Abstract
This study aimed to develop an efficient Saccharomyces cerevisiae-based cell factory for heme production. The industrial strain S. cerevisiae KCCM 12638, a starter strain used in American whisky production, was selected for its naturally high heme concentration, and its medium composition was optimized. Next, CRISPR/Cas9-based genome editing was employed to enhance carbon flux through the heme biosynthetic pathway by overexpressing HEM2, HEM3, HEM12, and HEM13 genes, which encode key enzymes involved in heme synthesis. Additionally, the HMX1 gene, which encodes heme oxygenase 1, was inactivated to prevent heme degradation. The resulting ΔHMX1_H2/3/12/13 strain achieved a heme titer of 9 mg/L in batch fermentation, a 1.7-fold improvement over the wild-type KCCM 12638 strain. In a glucose-limited fed-batch fermentation, the ΔHMX1_H2/3/12/13 strain produced 67 mg/L heme. This study demonstrates that engineering industrial yeast strains can significantly enhance heme production, with promising applications in food, pharmaceuticals, and bioenergy.
Similar content being viewed by others
Introduction
Heme, an important porphyrin derivative involved in cellular respiration, signal transduction, and iron metabolism, has widespread applications in dietary supplements, healthcare, and the alternative protein industry1,2,3. In recent years, the biological production of heme has garnered significant attention due to its simple and sustainable production process and the rapid growth of the global market for plant-based meat alternatives2,4. Notably, Saccharomyces cerevisiae has been employed as a chassis for heme production because it is generally recognized as safe (GRAS) and can produce heme cost-effectively. Moreover, yeast extracts from S. cerevisiae with high heme content often serve as flavor enhancers in plant-based meat alternatives5. However, the titer, yield, and productivity of heme produced by S. cerevisiae were not comparable to those produced by bacteria. For instance, the highest heme titer (380.5 mg/L) produced by engineered S. cerevisiae with enhanced resistance to heme toxicity6 was significantly lower than the titer (1.03 g/L) produced by engineered Escherichia coli7.
Traditionally, laboratory strains of haploid S. cerevisiae have been employed for heme production due to the limited genetic tools available for engineering industrial polyploid S. cerevisiae, which typically demonstrates greater tolerance to harsh fermentation conditions and achieves higher yields of target molecules8,9. However, conventional genetic engineering methods, involving sporulation and inactivation of the HO gene, convert polyploid S. cerevisiae into haploids, resulting in the loss of several advantageous traits10. The clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9-based genome editing system effectively addresses this challenge, enabling precise gene modifications without the drawbacks of traditional methods, thereby enhancing the potential for engineering industrial strains11.
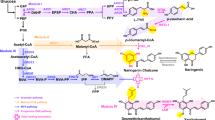
In this study, an edible industrial S. cerevisiae strain with high heme concentration (KCCM 12638) was selected, and the medium composition was optimized. Furthermore, the heme biosynthetic pathway of the selected strain was optimized using the CRISPR/Cas9-based genome editing system to maximize heme production. The heme biosynthetic pathway in S. cerevisiae consists of eight reactions, starting with the synthesis of 5-aminolevulinic acid and ending with heme production (Fig. 1). The expression of rate-limiting enzymes—aminolevulinate dehydratase (HEM2), porphobilinogen deaminase (HEM3), uroporphyrinogen decarboxylase (HEM12), and coproporphyrinogen III oxidase (HEM13)12,13,14—was optimized in the KCCM 12638 strain. Additionally, the gene encoding protoporphyrinogen oxidase (HEM14) was overexpressed and the HMX1 gene (encoding heme oxygenase 1, involved in heme degradation) was knocked out to further improve the efficiency of heme production in the KCCM 12638 strain. Taken together, the results of this study provide a foundation for the future commercial development of yeast extract with high heme content.
Flux in the heme biosynthetic pathway was enhanced by overexpressing genes encoding rate-limiting enzymes (HEM2, HEM3, HEM12, and HEM13), as well as enzymes involved in the final stages of the pathway (HEM14 and HEM15). Disruption of the ROX1 and HAP1 genes derepressed HEM13 expression, and inactivation of the HMX1 gene prevented heme degradation. Genes encoding key pathway enzymes: HEM1, 5-aminolevulinate synthase; HEM2, aminolevulinate dehydratase; HEM3, porphobilinogen deaminase; HEM4, uroporphyrinogen III synthase; HEM12, uroporphyrinogen decarboxylase; HEM13, coproporphyrinogen III oxidase; HEM14, protoporphyrinogen oxidase; HEM15, protoporphyrin ferrochelatase; ROX1, repressor of hypoxic functions; HAP1, heme activator protein; HMX1, heme oxygenase 1.
Results and discussion
Selection of a wild-type Saccharomyces cerevisiae with high heme concentration and optimization of medium composition
Heme, a porphyrin/iron complex cofactor, is naturally produced by various organisms, including yeast15. Even among strains of S. cerevisiae, fermentation capacity varies significantly16. Therefore, to identify a potential high-heme-producing strain, the heme concentrations produced by 31 edible S. cerevisiae strains (Supplementary Table S1) were compared. Overall, the wild-type strains exhibited higher heme production than the laboratory strain S. cerevisiae D452-2. Among the 31 wild-type strains, KCCM 12638, a starter strain used in American whisky production, exhibited the highest heme production, which was 3.3-fold greater than that observed in the D452-2 strain (Fig. 2). Therefore, KCCM 12638 was selected as the chassis for heme production.
The relative fold change in heme concentration was calculated by comparing the sample strains to the S. cerevisiae D452-2 strain. The yeast strains were cultured at 25 °C and 250 rpm for 48 h. Results represent the mean of two experiments, with error bars indicating standard deviation. Asterisk (*) indicates a significant difference compared to the control group, as determined by Student’s t test at p < 0.05.
Next, the composition of the complex medium was optimized to maximize heme production in KCCM 12638. While minimal media are typically preferred for the cost-effective production of target compounds, complex media containing byproduct-derived components, such as molasses and corn steep liquor (byproducts of the sugar and corn wet milling industries, respectively), are widely used for economical large-scale fermentation17,18. In this study, optimization was performed using the YP medium, with the potential for future substitution with byproducts to improve cost efficiency.
To assess the influence of nitrogen sources on heme production, various organic and inorganic nitrogen sources were tested by replacing 75% of the original nitrogen content in the YP50D medium (10 g/L yeast extract, 20 g/L peptone, and 50 g/L glucose). Specifically, different nitrogen sources were added at a concentration of 22.5 g/L, while yeast extract (2.5 g/L) and peptone (5 g/L) were maintained at one-fourth of the original concentrations. For example, 25 g/L of yeast extract, along with 5 g/L of peptone, was added to assess the impact of yeast extract on heme production (condition 1 in Figure S1A). Contrary to expectations, all nitrogen sources, except yeast extract, negatively affected heme production (Supplementary Fig. 1A). Notably, supplementing with yeast extract resulted in a 24% increase in heme concentration.
Further optimization of the yeast extract-to-peptone ratio revealed that the heme-enhancing effect of yeast extract was observed only in the presence of peptone (Supplementary Fig. 1B). The optimal combination of 40 g/L yeast extract and 20 g/L peptone increased heme production in the KCCM 12638 strain by 2.3-fold compared to the YP50D medium (Supplementary Fig. 1B). Although galactose as a carbon source increased heme production by 13% compared to glucose (Supplementary Fig. 2), glucose was selected due to the significantly higher cost of galactose.
Enhancing heme production in Saccharomyces cerevisiae KCCM 12638 through systematic metabolic engineering
To enhance heme production in the KCCM 12638 strain, four key genes (HEM2, HEM3, HEM12, and HEM13) encoding rate-limiting enzymes in heme biosynthesis (Fig. 1) were overexpressed, both individually and in combination. As expected, overexpression of these genes individually led to increased heme production. Specifically, overexpression of HEM2 or HEM13 resulted in 38% and 39% higher heme concentrations, respectively, compared to the wild-type KCCM 12638 strain (Fig. 3). Among strains overexpressing combinations of two or three of the four genes, the H2/13 strain (overexpressing HEM2 and HEM13) showed the highest increase, with a 63% boost in heme production. However, the H2/3/12/13 strain, which overexpressed all four genes overexpressed, achieved the highest overall heme concentration, with a 78% increase compared to the KCCM 12638 strain (Fig. 3).
The relative fold change in maximum heme concentration was calculated by comparing the sample strains to the S. cerevisiae KCCM 12638 strain. Results represent the mean of n ≥ 2 experiments, with error bars indicating standard deviation. Different letters indicate significantly different means (Tukey’s HSD tests, p < 0.05).
HAP1, a zinc finger DNA-binding protein, regulates heme levels in yeast19 by activating the transcription of a repressor of hypoxic functions (ROX1), which in turn suppresses the expression of various hypoxic genes, including HEM1320. Derepression of HEM13 transcription through the inactivation of HAP121 or ROX122 resulted in a significant increase in heme production. However, contrary to the results of previous studies, knockout of the HAP1 or ROX1 genes in the H2/3/12/13 strain led to decreased heme production (Fig. 4). This suggests that the effects of transcription factor inactivation are strain-dependent, and genetic background plays a significant role in metabolic engineering outcomes.
Additionally, yeast maintains heme homeostasis by regulating intracellular heme degradation. Heme oxygenase 1 (encoded by HMX1) plays a key role in heme degradation, facilitating its use as a source of nutritional iron23. Therefore, the HMX1 gene was selected as a knockout target to enhance heme production by preventing heme degradation. In wild-type yeast, the final steps of heme synthesis, catalyzed by two consecutive enzymes [HEM14 and protoporphyrin ferrochelatase (HEM15)] in the mitochondria, are not rate-limiting. However, it was hypothesized that the enhanced carbon flux in the H2/3/12/13 strain may outpace the activity of these enzymes. To validate this hypothesis, HEM14 and HEM15 were overexpressed individually and in combination in the HMX1-deficient H2/3/12/13 strain. Notably, the highest heme concentration (10 mg/L) was achieved in the ΔHMX1_H2/3/12/13/14 strain, which overexpressed HEM2, HEM3, HEM12, HEM13, and HEM14, producing 90% more heme than the wild-type KCCM 12638 strain in batch fermentation (Fig. 4). This result aligns with a previous study showing that rate-limiting enzymes may differ between strains with high and low heme production efficiency24.
Next, the role of 5-aminolevulinate synthase (HEM1), which synthesizes 5-aminolevulinic acid (5-ALA) from succinyl-CoA and glycine, was investigated. While HEM1 is typically not a rate-limiting enzyme in low-heme-producing yeast strains12,14, it may become limiting in strains engineered for higher heme production24. To determine whether 5-ALA availability limits heme production in the ΔHMX1_H2/3/12/13/14 strain, heme production was compared with and without 5-ALA supplementation. As controls, heme production in the wild-type KCCM 12638 strain and the ΔHMX1_H2/3/12/13 strain (lacking the HEM14 overexpression cassette) was also evaluated under the same conditions. As expected, 5-ALA supplementation did not enhance heme production in the low-heme-producing KCCM 12638 strain (Supplementary Fig. 3). Similarly, 5-ALA supplementation did not affect heme production in the ΔHMX1_H2/3/12/13 strain. However, the ΔHMX1_H2/3/12/13/14 strain exhibited a 25% increase in heme concentration compared to the KCCM 12638 strain when supplemented with 2.5 mM 5-ALA (Supplementary Fig. 3). These findings suggest that in metabolic engineering using a “push-pull-block” strategy, the introduction of the HEM14 overexpression cassette enhanced the “pull” capacity of the ΔHMX1_H2/3/12/13/14 strain, enabling efficient conversion of accumulated precursors into heme in response to increased 5-ALA levels. However, as the exogenous addition of 5-ALA is economically unfeasible, efforts to enhance 5-ALA availability are essential to maximize the heme production efficiency of the ΔHMX1_H2/3/12/13/14 strain.
Enhancing heme production via high-cell density culture
Initial experiments to increase heme concentration in fed-batch fermentation were performed using the wild-type KCCM 12638 strain. With intermittent additions of 20 g/L of carbon source, the maximum dry cell weight (DCW) reached only 17 g/L, resulting in heme production at a concentration of 8 mg/L (data not shown). In Crabtree-positive S. cerevisiae, excess glucose promotes ethanol production, reducing energy efficiency and biomass yield25. To mitigate this, a feeding solution consisting of 350 g/L glucose, 280 g/L yeast extract, and 140 g/L peptone was continuously added to match the glucose consumption rate. This approach helped limit ethanol production to below 15 g/L while promoting both cell growth and heme production. After the depletion of glucose (20–25 g/L) present in the 40YP medium (containing 40 g/L yeast extract and 20 g/L peptone), the feeding solution was introduced at rates of 9.6−16.0 mL/h for medium pH of 5.5, and 4.1–24.1 mL/h for medium pH of 4.5, and 4.6–15.2 mL/h for medium pH of 3.5 to optimize cell growth and heme production (Supplementary Fig. 4). Notably, the highest maximum DCW of 123 g/L and heme concentration of 49 mg/L were achieved when the medium pH was maintained at 4.5 (Fig. 5).
Fed-batch heme production using the Saccharomyces cerevisiae KCCM 12638 strain at pH 5.5 (A), pH 4.5 (B), and pH 3.5 (C). After the initially added glucose was depleted, a feeding solution containing 350 g/L glucose, 280 g/L yeast extract, and 140 g/L peptone was added to the bioreactor (black dot arrow). The detailed data are presented in Supplementary Fig. 4. Throughout the cultivation period, the medium temperature, agitation speed, and aeration rate were maintained at 25 °C, 700−1200 rpm, and 2.0−5.0 vvm, respectively. Measurements were performed in duplicate, and average values with standard deviations of less than 5% are presented.
The medium composition optimized for heme production in the KCCM 12638 strain requires a high concentration of yeast extract, which is not cost-effective for large-scale heme production. To address this issue, heme production by the ΔHMX1_H2/3/12/13 and ΔHMX1_H2/3/12/13/14 strains was compared in two different media: YP50D medium (containing 10 g/L yeast extract, 20 g/L peptone, and 50 g/L glucose) and 40YP50D medium (containing 40 g/L yeast extract, 20 g/L peptone, and 50 g/L glucose). Surprisingly, both strains produced 5.9- and 6.6-fold more heme in the YP50D medium than in the 40YP50D medium (Supplementary Fig. 5). This result suggests that the optimal medium composition is strain-dependent. Since YP50D contains four times less yeast extract than 40YP50D, the ΔHMX1_H2/3/12/13 and ΔHMX1_H2/3/12/13/14 strains benefit from high heme production efficiency in YP50D, offering a more cost-effective approach to heme production. After depleting the glucose (~25 g/L) in the YP medium, the feeding solution was introduced at a rate of 3.0−26.1 mL/h for the ΔHMX1_H2/3/12/13 strain and 2.4–24.2 mL/h for the ΔHMX1_H2/3/12/13/14 strain (Supplementary Fig. 6). Notably, while both strains produced similar concentrations of heme in batch fermentation, the ΔHMX1_H2/3/12/13 strain achieved a higher maximum DCW of 93 g/L and heme production of 67 mg/L in glucose-limited fed-batch fermentation, 20% and 52% higher than the ΔHMX1_H2/3/12/13/14 strain, respectively (Fig. 6). These results indicate that the additional overexpression of HEM14 in the ΔHMX1_H2/3/12/13/14 strain likely imposed a metabolic burden, impairing both cell growth and fermentation capacity. This result aligns with a previous study demonstrating a positive correlation between intracellular spermidine content and resistance to lignocellulose-derived inhibitors26. However, the same study also reported that S. cerevisiae strains with excessive gene overexpression reduced resistance despite elevated spermidine levels26. These findings underscore the need to balance gene overexpression with metabolic burden to achieve the desired phenotype. In future studies, we aim to determine the threshold at which gene overexpression induces metabolic burden in heme-producing strains.
Fed-batch heme production using the ΔHMX1_H2/3/12/13 (A) and ΔHMX1_H2/3/12/13/14 (B) strains. After the initially added glucose was depleted, a feeding solution containing 600 g/L glucose, 100 g/L yeast extract, and 200 g/L peptone was added to the bioreactor (black dot arrow). The detailed data are presented in Supplementary Fig. 6. Throughout the cultivation period, the medium acidity, temperature, agitation speed, and aeration rate were maintained at pH 4.5, 25 °C, 1000−1200 rpm, and 2.0 vvm, respectively. Measurements were performed in duplicate, and average values with standard deviations of less than 5% are presented.
Previously, we reported that a similar metabolic engineering approach in a laboratory S. cerevisiae strain yielded a lower heme titer (28 mg/L) in fed-batch fermentation14. This underscores the advantage of using industrial yeast strains for heme production. Nevertheless, despite these advancements, heme production in yeast remains significantly lower than that in E. coli, likely due to the bifurcation of the yeast heme biosynthetic pathway between the cytosol and mitochondria, which limits precursor accessibility. Additionally, yeast’s protoporphyrin-dependent heme biosynthesis pathway is thermodynamically less favorable than the coproporphyrin-dependent pathway used by bacteria. These limitations could potentially be mitigated by introducing the coproporphyrin-dependent pathway or compartmentalizing heme biosynthesis within the mitochondria. Guo et al.6 demonstrated that an engineered S. cerevisiae strain with an optimized heme biosynthetic pathway and enhanced heme tolerance achieved the highest reported yeast-based heme production (380.5 mg/L) in fed-batch fermentation. In contrast, a strain with only heme biosynthetic pathway optimization, without improved heme tolerance, produced just 5.9 mg/L of heme in batch fermentation6. Therefore, future studies should explore laboratory evolution approaches to enhance heme tolerance, ultimately maximizing heme production in the ΔHMX1_H2/3/12/13 strain.
This study aimed to develop a yeast-based microbial cell factory for efficient heme production. The industrial S. cerevisiae strain KCCM 12638 was selected as a potential heme producer. Using the CRISPR/Cas9-based genome editing system, the heme biosynthetic pathway in KCCM 12638 was reconstructed. Specifically, heme production efficiency was improved by overexpressing enzymes involved in the heme biosynthetic pathway and by preventing heme degradation. High-cell density cultivation of the ΔHMX1_H2/3/12/13 strain, which is HMX1-deficient and overexpresses HEM2, HEM3, HEM12, and HEM13, resulted in a heme concentration of 67 mg/L. Given the widespread concerns associated with the use of genetically modified organisms in the food industry, the ΔHMX1_H2/3/12/13 strain, developed using precise genome editing without the introduction of heterologous genes, shows promise for efficient heme production in this sector.
Methods
Strains and plasmids
The Escherichia coli TOP10 (Invitrogen, Carlsbad, CA, USA) strain was used as the host for plasmid construction. A laboratory S. cerevisiae strain, D452-227, and 31 wild-type S. cerevisiae strains from the Korean Agricultural Culture Collection (KACC) and the Korean Culture Center of Microorganisms (KCCM) were used as hosts for heme production. The S. cerevisiae strains and plasmids used in this study are listed in Supplementary Tables S1, S2.
Genetic manipulation
The primer sets used for the amplification of the HEM14 and HEM15 genes, intermediate plasmids, and guide RNA (gRNA) plasmids are listed in Supplementary Table S3. The resulting PCR products were assembled using NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s instructions to construct gene expression cassettes and gRNA plasmids for CRISPR/Cas9-based genome editing system.
The HEM2 (tHXT7p-HEM2-CYC1t), HEM3 (tHXT7p-HEM2-CYC1t), HEM12 (tHXT7p-HEM2-CYC1t), HEM13 (tHXT7p-HEM2-CYC1t), HEM14 (tHXT7p-HEM2-CYC1t), and HEM15 (tHXT7p-HEM2-CYC1t) expression cassettes were introduced into the CS10, CS828, CS628, CS12, INT1129, and INT12 loci, respectively, using the CRISPR/Cas9-based genome editing system as described previously30. In the S. cerevisiae KCCM 12638 strain, CS10, CS8, CS6, CS12, INT11, and INT12 are intergenic regions located at the following sites: (1) CS10 and CS8, between YPR015C and YPR014C on chromosome XVI; (2) CS6, between YHR19W and YNCH0042C on chromosome VII; (3) CS12, between LYS14 and YDR034C-D on chromosome IV; (4) INT11, between SYC1 and DCI1 on chromosome XV; (5) INT12, between MSC6 and GDS1 on chromosome XV. The knockout of the ROX1, HMX1, and HAP1 genes was achieved by integrating the HEM3 (tHXT7p-HEM2-CYC1t) expression cassette into the middle of these genes. Briefly, gRNA plasmids and repair DNA fragments, amplified with primers listed in Supplementary Table S3, were co-transformed into the S. cerevisiae KCCM 12638 strain harboring pCas9_AUR to introduce the gene expression cassettes.
Media and culture conditions
E. coli was grown in Luria–Bertani medium (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl) supplemented with 50 μg/mL of ampicillin for gene cloning. All S. cerevisiae strains were pre-cultured at 25 °C and 250 rpm for 48 h in YP20D medium (10 g/L yeast extract, 20 g/L Bacto Peptone, and 20 g/L glucose). The pre-cultured cells were harvested and inoculated into the main cultures to achieve an initial optical density at 600 nm (OD600) of 1.0. The main fermentation experiments were conducted in baffled flasks containing 100 mL of YP50D medium (10 g/L yeast extract, 20 g/L Bacto Peptone, and 50 g/L glucose). The temperature and agitation speed were maintained at 25 °C and 250 rpm throughout the fermentation.
Fed-batch fermentation was performed in a 2.5 L bioreactor (Kobiotech Co., Incheon, Republic of Korea) with an initial OD600 of 1.0, containing 1 L of 40YP20D medium for the KCCM 12638 strain and YP20D medium for the ΔHMX1_H2/3/12/13 and ΔHMX1_H2/3/12/13/14 strains. For the KCCM 12638 strain, a feeding solution containing 350 g/L glucose, 280 g/L yeast extract, and 140 g/L peptone was introduced at a rate from 4.1 to 24.1 mL/h after depletion of the initially added glucose. The medium pH, temperature, agitation speed, and air supply were maintained at 3.5–5.5, 25 °C, 700–1,200 rpm, and 2.0−5.0 vvm, respectively. For the ΔHMX1_H2/3/12/13 and ΔHMX1_H2/3/12/13/14 strains, a feeding solution containing 600 g/L glucose, 100 g/L yeast extract, and 200 g/L peptone was introduced at rates ranging from 2.4 to 26.1 mL/h after depletion of the initially added glucose. The medium pH, temperature, agitation speed, and air supply were maintained at 4.5, 25 °C, 1000–1200 rpm, and 2.0 vvm, respectively.
Analytical methods
A spectrophotometer (OPTIZEN POP, Mecasys Co., Ltd., Yuseong-gu, Daejeon, Republic of Korea) was used to measure cell growth at OD600. The concentrations of glucose, glycerol, acetic acid, and ethanol were determined using a high-performance liquid chromatography (HPLC) system (Ultimate 3000; Thermo Fisher Scientific Inc., Waltham, MA, USA) equipped with a Rezex ROA-organic acid H+ column (Phenomenex, Torrance, CA, USA). Metabolites were separated isocratically at 60 °C temperature and a flow rate of 0.6 mL/min in 5 mM sulfuric acid (H2SO4) and detected using a reflective index detector.
To determine intracellular heme concentrations, the culture broth was centrifuged at 15,000 rpm for 10 min. Cells were harvested at OD600 × mL of culture = 60 (e.g., for OD600 = 150, 0.4 mL of culture was harvested), resuspended in 0.4 mL of Y-PER reagent (Thermo Fisher Scientific Inc., Waltham, MA, USA), and lysed following the manufacturer’s instructions. Subsequently, 1.2 mL of acetonitrile was added to the cell lysate, and the mixture was incubated at room temperature for 5 min with occasional shaking. The precipitates were collected by centrifugation at 4000 rpm for 5 min and reacted with 1.6 mL of a solution containing acetonitrile and 1.7 M hydrochloric acid (8:2, v/v) at 25 °C and 250 rpm for 20 min. For neutralization, 0.4 mL of saturated magnesium sulfate solution containing 100 µg/L of sodium chloride was added to 1.6 mL of the reaction mixture, which was then incubated at 25 °C and 250 rpm for 20 min. Subsequently, the mixture was centrifuged at 4000 rpm for 5 min, and the acetonitrile layer was filtered through a 0.2 μm filter before HPLC analysis.
Heme concentrations were measured using a Thermo Fisher Ultimate 3000 HPLC system equipped with a YMC-Pack ODS-A column (YMC Co., Ltd., Kyoto, Japan). Heme was separated at a constant temperature of 50 °C using a gradient mode with mobile phases A and B as follows: 0 min, A:B = 80:20; 10–11 min, A:B = 0:100; 12–25 min, A:B = 80:20 (v/v). The flow rate was set to 0.4 mL/min, and detection was performed using an ultraviolet (UV) detector at 400 nm. Mobile phase A consisted of acetonitrile and water (5:95, v/v) containing 1.2 g/L of formic acid, whereas mobile phase B consisted of acetonitrile and water (95:5, v/v) containing 1.2 g/L of formic acid.
Statistical analysis
Statistical analyses were conducted using SPSS Statistics software (v.28.0, IBM Corp., Armonk, NY, USA). Data are presented as the mean ± standard deviation. One-way analysis of variance was performed, and statistical significance was assessed using Tukey’s honestly significant difference (HSD) test at a significance level of p < 0.05.
Data availability
Data are available upon request to the corresponding author.
References
Phillips, J. D. Heme biosynthesis and the porphyrias. Mol. Genet. Metab. 128, 164–177 (2019).
Su, H., Chen, X., Chen, S., Guo, M. & Liu, H. Applications of the whole-cell system in the efficient biosynthesis of heme. Int. J. Mol. Sci. 24, 8384 (2023).
Dailey, H. A. & Medlock, A. E. A primer on heme biosynthesis. Biol. Chem. 403, 985–1003 (2022).
Andreani, G. et al. Plant-based meat alternatives: technological, nutritional, environmental, market, and social challenges and opportunities. Nutrients 15, 452 (2023).
Tao, Z. et al. Yeast extract: characteristics, production, applications and future perspectives. J. Microbiol. Biotechnol. 33, 151–166 (2023).
Guo, Q. et al. Multidimensional engineering of Saccharomyces cerevisiae for the efficient production of heme by exploring the cytotoxicity and tolerance of heme. Metab. Eng. 85, 46–60 (2024).
Choi, K. R., Yu, H. E., Lee, H. & Lee, S. Y. Improved production of heme using metabolically engineered Escherichia coli. Biotechnol. Bioeng. 119, 3178–3193 (2022).
Akada, R. Genetically modified industrial yeast ready for application. J. Biosci. Bioeng. 94, 536–544 (2002).
Lee, Y. G. & Seo, J. H. Production of 2,3-butanediol from glucose and cassava hydrolysates by metabolically engineered industrial polyploid Saccharomyces cerevisiae. Biotechnol. Biofuels 12, 204 (2019).
Hu, X. H. et al. Genetic dissection of ethanol tolerance in the budding yeast Saccharomyces cerevisiae. Genetics 175, 1479–1487 (2007).
Zhang, G. C. et al. Construction of a quadruple auxotrophic mutant of an industrial polyploid saccharomyces cerevisiae strain by using RNA-guided Cas9 nuclease. Appl. Environ. Microbiol. 80, 7694–7701 (2014).
Hoffman, M., Gora, M. & Rytka, J. Identification of rate-limiting steps in yeast heme biosynthesis. Biochem. Biophys. Res. Commun. 310, 1247–1253 (2003).
Liu, L., Martinez, J. L., Liu, Z., Petranovic, D. & Nielsen, J. Balanced globin protein expression and heme biosynthesis improve production of human hemoglobin in Saccharomyces cerevisiae. Metab. Eng. 21, 9–16 (2014).
Lee, H. J., Shin, D. J., Nho, S. B., Lee, K. W. & Kim, S. K. Metabolic engineering of Saccharomyces cerevisiae for fermentative production of heme. Biotechnol. J. 19, e202400351 (2024).
Gallio, A. E., Fung, S. S., Cammack-Najera, A., Hudson, A. J. & Raven, E. L. Understanding the logistics for the distribution of heme in cells. JACS Au 1, 1541–1555 (2021).
Jeon, G.-B., Lee, Y.-O., Joo, B., Chin, Y.-W. & Kim, S.-K. Selection of wild type Saccharomyces cerevisiae with high protein content and improving its protein content via random mutagenesis. Food Eng. Prog. 27, 199–205 (2023).
Najafpour, G. D. & Shan, C. P. Enzymatic hydrolysis of molasses. Bioresour. Technol. 86, 91–94 (2003).
Do, S. H., Lee, T. G. & Kim, S. K. Enhancing protein content in wild-type Saccharomyces cerevisiae via random mutagenesis and optimized fermentation conditions. J. Microbiol. Biotechnol. 34, 1–7 (2024).
Zhang, L. & Guarente, L. The yeast activator HAP1-a GAL4 family member-binds DNA in a directly repeated orientation. Genes Dev. 8, 2110–2119 (1994).
Keng, T. HAP1 and ROX1 form a regulatory pathway in the repression of HEM13 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 2616–2623 (1992).
Tian, T. et al. High-level expression of leghemoglobin in Kluyveromyces marxianus by remodeling the heme metabolism pathway. Front. Bioeng. Biotech. 11, 1329016 (2024).
Zhang, T., Bu, P., Zeng, J. & Vancura, A. Increased heme synthesis in yeast induces a metabolic switch from fermentation to respiration even under conditions of glucose repression. J. Biol. Chem. 292, 16942–16954 (2017).
Protchenko, O. & Philpott, C. C. Regulation of intracellular heme levels by HMX1, a homologue of heme oxygenase, in Saccharomyces cerevisiae. J. Biol. Chem. 278, 36582–36587 (2003).
Ishchuk, O. P. et al. Genome-scale modeling drives 70-fold improvement of intracellular heme production in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 119, e2108245119 (2022).
Zhang, Y. M., Su, M., Wang, Z., Nielsen, J. & Liu, Z. H. Rewiring regulation on respiro-fermentative metabolism relieved Crabtree effects in Saccharomyces cerevisiae. Synth. Syst. Biotechnol. 7, 1034–1043 (2022).
Kim, S. K., Jin, Y. S., Choi, I. G., Park, Y. C. & Seo, J. H. Enhanced tolerance of Saccharomyces cerevisiae to multiple lignocellulose-derived inhibitors through modulation of spermidine contents. Metab. Eng. 29, 46–55 (2015).
Hosaka, K., Nikawa, J., Kodaki, T. & Yamashita, S. A dominant mutation that alters the regulation of INO1 expression in Saccharomyces cerevisiae. J. Biochem. 111, 352–358 (1992).
Kwak, S. et al. Enhanced isoprenoid production from xylose by engineered Saccharomyces cerevisiae. Biotechnol. Bioeng. 114, 2581–2591 (2017).
Park, S. et al. Functional expression of RuBisCO reduces CO2 emission during fermentation by engineered Saccharomyces cerevisiae. Process Biochem 134, 286–293 (2023).
Lee, Y. G., Jin, Y. S., Cha, Y. L. & Seo, J. H. Bioethanol production from cellulosic hydrolysates by engineered industrial Saccharomyces cerevisiae. Bioresour. Technol. 228, 355–361 (2017).
Acknowledgements
This study was financially supported by the National Research Foundation of Korea Grant (2022R1C1C1003154 and RS-2024-00440975), funded by the Korean Ministry of Science, ICT, and Future Planning. Additional support was provided by the Chung-Ang University Graduate Research Scholarship (Academic Scholarship for the College of Biotechnology and Natural Resources) in 2024.
Author information
Authors and Affiliations
Contributions
H.L.: Methodology, Investigation. T.L.: Investigation, Visualization. C.C.: Supervision. G.J.: Formal analysis. S.O.: Visualization. S.K.: Conceptualization, Writing - original draft preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lee, HJ., Lee, TG., Cho, C. et al. Enhanced heme production in industrial Saccharomyces cerevisiae through metabolic engineering. npj Sci Food 9, 244 (2025). https://doi.org/10.1038/s41538-025-00618-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41538-025-00618-1