Abstract
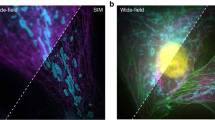
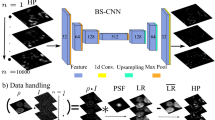
Blind structured illumination microscopy (blind-SIM) is a valuable tool for achieving super-resolution without the need for known illumination patterns. However, in its current formulation the algorithm requires many iterations to converge, leading to long inference times and limited use for real-time or video-rate imaging. We present unrolled blind-SIM (UBSIM), an algorithm which integrates a learnable neural network inside the unrolled iterations of the blind-SIM algorithm. UBSIM delivers a reconstruction speed two to three orders of magnitude faster than that of current iterative blind-SIM methods, while achieving similar resolution and image quality. Furthermore, we demonstrate that UBSIM can be trained in an unsupervised manner that reduces hallucinations and produces superior generalization capability when compared to benchmark super-resolution networks. We test UBSIM experimentally on live cells and present video-rate super-resolution imaging up to 50 Hz. Using our method, we observe dynamic remodeling of the endoplasmic reticulum with high spatiotemporal resolution.
Similar content being viewed by others
Data availability
The simulated datasets used for training and testing the models presented in this work, along with the experimental data used in the figures, are available on Zenodo at https://zenodo.org/records/17852915.
Code availability
The code for UBSIM is available on GitHub at https://github.com/Zach-T-Burns/Unrolled-blind-SIM. MATLAB code for generating the simulated datasets is available on Zenodo at https://zenodo.org/records/17852915.
References
Gustafsson, M. G. L. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82–87 (2000).
Wu, Y. & Shroff, H. Faster, sharper, and deeper: structured illumination microscopy for biological imaging. Nat. Methods 15, 1011–1019 (2018).
Kner, P., Chhun, B. B., Griffis, E. R., Winoto, L. & Gustafsson, M. G. L. Super-resolution video microscopy of live cells by structured illumination. Nat. Methods 6, 339–342 (2009).
Keller, P. J. et al. Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nat. Methods 7, 637–642 (2010).
Markwirth, A. et al. Video-rate multi-color structured illumination microscopy with simultaneous real-time reconstruction. Nat. Commun. 10, 4315 (2019).
York, A. G. et al. Resolution doubling in live, multicellular organisms via multifocal structured illumination microscopy. Nat. Methods 9, 749–754 (2012).
Lee, Y. U. et al. Metamaterial-assisted illumination nanoscopy via random super-resolution speckles. Nat. Commun. 12, 1559 (2021).
Karras, C. et al. Successful optimization of reconstruction parameters in structured illumination microscopy—a practical guide. Opt. Commun. 436, 69–75 (2019).
Demmerle, J. et al. Strategic and practical guidelines for successful structured illumination microscopy. Nat. Protoc. 12, 988–1010 (2017).
Mudry, E. et al. Structured illumination microscopy using unknown speckle patterns. Nat. Photonics 6, 312–315 (2012).
Yeh, L.-H., Tian, L. & Waller, L. Structured illumination microscopy with unknown patterns and a statistical prior. Biomed. Opt. Express 8, 695–711 (2017).
Labouesse, S. et al. Joint reconstruction strategy for structured illumination microscopy with unknown illuminations. IEEE Trans. Image Process. 26, 2480–2493 (2017).
Min, J. et al. Fluorescent microscopy beyond diffraction limits using speckle illumination and joint support recovery. Sci. Rep. 3, 2075 (2013).
Mangeat, T. et al. Super-resolved live-cell imaging using random illumination microscopy. Cell Rep. Methods 1, 100009 (2021).
Jin, L. et al. Deep learning enables structured illumination microscopy with low light levels and enhanced speed. Nat. Commun. 11, 1934 (2020).
Qiao, C. et al. Evaluation and development of deep neural networks for image super-resolution in optical microscopy. Nat. Methods 18, 194–202 (2021).
Ward, E. N. et al. Machine learning assisted interferometric structured illumination microscopy for dynamic biological imaging. Nat. Commun. 13, 7836 (2022).
Chen, R. et al. Single-frame deep-learning super-resolution microscopy for intracellular dynamics imaging. Nat. Commun. 14, 2854 (2023).
Hoffman, D. P., Slavitt, I. & Fitzpatrick, C. A. The promise and peril of deep learning in microscopy. Nat. Methods 18, 131–132 (2021).
Belthangady, C. & Royer, L. A. Applications, promises, and pitfalls of deep learning for fluorescence image reconstruction. Nat. Methods 16, 1215–1225 (2019).
Deng, M. et al. On the interplay between physical and content priors in deep learning for computational imaging. Opt. Express 28, 24152–24170 (2020).
Barbastathis, G., Ozcan, A. & Situ, G. On the use of deep learning for computational imaging. Optica 6, 921–943 (2019).
Wijesinghe, P. & Dholakia, K. Emergent physics-informed design of deep learning for microscopy. J. Phys. Photonics 3, 021003 (2021).
Li, Y. et al. Incorporating the image formation process into deep learning improves network performance. Nat. Methods 19, 1427–1437 (2022).
Qiao, C. et al. Rationalized deep learning super-resolution microscopy for sustained live imaging of rapid subcellular processes. Nat. Biotechnol. 41, 367–377 (2023).
Burns, Z. & Liu, Z. Untrained, physics-informed neural networks for structured illumination microscopy. Opt. Express 31, 8714–8724 (2023).
Monga, V., Li, Y. & Eldar, Y. C. Algorithm unrolling: Interpretable, efficient deep learning for signal and image processing. IEEE Signal Process. Mag. 38, 18–44 (2021).
Monakhova, K. et al. Learned reconstructions for practical mask-based lensless imaging. Opt. Express 27, 28075–28090 (2019).
Gilton, D., Ongie, G. & Willett, R. Deep equilibrium architectures for inverse problems in imaging. IEEE Trans. Comput. Imaging 7, 1123–1133 (2021).
Li, Y. et al. Efficient and interpretable deep blind image deblurring via algorithm unrolling. IEEE Trans. Comput. Imaging 6, 666–681 (2020).
Polak, E. and Ribière, G. Note sur la convergence de mèthodes de directions conjuguées. Rev. Francaise Informat. Recherche Opérationnelle 3, 35–43 (1969).
Nesterov, Y. A method of solving a convex programming problem with convergence rate O(1/k^2). Sov. Math. Dokl. 27, 372–376 (1983).
Benzi, M. Preconditioning techniques for large linear systems: a survey. J. Comput. Phys. 182, 418–477 (2002).
Pock, T. & Chambolle, A. Diagonal preconditioning for first order primal-dual algorithms in convex optimization. In 2011 International Conference on Computer Vision, 1762−1769 (2011).
Nocedal, J., & Wright, S. Numerical Optimization. (Springer, New York, 2006).
Li, Y., Chen, P.Y., Du, T., & Matusik, W. Learning preconditioners for conjugate gradient PDE solvers. In International Conference on Machine Learning, 19425−19439 (2023).
Azulay, Y. & Treister, E. Multigrid-augmented deep learning preconditioners for the Helmholtz equation. SIAM J. Sci. Comput. 45, S127–S151 (2022).
Andrychowicz, M. et al. Learning to learn by gradient descent by gradient descent. Adv. Neural Info. Process. Syst. 29, (2016).
Hauptmann, A. et al. Model-based learning for accelerated, limited-view 3-D photoacoustic tomography. IEEE Trans. Med. Imaging 37, 1382–1393 (2018).
Gong, D. et al. Learning deep gradient descent optimization for image deconvolution. IEEE Trans. Neural Netw. Learn Syst. 31, 5468–5482 (2020).
Adler, J. & Öktem, O. Solving ill-posed inverse problems with iterative deep neural networks. Inverse Problems 33, 124007 (2017).
Ronneberger, O., Fischer, P. & Brox, T. U-net: convolutional networks for biomedical image segmentation. Med. Image Comput. Comput. Assist. Interv. 9351, 234–241 (2015).
Qiao, C. & Li, D. BioSR: a biological image dataset for super-resolution microscopy. https://doi.org/10.6084/m9.figshare.13264793.v7 (2020).
Descloux, A., Grußmayer, K. S. & Radenovic, A. Parameter-free image resolution estimation based on decorrelation analysis. Nat. Methods 16, 918–924 (2019).
Lim, B., Son, S., Kim, H., Nah, S., & Mu Lee, K. Enhanced deep residual networks for single image super-resolution. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops 136–144 (2017).
Zhang, Y. et al. Image super-resolution using very deep residual channel attention networks. In Proceedings of the European Conference on Computer Vision (ECCV) 286–301 (2018).
LeCun, Y. & Cortes, C. MNIST handwritten digit database. http://yann.lecun.com/exdb/mnist (2010).
Zhang, R., Isola, P., Efros, A. A., Shechtman, E. & Wang, O. The unreasonable effectiveness of deep features as a perceptual metric. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 586–595 (2018).
Simonyan, K. & Zisserman, A. Very deep convolutional networks for large-scale image recognition. Preprint at http://arXiv.org/abs/1409.1556 (2014).
Riedl, J. et al. Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5, 605–607 (2008).
Schwarz, D. S. & Blower, M. D. The endoplasmic reticulum: structure, function and response to cellular signaling. Cell. Mol. Life Sci. 73, 79–94 (2016).
Pain, C. & Kriechbaumer, V. Defining the dance: quantification and classification of endoplasmic reticulum dynamics. J. Exp. Bot. 71, 1757–1762 (2020).
Shibata, Y., Voeltz, G. K. & Rapoport, T. A. Rough sheets and smooth tubules. Cell 126, 435–439 (2006).
Costantini, L. & Snapp, E. Probing endoplasmic reticulum dynamics using fluorescence imaging and photobleaching techniques. Curr. Protoc. Cell Biol. 60, 21–27 (2013).
Mo, G. C. H., Posner, C., Rodriguez, E. A., Sun, T. & Zhang, J. A rationally enhanced red fluorescent protein expands the utility of FRET biosensors. Nat. Commun. 11, 1848 (2020).
Snapp, E. L. et al. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163, 257–269 (2003).
Huang, X. et al. Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy. Nat. Biotechnol. 36, 451–459 (2018).
Zhao, W. et al. Sparse deconvolution improves the resolution of live-cell super-resolution fluorescence microscopy. Nat. Biotechnol. 40, 606–617 (2022).
Jang, W. & Haucke, V. ER remodeling via lipid metabolism. Trends Cell Biol (2024).
Chertkova, A. O. et al. Robust and Bright Genetically Encoded Fluorescent Markers for Highlighting Structures and Compartments in Mammalian Cells https://www.biorxiv.org/content/10.1101/160374v2.full Preprint at (2020).
Acknowledgements
This work was supported by the National Science Foundation (CBET-2348536 to Z.L.) and the National Institutes of Health (R35 CA197622 to J.Zhang). Z.B. was supported by the NSF graduate research fellowship program (DGE-2038238 to Z.B.), and A.Z.S. was supported by a fellowship from the National Institute of Dental and Craniofacial Research (1F31DE032886-01A1 to A.Z.S.).
Author information
Authors and Affiliations
Contributions
Z.B. conceived the study, designed the code, and trained the models. Z.B., J.Zhao., and A.Z.S. designed and conducted the experiments. Z.B. processed experimental and simulated data. Z.B. prepared the figures. Z.B., J.Zhao, and A.Z.S. wrote the manuscript. J.Zhang and Z.L. supervised the work and contributed to the discussion.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Simon Labouesse and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Burns, Z., Zhao, J., Sahan, A.Z. et al. High-speed blind structured illumination microscopy via unsupervised algorithm unrolling. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68693-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68693-w