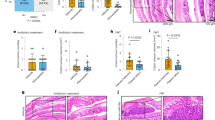
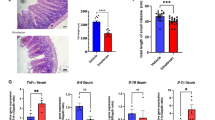
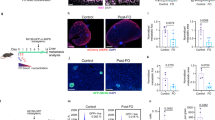
Abstract
Chemotherapy-induced intestinal toxicity is a major dose-limiting complication, but the underlying mechanisms linking systemic metabolism to localized gut damage are poorly understood. Here we show that serum L-kynurenine, a tryptophan metabolite, is elevated in patients with severe oxaliplatin-induced intestinal toxicity. Accumulation of L-kynurenine is driven by IFNγ-mediated induction of indoleamine 2,3-dioxygenase 1 (IDO1) in myeloid cells. Using scRNA-seq and myeloid cell-specific knockout models, we confirm that myeloid cell-derived L-kynurenine exacerbates toxicity. Critically, L-kynurenine accumulation drives gut dysbiosis, characterized by the loss of Lactobacillus johnsonii, and subsequently activates the TNFα/JNK pathway, leading to intestinal epithelial apoptosis. Pharmacological inhibition or engineered reduction of L-kynurenine mitigates chemotherapy-induced intestinal injury. Our findings reveal an important role of L-kynurenine from myeloid cells in chemotherapy tolerance and propose its targeting as a potential therapeutic strategy.
Similar content being viewed by others
Data availability
The data that support the findings of this study are available in the main text, the Supplementary Information, and Source Data file. The RNA-Seq data generated have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database under the accession code PRJNA1239919 and PRJNA1337323. The metabolomics data to the OMIX repository (accession number: OMIX013390 and OMIX013372). Any additional information required to reanalyze the data reported in this work is available from the lead contact upon request. Source data are provided with this paper.
References
Anand, U. et al. Cancer chemotherapy and beyond: current status, drug candidates, associated risks and progress in targeted therapeutics. Genes Dis. 10, 1367–1401 (2023).
Wu, W., Pu, Y. & Shi, J. Nanomedicine-enabled chemotherapy-based synergetic cancer treatments. J. Nanobiotechnol. 20, 4 (2022).
Boussios, S., Pentheroudakis, G., Katsanos, K. & Pavlidis, N. Systemic treatment-induced gastrointestinal toxicity: incidence, clinical presentation and management. Ann. Gastroenterol. 25, 106–118 (2012).
Cheng, F. et al. Oxaliplatin-induced peripheral neurotoxicity in colorectal cancer patients: mechanisms, pharmacokinetics and strategies. Front. Pharmacol. 14, 1231401 (2023).
Xu, X. et al. Metabolic reprogramming and epigenetic modifications in cancer: from the impacts and mechanisms to the treatment potential. Exp. Mol. Med. 55, 1357–1370 (2023).
Tintelnot, J. et al. Microbiota-derived 3-IAA influences chemotherapy efficacy in pancreatic cancer. Nature 615, 168–174 (2023).
Jia, D. et al. Microbial metabolite enhances immunotherapy efficacy by modulating T cell stemness in pan-cancer. Cell 187, 1651–1665 (2024).
He, Z. et al. Chemotherapy-induced microbiota exacerbates the toxicity of chemotherapy through the suppression of interleukin-10 from macrophages. Gut Microbes 16, 2319511 (2024).
Zou, S., Fang, L. & Lee, M. H. Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterol. Rep. 6, 1–12 (2018).
De Pietri, S. et al. Gastrointestinal toxicity during induction treatment for childhood acute lymphoblastic leukemia: the impact of the gut microbiota. Int. J. Cancer 147, 1953–1962 (2020).
Guo, H. et al. Multi-omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science. 370, eaay9097 (2020).
Golkhalkhali, B. et al. Strain-specific probiotic (microbial cell preparation) and omega-3 fatty acid in modulating quality of life and inflammatory markers in colorectal cancer patients: a randomized controlled trial. Asia Pac. J. Clin. Oncol. 14, 179–191 (2018).
Osterlund, P. et al. Lactobacillus supplementation for diarrhoea related to chemotherapy of colorectal cancer: a randomised study. Br. J. Cancer 97, 1028–1034 (2007).
Huai, M. et al. Intelligent nanovesicle for remodeling tumor microenvironment and circulating tumor chemoimmunotherapy amplification. J. Nanobiotechnol. 22, 257 (2024).
Mulder, K. et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity 54, 1883–1900 (2021).
Tan, X. X., Actor, J. K. & Chen, Y. Peptide nucleic acid antisense oligomer as a therapeutic strategy against bacterial infection: proof of principle using mouse intraperitoneal infection. Antimicrob. Agents Chemother. 49, 3203–3207 (2005).
Martinez, M. A., de Mendoza, D. & Schujman, G. E. Transcriptional and functional characterization of the gene encoding acyl carrier protein in Bacillus subtilis. Microbiology 156, 484–495 (2010).
Britton, T. A. et al. Inactivation of the Fusobacterium nucleatum Rnf complex reduces FadA-mediated amyloid formation and tumor development. Mbio 16, e103225 (2025).
Smith, P. A. et al. Optimized arylomycins are a new class of Gram-negative antibiotics. Nature 561, 189–194 (2018).
Craven, T. W. et al. Computational design of cyclic peptide inhibitors of a bacterial membrane lipoprotein peptidase. ACS Chem. Biol. 19, 1125–1130 (2024).
Zhang, P. & Liu, Z. Structural insights into the transporting and catalyzing mechanism of DltB in LTA D-alanylation. Nat. Commun. 15, 3404 (2024).
Park, J. & Wang, H. H. Systematic dissection of σ(70) sequence diversity and function in bacteria. Cell Rep. 36, 109590 (2021).
Jones, R. M. et al. Flagellin administration protects gut mucosal tissue from irradiation-induced apoptosis via MKP-7 activity. Gut 60, 648–657 (2011).
Pallotta, M. T. et al. Indoleamine 2,3-dioxygenase 1 (IDO1): an up-to-date overview of an eclectic immunoregulatory enzyme. FEBS J. 289, 6099–6118 (2022).
Zhai, L. et al. Immunosuppressive IDO in cancer: mechanisms of action, animal models, and targeting strategies. Front. Immunol. 11, 1185 (2020).
Yamashita, N. et al. MUC1-C integrates activation of the IFN-γ pathway with suppression of the tumor immune microenvironment in triple-negative breast cancer. J. Immunother. Cancer. 9, e002115 (2021).
Yan, J. et al. Molecular mechanisms and therapeutic significance of Tryptophan Metabolism and signaling in cancer. Mol. Cancer 23, 241 (2024).
Dorcas, A. T. et al. Modulation of brain kynurenic acid by N-acetylcysteine prevents cognitive impairment and muscular weakness induced by cisplatin in female rats. Cells. 13, 1989 (2024).
Nguyen, D. et al. Targeting the kynurenine pathway for the treatment of cisplatin-resistant lung cancer. Mol. Cancer Res. 18, 105–117 (2020).
Wu, J. et al. Microbiota-induced alteration of kynurenine metabolism in macrophages drives formation of creeping fat in Crohn’s disease. Cell Host Microbe 32, 1927–1943 (2024).
Haq, S., Grondin, J. A. & Khan, W. I. Tryptophan-derived serotonin-kynurenine balance in immune activation and intestinal inflammation. FASEB J. 35, e21888 (2021).
Chrysostomou, D., Roberts, L. A., Marchesi, J. R. & Kinross, J. M. Gut microbiota modulation of efficacy and toxicity of cancer chemotherapy and immunotherapy. Gastroenterology 164, 198–213 (2023).
Wallace, B. D. et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330, 831–835 (2010).
Anderson, C. J. et al. Metabolite-based inter-kingdom communication controls intestinal tissue recovery following chemotherapeutic injury. Cell Host Microbe 32, 1469–1487 (2024).
Alvarado, D. M. et al. Epithelial indoleamine 2,3-dioxygenase 1 modulates aryl hydrocarbon receptor and notch signaling to increase differentiation of secretory cells and alter mucus-associated microbiota. Gastroenterology 157, 1093–1108 (2019).
Sun, P. et al. High-fat diet-disturbed gut microbiota-colonocyte interactions contribute to dysregulating peripheral tryptophan-kynurenine metabolism. Microbiome 11, 154 (2023).
Agus, A., Planchais, J. & Sokol, H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 23, 716–724 (2018).
Qu, C. et al. Patchouli alcohol ameliorates dextran sodium sulfate-induced experimental colitis and suppresses tryptophan catabolism. Pharmacol. Res. 121, 70–82 (2017).
Sharaf, G., El Morsy, E. M. & El-Sayed, E. K. Augmented nephroprotective effect of liraglutide and rabeprazole via inhibition of OCT2 transporter in cisplatin-induced nephrotoxicity in rats. Life Sci. 321, 121609 (2023).
Meng, X. et al. Nanoplastics induced health risk: insights into intestinal barrier homeostasis and potential remediation strategy by dietary intervention. J. Hazard. Mater. 472, 134509 (2024).
Wang, C. et al. Maternal consumption of a fermented diet protects offspring against intestinal inflammation by regulating the gut microbiota. Gut Microbes 14, 2057779 (2022).
Mishra, S., Jain, S., Agadzi, B. & Yadav, H. A cascade of microbiota-leaky gut-inflammation- is it a key player in metabolic disorders?. Curr. Obes. Rep. 14, 32 (2025).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Acknowledgements
Guangdong S&T Program (Z.H., 2024B1111150001); Shenzhen Medical Research Special Fund Project Target disease (Z.H., B2302036); National Key R&D Program of China (P.L., 2022YFA1304000); the program of Guangdong Provincial Clinical Research Center for Digestive Diseases (P.L., 2020B1111170004), National Key Clinical Discipline; National Natural Science Foundation of China (P.L., U21A20344; Z.H., 82273346); Science and Technology Program of Guangdong Province, China (Z.H., 2021B1212040017, J.W., 2024A04J4086); Key laboratory start-up project (Sixth Affiliated Hospital of Sun Yat-sen University) (Z.H., 2023WST03); Guangdong Medical Development Foundation (H.X., B2025247). This project was also supported by Guang Dong Cheung Kong Philanthropy Foundation.
Author information
Authors and Affiliations
Contributions
Z.H., P.L. and S.Q.J. supervised the study and designed the experiments. H.Y.Xie, J.Y.Y., J.J.W., W.H.M., H.Y.Xu, S.G., Y.C.X., Z.H.L., D.Y.L., M.J.C. and D.L.L. performed the experiments. H.Y.Xie and J.J.W. collected, analyzed and interpreted the data. H.Y.Xie and J.Y.Y. wrote the manuscript. Z.H., P.L. and S.Q.J. revised the manuscript. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Hamid Akbarali, Eiji Hara and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xie, H., Yang, J., Wu, J. et al. Kynurenine mediates the chemotherapy-induced intestinal toxicity through modulation of gut microbiota. Nat Commun (2026). https://doi.org/10.1038/s41467-026-68741-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-026-68741-5