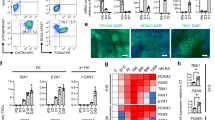
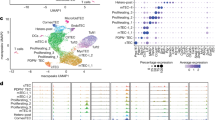
Abstract
The thymus educates thymocytes through a selection process mediated by thymic epithelial cells (TECs). Recent advances have made the generation of T lymphocytes from induced pluripotent stem cells (iPSc) a promising therapeutic strategy. However, current approaches often fail to replicate the thymic niche, leading to impaired T cell generation. Here we address the production of functional mature iPSc-derived TECs supporting in vitro T cell generation. We optimize thymic lineage differentiation through an unbiased multifactorial experimental design. By modulating specific signaling pathways, we generate progenitors that mature into medullary and cortical TECs. Co-culture with primary hematopoietic progenitors in a 3D thymic organoid setup induces their differentiation into CD4+ and CD8+ T cells. Importantly, thymic organoids support multilineage differentiation, with dendritic cell populations also emerging. Thus, the presented thymic organoid model provides a practical platform for studying thymic cellular interactions and thymopoiesis in vitro, and opens further research perspectives towards cell-based therapies.
Similar content being viewed by others
Data availability
The DGE-seq RNA sequencing data related to the DOE-based factor screening and generated in this study have been deposited in the GEO database under accession code GSE302942. The DGE-seq data related to the iPSc to TEP differentiation and generated in this study have been deposited in the GEO database under the accession code GSE304598. The CD205 TEP RNA-seq data generated in this study have been deposited in the GEO database under accession code GSE294118. The D7 hTO TEC scRNA-seq data generated in this study have been deposited in the GEO database under accession code GSE293928. The RNA-seq data of HLA-DRhi, HLA-DRlo TECs and thymic DCs in D28 hTOs generated in this study have been deposited in the GEO database under accession code GSE294061. The scRNA-seq data of D28 hTO detaching cells generated in this study have been deposited in the GEO database under accession code GSE293930. The scRNA-seq data of early pharyngeal development (Han) reused in this study are available in the GEO database under accession code GSE136689. The scRNA-seq data of late pharyngeal development (Magaletta) reused in this study are available in the GEO database under accession code GSE182135. Park’s scRNA-seq data of human thymic cells reused in this study are available at [https://cellatlas.io/studies/thymus-development#resources]. Bautista’s scRNA-seq data of human TECs reused in this study are available in the GEO database under accession code GSE147520. RNA-seq data from human mTEChi, mTEClo, and cTEC populations, used as controls in this study, are available in the GEO database under accession code GSE176445. All other data are available in the article and its Supplementary files or from the corresponding author upon request. Source data are provided with this paper.
Code availability
We have provided the documented R code that we developed for the calculation of gene tissue specificity scores93 as a Git repository available from https://github.com/TeamGiraud/GeneTissueSpe and as a Shiny application at https://nathanprovin.shinyapps.io/GeneTissueSpe.
References
Miller, J. F. A. P. The function of the thymus and its impact on modern medicine. Science 369, eaba2429 (2020).
Abramson, J. & Anderson, G. Thymic epithelial cells. Annu Rev. Immunol. 35, 1–34 (2016).
Bautista, J. L. et al. Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla. Nat. Commun. 12, 1096–15 (2021).
Kadouri, N., Nevo, S., Goldfarb, Y. & Abramson, J. Thymic epithelial cell heterogeneity: TEC by TEC. Nat. Rev. Immunol. 20, 239–253 (2019).
Bornstein, C. et al. Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature 559, 622–626 (2018).
Carpenter, A. C. & Bosselut, R. Decision checkpoints in the thymus. Nat. Immunol. 11, 666–673 (2010).
Deng, Y. et al. Leaving no one behind: tracing every human thymocyte by single-cell RNA-sequencing. Semin. Immunopathol. 43, 29–43 (2021).
Chopp, L. B. et al. An integrated epigenomic and transcriptomic map of mouse and human αβ T cell development. Immunity 53, 1182–1201.e8 (2020).
Klein, L., Klugmann, M., Nave, K. A., Tuohy, V. K. & Kyewski, B. Shaping of the autoreactive T-cell repertoire by a splice variant of self protein expressed in thymic epithelial cells. Nat. Med. 6, 56–61 (2000).
Klein, L., Kyewski, B., Allen, P. M. & Hogquist, K. A. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat. Rev. Immunol. 14, 377–391 (2014).
Anderson, M. S. et al. Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401 (2002).
Cumano, A. et al. New molecular insights into immune cell development. Annu. Rev. Immunol. 37, 497–519 (2019).
Cosway, E. J., James, K. D., Lucas, B., Anderson, G. & White, A. J. The thymus medulla and its control of αβT cell development. Semin. Immunopathol. 43, 15–27 (2021).
Liang, K. L. et al. Intrathymic dendritic cell-biased precursors promote human T cell lineage specification through IRF8-driven transmembrane TNF. Nat. Immunol. 24, 474–486 (2023).
Spidale, N. A., Wang, B. & Tisch, R. Cutting edge: antigen-specific thymocyte feedback regulates homeostatic thymic conventional dendritic cell maturation. J. Immunol. 193, 21–25 (2014).
Nusser, A. et al. Developmental dynamics of two bipotent thymic epithelial progenitor types. Nature 606, 165–171 (2022).
Bleul, C. C. et al. Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature 441, 992–996 (2006).
Ulyanchenko, S. et al. Identification of a bipotent epithelial progenitor population in the adult thymus. Cell Rep. 14, 2819–2832 (2016).
Alves, N. L. et al. Serial progression of cortical and medullary thymic epithelial microenvironments. Eur. J. Immunol. 44, 16–22 (2014).
Baik, S., Jenkinson, E. J., Lane, P. J. L., Anderson, G. & Jenkinson, W. E. Generation of both cortical and Aire+ medullary thymic epithelial compartments from CD205+ progenitors. Eur. J. Immunol. 43, 589–594 (2013).
Li, J. et al. NOTCH1 signaling establishes the medullary thymic epithelial cell progenitor pool during mouse fetal development. Development 147, dev178988 (2020).
Liu, D. et al. Canonical Notch signaling controls the early thymic epithelial progenitor cell state and emergence of the medullary epithelial lineage in fetal thymus development. Development 147, dev178582 (2020).
Irla, M. RANK signaling in the differentiation and regeneration of thymic epithelial cells. Front. Immunol. 11, 623265 (2021).
Irla, M. et al. Autoantigen-specific interactions with CD4+ thymocytes control mature medullary thymic epithelial cell cellularity. Immunity 29, 451–463 (2008).
Lopes, N. et al. Thymocytes trigger self-antigen-controlling pathways in immature medullary thymic epithelial stages. eLife 11, e69982 (2022).
Park, J.-E. et al. A cell atlas of human thymic development defines T cell repertoire formation. Science 367, eaay3224 (2020).
Gao, H. et al. The lineage differentiation and dynamic heterogeneity of thymic epithelial cells during thymus organogenesis. Front. Immunol. 13, 805451 (2022).
Zeng, Y. et al. Single-cell RNA sequencing resolves spatiotemporal development of pre-thymic lymphoid progenitors and thymus organogenesis in human embryos. Immunity 51, 930–948.e6 (2019).
Bredenkamp, N. et al. An organized and functional thymus generated from FOXN1-reprogrammed fibroblasts. Nat. Cell Biol. 16, 902–908 (2014).
Nowell, C. S. et al. Foxn1 regulates lineage progression in cortical and medullary thymic epithelial cells but is dispensable for medullary sublineage divergence. PLoS Genet. 7, e1002348 (2011).
Blackburn, C. C. et al. The nu gene acts cell-autonomously and is required for differentiation of thymic epithelial progenitors. Proc. Natl. Acad. Sci. USA 93, 5742–5746 (1996).
Magaletta, M. E. et al. Integration of single-cell transcriptomes and chromatin landscapes reveals regulatory programs driving pharyngeal organ development. Nat. Commun. 13, 457 (2022).
Farley, A. M. et al. Dynamics of thymus organogenesis and colonization in early human development. Development 140, 2015–2026 (2013).
Gordon, J., Bennett, A. R., Blackburn, C. C. & Manley, N. R. Gcm2 and Foxn1 mark early parathyroid- and thymus-specific domains in the developing third pharyngeal pouch. Mech. Dev. 103, 141–143 (2001).
Gordon, J. & Manley, N. R. Mechanisms of thymus organogenesis and morphogenesis. Development 138, 3865–3878 (2011).
Figueiredo, M., Zilhão, R. & Neves, H. Thymus inception: molecular network in the early stages of thymus organogenesis. Int. J. Mol. Sci. 21, 5765 (2020).
Han, L. et al. Single cell transcriptomics identifies a signaling network coordinating endoderm and mesoderm diversification during foregut organogenesis. Nat. Commun. 11, 4158 (2020).
Nowotschin, S. et al. The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature 569, 361–367 (2019).
Chhatta, A., Mikkers, H. M. M. & Staal, F. J. T. Strategies for thymus regeneration and generating thymic organoids. J. Immunol. Regen. Med. 14, 100052 (2021).
Parent, A. V. et al. Generation of functional thymic epithelium from human embryonic stem cells that supports host T cell development. Cell Stem Cell 13, 219–229 (2013).
Sun, X. et al. Directed differentiation of human embryonic stem cells into thymic epithelial progenitor-like cells reconstitutes the thymic microenvironment in vivo. Cell Stem Cell 13, 230–236 (2013).
Ramos, S. A. et al. Generation of functional human thymic cells from induced pluripotent stem cells. J. Allergy Clin. Immun. 149, 767–781.e6 (2022).
Gras-Peña, R. et al. Human stem cell-derived thymic epithelial cells enhance human T-cell development in a xenogeneic thymus. J. Allergy Clin. Immun. 149, 1755–1771 (2022).
Soh, C.-L. et al. FOXN1 (GFP/w) reporter hESCs enable identification of integrin-β4, HLA-DR, and EpCAM as markers of human PSC-derived FOXN1(+) thymic epithelial progenitors. Stem Cell Rep. 2, 925–937 (2014).
Provin, N. & Giraud, M. Differentiation of pluripotent stem cells into thymic epithelial cells and generation of thymic organoids: applications for therapeutic strategies against APECED. Front. Immunol. 13, 930963 (2022).
Callaghan, N. I. et al. Harnessing conserved signaling and metabolic pathways to enhance the maturation of functional engineered tissues. NPJ Regen. Med. 7, 44 (2022).
Toms, D., Deardon, R. & Ungrin, M. Climbing the mountain: experimental design for the efficient optimization of stem cell bioprocessing. J. Biol. Eng. 11, 35 (2017).
Yasui, R., Sekine, K. & Taniguchi, H. Clever experimental designs: shortcuts for better iPSC differentiation. Cells 10, 3540 (2021).
Besnard, M., Padonou, F., Provin, N., Giraud, M. & Guillonneau, C. AIRE deficiency, from preclinical models to human APECED disease. Dis. Model Mech. 14, dmm046359 (2021).
Chaudhry, M. S., Velardi, E., Dudakov, J. A. & Brink, M. R. M. Thymus: the next (re)generation. Immunol. Rev. 271, 56–71 (2016).
Bosticardo, M. et al. Artificial thymic organoids represent a reliable tool to study T-cell differentiation in patients with severe T-cell lymphopenia. Blood Adv. 4, 2611–2616 (2020).
Montel-Hagen, A. et al. In vitro recapitulation of murine thymopoiesis from single hematopoietic stem cells. Cell Rep. 33, 108320–108320 (2020).
Seet, C. S. et al. Generation of mature T cells from human hematopoietic stem and progenitor cells in artificial thymic organoids. Nat. Methods 14, 521–530 (2017).
Iriguchi, S. et al. A clinically applicable and scalable method to regenerate T-cells from iPSCs for off-the-shelf T-cell immunotherapy. Nat. Commun. 12, 430 (2021).
Canté-Barrett, K. et al. Loss of CD44dim expression from early progenitor cells marks T-cell lineage commitment in the human thymus. Front. Immunol. 8, 32 (2017).
Lavaert, M. et al. Integrated scRNA-Seq identifies human postnatal thymus seeding progenitors and regulatory dynamics of differentiating immature thymocytes. Immunity 52, 1088–1104.e6 (2020).
Li, L.-C. et al. Single-cell patterning and axis characterization in the murine and human definitive endoderm. Cell Res. 31, 326–344 (2021).
D’Amour, K. A. et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 23, 1534–1541 (2005).
Davenport, C., Diekmann, U., Budde, I., Detering, N. & Naujok, O. Anterior–posterior patterning of definitive endoderm generated from human embryonic stem cells depends on the differential signaling of retinoic acid, Wnt-, and BMP-signaling. Stem Cells 34, 2635–2647 (2016).
Balciunaite, G. et al. Wnt glycoproteins regulate the expression of FoxN1, the gene defective in nude mice. Nat. Immunol. 3, 1102–1108 (2002).
Barbarulo, A. et al. Hedgehog signalling in the embryonic mouse thymus. J. Dev. Biol. 4, 22 (2016).
Bleul, C. C. & Boehm, T. BMP signaling is required for normal thymus development. J. Immunol. 175, 5213–5221 (2005).
Lucas, B. et al. Progressive changes in CXCR4 expression that define thymocyte positive selection are dispensable for both innate and conventional αβT-cell development. Sci. Rep. 7, 5068 (2017).
Farley, A. M. et al. Thymic epithelial cell fate and potency in early organogenesis assessed by single cell transcriptional and functional analysis. Front. Immunol. 14, 1202163 (2023).
Fang, X. et al. MIR148A family regulates cardiomyocyte differentiation of human embryonic stem cells by inhibiting the DLL1-mediated NOTCH signaling pathway. J. Mol. Cell. Cardiol. 134, 1–12 (2019).
Ivey, K. N. et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell 2, 219–229 (2008).
Lucas, B. et al. Embryonic keratin19+ progenitors generate multiple functionally distinct progeny to maintain epithelial diversity in the adult thymus medulla. Nat. Commun. 14, 2066 (2023).
Lopes, N., Sergé, A., Ferrier, P. & Irla, M. Thymic crosstalk coordinates medulla organization and T-cell tolerance induction. Front. Immunol. 6, 365 (2015).
Pinto, S. et al. An organotypic coculture model supporting proliferation and differentiation of medullary thymic epithelial cells and promiscuous gene expression. J. Immunol. 190, 1085–1093 (2013).
Campinoti, S. et al. Reconstitution of a functional human thymus by postnatal stromal progenitor cells and natural whole-organ scaffolds. Nat. Commun. 11, 6372 (2020).
Hun, M. et al. Native thymic extracellular matrix improves in vivo thymic organoid T cell output, and drives in vitro thymic epithelial cell differentiation. Biomaterials 118, 1–15 (2017).
Fan, Y. et al. Bioengineering thymus organoids to restore thymic function and induce donor-specific immune tolerance to allografts. Mol. Ther. 23, 1262–1277 (2015).
Yanai, I. et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21, 650–659 (2005).
Williams, J. A., Tai, X. & Hodes, R. J. CD28-CD80/86 and CD40-CD40L interactions promote thymic tolerance by regulating medullary epithelial cell and thymocyte development. Crit. Rev. Immunol. 35, 59–76 (2015).
Yang, S., Fujikado, N., Kolodin, D., Benoist, C. & Mathis, D. Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science 348, 589–594 (2015).
Mellins, E. D. & Stern, L. J. HLA-DM and HLA-DO, key regulators of MHC-II processing and presentation. Curr. Opin. Immunol. 26, 115–122 (2014).
Lepletier, A. et al. Interplay between follistatin, activin A, and BMP4 signaling regulates postnatal thymic epithelial progenitor cell differentiation during aging. Cell Rep. 27, 3887–3901.e4 (2019).
Ramos, S. A. et al. Generation of functional thymic organoids from human pluripotent stem cells. Stem Cell Rep. 18, 829–840 (2023).
Hosaka, N., Kanda, S., Shimono, T. & Nishiyama, T. Induction of γδT cells from HSC-enriched BMCs co-cultured with iPSC-derived thymic epithelial cells. J. Cell Mol. Med. 25, 10604–10613 (2021).
Azzam, H. S. et al. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188, 2301–2311 (1998).
von Boehmer, H. & Fehling, H. J. Structure and function of the pre-T cell receptor. Immunology 15, 433–452 (1997).
Staal, F. J. T., Weerkamp, F., Langerak, A. W., Hendriks, R. W. & Clevers, H. C. Transcriptional control of T lymphocyte differentiation. Stem Cells 19, 165–179 (2001).
Douagi, I., André, I., Ferraz, J. & Cumano, A. Characterization of T cell precursor activity in the murine fetal thymus: evidence for an input of T cell precursors between days 12 and 14 of gestation. Eur. J. Immunol. 30, 2201–2210 (2000).
Kraft, D. L., Weissman, I. L. & Waller, E. K. Differentiation of CD3-4-8- human fetal thymocytes in vivo: characterization of a CD3-4+8- intermediate. J. Exp. Med. 178, 265–277 (1993).
Flippe, L. et al. Rapid and reproducible differentiation of hematopoietic and t cell progenitors from pluripotent stem cells. Front. Cell Dev. Biol. 8, 577464 (2020).
Inami, Y. et al. Differentiation of induced pluripotent stem cells to thymic epithelial cells by phenotype. Immunol. Cell Biol. 89, 314–321 (2011).
Gaignerie, A. et al. Urine-derived cells provide a readily accessible cell type for feeder-free mRNA reprogramming. Sci. Rep. 8, 14363 (2018).
Kilens, S. et al. Parallel derivation of isogenic human primed and naive induced pluripotent stem cells. Nat. Commun. 9, 360 (2018).
Charpentier, E. et al. 3’ RNA sequencing for robust and low-cost gene expression profiling. https://doi.org/10.21203/rs.3.pex-1336/v1 (2021).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Xu, J. et al. Genotype-free demultiplexing of pooled single-cell RNA-seq. Genome Biol. 20, 290 (2019).
Pan, J.-B., Hu, S.-C., Wang, H., Zou, Q. & Ji, Z.-L. PaGeFinder: quantitative identification of spatiotemporal pattern genes. Bioinformatics 28, 1544–1545 (2012).
Provin, N. GeneTissueSpe: combinatory differentiation of human induced pluripotent stem cells generates functional thymic epithelium driving dendritic- and CD4/CD8 T-cell development. https://doi.org/10.5281/zenodo.17674360 (2025).
Acknowledgements
This work was supported by the EJP-Rare Disease JTC2019 program TARID project (ANR-19-RAR4-0011-05), “la Région Pays de la Loire” through the RFI Bioregate grant (ThymIPS), the NORD (The APS Type 1 Foundation grant) ID: 22001 and the “SATT Ouest Valorisation” project OrgaTreg to M.G., as well as “la Fondation pour la Recherche Médicale” (FRM) under grant Equipe FRM EQU202303016266 to C.G. and M.G. This work was also partially supported by the ANR SelfExpress (ANR-22-CE15-0045-01) and by "la Région Pays de la Loire" through the Trajectoire Nationale program (2023_01170) to M.G. N.P. was supported by “la Fondation d’entreprise ProGreffe”. M.d.A. received PhD fellowship support from the FRM (grant no. 17366). This work was partially funded by the Labex IGOprogram supported by the National Research Agency via the investment program ANR-11-LABX-0016-01. We thank the members of the “Genomic’IC” core facility headed by Franck Letourneur at Cochin Institute, Paris, France, for bulk RNA-seq data production, as well as, Drs Pärt Peterson, Julia Maslovskaja and Kai Kisand from Tartu University, Estonia, for human TEC RNA-seq datasets. We thank the iPSC and the MicroPICell core facilities of Nantes supported by IBiSA and Biogenouest, for the use of their resources and technical support. We specifically thank Caroline Chariau and Isabelle Leray (iPSC facility), as well as Anne Gaignerie (MicroPICell) for her advice on iPSc-to-HSPC differentiation, and Laurence Delbos for her advice on flow cytometry. We also thank Ella Beaumann for bioinformatics assistance and Léana Lecompte and Teresa Rodriguez-Trenchs for their help in iPSc culture and differentiation.
Author information
Authors and Affiliations
Contributions
N.P. and M.G. designed the study and wrote the manuscript; N.P., M.d.A. performed most of the experimental work; A.L.B. performed key HSPC experiments; A.B., L.B., C.F., J.P., and P.H. performed experiments; N.P., E.K., and M.G. performed bioinformatics analyses; P.M., O.B., and X.S. provided key material; C.G. and L.D. contributed to the study design and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Kaomei Guan and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Provin, N., d’Arco, M., Le Bozec, A. et al. Combinatory differentiation of human induced pluripotent stem cells generates functional thymic epithelium driving dendritic cell and CD4/CD8 T cell development. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68675-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68675-y