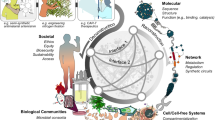
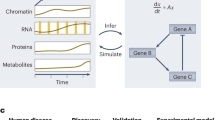
Abstract
While reductionism has advanced biology and medicine, it fosters a fragmented understanding of health, ill-suited to modern challenges like chronic and systemic diseases. Systems biology offers a new perspective, framing biological entities within interconnected networks. Using French medical education as an example, we argue that systems thinking should be foundational, not optional. Integrating systems biology early in medical education can better prepare future physicians for biomedical complexity and precision medicine.
Similar content being viewed by others
Introduction
Both biology and medicine are undergoing profound transformations1. Advances in omics technologies, systems-level modeling, and precision medicine have revealed that diseases rarely confine themselves to single organs, that molecular pathways interact in complex and non-linear ways, and that patient outcomes depend as much on systemic factors as on localized lesions2,3,4,5. Yet, medical education remains largely rooted in 20th-century reductionist paradigm, presenting knowledge in discipline-specific silos and introducing systemic perspectives only late in training. Reductionism often leads to an overly mechanistic understanding of biology, prioritizing isolated molecular mechanisms over the interconnected systems that define health and disease. The epistemological challenge of symbolic substitution in medicine, where organs, cells, molecular pathways, or computational models replace the irreducible complexity of living organisms, highlights the limits of reductionist frameworks. As Georges Canguilhem argued, the normative and adaptive dimensions of life and care cannot be fully captured by static or fragmented models6. Using French medical training as a case study, we propose that embracing a systemic paradigm is essential for preparing physicians to address the biomedical complexity of 21st‑century healthcare.
Revisiting the reductionist legacy in biology and medicine
The traditional model of organ-based medicine, which has dominated clinical practice for a century, shares deep conceptual roots with the reductionist paradigm in physics and biology7,8. Both approaches follow a Cartesian logic of decomposition: to understand the whole, one must first isolate and analyze its constituent parts9. In medicine, this led to segment the human body into discrete functional units such as heart, lungs, liver, or kidneys, each governed by a specialized medical discipline. Similarly, in biology, reductionism has fostered the belief that biological phenomena can be explained by tracing them back to individual genes, proteins, or signaling pathways, each associated with a specific function10.
This organ-based logic has been remarkably effective, enabling highly specialized diagnostics and therapeutic interventions, supported by increasingly sophisticated tools in imaging, surgery, and pharmacology. In parallel, reductionist molecular biology has driven transformative discoveries as well as exponential growth of knowledge in genetics, cell biology, physiology, genetics, enzymology, and physiopathology.
However, given the complexity of contemporary health challenges11, both paradigms reveal their limitations. In medicine, the fragmentation inherent to organ-based approaches often preclude to adequately manage patients with multiple chronic conditions12, functional disorders13, or syndromes that transcend anatomical boundaries, leading to partial treatments, delayed diagnoses, or failure to address underlying etiologies. In biology, the central dogma of molecular biology, “one gene, one protein, one function”, is at least incomplete (if not incorrect) to explain pleiotropy, gene-environment interactions, or epigenetic modifications14,15.
These limitations are particularly evident in common chronic conditions, such as asthma and hypertension, where reductionist views have long obscured their systemic nature. Asthma, once considered a purely bronchial disorder, is now recognized as a heterogeneous disease involving systemic immune dysregulation, obesity, and microbiome alterations16 and thus multiple medical specialities. The identification of distinct inflammatory phenotypes (eosinophilic, neutrophilic, IgE-mediated) has led to the development of targeted biologics, calling for a personalized, system-level approach beyond bronchial mechanics. Hypertension, often reduced to a blood pressure threshold, is defined as a dysfunction of multiple regulatory systems, linked initially to renal, endocrine, neural, and vascular mechanisms, and now also to inflammation, immunity, microbiome, oxidative stress, and behavioral factors17. Its management extends far beyond pharmacological control of one target molecule, requiring synergistic pharmacological strategies, combined with lifestyle interventions, psychosocial evaluation and support, and continuous monitoring of comorbidities and target-organ damages18. These examples illustrate how diseases traditionally defined by a single organ are, in fact, systemic disorders with localized manifestations.
Similar concerns apply to the field of therapeutics. Molecularly targeted therapies have revolutionized the treatment of several malignancies19. Designed to act on specific molecular alterations (e.g., oncogenic mutations, overexpressed receptors, or aberrant signaling pathways), these therapies embody the promise of precision medicine: offering treatments tailored to the patient tumor molecular profile. However, this therapeutic precision promoted a reductionist vision of diseases, that equates complex pathologies with discrete, targetable molecular entities. The conceptual elegance of the “one gene, one target, one drug” model, mirroring the earlier “one gene, one protein, one function” paradigm, provides clarity at the expense of obscuring the broader systemic context in which these molecular defects operate, including network-level interactions, compensatory mechanisms, and patient variability. Understanding this context is particularly critical in oncology, where tumors are not isolated entities defined solely by their genomic landscape; they exist within dynamic microenvironments shaped by immune interactions, metabolic flux, stromal remodeling, inflammation, and even psychosocial and circadian influences20,21,22.
Moreover, this focused approach can lead to unexpected adverse effects, since many molecular targets are not entirely tumor-specific and systemic regulatory feedback loops may be significantly disrupted. Examples include inhibitors of angiogenesis (e.g., VEGF-targeting agents), which can cause hypertension, thrombosis, and impaired wound healing through both direct VEGF inhibition and off-target effects on other tyrosine kinases23; immune checkpoint inhibitors, which can trigger immune-related adverse events in organs such as the gut, liver, skin, endocrine glands, or lungs due to widespread immune activation beyond the tumor site24; and BRAF inhibitors in melanoma, which may paradoxically activate the MAPK pathway in normal cells, leading to secondary neoplasms25.
A particularly illustrative case is imatinib, often celebrated as the first major success of rationally designed targeted therapy26. Developed to inhibit the constitutively active tyrosine kinase produced by the BCR-ABL fusion protein in chronic myeloid leukemia, imatinib dramatically improved survival in patients with Philadelphia chromosome-positive disease. Yet the clinical reality is more nuanced. Some patients develop primary or secondary resistance due to kinase domain mutations, activation of alternative pathways, or host-related factors such as pharmacogenetic variation27. Moreover, in addition to BCR-ABL, imatinib inhibits other tyrosine kinases, including c-KIT and PDGFRs, making it effective in unrelated malignancies such as gastrointestinal stromal tumors and in non-malignant disorders such as hypereosinophilic syndrome26,28,29. What began as a highly selective drug revealed a broader, multi-targeted profile, demonstrating that even the most “targeted” therapies can act systemically. In some cases, these systemic effects may be beneficial, but in others, they give rise to unexpected toxicities such as fluid retention, hepatotoxicity, or myelosuppression, reflecting the complex, network-level interactions of the human organism30.
These insights challenge the notion that a single molecular change can fully explain a disease or predict treatment response, urging the need to view the human body as a dynamic network of interacting physiological systems14. Although systems biology, for some authors31, is a notion that, in spite of its broad appeal, is still lacking a definition, it can be characterized as an integrative research strategy designed to tackle the complexity of biological systems and their behavior at all levels of organization (from molecules, cells, and organs to organisms and ecosystems) in normal and perturbed conditions32. Systems biology is evolving to systems medicine as a new discipline that aims to offer new approaches for addressing the diagnosis and treatment of major human diseases uniquely, effectively, and with personalized precision33. Systems medicine and systems biology integrate inter-organ communication, compensatory signaling, immune responses, individual variability, and broader social and environmental determinants, while examining interactions across molecular, cellular, tissue, and organismal scales34,35. Advances in computational modeling, high-throughput omics, and artificial intelligence now enable predictive models that move beyond descriptive biology to uncover mechanisms underlying health and disease36,37. Systems biology has transformed research in neuroscience, molecular biology, pharmacology, and translational medicine through data integration, modeling, and predictive analytics38.
As shown in Table 1, reductionist approaches and systemic paradigm are complementary. While reductionism isolates variables for precise analysis, the systemic paradigm emphasizes interdependence, feedback mechanisms, and multiscale complexity. Importantly, embracing systems biology does not replace reductionist methods; rather, it integrates them into a broader framework that accounts for interactions within the environment sustaining the entire system31. This integrated view is crucial for the future of precision medicine, requiring more than molecular profiling to achieve a systems-level understanding of how individual variability shapes outcomes within complex pathophysiological networks32,39.
From fragmented knowledge to integrated thinking: the case of French medical education
Medical education critically shapes how future physicians understand the human body, reason through clinical problems, and engage with biomedical complexity. Yet in many countries, including France, training remains rooted in 20th-century reductionist paradigms. While academically rigorous, it reinforces compartmentalized learning and organ-based reasoning. Introducing systems biology early in the curriculum could provide a crucial conceptual shift toward holistic care and systems thinking.
The first two years of medical school are highly selective and intensive, with limited basic science taught in fragmented, discipline-specific blocks. This structure emphasizes memorization over conceptual synthesis, leaving students with little sense of how molecular mechanisms connect to physiological and pathological processes. Furthermore, French medical education is strongly influenced by Claude Bernard’s “méthode expérimentale”, a hypothesis-driven approach that emphasizes the formulation of testable ideas, their evaluation through observable clinical signs or biological mechanisms, and the drawing of diagnostic conclusions40. While invaluable for diagnosis, this linear, cause-effect model can leave students ill-prepared to interpret the multidimensional datasets central to modern research and precision medicine31.
The organization of medical curricula further reinforces this reductionist inclination. Teaching remains largely structured around organ-based disciplines. Beyond internal medicine, cross-disciplinary fields like geriatrics, general practice, mental health, or gender medicine, often remain peripheral. This siloed approach, which mirrors that of biological sciences, tends to fragment knowledge and may hinder the development of an integrated understanding of the human body. As a result, it can be challenging for students to understand the human body as a dynamic interconnected system and acquire a holistic view of the patient, which is essential for comprehensive and person-centered care.
In this context, systems thinking offers a much-needed conceptual shift. It emphasizes the interdependence of biological, psychological, social, and environmental factors in health and disease, and encourages recognition of feedback loops, emergent properties, and the dynamic nature of complex systems. Although systems thinking is increasingly promoted by public health agencies (see for ref. 41), educators, and healthcare quality organizations, its implementation into the early years of medical school remains limited. Where present, courses are mostly restricted to global health or epidemiology courses42,43.
Systems biology and systems thinking share conceptual foundations: both focus on integration across levels, networks, regulatory feedback, and emergent properties of complex systems. Introducing systems biology early in medical education offers far more than a technical or research-oriented perspective. It provides a conceptual framework that prepares students to think systemically, considering systems biology as a unifying paradigm for medical education44. By learning how cells and molecules operate within dynamic, interconnected networks, students develop reasoning skills applicable across clinical care, public health, and health system domains.
Teaching cell biology through systems thinking
We illustrate this approach with an educational programme45 that incorporates core principles of systems biology into medical training. Designed to demonstrate both feasibility and benefits, this programme consists of three activities spread across several years to promote progressive learning and reinforcement of systems biology concepts. The first, delivered at the very beginning of the first year, introduces foundational principles and establishes an epistemological framework for subsequent learning. The second, in the second year, provides a detailed example of systems biology in practice. The third, in the clinical phase, embeds systems thinking within organ-based teaching, linking basic science and clinical decision-making.
Activity 1: Introducing the principles of systems biology
This activity is built as an introductory class establishing systems biology as the foundational framework for teaching cell biology to first-year medical students. Unlike traditional curricula, which often begin with a year of reductionist teaching (e.g., one ligand, one receptor, one function) and only later introduce the complexity of networks, we present systems biology concepts from the outset. This early exposure positions molecular biology as a remarkable achievement of reductionist science that now requires extension into integrative, systems-level models31. The class begins by defining the concepts of system structure and organization, explores internal interactions within the system, expands to view the system as a whole, and finally examines its relationships with other systems44. The class then builds on the presentation of the core Principles of Systems Biology, using seven principles inspired by Denis Noble in his influential assays, particularly his seminal 2006 article14,46. These principles were originally conceived to counteract genetic determinism, a form of reductionism prevalent at the time. As Nobel laureate Sydney Brenner famously stated47, and as Denis Noble emphasized: “The correct level of abstraction is the cell. The cell is the fundamental unit of structure, function, and organization.” Building on this perspective, we refer to the cell as the correct level of functional integration. In line with this, we have adapted several of these principles to the cellular scale and illustrated them with concrete examples drawn from the annual course in cell biology. This includes pathological processes and therapeutic strategies such as cancer or gene therapy for type 1 diabetes9 (see Box 1 and supplemental data 1 for further details and references). Recognizing that most 1st-year medical students have limited prior exposure to molecular and cellular biology, we adopted a pedagogical strategy aimed at making complex systems more accessible. Like Noble, we rely extensively on metaphors and analogies to build intuitive understanding14. For example, we compare the cell to a networked society, rather than a simple machine, in order to emphasize its dynamic, adaptive, and context-sensitive nature. This approach encourages students to move beyond static, linear representations and to appreciate the emergent properties and systemic interdependence that characterize living organisms. This conceptual grounding is then complemented in subsequent classes by classical content of cell biology (cell structures, molecular pathways), but with a deliberate emphasis on interconnections, regulatory feedback, and biological context.
Conclusion: preparing clinicians for complexity through systems thinking
Integrating systems biology from the outset represents a vital paradigm shift to prepare future clinicians for the complexity of modern medicine52,53. Moving beyond the traditional reductionist, organ-based model, systems thinking offers a framework linking molecular, cellular, organismal, populational and environmental levels, fostering a deep understanding of biological networks and their dynamic regulation54. This approach aligns education with current scientific advances, strengthens interdisciplinary collaboration, and supports the principles of precision medicine. Embedding systems biology early and continuously within the curriculum can cultivate critical reasoning skills, cognitive flexibility, and an integrative mindset in medical students, enabling them to navigate multifaceted disease processes, interpret multidimensional data, and deliver personalized, context-aware patient care. Ultimately, embracing systems thinking is essential to train the next generation of physicians addressing the challenges of modern healthcare, where complexity and individual variability are the norm rather than the exception.
Data availability
No datasets were generated or analysed during the current study.
References
Hood, L., Balling, R. & Auffray, C. Revolutionizing medicine in the 21st century through systems approaches. Biotechnol. J. 7, 992–1001 (2012).
Comte, B. et al. Network and Systems Medicine: Position Paper of the European Collaboration on Science and Technology Action on Open Multiscale Systems Medicine. Netw. Syst. Med. 3, 67–90 (2020).
Denny, J. C. & Collins, F. S. Precision medicine in 2030—seven ways to transform healthcare. Cell 184, 1415–1419 (2021).
Manicka, S., Johnson, K., Levin, M. & Murrugarra, D. The nonlinearity of regulation in biological networks. npj Syst. Biol. Appl 9, 10 (2023).
Shen, X. et al. Nonlinear dynamics of multi-omics profiles during human aging. Nat. Aging 4, 1619–1634 (2024).
Canguilhem, G. Le statut épistémologique de la médecine. Hist. Philos. Life Sci. 10, 15–29 (1988).
Schmidt, H. H. H. W. Organ-Based Medicine. In The end of medicine as we know it - and why your health has a future (ed. Schmidt, H. H. H. W.) 119–128 (Springer International Publishing, Cham, 2022). https://doi.org/10.1007/978-3-030-95293-8_9.
Von Bertalanffy, L. General System Theory. 40 (1968).
Ahn, A. C., Tewari, M., Poon, C.-S. & Phillips, R. S. The Limits of Reductionism in Medicine: Could Systems Biology Offer an Alternative?. PLOS Med. 3, e208 (2006).
Karsenti, E. A journey from reductionist to systemic cell biology aboard the schooner Tara. MBoC 23, 2403–2406 (2012).
Rocca, E. & Anjum, R. L. Complexity, Reductionism and the Biomedical Model. In Rethinking Causality, Complexity and Evidence for the Unique Patient: A CauseHealth Resource for Healthcare Professionals and the Clinical Encounter (eds Anjum, R. L., Copeland, S. & Rocca, E.) 75–94 (Springer International Publishing, Cham, 2020). https://doi.org/10.1007/978-3-030-41239-5_5.
Smith, S. M. & O’Dowd, T. Chronic diseases: what happens when they come in multiples? Br. J. Gen. Pr. 57, 268–270 (2007).
Wessely, S., Nimnuan, C. & Sharpe, M. Functional somatic syndromes: one or many? Lancet 354, 936–939 (1999).
Noble, D. Claude Bernard, the first systems biologist, and the future of physiology. Exp. Physiol. 93, 16–26 (2008).
Crick, F. Central Dogma of Molecular Biology. Nature 227, 561–563 (1970).
Pavord, I. D. et al. After asthma: redefining airways diseases. Lancet 391, 350–400 (2018).
Harrison, D. G., Coffman, T. M. & Wilcox, C. S. Pathophysiology of Hypertension: The Mosaic Theory and Beyond. Circ. Res. 128, 847–863 (2021).
McEvoy, J. W. et al. 2024 ESC Guidelines for the management of elevated blood pressure and hypertension. Eur. Heart J. 45, 3912–4018 (2024).
Lee, Y. T., Tan, Y. J. & Oon, C. E. Molecular targeted therapy: Treating cancer with specificity. Eur. J. Pharm. 834, 188–196 (2018).
Li, Z., Li, J., Bai, X., Huang, X. & Wang, Q. Tumor microenvironment as a complex milieu driving cancer progression: a mini review. Clin. Transl. Oncol. 27, 1943–1952 (2025).
Lee, Y. Roles of circadian clocks in cancer pathogenesis and treatment. Exp. Mol. Med. 53, 1529–1538 (2021).
Yuan, A., Wang, S., Li, Z. & Huang, C. Psychological aspect of cancer: From stressor to cancer progression. Exp. Ther. Med. 1, 13–18 (2010).
Dobbin, S. J. H., Petrie, M. C., Myles, R. C., Touyz, R. M. & Lang, N. N. Cardiotoxic effects of angiogenesis inhibitors. Clin. Sci. (Lond.) 135, 71–100 (2021).
Yin, Q. et al. Immune-related adverse events of immune checkpoint inhibitors: a review. Front. Immunol. 14, 1167975 (2023).
Thompson, E. L., Hu, J. J. & Niedernhofer, L. J. The Role of Senescent Cells in Acquired Drug Resistance and Secondary Cancer in BRAFi-Treated Melanoma. Cancers 13, 2241 (2021).
Iqbal, N. & Iqbal, N. Imatinib: A Breakthrough of Targeted Therapy in Cancer. Chemother. Res. Pract. 2014, 357027 (2014).
Ramirez, P. & DiPersio, J. F. Therapy options in imatinib failures. Oncologist 13, 424–434 (2008).
Gleich, G. J., Leiferman, K. M., Pardanani, A., Tefferi, A. & Butterfield, J. H. Treatment of hypereosinophilic syndrome with imatinib mesilate. Lancet 359, 1577–1578 (2002).
Demetri, G. D. et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N. Engl. J. Med. 347, 472–480 (2002).
Guilhot, F. Indications for imatinib mesylate therapy and clinical management. Oncologist 9, 271–281 (2004).
Kohl, P., Crampin, E. J., Quinn, T. A. & Noble, D. Systems Biology: An Approach. Clin. Pharmacol. Therapeutics 88, 25–33 (2010).
Auffray, C., Chen, Z. & Hood, L. Systems medicine: the future of medical genomics and healthcare. Genome Med. 1, 2 (2009).
Wang, R.-S., Maron, B. A. & Loscalzo, J. Systems Medicine: Evolution of Systems Biology From Bench To Bedside. Wiley Interdiscip. Rev. Syst. Biol. Med. 7, 141–161 (2015).
Williams, E. G. & Auwerx, J. The Convergence of Systems and Reductionist Approaches in Complex Trait Analysis. Cell 162, 23–32 (2015).
Gatherer, D. So what do we really mean when we say that systems biology is holistic? BMC Syst. Biol. 4, 22 (2010).
Puniya, B. L., Verma, M., Damiani, C., Bakr, S. & Dräger, A. Perspectives on computational modeling of biological systems and the significance of the SysMod community. Bioinforma. Adv. 4, vbae090 (2024).
Yue, R. & Dutta, A. Computational systems biology in disease modeling and control, review and perspectives. NPJ Syst. Biol. Appl 8, 37 (2022).
Chuang, H.-Y., Hofree, M. & Ideker, T. A Decade of Systems Biology. Annu. Rev. Cell Dev. Biol. 26, 721–744 (2010).
Vogt, H., Hofmann, B. & Getz, L. The new holism: P4 systems medicine and the medicalization of health and life itself. Med Health Care Philos. 19, 307–323 (2016).
Ayala, R. A. Welcome to the New Age. Claude Bernard’s “Introduction to the Study of Experimental Medicine” and the Shift of Medical Thought Towards Science: 150 Years Later. Arch. Med. Res. 48, 393–396 (2017).
de Savigny, D. & Adam, T. Systems Thinking for Health Systems Strengthening. https://wkc.who.int/resources/publications/i/item/2009-11-13-systems-thinking-for-health-systems-strengthening (2009).
Leischow, S. J. et al. Systems Thinking to Improve the Public’s Health. Am. J. Preventive Med. 35, S196–S203 (2008).
Earp, B. D. Systems thinking in gender and medicine. https://philpapers.org/rec/EARSTI (2020).
Momsen, J., Speth, E. B., Wyse, S. & Long, T. Using Systems and Systems Thinking to Unify Biology Education. CBE Life Sci. Educ. 21, es3 (2022).
Educational programme. https://uis.unesco.org/en/glossary-term/educational-programme (2020).
Noble, D. The Music of Life: Biology beyond the Genome (Oxford University Press, 2006). https://doi.org/10.1093/oso/9780199295739.001.0001.
Brenner, S. Sequences and consequences. Philos. Trans. R. Soc. B: Biol. Sci. 365, 207–212 (2010).
Huyghe, A., Trajkova, A. & Lavial, F. Cellular plasticity in reprogramming, rejuvenation and tumorigenesis: a pioneer TF perspective. Trends Cell Biol. 34, 255–267 (2024).
Huyghe, A. et al. Comparative roadmaps of reprogramming and oncogenic transformation identify Bcl11b and Atoh8 as broad regulators of cellular plasticity. Nat. Cell Biol. 24, 1350–1363 (2022).
Grimwade, D., Mistry, A. R., Solomon, E. & Guidez, F. Acute promyelocytic leukemia: a paradigm for differentiation therapy. Cancer Treat. Res. 145, 219–235 (2010).
Abou-Alfa, G. K. et al. Ivosidenib in IDH1-mutant, chemotherapy-refractory cholangiocarcinoma (ClarIDHy): a multicentre, randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 21, 796–807 (2020).
Auffray, C., Noble, D., Nottale, L. & Turner, P. Progress in integrative systems biology, physiology and medicine: towards a scale-relative biology. Eur. Phys. J. A 56, 88 (2020).
Qiao, L., Khalilimeybodi, A., Linden-Santangeli, N. J. & Rangamani, P. The Evolution of Systems Biology and Systems Medicine: From Mechanistic Models to Uncertainty Quantification. Annu. Rev. Biomed. Eng. 27, 425–447 (2025).
Didier, J. et al. Challenges and opportunities in systems biology education. Endocr. Relat. Cancer 32, e250024 (2025).
Yang, C. et al. Regulation of insulin secretion by the post-translational modifications. Front Cell Dev. Biol. 11, 1217189 (2023).
Pauklin, S. & Vallier, L. The cell-cycle state of stem cells determines cell fate propensity. Cell 155, 135–47 (2013).
Kruiswijk, F., Labuschagne, C. F. & Vousden, K. H. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 16, 393–405 (2015).
Harris, S. L. & Levine, A. J. The p53 pathway: positive and negative feedback loops. Oncogene 24, 2899–2908 (2005).
Syrovatkina, V., Alegre, K. O., Dey, R. & Huang, X.-Y. Regulation, Signaling, and Physiological Functions of G-Proteins. J. Mol. Biol. 428, 3850–3868 (2016).
Ye, K., Lu, J., Ma, F., Keinan, A. & Gu, Z. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc. Natl. Acad. Sci. USA 111, 10654–10659 (2014).
Wheat, J. C. et al. Single-molecule imaging of transcription dynamics in somatic stem cells. Nature 583, 431–436 (2020).
Acknowledgements
The authors used AI assisted copy editing tools (Mistral, ChatGPT (OpenAI) and Perplexity AI) accordingly to journal guidelines.
Author information
Authors and Affiliations
Contributions
L.D. drafted the original version, and was a major contributor in writing the manuscript. P.A.G. and G.L. were substantial contributors in writing and editing the manuscript. P.L. contributed to the conception and design of the manuscript, and was a major contributor in writing the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
L.D., G.L., and P.L. declare no conflicts of interest.PA Gourraud is the founder of Methodomics (2008) and its spin-off Big data Santé (2018- “Octopize” brand). He consults for major pharmaceutical companies, and start-ups, all of which are handled through academic pipelines (AstraZeneca, Amgen, Biogen, Boston Scientific, Cemka, Cook, Docaposte/Heva, Edimark, Ellipses, Elsevier, Grunenthal, Hemovis, Janssen, IAGE, Lek, Methodomics, Merck, Mérieux, Novartis, Octopize, Sanofi-Genzyme, Lifen, TuneInsight, Aspire UAE). PA Gourraud is a volunteer board member at AXA not-for-profit mutual insurance company (2021). He has no prescription activity with either drugs or devices. He receives no wages from these activities.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
David, L., Gourraud, PA., Lamirault, G. et al. Rethinking medical education through systems biology to address complexity. npj Syst Biol Appl 12, 12 (2026). https://doi.org/10.1038/s41540-025-00636-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41540-025-00636-5