Abstract
Embryo development undergoes critical morphological transformations post-implantation, largely driven by the complex and dynamic microenvironment of the uterus. Despite advances, current 3D culture models inadequately recapitulate the uterine environment necessary for studying embryo-uterus interactions. In this work, we engineer a hydrogel inspired by the properties of the decidua, incorporating Matrigel to support blastocyst implantation and embryo development in vitro. Our findings reveal that embryos cultured within this hydrogel system successfully progress to an early organogenesis-like stage, including the development of first and second heart fields, mimicking natural embryogenesis. Moreover, we identify that the mechanical properties, particularly stress relaxation, play a crucial role in facilitating focal adhesion (FA) formation between the trophoblast and the hydrogel. Additionally, the degradation of the hydrogel by embryo-secreted metalloproteinases (MMP2 and MMP9) creates a favorable environment for continued embryonic growth and development. These insights contribute to a deeper understanding of how the external environment regulates embryo development and offer an enhanced approach for in vitro embryo culture.
Similar content being viewed by others
Introduction
Embryonic development is a complex and highly regulated process1,2,3,4. However, studying the in vivo development of mammalian embryos post-implantation remains a significant challenge, due to their small size and the limitations of current observation techniques. The transformation from a blastocyst invading the uterine stroma to a complete organism involves numerous intricate morphogenetic and signaling events that are still poorly understood. Therefore, innovative techniques and approaches are essential to uncover the mechanisms governing post-implantation embryonic development5.
In vivo, embryonic development is closely orchestrated by the dynamic physical conditions within the uterus, coupled with chemical signals. Embryo implantation triggers rapid and extensive changes in the endometrium and the ECM6, while the trophoblast cells interact closely with the uterine ECM7. Throughout development, the environment surrounding the embryo transitions from a 2D to a 3D state, and from an ECM-free to an ECM-encapsulated state. Trophoblastic cells engage in highly integrated physicochemical interactions with the uterine ECM and endometrial cells7. This indicates significant correlations between the developmental stage of the blastocyst and the remodeling of its physical microenvironment, as well as interactions between blastocysts and the uterus that involve factors such as viscoelasticity (e.g., stress relaxation, creep), modulus (e.g., Young’s modulus), the ECM fibrous network (e.g., pore size, fiber diameter), cell adhesion (e.g., focal adhesions, integrins), and cellular tension. However, detailed studies exploring the specific effects of the viscoelastic microenvironment and ECM fibrous network on embryonic development and embryo-uterine interactions remain insufficient.
Mechanical forces are critical during the various stages of embryo morphogenesis and organ formation8,9,10,11, including embryo hatching from the zona pellucida12, gastrulation13, and organ size determination10. Despite ample evidence demonstrating the profound influence of mechanical forces on cell behavior, the precise mechanisms by which these forces impact embryonic tissue and organ development remain elusive. Further, a deep understanding of the mechanics between the early uterine decidua and embryonic structure is also lacking.
The challenges associated with replicating the intricate processes of implantation and post-implantation development have long been recognized, dating back to the early proposals of embryonic in vitro culture techniques in 196314,15. Despite the progress made in this field16,17,18,19,20, current in vitro embryo culture systems continue to face significant hurdles, including abnormal trophectoderm development and developmental blocks during implantation. These limitations underscore the need for a model that can better replicate the complex interactions between the embryo and its physical environment within the uterus. Our findings, along with recent studies, have reinforced the idea that both biochemical and physical factors within the microenvironment play pivotal roles in embryonic development21,22. Specifically, factors such as tissue stiffness, viscoelasticity, and cellular tension have emerged as crucial determinants of cell fate, embryonic patterning, and tissue morphogenesis23,24,25,26,27. However, the specific role of these physical microenvironmental factors in modulating embryo-uterus interactions has not been fully explored.
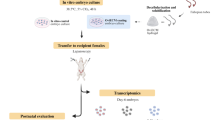
The impact of the viscoelastic microenvironment on embryonic development and its underlying mechanisms remains largely unexplored in current studies of blastocyst in vitro culture. Therefore, developing an in vitro embryo culture method conducive to blastocyst implantation and subsequent development is crucial. To address this, we establish a three-dimensional (3D) embryo culture system (Fig. 1A) inspired by the characteristics of the decidual ECM. Within this model, embryos progress to the early organogenesis stage in vitro. Using this innovative culture system, we successfully conduct real-time monitoring and observation of embryo development. We then use the model to examine how stress relaxation affects embryo development and how ECM spatial structure influences embryo growth. Consequently, this 3D system introduces a approach for examining in vitro embryo-ECM interactions, with potential applications in diverse realms, including reproductive biology and regenerative medicine.
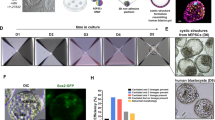
A Schematics of in vivo and in vitro environments. Upper: positions of embryonic and decidual tissues. Lower: in vitro 3D culture system showing embryo and material positions, with the culture protocol on the right. B SEM images of Matrigel at different concentrations (n = 3) and decellularized E5.5 decidua (n = 8). Scale bars, 1 μm. C Pore sizes of decellularized E5.5 decidua and Matrigel at different concentrations were quantified based on SEM images. Data were plotted as mean ± s.d. n = 3. P-values were calculated using one-way ANOVA test. ns, not significant; ****P < 0.0001. D Representative bright-field images of in-vitro-cultured (IVC) mouse embryos from days 0–9. Scale bars, 100 μm. Day, IVC day; EXE, extraembryonic ectoderm; Epi, epiblast; PS, primitive streak; VE, visceral endoderm; Am, amnion; PAC, proamniotic cavity; AC, amniotic cavity; EPC, ectoplacental cone; EB, embryonic body; TB, tail bud; YS, yolk sac; H, heart; YSV, yolk sac vessel. n = 3. E Percentages of embryos in 40-MG with two cavities (IVC day 3), an egg cylinder (IVC day 4), a yolk sac (IVC day 6), and a heartbeat-like structure (IVC days 8–10). Data were plotted as mean ± s.d. n = 7. F IVC day 8 and natural E8.5 embryos stained for FOXA2 (green) and OTX2 (red). Scale bars, 100 μm. n = 3. G IVC day 9 embryos in 40-MG stained for cardiomyocyte markers MHC (green) and NKX2.5 (red), compared with E8.5 embryos. Scale bars, 100 μm. n = 3. H IVC day 9 embryos in 40-MG stained for cardiomyocyte markers MHC (green), GATA4 (gray), and MLC (red), compared with E8.5 embryos. Dotted boxes indicate magnified regions. Scale bars, 200 μm. A-P axis formation, dorsal (D), ventral (V), Distal (Di), and proximal (Pr) orientations in the mouse embryo are labeled. n = 3.
Results
Constructing a decidual-ECM-mimicking 3D system for in vitro culture of mouse embryos
Mouse embryo implantation occurs in vivo at E4.5, triggering decidualization of the uterus and rapid proliferation of the decidua tissue, which forms a 3D growth environment encapsulating the embryo. Consequently, an in vitro culture environment that mimics this 3D metamorphosis may be more conducive to optimal embryo development. Therefore, there is an urgent need to establish a 3D environment that serves as a substitute for in vivo uterine tissue. To better simulate the decidua-embryo interaction compared to existing methods, we constructed a 3D environment designed to match the characteristics of the decidua. We selected Matrigel as it is widely employed in embryo culture and, importantly, as its mechanical properties are comparable to those of the decidua, making it a suitable biomaterial for mimicking the in vivo environment28,29,30. We prepared Matrigel at concentrations of 20% to 50% (termed 20-MG, 30-MG, 40-MG, and 50-MG, respectively) and measured their moduli. The moduli of 20 to 50-MG (83.4 Pa, 85.5 Pa, 89.1 Pa, 104.6 Pa, respectively) were comparable to those of E5.5 to E8.5 decidua (121.6 Pa, 133.2 Pa, 138.7 Pa, 138 Pa, respectively) (Fig. S1A and B). Furthermore, the pore area of 40-MG was most similar to that of the E5.5 decidua (Fig. 1B, C, and Supplementary Movies 1–5). Thus, 40-MG was selected as the material for constructing the 3D environment.
The 3D culture system production process is described in Fig. S1C. The developmental rate of five E3.5 embryos cultured in a 5 μL volume was higher than that of other groups (Fig. S2A). Additionally, embryos developed more readily in 40-MG than in 20-MG, 30-MG, and 50-MG, consistent with our modulus data (Fig. S2B). Finally, we found that the developmental efficiency of embryos with two cavities and an egg cylinder was greater in the 3D environment than in the 2D environment (Fig. S2C). These results suggest that the 3D environment is more suitable for in vitro embryo culture and better supports the study of ECM-embryo interactions.
To evaluate the impact of the 3D environment on embryo development, we quantified various developmental stages. On IVC day 1, embryos maintained a blastocyst-like structure with a developmental rate of 92.9 ± 0.8% (Fig. S3A and C). By IVC day 2, the embryo cells had aggregated, resulting in a developmental rate of 92.3 ± 0.7% (Fig. S3B and C). A cavity, either epiblast (EPI) or extra-embryonic ectoderm (ExE), appeared on IVC day 3, with developmental rate of 69.7 ± 17.7% (Figs. 1D, E, S3D and E). The EPI and ExE cavities then fused to form an egg cylinder by IVC day 4, with a developmental rate of 59.8 ± 18.9% (Fig. 1D and E). The yolk sac began to grow on IVC days 5–6, with a developmental rate of 28.2 ± 2.3% (Fig. 1D and E). Early neural structures of the embryo appeared on IVC day 7. By IVC day 8, the head and tail of the embryo were clearly observed, and the early heart-like structures were visible, with a developmental rate of 23.6 ± 0.6% (Fig. 1D, E, and Supplementary Movie 6).
To characterize the molecular properties of 3D IVC cultured embryos, we analyzed the expression of critical developmental genes in embryos from IVC day 2 to day 9, as well as natural E4.5–E8.5 embryos. On IVC day 1, we found that the embryos expressed OCT4 and CDX2 (Fig. S3A) and formed a rosette-like structure in the ectoderm region by IVC day 2, similar to natural in vivo embryos (Figs. S3B, S4A and B). By IVC day 3, the EOMES+ segment of the ExE cavity and the SOX2+/OCT4+ segment of the EPI cavity were observed, aligning with natural embryos (Fig. S4C, D, and E). On IVC day 4, the amniotic cavity was formed by the fusion of the ExE and EPI cavities, again reflecting the development seen in natural embryos (Fig. S4F and G). Additionally, T+ cells migrated anterolaterally during development, indicating the onset of gastrulation (Fig. S4H, K and L).
On IVC day 5, a few FOXA2+ cells began to appear in the embryos, showing a developmental stage comparable to that of natural embryos (Fig. S4I). By IVC day 7, the number of FOXA2+ cells had increased and their expression patterns matched those seen in E7.5 embryos (Fig. S4J). Early organogenesis initiated on IVC day 8, with the expression of the early brain and nerve development markers, OTX2 and PAX6, corresponding to the neural structures in E8.5 embryos (Fig. S5A). Notably, OTX2+ and FOXA2+ cells, which are markers for early gut, brain, and notochord development, were observed in both natural and in vitro embryos (Figs. 1F, S5B, and C). To further confirm this, we detected the expression of MHC, NKX2.5, MLC, and GATA4 in the early heart-like structures. Additionally, the formation and morphology of the early heart field were similar to those of E8.5 embryos (Fig. 1G and H). These findings validate the successful establishment of a 3D culture system with decidual ECM characteristics.
Application of the decidual-ECM-mimicking 3D system
To validate the effectiveness of our embryo culture platform, we first assessed whether the heart-like structures of IVC day 8 embryos exhibited early functional heart activity. Tissue staining with a calcium (Ca2+)-binding fluorescent dye revealed transient Ca2+ contractions in the heart-like structure of IVC day 9 embryos (Figs. 2A , B, C, S6A, and Supplementary Movie 7). Additionally, administration of isoprenaline increased the beat rate of these heart-like structures (Figs. 2A, B, C, S6B, and Supplementary Movie 8), similar to the response observed in natural E8.5 embryos upon cardiac β-adrenaline stimulation. For live imaging, embryos were cultured and photographed at two-hour intervals over a period of nine days. On IVC day 0.5, the embryo size exhibited fluctuations, initially decreasing before expanding. By IVC day 1, the embryos began penetrating the zona pellucida. By IVC day 1.5, they displayed mobility within the hydrogel. On IVC day 3, distinct cavities corresponding to EPI and EXE formed. On IVC day 4, the egg cylinder was observed. On IVC day 5, the yolk sac started to expand. On IVC day 6, the embryonic body structure became visible. This developmental progression culminated on IVC day 9 with the first detection of the early heartbeat (Fig. 2D and Supplementary Movie 9). These results suggest that the culture system with the decidua ECM properties is capable of supporting in vitro embryo development and can serve as a platform for early embryonic developmental tracing.
A Representative images showing Ca2+ dynamics in IVC day 9 embryos, before and after treatment with the β-adrenergic agonist isoproterenol (1 μM). The Ca2+-binding dye Fluo8-AM was used to visualize calcium fluxes, highlighting changes in intracellular calcium concentrations. Scale bars, 50 μm. n = 8 independent experiments. B Graphical representation of Ca2+ signal changes (F/F0) in IVC day 9 embryos, captured before and after isoproterenol treatment. The traces depict a clear enhancement in calcium transients post-treatment, indicative of increased cardiac activity. C Heart rate (HR) response to isoproterenol: Quantification of the beating rate of embryos, illustrating a significant increase in HR (beats per minute, BPM) following isoproterenol treatment. Data were plotted as mean ± s.d., n = 8. P-values were calculated using unpaired, two-sided Student’s t-test, ****P < 0.0001. D Time-lapse light-field imaging of E3.5 embryo development: Sequential light-field images showing the development of an E3.5 embryo over a nine-day IVC period. Images were acquired every two hours during culture, capturing the progression of embryonic development from the blastocyst stage to more advanced stages. See also Supplementary Movie 9 for the complete time-lapse sequence. Scale bars, 500 μm. n = 3.
Transcriptomic comparison of IVC and natural embryos
We performed single-cell RNA sequencing (scRNA-seq) of three IVC day 9 embryos, three natural E6.5 embryos and three natural E8.5 embryos to investigate how the 3D culture system, designed with decidua ECM properties, affects gene expression during embryo development. After stringent quality control, 14,315 E6.5 cells, 14,761 E8.5 cells, and 10,756 cells from IVC day 9 embryos were analyzed (Fig. S7A and B). UMAP analyses revealed a similar contribution of cells to the developing lineages in both the E8.5 and IVC day 9 embryos (Figs. 3A and S7C). Using Seurat and published datasets, we identified 19 distinct cell types based on gene expression patterns, which were clearly represented in both E8.5 and IVC day 9 embryo datasets (Fig. 3B).
A Uniform manifold approximation and projection (UMAP) analysis of scRNA-seq data at the indicated timepoints for E6.5 and E8.5 embryos and IVC day 9 (D9) embryos. B UMAP plot showing the cell atlas. Cells are colored according to their cell-type (see legend below). E6.5 (14,315 cells), E8.5 (14,761 cells), D9 (10,756 cells). A comparison of cell-type proportions between E8.5 embryos and IVC day 9 embryos. D9: IVC day 9. C Heatmap showing the global level of similarity across all identified tissues between D9 and E8.5 embryos. D Cell type proportions for E6.5 and E8.5 embryos and IVC day 9 embryos. E Violin plots depicting genes that were upregulated and downregulated across 19 fractions in D9 embryos relative to E8.5 embryos. F UMAP analysis of scRNA-seq data for the heart regions of E8.5 and D9 embryos. G Localization of first heart field (FHF) and second heart field (SHF) on the UMAP in (A) based on expression of TBX5, HCN4, NKX2.5, HAND1 for FHF and TBX1, SIX1, SIX2 for SHF. H Bubble plot showing a curated list of marker genes that were used to identify the first heart field and second heart field. I UMAP plot of reclustered multipotent progenitor populations, colored by cluster. AHF, anterior heart field; pSHF, posterior second heart field; BMP, branchiomeric muscle progenitors. J Bubble plot showing a curated list of marker genes that were used to identify the posterior second heart field, anterior heart field, and branchiomeric muscle progenitors.
A Pearson correlation matrix indicated a high level of congruence in gene expression between the cell-type clusters of the E8.5 and IVC day 9 embryos (Fig. 3C). The comparison of cell-type proportions between E8.5 embryos and IVC day 9 embryos showed close similarities between individual clusters, with some variability (Fig. 3D). In IVC day 9 embryos, GPX3, GSTM1, and CD63 were upregulated across all 19 gene clusters compared to E8.5 embryos, while HSPH1, PPA1, and TCOF1 were downregulated (Figs. 3E and S8). PPA1, GSTM1, GPX3, and HSPH1 play key roles in intracellular metabolism and in mitigating external stress, thereby maintaining intracellular stability. This suggests that embryos cultured in vitro primarily rely on nutrients supplied by the culture medium, and metabolism may become dysregulated in IVC day 9–10. The reduced expression of TCOF1 points to potential developmental abnormalities in the head and facial structures during the IVC day 9–10 stage31. Extracellular vesicles (EVs) serve as significant microenvironmental regulators, with CD63 being recognized as an EV marker. Higher levels of released EVs may reflect poorer embryo quality assessment32.
On IVC day 9, the embryos displayed expression profiles similar to E8.5 embryos, including the expression of hematopoietic-related genes and the presence of blood cells in the yolk sac (Fig. S9). Additionally, genes related to neural and early gut development were observed (Fig. S9). We also identified the expression of key genes associated with cardiac development, indicating the emergence of the first heart field (FHF) and second heart field (SHF), progenitor cells (Fig. 3F–J). Additionally, scRNA-seq analysis confirmed that endothelial cells exhibit similar expression profiles both in vivo and in vitro (Fig. S9).
Stress relaxation regulates embryonic development in vitro
The stiffness and viscoelasticity of the ECM significantly influence cellular behaviors such as spreading, growth, proliferation, migration, differentiation, and organoid formation33,34. We observed that the stress relaxation properties of 40-MG were similar to that of E5.5 decidua (Fig. 4A). To investigate whether viscoelasticity regulates embryonic development, we modified our 3D culture system by incorporating two cross-linking agents commonly used in bioengineering: glutaminase and PEG-NHS, to further covalently cross-link the Matrigel35,36,37,38,39. After optimizing the concentrations of glutaminase and PEG-NHS in ES and TS cells and drawing on previous reports38,40,41, we determined that 50 U/g glutaminase and 1 mg/mL PEG-NHS provided the optimal environment for cross-linking Matrigel (Fig. S10A–D). We measured the hydrogel modulus and found no significant differences in the resulting glutaminase cross-linked (40-MGT) and the PEG-NHS cross-linked (40-MGN) hydrogels (Figs. 4B, C, S10E and F). The pore sizes on the surface of 40-MG, 40-MGT, and 40-MGN remained largely unchanged (Fig. S10G, H, Supplementary Movies 10 and 11). However, stress relaxation rates e decreased (Figs. 4D, S10I, and J).
A Characteristic relaxation timescale (τ1/2) quantifying the time required for stress to relax to half of its original value in the E5.5 De and 40-MG. Data were plotted as mean ± s.d. n = 10, 9 independent experiments E5.5 De and 40-MG, respectively. P-values were calculated using unpaired, two-sided Student’s t-test. B Design and characterization of 40-MG, 40-MGT, and 40-MGN hydrogels. Schematics illustrate 40-MG formed by physical crosslinking, 40-MGT by TG-mediated covalent bonding, and 40-MGN by PEG-NHS covalent bonding. C Quantification of the Young’s modulus (E) of 40-MG, 40-MGT, and 40-MGN, as detected using a nanoindenter. Data were plotted as mean ± s.d. n = 10. P-values were calculated using one-way ANOVA test. ns, not significant. D Characteristic relaxation timescale (τ₁/₂) of 40-MG, 40-MGT, and 40-MGN, measured by nanoindentation. Data were plotted as mean ± s.d. n = 5. P-values were calculated using one-way ANOVA test. **P < 0.01, ***P < 0.001. E, F Percentages of embryos with two cavities (IVC day 3) and an egg cylinder (IVC day 4) in 40-MG, 40-MGT, and 40-MGN. Data were plotted as mean ± s.d. n ≥ 4. P-values were calculated using one-way ANOVA test. ****P < 0.0001. G–I IVC day 2–4 embryos in 40-MG, 40-MGT, and 40-MGN labeled for OCT4 (gray), CDX2 (green), and Phall (red). Scale bars, 50 μm. n = 3. J Schematic showing how altering polymer molecular weight changes stress relaxation: LA + 40-MG (1% low MW alginate + 40-MG) and HA + 40-MG (1% high MW alginate + 40-MG). K, L Percentages of embryos with two cavities (IVC day 3) and an egg cylinder (IVC day 4) in LA + 40-MG and HA + 40-MG. Data were plotted as mean ± s.d. n ≥ 4 P-values were calculated using one-way ANOVA test. ns, not significant, *P < 0.05, ****P < 0.0001. M, N Percentages of embryos with two cavities (IVC day 3) and an egg cylinder (IVC day 4) in 20-MG and 20-MGT. Data were plotted as mean ± s.d. n = 3. P-values were calculated using unpaired, two-sided Student’s t-test. **P < 0.01, ****P < 0.0001.
We found that embryo development was markedly inhibited in the 40-MGT, with a developmental rate of 31.5 ± 13.0% on IVC day 3 and 27.7 ± 13.0% on IVC day 4. In the 40-MGN, the rates were slightly higher at 37.4 ± 14.1% and 34.7 ± 13.3% (Fig. 4E and F). Additionally, embryos cultured in 40-MGT and 40-MGN did not form rosette-like structures with OCT4+ cells by IVC day 2, and the number of OCT4+ cells remained low (Figs. 4G, S11A and B). By IVC day 3, only a limited number of OCT4+ cells formed a cavity (Fig. 4H), and by IVC day 4, CDX2+ cells were predominantly observed (Fig. 4I). Bright field images also revealed abnormal embryo morphologies (Fig. S11C). Atomic force microscopy (AFM) measurements showed that the modulus difference of the E5.5–E7.5 embryos decidua was small (Fig. S1A). Additionally, we observed that the stress relaxation of the E5.5–E8.5 embryo decidua gradually slowed down (Fig. S11D and E). Our investigation revealed a gradual deceleration in stress relaxation within the 3D culture system over time (Fig. S11F); notably, 40-MG itself underwent these changes more rapidly (Fig. S11F). These findings suggested that stress relaxation played a key role in embryo development during in vitro culture.
To further validate the impact of stress relaxation on embryo development, we utilized the widely used alginate plus Matrigel system (Figs. 4J and S12A). We combined both high and low molecular weight alginates with 40% Matrigel, resulting in comparable moduli (Fig. S12B and C). Notably, the low molecular weight alginate mixed with 40% Matrigel exhibited faster stress relaxation (Fig. S12D and E) and superior developmental efficiency (Figs. 4K, L and S12F). Furthermore, the addition of glutaminase to 20-MG maintained an unaltered modulus while showing slower stress relaxation (Fig. S12G and H), which corresponded with an increased embryo development rate (Fig. 4M and N). These findings further confirm that stress relaxation plays a critical role in embryo development.
Enhancement of integrin and FA aggregation in TE cells facilitates embryo-hydrogel interactions
Integrins are key adhesion molecules present at the embryo-uterine interface during implantation, playing a critical role in the adhesion and migration of trophoblast cells42,43. The interaction between the ECM and integrins is crucial for mechanosensing during early embryo development19,44. Stress relaxation significantly affects the cell morphology of the implanted embryos. Specifically, under 40-MG conditions, we observed more pseudopods compared to 40-MGT or 40-MGN conditions (Fig. 5A), indicating the development of a more invasive trophectoderm (TE)19. Additionally, integrin subunit expression in embryos cultured in 40-MG was higher than in those cultured in 40-MGT or 40-MGN (Fig. S13A), suggesting that elevated integrin expression is a significant factor in promoting embryo implantation in vitro. The aggregation of integrins subsequently led to vinculin recruitment and the formation of stable FAs in embryos cultured in 40-MG (Fig. 5B, C, and D). The 3D environment modulates cell contraction, thereby regulating FA formation24. Consequently, enhanced integrin expression and FA formation under 40-MG conditions promote better embryo implantation and development, highlighting the critical role of stress relaxation in the 3D system.
A IVC day 2 embryos in 40-MG and 40-MGT labeled with Phalloidin (red, cytoskeleton) and Hoechst (blue, nuclei). TE cells in 40-MG embryos show filopodia, while those in 40-MGT appear smoother. Scale bars, 50 μm. n = 3. B IVC day 2 embryos in 40-MG, 40-MGT, and 40-MGN labeled for Vinculin (green, FAs) and TFAP2C (red, TE). TE cells in 40-MG embryos show more FAs. Dotted boxes indicate magnified regions. Scale bars, 50 μm. n = 3. C, D Quantification of FA size (C) and number (D) via vinculin staining on IVC day 2, showing larger and more FAs in 40-MG embryos than in 40-MGT and 40-MGN. FA number per TE cell determined using TFAP2C localization. Data were plotted as mean ± s.d. n = 3. P-values were calculated using one-way ANOVA test. ****P < 0.0001. E IVC day 2 embryos in 40-MG, 40-MGT, and 40-MGN labeled for pMLC (green). pMLC is cytoplasmic in 40-MG and nuclear in 40-MGT and 40-MGN. Scale bars, 50 μm. n = 3. F Percentage of trophoblast cells with nuclear pMLC in embryos cultured in 40-MG, 40-MGT, and 40-MGN. Data were plotted as mean ± s.d. n = 3. P-values were calculated using one-way ANOVA test. ****P < 0.0001. G Dot plot of pathway enrichment between 40-MG and 40-MGT. Vertical axis: pathways; horizontal axis: Gene Ratio. Dot color: p-value (red smaller); dot size: gene count. Up/down arrows indicate higher/lower expression in 40-MGT versus 40-MG. H Schematic showing RGD concentrations in HA + 40-MG achieved by adjusting RGD levels and mixing with different components. (I-J) Percentages of embryos with two cavities (IVC day 3) and egg cylinder (IVC day 4) in HA-40-MG, HA/HARGD-40-MG, and HARGD-40-MG. Data were plotted as mean ± s.d. n ≥ 4. P-values were calculated using one-way ANOVA test. *P < 0.05, **P < 0.01. K Diagram summarizing the study: On IVC day 2, rapid stress relaxation (40-MG, LA-40-MG) promoted FA aggregation, while slow relaxation (40-MGT, 40-MGN, HA-40-MG) reduced FAs and hindered embryo development; RGD in HA-40-MG alleviated these effects.
It is known that hydrogels regulate the formation of adhesive patches by modulating cell contraction25. To investigate how 40-MG promotes FA formation, we analyzed the localization of phosphorylated myosin light chain (pMLC) in TE cells. In embryos cultured in 40-MGT and 40-MGN, pMLC was predominantly localized in the nuclei, whereas in 40-MG, it was found at the periphery of embryos (Fig. 5E and F). To further investigate the interaction between TE cells and the hydrogel, we cultured trophospheres (spheroids derived from trophoblast stem cells, TS cells) in 40-MG, 40-MGT, and 40-MGN. The localization patterns of FAs and pMLC in these trophospheres were consistent with those observed in embryos (Fig. S13B–F). Inhibiting myosin activity with blebbistatin on IVC days 1–2 reduced FA formation in embryos cultured in 40-MG and increased nuclear localization of pMLC in TE cells, confirming the importance of TE cell contraction in this process (Fig. S14A and B). This inhibition also led to decreased embryonic development rates in the 40-MG environment (Fig. S14C–E).
On IVC day 2, we sequenced the hydrogel-contact regions of embryos from 40-MG, 40-MGT, and 40-MGN groups. Analysis of differentially expressed genes revealed that the 40-MG group exhibited elevated levels of cellular processes, including cell migration, cell adhesion, regulation of actin cytoskeleton reorganization, compared to 40-MGT or 40-MGN groups (Fig. 5G). Since stress relaxation influences FA formation, we explored whether altering the RGD content in the material could enhance FA formation and mitigate the adverse effects of slow stress relaxation on embryo development (Fig. 5H). To do this, we created a group with intermediate RGD content by mixing equal proportions of RGD-free and RGD containing high molecular weight sodium alginate, followed by assessment of embryo development. We found that as the RGD content increased, embryo development initially improved and then decreased, though it remained significantly higher than the group without RGD (Figs. 5I, J, and S15).
Overall, our findings indicate that rapid stress relaxation facilitates swift FA aggregation, while slow stress relaxation combined with RGD inclusion enhances embryo development (Fig. 5K). Furthermore, 40-MG promotes TE cell contraction, integrin aggregation, and FA formation, thereby establishing effective mechanotransduction between the embryo and the ECM.
The fibrous network in hydrogel regulates embryonic development in vitro
Another crucial property of ECM is its fibrous network structure. Pore size and density are pivotal in defining the microenvironment for cell growth. Extensive evidence suggests that the porous architecture of tissues can offer guidance for cell behaviors45. To investigate whether the spatial network of the ECM plays a regulatory role in embryo development, we initially assessed alterations in the spatial network of decellularized natural decidual tissues from E5.5 to E7.5 (Fig. 6A, Supplementary Movies 12 and 13). Our observations revealed that the spatial network of these tissues exhibited a compact structure at E5.5, which gradually expanded from E6.5 onwards (Fig. 6A). This expansion was characterized by an increase in pore size (Fig. 6B) and fiber diameter thickness (Fig. S16A). Similarly, SEM images of the interface between the embryo and 40-MG showed that the fibrous network around the embryo became loosened over time (Fig. 6C and D), accompanied by an increase in fiber diameter thickness (Fig. S16B).
A SEM images of decellularized E5.5, E6.5, and E7.5 decidua surfaces, with inset showing a magnified ECM region. Scale bars, 1 μm. B Pore size quantification on decellularized E5.5, E6.5, and E7.5 decidua. Data were plotted as mean ± s.d. n = 3. P-values were calculated using one-way ANOVA test. ***P < 0.001, ****P < 0.0001. C, E SEM images of ECM regions (yellow) showing the fibrous network in 40-MG and 40-MG + GM6001. Scale bars, 1 μm. D, F Pore size quantification on 40-MG and 40-MG + GM6001 surfaces from Day 0 to Day 4. Data were plotted as mean ± s.d. n = 3. P-values were calculated using one-way ANOVA test. *P < 0.05, ****P < 0.0001. G, H Percentages of embryos with two cavities (IVC day 3) and an egg cylinder (IVC day 4), embedded in 40-MG treated with DMSO or GM6001. Data were plotted as mean ± s.d. n = 3. P-values were calculated using unpaired, two-sided Student’s t-test. *P < 0.05, ***P < 0.001. (I) IVC day 2 embryos in 40-MG or 40-MG + FI 14, stained for pFAK, showing higher activation than 40-MGT. Yellow arrowheads mark focal adhesions. Scale bars, 50 μm. n = 3. J, K Percentages of embryos with two cavities on IVC day 3 and an egg cylinder on IVC day 4, embedded in 40-MG or 40-MG + FI 14. Data were plotted as mean ± s.d. n = 3. P-values were calculated using unpaired, two-sided Student’s t-test. **P < 0.01. L, M Percentages of embryos with two cavities on IVC day 3 and an egg cylinder on IVC day 4, embedded in 50-MG treated with DMSO or collagenase IV. Data were plotted as mean ± s.d. n = 3. P-values were calculated using unpaired, two-sided Student’s t-test. **P < 0.01, ****P < 0.0001. N A summary diagram illustrating how the hydrogel network promotes in vitro embryo development. MMP2 and MMP9 expression facilitates remodeling of the ECM, creating an environment conducive to embryo development.
To investigate the impact of MMP on this process, we added MMP inhibitor GM600124,46 to IVC embryos from day 0 to day 4, which slowed down the loosening process (Figs. 6E, F and S16C). Moreover, GM6001 also reduced the expression levels of MMP2 and MMP9 in the embryos cultured in 40-MG (Fig. S16D), indicating that MMP secretion by TE cells regulates ECM. The addition of GM6001 decreased the developmental rates of embryos in 40-MG on IVC days 3 and 4 (38.6 ± 9.5% and 34.2 ± 11.0%, respectively) (Fig. 6G and H), suggesting that the proper regulation of ECM spatial network is crucial for optimal embryonic development.
FAK is known to regulate the expression of MMP2 and MMP9, which are crucial for various cellular processes47. Our results showed higher levels of FAK activation in TE cells of embryos in 40-MG compared to those treated with the FAK inhibitor FI 14 (Fig. 6I). The inhibitor significantly reduced the developmental rate of the embryos on IVC days 3 and 4 to18.3 ± 12.3% and 10.8 ± 11.7%, respectively (Figs. 6J, K and S16E). Following the addition of FI 14, p-FAK was undetectable (Fig. 6I). To further validate the impact of spatial structure on embryonic development in 3D system, we added collagenase IV to 50-MG, which significantly increased developmental rate of embryos on IVC days 3 and 4 to 53.74 ± 12.0% and 50.11 ± 11.3%, respectively, compared to embryos cultured in 50-MG alone (Fig. 6L and M). This suggests that the controlled degradation of ECM components can be beneficial for in vitro embryonic development by providing additional space for growth and by facilitating cellular interactions. In conclusion, our findings indicate that in vitro embryos degrade surrounding materials through the secretion of MMP2 and MMP9, thereby creating additional growth space conducive to in vitro embryonic development (Fig. 6N).
Discussion
Our findings highlight the limitations of conventional embryo culture models in replicating the 3D physical microenvironment of the uterus, which is critical for proper embryo development. In this study, we reported a 3D embryo culture system that supported blastocyst development up to an E8.5-like stage. Gu et al. (2022) reported an egg cylinder formation efficiency of 47 ± 15% and a heartbeat-like stage formation rate of 11 ± 5% using the PDCO system27. In comparison, our 3D system achieved an egg cylinder formation efficiency of 69.7 ± 17.7% and a heartbeat-like stage formation rate of 23.6 ± 0.6%. However, we would like to emphasize that our culture system offers a simplified and more efficient model compared to static cultures. The 3D system enabled the detection of key markers such as FOXA2, T, OTX2, MHC, MLC, and GATA4 within various embryonic structures. Additionally, our data demonstrate that the dynamic properties of the 3D system, particularly its ability to modulate stress relaxation, significantly influence FA formation between the trophoblast cells and the hydrogel, underscoring the importance of physical characteristics in embryonic development.
This study establishes a platform for observing the early stages of life outside the uterus and provides a theoretical framework for extending in vitro embryo culture by elucidating the impact of the physical microenvironment on development. However, while our 3D culture system addresses some challenges of in vitro modeling, it cannot substitute for the placenta, which is crucial for supplying nutrients during later development stages. As current models, including ours, rely on nutrient infiltration, they are limited to supporting early embryonic stages. Therefore, although our findings are a significant step forward, they also highlight the need for continued research to develop methods that can sustain embryonic development through organogenesis without a placenta—a complex challenge that future research must address.
Our study contributes to this effort by demonstrating the critical role of a dynamic hydrogel network in supporting embryonic development in vitro. The 3D culture system we developed closely mimics the properties of the natural decidua, successfully replicating the early stages of in vivo embryonic development. By integrating principles from both developmental biology and tissue engineering, we have created a model that not only advances our understanding of hydrogel network dynamics but also provides a robust platform for further exploration of embryogenesis. This model serves as a foundational tool for future studies aimed at unraveling the complex processes governing embryonic development and implantation, while acknowledging that the road to fully replicating in vivo conditions remains challenging and incomplete.
Methods
Animal and ethical approval
ICR 8-week-old females and 8-week-old males mouse, SD 8-week-old females and 8-week-old males rats were purchased from SPF (Beijing) Biotechnology. All animals were maintained under specific-pathogen-free conditions at the same facility, with ad libitum access to food and water, a 12 h light/12 h dark cycle, and controlled temperature (18–23 °C) and humidity (40–60%). At E3.5, pregnant mice were humanely euthanized, and embryos were randomly collected without sex selection. The sex of mouse embryos cannot be determined prior to or during experiments, and there is no evidence that early embryo development is sex-dependent. Therefore, sex-based analysis was not applicable to this study.
Atomic force microscopy (AFM) testing
The Young’s moduli of deciduae at different developmental stages (E5.5 to E8.5) and various concentrations of Matrigel were measured using atomic force microscopy (AFM, Nanowizard4, Bruker, USA). AFM measurements were conducted with silicon nitride cantilevers (NP-O10, Nanoworld, USA) with a nominal spring constant of 0.12 N/m and a 30-μm-diameter silica bead (Thermo Fisher Scientific). Each sample was measured at five distinct points, and five force curves were collected at each point.
The applied mechanics model and Hertz model could be used for data fitting and calculation with the spherical indenters involved in the indentation, and the key factors have also been specified in eqn. Following the Hertz model describing the indentation of elastic bodies, the force-to-penetration equation is:
here, F is the applied force, R is the radius of the indenter tip, δ is the indentation depth, and E* is the reduced modulus, which can be defined by the combination of the plane strain moduli of the indenter (E1) and the substrate modulus (E2)48:
where v1 and v2 are the Poisson’s ratios of the indenter and the substrate, respectively, and the indenter is much stiffer than the substrate (E1 ≫ E2), so the reduced modulus E* is simply:
For decidua, the Poisson ratio (v2) is assumed to be 0.549 Eq. (1), Eq. (2) and Eq. (3) become:
The data in the loading section of the load-displacement curve were used to determine Young’s modulus using a fit of all data points from the contact point to the maximum load point.
Decellularization of decidua
Decidua was decellularized by shaking in solutions of SDS (1%, 0.1%, and 0.01%) and Triton X-100 (1%, 0.1%, and 0.01%), each solution for 3 h. This was followed by washing with water for 1 h. The decellularized decidua was then fixed for further use.
Scanning electron microscopy (SEM) observations
Decellularized decidua, varying concentrations of Matrigel, and embryos embedded into 40-MG were fixed in 2.5% glutaraldehyde at room temperature for 4 h. The samples were then dehydrated in a graded ethanol series (20%, 30%, 40%, 50%, 60%, 70%, 80%, 90% ethanol for 15 min each), followed by 100% ethanol for 15 min. After supercritical drying (CPD300, Leica), the samples were coated with platinum using a sputter coater (JEC-3000FC, JEOL) and observed using scanning electron microscopy (SEM, SU8010, Hitachi, Japan).
Contact angle measurement
Contact angle measurements were performed using a KRÜSS Contact Angle machine with KRÜSS ADVANCE software version 1.11.0.15801. Prior to testing, samples, such as 40% Matrigel (354230, BD Biosciences), were stored at 4 °C. For the sessile drop technique, a 2 μL droplet of the sample was deposited on a PDMS surface that had been treated with plasma for varying durations. Measurements were taken within 5 s of droplet deposition. Contact angles were analyzed from recorded videos using ImageJ software.
Culture system
The PDMS prepolymer (Sylgard 184, Dow Corning, United States) was prepared with a mass ratio of 10:1 between the matrix and crosslinker. The prepolymer was mixed thoroughly and poured into a four-well plate (176740, Thermo Fisher Scientific). A total of 0.05 g of PDMS prepolymer was added to each well, followed by curing at 80 °C for 2 h. The PDMS wells were then treated with plasma at 30 W for 5 s. After treatment, 5 μL of 40-MG was added to each well, and 5 embryos were embedded in the 40-MG. The plates were cured at 37 °C for 8-10 min, after which culture medium was added. Embryos were cultured at 37 °C in a 5% CO2 atmosphere. For 40-MGT, 50 U/g Transglutaminase (G8661-5g, Solarbio) was added to the 40-MG. For 40-MGN, 1 mg/mL PEG-NHS (N164064-1g, Aladdin) was added to the 40-MG.
Embryo recovery and culture
Pregnant mice were humanely euthanized at 3.5 days post coitum by cervical dislocation. Embryos were then flushed from the uterus using a flush medium and seeded onto a designated well a plate. This time point was recorded as IVC day 0. Embryo culture was conducted at 37 °C in a 5% CO2 atmosphere.
In vitro culture medium
Flush medium: CMRL 1066 (11530037, Invitrogen) + 5 × penicillin-streptomycin (60162ES76, YEASEN) + 10% fetal bovine serum (FBS, SE200-ES, Vistech). Medium for IVC days 0-1: CMRL 1066 + 1 × penicillin-streptomycin+ 1 × GlutaMAX™ Supplement (35050061, Thermo Fisher Scientific) + 1 × MEM Non-Essential Amino Acids Solution (11140050, Thermo Fisher Scientific) + 0.5 × N-2 Supplement (17502048, Gibco) + 0.5 × B-27™ Supplement (17504044, Gibco) + 10% FBS.Medium for IVC day 2: CMRL 1066 + 1 × penicillin-streptomycin + 1 × GlutaMAX™ Supplement+ 1 × MEM Non-Essential Amino Acids Solution + 0.5 × N-2 Supplement + 0.5 × B-27™ Supplement + 20% FBS. Medium for IVC day 3: CMRL 1066 + 1 × penicillin-streptomycin + 1 × GlutaMAX™ Supplement + 1 × MEM Non-Essential Amino Acids Solution + 30% KnockOut™ Serum Replacement (10828028, Gibco). Medium for IVC day 4: CMRL 1066 + 1 × penicillin-streptomycin+ 1 × GlutaMAX™ Supplement + 1 × MEM Non-Essential Amino Acids Solution + 30% rat serum (RS) + 0.5 mg/mL glucose (Sigma, D9434). Medium for IVC day 5: CMRL 1066 + 1 × penicillin-streptomycin + 1 × GlutaMAX™ Supplement + 1 × MEM Non-Essential Amino Acids Solution + 40% RS + 1 mg/mL glucose. Medium for IVC day 6–7: CMRL 1066 + 1 × penicillin-streptomycin + 1 × GlutaMAX™ Supplement + 1 × MEM Non-Essential Amino Acids Solution + 50% RS + 1 mg/mL glucose. Medium for IVC days 8-10: CMRL 1066 + 1 × penicillin-streptomycin + 1 × GlutaMAX™ Supplement + 1 × MEM Non-Essential Amino Acids Solution + 50% RS + 2 mg/mL glucose.
Inhibitor treatment
A concentration of 10 µM of each inhibitor, including Fak-specific inhibitor (FI 14, SML0837-10MG, Sigma), MMP inhibitor (GM6001, HAT08408, TargetMol), and blebbistatin inhibitor (S7099, Selleck), was added per well. Vehicle control (DMSO, D2650-100ML, Sigma) was also added per well. 0.02 mg/mL Collagenase IV (07909, Stemcell) was also added per well.
Immunofluorescence staining
-
(1)
Characterization of embryos from IVC day 1 to day 5 and natural embryos (E4.5-E6.5):
Embryos were fixed with 4% paraformaldehyde (PFA, 60536ES76, Yeasen) at 4 °C for 6 h, then permeabilized with 1% Triton X-100 (9002-93-1, Sigma) in phosphate buffered saline (PBS, PM5090-50, Coolaber) at 4 °C for 6 h. Blocking was performed with 0.1% Tween-20 (P1379-25, Sigma), 0.01% Triton X-100, and 1% bovine serum albumin (BSA, B09354-100g, Innochem) in PBS at 4 °C for 6 h. Embryos were incubated overnight at 4 °C with primary antibodies diluted in 0.1% Tween-20, 0.01% Triton X-100, in PBS. The primary antibodies used were CDX2 (1:100, 3977S, Cell Signaling Technology); OCT4 (1:100, sc-5279, Santa); SOX2 (1:100, ab97959, Abcam); EOMES (1:100, ab23345, Abcam); FOXA2 (1:100, 8186S, Cell Signaling Technology); T (1:100, ab209665, Abcam); Phalloidin (1:500, 40737ES75, Yeasen); VINCULIN (1:100, 26520-1-AP, Proteintech); p-MLC (1:100, 3674S, Cell Signaling Technology); p-FAK (1:100, PA5-17084, Thermo Fisher Scientific). The embryos were then incubated with the following secondary antibodies at 4 °C for 2 h: Alexa Fluor 647 goat anti-mouse (1:500, A21235, Invitrogen), Alexa 488 goat anti-rabbit (1:500, A11034, Invitrogen) and Cyanine3 goat anti-rabbit (1:500, A10520, Invitrogen). Nuclear staining was performed using 10 μg/mL Hoechst 33342 (H3570, Invitrogen) at room temperature for 10 minutes. Imaging was conducted using a Zeiss LSM 880 confocal microscope.
-
(2)
Characterization of embryos from IVC day 6 to day 10 and natural embryos (E7.5-E8.5).
The embryos were fixed with 4% PFA at 4 °C for 12 h, then permeabilized with 2% Triton X-100 in PBS at 4 °C for 12 h. Blocking was performed with 0.1% Tween-20, 0.01% Triton X-100 and 1% BSA in PBS at 4 °C for 12 h. Embryos were incubated overnight at 4 °C with primary antibodies diluted in 0.1% Tween-20, 0.01% Triton X-100 in PBS. The primary antibodies used were FOXA2 (1:100, 8186S, Cell Signaling Technology); T (1:100, ab209665, Abcam); SOX2 (1:100, ab97959, Abcam); PAX6 (1:100, AF8150, R&D Systems); OTX2 (1:100, AF1979, R&D Systems); GATA4 (1:100, sc-25310, Santa Cruz); myosin light chain (1:100, ab79935, Abcam); myosin heavy chain (1:100, 53-6503-82, eBioscience). The embryos were then incubated overnight at 4 °C with the following secondary antibodies: Alexa Fluor 647 goat anti-mouse (1:500, A21235, Invitrogen); Cyanine3 donkey anti-goat (1:500, A21432, Invitrogen); Cyanine3 goat anti-rabbit (1:500, A10520, Invitrogen) and Alexa Fluor 647 donkey anti-sheep (1:500, A21448, Invitrogen). Nuclear staining was performed 10 μg/mL Hoechst 33342 (H3570, Invitrogen) at room temperature for 10 minutes. Imaging was conducted using a Zeiss LSM 880 confocal microscope.
Live imaging of IVC embryos
Pregnant mice were humanely euthanized at 3.5 days post coitum by cervical dislocation. Embryos were then flushed from the uterus using a flush medium and seeded onto a designated well of a plate. This time point was recorded as IVC day 0. Embryo brightfield images were captured every 2 h over a 9-day period using a real-time dynamic cell monitoring and functional analysis system (JS170405-102, Juli-stage) with a 4× objective at 37 °C and 5% CO2.
Ca2+ measurement
The embryos were washed twice with PBS, incubated at 37 °C for 30 min with 8 μM of the green fluorescent intracellular Ca2+-binding dye Fluo8-AM (21080, AAT Bioquest) and 0.04% Pluronic F127 (P3000MP, Invitrogen) in HBSS solution (14025092, Gibco). After incubation, the embryos were washed twice again with PBS. They were then mounted in glass-bottom dishes with HBSS solution and observed under a confocal microscope (Zeiss 880). Isoproterenol was added to achieve a final concentration of 1 μM. The images obtained were analyzed with ImageJ software, and the time constant of Ca2+ decay was calculated by a single exponential fit using Origin software (OriginLab).
Rheological test
Rheological experiments are conducted on a rheometer (MCR-302, Anton Paar GmbH, Austria) equipped with a parallel plate (25 mm diameter, 3882). For the time sweep model, 200 μL of hydrogel solution was applied to the measuring stage at 4 °C. After trimming the sample to match the geometry, the temperature was elevated to 37 °C for 90 min. The shear storage (G’) and loss (G”) moduli were measured at a fixed frequency and strain (1 Hz, 1%) to monitor the hydrogel moduli. For the stress relaxation test model, the hydrogel solution was added at 4 °C and maintained at 37 °C for 60 min. The stress relaxation test was subsequently initiated at a fixed strain (1%). The stress profiles were normalized to the initial maximum stress and fitted using a generalized Maxwell–Weichert model with three parallel elements (n = 3)50:
Here, σ represents the normalized stress, while\({{{\rm{\sigma }}}}_{0}\), \({{{\rm{\sigma }}}}_{1}\), and \({{{\rm{\sigma }}}}_{2}\) are the normalized stresses in the spring and dashpot 1 and 2 models, respectively. The constants \({{{\rm{\tau }}}}_{1}\) and \({{{\rm{\tau }}}}_{2}\) describe the stress decay in the dashpot 1 and 2 models. All tests are repeated at least three times.
Nanoindentation test
To determine stress relaxation properties, a nanoindenter (Optics11Piuma, The Netherlands) was used. The generalized Maxwell model, which is mathematically expressed using the Prony series, was selected. The following analytical formulation describes the load decay during the holding phase as a function of time51:
where P(t = ∞)=\({{{\rm{P}}}}_{\infty }\) represents the equilibrium load, which is the value reached at the end of the relaxation time. All samples were measured using a probe with a 3 µm radius and a cantilever stiffness of 0.024 N/m. Cantilever bending calibrations were performed before each series of experiments by indenting a rigid surface beneath the probe. Each sample was tested over a 400 × 600 μm grid scan with 200 µm spacing between measurements. The indentation protocol consisted of a 4-second loading phase to a 15,000 nm indentation depth, held for 40 seconds, followed by a 4-second unloading phase. Three experiments were conducted on each tested sample to ensure reliability. All single indentation values were calculated using GraphPad Prism 9 software.
Trophosphere formation
Mouse trophoblast stem (TS) cells were obtained from the Zhou laboratory (Institute of Zoology, Chinese Academy of Sciences, Beijing, China), which were isolated from mouse blastocysts. TS cell lines tested negative for mycoplasma contamination. TS cells were cultured at 37 °C and 5% CO2 in RPMI 1640 (61870036, Gibco) supplemented with 20% FBS, 2 mM L-glutamine (TMS-002-C, Sigma), 0.1 mM 2-mercaptoethanol (2-ME, 21985023, Gibco), 1 mM sodium pyruvate (11360070, Gibco), and 1% penicillin-streptomycin, plus FGF4 (25 ng/mL, 100-31, PeproTech), FGF2 (25 ng/mL, 450-33, PeproTech) and Heparin (1 μg/mL, 07980, STEMCELL Technologies) in the presence of inactivated ICR MEFs. Trophosphere were obtained by seeding TS cells (approximately 12 cells per microwell) into a 96-well ultra-low attachment microplate (7007, Corning) using the same medium as for TS cell culture52.
Preparation of sodium alginate hydrogel
Sodium alginate with a high molecular weight (209.97 kDa) was purchased from Sigma-Aldrich (A2033), Low-molecular-weight sodium alginate (135.2 kDa) was prepared as follows: high molecular weight alginate was irradiated with a 3 or 8 Mrad Cobalt source, followed by dialysis and purification. Arginine-Glycine-Aspartate peptides (RGD, SynthBio) were coupled to alginate using carbodiimide chemistry53,54. To prepare the mixed hydrogels, the appropriate 1% (w/v) alginate and 40-MG were added to a Luer lock syringe (Cole-Parmer). In a separate syringe, appropriate volumes of calcium sulfate and 1066 medium were added (high molecular weight alginate required 19.52 mM calcium sulfate; low molecular weight alginate required 122 mM calcium sulfate). The two syringes were connected with a coupler, and the solutions were mixed by passing them back and forth six times. A 10 μL volume of the hydrogel was used to culture five embryos.
Real-time RT-PCR
Total RNA was extracted from 10 embryos collected into a 0.2 mL tube, followed by reverse transcription using the CellAmp™ Whole Transcriptome Amplification Kit (Real Time) Ver.2 (3734, Takara). Quantitative PCR was then performed using TB Green® Premix Ex Taq™ (Tli RNaseH Plus, RR420, Takara) on a QuantStudio™ 6 Flex system (ABI). INTEGRIN α3, INTEGRIN α4, INTEGRIN α5, INTEGRIN α6, INTEGRIN α7, INTEGRIN αV, INTEGRIN β1, and INTEGRIN β8 primer sequences were obtained from the referenced publications55. Detailed sequence information can be found in Supplementary Table 1.
RNA-seq analysis (SmartSeq2)
RNA was extracted from embryos cultured in 40-MG and 40-MGT on IVC day 2 for sequencing. Embryos were removed from 40-MG and 40-MGT cultures, washed three times with PBS, and then placed under a microscope for lysis. Embryos cultured under 40-MG and 40-MGT conditions on day 2 of in vitro culture (IVC) were sectioned as shown in Fig. 5G. Trophoblast cells from the deposited regions were then isolated and lysed for subsequent sequencing. Trophoblast cells from the deposited regions of embryos from three separate embryos were sequenced for both 40-MG and 40-MGT conditions. The raw sequencing data were processed with Cell Ranger using the default mapping arguments. Gene expression analyses and cell-type identifications were conducted using Seurat V3.1. IntegrateData is a function in the Seurat package used to integrate multiple single-cell datasets into a unified dataset. The data are processed to remove batch effects through the IntegrateData function. In this process, the dim parameter is set to 1:20, and other parameters are set to their default values. The Nonlinear dimensionality reduction was performed using t-SNE. Figures were generated in Seurat using the DimPlot and VlnPlot functions.
Single-cell RNA sequencing (scRNA-seq) library generation
Three E8.5 embryos from three different pregnant mice and three IVC day 9 embryos were selected for sequencing. Embryos were washed three times in PBS containing 0.4% BSA, then dissociated with 3 mL Tryple (12605010, Gibco) with 0.02% DNase (R1600, Solarbio) at 37 °C in 5% CO2 for 12 minutes with gentle agitation. Following dissociation, 2 mL of PBS containing 20% FBS was added, then the sample was centrifuged at 12,000 × g for 2 min. Cell pellets were resuspended in PBS containing 0.4% BSA and then filtered through a 40-μm cell strainer. The cell count was determined using a hemocytometer. Single-cell suspensions (1,000,000 cells/mL) were loaded into a 10x Genomics Chromium Chip according to the manufacturer’s instructions (10x Genomics, Chromium Next GEM Single Cell 3’ GEM, Library and Gel Bead Kit v.3.1). The 10x Genomics scRNA-Seq libraries were generated by Annoroad Gene Tech. (Beijing) Co. Ltd.
Video editing
All videos presented in the manuscript were processed and edited using VideoStudio to ensure clarity, proper sequencing, and high-quality visualization of the experimental results.
Quantification and statistical analysis
Data were statistically analyzed using One-way ANOVA with Welch’s correction for unequal sample sizes, followed by the Bonferroni post hoc test, or two-tailed Student’s t-tests, as appropriate. Analyses were performed using GraphPad Prism 7 for Windows. Data are presented as the mean ± standard deviation (s.d.). The number of independent experiments conducted for each analysis is indicated in the figure legends. All experiments were repeated at least three times unless otherwise stated. The following software were used in this study: Imaris 8.2, ImageJ 1.47, GraphPad Prism 7, Excel 2019, and ZEN Blue Edition (Zeiss).
Ethical statement
All animal procedures were approved by the Animal Care and Use Committee of the Institute of Zoology, Chinese Academy of Sciences (Ethical approval No. IOZ20180058).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The scRNA-Sequence data generated in this study have been deposited in the Chinese Academy of Sciences database under accession code PRJCA004049 [http://gsa.big.ac.cn/]. The SmartSeq2 data generated in this study have been deposited in the Chinese Academy of Sciences database under accession code PRJCA052100 [http://gsa.big.ac.cn/]. Source data are provided with this paper.
Code availability
The data analysis customized scripts are available to download from GitHub (https://github.com/dongjingxi/embryo-development-in-vitro.git).
References
Aguilera-Castrejon, A. et al. Ex utero mouse embryogenesis from pre-gastrulation to late organogenesis. Nature 593, 119 (2021).
Peng, G. et al. Molecular architecture of lineage allocation and tissue organization in early mouse embryo. Nature 572, 528–532 (2019).
Pijuan-Sala, B. et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490 (2019).
Zheng, Y. et al. Controlled modelling of human epiblast and amnion development using stem cells. Nature 573, 421 (2019).
Hantak, A. M., Bagchi, I. C. & Bagchi, M. K. Role of uterine stromal-epithelial crosstalk in embryo implantation. Int. J. Dev. Biol. 58, 139–146 (2014).
Jones-Paris, C. R. et al. Embryo implantation triggers dynamic spatiotemporal expression of the basement membrane toolkit during uterine reprogramming. Matrix Biol. 57-58, 347–365 (2017).
D’Occhio, M. J., Campanile, G., Zicarelli, L., Visintin, J. A. & Baruselli, P. S. Adhesion molecules in gamete transport, fertilization, early embryonic development, and implantation-role in establishing a pregnancy in cattle: A review. Mol. Reprod. Dev. 87, 206–222 (2020).
Campas, O. et al. Quantifying cell-generated mechanical forces within living embryonic tissues. Nat. Methods 11, 183 (2014).
Chan, C. J. et al. Hydraulic control of mammalian embryo size and cell fate. Nature 571, 112 (2019).
Mammoto, T. & Ingber, D. E. Mechanical control of tissue and organ development. Development 137, 1407–1420 (2010).
Valet, M., Siggia, E. D. & Brivanlou, A. H. Mechanical regulation of early vertebrate embryogenesis. Nat. Rev. Mol. Cell Biol. 23, 169–184 (2022).
Leonavicius, K. et al. Mechanics of mouse blastocyst hatching revealed by a hydrogel-based microdeformation assay. Proc. Natl. Acad. Sci. USA 115, 10375–10380 (2018).
Saadaoui, M., Rocancourt, D., Roussel, J., Corson, F. & Gros, J. A tensile ring drives tissue flows to shape the gastrulating amniote embryo. Science 367, 453–458 (2020).
New, D. A. T. & Stein, K. F. Cultivation of mouse embryos in vitro. Nature 199, 297 (1963).
Jenkinson, E. J. & Wilson, I. B. In-vitro support system for study of blastocyst differentiation in mouse. Nature 228, 776–778 (1970).
Bedzhov, I., Leung, C. Y., Bialecka, M. & Zernicka-Goetz, M. In vitro culture of mouse blastocysts beyond the implantation stages. Nat. Protoc. 9, 2732–2739 (2014).
Bedzhov, I. & Zernicka-Goetz, M. Self-Organizing Properties of Mouse Pluripotent Cells Initiate Morphogenesis upon Implantation. Cell 156, 1032–1044 (2014).
Ichikawa, T. et al. An ex vivo system to study cellular dynamics underlying mouse peri-implantation development. Dev. Cell 57, 373–386 (2022).
Govindasamy, N. et al. 3D biomimetic platform reveals the first interactions of the embryo and the maternal blood vessels. Dev. Cell 56, 3276–3287 (2021).
Ozguldez, H. O. & Bedzhov, I. In vitro culture of mouse blastocysts to the egg cylinder stage via mural trophectoderm excision. Methods Mol. Biol. 2214, 31–40 (2021).
Shao, Y. et al. Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche. Nat. Mater. 16, 419–427 (2017).
Fu, J. et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods 7, 733–736 (2010).
Panciera, T. et al. Reprogramming normal cells into tumour precursors requires ECM stiffness and oncogene-mediated changes of cell mechanical properties. Nat. Mater. 19, 797 (2020).
Wei, Z., Schnellmann, R., Pruitt, H. C. & Gerecht, S. Hydrogel network dynamics regulate vascular morphogenesis. Cell Stem Cell 27, 798–812 (2020).
Adebowale, K. et al. Enhanced substrate stress relaxation promotes filopodia-mediated cell migration. Nat. Mater. 20, 1290–1299 (2021).
Guo, J. et al. 3D printed controllable microporous scaffolds support embryonic development in vitro. J. Cell. Physiol. 8, 3408–3420 (2022).
Gu, Z. et al. A uterus-inspired niche drives blastocyst development to the early organogenesis. Adv. Sci. 28, e2202282 (2022).
Amadei, G. et al. Embryo model completes gastrulation to neurulation and organogenesis. Nature 610, 143–153 (2022).
Xiang, L. et al. A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature 577, 537–542 (2020).
Oldak, B. et al. Complete human day 14 post-implantation embryo models from naive ES cells. Nature 622, 562–573 (2023).
Sakai, D. & Trainor, P. A. Face off against ROS: Tcof1/Treacle safeguards neuroepithelial cells and progenitor neural crest cells from oxidative stress during craniofacial development. Dev. Growth Differ. 58, 577–585 (2016).
Dissanayake, K. et al. Individually cultured bovine embryos produce extracellular vesicles that have the potential to be used as non-invasive embryo quality markers. Theriogenology 149, 104–116 (2020).
Chaudhuri, O., Cooper-White, J., Janmey, P. A., Mooney, D. J. & Shenoy, V. B. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature 584, 535–546 (2020).
Ma, Y. F. et al. Viscoelastic cell microenvironment: hydrogel-based strategy for recapitulating dynamic ECM mechanics. Adv. Funct. Mater. 31, 2100848 (2021).
Arkenberg, M. R., Moore, D. M. & Lin, C. C. Dynamic control of hydrogel crosslinking via sortase-mediated reversible transpeptidation. Acta Biomater. 83, 83–95 (2019).
Chau, D. Y. S., Collighan, R. J., Verderio, E. A. M., Addy, V. L. & Griffin, M. The cellular response to transglutaminase-cross-linked collagen. Biomaterials 26, 6518–6529 (2005).
Cui, J. et al. Super-soft hydrogel particles with tunable elasticity in a microfluidic blood capillary model. Adv. Mater. 26, 7295–7299 (2014).
Fernandes-Cunha, G. M. et al. In situ-forming collagen hydrogel crosslinked via multi-functional PEG as a matrix therapy for corneal defects. Sci. Rep. 10, 16671 (2020).
Orban, J. M. et al. Crosslinking of collagen gels by transglutaminase. J. Biomed. Mater. Res. Part A 68, 756–762 (2004).
Na, K. S. et al. Effect of mesenchymal stromal cells encapsulated within polyethylene glycol-collagen hydrogels formed in situ on alkali-burned corneas in an ex vivo organ culture model. Cytotherapy 23, 500–509 (2021).
Zhao, L. L. et al. A novel smart injectable hydrogel prepared by microbial transglutaminase and human-like collagen: Its characterization and biocompatibility. Mater. Sci. Eng. C.-Mater. Biol. Appl. 68, 317–326 (2016).
Burghardt, R. C. et al. Enhanced focal adhesion assembly reflects increased mechanosensation and mechanotransduction at maternal-conceptus interface and uterine wall during ovine pregnancy. Reproduction 137, 567–582 (2009).
Sutherland, A. E., Calarco, P. G. & Damsky, C. H. Developmental regulation of integrin expression at the time of implantation in the mouse embryo. Development 119, 1175–1186 (1993).
Vining, K. H. & Mooney, D. J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 18, 728–742 (2017).
Jana, S., Cooper, A. & Zhang, M. Chitosan scaffolds with unidirectional microtubular pores for large skeletal myotube generation. Adv. Health. Mater. 2, 557–561 (2013).
Blatchley, M. R., Hall, F., Wang, S., Pruitt, H. C. & Gerecht, S. Hypoxia and matrix viscoelasticity sequentially regulate endothelial progenitor cluster-based vasculogenesis. Sci. Adv. 5, eaau7518 (2019).
Chen, J. S. et al. FAK is involved in invasion and metastasis of hepatocellular carcinoma. Clin. Exp. Metastasis 27, 71–82 (2010).
Fiore, V. F. et al. Mechanics of a multilayer epithelium instruct tumour architecture and function. Nature 585, 433–439 (2020).
Qi, Y. et al. Membrane stiffening by STOML3 facilitates mechanosensation in sensory neurons. Nat. Commun. 6, 8512 (2015).
Babaei, B., Davarian, A., Pryse, K. M., Elson, E. L. & Genin, G. M. Efficient and optimized identification of generalized Maxwell viscoelastic relaxation spectra. J. Mech. Behav. Biomed. Mater. 55, 32–41 (2016).
Tosini, M. et al. A methodological approach for interpreting and comparing the viscoelastic behaviors of soft biological tissues and hydrogels at the cell-length scale. Appl. Sci. 14, 1093 (2024).
Rivron, N. C. et al. Blastocyst-like structures generated solely from stem cells. Nature 557, 106–111 (2018).
Bouhadir, K. H. et al. Degradation of partially oxidized alginate and its potential application for tissue engineering. Biotechnol. Prog. 17, 945–950 (2001).
Chaudhuri, O. et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater. 15, 326–334 (2015).
Gao, Z. et al. Microcarriers promote the through interface movement of mouse trophoblast stem cells by regulating stiffness. Bioact. Mater. 24, 196–205 (2023).
Acknowledgements
Q.G.’s work is supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB0820000), National Natural Science Foundation of China (T2222029 and U21A20396), the Joint project of Chongqing Health Commission and Science and Technology Bureau (2025DBXM001), CAS Project for Young Scientists in Basic Research (YSBR-012), Beijing Institute for Stem Cell and Regenerative Medicine Project Incubation Fund (2022FH107), CAS Pioneer Hundred Talents Program (Y829F11102), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16020802), and K. C. Wong Education Foundation (GJTD-2019-06). J.G.’s work is supported by the National Natural Science Foundation of China (32400684). Z.L.G.’s work is supported by the National Natural Science Foundation of China (32501207), and China Postdoctoral Science Foundation (2025M772889). S.J.’s work is supported by Natural Science Foundation of Beijing Municipality (7254546). We are grateful to Shiwen Li and Xili Zhu of the imaging platform of CAS for their outstanding support, and Chunli Li for help with a scanning electron microscope.
Author information
Authors and Affiliations
Contributions
Q.G., Q.Z., G.H.F., L.Q.Y., J.L.Z., and J.G. designed the experiments. J.G., J.W.L., Z.L.G., L.Y.W., and T.J. performed the experiments and analyzed the results. J.G. wrote the manuscript. J.X.D., Z.G., W.L., H.M.W., and S.J. provided assistance in carrying out experiments and discussed results. Q.G. supervised the work and revised the manuscript. J.G., J.W.L., Z.L.G., L.Y.W., and T.J. together with J.L.Z. and Q.G. discussed results and prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Guo, J., Lyu, J., Gao, Z. et al. 3D biomimetic niche modulates embryo development in vitro. Nat Commun 17, 1279 (2026). https://doi.org/10.1038/s41467-025-68039-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-68039-y