Abstract
Parkinson’s disease (PD) is the second most common neurodegenerative disorder, characterized by motor dysfunction coupled with gastrointestinal disturbances. Recent studies implicate the gut microbiome with the development of PD, yet pinpointing the exact microbial players is still to be determined. This meta-analysis is the first to consolidate five homogenous case-control studies, covering the same variable regions of the 16S rRNA of 1007 fecal samples. Utilizing our unified pipeline, we identified several key players potentially contributing to PD. Our findings reveal higher microbial diversity characterized by elevated levels GABA consuming species particularly Evtepia gabavorous, contributing to neuronal excitability. We also report the abundance of the proinflammatory Klebsiella variicola and the H2S-producing Streptococcus anginosus bacteria, potentially promoting α-synuclein accumulation in the brain. This comprehensive analysis highlights the potential of gut microbiota as a biomarker and a therapeutic strategy to mitigate the progression of PD, possibly facilitating diagnosis and enhancing patient outcomes.
Similar content being viewed by others
Introduction
Parkinson’s disease (PD) is the second most common age-related neurodegenerative disorder (NDD), affecting over 10 million people worldwide as of 2020, which is expected to increase by 56% by 20301,2. It is primarily characterized by motor dysfunction, often initially manifesting as tremor, balance issues, rigidity, and bradykinesia3. As the disease progresses, symptoms expand to gastrointestinal disturbances such as gastroparesis, gut inflammation, increased intestinal permeability, and constipation4. These symptoms are largely attributed to the accumulation of Lewy bodies in addition to misfolded α-synuclein (αSyn) protein aggregates, which is one of the PD hallmarks and a leading cause of neuronal toxicity and death5. This is accompanied by the degeneration of dopaminergic neurons and a subsequent reduction in dopamine levels, which are the primary molecular mechanisms responsible for the motor impairments in PD6. Additionally, mitochondrial dysfunction, oxidative stress, neuroinflammation, and impaired protein degradation pathways further contribute to disease progression7.
Given the early involvement of the gastrointestinal tract, which often precedes motor symptoms by several years, alterations in gut microbiota composition have been widely investigated in the context of PD pathophysiology8. Studies support the potential role of gut microbiota in PD, demonstrating that the microbiota can influence αSyn deposition, which may begin in the neurons of the intestinal submucosa up to 8 years before motor symptoms emerge9,10. Altered gut microbiota composition may trigger immune activation through a compromised gut barrier, leading to systemic inflammation, which can disrupt the blood-brain barrier (BBB), promote neuroinflammation, and contribute to neuronal degeneration11. Intestinal inflammation triggered by bacterial pathobionts may also initiate αSyn misfolding, which can reach the brain through the vagus nerve12.
Another pivotal player in PD is the Gamma-aminobutyric acid (GABA), the brain’s primary inhibitory neurotransmitter, which plays a critical role in the disease pathophysiology13,14,15. Increasing evidence highlights a dynamic interplay between gut-derived GABA and its circulating and cerebral levels, underscoring the microbiome’s potential influence on neurological health16,17,18. Certain gut bacterial species can consume or produce GABA, thereby modulating its systemic availability. The work of Strandwitz et al. has identified Evtepia gabavorous as a GABA-consuming species that thrives in the presence of Bacteroides fragilis, a potent GABA producer16. Their study also revealed numerous candidate GABA-producing and -consuming species, underscoring the complex microbial networks involved in GABA metabolism. Another interesting work by Wu et al. has demonstrated that supplementation with GABA-producing bacterial strains can significantly alleviate essential tremors via the gut-brain axis, particularly through the enteric nervous system and vagus nerve pathways19. These findings suggest the microbiota-derived GABA may influence brain function and potentially contribute to PD progression. The growing interest in the gut–brain axis has led to extensive research into gastrointestinal pathology and gut microbiota alterations in patients with PD. Numerous studies have linked the disease with an increased abundance of specific gut microbiota, such as the Akkermansia genus and the Verrucomicrobiaceae family12. However, some findings remain contested, such as the reported enrichment of Lactobacillaceae in Western cohorts but not in Chinese populations, along with inconsistent results regarding the role of Prevotellaceae20,21,22. Moreover, there remains limited consensus on the specific bacterial species and their underlying mechanisms that may contribute to the PD pathophysiology23. Discrepancies among studies may arise from variations in study designs and methods used for the data analysis, as well as the inherent variability of gut microbiota across populations, lifestyles, and diets. These factors make it challenging to identify specific intestinal bacteria contributors to the pathogenesis of PD.
This variability has underscored the necessity for a comprehensive meta-analysis to synthesize and reconcile the diverse findings across studies4,24,25,26,27,28,29. Several meta-analyses have explored the gut microbiome in PD, and variations in alpha and beta diversity across studies have been noted. However, these analyses have not adequately resolved the inconsistencies or identified the specific microbial species that may contribute to the disease23,30,31,32,33,34,35,36. Examples of heterogeneity in previous meta-analyses include variations in sample collection, variable sections of 16S rRNA regions, and sequencing strategies used in these meta-analyses23,30,31,32,35. Some meta-analyses only presented summary statistics without analyzing raw data, potentially overlooking methodological biases36. Selecting the V3 region over V4 has been questioned due to heterogeneity, even though combining regions V3-V4 tends to yield more accurate results30,32,37. Furthermore, integrating shotgun and amplicon sequencing in meta-analyses has been problematic, as few studies have successfully merged the advantages of both technologies due to a lack of dataset comparability and standardized protocols30. Challenges in accessing raw data have led researchers to resort to tools like GetData Graph Digitizer, which may introduce bias35.
Our meta-analysis aims to address study heterogeneity by standardizing methodology and minimizing potential biases arising from varied data processing techniques. This was achieved by applying a standardized workflow across all samples, incorporating a compositionality-aware method to correct for artefactual variations in the relative abundances of bacterial species within the microbiome. By carefully selecting studies with the highest degree of homogeneity, we focused on paired-end 16S rRNA sequencing of the V3-V4 regions of the gut microbiota from PD patients without prior antibiotic administration. This stringent selection strategy ensured consistency across the dataset and minimized variability while confirming the PD diagnosis. As a result, our meta-analysis is uniquely comprehensive, offering a robust foundation for early PD detection through gut microbiome profiling.
Results
Systematic search results and quality control on the selected studies
Our comprehensive initial search yielded 57 records from the SRA database, which were then carefully screened for relevance and quality (Supplementary Table 1). We identified 15 primary case-control studies addressing the PD-associated gut microbiome using 16S rRNA gene amplicon sequencing data. Upon further review, 10 articles were excluded due to various inconsistencies, ultimately leaving us with the five most homogenous studies, which were included in our meta-analysis (Fig. 1a). These studies covered cohorts from Japan, Malaysia, Italy, South Korea, and China.
a PRISMA chart illustrating studies removed for being irrelevant or heterogeneous for the meta-analysis. b PCA biplot of QC measures (Internal Quality Control (IQC), Accuracy Quality Control (AQC), Consistency Quality Control (CQC), and Standardized Mean Rank (SMR)) demonstrating that the 5 studies are conceding with at least one of these quality measures of MetaQC. c Summary table of MetaQC results covering IQC, AQC, CQC, and SMR where the high score represents high consistency and the low one represents low consistency.
In order to assess the level of the heterogeneity among the included studies and to assess the presence of any outlier studies, we performed rigorous quality control using MetaQC38. From the results, all studies were consistent with at least one of the consistency measurements, namely IQC, AQC, and CQC. This was also reflected in the overall Standardized Mean Rank (SMR), which measures overall consistency, indicating that none of the cohorts showed inconsistency (Fig. 1b, c).
Collectively, the five cohorts provided a total of 1007 samples after filtration, all derived from case–control studies (Table 1). Sequencing technologies used across these studies included MiSeq, HiSeq, and NovaSeq platforms, ensuring comprehensive microbial profiling. Cases had an average age ranging between 55.65 and 78.1 years across all studies, while controls’ ages ranged from 55.4 to 78.1 years. To assess the microbial difference between the cohorts, we used the control samples within each study. The results showed a significant difference between the cohorts’ alpha diversity in both inverse simpson and observed richness indices (P < 0.05; Kruskal-Wallis) among their baselines. Also, beta-diversity analysis revealed a significant difference between the cohorts (P < 0.05; Kruskal-Wallis). However, when observing the over-represented species as well as under-represented species taking the China sample as baseline, we reported 24 differentially abundant OTUs, as shown in (Supplementary Fig. 1). This approach was also applied to the PD samples to measure the differences across the studies, where a significant difference was observed between the cohorts’ alpha diversity (P < 0.05; Kruskal-Wallis), and the beta-diversity analysis was also significant, but only 11 significant OTUs were identified among the PD samples (Supplementary Fig. 2).
Variation in microbial composition and increased diversity in Parkinson’s disease versus controls
Our in-depth analysis of gut microbiomes from PD patients and Healthy Control (HC) included comprehensive assessments of alpha and beta diversities along with microbial abundance across the five included studies. The alpha diversity, measured by the inverse simpson index, showed a significant increase (P < 0.05) in microbial diversity within the PD gut microbiome compared to HC. On the other hand, the observed richness index did not show significant differences between the two groups (Fig. 2a). When examined across individual studies, significant differences for both the inverse simpson and observed richness indices were also observed in the studies from Italy and Japan (Fig. 2b).
a Observed Richness and Inverse Simpson Index, assessing species diversity across different classes. b Observed Richness and Inverse Simpson Index, depicting the estimated variety of microbial species across groups and within individual studies. c Beta diversity analysis, comparing the overall composition of microbial communities between PD and HC samples, both across all studies and within each individual study. d A cladogram showing significant bacteria across various taxonomic levels, offering a hierarchical perspective of microbial differences.
The beta diversity analysis was first conducted using the Rhea pipeline, combining data from all studies using unweighted UniFrac distances. Although the combined analysis of the five studies indicated significant results (P < 0.05; PERMANOVA; Fig. 2c), no clear separation between both clusters appeared. This might be attributed to the fact that the inter-study variability overshadowed the differences between PD and HC. To address this, each study was analyzed separately, revealing clear distinctions between the two conditions. This was further confirmed using RCM to account for the influence of study-specific effects (Supplementary Fig. 3a, b) and when accounting for the study effect (Supplementary Fig. 3c, d).
In our analysis, microbial abundance was quantified, identifying different taxonomic levels with significant differences between PD samples and HC, as determined by ANCOM-BC with a False Discovery Rate (FDR) below 0.05. The cladogram showed that the phyla Bacteroidetes and Synergistetes were significantly upregulated. In the phylum Firmicutes, the class Negativicutes was found to be downregulated, whereas Bacilli and Clostridia showed upregulation. The phylum Verrucomicrobia, including its class Verrucomicrobiae, as well as the phylum Actinobacteria and its class Coriobacteriia, experienced upregulation. Furthermore, the classes Betaproteobacteria and Alphaproteobacteria within the phylum Proteobacteria and the class Synergistia within the phylum Synergistetes were also upregulated (Fig. 2d, detailed table available in Supplementary Table 2).
Identification of bacterial profiles as potential biomarkers in the Parkinson’s disease microbiome
Through our in-depth analysis, species that were differentially abundant between PD patients and HC were confirmed by both ANCOM-BC and MetaDE analyses. ANCOM-BC is particularly adept at handling the compositional nature of microbiome data, accounting for potential biases that traditional differential abundance methods might overlook. Using ANCOM-BC, we were able to identify 138 species, highlighting its robust sensitivity in detecting differentially abundant taxa. MetaDE, on the other hand, aggregates data across multiple studies, enhancing the statistical power and reliability of the findings through combined effect sizes (detailed list available in Supplementary Table 3). Ten species showed significant upregulation in PD, including Bifidobacterium longum, Bifidobacterium bifidum, Bifidobacterium pseudocatenulatum, Streptococcus anginosus, Lactobacillus acidophilus, Evtepia gabavorous, Mordavella massiliensis, Eubacterium siraeum, Klebsiella variicola, and Parabacteroides goldsteinii. Additionally, nine species were significantly downregulated in PDs, including Lachnoclostridium edouardi, Clostridium fessum, Dialister invisus, Coprococcus eutactus, and Lactobacillus rogosae (FDR < 0.05; Fig. 4a, b & c).
Building upon these species-specific findings, further analysis revealed that these changes extended to higher taxonomic levels, impacting entire classes and phyla. The species C. eutactus, L. edouardi, and C. fessum within the Clostridia class of the Firmicutes phylum were significantly downregulated. Conversely, the species A. butyriciproducens from the Ruminococcaceae family in the same phylum was upregulated. Species within the genus Bifidobacterium of the Actinobacteria phylum were consistently upregulated across all levels, from species to phylum. Similarly, in the Bacteroidetes phylum, species such as P. goldsteinii within the Bacteroidia class and Bacteroidales order were notably upregulated. Furthermore, within the Proteobacteria phylum, the species K. variicola in the Gammaproteobacteria class and Enterobacteriaceae family were significantly upregulated. In contrast, the Firmicutes phylum showed a decrease, especially notable within the Clostridia class where numerous families, including Lachnospiraceae and Clostridiaceae, were markedly downregulated.
Among the detected species, we have identified several GABA producers, consumers, and common species capable of both consuming and producing GABA from the collection reported in the work of Strandwitz et al.16. When looking into the 23 identified GABA producers, we are not able to report any significant difference between PD and HC neither on the overall producer load (Fig. 3c, P > 0.18, Mann-Whitney) nor on the individual species abundance (with exception of the P. goldsteinii species; FDR = 0.049; meta-DE). Regarding the 18 GABA consumers detected, PD samples demonstrated a significant increase in the overall microbial load (Fig. 3b, P < 0.006, Mann-Whitney), particularly E. gabavorus a potent GABA eating species (FDR = 0.003; meta-DE). This was further confirmed when correlating over all GABA-consuming levels with other GABA-producing species within our cohorts. The results demonstrated an increase in the microbial load of GABA consumers with an increase in the GABA producers within HC samples (P < 0.05; lm; Fig. 3d). Yet, the GABA consumers are present at a higher abundance in the PD samples throughout the different levels of the GABA producers (P < 0.001; lm; Fig. 3d). This was also confirmed when modeling E. gabavorus levels to the overall GABA producers (P < 0.0007; lm; Fig. 3d) and with each one independently (Supplementary Figure 4).
a Heatmap illustrating the distribution of GABA-producing, GABA-eating, and common (both producing and eating) species. b Violin plots comparing the abundance of GABA-eating species between Parkinson’s disease (PD) and Healthy controls (HC). c Violin plots comparing the abundance of GABA-producing bacterial species between Parkinson’s disease (PD) and Healthy control (HC). d Linear model showing the relationship between the total GABA-eating species and the total GABA-producing species.
Gut microbiome functional pathways associated with Parkinson’s disease in patients and controls
Our functional analysis, performed using STAMP, revealed five significantly enriched KOs in PD samples (Fig. 4e). Among these KOs, fatty acid synthase is involved in multiple metabolic pathways such as fatty acid biosynthesis, mycolic acid biosynthesis, fatty acid metabolism, and insulin resistance. The peptide/nickel transport system substrate-binding protein was linked to quorum sensing, while the cobalt/nickel transport system ATP-binding protein was associated with ABC transporter pathways. Lastly, ribonuclease E was identified as a key player in RNA degradation.
a heatmap illustrating the significant bacterial species with differential abundance between Parkinson’s Disease (PD) and Healthy Control (HC), providing a clear visual representation of microbial alterations associated with PD. b Venn diagram showing the overlap of significant species identified as both upregulated and downregulated in PD patients by ANCOM-BC and MetaDE. c Volcano plot of significant species in PD as identified by ANCOM-BC, with green labels denoting species common between ANCOM-BC and MetaDE analyses. d Confusion matrices and Receiver Operating Characteristic (ROC) curves for machine learning models. e Results from STAMP, highlighting key functional pathways affected in PD, which may offer insights into potential mechanisms of disease progression. f Table of performance metrics, showcasing the predictive accuracy and effectiveness of these models in differentiating between PD and HC samples.
Further association analysis using MicrobiomeAnalyst revealed substantial differences between PD patients and HC in several metabolic pathways. Notably, several pathways such as Glycerolipid metabolism, Valine, Leucine, and Isoleucine degradation, Glycine, Serine, and Threonine metabolism, and Sulfur metabolism were significantly enriched in PD (FDR < 0.05, detailed list available in Supplementary Table 4).
Effectiveness of machine learning models in differentiating Parkinson’s disease patients from healthy individuals using gut microbiota
We developed a machine learning model for classifying samples into two diagnostic categories: PD and HC. The feature selection was based on correlation with these categories, resulting in an optimized and regularized XGBoost model that demonstrated high accuracy and generalizability. A final set of 40 features at the genus level, 18 at the species level, 13 at the family level, 10 at the order level, 4 at the class level, and 1 at the phylum level. Additionally, some geographic and demographic factors were incorporated to ensure a comprehensive representation of the dataset. The accuracy scores of the XGBoost model achieved 77.0% for training accuracy and 77.2% for testing accuracy, indicating better generalizability in predicting PD. Furthermore, grid search cross-validation was conducted on the XGBoost model to optimize its hyperparameters. Using the best parameters, with the L1 regularization parameter set to 0.9 and the L2 regularization parameter set to 0.6, the model achieved a training accuracy of 86.5% and a testing accuracy of 83.2% (Fig. 4d). These results suggest that the optimized and regularized XGBoost model performed well on both the training and test datasets (Fig. 4f).
Discussion
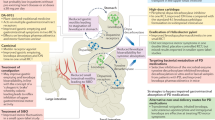
The intricate relationship between gut microbiota and the role it plays in the development of Parkinson's Disease (PD) has been an active research topic for the past couple of years. While considerable research has focused on elucidating this relationship, a consensus identification of the microbial species and their contributions remains unclear. From our meta-analysis of fifteen primary studies exploring this relationship, we have identified five homogeneous case-control studies of 16S rRNA V3-V4 regions comprising 1007 fecal samples that were selected for further analysis. This meta-analysis addresses the inconsistency of primary studies and other meta-analyses attributed to different regions and study designs, where we attempted to pinpoint the microbial players involved and their potential contributions to PD pathogenesis (Fig. 5).
Evtepia gabavorous reduces gamma-aminobutyric acid (GABA) production by consuming it, impacting neuronal excitability in brain regions, thereby exacerbating Parkinson’s Disease (PD) symptoms. Streptococcus anginosus generates H2S, which disrupts mitochondrial functions, elevating ROS levels and encouraging alpha-synuclein (αSyn) aggregation and prion-like spread via the vagal nerve. Moreover, Klebsiella variicola targets microglial cells, augmenting beta-amyloid and αSyn production, potentially intensifying PD progression through increased pro-inflammatory responses mediated by toll-like receptors. Created with BioRender.com.
In this work, we characterized a gut microbial composition consistently associated with PD across all of the included studies, a finding that is concordant with earlier research20,23,31,33,39,40. A significant increase in alpha diversity was observed in PD patients compared to Healthy Control (HC), which is indicative of a broader range of microbial species. Moreover, differences in beta diversity were identified in our analysis, indicating significantly distinct microbial community compositions consistently observed across other studies as well20,21,23,31,41,42,43,44. This was emphasized by notable bacterial shifts in PD patients compared to HC, collectively observed in other studies, where several genera showed an increase in abundance, namely Desulfovibrio, Enterococcus, Bifidobacterium, Hungatella, Lactobacillus, Parabacteroides, Akkermansia, and Alistipes12,21,32,45,46,47,48. Additionally, according to our knowledge, we report for the first time the abundance of the Weissella genus, which has been found to reduce pro-inflammatory markers49, and that of the Neglectibacter genus which has a fatty acid profile that may affect PD progression50.
Confirming previous findings, the involvement of the gut microbiome in the etiology of PD may involve alterations of short-chain fatty acids (SCFAs)-producing bacteria where some showed elevation and others depletion12. Some of these taxa namely, Bifidobacterium, Escherichia, Desulfovibrio, and Akkermansia, were elevated, potentially affecting intestinal barrier integrity and permeability12,23,51. Conversely, other genera such as Blautia, Faecalibacterium, Fusicatenibacter, Lachnospira, Roseburia, Prevotella, and Coprococcus were reduced, potentially affecting the gut health and reducing inflammation23,45,46,47. Furthermore, we report the abundance of a novel SCFA-producing genus in PD, namely Agathobaculum, which has been linked to gut health, reduced inflammation, and cholesterol levels, triglycerides, and TNF-α production52,53. Based on our results, we report the depletion of Agathobaculum butyriciproducens, which was found to have neuroprotective effects in a PD model54. Of interest, our functional analysis demonstrated that threonine, serine, and glycerolipid metabolism were enriched in PD samples, contributing to SCFAs production55,56,57. This is complemented by the enrichment of pathways involved in the degradation of branched-chain amino acids such as valine, leucine, and isoleucine, which can also be converted into branched-chain SCFAs58. This overall shift in microbial composition and SCFAs altered production suggests a potential link to PD progression, highlighting the need for further exploration of this area as that could serve as a biomarker or a therapeutic target for PD.
In addition to the microbial shift linked to PD and consequent alteration in SCFA, we have characterized several species likely to be implicated in the disease pathophysiology. Klebsiella variicola, a pro-inflammatory bacterium, emerged as one of the most distinctive and significantly enriched bacteria in PD patients across all five studies, with a relative abundance of 1.2%. K. variicola has been shown to significantly elevate the expression of pro-inflammatory cytokines, including TNF-α, IL-1β, and IL-6, as well as toll-like receptors (TLR2 and TLR9) in microglial cells59,60, likely by reaching the brain through blood circulation59,61,62. This elevation was reportedly correlated with a pro-inflammatory state in microglia, which subsequently enhances their phagocytic activity towards beta-amyloid (Aβ), facilitated by a 33-fold increase in the expression of the phagocytic receptor MSR159. Accumulation of Aβ was suggested to promote the aggregation of α-synuclein (αsyn), which may exacerbate the pathology and progression of PD63,64. The survival and pathogenesis mechanisms of K. variicola were proposed to be linked to adaptive metabolism, biosynthesis pathways, stress resistance, and various virulence regulatory systems, such as the ABC transporter, and quorum sensing systems65. Next to K. variicola, the pro-inflammatory bacterium E. siraeum was also observed to be significantly abundant in PD patients. The inflammaging E. siraeum may disrupt gut barrier integrity and promote intestinal inflammation, which is linked to PD pathogenesis by exacerbating gut-brain axis dysfunction and contributing to disease progression66.
Our results also revealed elevated levels of hydrogen sulfide (H2S)-producing bacteria, such as Streptococcus anginosus and Mordavella massiliensis, in PD patients, which may result in excess H2S production and elevated reactive oxygen species (ROS) levels67,68,69,70,71,72. Increased ROS levels in the gut may promote α-syn aggregation into oligomers and fibrils, facilitating prion-like spreading from the gut to the brainstem and accelerating PD progression73. Moreover, H2S entering the bloodstream and reaching the brain might inhibit the cytochrome c oxidase enzyme—crucial for ATP generation in neurons—by binding to the CuB site and disrupting oxygen kinetics, ultimately impairing ATP production70, exacerbating neurotoxicity, and further advancing PD pathology74.
Our analysis further identified the novel gamma-aminobutyric acid (GABA) modulating species, Evtepia gabavorous, that is significantly more abundant in PD patients compared to HC. Strandwitz et al. identified Evtepia gabavorous as a GABA-eating bacterium that thrives in the presence of Bacteroides fragilis, a potent GABA producer16. Consistent with this, we observed an overall increase in the abundance of GABA-consuming species in PD samples relative to HC, reinforcing the role of GABA-modulating bacteria in PD. This aligns with emerging evidence linking gut dysbiosis to altered GABAergic signaling in PD. For instance, Barichella et al. (2021) observed dysregulation of microbial GABA metabolism pathways, particularly an increase in the GABA degradation pathway, in PD fecal metagenomes, suggesting a systemic imbalance in GABA homeostasis driven by gut microbiota29. The malfunction of the GABA pathway due to the increase of this GABA-eating bacteria contributes to mitochondrial dysfunction and oxidative stress, integral to PD’s pathology75. Reduced levels of GABA, which reach the brain through either enteric nerves or the vagus nerve, were suggested to be linked to the motor and non-motor symptoms in PD76. This could be explained by promoting mitochondrial dysfunction, oxidative stress, disruption of calcium homeostasis, increasing neuronal excitability, and contributing to dopaminergic neuron loss, which are highly related to PD neuropathology77,78,79,80. Notably, therapeutic interventions targeting microbial GABA metabolism show promise. Wu et al. (2023) demonstrated that supplementation with GABA-producing bacterial strains alleviates motor deficits such as tremors, likely by restoring GABAergic tone through gut-brain axis signaling19, and an illustrative figure for the role of the GABA is available in (Supplementary Fig. 5).
On the functional level, several PD-associated pathways were predicted to be significantly enriched in PD patients, including fatty acid metabolism, insulin resistance, RNA degradation, and the quorum sensing pathway. Fatty acid metabolism, a key pathway influencing PD pathophysiology, was found significantly enriched in PD patients, which was reported to affect neuronal differentiation and function81,82,83. Insulin resistance is another crucial pathway influencing PD by promoting αSyn aggregation, dopaminergic neuron loss, and heightened neuroinflammation, alongside mitochondrial dysfunction and disrupted autophagy due to PI3K/AKT/mTOR pathway hyperactivation, all of which exacerbate PD symptoms84. RNA degradation, particularly of circular RNAs, was also implicated in PD pathology. The degradation mediated by RNase L activates protein kinase R, linked to neuroinflammation and systemic responses in PD85.
Ultimately, this meta-analysis identified bacterial species that are significant in PD patients, pinpointing their exact roles in PD pathogenesis. Notably, S. anginosus, K. variicola, and E. gabavorous emerged as key species due to their potential involvement in mechanisms contributing to PD progression. This study represents the most robust meta-analysis conducted to date, offering novel insights into the relationship between the gut microbiome and PD at a deeper level. However, the inclusion of more studies with identical sequencing technology could further increase the power. To fully comprehend the complexity of these interactions and their clinical implications, further comprehensive research is essential. Future studies should include larger, more diverse study populations and incorporate transcriptomics and proteomics analyses to provide a deeper understanding of gut microbiota function. These aspects need to be considered and validated through additional cohort-based wet lab investigations. By expanding knowledge in this area, we can pave the way for more personalized and effective approaches for the early diagnosis of PD and novel therapeutic interventions.
Methods
Systematic search
A comprehensive search was initiated following the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines on January 15, 2023, on the Sequence Read Archive (SRA) database86, which was selected as the primary source to identify any relevant research related to PD. This search employed specific keywords, including ‘Parkinson’ combined with at least one of the terms “metagenome,” “microbiome,” “amplicon sequence,” or ‘metagenomics,’ to identify 16S rRNA datasets. Studies included: (1) human participants, (2) case-control design comparing the gut microbiota of PD patients to HC, (3) 16S rRNA gene sequencing, (4) raw data is available, and (5) stool samples. Studies were excluded during the screening and eligibility stages if they: (1) lacked accompanying scientific papers, (2) utilized shotgun sequencing, (3) did not focus on V3-V4 regions of 16S rRNA, (4) lacked necessary metadata, or (5) included participants with recent antibiotic use.
16S rRNA gene data pre-processing
The raw 16S rRNA amplicon sequencing data in this study were obtained from the open-access database: SRA86, and initial processing was conducted using OCToPUS pipeline (v.1.087). The reads were denoised using SPAdes (v.4.0.088) and then merged using Mothur (v.1.48.189), and then the contigs with ambiguities exceeding 8 homopolymers of the SILVA database (v.138.290) were excluded. Additional denoising using IPED algorithm (v.1.091) was performed, followed by de novo chimera removal by CATCh (v.1.092). Operational taxonomic unit (OTU) clustering was conducted at a 97% identity using USEARCH (v.8.2.1893)) and classified against the Ribosomal Database Project dataset (v.1694). This approach enabled a more accurate and comprehensive understanding of microbial diversity. Quality control was conducted using the MetaQC module from the MetaOmics R package to ensure the reliability and consistency of the meta-analysis38. This includes Internal Quality Control (IQC), calculating the correlation of OTU pairs correlations across studies; Accuracy Quality Control (AQC), comparing the differentially abundant OTUs of each study to the remainder of the studies. Consistency Quality Control (CQC) examined the consistency of the ranking of differentially abundant OTUs from individual study analyses compared to the rest, thus evaluating the robustness of the results. The outcomes of these three quality control measures were then summarized using the Standardized Mean Rank (SMR), which provided a comprehensive indication of the studies’ overall level of consistency and reliability.
Diversity analysis
Hypothesis testing for group differences in taxonomy and gene profile was conducted using the car package in R95 with t.test function to implement Mann-Whitney U test96 as Levene’s test with leveneTest indicated heterogeneous variances, and the Shapiro-Wilk test by shapiro.test function revealed non-normal data distribution. All statistically significant outcomes in this study were identified with p-values less than 0.05. Using the phyloseq R package97 alpha diversity indices were conducted by plot_richness function including the observed richness index (i.e., species richness, including rare species) and inverse simpson index (i.e., species richness and evenness98). To assess beta diversity, two different dimensionality reduction techniques were applied first using Rhea pipeline(v.1.199), with calculations based on an unweighted UniFrac distance matrix. Analyses were performed separately for each study, and Ward’s hierarchical clustering along with NMDS plots were used for visualization. Secondly, Residuals and Covariance Modelling (RCM)100 was applied to account for inter-study differences. Additionally, accounting for cohort difference as a confounder was also applied for beta diversity calculations, which were further examined through PERMANOVA (Permutational Multivariate Analysis of Variance)101 using the vegan package’s adonis2 function (v.2.6-4102) for both NMDS and RCM approaches.
Biomarkers detection through meta-analysis
To identify taxa that differed significantly in abundance between groups PD and HC, Analysis of Composition of Microbiomes with Bias Correction (ANCOM-BC)103 was used, addressing challenges of compositionality, study difference, and sampling bias. For clearer representation, we constructed a cladogram showing phylogenetic relationships and abundance variations. MetaDE module of MetaOmics38 was then utilized performing a meta-analysis from the five studies with the random-effects model (REM)104. OTUs with false discovery rate (FDR) values below 0.05 were identified as significantly different, and the detected bacteria were classified using the NCBI Nucleotide BLAST, achieving approximately 97% identity and 100% coverage by rRNA/ITS database105. Another mapping was conducted to map the identified OTUs to the GABA-producing and GABA-consuming species presented in the work of Strandwitz et al.16; for this purpose, we applied BLAST with 100% coverage and at least 99% sequence identity. Then, to further assess the correlation between the GABA producers and consumers, in particular those that have been identified to be significant, we performed a linear model using the (linear model) lm function from the stats package106, coupled with Mann-Whitney U test96.
Functional prediction
The metabolic and functional capabilities of the microbial communities were inferred through functional prediction using Tax4Fun function from themetagenomics package107. In addition, STAMP (Statistical Analysis of Metagenomic Profiles) was employed108 to conduct statistical hypothesis tests and generate visual comparisons of taxonomic and functional profiles between PD and HC. For this comparison, we applied Welch’s t-test, with alpha of 0.05 and an effect size of >0.1. To further explore the metabolic potential, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database109 was used to determine the relevant pathways. Moreover, another pathway association analysis was conducted using MicrobiomeAnalyst110 to draw more connections between microbial functions.
Machine learning
Machine learning techniques were employed to build a robust predictive model. The data used in this investigation were pre-processed using significant taxa across taxonomy level, and available metadata, namely the center name, city, and instrument. Prior to model training, categorical features were encoded with label encoding and one-hot encoding. A correlation matrix was computed, and features with a correlation > 0.1 were selected. The dataset was divided into two sets: training (80%), and testing (20%), with a random state of 50. Several machine learning models were compared, including Random Forest, XGBoost, and Decision tree111,112,113. The XGBoost classifier was selected due to its efficacy and robustness. Then it was initialized using grid search with the following hyperparameters: number of estimators set to 50, maximum depth at 3, column sample by tree and subsample at 0.6, use label encoder at False, and learning rate at 0.1. To avoid overfitting, the lasso regularization parameter set to 0.9 and the rigid regularization parameter set to 0.6.
Data availability
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Code availability
The underlying code for this study is not publicly available but may be made available to researchers on reasonable request from the corresponding author.
References
Kouli, A., Torsney, K. M. & Kuan, W. L. Parkinson’s Disease: Etiology, Neuropathology, and Pathogenesis, in Parkinson’s Disease: Pathogenesis and Clinical Aspects, T. B. Stoker and J. C. Greenland, Editors. Brisbane (AU, 2018).
Bayen, S. et al. Parkinson’s disease: content analysis of patient online discussion forums. A prospective observational study using Netnography. Patient Educ. Couns. 104, 2060–2066 (2021).
Simonet, C. et al. The motor prodromes of parkinson’s disease: from bedside observation to large-scale application. J. Neurol. 268, 2099–2108 (2021).
Jo, S. et al. Oral and gut dysbiosis leads to functional alterations in Parkinson’s disease. NPJ Parkinsons Dis. 8, 87 (2022).
Stefanis, L. α-Synuclein in Parkinson’s disease. Cold Spring Harb. Perspect. Med 2, a009399 (2012).
Kalia, L. V. & Lang, A. E. Parkinson’s disease. Lancet 386, 896–912 (2015).
Dong-Chen, X. et al. Signaling pathways in Parkinson’s disease: molecular mechanisms and therapeutic interventions. Signal Transduct. Target Ther. 8, 73 (2023).
Sampson, T. R. et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469–1480.e12 (2016).
Hilton, D. et al. Accumulation of alpha-synuclein in the bowel of patients in the pre-clinical phase of Parkinson’s disease. Acta Neuropathol. 127, 235–241 (2014).
Sanchez-Ferro, A. et al. In vivo gastric detection of alpha-synuclein inclusions in Parkinson’s disease. Mov. Disord. 30, 517–524 (2015).
Quigley, E. M. M. Microbiota-brain-gut axis and neurodegenerative diseases. Curr. Neurol. Neurosci. Rep. 17, 94 (2017).
Li, Z. et al. Gut bacterial profiles in Parkinson’s disease: a systematic review. CNS Neurosci. Ther. 29, 140–157 (2023).
Blaszczyk, J. W. Parkinson’s disease and neurodegeneration: GABA-collapse hypothesis. Front. Neurosci. 10, 269 (2016).
Yadav, D. & Kumar, P. Restoration and targeting of aberrant neurotransmitters in Parkinson’s disease therapeutics. Neurochem. Int. 156, 105327 (2022).
Gong, T. et al. Inhibitory motor dysfunction in parkinson’s disease subtypes. J. Magn. Reson Imaging 47, 1610–1615 (2018).
Strandwitz, P. et al. GABA-modulating bacteria of the human gut microbiota. Nat. Microbiol. 4, 396–403 (2019).
Braga, J. D., Thongngam, M. & Kumrungsee, T. Gamma-aminobutyric acid as a potential postbiotic mediator in the gut–brain axis. npj Sci. Food 8, 16 (2024).
Lyte, M. Microbial endocrinology: Host-microbiota neuroendocrine interactions influencing brain and behavior. Gut Microbes 5, 381–389 (2014).
Zhong, H.-J. et al. Supplementation with high-GABA-producing Lactobacillus plantarum L5 ameliorates essential tremor triggered by decreased gut bacteria-derived GABA. Transl. Neurodegener. 12, 58 (2023).
Qian, Y. et al. Alteration of the fecal microbiota in Chinese patients with Parkinson’s disease. Brain Behav. Immun. 70, 194–202 (2018).
Li, C. et al. Gut microbiota differs between Parkinson’s disease patients and healthy controls in Northeast China. Front. Mol. Neurosci. 12, 171 (2019).
Jin, M. et al. Analysis of the gut microflora in patients with Parkinson’s disease. Front. Neurosci. 13, 1184 (2019).
Romano, S. et al. Meta-analysis of the Parkinson’s disease gut microbiome suggests alterations linked to intestinal inflammation. NPJ Parkinsons Dis. 7, 27 (2021).
Cerroni, R. et al. Not just a snapshot: an Italian longitudinal evaluation of stability of gut microbiota findings in Parkinson’s disease. Brain Sci. 12, 739 (2022).
Nishiwaki, H. et al. Short-chain fatty acid-producing gut microbiota is decreased in parkinson’s disease but not in rapid-eye-movement sleep behavior disorder. mSystems. 5, e00797-20 (2020).
Tan, A. H. et al. Gut microbial ecosystem in Parkinson disease: new clinicobiological insights from multi-omics. Ann. Neurol. 89, 546–559 (2021).
Zhang, P. et al. Specific gut microbiota alterations in essential tremor and its difference from Parkinson’s disease. NPJ Parkinsons Dis. 8, 98 (2022).
Chiang, H. L. & Lin, C. H. Altered gut microbiome and intestinal pathology in Parkinson’s disease. J. Mov. Disord. 12, 67–83 (2019).
Wallen, Z. D. et al. Metagenomics of Parkinson’s disease implicates the gut microbiome in multiple disease mechanisms. Nat. Commun. 13, 6958 (2022).
Nie, S. et al. Inflammatory microbes and genes as potential biomarkers of Parkinson’s disease. npj Biofilms Microbiomes 8, 101 (2022).
Toh, T. S. et al. Gut microbiome in Parkinson’s disease: new insights from meta-analysis. Parkinsonism Relat. Disord. 94, 1–9 (2022).
Nishiwaki, H. et al. Meta-analysis of gut dysbiosis in Parkinson’s disease. Mov. Disord. 35, 1626–1635 (2020).
Nishiwaki, H. et al. Meta-analysis of shotgun sequencing of gut microbiota in Parkinson’s disease. NPJ Parkinsons Dis. 10, 106 (2024).
Romano, S. et al. Machine learning-based meta-analysis reveals gut microbiome alterations associated with Parkinson’s disease. bioRxiv, 2023: p. 2023.12.05.569565.
Shen, T. et al. The association between the gut microbiota and Parkinson’s disease, a meta-analysis. Front. Aging Neurosci. 13, 636545 (2021).
Zuo, S. et al. High levels of Bifidobacteriaceae are associated with the pathogenesis of Parkinson’s disease. Front. Integr. Neurosci. 16, 1054627 (2022).
Claesson, M. J. et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38, e200 (2010).
Ma, T. et al. MetaOmics: analysis pipeline and browser-based software suite for transcriptomic meta-analysis. Bioinformatics 35, 1597–1599 (2019).
Barichella, M. et al. Unraveling gut microbiota in Parkinson’s disease and atypical Parkinsonism. Mov. Disord. 34, 396–405 (2019).
Keshavarzian, A. et al. Colonic bacterial composition in Parkinson’s disease. Mov. Disord. 30, 1351–1360 (2015).
Lin, A. et al. Gut microbiota in patients with Parkinson’s disease in Southern China. Parkinsonism Relat. Disord. 53, 82–88 (2018).
Lin, C. H. et al. Altered gut microbiota and inflammatory cytokine responses in patients with Parkinson’s disease. J. Neuroinflammation 16, 129 (2019).
Qian, Y. et al. Gut metagenomics-derived genes as potential biomarkers of Parkinson’s disease. Brain 143, 2474–2489 (2020).
Cosma-Grigorov, A. et al. Changes in gastrointestinal microbiome composition in PD: a pivotal role of covariates. Front. Neurol. 11, 1041 (2020).
Mirzaei, R. et al. Role of microbiota-derived short-chain fatty acids in nervous system disorders. Biomed. Pharmacother. 139, 111661 (2021).
Li, W. et al. Structural changes of gut microbiota in Parkinson’s disease and its correlation with clinical features. Sci. China Life Sci. 60, 1223–1233 (2017).
Hester, C. M. et al. Fecal microbes, short chain fatty acids, and colorectal cancer across racial/ethnic groups. World J. Gastroenterol. 21, 2759–2769 (2015).
Weis, S. et al. Effect of Parkinson’s disease and related medications on the composition of the fecal bacterial microbiota. NPJ Parkinsons Dis. 5, 28 (2019).
Ahmed, S. et al. The Weissella Genus: clinically treatable bacteria with antimicrobial/probiotic effects on inflammation and cancer. Microorganisms. 10, 2427 (2022).
Zgheib, R. et al. Neglectibacter timonensis gen. nov., sp. nov. and Scatolibacter rhodanostii gen. nov., sp. nov., two anaerobic bacteria isolated from human stool samples. Arch. Microbiol. 204, 45 (2021).
Kim, C. H. Complex regulatory effects of gut microbial short-chain fatty acids on immune tolerance and autoimmunity. Cell Mol. Immunol. 20, 341–350 (2023).
Akinsuyi, O. S. & Roesch, L. F. W. Meta-analysis reveals compositional and functional microbial changes associated with osteoporosis. Microbiol. Spectr. 11, e0032223 (2023).
Eom, J. A. et al. Diet-regulating microbiota and host immune system in liver disease. Int. J. Mol. Sci. 22, 6326 (2021).
Lee, D. W. et al. Agathobaculum butyriciproducens shows neuroprotective effects in a 6-OHDA-induced mouse model of Parkinson’s disease. J. Microbiol. Biotechnol. 32, 1168–1177 (2022).
Hampson, R. K., Barron, L. L. & Olson, M. S. Stimulation of the glycine cleavage system by short-chain fatty acids in isolated rat liver mitochondria. Biochemistry 23, 4604–4610 (1984).
Kalyanaraman, B., Cheng, G. & Hardy, M. Gut microbiome, short-chain fatty acids, alpha-synuclein, neuroinflammation, and ROS/RNS: Relevance to Parkinson’s disease and therapeutic implications. Redox Biol. 71, 103092 (2024).
Peterson, C. T. et al. Short-chain fatty acids modulate healthy gut microbiota composition and functional potential. Curr. Microbiol. 79, 128 (2022).
Rios-Covian, D. et al. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 7, 185 (2016).
Almarhoumi, R. et al. Microglial cell response to experimental periodontal disease. J. Neuroinflammation 20, 142 (2023).
Gong, J. et al. Mesenteric adipose tissue-derived Klebsiella variicola disrupts intestinal barrier and promotes colitis by type VI secretion system. Adv. Sci. 10, e2205272 (2023).
Akine, D. et al. Post-surgical meningitis caused by Klebsiella variicola. IDCases 18, e00622 (2019).
Long, D. L. et al. Fatal community-acquired bloodstream infection caused by Klebsiella variicola: a case report. World J. Clin. Cases 10, 2474–2483 (2022).
Clinton, L. K. et al. Synergistic Interactions between Abeta, tau, and alpha-synuclein: acceleration of neuropathology and cognitive decline. J. Neurosci. 30, 7281–7289 (2010).
Marsh, S. E. & Blurton-Jones, M. Examining the mechanisms that link beta-amyloid and alpha-synuclein pathologies. Alzheimers Res. Ther. 4, 11 (2012).
Yin, L. et al. Transcriptome response of a new serotype of avian type Klebsiella varicella strain to chicken sera. Res. Vet. Sci. 145, 222–228 (2022).
Hall, D. A. et al. An open label, non-randomized study assessing a prebiotic fiber intervention in a small cohort of Parkinson’s disease participants. Nat. Commun. 14, 926 (2023).
Pilarczyk-Zurek, M., Sitkiewicz, I. & Koziel, J. The clinical view on Streptococcus anginosus group—opportunistic pathogens coming out of hiding. Front. Microbiol. 13, 956677 (2022).
Madathil, S. et al. Central nervous system infections due to streptococcus anginosus group: a single-center case series. J. Child Neurol. 37, 210–217 (2022).
Deutschmann, M. W. et al. The significance of Streptococcus anginosus group in intracranial complications of pediatric rhinosinusitis. JAMA Otolaryngol. Head. Neck Surg. 139, 157–160 (2013).
Verma, S. et al. Presumed hydrogen sulfide-mediated neurotoxicity after Streptococcus anginosus group meningitis. Pediatr. Infect. Dis. J. 32, 189–191 (2013).
Yoshida, Y. et al. Differences in the betaC-S lyase activities of viridans group streptococci. Biochem. Biophys. Res. Commun. 300, 55–60 (2003).
Ndongo, S. et al. ‘Collinsella phocaeensis’ sp. nov., ‘Clostridium merdae’ sp. nov., ‘Sutterella massiliensis’ sp. nov., ‘Sutturella timonensis’ sp. nov., ‘Enorma phocaeensis’ sp. nov., ‘Mailhella massiliensis’ gen. nov., sp. nov., ‘Mordavella massiliensis’ gen. nov., sp. nov. and ‘Massiliprevotella massiliensis’ gen. nov., sp. nov., 9 new species isolated from fresh stool samples of healthy French patients. N. Microbes N. Infect. 17, 89–95 (2017).
Murros, K. E. Hydrogen sulfide produced by gut bacteria may induce Parkinson’s disease. Cells. 11, 978 (2022).
Rumbeiha, W. et al. Acute hydrogen sulfide-induced neuropathology and neurological sequelae: challenges for translational neuroprotective research. Ann. N. Y Acad. Sci. 1378, 5–16 (2016).
Alharbi, B. et al. Role of GABA pathway in motor and non-motor symptoms in Parkinson’s disease: a bidirectional circuit. Eur. J. Med. Res. 29, 205 (2024).
Chen, Y., Xu, J. & Chen, Y. Regulation of neurotransmitters by the gut microbiota and effects on cognition in neurological disorders. Nutrients. 13, 2099 (2021).
van Nuland, A. J. M. et al. GABAergic changes in the thalamocortical circuit in Parkinson’s disease. Hum. Brain Mapp. 41, 1017–1029 (2020).
Gao, J. et al. Glutamate and GABA imbalance promotes neuronal apoptosis in hippocampus after stress. Med Sci. Monit. 20, 499–512 (2014).
Mele, M., Costa, R. O. & Duarte, C. B. Alterations in GABA(A)-receptor trafficking and synaptic dysfunction in brain disorders. Front Cell Neurosci. 13, 77 (2019).
Lakhani, R. et al. Defects in GABA metabolism affect selective autophagy pathways and are alleviated by mTOR inhibition. EMBO Mol. Med 6, 551–566 (2014).
Tong, B. et al. Targeting dysregulated lipid metabolism for the treatment of Alzheimer’s disease and Parkinson’s disease: Current advancements and future prospects. Neurobiol. Dis. 196, 106505 (2024).
Bogie, J. F. J. et al. Fatty acid metabolism in the progression and resolution of CNS disorders. Adv. Drug Deliv. Rev. 159, 198–213 (2020).
Calder, P. C. Sir David Cuthbertson Medal Lecture. Immunomodulatory and anti-inflammatory effects of n-3 polyunsaturated fatty acids. Proc. Nutr. Soc. 55, 737–774 (1996).
Ruiz-Pozo, V. A. et al. The molecular mechanisms of the relationship between insulin resistance and Parkinson’s disease pathogenesis. Nutrients. 15, 3585 (2023).
Whittle, B. J. et al. Early-stage idiopathic Parkinson’s disease is associated with reduced circular RNA expression. NPJ Parkinsons Dis. 10, 25 (2024).
Katz, K. et al. The Sequence Read Archive: a decade more of explosive growth. Nucleic Acids Res. 50, D387–D390 (2022).
Mysara, M. et al. From reads to operational taxonomic units: an ensemble processing pipeline for MiSeq amplicon sequencing data. Gigascience 6, 1–10 (2017).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Pruesse, E. et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196 (2007).
Mysara, M. et al. IPED: a highly efficient denoising tool for Illumina MiSeq Paired-end 16S rRNA gene amplicon sequencing data. BMC Bioinforma. 17, 192 (2016).
Mysara, M. et al. CATCh, an ensemble classifier for chimera detection in 16S rRNA sequencing studies. Appl. Environ. Microbiol. 81, 1573–1584 (2015).
Edgar, R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998 (2013).
Wang, Q. et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007).
Fox, J. & Fox, M. The car Package. 2001.
Mann, H. B. & Whitney, D. R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 18, 50–60 (1947).
McMurdie, P. J. & Holmes, S. Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. Pac. Symp. Biocomput. 235-246 (2012).
Simpson, E. H. Measurement of diversity. Nature 163, 688–688 (1949).
Lagkouvardos, I. et al. Rhea: a transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. PeerJ 5, e2836 (2017).
De Faveri, J. et al. Residual variance–covariance modelling in analysis of multivariate data from variety selection trials. J. Agric. Biol. Environ. Stat. 22, 1–22 (2017).
Anderson, M. J., Permutational Multivariate Analysis of Variance (PERMANOVA), in Wiley StatsRef: Statistics Reference Online. p. 1-15.
Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003).
Lin, H. & Peddada, S. D. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 11, 3514 (2020).
Bollen, K. A. & Brand, J. E. A general panel model with random and fixed effects: a structural equations approach. Soc. Forces 89, 1–34 (2010).
Cole, J. R. et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642 (2014). (Database issue).
Team, R. C., R: A language and environment for statistical computing. 2022, Vienna, Austria: R Foundation for Statistical Computing.
Asshauer, K. P. et al. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31, 2882–2884 (2015).
Parks, D. H. et al. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124 (2014).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Dhariwal, A. et al. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 45, W180–W188 (2017).
Chen, T. & Guestrin, C. XGBoost: A Scalable Tree Boosting System, in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2016, Association for Computing Machinery: San Francisco, California, USA. p. 785–794.
Rokach, L. & Maimon, O. Decision Trees, in Data Mining and Knowledge Discovery Handbook, O. Maimon and L. Rokach, Editors. 2005, Springer US: Boston, MA. p. 165-192.
Tin Kam, H. Random decision forests. In: Proc. of 3rd International Conference on Document Analysis and Recognition. (1995).
Acknowledgements
I would like to acknowledge M Oweda, O. Saeed, Marawan Yasser, MK Khaled, Omar M El-Habbak, Mohamed Yousry, and Omar Khaled for their collaboration and significant contributions to this research.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
Conceptualization—M.M. & M.E., Design of work —M.M. & M.E. Data analysis—N.H. & H.H. Data visualization—N.H. Data interpretation—R.R., M.M., H.H. & N.H. Writing—N.H., R.R. & M.M. Review—H.H., R.R., M.M. & M.E. All authors read, gave input, and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Marzouk, N.H., Rashwan, H.H., El-Hadidi, M. et al. Proinflammatory and GABA eating bacteria in Parkinson's disease gut microbiome from a meta-analysis perspective. npj Parkinsons Dis. 11, 145 (2025). https://doi.org/10.1038/s41531-025-00950-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41531-025-00950-z
This article is cited by
-
Insights into the tripartite relationship between cervical cancer, human papillomavirus, and the vaginal microbiome: a mega-analysis
Human Genomics (2025)
-
Unravelling novel microbial players in the breast tissue of TNBC patients: a meta-analytic perspective
npj Biofilms and Microbiomes (2025)