. Marker selection proves critical for characterizing distinct PNEC states during injury response. While CGRP reliably identifies terminally differentiated PNECs and ASCL1 marks developmental progenitors, PGP9.5 provides superior utility by detecting both constitutive PNECs and activated states, including potential epithelial-to-neuroendocrine reprogrammed cells11. This broad detection capability reveals the full spectrum of neuroendocrine involvement in repair processes. Supporting this approach, CGRP ablation studies show no impairment of homeostasis or repair10, while ASCL1+ cells exhibit limited proliferation capacity14. These findings suggest that PGP9.5+ populations likely encompass transitional states essential for tissue regeneration. Notably, we observed that post-injury PGP9.5+ cells lose their characteristic neuroepithelial body (NEB) clustering and expand diffusely across the airway epithelium, consistent with PGP9.5’s pan-neuroendocrine labeling. These dispersed PGP9.5+ population may contribute to repair through non-canonical mechanisms distinct from the classical Ascl1-dependent proliferation observed in intact NEBs.
The temporal expression pattern of S100B in PNECs, coinciding with their proliferation beginning at day 2 post-injury, suggests an important role for this protein in PNEC stem cell functions. Pharmacological inhibition of S100B (using PTM) significantly reduces both PNEC proliferation and the population of CC10+/PGP9.5+ transitional cells, demonstrating S100B’s involvement in PNEC proliferation and reprogramming. Interestingly, while we observe a substantial increase in S100B+ PNECs following NAPH-induced injury, a distinct subset of S100B-negative PNECs persists. This finding aligns with single-cell analyses by Ouadah et al., which revealed that only a minor subpopulation of differentiated PNECs possesses stem cell capabilities14. The proliferation of PGP9.5+ cells alone further underscores the heterogeneity within the PNEC stem cell population. Current evidence identifies at least three injury-induced signals that regulate PNEC renewal, dispersal and transdifferentiation14. To fully elucidate the behavior and functions of specific PNEC subpopulations during injury response, future studies should focus on identifying additional biomarkers that can distinguish these functionally distinct cell groups.
Numerous studies have indicated that PNECs play a role in oxygen sensing and immune responses7. The neuroimmune recognition and regulation within the respiratory system is a sophisticated and highly coordinated process to detect and respond to pathogens, pollutants and other potential hazards. Sensory neurons can sense pathogen directly by expressing pathogen receptors and immune factors, or indirectly by forming recognition units in collaboration with respiratory epithelial cells34. The sensory capacity of PNECs is demonstrated by their release of CGRP to mitigate lung damage in response to hypoxia6. Additionally, CGRP secretion from PNECs induces the recruitment of macrophages, thereby regulating the immune environment within the lungs. Dysregulation of PNECs and excessive CGRP lead to increased immune infiltrates8. However, genetic ablation of CGRP did not affect homeostasis or the ability to repair airway10, suggesting CGRP-independent mechanisms govern PNEC stem cell function and their interaction with the immune system. Our results establish S100B as a major factor controlling the PNEC-macrophage response by interacting with the ARG1 protein. Dysfunction of S100B leads to fewer ARG1+ macrophages and transitional cells (CC10+/PGP9.5+), indicating that the S100B-ARG1 axis is necessary for PNEC function as stem cell in club cell regeneration. However, the exact role of ARG1+ macrophage, whether as signal transmitters to PNEC proliferation or as part of the stem cells niche, remains to be elucidated.
A multitude of immune cells resident in the respiratory tract, including mast cells35, dendritic cells36, and interstitial macrophages37. Among these, macrophages initiate the immune response to injury by secreting a variety of soluble mediators, chemokines, and growth factors that recruit and stimulate diverse cell types involved in inflammation and lung repair38. Macrophages are typically categorized into two functional phenotypes: pro-inflammatory (M1) and anti-inflammatory (M2) macrophages39. Studies have shown that platelets are recruited into the lung alongside regulatory T (Treg) cells during the resolution phase of lung injury40. In NAPH-induced lung injury, M2 macrophages and T reg cells work synergistically to suppress inflammation and promote epithelial regeneration. M2 macrophages, polarized by Treg-derived IL-10 and TGF-β, secrete anti-inflammatory cytokines and growth factors that facilitate airway epithelial repair41. Conversely, M2 macrophages enhance Treg cell stability and expansion by producing retinoic acid and PGE2, reinforcing a feedback loop that sustains immune tolerance and injury resolution42. The association between S100B and ARG1, a marker of M2 macrophages, suggests that the PNEC-M2 macrophage interaction is more critical for tissue repair than pathogen clearance following NAPH injury. M2-derived cytokines and growth factors may also promote the proliferation of PNECs and MLE-15 cells. Notably, disruption of S100B primarily affects M2 macrophages at 3 days post-injury, highlighting its dual role in PNEC proliferation and reprogramming. However, whether S100B modulates the Treg-M2 axis in post-NAPH lung homeostasis remains to be investigated. Given that small cell lung cancer (SCLC) is hypothesized to originate from PNECs14, further studies are needed to molecularly characterize M2 macrophage-promoted PNECs and determine if they serve as the primary cell of origin for SCLC.
Our study demonstrates that P110α activation in P110*/*; Gata5-cre lungs drives a stepwise regeneration program, promoting both PNEC proliferation and their differentiation into club cells (Fig. 5C, D). Although P110α is broadly expressed in airway epithelium, PNECs serve as the primary regenerative mediators, while ciliated cells contribute minimally, as shown by their low proliferation during repair (Supplementary Fig. 17). We identified a P110α-S100B-ARG1+ macrophage axis wherein P110α upregulates S110B expression to recruit ARG1+ macrophages and enhance club cell regeneration. Notably, S100B expression is injury-dependent, as absent in uninjured P110*/*; Gata5-cre PNECs, suggesting additional regulatory factors. Furthermore, PTM-mediated S100B inhibition reduces P110α levels, revealing a bidirectional regulatory relationship. Specifically, P110α acts upstream of S100B to promote regeneration, while S100B feedback is required for maintaining P110α activity. These findings uncover a sophisticated P110α-S100B interplay in lung repair that warrants further mechanistic investigation.
The absence of significant alterations in airway epithelial composition, including club cells, following early P110α activation in P110*/*; Gata5-cre airways implies that P110α signaling may function redundantly or in a context-dependent manner during lung epithelial fate determination. This resilience likely stems from the robust transcriptional networks governing airway development, which appear resistant to moderate perturbations in PI3K signaling. Notably, the failure of P110α activation to enhance AKT phosphorylation (Fig. 5B) further supports the existence of stringent regulatory mechanisms that constrain constitutive PI3K-AKT pathway activity. These findings suggest that P110α activation alone is insufficient to disrupt established developmental programs, potentially due to compensatory adaptations such as PI3K feedback inhibition or activation of parallel pathways, such as Wnt/β-catenin, that preserve epithelial homeostasis.
Collectively, our study provides a novel insight into the role of S100B in the regulatory mechanisms governing PNEC stemness and the interaction between PNEC and M2 macrophage, placing S100B as a crucial factor in the repair and regeneration processes following lung airway injury.
Methods
This study was conducted in full compliance with the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines 2.0. All experiments complied with relevant ethical regulations and were approved by China Agricultural University under protocol number AW81801202-3-2.
Mouse lines
P110*/* mice were purchased from Jackson Laboratory (strain name: C57BL/6-Gt (ROSA)26Sortm7 (Pik3ca*, EGFP) Rsky/J, stock number: 012343, also known as R26StopFLP110*), allowing inducible expression of activated PI3K heterodimer activity43. Gata5-Cre mice were kindly provided by Dr. Parviz Minoo (University of Southern California, USA). P110*/*; Gata5-Cre mice were generated by crossing P110*/* mice with Gata5-Cre mice. C57BL/6 mice served as control animals. All animals were housed in accordance with protocol approved by the Beijing Association of Laboratory Animal Care (Beijing, China), and experiments were conducted under protocol AW81801202-3-2 from China Agricultural University. Mice were housed in individually ventilated units within a specific-pathogen-free facility, maintained on a 12 h light/dark cycle, with ad libitum access to water and standard chow. Ambient temperature and humidity were kept within standard laboratory conditions.
Single-cell RNA-seq analysis
Single-cell RNA-seq analysis was performed on the GSE202325 dataset (young and aged mouse lungs at days 3/9 post-influenza) using Seurat v5.3.0. After quality control (retaining cells with 200-6000 genes and <15% mitochondrial content; removing genes in <3 cells), data were ln (CPM/100 + 1) normalized. We identified variable features, performed PCA (selecting 17 PCs via elbow/90% variance/maximum curvature methods), and clustered cells (Louvain algorithm, resolution = 0.5) following SNN graph construction. Cell types were annotated using SingleR v2.8.0 with celldex v1.16.0’s MouseRNAseq reference, assigning cluster identities by majority type. Marker genes were identified via the Wilcoxon rank-sum test with Benjamini-Hochberg correction, visualized using t-SNE. For S100B analysis, expression outliers (mean ± SD) were excluded from dot/violin plots.
Naphthalene exposure
Adult male and female mice were intraperitoneally injected with 300 mg/kg NAPH (Sigma, USA) dissolved in corn oil. No significant sex-related differences were observed in injury repair. Therefore, all data presented in this study were obtained from 6-week-old female mice for consistency. Animals were sacrificed at 0, 1, 2, 3, 5, and 7 days after NAPH administration. At each time point, five mice per group (n = 5) were analyzed, and all experiments were independently repeated at least three times. Anesthesia and euthanasia. During experimental procedures, animals were monitored at least twice daily or more frequently depending on the intervention and anticipated level of distress. Humane euthanasia was performed using CO₂ inhalation (20–30% chamber displacement/min).
Histology and immunohistochemistry (IHC)
Lungs were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS, pH 7.0) and processed into serial paraffin sections using standard protocols. Tissue sections (5 μm) were prepared for hematoxylin-eosin (H&E) staining and immunohistochemical (IHC) analysis. H&E staining was performed according to standard procedures for morphological evaluation. For IHC, a commercial kit (ZSGB-BIO, China) was used. To ensure staining specificity and reliability, appropriate controls were systematically included for each antibody and marker, including no-primary antibody controls, positive tissue controls, and internal controls within experimental samples. These controls consistently confirmed that the observed staining in experimental samples was antigen-specific and not due to nonspecific antibody binding, cross-reactivity of secondary antibodies, or artifacts of the detection system. The primary antibodies used in the experiments are listed in Supplementary Table 1.
RNA extraction and qRT-PCR
Total RNA was extracted from cells and tissues using TRIzol reagent (Invitrogen, Life Technologies, China). cDNA was synthesis with the StarScript III RT Kit (Genstar, China) following the manufacturer’s instruction. Quantitative real-time PCR was performed on a LightCycler 480 System (Roche). Primers were designed using Primer3 software and are listed in Supplementary Table 2. Relative gene expression was determined by the 2^(−ΔΔCt) method. ΔCt values were calculated as (Ct_target—Ct_18S) for both treated and control groups, and ΔΔCt as (ΔCt_treated—ΔCt_control). Expression levels were normalized to the control group (set to 1).
Western blot analysis
Lung tissues were harvested and frozen in liquid nitrogen. Protein extracts were prepared in RIPA buffer containing 1 mM PMSF (Beyotime, China) and phosphatase inhibitor (Roche, Switzerland). Equal amounts of protein were separated on 4–12% SDS-PAGE gel, transferred to Immobilon-P membrane, and hybridized to selected primary antibody and HRP-conjugated second antibody for subsequent detection by SuperSignal* West Pico Chemiluminescent Substrate (Pierce). The intensity of bands was analyzed by ImageJ. Full, uncropped blot scans are provided in the Source Data file.
Bronchoalveolar lavage (BAL)
After sacrificing the mice by cervical dislocation, the trachea was exposed and intubated to facilitate the lavage process. PBS was instilled into the lungs in three aliquots, and each aliquot of PBS was retained in the lungs for approximately 30 s, during which the lung tissue was gently massaged to facilitate the distribution of fluid and interaction with the cells. The PBS was then aspirated, and the resulting lavage fluid was collected for counting and analyzing the presence of macrophages.
Immunoprecipitation-mass spectrometry (IP-MS)
Proteins were extracted from lung tissues 3 days post-NAPH injury, and specific antibodies was used to capture S100B and interacting proteins. The antibody-antigen complex was immunoprecipitated and subjected to mass spectrometry analysis (n = 5) following the manufacture’s protocol (Beyotime, China). Proteins were digested according to the FASP method. Nano LC-MS analysis was performed on Q-Exactive high-resolution mass spectrometer (Thermo Scientific) and Nano-Acquity nano HPLC (Waters, MA, USA). Peak picking and alignment were performed by using Progenesis QI for Proteomics (build 2.0, Nonlinear Dynamics, Newcastle, UK). Protein interactions were analyzed by using STRING10 (https://cn.string-db.org/), and pathway enrichment was assessed via KEGG analysis.
Macrophage depletion in the mouse NAPH injury model
For macrophage depletion, mice were administered clodronate liposomes (5 mg/mL, 50 µL per dose; YEASEN, Shanghai, China) via intratracheal instillation 48 h prior to NAPH treatment. Subsequent doses were administered every 48 h until the end of the study.
Flow cytometry
To analyze anti-inflammatory macrophages in lung tissue and bronchoalveolar lavage (BAL) samples, cells were first incubated with an Fc receptor blocking agent (anti-mouse CD16/32) to prevent nonspecific binding, and then identified using a standardized gating strategy wherein nucleated cells were selected by FSC-A/SSC-A, doublets were excluded by FSC-A/FSC-W, dead cells were removed with 7-AAD staining, leukocytes were gated on CD45 positivity, and M2 macrophages were ultimately defined by high F4/80 and ARG1 expression. This gating strategy was also applied to evaluate macrophage depletion following clodronate liposome treatment. Finally, after washing with flow cytometry staining buffer (Proteintech, China), cells were incubated with the appropriate secondary antibody, centrifuged, and resuspended for analysis on BD FACSVerse™ and BD FACSAria Fusion flow cytometer.
3D cell culture
AGR1+ macrophages from injured mouse were mixed with Matrigel and plated in Transwell chambers (Corning, USA). MLE-15 cells, a mouse lung epithelial cell line, were added to the lower chamber wells to co-culture with AGR1+ macrophages in a 3D format.
Cell counting and image analysis
Systematic random sampling was used for histomorphometric quantification. Each group included lung tissues from five mice, with more than three cross-sections from proximal airways and five from distal airways per mouse. Epithelial cells were quantified in ImageJ (v1.53), with positive cells defined by epithelial localization, distinct nuclear morphology, and fluorescence intensity >3× background. Data were normalized to the number of positive cells per unit length of basement membrane. All analyses were performed independently, showing intra-group variation <15% and inter-observer reliability (ICC) > 0.85. Images were acquired with an Olympus BX53 microscope (20x objective). Cell counts were obtained manually based on DAPI-stained nuclei and cell-type-specific markers across more than 30 randomly selected fields per sample. Each group contained 5–8 biological replicates, and cell population percentages were calculated from pooled counts.
Statistical analysis
Each experimental group contained a minimum sample size of n = 3 biologically independent replicates. All measurements were derived from distinct biological specimens (one data point per sample). Technical replicates (e.g., triplicate qPCR runs per RNA sample) were averaged prior to analysis. Statistical analyses were conducted using SPSS 16.0 software, with between-group comparisons performed using two-sided Student’s t tests assuming equal variance. Data are presented as mean ± standard error of the mean (SEM), with statistical significance defined as P < 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors confirm that all data supporting the findings of this study are available within the article and its supplementary information. The single-cell RNA sequencing analysis uses the publicly available dataset GSE202325. The raw mass spectrometry data for the IP-MS analyses are included in Supplementary Table 3. Full length uncropped blots and other source data are provided in the accompanying Source Data file. Source data are provided with this paper.
References
Han, S., Budinger, G. R. S. & Gottardi, C. J. Alveolar epithelial regeneration in the aging lung. J. Clin. Investig. 133, e170504 (2023).
Bin, N. R. et al. An airway-to-brain sensory pathway mediates influenza-induced sickness. Nature 615, 660–667 (2023).
Han, X. et al. Long-term radiological and pulmonary function abnormalities at 3 years after COVID-19 hospitalisation: a longitudinal cohort study. Eur. Respir. J. 64, 2301612 (2024).
Prescott, S. L., Umans, B. D., Williams, E. K., Brust, R. D. & Liberles, S. D. An airway protection program revealed by sweeping genetic control of vagal afferents. Cell 181, 574–589.e514 (2020).
Boers, J. E., den Brok, J. L., Koudstaal, J., Arends, J. W. & Thunnissen, F. B. Number and proliferation of neuroendocrine cells in normal human airway epithelium. Am. J. Respir Crit. Care Med. 154, 758–763 (1996).
Shivaraju, M. et al. Airway stem cells sense hypoxia and differentiate into protective solitary neuroendocrine cells. Science 371, 52–57 (2021).
Cutz, E., Pan, J., Yeger, H., Domnik, N. J. & Fisher, J. T. Recent advances and contraversies on the role of pulmonary neuroepithelial bodies as airway sensors. Semin. Cell Dev. Biol. 24, 40–50 (2013).
Branchfield, K. et al. Pulmonary neuroendocrine cells function as airway sensors to control lung immune response. Science 351, 707–710 (2016).
Shan, L., Aster, J. C., Sklar, J. & ME, S. unday Notch-1 regulates pulmonary neuroendocrine cell differentiation in cell lines and in transgenic mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 292, L500–L509 (2007).
Song, H. et al. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc. Natl. Acad. Sci. USA 109, 17531–17536 (2012).
Xing, Y., Li, A., Borok, Z., Li, C. & Minoo, P. NOTCH1 is required for regeneration of Clara cells during repair of airway injury. Stem Cells 30, 946–955 (2012).
Stevens, T. P., McBride, J. T., Peake, J. L., Pinkerton, K. E. & Stripp, B. R. Cell proliferation contributes to PNEC hyperplasia after acute airway injury. Am. J. Physiol. 272, L486–L493 (1997).
Peake, J. L., Reynolds, S. D., Stripp, B. R., Stephens, K. E. & Pinkerton, K. E. Alteration of pulmonary neuroendocrine cells during epithelial repair of naphthalene-induced airway injury. Am. J. Pathol. 156, 279–286 (2000).
Ouadah, Y. et al. Rare pulmonary neuroendocrine cells are stem cells regulated by Rb, p53, and Notch. Cell 179, 403–416.e423 (2019).
Kuo, C. S. & Krasnow, M. A. Formation of a neurosensory organ by epithelial cell slithering. Cell 163, 394–405 (2015).
Hong, K. U., Reynolds, S. D., Giangreco, A., Hurley, C. M. & Stripp, B. R. Clara cell secretory protein-expressing cells of the airway neuroepithelial body microenvironment include a label-retaining subset and are critical for epithelial renewal after progenitor cell depletion. Am. J. Respir. Cell Mol. Biol. 24, 671–681 (2001).
Giangreco, A., Reynolds, S. D. & Stripp, B. R. Terminal bronchioles harbor a unique airway stem cell population that localizes to the bronchoalveolar duct junction. Am. J. Pathol. 161, 173–182 (2002).
Kim, C. F. et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121, 823–835 (2005).
Sorci, G. et al. The many faces of S100B protein: when an extracellular factor inactivates its own receptor and activates another one. Ital. J. Anat. Embryol. 115, 147–151 (2010).
Michetti, F. et al. The S100B story: from biomarker to active factor in neural injury. J. Neurochem 148, 168–187 (2019).
Henze, G., Dummer, R., Joller-Jemelka, H. I., Böni, R. & Burg, G. Serum S100-a marker for disease monitoring in metastatic melanoma. Dermatology 194, 208–212 (1997).
Michetti, F., Massaro, A., Russo, G. & Rigon, G. The S-100 antigen in cerebrospinal fluid as a possible index of cell injury in the nervous system. J. Neurol. Sci. 44, 259–263 (1980).
Choi, J., Kim, D. I., Kim, J., Kim, B. H. & Kim, A. Hornerin is involved in breast cancer progression. J. Breast Cancer 19, 142–147 (2016).
Kondrup, M., Nygaard, A. D., Madsen, J. S. & Bechmann, T. S100B as a biomarker for brain metastases in patients with non-small cell lung cancer. Biomed. Rep. 12, 204–208 (2020).
Mu, S., Ma, H., Shi, J. & Zhen, D. The expression of S100B protein in serum of patients with brain metastases from small-cell lung cancer and its clinical significance. Oncol. Lett. 14, 7107–7110 (2017).
Smith, J. et al. The effect of pentamidine on melanoma ex vivo. Anticancer Drugs 21, 181–185 (2010).
Arcuri, C., Bianchi, R., Brozzi, F. & Donato, R. S100B increases proliferation in PC12 neuronal cells and reduces their responsiveness to nerve growth factor via Akt activation. J. Biol. Chem. 280, 4402–4414 (2005).
Hartman, K. G., McKnight, L. E., Liriano, M. A. & Weber, D. J. The evolution of S100B inhibitors for the treatment of malignant melanoma. Future Med. Chem. 5, 97–109 (2013).
Xing, Y. et al. Signaling via Alk5 controls the ontogeny of lung Clara cells. Development 137, 825–833 (2010).
Adami, C. et al. S100B expression in and effects on microglia. Glia 33, 131–142 (2001).
Mittl, P. R. et al. Metal-free MIRAS phasing: structure of apo-S100A3. Acta Crystallogr. D Biol. Crystallogr. 58, 1255–1261 (2002).
Garg, A., Sui, P., Verheyden, J. M., Young, L. R. & Sun, X. Consider the lung as a sensory organ: a tip from pulmonary neuroendocrine cells. Curr. Top. Dev. Biol. 132, 67–89 (2019).
Reynolds, S. D. et al. Conditional Clara cell ablation reveals a self-renewing progenitor function of pulmonary neuroendocrine cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 278, L1256–L1263 (2000).
Chen, J., Lai, X., Song, Y. & Su, X. Neuroimmune recognition and regulation in the respiratory system. Eur. Respir. Rev.33, 240008 (2024).
Xu, H. et al. Neurotransmitter and neuropeptide regulation of mast cell function: a systematic review. J. Neuroinflammation 17, 356 (2020).
Veres, T. Z. et al. Spatial interactions between dendritic cells and sensory nerves in allergic airway inflammation. Am. J. Respir. Cell Mol. Biol. 37, 553–561 (2007).
Ural, B. B. et al. Identification of a nerve-associated, lung-resident interstitial macrophage subset with distinct localization and immunoregulatory properties. Sci. Immunol. 5, eaax8756 (2020).
Cox, N., Pokrovskii, M., Vicario, R. & Geissmann, F. Origins, biology, and diseases of tissue macrophages. Annu Rev. Immunol. 39, 313–344 (2021).
Ge, Z., Chen, Y., Ma, L., Hu, F. & Xie, L. Macrophage polarization and its impact on idiopathic pulmonary fibrosis. Front. Immunol. 15, 1444964 (2024).
Rossaint, J. et al. Platelets orchestrate the resolution of pulmonary inflammation in mice by T reg cell repositioning and macrophage education. J. Exp. Med. 218, e20201353 (2021).
D’Alessio, F. R. et al. CD4+CD25+Foxp3+ Tregs resolve experimental lung injury in mice and are present in humans with acute lung injury. J. Clin. Investig. 119, 2898–2913 (2009).
Shirey, K. A. et al. Role of the lipoxygenase pathway in RSV-induced alternatively activated macrophages leading to resolution of lung pathology. Mucosal Immunol. 7, 549–557 (2014).
Dai, H., Zhu, M., Li, W., Si, G. & Xing, Y. Activation of PI3K/p110α in the lung mesenchyme affects branching morphogenesis and club cell differentiation. Front. Cell Dev. Biol. 10, 880206 (2022).
Acknowledgements
This work was supported by Biological Breeding-National Science and Technology Major Project (2023ZD0407106) and the 2115 Talent Development Program of China Agricultural University.
Author information
Authors and Affiliations
Contributions
Yiming Xing: conceptualization, investigation, supervision, formal analysis, writing-review and editing, funding acquisition. Bing Sun, Haiting Dai, and Tiemei Zhao: investigation, design and execution of molecular biology and biochemical experiments. Yun Guo: data analysis. Yujia Liu, Wenya Li, Mingli Zhu, Yilina Bai, and Hanyuhui Yang: animal experiments. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sun, B., Dai, H., Zhao, T. et al. S100B triggers neuroendocrine macrophage networks to drive airway regeneration in mice. Nat Commun 17, 965 (2026). https://doi.org/10.1038/s41467-025-67691-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-67691-8