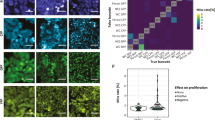
Abstract
Active, responsive, non-equilibrium materials–at the forefront of materials engineering–offer dynamical restructuring, mobility and other complex life-like properties. Yet, this enhanced functionality comes with significant amplification of the size and complexity of the datasets needed to characterize their properties, thereby challenging conventional approaches to analysis. To meet this need, we present BARCODE: Biomaterial Activity Readouts to Categorize, Optimize, Design and Engineer, an open-access software that automates high throughput screening of microscopy video data to enable non-equilibrium material optimization and discovery. BARCODE produces a unique fingerprint or ‘barcode’ of performance metrics that visually and quantitatively encodes dynamic material properties with minimal file size. Using three complementary material-agnostic analysis branches, BARCODE significantly reduces data dimensionality and size, while providing rich, multiparametric outputs and rapid tractable characterization of activity and structure. We analyze a series of datasets of cytoskeleton networks and cell monolayers to demonstrate BARCODE’s abilities to accelerate and streamline screening and analysis, reveal unexpected correlations and emergence, and enable broad non-expert data access, comparison, and sharing.
Similar content being viewed by others
Data availability
The source data for all plots shown in Figs. 2–5 have been deposited in the Dryad repository and can be accessed at: https://doi.org/10.5061/dryad.pc866t235. Source data are provided with this paper.
Code availability
BARCODE and supporting documentation are available on the GitHub repository58 at https://github.com/softmatterdb/barcode and can be accessed at: https://doi.org/10.5281/zenodo.17585069.
References
Ziepke, A., Maryshev, I., Aranson, I. S. & Frey, E. Multi-scale organization in communicating active matter. Nat. Commun. 13, 6727 (2022).
Shaebani, M. R., Wysocki, A., Winkler, R. G., Gompper, G. & Rieger, H. Computational models for active matter. Nat. Rev. Phys. 2, 181–199 (2020).
Ghanbari, F., Sgarrella, J. & Peco, C. Emergent dynamics in slime mold networks. J. Mech. Phys. Solids 179, 105387 (2023).
Hallatschek, O. et al. Proliferating active matter. Nat. Rev. Phys. 5, 407–419 (2023).
Banerjee, S., Gardel, M. L. & Schwarz, U. S. The actin cytoskeleton as an active adaptive material. Annu. Rev. Condens. Matter Phys. 11, 421–439 (2020).
Hueschen, C. L. et al. Emergent actin flows explain distinct modes of gliding motility. Nat. Phys. 20, 1989–1996 (2024).
Maroudas-Sacks, Y., Garion, L., Suganthan, S., Popović, M. & Keren, K. Confinement modulates axial patterning in regenerating hydra. PRX Life 2, 043007 (2024).
Fletcher, D. A. & Geissler, P. L. Active biological materials. Annu. Rev. Phys. Chem. 60, 469–486 (2009).
Needleman, D. & Dogic, Z. Active matter at the interface between materials science and cell biology. Nat. Rev. Mater. 2, 17048 (2017).
Alvarado, J., Sheinman, M., Sharma, A., MacKintosh, F. C. & Koenderink, G. H. Molecular motors robustly drive active gels to a critically connected state. Nat. Phys. 9, 591–597 (2013).
Linsmeier, I. et al. Disordered actomyosin networks are sufficient to produce cooperative and telescopic contractility. Nat. Commun. 7, 12615 (2016).
Lee, G. et al. Myosin-driven actin-microtubule networks exhibit self-organized contractile dynamics. Sci. Adv. 7, eabe4334 (2021).
Nguyen, D. T. & Saleh, O. A. Tuning phase and aging of DNA hydrogels through molecular design. Soft Matter 13, 5421–5427 (2017).
Bate, T. E. et al. Self-mixing in microtubule-kinesin active fluid from nonuniform to uniform distribution of activity. Nat. Commun. 13, 6573 (2022).
Zhang, M., Pal, A., Lyu, X., Wu, Y. & Sitti, M. Artificial-goosebump-driven microactuation. Nat. Mater. 23, 560–569 (2024).
Daly, M. L. et al. Designer peptide–DNA cytoskeletons regulate the function of synthetic cells. Nat. Chem. 16, 1229–1239 (2024).
Schwartz, M. & Lagerwall, J. P. F. Embedding intelligence in materials for responsive built environment: a topical review on liquid crystal elastomer actuators and sensors. Build. Environ. 226, 109714 (2022).
Wang, J., Zhang, M., Han, S., Zhu, L. & Jia, X. Multiple-stimuli-responsive multicolor luminescent self-healing hydrogel and application in information encryption and bioinspired camouflage. J. Mater. Chem. C. 10, 15565–15572 (2022).
Hua, M. et al. Strong tough hydrogels via the synergy of freeze-casting and salting out. Nature 590, 594–599 (2021).
McGorty, R. J. et al. Kinesin and myosin motors compete to drive rich multiphase dynamics in programmable cytoskeletal composites. PNAS Nexus 2, pgad245 (2023).
Ross, T. D. et al. Controlling organization and forces in active matter through optically defined boundaries. Nature 572, 224–229 (2019).
Schuppler, M., Keber, F. C., Kröger, M. & Bausch, A. R. Boundaries steer the contraction of active gels. Nat. Commun. 7, 13120 (2016).
Sheung, J. Y. et al. Motor-driven restructuring of cytoskeleton composites leads to tunable time-varying elasticity. ACS Macro Lett. 10, 1151–1158 (2021).
Foster, P. J., Fürthauer, S., Shelley, M. J. & Needleman, D. J. Active contraction of microtubule networks. eLife 4, e10837 (2015).
Lee, G. et al. Active cytoskeletal composites display emergent tunable contractility and restructuring. Soft Matter 17, 10765–10776 (2021).
Chaudhuri, O., Parekh, S. H. & Fletcher, D. A. Reversible stress softening of actin networks. Nature 445, 295–298 (2007).
Jorgenson, T. D. et al. Highly flexible PEG-LifeAct constructs act as tunable biomimetic actin crosslinkers. Soft Matter 20, 971–977 (2024).
Lemma, L. M. et al. Multiscale microtubule dynamics in active nematics. Phys. Rev. Lett. 127, 148001 (2021).
Henkin, G., DeCamp, S. J., Chen, D. T. N., Sanchez, T. & Dogic, Z. Tunable dynamics of microtubule-based active isotropic gels. Philos. Trans. R. Soc. A Math., Phys. Eng. Sci. 372, 20140142 (2014).
Berezney, J., Goode, B. L., Fraden, S. & Dogic, Z. Extensile to contractile transition in active microtubule–actin composites generates layered asters with programmable lifetimes. Proc. Natl. Acad. Sci. 119, e2115895119 (2022).
López Barreiro, D. et al. Sequence control of the self-assembly of elastin-like polypeptides into hydrogels with bespoke viscoelastic and structural properties. Biomacromolecules 24, 489–501 (2023).
Conboy, J. P., Istúriz Petitjean, I., van der Net, A. & Koenderink, G. H. How cytoskeletal crosstalk makes cells move: bridging cell-free and cell studies. Biophys. Rev. 5, 021307 (2024).
Tan, T. H. et al. Self-organized stress patterns drive state transitions in actin cortices. Sci. Adv. 4, eaar2847 (2018).
Luo, Y. et al. Molecular-scale substrate anisotropy, crowding and division drive collective behaviours in cell monolayers. J. R. Soc. Interface 20, 20230160 (2023).
Zhang, G. & Yeomans, J. M. Active forces in confluent cell monolayers. Phys. Rev. Lett. 130, 038202 (2023).
Ladoux, B. & Mège, R.-M. Mechanobiology of collective cell behaviours. Nat. Rev. Mol. Cell Biol. 18, 743–757 (2017).
Malinverno, C. et al. Endocytic reawakening of motility in jammed epithelia. Nat. Mater. 16, 587–596 (2017).
Palamidessi, A. et al. Unjamming overcomes kinetic and proliferation arrest in terminally differentiated cells and promotes collective motility of carcinoma. Nat. Mater. 18, 1252–1263 (2019).
Angelini, T. E., Hannezo, E., Trepat, X., Fredberg, J. J. & Weitz, D. A. Cell migration driven by cooperative substrate deformation patterns. Phys. Rev. Lett. 104, 168104 (2010).
Farnebäck, G. Two-Frame Motion Estimation Based on Polynomial Expansion. In: Image Analysis (eds Bigun, J., Gustavsson, T.). Lecture Notes in Computer Science, vol. 2749, p. 363–370 (Springer, Berlin, Heidelberg, 2003).
Pandey, S. & Bharti, J. Review of Different Binarization Techniques Used in Different Areas of Image Analysis. In: Evolution in Signal Processing and Telecommunication Networks. (eds Chowdary, P.S.R., Anguera, J., Satapathy, S.C., Bhateja, V.) Lecture Notes in Electrical Engineering, vol. 839, p. 249–268 (Springer, Singapore, 2022).
Robertson-Anderson, R. M. Biopolymer Networks: Design, dynamics and discovery. (Institute of Physics Publishing, 2024).
Tran, P. N. et al. Deep-learning optical flow for measuring velocity fields from experimental data. Soft Matter 20, 7246–7257 (2024).
Gurmessa, B. et al. Counterion crossbridges enable robust multiscale elasticity in actin networks. Phys. Rev. Res. 1, 013016 (2019).
Krauss, P., Metzner, C., Lange, J., Lang, N. & Fabry, B. Parameter-Free Binarization and Skeletonization of Fiber Networks from Confocal Image Stacks. PLOS ONE 7, e36575 (2012).
LaTorre, A., Alonso-Nanclares, L., Muelas, S., Peña, J.-M. & DeFelipe, J. 3D segmentations of neuronal nuclei from confocal microscope image stacks. Front. Neuroanat. 7, 49 (2013).
Delpiano, J. et al. Performance of optical flow techniques for motion analysis of fluorescent point signals in confocal microscopy. Mach. Vis. Appl. 23, 675–689 (2012).
Living Biotic-Abiotic Materials: BARCODE Tutorial https://www.livingbam.org/barcode-tutorial (2025).
Cerbino, R. & Trappe, V. Differential Dynamic Microscopy: Probing Wave Vector Dependent Dynamics with a Microscope. Phys. Rev. Lett. 100, 188102 (2008).
Verwei, H. N. et al. Quantifying Cytoskeleton Dynamics Using Differential Dynamic Microscopy. Vol. 184 (1940-087X, 2022).
Cerbino, R., Giavazzi, F. & Helgeson, M. E. Differential dynamic microscopy for the characterization of polymer systems. J. Polym. Sci. 60, 1079–1089 (2022).
Robertson, C. & George, S. Theory and practical recommendations for autocorrelation-based image correlation spectroscopy. J. Biomed. Opt. 17, 080801 (2012).
Crocker, J. C. & Grier, D. G. Methods of Digital Video Microscopy for Colloidal Studies. J. Colloid Interface Sci. 179, 298–310 (1996).
Valentine, M. T. et al. Investigating the microenvironments of inhomogeneous soft materials with multiple particle tracking. Phys. Rev. E 64, 061506 (2001).
Nédélec, F. J., Surrey, T., Maggs, A. C. & Leibler, S. Self-organization of microtubules and motors. Nature 389, 305–308 (1997).
Young, E. C., Berliner, E., Mahtani, H. K., Perez-Ramirez, B. & Gelles, J. Subunit interactions in dimeric kinesin heavy chain derivatives that lack the Kinesin Rod. J. Biol. Chem. 270, 3926–3931 (1995).
Ricketts, S. N. et al. Varying crosslinking motifs drive the mesoscale mechanics of actin-microtubule composites. Sci. Rep. 9, 12831 (2019).
Chen, Q. et al. BARCODE: high throughput screening and analysis of soft active materials, Zenodo, https://doi.org/10.5281/zenodo.17585069 (2025).
Acknowledgements
BARCODE was developed through Hackathon events held at University of California, Santa Barbara and supported by the US National Science Foundation (NSF) Designing Materials to Revolutionize and Engineer our Future (DMREF) program with contributions from the following participants: Jonathan Michel, Mengyang Gu, Prashali Chauhan, Laura Morocho, Nimisha Krishnan, Anindya Chowdhury, Lauren Melcher, JJ Siu, Gregor Leech, Mehrzad Sasanpour, and Karthik Peddireddy. We acknowledge funding from the US National Science Foundation DMREF program through following grants: NSF DMR-2119663 (to RMRA), NSF DMR-2118403 (to JLR), NSF DMR-2118449 (to MD), NSF DMR-2118497 (to MTV), Research Corporation for Science Advancement award no. CS-PBP-2023-019 (to RMRA, MH), Arnold and Mabel Beckman Foundation Beckman Scholars Program (to RMRA, KM) and the NSF BioPACIFIC Materials Innovation Platform NSF DMR-1933487 for personnel support (QC) and access to research infrastructure. We thank Christopher Dunham, BioPACIFIC MIP, Emmie Kao and Christopher Tao for helpful discussions and preliminary research development, and Eric Feng for assistance in updating the graphical user interface. We thank Jose Alvarado, Gijsje Koenderink, and Yimin Luo for providing data10,34 and for helpful discussions. We thank Roberto Cerbino and Jasmin Di Franco for providing unpublished data and for helpful discussions.
Author information
Authors and Affiliations
Contributions
M.T.V. and R.M.R.A. conceived and designed the research. Q.C., A.S., A.D., K.M., M.H., K.T., R.J.M., R.M.R.A. and M.T.V. designed BARCODE algorithms, pipeline, and documentation. K.M. and M.H. performed experimental work and curated data. Q.C., A.S., A.D., R.M.R.A. and M.T.V. analyzed data. Q.C., A.S., R.M.R.A., and M.T.V. prepared figures and wrote the manuscript. M.D., R.J.M., R.M.R.A., and M.T.V. supervised the research. All authors interpreted data and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Anne Bernheim-Groswasser, Kelly Schultz and the other anonymous reviewer(s) for their contribution to the peer review of this work. [A peer review file is available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chen, Q., Sriram, A., Das, A. et al. BARCODE: high throughput screening and analysis of soft active materials. Nat Commun (2025). https://doi.org/10.1038/s41467-025-67963-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-67963-3