Abstract
Oligodendroglial pathology is a hallmark of multiple system atrophy (MSA), yet it remains unclear whether MRI-detected atrophy reflects underlying biological mechanisms. This study investigated whether regional atrophy aligns with gene expression and neurotransmitter systems. We recruited 65 MSA patients and derived brain atrophy measures from T1-weighted MRIs. Using postmortem data from the Allen Human Brain Atlas, partial least squares (PLS) regression identified gene expression components associated with brain atrophy. Gene enrichment analyses explored biological processes, and annotation mapping identified neurotransmitter systems matching atrophy patterns. Specificity was tested against 57 patients with Parkinson’s disease. Atrophy primarily affected the cerebellar white matter, pons, putamen, olive, and substantia nigra. PLS revealed two latent variables explaining 27.5% of the covariance. Atrophic regions overexpressed genes linked to mitochondrial function and oligodendrocytes, showing patterns distinct from Parkinson’s disease. These regions also exhibited lower serotonin and GABA levels, and higher acetylcholine and noradrenaline receptor densities. MRI-derived atrophy in MSA is biologically grounded and may inform future therapeutic studies.
Similar content being viewed by others
Introduction
Multiple system atrophy (MSA) is a rare, rapidly progressing synucleinopathy with a poor prognosis, characterized by autonomic failure and varying degrees of motor impairment, including poorly levodopa-responsive parkinsonism in the parkinsonian variant (MSAp) and predominant cerebellar symptoms in the cerebellar variant (MSAc)1. α-synuclein aggregates mainly within the cytoplasm of oligodendrocytes, forming glial cytoplasmic inclusions2,3. Although mostly sporadic, some rare familial cases linked MSA to SNCA, MAPT, and COQ2 gene variants4,5. However, the biological mechanisms underlying selective brain vulnerability in MSA remain poorly understood.
Neuroimaging techniques, such as magnetic resonance imaging (MRI) and positron emission tomography (PET) provide sensitive, non-invasive biomarkers of disease-related brain changes6,7. Yet, these imaging-derived phenotypes remain largely descriptive and offer limited mechanistic insight into why certain brain regions are more susceptible to degeneration. The emerging field of imaging transcriptomics addresses this gap by linking spatial patterns of neuroimaging abnormalities to normative gene expression profiles from healthy postmortem human brains8,9,10,11. The core rationale of imaging transcriptomics is to move beyond purely descriptive neuroimaging findings and gain insight into the biological underpinnings of regional vulnerability in neurodegenerative diseases. In this context, MRI-derived atrophy patterns are not viewed as endpoints in themselves, but as in vivo phenotypic readouts of latent molecular predispositions. If regions showing pronounced atrophy overexpress genes involved in specific biological pathways, this suggests that neurodegeneration targets pre-existing transcriptomic landscape, thus positioning neuroimaging findings as biologically grounded and mechanistically interpretable markers of disease.
This approach has already yielded disease-specific insights into synucleinopathies and other disorders. In Parkinson’s disease (PD), regional brain iron deposition has been associated with genes involved in metal detoxification and synaptic function12 while cortical atrophy progression was associated with mitochondrial13 and synaptic gene expression14 and was reduced in regions enriched with oligodendrocyte and endothelial cell markers14. In isolated rapid eye movement sleep behavior disorder (iRBD), a prodromal synucleinopathy15, regions showing cortical thinning overexpressed genes related to mitochondrial function and macroautophagy16. Imaging transcriptomics has also demonstrated specificity: in Alzheimer’s disease, atrophy correlates with regions overexpressing genes of the protein remodeling complex, with APOE (coding for apolipoprotein E) emerging as a key contributor16. Similarly, kidney-brain axis-related neurodegeneration was shown to follow spatial gene expression patterns, including AGT (coding for angiotensinogen)17, involved in vascular regulation18.
Beyond transcriptomics, molecular annotation mapping enables the comparison of neuroimaging patterns with PET-based receptor density maps19. In iRBD, atrophy maps overlap with neurotransmitter systems, such as dopamine, serotonin, and noradrenaline16. These complementary approaches provide a framework for elucidating the molecular and neurochemical architecture of regional vulnerability. However, they have not been applied to MSA, which remains poorly understood at the mechanistic level and urgently requires novel therapeutic targets.
In this study, our objective was to quantify the spatial pattern of brain atrophy in MSA and its subtypes and apply imaging transcriptomics and PET-based annotation mapping to uncover the molecular and neurochemical signatures of vulnerable brain regions. Using the Allen Human Brain Atlas, we identified gene expression components aligned with MSA-related atrophy and performed gene enrichment analyses to characterize the underlying biological processes. We further assessed which neurotransmitter systems best matched the spatial distribution of atrophy. To evaluate disease specificity, we repeated the same analyses in PD. We hypothesized that atrophy in MSA would primarily involve regions enriched in oligodendrocyte-related gene expression, but also reveal additional disease-relevant molecular and neurochemical mechanisms contributing to neurodegeneration beyond oligodendroglial dysfunction.
Results
Participants
The study included 69 patients with MSA, 57 with PD and 190 HCs. Of these, 13 (4 MSA, 9 HCs) failed image processing or quality control, resulting in 303 participants, namely 65 MSA patients, 57 PD patients, and 181 HCs. The MSA group included 34 MSAp, 22 MSAc, and 9 mixed. Average disease duration was 4.0 ± 2.2 years. There were no significant between-group differences in age, sex, disease duration or UPDRS scores. MSA patients had lower LEDD values (p < 0.0001) and higher Hoehn and Yahr stages (p < 0.0001) compared to PD patients. As expected, patients with MSAc (10.8 ± 6.2) and mixed MSA (16.0 ± 4.2) had higher cerebellar Parkinson Plus Scale (PPS) subscores than those with MSAp (0.8 ± 1.2, p = 0.002). No other group differences were seen (Table 1).
MSA patients show subcortical and cortical brain atrophy
Compared to HCs, MSA patients had severe deep brain atrophy, particularly in the cerebellar white matter and cortex, pons, putamen, superior and inferior olive, and SN (all PFDR < 0.0001, Table S1). Asymmetry was only observed in the inferior olive (PFDR < 0.0001). Cortical atrophy was seen in 46 (68%) regions, particularly in the frontal and parietal cortices. The most atrophic regions were the bilateral precentral and caudal middle frontal cortices, the right pars opercularis and supramarginal cortices, the left precuneus, inferior parietal, paracentral, and rostral middle frontal cortices (PFDR < 0.0001, Fig. 1, Table S1). No asymmetry was found in cortical atrophy.
Average regional W-scores for cortical (top row) and deep brain regions (bottom row) were projected onto a brain volume for all MSA patients (a), MSAp (b), and MSAc (c). Red represent lower W-scores relative to HCs (greater atrophy); white indicate no significant atrophy. Only regions with significant W-score differences are shown. The same color scale was used for cortical and deep brain regions to ensure comparability of the magnitude of atrophy. HC healthy control, MSA multiple system atrophy, MSAp parkinsonian variant of MSA, MSAc cerebellar variant of MSA.
When comparing MSAc and MSAp groups, as expected, MSAp patients exhibited more atrophy in the putamen (PFDR < 0.0001) and caudate (left: PFDR = 0.006, right: PFDR = 0.04, Table S2). MSAc patients had greater atrophy in the cerebellar white matter (PFDR < 0.0001) and cortex (PFDR < 0.0001), followed by the pons (PFDR < 0.0001), midbrain (PFDR < 0.02), superior cerebellar peduncles (PFDR = 0.02), and superior olive (PFDR ≤ 0.008). No differences were found in the SN, inferior olive, or cortical regions between MSAp and MSAc patients (Table S2).
MRI subcortical atrophy correlates with disease severity
We found significant negative correlation after adjusting for age and sex between UPDRS III scores and atrophy in the bilateral putamen (right: r = -0.49, PFDR = 0.02, left: r = -0.48, PFDR = 0.02), left pallidum (r = -0.53, PFDR = 0.01), left caudate (r = -0.38, PFDR = 0.049), bilateral inferior olive (right: r = -0.46, PFDR = 0.02, left r = -0.43, PFDR = 0.02), and medulla (r = -0.45, PFDR = 0.02) (Fig. 2a). When further adjusting for disease duration and LEDD values, four correlations remained significant, including the bilateral putamen (right: r = -0.51, PFDR = 0.01, left: r = -0.51, PFDR = 0.01), left pallidum (r = -0.51 PFDR = 0.01) and right inferior olive (r = -0.43, PFDR = 0.04).
a Scatterplots with linear regression lines showing the associations between individual regional W-scores in deep brain regions and UPDRS III scores. Pearson’s partial correlation coefficients (r) after adjustment by age and sex and p values after FDR correction are reported. Only significant correlations are plotted. b Heatmaps showing correlations between individual regional W-scores in deep brain regions and PPS total scores and autonomic, cerebellar, oculomotor and motor subscores. Pearson’s partial correlation coefficients (r) adjusted for age and sex and p values after FDR correction are reported. The color scale indicates the strength and direction of the correlation (blue for negative and red for positive correlations). Of note, PPS scores were only available for a subset of participants (n = 20). **0.001 < PFDR ≤ 0.01; *0.01 < PFDR ≤ 0.05; °PFDR ≤ 0.10. FDR false discovery rate, ION inferior olive nucleus, MSA multiple system atrophy, PPS Parkinson Plus Scale, SN substantia nigra, SON superior olive nucleus, UPDRS Unified Parkinson’s Disease Rating Scale.
The total PPS scores negatively correlated with atrophy in the bilateral SN (right: r = -0.61, PFDR = 0.048, left: r = -0.58, PFDR = 0.03) and right inferior olive (right: r = -0.64, PFDR = 0.04). In addition, the cerebellar PPS subscore strongly correlated with the pons (r = -0.75, PFDR = 0.01) cerebellar white matter (right: r = -0.80, PFDR = 0.007, left: r = -0.74, PFDR = 0.01), bilateral SN (right: r = -0.61, PFDR = 0.04, left: r = -0.66, PFDR = 0.04), inferior olive (right: r = -0.69, PFDR = 0.02, left: r = -0.63, PFDR = 0.04), and superior olive (right: r = -0.72, PFDR = 0.02, left: r = -0.71, PFDR = 0.02). The motor subscore also correlated with the left putamen (r = -0.67, PFDR = 0.03) and left SN (r = -0.66, PFDR = 0.03; Fig. 2b). After adjusting for disease duration and LEDD values, only two associations remained significant: between the cerebellar subscore and right cerebellar white matter atrophy (r = -0.74, PFDR = 0.0498), and between the motor subscore and left substantia nigra atrophy (r = -0.77, PFDR = 0.045).
No significant correlations were found between subcortical atrophy and either disease duration or LEDD values. Similarly, cortical W-scores did not correlate significantly with any clinical variables, including MMSE scores.
The brain’s gene expression spatial distribution predicts atrophy in MSA
We investigated the relationship between deep brain atrophy and the brain’s spatial gene expression distribution. Two LVs were significant, explaining 28% of the covariance in gene-atrophy compared to random (LV3: 19.8% of covariance explained vs. 11.3% in null models, P = 0.03; LV5: 7.6 vs. 3.7%, P = 0.02) and spatial null models (LV3: 12.1%, P = 0.04; LV5: 3.7%, P = 0.01; Fig. 3a). For both variables, regions with more atrophy had more negative weights (LV3: r = 0.44, P < 0.001; LV5: r = 0.28, P = 0.05), meaning that negatively-weighted genes were more expressed in atrophic regions (Fig. 3b, d; Fig. S1).
a PLS regression results on deep brain regions with violin plots representing the covariance percentage in W-scores explained by gene expression (y-axis). Black dots indicate empirical covariance, asterisks indicate significant latent variables (LV) against random (rd) and spatial (sp) null models (x-axis). b, d Scatterplots of the correlation between W-scores associated with deep brain regions (x-axis) and regional weights of LV3 and LV5 (y axis). c, e Histograms of bootstrapped gene weights on LV3 and LV5, with red weights indicating robust association with regions with greater atrophy (below -3.29). LV latent variable, PLS partial least square rd random null models, sp spatial null models.
MRI atrophy in MSA is primarily associated with mitochondrial function
Next, we performed GSEA to identify biological processes, cellular components, and human disease terms enriched among genes most strongly associated with brain atrophy in MSA. For genes negatively weighted on LV3, 652 genes (4.2%) were significantly associated with atrophy after bootstrapping (Fig. 3c, e). Strikingly, the most significantly enriched biological processes in atrophic regions were related to mitochondrial function. These included proton transmembrane transport, complex I assembly, electron transport chain, mitochondrial transport, and mitochondrion organization (all PFDR < 0.0001). Other key processes included energy derivation by oxidation of organic compounds (PFDR < 0.001) and nucleoside triphosphate metabolic process (PFDR < 0.001) (Table S3, Fig. 4a). Among biological processes, ensheathment of neurons (a function primarily mediated by oligodendrocytes) was also significantly enriched, but ranked twelfth among all significant terms, indicating that although oligodendrocyte-related functions were involved in regions showing neurodegeneration, mitochondrial processes are more prominently associated with regional vulnerability in MSA. Regarding cellular components, atrophic regions were enriched for genes associated with the mitochondrial protein-containing complex, respirasome, oxidoreductase complex, mitochondrial inner membrane, and myelin sheath (all PFDR < 0.0001; Fig. 4b, Table S3). Atrophic regions also overexpressed terms related to mitochondrial diseases (Table S3). Similar findings were obtained when performing GSEA using the PANTHER gene enrichment platform (Table S4).
Significant biological processes (a), cellular components (b), and tissue cell types (c) enriched in the negatively-weighted genes associated with deep brain atrophy in MSA. Terms are ranked based on the normalized enrichment score; all enriched terms were significant after FDR correction. Only the top 15 terms are shown for visualization purposes. FDR false discovery rate, NES normalized enrichment score.
For genes negatively weighted on LV5, 219 (1.4%) were associated with greater atrophy (Fig. 3e). GSEA revealed similar findings as in LV3, with processes related to energy production and metabolism. Additional sub-analyses on the MSA subgroups separately also identified terms related to mitochondrial functions in both subgroups (Table S5).
MRI atrophy in MSA relates to cell-type vulnerability
Using GSEA, cell type-related genes were overexpressed in relation to atrophy in MSA, including brain-related oligodendrocytes and endothelial cells (both PFDR < 0.001) (Fig. 4c). Similarly, using single-cell RNA sequencing specific to neural cells, we found a negative correlation between W-scores in deep brain regions and oligodendrocyte gene expression (r = -0.39, P = 0.0006; PFDR spatial=0.002), suggesting higher gene expression of these cell types in atrophic areas (Table S6). These results support that atrophic regions overexpress gene expression profiles related to oligodendrocytes. The same enrichment profile was seen in MSAp and MSAc subgroups.
Using Webgestalt, we next investigated the genes driving the cell-type enrichment in atrophic regions. For oligodendrocyte-related genes, among the eight top genes identified, several were implicated in myelin synthesis, assembly and regulation, including Leucine-rich repeat LGI family member 3 (LGI3), proteolipid protein 1 (PLP1), and plasmolipin (PLLP) from LV3; and myelin-associated glycoprotein (MAG), gap junction protein beta 1 (GJB1), and myelin-associated oligodendrocyte basic protein (MOBP) from LV5 (Table S7).
Specificity analysis: comparison with PD
To assess the specificity of these findings to MSA, we repeated the PLS regression in 57 PD patients age- and sex-matched to the MSA patients. We found two significant LVs explaining in 20.0% of the gene expression-atrophy covariance in PD. Regions associated with negatively-weighted genes (atrophy) in PD were enriched for synaptic functions. In terms of cellular components, terms related to synapses were significant, including postsynaptic specialization, neuron spine, neuron-to-neuron synapse, and GABA-ergic synapse (Fig. S2). There was no significant enrichment of cell types, including oligodendrocytes. These results suggest that atrophy patterns in MSA and PD may be associated with distinct underlying molecular profiles in the healthy brain.
Brain atrophy maps onto specific neurotransmitter systems
We further tested whether the pattern of cortical and deep brain regions atrophy in MSA mapped onto specific neurotransmitter systems. Atrophic regions had lower density of serotonin receptors, namely 5-HT1A (r = 0.75, P < 0.0001; PFDR spatial<0.0001) and 5-HT2A (r = 0.36, P = 0.001; PFDR spatial =0.046), and GABAA/BZ receptors (r = 0.42, P < 0.0001; PFDR spatial=0.046), as well as a higher density of α4β2 acetylcholine (r = -0.61, P < 0.0001; PFDR spatial <0.0001) and NET noradrenaline transporters (r = -0.36, P = 0.001; PFDR spatial=0.046) (Fig. S3, Table S8).
Discussion
Our study provides novel evidence that MRI-derived brain atrophy patterns observed in MSA reflect specific underlying biological mechanisms. The pathological involvement of oligodendrocytes is well established, given their role in glial cytoplasmic inclusions, but the molecular underpinnings of MRI-derived atrophy patterns have remained poorly characterized. Here, we showed that regions most affected by atrophy in MSA, including the putamen, cerebellar white matter, and pons, are not only enriched for oligodendrocyte-related gene expression, but also display strong overexpression of genes involved in mitochondrial respiratory chain function. This finding suggests that MRI atrophy in MSA is not merely a structural readout, but a phenotypic manifestation of region-specific molecular vulnerability. In addition, these atrophic regions exhibited a distinct neurochemical profile, with increased densities of acetylcholine and noradrenaline receptors/transporters and decreased densities of serotonin and GABA receptors. This extends current understanding of selective vulnerability in MSA by integrating transcriptomic and neurochemical context and supports imaging transcriptomics as an approach to anchor neuroimaging observations in both established and novel disease-relevant biology.
As expected, atrophy in MSA was more severe in deep brain regions, mainly involving the putamen in the parkinsonian variant, and the cerebellum, pons, and olive in the cerebellar subtype6,7. Subcortical atrophy was significantly associated with disease severity scores of motor and cerebellar dysfunction, supporting the functional relevance of these structural changes. In contrast, no significant correlation was found with autonomic dysfunction scores. This lack of association may be due to limited statistical power. Notably, a recent study linked gray matter loss in the medulla and cerebellum (cerebellar cortex and deep cerebellar nuclei) with cardiovascular autonomic failure20, suggesting that more targeted or higher-resolution analyses may be required to detect such associations. Atrophy was also present in specific cortical areas, notably in the bilateral precentral and caudal middle frontal cortices, the right pars opercularis region, and the left precuneus, in line with previous studies21. The involvement of the precentral cortices, more prominent in MSAp than MSAc, may reflect extrapyramidal and corticospinal involvement. Subcortical degeneration may alter cortico-subcortical loops, potentially leading to structural reorganization in connected cortical structures, and contributing to cortical atrophy21. Moreover, MSA patients often present with pyramidal signs1, reflecting corticospinal tract damage, as reported in neuropathological22 and MRI studies23.
Using imaging transcriptomics, we found that atrophic regions overexpressed genes involved in mitochondrial assembly and functioning, suggesting that mitochondrial dysfunction represents an important factor of selective vulnerability to neurodegeneration. Mitochondria play a crucial role in several neurodegenerative diseases24. As for PD25, there is evidence of mitochondrial dysfunction in the pathogenesis of MSA based on genome-wide association studies4, postmortem studies25,26, and cell27,28 and animal models24. Mutations in the COQ2 gene, encoding an enzyme involved in Coenzyme Q10 (CoQ10) biosynthesis, have been linked to familial and sporadic cases of MSA4. CoQ10 is found in the inner mitochondrial membrane and is involved in the transfer of electrons from complexes I/II to complex III, thus playing an essential role in the functioning of the respiratory chain and the ATP production. Impaired CoQ10 activity increases vulnerability to oxidative stress24,25. Autopsy studies have shown reduced CoQ10 levels in the cerebrospinal fluid29 and blood30 of MSA patients as well as in cerebellar25,26 and motor cortex postmortem samples26. A study investigating mitochondrial function within dopaminergic neurons derived from induced pluripotent stem cells (iPSCs) of MSA patients found evidence of impaired respiratory chain activity and up-regulation of CoQ10 biosynthesis, the latter indicating possible compensatory mechanism27. Another study found that oxidative stress exposure in neural progenitor cells derived from MSA patient iPSCs triggered excessive generation of reactive oxygen species and cell damage28. In line with these findings, a recent placebo-controlled phase 2 trial tested the efficacy of Ubiquinol, a coenzyme Q10 supplementation, in MSA patients and showed a significantly smaller decline of the Unified MSA Rating Scale (UMSARS) in the treated group31, further supporting the evidence of mitochondrial dysfunction in MSA.
An expected finding was the overexpression of oligodendrocyte-related genes in regions showing greater atrophy in MSA patients, aligning with the fact that oligodendrocytes are the primary target of α-synuclein aggregates in MSA2,3. Several of the oligodendrocyte-related genes identified by our GSEA are implicated in myelin synthesis, assembly and regulation. Interestingly, the PLP1 gene encodes the predominant component of myelin, which is expressed almost exclusively in mature oligodendrocytes. PLP1 plays a critical role in myelin sheath stability, oligodendrocyte development, and axonal survival. Because the PLP1 promoter is active only in oligodendrocytes, driving expression of the human wild-type α-synuclein gene under this promoter in transgenic mice restricts α-synuclein production to oligodendrocytes. These PLP-α-synuclein mice recapitulate the oligodendroglial α-synuclein accumulation seen in MSA and are therefore used as models to study MSA mechanisms. These mice show glial cell inclusions as well as mild, progressive striatonigral degeneration, microgliosis, motor impairment, and features of autonomic failure32,33. Another gene of interest is LGI3, which encodes a secreted oligodendrocyte protein that binds the neuronal receptor ADAM23 to regulate potassium channels (Kv1) at the juxtaparanodes of myelinated axons. In mice, LGI3 deficiency disrupts juxtaparanodal clustering of ADAM23 and Kv1 channels and suppresses Kv1-channel-mediated short-term synaptic plasticity, highlighting a mechanism of glia-neuron interaction34. GSEA revealed other genes not directly linked to known MSA, which warrant further investigation. Thus, imaging transcriptomics may allow generating a set of hypotheses about novel mechanisms potentially involved in disease pathophysiology.
The overexpression of oligodendrocyte-related genes in atrophic regions mirrors the association with genes enriched for mitochondrial functions given the energetic need of oligodendrocytes, especially during the process of myelination. Whether mitochondrial dysfunction is a primary mechanism underlying oligodendropathy or is secondary to other pathological processes, especially α-synuclein accumulation, remains to be determined. Indeed, α-synuclein-mediated oligodendroglial pathology contributes to neuronal damage, as supported by the positive correlation between neuronal loss and glial cytoplasmic inclusion density3. Oligodendrocytes are crucial not only in the formation of the myelin sheath, but also for providing trophic support to neurons. Factors released by oligodendroglial precursors and mature oligodendrocytes are essential for neuronal survival35. Consequently, the deficiency of oligodendroglia-derived neurotrophic factors resulting from oligodendroglial impairment may account for the neuronal loss24. In sum, our results support the theory that neurodegeneration in MSA may derive from neuronal energy failure and lack of trophic support from oligodendroglia. On the other hand, we also found enrichment for genes related to endothelial cells, consistent with pathological reports of microvascular involvement in MSA and suggesting glial-vascular interactions contributing to regional vulnerability36,37.
Our results resemble those seen in other synucleinopathies, namely PD and iRBD, showing that cortical brain atrophy occurs in regions overexpressing genes involved in mitochondrial function or synaptic functioning13,16. However, when repeating the same analyses on deep brain regions in PD, we found an overexpression of synapse-related genes, as in previous studies14. Interestingly, no mitochondria-related terms were associated with deep brain regions in PD. These findings suggest that the regional vulnerability to neurodegeneration in MSA and PD may be linked to distinct underlying molecular profiles in the healthy brain, highlighting potential mechanisms of differential vulnerability associated with these disorders, with the relationship between deep brain atrophy, mitochondrial function and oligodendrocytes being specific to MSA. This divergence between MSA and PD is biologically plausible given their distinct biology and our study design. A previous study found modest negative associations between longitudinal atrophy progression over 4 years and oligodendrocytes and endothelial cells (r = -0.15 and r = -0.11 respectively) in PD14, which was interpreted as possible neuroprotective role of oligodendrocytes via the synthesis of neurotrophic factors35. Thus, in PD, regions richer in oligodendrocytes may be relatively protected over the short to midterm, whereas oligodendroglia are a primary lesion site and track atrophy in MSA.
Furthermore, regions most vulnerable to atrophy in MSA colocalized with areas with higher distribution of NET (noradrenaline) and α4β2 (acetylcholine), and lower distribution of serotonin (5-HT1A/5-HT2A) and GABA receptors. These associations may provide clues about the neurochemical environment of regions that are preferentially vulnerable in MSA, supporting the idea that neurotransmitter systems may contribute to a biological substrate of local selective vulnerability. These findings may also support the involvement of multi-neurochemical systems related to neuronal cell loss across several neurotransmitter projection systems, as demonstrated by postmortem and PET studies38. To date, only one study applied the spatial mapping approach in MSA to investigate the neurochemical underpinnings of iron deposition patterns, demonstrating that regions with higher cortical iron content overlapped with areas of higher density of noradrenaline and acetylcholine receptors39. Central autonomic pathways, crucial for autonomic cardiovascular and respiratory control, are impaired in MSA due to central noradrenergic deficiency40. This denervation is reflected by reduced cerebrospinal fluid levels of noradrenaline, coupled with decreased noradrenaline density in the frontal cortex and putamen in post-mortem tissue analyses41. The locus coeruleus (LC) is the main source of noradrenaline in the brain. Noradrenergic neurons project throughout the brain and spinal cord, regulating arousal, attention, and stress responses42. Central noradrenergic deficiency, possibly due to LC damage, may contribute to baroreflex dysfunction and orthostatic hypotension in MSA. The LC is also known to project to the nucleus of the solitary tract, where all baroreceptor afferents initially synapse in the brain, and to the rostral ventrolateral medulla, a major source of descending projections to sympathetic pre-ganglionic neurons, crucial for tonic maintenance of sympathetic vasomotor tone and blood pressure control43. The association between atrophy and higher cholinergic receptor density shown here aligns with postmortem studies demonstrating severe cholinergic neurons depletion in the pedunculopontine and laterodorsal tegmental nuclei and LC42. Similarly, using 1-[11C]Methylpiperidin-4-yl propionate, a PET radiotracer targeting acetylcholinesterase, a cholinergic activity reduction was shown in MSAp patients, similar to PD in the cortex, but more pronounced in subcortical regions, potentially accounting for the gait and cognitive disturbances observed in MSAp44. Furthermore, there is evidence of serotonergic neuronal loss in the caudal brainstem raphe nucleus in MSA, as shown by neuropathological38 and PET imaging studies reporting decreased cortical serotonin transporter binding45. This may contribute to impaired autonomic and respiratory control.
Our study has limitations. First, we lacked pathological confirmation of diagnoses. Instead, we only included patients meeting criteria for clinically established MSA, which is currently the highest level of diagnostic certainty. Second, while the sample size was relatively small given the rarity of the disease, it still provides valuable insights into brain abnormalities. Third, the Unified MSA Rating scale was not available for our cohort; instead, we used UDPRS III scores and the PPS scale in a subset of patients. Although the UPDRS III is not MSA-specific and does not include dedicated items for cerebellar dysfunction, it provided a standardized measure of motor severity across the entire cohort. Furthermore, we performed a cross-sectional analysis on a cohort with variable disease duration and severity. Future longitudinal analyses will help investigate whether these molecular correlates are stage-dependent and how the spatial and biological landscape of atrophy evolves over time. Finally, gene expression data came from six healthy donors with varying ages and medical histories. Future studies should use transcriptomic atlases from a larger number of HCs closely matched to the MSA population.
To conclude, we showed that brain atrophy in MSA, which is associated with clinical disease severity, affects deep regions enriched in genes related to mitochondrial function and oligodendrocytes and aligns with specific neurochemical systems. Using only MRI-derived morphological markers, we identified biologically meaningful correlates of neurodegeneration in MSA. These results strengthen the link between imaging-derived phenotypes and underlying molecular mechanisms in MSA, and may open up new avenues for further investigating disease pathophysiology and therapeutic targets.
Methods
Participants
This is a retrospective single-center study of patients with clinically established MSA prospectively enrolled: 1) between 2007 and 2012 at the Paris Brain Institute-ICM as part of two research protocols (Genepark (LSHB-CT-2006-037544) and BBBIPPS (DGS 2006/0524)) and 2) between 2013 and 2020 in the movement disorders clinic at the Pitié-Salpêtrière Hospital. All cases were retrospectively reassessed by neurologists expert in movement disorders according to the most recent MDS criteria1. MRI findings (including atrophy, signal abnormalities or increased diffusivity of the putamen and/or infratentorial regions -pons, cerebellum, middle cerebellar peduncles) had to be consistent with the clinical diagnosis of MSA6,7. Participants were excluded if they had other neurological or psychiatric disorders, in case of evidence of vascular lesions on MRI (stroke, lacunar infarcts or Fazekas grade 3 leukopathy) or if MRI findings contradicted the clinical diagnosis (e.g., midbrain atrophy suggestive of PSP in a clinically diagnosed MSA patient). Based on neurological examination, patients were clinically classified as the parkinsonian subtype (MSAp), cerebellar subtype (MSAc), or mixed subtype. The following clinical variables were collected: disease onset (defined as the first occurrence of either motor symptoms or autonomic dysfunction), levodopa equivalent daily dose (LEDD), Unified PD Rating Scale (UPDRS)46 part III scores, Hoehn and Yahr stage47, Mini Mental State Examination (MMSE)48, and Parkinson Plus Scale (PPS)49, the latter being available for a subset of participants (n = 20).
We recruited age- and sex-matched healthy controls (HC) without a history of neurological or psychiatric disorders from the same sites and the Parkinson’s Progression Markers Initiative (PPMI) Database (RRID:SCR 006431, February 2024)50.
Additionally, patients with PD, diagnosed according to the current diagnostic clinical criteria51, were recruited through the Quebec Parkinson Network (QPN) at the Montreal Neurological Institute-MNI52 as a second control group. They were age- and sex-matched to the MSA patients.
The study was performed in accordance with the Declaration of Helsinki and was approved by the institutional ethical standard committee (Genepark: CPP Paris II, 2007-A00208-45; BBBIPPS: CPP Paris VI, P040410–65-06; Parkatypique: CPP Ile-de-France VI08012015; C-BIG general protocol: 2017-330, 15-944-MUHC; C-BIG imaging protocol: 2019-4759; QPN protocol: 2015-143, MP-CUSM-NEU-14-053, MP-37-2015-143). Participants gave written informed consent.
MRI acquisition
Participants were scanned on three 3 T MRI scanners (Siemens TRIO, Siemens SKYRA, General Electric SIGNA, 1.5 T General Electric OPTIMA) using a three-dimensional gradient-recalled echo T1-weighted sequence (Tables S9−S10, PPMI imaging protocols).
MRI processing
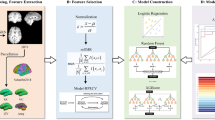
Figure 5 outlines the pipeline. FreeSurfer (v7.1.1) was used for cortical parcellation and volumetric segmentation of T1-weighted images53. Maps passing quality control were segmented to derive 68 bilateral cortical thickness measurements from the Desikan-Killiany atlas and 14 volume measurements from the bilateral subcortical structures (putamen, caudate, pallidum, thalamus, nucleus accumbens, amygdala, hippocampus). The brainstem was segmented into subregions (midbrain, pons, medulla, superior cerebellar peduncles) using FreeSurfer’s brainstem toolbox53. The cerebellum was segmented into 28 volumes with CerebNet(v.2.1.2)54 and the Schmahmann atlas55. All volumes were normalized by total intracranial volume.
a T1-weighted images from MSA patients and HCs were processed to derive cortical thickness, volume, and deformation metrics. Scan harmonization corrected inter-scanner variability, and regional values were converted to W-scores. For subsequent analyses, average W-scores across MSA patients were used for each brain region. b Regional gene expression values were extracted from postmortem brain data. Using PLS, the MSA brain atrophy vector was linked with gene expression, followed by gene set enrichment analysis to identify significant terms in MSA-relevant regions. HC healthy control, MSA multiple system atrophy, PLS Partial least squares regression, SD standard deviation, T1-w T1-weighted.
To extract morphological information from brainstem nuclei of interest, we performed deformation-based morphometry (DBM) on each subject’s T1-weighted image using the Computational Anatomy Toolbox (CAT12; r1742) in Statistical Parametric Mapping software (SPM12)56 and MATLAB (vR2019b). The Brainstem Navigator toolkit (v0.9)57,58 was applied to MNI-registered DBM maps to extract the extent of deformation in the substantia nigra (SN), inferior olive, and superior olive.
Bilateral regions were analyzed separately.
Processing of atrophy maps
Morphological values were harmonized using ComBat to remove scanner-related variability while preserving biological effects59. W-scoring was applied to ComBat-corrected values to remove the age and sex effects and derive deviations from what is expected for age and sex based on regressions generated within the HC group13,14,16. Negative W-scores indicate atrophy, whereas positive W-score indicate expansion.
Regional gene expression extraction
To characterize the gene expression patterns associated with atrophic regions, we performed an imaging transcriptomics analysis11. Regional expression values of >20,000 genes from the Allen Human Brain Atlas (AHBA)60 were extracted in atrophic regions for each post-mortem brain donor using abagen (v0.1.3)61 and then averaged across the six AHBA brains. Two cerebellar lobules which did not match with any AHBA region (vermis and vermis VII) were discarded, resulting in 26 cerebellar regions. The main analysis focused on deep brain regions using a region-by-gene expression matrix (50 regions, 15,611 genes), as atrophy in MSA is more prominent in subcortical than cortical regions, avoiding dilution of disease-specific components. Since only two brains had available right hemisphere gene expression values, measurements from the left hemisphere were mirrored onto the right hemisphere to ensure whole-brain transcriptomic coverage. These average gene expression values were used as predictors for the partial least squares regression.
Partial least squares regression
Partial least squares (PLS) regression was used to identify gene expression patterns associated with deep brain atrophy. PLS is a multivariate approach that identifies latent variables (LV) explaining maximal covariance between two matrices: average W-scores of atrophy (65 patients, 50 regions) and average gene expression values (15,611 genes, 50 regions). The matrices were multiplied, and the resulting correlation matrix was subjected to singular value decomposition. Significance of the LVs was assessed by comparing the empirical covariance explained by each LV to the covariance of 10,000 null models where atrophy was randomly permuted between regions (random null models). Given that the brain is characterized by a high degree of spatial autocorrelation between brain regions, the significance was also tested against 10,000 spatially-constrained null models generated with BrainSMASH62. A LV was considered significant if fewer than 5% of null models explained more covariance than the original LV (P < 0.05).
To identify the genes most robustly associated with each LV, we performed bootstrap resampling by randomly shuffling the matrix rows and repeating the PLS regression 5000 times to obtain bootstrap ratio weights. The ranked gene lists were used as inputs for gene set enrichment analysis.
Gene set enrichment analysis
To identify the biological processes, cellular components, and human diseases gene terms overexpressed in association with atrophy in MSA, we performed gene set enrichment analysis (GSEA) in WebGestalt 202463. Gene Ontology terms64 were used for biological processes and cellular components, and DisGeNET terms65 for human diseases. GSEA assessed whether negatively-weighted genes (associated with atrophy) were found more frequently within certain gene terms63. Only gene terms with a minimum of 20 and maximum of 2000 genes were considered. Significance was determined using 1000 random permutations with false discovery rate (FDR) correction for multiple comparisons. To ensure that our results were independent of the enrichment platform used, we repeated the GSEA using PANTHER (version 19.0, 20240619)66.
To identify which terms were specifically overexpressed in each MSA subgroup, we ran additional sub-analyses in each subgroup.
Cell type analysis
Two transcriptomic-based approaches were performed to identify the cell types whose genes were expressed in relation to atrophy. First, we used a GSEA as described above using WebGestalt 202463 and the Human Cell Landscape67, which analyzes cells from >50 human tissues. We further examined which genes were driving the cell-type enrichments in atrophic regions. Second, using single-cell RNA sequencing performed on cortical samples68, we extracted the gene expression associated with oligodendrocytes, oligodendrocyte progenitor cells, microglia, astrocytes, excitatory neurons, inhibitory neurons, and endothelial cells19. For each cell type, we computed the regional average expression across all genes and calculated Pearson’s correlations between the average gene expression and atrophy maps. Correlations were corrected for 7 comparisons with Bonferroni (P < 0.007), tested against 10,000 spatial null models, and resulting P values were FDR-corrected62.
Specificity analysis: comparison with PD
To test whether the gene enrichment patterns in MSA were disease-specific, we replicated the gene set enrichment analysis on a sample of PD patients.
Neurotransmitter mapping
We investigated whether atrophy in MSA occurred in regions overexpressing certain neurochemical systems. We parcellated and Z-scored the average regional density maps of 19 receptors, transporters, and binding sites collected from 1238 healthy individuals.These included dopamine (D1, D2, dopamine transporter [DAT]), serotonin (5-HT1A, 5-HT1B, 5-HT2A, 5-HT4, 5-HT6, serotonin transporter [5-HTT]), noradrenaline (noradrenaline transporter [NET]), acetylcholine (α4β2, vesicular acetylcholine transporter [VAChT], M1), GABA (GABAA/BZ), glutamate (mGluR5, NMDA), histamine (H3), endocannabinoids (CB1), and opioids (μ)19. We quantified receptor densities in the same regions where atrophy was measured, using neuromaps (Table S11)69. Cerebellar regions were excluded since PET images were normalized to the cerebellum for most tracers. Spearman’s correlation was calculated between each neurochemical regional density map and the average W-scores for all regions considered in the analysis. Correlations were corrected for 19 comparisons with Bonferroni (P < 0.003), tested against 10,000 spatial null models, and resulting P values were further FDR-corrected.
Specifics on the imaging and statistical methods are described in previous studies13,14,16,19.
Statistical analysis
Differences in sex proportion, age, and clinical scores were compared between groups using chi-squared test and Kruskal-Wallis test, respectively. Two-tailed one-sample t-tests with FDR correction were conducted on the W-scores to characterize the regions where MSA patients differed from HCs. Effects of laterality on the W-scores were investigated in MSA using two-tailed paired t-tests and FDR correction. Comparisons were also performed between the MSAp and MSAc subgroups.
Associations between clinical variables and individual atrophy W-scores in MSA patients were investigated using partial Pearson’s correlation coefficients, adjusting for age and sex with FDR correction. To account for the potential effects of disease duration and LEDD, we repeated sensitivity analyses including these variables as covariates.
Data availability
The data that support the findings of this study are not publicly available, but may be made available to qualified researchers on reasonable request from the corresponding author.
Code availability
The underlying code for this study is not publicly available, but may be made available to qualified researchers on reasonable request from the corresponding author.
References
Wenning, G. K. et al. The movement disorder society criteria for the diagnosis of multiple system atrophy. Movement Disorders mds.29005 https://doi.org/10.1002/mds.29005 (2022).
Dickson, D. W. Parkinson’s disease and parkinsonism: neuropathology. Cold Spring Harb. Perspect. Med. 2, a009258 (2012).
Jellinger, K. A. Neuropathology of multiple system atrophy: new thoughts about pathogenesis. Mov. Disord. 29, 1720–1741 (2014).
Multiple-System Atrophy Research Collaboration Mutations in COQ2 in familial and sporadic multiple-system atrophy. N. Engl. J. Med. 369, 233–244 (2013).
Sailer, A. et al. A genome-wide association study in multiple system atrophy. Neurology 87, 1591–1598 (2016).
Chougar, L., Pyatigorskaya, N., Degos, B., Grabli, D. & Lehéricy, S. The role of magnetic resonance imaging for the diagnosis of atypical Parkinsonism. Front. Neurol. 11, 665 (2020).
Chougar, L., Pyatigorskaya, N. & Lehéricy, S. Update on neuroimaging for categorization of Parkinson’s disease and atypical parkinsonism. Curr. Opin. Neurol. 54, 514–524 (2021).
Martins, D. et al. Imaging transcriptomics: convergent cellular, transcriptomic, and molecular neuroimaging signatures in the healthy adult human brain. Cell Rep. 37, 110173 (2021).
Arnatkeviciute, A., Fulcher, B., Bellgrove, M. & Fornito, A. Imaging transcriptomics of brain disorders. https://doi.org/10.31234/osf.io/4exug (2021).
Mroczek, M., Desouky, A. & Sirry, W. Imaging transcriptomics in neurodegenerative diseases. J. Neuroimag. 31, 244–250 (2021).
Arnatkeviciute, A., Markello, R. D., Fulcher, B. D., Misic, B. & Fornito, A. Toward best practices for imaging transcriptomics of the human brain. Biol. Psychiatry 93, 391–404 (2023).
Thomas, G. E. C. et al. Regional brain iron and gene expression provide insights into neurodegeneration in Parkinson’s disease. Brain 144, 1787–1798 (2021).
Vo, A. et al. Network connectivity and local transcriptomic vulnerability underpin cortical atrophy progression in Parkinson’s disease. NeuroImage: Clin. 40, 103523 (2023).
Tremblay, C. et al. Brain atrophy progression in Parkinson’s disease is shaped by connectivity and local vulnerability. Brain Commun. 3, fcab269 (2021).
Galbiati, A., Verga, L., Giora, E., Zucconi, M. & Ferini-Strambi, L. The risk of neurodegeneration in REM sleep behavior disorder: a systematic review and meta-analysis of longitudinal studies. Sleep. Med. Rev. 43, 37–46 (2019).
Rahayel, S. et al. Mitochondrial function-associated genes underlie cortical atrophy in prodromal synucleinopathies. Brain 146, 3301–3318 (2023).
Rahayel, S. et al. Lower estimated glomerular filtration rate relates to cognitive impairment and brain alterations. Alzheimer's Dement. 16, e70044 (2024).
Nakagawa, P. & Sigmund, C. D. How is the brain renin-angiotensin system regulated? Hypertension 70, 10–18 (2017).
Hansen, J. Y. et al. Mapping neurotransmitter systems to the structural and functional organization of the human neocortex. Nat. Neurosci. 25, 1569–1581 (2022).
Georges, C. et al. Structural alterations associated with cardiovascular autonomic failure in multiple system atrophy. Eur. J. Neurol. 32, e70212 (2025).
Fiorenzato, E. et al. Brain structural profile of multiple system atrophy patients with cognitive impairment. J. Neural Transm. 124, 293–302 (2017).
Lin, C. R. et al. Clinicopathological correlates of pyramidal signs in multiple system atrophy. Ann. Clin. Transl. Neurol. 9, 988–994 (2022).
Ogawa, T. et al. White matter and nigral alterations in multiple system atrophy-parkinsonian type. npj Parkinsons Dis. 7, 1–12 (2021).
Compagnoni, G. M. & Fonzo, A. D. Understanding the pathogenesis of multiple system atrophy: state of the art and future perspectives. Acta Neuropathologica Commun. 7, 113 (2019).
Foti, S. C. et al. Cerebral mitochondrial electron transport chain dysfunction in multiple system atrophy and Parkinson’s disease. Sci. Rep. 9, 6559 (2019).
Hsiao, J.-H. T., Purushothuman, S., Jensen, P. H., Halliday, G. M. & Kim, W. S. Reductions in COQ2 expression relate to reduced ATP levels in multiple system atrophy brain. Front. Neurosci. 13, 1187 (2019).
Monzio Compagnoni, G. et al. Mitochondrial dysregulation and impaired autophagy in iPSC-derived dopaminergic neurons of multiple system atrophy. Stem Cell Rep. 11, 1185–1198 (2018).
Herrera-Vaquero, M. et al. Signs of early cellular dysfunction in multiple system atrophy. Neuropathol. Appl. Neurobiol. 47, 268–282 (2021).
Compta, Y. et al. Cerebrospinal fluid levels of coenzyme Q10 are reduced in multiple system atrophy. Parkinsonism Relat. Disord. 46, 16–23 (2018).
Kasai, T. et al. Serum levels of coenzyme Q10 in patients with multiple system atrophy. PLoS One 11, e0147574 (2016).
Mitsui, J. et al. High-dose ubiquinol supplementation in multiple-system atrophy: a multicentre, randomised, double-blinded, placebo-controlled phase 2 trial. eClinicalMedicine 59, 101920 (2023).
Kahle, P. J. et al. Hyperphosphorylation and insolubility of α-synuclein in transgenic mouse oligodendrocytes. EMBO Rep. 3, 583–588 (2002).
Stemberger, S., Poewe, W., Wenning, G. K. & Stefanova, N. Targeted overexpression of human alpha-synuclein in oligodendroglia induces lesions linked to MSA-like progressive autonomic failure. Exp. Neurol. 224, 459–464 (2010).
Miyazaki, Y. et al. Oligodendrocyte-derived LGI3 and its receptor ADAM23 organize juxtaparanodal Kv1 channel clustering for short-term synaptic plasticity. Cell Rep. 43, 113634 (2024).
Wilkins, A., Majed, H., Layfield, R., Compston, A. & Chandran, S. Oligodendrocytes promote neuronal survival and axonal length by distinct intracellular mechanisms: a novel role for oligodendrocyte-derived glial cell line-derived neurotrophic factor. J. Neurosci. 23, 4967–4974 (2003).
Gabdulkhaev, R. et al. Blood–brain barrier dysfunction in multiple system atrophy: a human postmortem study. Neuropathology 45, 210–222 (2025).
Miller, V. M., Kalaria, R. N., Hall, R., Oakley, A. E. & Kenny, R. A. Medullary microvessel degeneration in multiple system atrophy. Neurobiol. Dis. 26, 615–622 (2007).
Benarroch, E. E., Schmeichel, A. M., Low, P. A. & Parisi, J. E. Involvement of medullary serotonergic groups in multiple system atrophy. Ann. Neurol. 55, 418–422 (2004).
Yan, S. et al. Quantitative susceptibility mapping of multiple system atrophy and Parkinson’s disease correlates with neurotransmitter reference maps. Neurobiol. Dis. 198, 106549 (2024).
Chelban, V. et al. An update on MSA: premotor and non-motor features open a window of opportunities for early diagnosis and intervention. J. Neurol. 267, 2754–2770 (2020).
Goldstein, D. S. et al. Differential abnormalities of cerebrospinal fluid dopaminergic versus noradrenergic indices in synucleinopathies. J. Neurochem 158, 554–568 (2021).
Benarroch, E. E., Schmeichel, A. M. & Parisi, J. E. Depletion of mesopontine cholinergic and sparing of raphe neurons in multiple system atrophy. Neurology 59, 944–946 (2002).
Coon, E. A., Cutsforth-Gregory, J. K. & Benarroch, E. E. Neuropathology of autonomic dysfunction in synucleinopathies. Mov. Disord. 33, 349–358 (2018).
Gilman, S. et al. Cerebral cortical and subcortical cholinergic deficits in parkinsonian syndromes. Neurology 74, 1416–1423 (2010).
Chou, K. L. et al. Serotonin transporter imaging in multiple system atrophy and Parkinson’s disease. Mov. Disord. 37, 2301–2307 (2022).
Fahn, S., Elton, R. & Members of the UPDRS Development Committee. Recent Developments in Parkinson’s Disease. Vol 2. Florham Park, NJ. (Macmillan Health Care Information, 1987).
Hoehn, M. M. & Yahr, M. D. Parkinsonism: onset, progression and mortality. Neurology 17, 427–442 (1967).
Folstein, M. F., Folstein, S. E. & McHugh, P. R. ‘Mini-mental state’. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 12, 189–198 (1975).
Payan, C. A. M. et al. Disease severity and progression in progressive supranuclear palsy and multiple system atrophy: validation of the NNIPPS – Parkinson plus scale. PLoS One 6, e22293 (2011).
Marek, K. et al. The Parkinson’s progression markers initiative (PPMI) – establishing a PD biomarker cohort. Ann. Clin. Transl. Neurol. 5, 1460 (2018).
Postuma, R. B. et al. MDS clinical diagnostic criteria for Parkinson’s disease. Mov. Disord. 30, 1591–1601 (2015).
Gan-Or, Z. et al. The Quebec Parkinson network: a researcher-patient matching platform and multimodal biorepository. J. Park. ’s Dis. 10, 301–313 (2020).
Iglesias, J. E. et al. Bayesian segmentation of brainstem structures in MRI. Neuroimage 113, 184–195 (2015).
Faber, J. et al. CerebNet: a fast and reliable deep-learning pipeline for detailed cerebellum sub-segmentation. NeuroImage 264, 119703 (2022).
Schmahmann, J. D. et al. Three-dimensional MRI atlas of the human cerebellum in proportional stereotaxic space. Neuroimage 10, 233–260 (1999).
Gaser, C. et al. CAT – a computational anatomy toolbox for the analysis of structural MRI data. bioRxiv 2022.06.11.495736. https://doi.org/10.1101/2022.06.11.495736 (2023).
García-Gomar, M. G., Singh, K., Cauzzo, S. & Bianciardi, M. In vivo structural connectome of arousal and motor brainstem nuclei by 7 Tesla and 3 Tesla MRI. Hum. Brain Mapp. 43, 4397–4421 (2022).
Singh, K. et al. Functional connectome of arousal and motor brainstem nuclei in living humans by 7 Tesla resting-state fMRI. NeuroImage 249, 118865 (2022).
Fortin, J.-P. et al. Harmonization of cortical thickness measurements across scanners and sites. NeuroImage 167, 104–120 (2018).
Hawrylycz, M. J. et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399 (2012).
Markello, R. D. et al. Standardizing workflows in imaging transcriptomics with the abagen toolbox. eLife 10, e72129 (2021).
Burt, J. B., Helmer, M., Shinn, M., Anticevic, A. & Murray, J. D. Generative modeling of brain maps with spatial autocorrelation. NeuroImage 220, 117038 (2020).
Elizarraras, J. M. et al. WebGestalt 2024: faster gene set analysis and new support for metabolomics and multi-omics. Nucleic Acids Res. 52, W415–W421 (2024).
Consortium, T. G. O. et al. The gene ontology knowledgebase in 2023. Genetics 224, iyad031 (2023).
Piñero, J. et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 48, D845 (2019).
Mi, H. et al. Protocol Update for large-scale genome and gene function analysis with PANTHER classification system (v.14.0). Nat. Protoc. 14, 703–721 (2019).
Han, X. et al. Construction of a human cell landscape at single-cell level. Nature 581, 303–309 (2020).
Seidlitz, J. et al. Transcriptomic and cellular decoding of regional brain vulnerability to neurogenetic disorders. Nat. Commun. 11, 3358 (2020).
Markello, R. D. et al. neuromaps: structural and functional interpretation of brain maps. Nat. Methods 19, 1472–1479 (2022).
Acknowledgements
This work was supported by grants from Agence Nationale de la Recherche (grant numbers ANR-11-INBS-0006 [France Life Imaging], ANR-11-INBS-0011 [NeurATRIS, Investissements d’Avenir], ANR-19-P3IA-0001 [PRAIRIE 3IA Institute], and ANR-10-IAIHU-06 [IHU–Paris Institute of Neurosciences]), the European Union (EU) Framework Project 6–GENEPARK: Genomic Biomarkers for PD, Action Line: LIFESCIHEALTH Life sciences, genomics and biotechnology for health (LSH-2005-1.2.2.2)—Development of Innovative methods for diagnosis of nervous system disorders, the Program Hospitalier de Recherche Clinique [grant Numbers: PHRC 2007-A00169-44 (LRRK) and PHRC 2004 (BBBIPPS), Association France Parkinson, Ecole Neuroscience de Paris, Electricité de France (Fondation d’Entreprise EDF), Institut National de la Santé et de la Recherche Médicale, Fondation Thérèse and René Planiol pour l’étude du Cerveau, Société Française de Radiologie (SFR) / Collège des Enseignants en Radiologie de France (CERF), and Société Française de Neuroradiologie (SFNR). The work was also supported by Fonds de recherche du Quebec – Santé (FRQS), the Canadian Institutes of Health Research (CIHR) and the Healthy Brains for Healthy Lives (HBHL) program of the Canada First Research Excellence Fund. This work was also supported by a donation in memoriam of Gaetan Boulianne and Pierre-Claude Durivage to the Foundation of Hopital du Sacré-Coeur de Montréal, in support of PD research at the Center for Advanced Research in Sleep Medicine.
Author information
Authors and Affiliations
Contributions
L.C. designed and conceptualized the study, collected and analyzed the data, drafted and revised the manuscript for intellectual content. C.T., A.D., M.F., A.V., J.Y.H., A.F., P.K., C.-E.C., G.R., and A.D. analyzed the data and revised the manuscript. B.M. assisted with the methodology and revised the manuscript. J.-C.C., M.V., B.D., D.G., N.P., C.P., and S.L. performed data collection and revised the manuscript. S.R. designed and conceptualized the study, collected and analyzed the data, drafted and revised the manuscript for intellectual content.
Corresponding authors
Ethics declarations
Competing interests
None of the authors report any competing interests related to the current work. J.C.C. has served in advisory boards for Alzprotect, Bayer, Ferrer, iRegene, Servier, UC; and received grants from the AXA and the ICM Foundations outside of this work. BD received honoraria for lectures from IPSEN, ORION, MERZ. SR holds a research scholar award from the Fonds de recherche du Québec –Santé (FRQS) and receive grants from Parkinson Canada, Alzheimer Society of Canada, and The Michael J. Fox Foundation.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chougar, L., Tremblay, C., Delva, A. et al. MRI-derived atrophy in multiple system atrophy aligns with mitochondrial and glial gene expression patterns. npj Parkinsons Dis. 12, 16 (2026). https://doi.org/10.1038/s41531-025-01227-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41531-025-01227-1