Abstract
A new Escherichia coli laboratory evolution screen for detecting plant ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) mutations with enhanced CO2-fixation capacity has identified substitutions that can enhance plant productivity. Selected were a large subunit catalytic (Met-116-Leu) mutation that increases the kcatc of varying plant Rubiscos by 25% to 40% and a solubility (Ala-242-Val) mutation that improves plant Rubisco biogenesis in E. coli 2- to 10-fold. Plastome transformation of either mutation into the tobacco plastome rbcL gene had no impact on leaf Rubisco production, photosynthesis or plant growth. However, tobacco transformed with low-abundance hybrid Arabidopsis Rubisco coding M116L improved plant exponential growth rate by ~75% relative to unmutated hybrid enzyme, with the A242V substitution increasing both hybrid Rubisco production and plant growth by ~50%. Our identification of mutations with the potential to enhance plant growth bodes well for broadening the survey of Rubisco sequence space for catalytic switches that can impart more substantive plant productivity improvements.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The materials generated as part of this study are available from the corresponding author upon request. Source data are provided with this paper.
Change history
24 September 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41477-025-02136-0
References
Furbank, R. T. et al. Genetic manipulation of key photosynthetic enzymes in the C-4 plant Flaveria bidentis. Aust. J. Plant Physiol. 24, 477–485 (1997).
Pettersson, G. Control properties of the Calvin photosynthesis cycle at physiological carbon dioxide concentrations. Biochim. Biophys. Acta Bioenerg. 1322, 173–182 (1997).
Woodrow, I. E. & Berry, J. A. Enzymatic regulation of photosynthetic CO2 fixation in C3 plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 533–594 (1988).
Iñiguez, C., Galmés, J. & Gordillo, F. J. L. Rubisco carboxylation kinetics and inorganic carbon utilization in polar versus cold-temperate seaweeds. J. Exp. Bot. 70, 1283–1297 (2018).
Sharwood, R. E. Engineering chloroplasts to improve Rubisco catalysis: prospects for translating improvements into food and fiber crops. New Phytol. 213, 494–510 (2017).
Bailey-Serres, J., Parker, J. E., Ainsworth, E. A., Oldroyd, G. E. D. & Schroeder, J. I. Genetic strategies for improving crop yields. Nature 575, 109–118 (2019).
Whitney, S. M. et al. Isoleucine 309 acts as a C4 catalytic switch that increases ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) carboxylation rate in Flaveria. Proc. Natl Acad. Sci. USA 108, 14688–14693 (2011).
Lin, M. T., Salihovic, H., Clark, F. K. & Hanson, M. R. Improving the efficiency of Rubisco by resurrecting its ancestors in the family Solanaceae. Sci. Adv. 8, abm6871 (2022).
Walker, B. J., VanLoocke, A., Bernacchi, C. J. & Ort, D. R. The costs of photorespiration to food production now and in the future. Annu. Rev. Plant Biol. 67, 107–129 (2016).
Eisenhut, M., Roell, M.-S. & Weber, A. P. M. Mechanistic understanding of photorespiration paves the way to a new green revolution. New Phytol. 223, 1762–1769 (2019).
Adler, L. et al. New horizons for building pyrenoid-based CO2-concentrating mechanisms in plants to improve yields. Plant Physiol. 190, 1609–1627 (2022).
Furbank, R., Kelly, S. & von Caemmerer, S. Photosynthesis and food security: the evolving story of C4 rice. Photosynth. Res. 158, 121–130 (2023).
Hennacy, J. H. & Jonikas, M. C. Prospects for engineering biophysical CO2 concentrating mechanisms into land plants to enhance yields. Annu. Rev. Plant. Biol. 71, 461–485 (2020).
Nguyen, N. D. et al. A carboxysome-based CO concentrating mechanism for C crop chloroplasts: advances and the road ahead. Plant J. 118, 940–952 (2024).
Erb, T. J. In Curious Future Insight: Science for a Better Tomorrow (ed. Betz, U. A. K.) 49–64 (Springer, 2024).
Salesse-Smith, C. E. et al. Overexpression of Rubisco subunits with RAF1 increases Rubisco content in maize. Nat. Plants 4, 802–810 (2018).
Yoon, D.-K. et al. Transgenic rice overproducing Rubisco exhibits increased yields with improved nitrogen-use efficiency in an experimental paddy field. Nat. Food 1, 134–139 (2020).
Salesse-Smith, C. E. et al. Adapting C4 photosynthesis to atmospheric change and increasing productivity by elevating Rubisco content in sorghum and sugarcane. Proc. Natl Acad. Sci. USA 122, e2419943122 (2025).
Qu, Y., Mueller-Cajar, O. & Yamori, W. Improving plant heat tolerance through modification of Rubisco activase in C3 plants to secure crop yield and food security in a future warming world. J. Exp. Bot. 74, 591–599 (2022).
Andrews, T. J. & Whitney, S. M. Manipulating ribulose bisphosphate carboxylase/oxygenase in the chloroplasts of higher plants. Arch. Biochem. Biophys. 414, 159–169 (2003).
Prywes, N. et al. A map of the rubisco biochemical landscape. Nature 638, 823–828 (2025).
Wilson, R. H., Alonso, H. & Whitney, S. M. Evolving Methanococcoides burtonii archaeal Rubisco for improved photosynthesis and plant growth. Sci. Rep. 6, 22284 (2016).
Wilson, R. H., Martin-Avila, E., Conlan, C. & Whitney, S. M. An improved Escherichia coli screen for Rubisco identifies a protein–protein interface that can enhance CO2-fixation kinetics. J. Biol. Chem. 293, 18–27 (2018).
Zhou, Y., Gunn, L. H., Birch, R., Andersson, I. & Whitney, S. M. Grafting Rhodobacter sphaeroides with red algae Rubisco to accelerate catalysis and plant growth. Nat. Plants 9, 978–986 (2023).
Aigner, H. et al. Plant Rubisco assembly in E. coli with five chloroplast chaperones including BSD2. Science 358, 1272–1278 (2017).
Archer, J., Kathpalia, M., Lee, B., Li, S. & Wang, T. Effects of chaperone selectivity on the assembly of plant Rubisco orthologs in E. coli. J. Exp. Bot. 76, 2809–2820 (2025).
Buck, S. et al. Escherichia coli expressing chloroplast chaperones as a proxy to test heterologous Rubisco production in leaves. J. Exp. Bot. 74, 664–676 (2023).
Lin, M. T., Stone, W. D., Chaudhari, V. & Hanson, M. R. Small subunits can determine enzyme kinetics of tobacco Rubisco expressed in Escherichia coli. Nat. Plants 6, 1289–1299 (2020).
Oh, Z. G. et al. Unique biogenesis and kinetics of hornwort Rubiscos revealed by synthetic biology systems. Mol. Plant 17, 1833–1849 (2024).
Whitney, S. M., Birch, R., Kelso, C., Beck, J. L. & Kapralov, M. V. Improving recombinant Rubisco biogenesis, plant photosynthesis and growth by coexpressing its ancillary RAF1 chaperone. Proc. Natl Acad. Sci. USA 112, 3564–3569 (2015).
Conlan, B. & Whitney, S. Preparing Rubisco for a tune up. Nat. Plants 4, 12–13 (2018).
Gionfriddo, M., Rhodes, T. & Whitney, S. M. Perspectives on improving crop Rubisco by directed evolution. Semin. Cell Dev. Biol. 155, 37–47 (2024).
Tcherkez, G., Farquhar, G. & Andrews, T. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl Acad. Sci. USA 103, 7246–7251 (2006).
Farquhar, G. D., von Caemmerer, S. & Berry, J. A. A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 149, 78–90 (1980).
Whitney, S. M. & Sharwood, R. E. Construction of a tobacco master line to improve Rubisco engineering in chloroplasts. J. Exp. Bot. 59, 1909–1921 (2008).
Passioura, J. B. Viewpoint: the perils of pot experiments. Funct. Plant. Biol. 33, 1075–1079 (2006).
Poorter, H., Hler, J. B., van Dusschoten, D., Climent, J. & Postma, J. A. Pot size matters: a meta-analysis of the effects of rooting volume on plant growth. Funct. Plant. Biol. 39, 839–850 (2012).
Eckardt, N. A. & Portis, A. R. Jr. Heat denaturation profiles of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) and Rubisco activase and the inability of Rubisco activase to restore activity of heat-denatured Rubisco. Plant Physiol. 113, 243–248 (1997).
Rodermel, S., Haley, J., Jiang, C. Z., Tsai, C. H. & Bogorad, L. A mechanism for intergenomic integration: abundance of ribulose bisphosphate carboxylase small-subunit protein influences the translation of the large-subunit mRNA. Proc. Natl Acad. Sci. USA 93, 3881–3885 (1996).
Wostrikoff, K., Clark, A., Sato, S., Clemente, T. & Stern, D. Ectopic expression of Rubisco subunits in maize mesophyll cells does not overcome barriers to cell type-specific accumulation. Plant Physiol. 160, 419–432 (2012).
Durao, P. et al. Opposing effects of folding and assembly chaperones on evolvability of Rubisco. Nat. Chem. Biol. 11, 148–155 (2015).
Zhou, Y. & Whitney, S. Directed evolution of an improved Rubisco; in vitro analyses to decipher fact from fiction. Int. J. Mol. Sci. 20, 5019 (2019).
Nick Pace, C. & Martin Scholtz, J. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 75, 422–427 (1998).
McLure, R. J., Radford, S. E. & Brockwell, D. J. High-throughput directed evolution: a golden era for protein science. Trends Chem. 4, 378–391 (2022).
Mueller-Cajar, O., Morell, M. & Whitney, S. M. Directed evolution of Rubisco in Escherichia coli reveals a specificity-determining hydrogen bond in the form II enzyme. Biochemistry 46, 14067–14074 (2007).
Manning, T., Birch, R., Stevenson, T., Nugent, G. & Whitney, S. Bacterial form II Rubisco can support wild-type growth and productivity in Solanum tuberosum cv. Desiree (potato) under elevated CO2. PNAS Nexus 2, pgac305 (2023).
Nakazato, I. et al. Targeted base editing in the plastid genome of Arabidopsis thaliana. Nat. Plants 7, 906–913 (2021).
Whitney, S. M. & Sharwood, R. E. Rubisco engineering by plastid transformation and protocols for assessing expression. Methods Mol. Biol. 2317, 195–214 (2021).
Sharwood, R. E., Ghannoum, O., Kapralov, M. V., Gunn, L. H. & Whitney, S. M. Temperature responses of Rubisco from Paniceae grasses provide opportunities for improving C3 photosynthesis. Nat. Plants 2, 16186 (2016).
Kane, H. J., Wilkin, J. M., Portis, A. R. & Andrews, T. J. Potent inhibition of ribulose-bisphosphate carboxylase by an oxidized impurity in ribulose-1,5-bisphosphate. Plant Physiol. 117, 1059–1069 (1998).
Sharwood, R. E., von Caemmerer, S., Maliga, P. & Whitney, S. M. The catalytic properties of hybrid Rubisco comprising tobacco small and sunflower large subunits mirror the kinetically equivalent source Rubiscos and can support tobacco growth. Plant Physiol. 146, 83–96 (2008).
Sharwood, R. E., Sonawane, B. V., Ghannoum, O. & Whitney, S. M. Improved analysis of C4 and C3 photosynthesis via refined in vitro assays of their carbon fixation biochemistry. J. Exp. Bot. 67, 3137–3148 (2016).
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Eriksson, A. E. et al. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science 255, 178–183 (1992).
Acknowledgements
This research was supported by Australian Research Council grants CE140100015 (S.W.) and DP210101757 (S.W.) in addition to the Czech Science Foundation (GACR project number 24-10671S, I.A.) and the Johannes Amos Comenius Operational Programme (OPJAK project number SENDISO-CZ.02.01.01/00/22_008/0004596, I.A.). We thank N. Paul for technical assistance and acknowledge the use of the facilities of the Australian Plant Phenomics Network, which is supported by the Australian Government’s National Collaborative Research Infrastructure Strategy.
Author information
Authors and Affiliations
Contributions
M.G. and S.W. conceived the study. M.G., T.R. and S.B. designed the directed evolution system with M.G. performing the mutant screening experiments. M.G., R.B. and S.W. generated and analysed the Rubisco mutant tobacco lines with M.G. and S.W. performing the Rubisco kinetic analyses with assistance from T.S. and T.R. Structure–function interpretations were performed by I.A., and the paper was written by M.G., I.A. and S.W. with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors R.B. and I.A. declare no competing interests. A provisional patent application (AU2025902586) has been filed for aspects of the work reported in this paper with S.W., M.G., T.R., S.B. and T.S. listed as inventors.
Peer review
Peer review information
Nature Plants thanks Elizabete Carmo-Silva, Alistair McCormick and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Tobacco rbcL-rbcS mutagenic library construction and insertion of point mutations by golden gate (GG) cloning.
(a) Error prone PCR (epPCR) using the GeneMorph II Random Mutagenesis Kit, plasmid pBP-LS and primers 1555-F and 1568-R yielded 2075-bp products containing flanking BsaI sites to facilitate GG cloning into (b) BsaI cut pET16-GFP-NtRCA to create (c) a pET16-NtL*S*RCA (P2*) plasmid library comprising random mutations (*) in the Rubisco genes. Transformed E. coli containing pET16-GFP-NtRCA are visually identified as GFP expressing colonies while those expressing Rubisco are white. T7 promoter (P) and terminator (T). See Source data Figs. 1 and 5 and Extended Data Figs. 1 and 3 for full plasmid and primer sequences. The mutant tobacco Rubisco library was generated by error prone-PCR (ep-PCR) amplification of the rbcL-rbcS genes including their independent promoter, ribosome binding site and terminator elements) with primers 1555-F and 1568-R using Mutazyme manufacturer’s recommendations (Agilent Technologies, Santa Clara, CA, USA). Following agarose gel separation, the ep-PCR product (library) was purified using a Wizard® SV Gel and PCR Clean-Up System kit (Promega), digested with BsaI and ligated into pET16 (that is Golden Gate [GG] cloning) to generate the P2 plasmid library. Following dialysis by floating Millipore disc Filter Membranes on water for 2 hours the GG reactions were transformed into XL-1Blue E. coli by electroporation (2.5 V, 0.2 mm electro-cuvettes) and plated on LB-Amp at 37 °C. Once colonies appeared they were collected by scraping and the P2 plasmid library purified before transforming into BL21Star E. coli already transformed with plasmids P1 and P3 (step 3 of Fig. 1d) and then screening under selective (RDE) conditions (Fig. 2). The Rubisco library made contained ~1.5 × 106 different mutants.
Extended Data Fig. 2 PAGE analysis of tobacco Rubisco production in E. coli BL21-Star.
PAGE analysis of a single experiment examining the soluble protein from a tobacco (Nt) leaf and E. coli BL21-star expressing wildtype (WT) and mutant tobacco L8S8 Rubiscos (coded by plasmid P2, Fig. 1c) and tobacco chloroplast chaperones (plasmid P3) were separated by native-PAGE and (a) stained with Coomassie or (b) blotted onto nitrocellulose and immuno-probed with tobacco Rubisco antibody. In addition to the A242V substitution promoting L8S8 holoenzyme production, it facilitated the production of a larger RbcL intermediary complex (*), previously shown to contain RAF1 (R1) and/or BSD2 (B2) subunits (L8(R1/B2)n28. (c) Corresponding SDS PAGE separation of the same soluble (S) protein fractions and corresponding total (T) cellular protein (comprising soluble and insoluble membrane and aggregated/inclusion body proteins) showing the Cpn60 and Cpn20 produced are mostly fully soluble while only a small proportion ( < 20%) of the RbcL, RbcS and Rca produced in E. coli are soluble (+), more so in those containing the A242V mutation in RbcL (++). Asmbl refers to the expression of only P3 plasmid; EV, empty pCDF plasmid; GroE, E. coli GroELS chaperonin cages; CPN, tobacco Cpn60α/β/20 chaperonin complexes; CPN + L, CPN encaged RbcL (slightly visible in panel b). Uncropped images of the original gels and blot are provided in Source Data Fig. 2 and Extended Data Figs. 2, 4, 6 and 7.
Extended Data Fig. 3 Generating rbcL plastome transformation plasmids containing direct repeats to facilitate marker excision via homologous recombination within the plastome.
(a) The primers, PCR amplifications and plasmids required for (b) the (i) Golden Gate assembly of the (ii) 4 cloning fragments required to (iii) construct the final transformation vector pLEV-GG-rbcL that contain 405-bp direct repeats (DR) of duplicated regions of the 3’rbcL coding sequence (∆L) and 3’UTR (green T). P, tobacco rbcL promoter + 5′UTR; T, tobacco rps16 terminator; p, T7 promoter; t, T7 terminator.
Extended Data Fig. 4 Generation of gene edited rbcL tobacco plastome transformants.
(a) Plasmids pLEVNt-RbcL116/RbcL242 (see Extended Data Fig. 3 for method of synthesis) were (b) transformed into the 100 kDa L2 R. rubrum Rubisco producing TobRr35 and (c) independent spectinomycin (0.5 mg.ml−1) resistant (specR) T0 transplastomic Tob116 and Tob242 lines selected. Correctly transformed lines (that is those making L8S8 Rubisco) were (d) identified by native PAGE and three Tob116 and two Tob242 lines grown to maturity and fertilized with (e) wild-type (WT) pollen over the (f) T1 and (g) T2 generations. The seeds from each generation were screened for specR, with 12–16% white spectinomycin sensitive progeny detected in the T2 generation and assumed to be (h) aadA free lines resulting from aadA excision via recombination between the homologous direct repeat (DR) regions and thus restoring the plastome sequence to WT apart from single nucleotide gene edits in rbcL (as indicated in panel (a)). Growth of the aadA-free plants was restored once spectinomycin selection was removed and at maturity (i) fertilised with WT pollen to produce T3 plants that were all spectinomycin sensitive and used for growth analyses. (j) A DNA blot analysis of BamHI (B) digested DNA to confirm the T3 plants were aadA-free homoplasmic rbcL gene edited lines (that is contained no aadA-containing 2.7 kb plastome fragment) and the T1 plants had both aadA-containing and aadA-excised plastome copies (that is were heteroplasmic). Uncropped germination plate photos and PAGE images are provided in Source Data Fig. 2 and Extended Data Figs. 2, 4, 6 and 7.
Extended Data Fig. 5 Comparing the Ko and Kc of mutant tobacco and hybrid Arabidopsis Rubisco.
(a) Linear response in Km for CO2 (Kc) with elevating [O2] for the differing Rubiscos. Data shows a collation of measures made on n = 3 to 5 samples (each symbol indicating an independent biological sample). For comparison, the grey dashed line represents the linear fit to the Kc vs [O2] response for wild-type tobacco Rubisco from the top left panel. The mean ( ± SD) values of (b) Kc derived the y-axis (that is where [O2] = 0) and (c) Ko calculated as Kc/slope of the linear fit for each biological sample assayed (circles representing values for each sample). Shown are the mean ( ± SD) of individual values (white circles) from n = 4–6 biological replica measurements as indicated. Lower case letters indicate significant differences to p < 0.05 using a Tukey multiple comparison test with p-values and all data points provided in Source Data Figs. 4 and 5 and Extended Data Fig. 5.
Extended Data Fig. 6 Ambient CO2 growth experiments on tobacco and the RbcL gene edited tob116 (T3, T4) and tob242 (T3) lines.
(a) Summary detail of the experimental conditions for three temperature-controlled growth experiments (GE) under ambient (410 ppm) CO2 and (b) the corresponding individual measures of height increase over with destructive measures of leaf and stem dry biomass taken at differing time points (the number of plants (n) harvested at each time point indicated). Black lines represent sigmoidal growth fits to the height measurements (circles; see Source Data Fig. 6 and Extended Data Figs. 6 and 7 for measurements of each plant) with the wild-type tob growth fit for each GE shown as a dashed line in the lower panels. Shown are the mean ( ± SD) of the linear exponential growth rates (red text and line) measured for n = 6 to 10 plants during stem elongation as indicated. (c) Representative phenotype of the plants at the time of harvest indicated by the circled numbers. Timing of A-Ci gas exchange measurements performed during GE#3 are indicated by the horizontal yellow line (that is when plants at 35 ± 3 cm in height). Uncropped images plants are provided in Source Data Fig. 2 and Extended Data Figs. 2, 4, 6 and 7.
Extended Data Fig. 7 Comparative growth of tobacco and hybrid Arabidopsis Rubisco producing tobacco lines under elevated CO2 – Growth experiment 4.
Plants were grown in a greenhouse as in growth experiment 2 (Extended Data Fig. 6) during Oct 2023 – Feb 2024 in air containing 600 ppm CO2. (a) Individual measures of height increase over time were made for n = 12 plants with destructive measures of leaf and stem dry biomass for (b) n = 6 or7 plants when 66 ± 2 cm in height (indicated by purple horizontal line in panel a) at the times indicated for each genotype (numbers circles correlating to panel a). Black lines represent sigmoidal growth fits to the height measurements (see Source Data Fig. 6 and Extended Data Figs. 6 and 7 for measurements of each plant). Shown are the mean ( ± SD) of the linear exponential growth rates (red text and line) measured for each plant during stem elongation. Timing of A-Ci gas exchange measurements are indicated by the horizontal yellow line (that is, when plants were at 35 ± 3 cm in height). Uncropped plants images are provided in Source Data Fig. 2 and Extended Data Figs. 2, 4, 6 and 7.
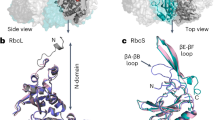
Extended Data Fig. 8 Contacts within van der Waals distance of residue M/L116.
Contacts (dashed red lines) within van der Waals distance of (a) the modelled M116 in wild type (PDB id 4RUB) and (b) mutant L116 along with (c) a full list of contacts around atoms Cβ and Cγ of Met116 and Leu116. The structure analysis is based on the crystal structure of Rubisco from tobacco (PDB 4RUB). The M116L and A242V substitutions were modeled in their most likely conformations from a library of rotamers53. The modelled residue conformations aligned with the corresponding residues in the structure of the highly CO2-specific non-green algae Griffithsia monilis (PDB id 8BDB)24. The effect of cavity-creating substitutions on protein structure has been discussed in detail in the literature54.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2 and Fig. 1.
Source data
Source Data Figs. 1 and 5 and Extended Data Figs. 1 and 3
Gene, plasmid, primer and protein sequence information.
Source Data Fig. 2 and Extended Data Figs. 2, 4, 6 and 7
Unprocessed micrographs and images.
Source Data Figs. 4 and 5 and Extended Data Fig. 5
E. coli-made Rubisco kinetic and biochemical raw data values and statistics for Fig. 4, plant-made Rubisco kinetic and biochemical raw data values and statistics for Fig. 5 and raw kinetic data of plant-made Rubisco that was used to determine Ko for Exended Data Fig. 5.
Source Data Fig. 6 and Extended Data Figs. 6 and 7
Plant growth, biomass, photosynthesis and leaf Rubisco/protein raw data values and statistics for Fig. 6 and plant growth raw data values for Extended Data Figs. 6 and 7.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gionfriddo, M., Birch, R., Rhodes, T. et al. Laboratory evolution of Rubisco solubility and catalytic switches to enhance plant productivity. Nat. Plants 11, 1939–1950 (2025). https://doi.org/10.1038/s41477-025-02093-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41477-025-02093-8