Abstract
mRNA vaccines have demonstrated significant potential in preventing human diseases and controlling livestock infections. However, the application of mRNA vaccines in aquaculture, especially on fish, remains limited. Infectious hematopoietic necrosis virus (IHNV) is an RNA virus that mainly affects rainbow trout (Oncorhynchus mykiss), leading to high mortality rates. In this study, we systematically engineered three UTR-optimized mRNA constructs, exhibiting comparable and sustained in vitro antigen expression. Following encapsulation, the mG1-LNP formulation, incorporating endogenous antigen-specifics, conferred robust relative protection against IHNV challenge, accompanied by enhanced levels of IgM and neutralizing antibodies. Furthermore, dose-response profiling identified 10 μg/dose as the immunologically optimized regimen, eliciting efficient immunogenicity. Moreover, biodistribution analyses revealed complete mG1-LNP clearance from injection sites and hepatic tissues by 28 dpv, confirming favorable biosafety. Collectively, our work demonstrates the successful development of mRNA-LNP vaccine against infectious IHNV in rainbow trout, providing the first empirical demonstration of mRNA-LNP vaccine efficacy in aquaculture.
Similar content being viewed by others
Introduction
As an emerging vaccine platform, messenger RNA (mRNA)-based vaccines have been developed to eliminate the existing bottleneck of conventional vaccines with distinct advantages in several areas, such as faster development cycles, favorable safety profiles, and the ability to elicit stronger cellular immune responses1,2. Since the onset of the COVID-19 pandemic, mRNA vaccines developed by companies such as Pfizer-BioNTech and Moderna have been widely used3,4, demonstrating significant potential in preventing and treating infectious diseases5. To cope with other respiratory viral diseases, such as influenza, mRNA vaccine candidates are also under different stages of development and possess a robust immune response with high levels of protective antibodies6. The development of mRNA vaccines thus provides new insights and methods for the prevention and control of infectious diseases7,8.
In addition to their applications in human health and disease, an increasing number of studies have focused on mRNA vaccines targeting various animal diseases, due to the potential to elicit stronger cellular immune responses, faster development cycles, and favorable safety profiles9,10. For instance, the mRNA vaccine encoding ASFV p30 elicited strong immune responses and provided partial protection in experimental swine11. For poultry disease prevention, self-amplifying mRNA vaccines efficiently induced robust neutralizing antibody titers and enhanced T-cell activation in broiler chickens12. Despite the promising achievements of mRNA vaccines in human and terrestrial animal health, mRNA vaccine application in aquatic species remains limited.
As a promising technology for disease prevention in aquaculture, mRNA vaccines have to address significant challenges, such as the delivery and stability of mRNA-based antigens, species-specific immune responses, and environmental impact. Unlike the constant temperature environment within mammals, fluctuating water and fish temperatures may cause alterations in the activity of ligands and lipid arrangements on lipid nanoparticle (LNP) surface, eventually impacting antigen stability and release, which has been considered as the most successful delivery system13. Compared to the mammalian immune system, fish possess distinct innate and adaptive immunity characteristics, such as different immune response mechanisms and antigen presentation pathways14. To ensure effective protection, mRNA vaccine formulations may be required to enhance the immunogenicity, rather than attenuating it as is done within human vaccines15. The release of mRNA or delivery components into aquatic ecosystems must be carefully evaluated to avoid unintended ecological consequences.
Generally, the conventional structure of mRNA-based antigen consists of five components: the 5’ cap, the 5’ untranslated region (5’ UTR), the antigen-coding sequence (CDS), the 3’ UTR (3’ UTR), and the 3’ poly (A) tail16. Appropriate sequence design and component optimization are required to elicit strong and durable immune responses in aquatic species17. The UTR serves as a critical component of mRNA, playing a pivotal role in determining mRNA stability, translational efficiency, and functionality. Current design strategies for UTR optimization primarily encompass: (1) direct utilization of UTRs derived from highly expressed genes, such as α-globin and β-globin UTRs, which significantly optimize mRNA expression efficiency and stability and have been extensively employed in COVID-19 mRNA vaccines18,19,20; (2) viral-derived UTR elements, which typically enable efficient initiation of viral protein translation17; (3) de novo computational design of UTR sequences, such as LinearDesign algorithm21.
Previously, we developed a DNA vaccine with self-designed CpG encoding glycoprotein of J genotype IHNV and conducted large-scale trials to evaluate its efficacy and safety22. IHNV is a single-stranded RNA virus capable of causing mortality rates of 80%-100% in salmonid fish, such as rainbow trout (Oncorhynchus mykiss), leading to significant losses in the global salmon and trout aquaculture industry23. Here, the optimized mRNA encoding glycoprotein of IHNV GS21 was engineered and encapsulated in commercial LNP. Following vaccination, the immune protective efficacy against IHNV, specific antibody titers, and immune response were evaluated, defining it as an effective mRNA vaccine candidate for rainbow trout against IHNV infection. Dose optimization and vaccine residue biodistribution analysis addressed aquaculture-specific safety concerns. Our results provide proof-of-principle evidence that mRNA-LNP vaccines can protect against aquatic viruses in a fish model.
Results
Optimization of different UTRs, ensuring mRNA constructs expression
The 5’ and 3’ UTRs and poly(A) tails play crucial roles in ensuring that mRNA vaccine is efficiently translated into the target protein, elicits a robust immune response, and remains stable long enough to achieve the immunoprophylaxis effects24. Albeit with a varied length of poly(A) tails, synthetic mRNA is engineered to contain poly(A) tails within a range of 100–150 adenosines to ensure stability and efficient translation25. In regard of UTR elements, the original virus UTRs are believed to be capable of efficiently initiating the translation of their corresponding CDS regions in the hosts17, moreover, the modified α-globin 5’ UTR and β-globin 3’ UTR have been commonly adopted in mRNA vaccine molecular design, especially in the successful implementation of COVID-19 mRNA vaccines18,19,20. To this end, three mRNAs with distinct UTRs were constructed using enhanced green fluorescent protein (EGFP) as a reporter, including the original 5’ and 3’ UTRs of IHNV G, modified α-globin 5’ UTR, and β-globin 3’ UTR (Fig. 1A).
A The construction diagram of mRNA encoding EGFP with different UTRs. B and C Representative images of the indicated mE constructs expressing green fluorescent signal (scale bar 30 µm; n = 6) (B). Statistical quantification of the corresponding green fluorescent signals (C). D and E EGFP protein productions by the indicated mE constructs were analyzed by Flow cytometry (n = 3) (D). Statistics of the relative fluorescence intensity of FITC are shown (E). F and G Western blotting analysis of EGFP yields by the indicated mE constructs (F). GADPH was used as the internal control. Statistical quantification of EGFP protein productions (n = 3) (G).
We subsequently incubated the naked three EGFP constructs consisting of different UTR combinations (mEs; Detailed in Fig. 1A) with HEK293T (a common mammalian cell line extensively utilized for exogenous nucleic acid expression) and CHSE-214 (a conventional fish cell line mimicking the physiological conditions in fish), using transfection reagents, respectively. In the cells transfected with all EGFP constructs, we observed apparent fluorescence signals compared to the absence of fluorescence signals in the mock group (Fig. 1B). Analysis of the mean fluorescence intensity (MFI) indicated that all mEs exhibited efficient protein expressions (Fig. 1C). Likewise, flow cytometry and Western blotting (WB) analysis indicated that each mE provided robust and consistent effectiveness of EGFP expression across different cell lines, in line with the aforementioned findings (Fig. 1D–G). Specifically, mE2 exhibited 15-fold greater fluorescence intensities in HEK293T and 9.7-fold greater fluorescence intensities in CHSE-214 cells.
Validation of mRNA vaccines encoding glycoprotein ex vivo
Antigens are particularly important for immunogenicity, and in vaccines against IHNV, glycoprotein has been widely utilized as an antigen due to its critical role in viral infection and immune response26. Therefore, we employed the indicated non-coding elements to generate mRNAs with the codon-optimized G protein sequence (mGs; Detailed in Fig. 2A). Considering that stability is a critical parameter for evaluating the performance of mRNA vaccines, directly influencing the duration of mRNA expression and overall expression efficacy, we initially assessed the stability of three mG constructs in HEK293T and CHSE-214 cells (Fig. 2B, C). Within 12 h post-transfection, all levels of mG constructs decreased by less than 50% and mG1 possessed the slowest degradation rate with no significant decline in intracellular levels. Remarkably, the degradation of mG constructs proceeded at a relatively faster pace in CHSE-214 cells.
A The construction diagram of mRNA encoding glycoprotein with different UTRs. B and C Cellular amount of mG constructs in HEK293T (B) and CHSE-214 (C) cells after transfection for 6, 12, 24, 48, and 72 h (n = 3). D and E Representative images of the indicated mG constructs expressing glycoprotein in HEK293T and CHSE-214 cells via indirect Immunofluorescence (scale bar 30 µm; n = 4) (D). Statistical quantification of the corresponding glycoprotein productions (E). F and G Western blotting analysis of glycoprotein yields by the indicated mG constructs in HEK293T and CHSE-214 cells (F). GADPH was used as the internal control. Statistical quantification of glycoprotein productions (n = 3) (G). H Representative images of glycoprotein expression encoded by mG1 construct in muscle via IHC analysis at 48 hpi (scale bar 100 µm; n = 2). All data were presented as mean ± SEM and analyzed by one-way ANOVA with Kruskal–Wallis test (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, ns, not significant).
Subsequently, all mGs constructs were transfected into both cell lines for indirect immunofluorescence (IIF) and WB analyses. IIF analysis revealed distinct green fluorescence was observed across all experimental groups (Fig. 2D, E), supporting the successful mGs expression in both HEK293T and CHSE-214 systems. WB results aligned well with these findings. All mG constructs generated glycoproteins in both cell types, while the most abundant glycoproteins were determined compared to those of mG2 and mG3 when the original UTRs of glycoprotein were employed (Fig. 2F, G). The combination of the original glycoprotein UTRs and the codon-optimized G protein sequence slightly improved glycoprotein production, probably due to the more natural conformation formed by the original non-coding elements. To further investigate the in vivo antigen expression, the muscle tissues of rainbow trout injected with 10 μg mG1 construct were sampled at 48 hours post-injection for immunohistochemical (IHC) analysis. Congruent with the ex vivo observation, substantial G-proteins were generated by mG1 construct compared to the PBS group (Fig. 2H).
Immune protection effect of G mRNA-LNPs against IHNV infection
We further explored whether these mG constructs can provide efficient protection against IHNV infection. To this end, mGs (capping efficiency: mG1 95.02%, mG2 93.93%, mG3 93.52%) were encapsulated using the commercial SM102 lipids to generate stable G mRNA-LNPs. We previously established a DNA vaccine encoding the same glycoprotein, which elicited robust protective immunity at a 10 μg dose per fish22. Additionally, unlike mammalian vaccination protocol27,28,29, the operation of booster immunization in aquaculture is challenging and often induces a physiological stress response in fish. Collectively, to achieve optimal immune protection under these constraints, we immunized rainbow trout weighing 6–10 g intramuscularly with a single dose (10 μg) mRNA-LNP strategy (Fig. 3A). At 2 and 4 weeks post-vaccination, serum samples were collected to determine the production of serum IgM antibody and neutralizing antibody. After intraperitoneal challenge with a highly virulent clinical isolate GS21, survival rates were monitored for additional 2 weeks and relative percent survival (RPS) was calculated. Considering that encapsulation efficiency and LNP usage may influence the immune responses30, FLUC-LNP, encapsulating mRNA molecules encoding firefly luciferase (mFLUC), was employed as the internal control supplement to PBS vaccination.
A Scheme of vaccination and challenge (Created with BioRender.com with permission). Rainbow trout (n = 40 per group) were immunized with 10 μg doses of mG-LNPs. The control groups received PBS or 10 μg doses of FLUC-LNP. At 14 dpv and 28 dpv, serum from rainbow trout was collected to detect IgM titers (n = 6) and neutralizing antibody levels (n = 4). At 28 dpv, rainbow trout (n = 30 per group) were challenged with 103 TCID50 GS21 virus and monitored for survival rates for 14 days. B Serum IgM antibody levels at 14 dpv and 28 dpv (n = 6). C Serum neutralizing antibody levels at 14 dpv and 28 dpv (n = 4). D Survival rates of vaccinated rainbow trout. Survival curves are from two independent experiments. E RPS of vaccinated groups and negative groups after challenge. All data were presented as mean ± SEM and analyzed by two-way ANOVA (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, ns, not significant).
Serum antibody and neutralizing antibody levels are key metrics for evaluating vaccine efficacy31. As demonstrated in previous studies on DNA vaccine encoding glycoprotein32, all G mRNA-LNPs stimulated the significant production of IgM antibodies and neutralizing antibodies in rainbow trout at 28 dpv, whereas the PBS- and FLUC- LNP- vaccinated groups did not yield considerable antibodies (Fig. 3B, C). In line with glycoprotein production in transfected HEK293T and CHSE-214 cells, mG1-LNPs resulted in the highest level of IgM and neutralizing antibodies. No apparent IgM and neutralizing antibodies were detected in all test rainbow trout at 14 dpv (Fig. 3B, C). Remarkably, only mG1-LNPs provided strong protection against IHNV infection, of which 83.3% of rainbow trout immunized with mG1-LNP survived the entire period, with the RPS of 75.0% (Fig. 3D, E).
mG1-LNP offers strong protection against IHNV infection
We thus decided to use mG1-LNP for further investigation due to its ability to induce high-level antigen expression and robust immune protection. Following the same immunological assessment (Fig. 4A), different doses of mG1-LNPs were employed for vaccination, including 1 μg, 10 μg, 20 μg, 30 μg, and 40 μg, respectively. Similarly, all doses of mG1-LNPs induced the production of IgM antibodies and neutralizing antibodies in rainbow trout, with antibody levels increasing from 14 to 28 dpv (Fig. 4B, C). Notably, a negative correlation was observed between immunization dosage and antibody level and the immunization of 10 μg mG1-LNPs resulted in the highest level of IgM antibodies and neutralizing antibodies, of which IgM level was four-fold higher than that of the PBS group (Fig. 4B) and the neutralizing antibody titer achieved 1:244 at 28 dpv (Fig. 4C).
A Scheme of vaccination and challenge (Created with BioRender.com with permission). Rainbow trout (n = 40 per group) were immunized with 1, 10, 20, 30, and 40 μg doses of mG1-LNP. The control groups received PBS or 40 μg doses of FLUC-LNP. At 14 dpv and 28 dpv, serum from rainbow trout was collected to detect IgM titers (n = 6) and neutralizing antibody levels (n = 4), spleen and head kidney tissues were collected to detect immune factor expression (n = 4), and liver and injection site muscle tissues were collected to detect mRNA retention (n = 4). At 28 dpv, rainbow trout (n = 30 per group) were challenged with 103 TCID50 GS21 virus and monitored for survival rates for 14 days. B Serum IgM antibody levels at 14 and 28 dpv (n = 6). C Serum neutralizing antibody levels at 14 and 28 dpv (n = 4). D Survival rates of vaccinated rainbow trout. Survival curves are from two independent experiments. E RPS of vaccinated groups and negative groups after challenge. F and G Retention of mG1-LNP in muscle (F) and liver (G) at 14 and 28 dpv (n = 4). All data were presented as mean ± SEM and analyzed by two-way ANOVA (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, ns, not significant).
After intraperitoneal injection challenges with 103 TCID50 GS21, all doses of mG1-LNPs exhibited considerable immune protection. All vaccinated groups significantly improved the survival rate, compared to less than 30% survival in the unvaccinated group (Fig. 4D). Consistent with the profiles of IgM antibodies and neutralizing antibodies, 10 μg mG1-LNP demonstrated the RPS of 84.61% and the RPS slightly declined as the vaccine dosage increased (Fig. 4E).
We then evaluated the persistence of mG1-LNP residues at the site of immunization to identify any potential for prolonged immune stimulation or adverse reactions. Typically, after intramuscular injection of mRNA vaccines, there is prolonged distribution at the injection site. It was found that the vaccinated rainbow trout still exhibited relatively high retention of mG1-LNP in muscle tissue at 14 dpv (Fig. 4F). Moreover, the liver-targeting nature of LNP generally results in higher mRNA exposure in the liver33,34. Consistent with this, a certain amount of mG1-LNP was detectable in the liver tissue (Fig. 4G). By 28 dpv, mG1-LNP was nearly undetectable in muscle and liver as in PBS- and FLUC-LNP- vaccinated groups. Furthermore, no side effects were observed during the 28-day immunization period with the increasing immune doses.
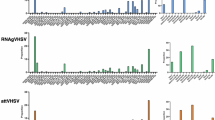
To explore the immune response stimulated by different doses of mG1-LNP vaccines, the transcriptional levels of Mx-1, IFN-γ, CD4, CD8, and IgM in the head kidney and spleen were assessed by RT-qPCR at indicated time points after vaccination. A dose-dependent escalation in antiviral gene transcription was observed and the corresponding expression profiles peaked at 14 dpv and gradually recovered to those in the PBS-vaccinated group (Fig. 5A, B). Notably, the vaccination by FLUC-LNP also triggered an apparent antiviral immune response, probably due to the immune recognition against exogenous nucleic acids. Similarly, the expression levels of adaptive immune-related genes in the mG1-LNP vaccinated group were significantly higher than those in the PBS-vaccinated group throughout the immune process (Fig. 5C, D). The transcriptional levels of these immune factors peaked at 14 dpv and exhibited only marginal attenuation by 28 dpv, indicative of sustained immune response. Considering neutralizing antibody titers reached maximal geometric mean titers at 28 dpv, a 14-day lag phase between immune system activation and immune response maturation was observed, conferring immunological precision that early IFN-γ-driven viral suppression gains time for high-affinity antibody development.
A and B Transcripts of anti-viral immune-related factors expression in the spleen (A) and head kidney (B) at 14 dpv and 28 dpv (n = 4). C and D Transcripts of adaptive immune-related factors expression in the spleen (C) and head kidney (D) at 14 dpv and 28 dpv (n = 4). All data were presented as mean ± SEM and analyzed by two-way ANOVA (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, ns, not significant).
Discussion
Since the initial testing of mRNA influenza vaccines in mice in 199035 and the subsequent global rollout of COVID-19 mRNA vaccine1, mRNA vaccines have undergone decades of development and are now extensively used in the prevention of infectious diseases5. Currently, the primary methods for preventing and controlling aquatic diseases rely on traditional inactivated vaccines, attenuated vaccines, and DNA vaccines, which have achieved considerable research progress22,23,24,25. Herein, we developed a mRNA vaccine encoding the glycoprotein with its specific UTRs. The mG1-LNP formulation conferred 84.61% relative protection against IHNV challenge, accompanied by robust immune responses. Dose optimization supported 10 μg/dose to ensure immunogenicity, and biodistribution analyses underscored its biosafety.
The 5’ UTR and 3’ UTR regions are crucial for efficient in vivo expression of antigens and reducing vaccine cost. The 5’ UTR of human α-globin and two serial 3’ UTRs of the β-globin have been widely and successfully employed in mammalian vaccine mRNA18,19,20. However, due to the distinct protein translation and immune microenvironment in fish compared to mammals, it is essential to select effective UTRs that drive high and uniform mRNA expression in fish. In our study, we utilized the original UTRs of IHNV G gene as well as human α-globin 5’ UTR and two duplicate β-globin 3’ UTRs. Preliminary intracellular results demonstrated that all UTR constructs mediated robust EGFP and glycoprotein protein production, with no statistically significant intergroup differences in protein yield across cellular models. Strikingly, quantitative analyses of the vaccination effect indicated that the vaccine encoding antigen-specific native UTRs elicited superior immunoprotection in vivo, as evidenced by the best survival rate post-challenge and highest neutralizing antibody titers, compared to heterologous UTR variants.
The divergence between in vitro and in vivo efficacy may reflect unique immunological and physiological constraints in fish. The enhanced performance of endogenous UTRs likely stems from their evolutionary optimization for temperature-resilient translational fidelity and mRNA stability maintenance within the poikilothermic host, where the environmental temperature fluctuations may impose distinct biomolecular demands36. Furthermore, species-specific ribosome loading kinetics and innate immune responses—potentially compromised by exogenous UTRs—may critically modulate antigen persistence required for robust adaptive immunity in fish37,38. Our framework aligns with emerging paradigms in comparative vaccinology, emphasizing that cross-species mRNA vaccine optimization requires the holistic integration of host-specific translational regulation and environmental adaptation networks37.
Although mRNA vaccines carry a lower risk of integration into the host genome than DNA vaccines30, safety considerations must address the possibility of tissue residue from mRNA constructs. Typically, after intramuscular injection of mRNA vaccines, there is prolonged distribution at the injection site, and the liver-targeting nature of LNP results in higher mRNA exposure in the liver33. Data released by BioNTech indicated that after intramuscular injection of LNP-encapsulated luciferase mRNA into the hind leg muscles of mice, strong fluorescence was observed in the liver from 8 to 48 h post-administration and then undetectable after 48 h34. In our study, we detected a significant presence of mRNA molecules at both the injection site and liver at 14 dpv. However, by 28 dpv, the levels were nearly indistinguishable from those in the non-immunized group. Considering that retention time reflects the durability of vaccine-induced immunoprotection to a certain extent39, our results confirmed that mG1-LNP conferred a prolonged immune duration and simultaneously demonstrated reliable biosafety in rainbow trout.
The immunoprotective efficacy of mRNA vaccines exhibits a dose-dependent relationship, wherein the administered mRNA quantity critically governs the efficacy and durability of adaptive immune responses40. Preclinical and clinical data demonstrate that suboptimal dosing (<10 μg in murine models) fails to achieve the threshold antigen expression required for robust germinal center activation, resulting in diminished neutralizing antibody titers41. Conversely, supratherapeutic doses may paradoxically induce innate immune overactivation, triggering interferon-mediated suppression of antigen translation and restraining immunogen exposure windows4. Additionally, the 10 μg vaccination dose (a relatively high antigen load on rainbow trout weighing 6–10 g) employed in our study diverges fundamentally from mammalian vaccination paradigms reliant on booster immunizations27,28,29. In aquaculture contexts, booster immunization protocols impose significant environmental and handling stress on fish, often precipitating physiological dysregulation that undermines vaccine efficacy. Therefore, our single high-dose mRNA-LNP strategy circumvents these limitations by achieving sterilizing immunity (84.61% of RPS) without requiring boosters. In the future, the dose dependency underscores the necessity for optimization in mRNA vaccine development to achieve the critical balance between immunoprotective efficacy and safety.
In summary, our study demonstrated a robust immune response provided by mRNA vaccination in rainbow trout following effective molecular design and dose optimization. Although the RPS of mG1-LNP was less than those of DNA vaccine encoding glycoprotein, we believe that the potential of mRNA vaccines to revolutionize disease prevention in aquaculture is significant due to its advantages in terms of biosafety and developmental potential in aquaculture. Future optimization of aquatic mRNA vaccines should focus on the delivery systems suitable for fish, the unique immune responses of fish, and large-scale clinical evaluations of efficacy and safety.
Material and methods
Cell lines, virus, and fish
The IHNV-sensitive Chinook salmon embryonic cell line (CHSE-214) was a kind gift from Northeast Agricultural University. The IHNV strain GS21 (CCTCC V202467) was isolated and characterized as the J genotype. Triploid rainbow trout larvae (6–10 g) were sourced from a fishery in Gansu, China.
mRNA design and synthesis
Three distinct models of 5’ UTR-egfp-6×his-3’ UTR-A(100) were designed and constructed. In mE1, both 5’ and 3’ UTRs were derived from the IHNV glycoprotein. The modified α-globin 5’ UTR designed by Pfizer/BioNTech18,19 was employed in mE2 and mE3. mE2 incorporated the IHNV glycoprotein 3’ UTR, while mE3 contained a tandem sequence of two human β-globin 3’ UTRs (Fig. 1A). The three segments were individually inserted into the cloning vector pVAX1 to create mRNA production plasmids. mG1, mG2, and mG3 were the corresponding mE1, mE2, and mE3 constructs, of which EGFP was replaced by optimized IHNV glycoprotein (Fig. 2A). The glycoprotein sequences were codon-optimized based on the codon usage bias within zebrafish (Geneart, Thermo-Fisher) and synthesized (GenScript).
After ligation into expression vectors, mRNAs were produced using T7 RNA polymerase (MEGAscript) on linearized plasmids. The 100nt poly(A) tail is synthesized based on the DNA template. m1Ψ-5’-triphosphate instead of UTP, was used to generate modified nucleoside-containing mRNA. Capping of the in vitro-transcribed mRNAs was performed co-transcriptionally using the trinucleotide cap1 analog CleanCap. mRNA was purified using cellulose chromatography, as described.
Cell cultivation
CHSE-214 was propagated at 20 °C in Medium 199 (Gibco) supplemented with 10% fetal bovine serum (FBS) (Biosera) and 1% streptomycin and penicillin. HEK293T was propagated at 37 °C with 5% CO2 in DMEM (OPM) supplemented with 10% FBS and 1% streptomycin and penicillin.
Detection for mRNA expression
The expression of mEs was evaluated in HEK293T and CHSE-214 cells, seeded in 24-well plates at a density of 500,000 cells/well. For mGs expression assessment, HEK293T cells were seeded in 24-well plates at a density of 500,000 cells/well, while CHSE-214 cells were seeded in 12-well plates at a density of 1,200,000 cells/well. Cells were transfected with 1 μg of mRNA per well via Lipofection after 24 h. RNAs were complexed with Lipofectamine MessengerMAX (Thermo-Fisher) at a ratio of 1:1.5 and transfected into cells according to the manufacturer’s protocol.
After transfecting the cells with mE for 24 h, observe the EGFP fluorescence signal under a fluorescence microscope. Select the appropriate fluorescence channel, typically using a 488 nm excitation light source, to visualize the green fluorescence in the cells. The MFI was analyzed using ImageJ. MFI is calculated by dividing the integrated density (IntDen), which represents the total fluorescence intensity within the region of interest (ROI), by the area of the ROI.
For FACS analysis, 24 h post-transfection of the three mE, the original culture medium was discarded, and the cells were washed twice with PBS to remove the residual medium. The final cell suspension was prepared in 1 ml of PBS. FACS analysis was then performed using the FITC channel. Fluorescence intensity was analyzed using FlowJo software.
For Western blotting, protein expression in whole cell lysates was analyzed. Protein samples were separated by 10% SDS-PAGE and transferred onto PVDF membranes using a wet transfer method. The PVDF membranes were blocked with 5% non-fat milk in 1× PBST buffer for 2 h at room temperature. Protein detection was performed using 6X His-tag Rabbit Polyclonal Antibody (ABclonal, AE086) for 2 h, followed by incubation with a secondary goat anti-rabbit IgG-HRP antibody (HUABIO, HA1001) for 2 h. GADPH was used as the reference gene, using Anti-GAPDH Recombinant Rabbit Monoclonal Antibody (HUABIO, ET1601-4), followed by incubation with a secondary goat anti-rabbit IgG-HRP antibody (HUABIO, HA1001). Membranes were imaged using the Tanon-5200 chemiluminescence imaging system with Super ECL detection reagent. The relative densities of protein bands were quantified using ImageJ. Protein intensity was normalized to GAPDH intensity from the blank control group, and relative expression was calculated by comparing the target protein intensity to GAPDH.
For IIF, 48 h post-transfection of mG, the upper culture medium was discarded, and the cells were washed three times with PBS. Then, 500 μl of immunofluorescence fixation solution was added to each well of a 24-well plate and incubated at room temperature for 20 min. Next, the cells were permeabilized using Triton X-100 permeabilization solution at room temperature for 10 min. The cells were then blocked with 5% BSA in PBST (PBS with 0.05% Tween 20) at room temperature for 30 min. After blocking, the cells were incubated with 6X His-tag Rabbit Polyclonal Antibody (ABclonal, AE086) at room temperature for 1 h, followed by incubation with FITC-Conjugated Goat anti-Rabbit lgG polyclonal Antibody (HUABIO, HA1004) at room temperature in the dark for 1 h. Finally, 300 μl of PBS was added, and the results were observed under a microscope. The MFI was analyzed using ImageJ by selecting the ROI and measuring the MFI of both green fluorescence and DAPI. The green fluorescence MFI was then normalized by dividing it by the DAPI MFI to account for variations in area.
To evaluate in vivo antigen expression, muscle tissue at the injection site was harvested and performed IHC analysis. Briefly, rainbow trout weighing 6–10 g were anesthetized by MS-222 and then intramuscularly injected with 10 μg mG1 construct encapsulated with the commercial SM102 lipids at the base of the dorsal fin. At 48 h post-injection, tissue samples were fixed in 4% paraformaldehyde at 4 °C and then embedded in paraffin. Sections (5 μm thick) were incubated with 6x-His Tag Rabbit Monoclonal Antibody (Invitrogen, MA5-33032) according to the manufacturer’s instructions. The immunoreactivity was visualized using 3,3’-diaminobenzidine as the chromogen, and sections were counterstained lightly with Mayer’s hematoxylin.
Detection for mRNA stability
The transfection method was performed as described above. Cell samples were collected at 6, 12, 24, 48, and 72 h post transfection (n = 3), and total RNA was extracted using Trizol (Invitrogen, 15596018CN). cDNA was synthesized using the FastQuant RT Kit (TransGen, AT341-01). Subsequently, RT-qPCR was performed in triplicate using SuperReal PreMix Plus (SYBR Green) (Monard, MQ10701S). The absolute quantification method was employed to measure the gene expression of glycoprotein, thereby assessing the degradation rate of mRNA within the cells. The primers used are presented in Table 1.
Detection for mRNA capping efficiency
The capping efficiency of mRNA was quantified via liquid chromatography-mass spectrometry (LC-MS). Briefly, a biotinylated probe featuring a 2’-O-methyl-modified RNA oligonucleotide with 4–6 cleavage sites at its 3’ terminus was designed to hybridize with the 5’ UTR region of target mRNA. Following streptavidin magnetic bead isolation of 5’ cleavage products digested by RNase H, capping efficiency was determined by LC-MS analysis and uncapped mRNA was served as the negative control. The analytical procedure was performed by OPM (China).
Package N1ψmRNA in LNPs
LNPs was incorporated based on its application in Moderna’s COVID-19 Spike mRNA-LNP vaccine. The lipid mixture includes the ionizable cationic amino-lipid SM-102 (CAS 2089251-47-6, AVT Pharmaceutical Tech Co., Ltd., Cat. O02010), the zwitterionic amino phospholipid DSPC (CAS 816-94-4, AVT Pharmaceutical Tech Co., Ltd, Cat. S01005), the stealth amino phospholipid DMG-PEG2000 (CAS 160743-62-4, AVT Pharmaceutical Tech Co., Ltd, Cat. O02005), and cholesterol (CAS 57-88-5, AVT Pharmaceutical Tech Co., Ltd, Cat. O01001) in molar ratios of 50:10:1.5:38.5, respectively. To prepare mRNA-LNPs, microfluidic mixing was employed using a NWDPS II 40 rapid nanodrug preparation system (Nanowetech Co., Ltd.). A mixture of 8 mg of lipids dissolved in ethanol and 100 μg of mRNA in a pH 4.0 buffer (50 mM citrate buffer) was combined at a 3:1 volume ratio with a flow rate of 12 mL/min using Nanowetech’s microfluidic chips. Following synthesis, the mRNA-lipid complexes were dialyzed into 100 volumes of pH 7.4 TBS buffer (Servicebio, Cat.#G0001). Dialysis in a neutral buffer facilitates the formation of closed, spherical LNPs.
Virus replication
The IHNV isolate GS21 from the Gansu fishery was added to 24-hour-old CHSE-214 cell monolayers and incubated at 15 °C until a pronounced cytopathic effect (CPE) developed42. The cells were collected and centrifuged at 10,000 × g for 10 min at 4 °C. The pellet was resuspended in the culture medium containing dual antibiotics and 2% serum, and subsequently stored at −20 °C, frozen and thawed three times, followed by centrifugation at 10,000 × g for 10 min. The supernatant was transferred into aliquots and stored at −80 °C for future use.
Determination of virus titers
The virus stock was serially diluted in Medium 199 using 10-fold dilutions and added to a 96-well cell culture plate containing a confluent monolayer of CHSE-214 cells. Each well received 100 μl of the diluted virus, with eight replicate wells per dilution. Concurrently, eight wells were designated as normal cell controls, each receiving 100 μl of Medium 199. The plate was incubated at 15 °C for 7 days. The number of wells exhibiting CPE was recorded. The 50% TCID50 was calculated using the karber method.
Vaccination and challenge
Rainbow trout weighing 6–10 g were randomly divided into groups of 40 fish each. The fish were acclimated for 14 days in recirculating water tanks (65 × 65 × 70 cm) maintained at a temperature of (15 ± 1) °C. Following acclimation, the fish were anesthetized by MS-222 and then intramuscularly injected with 50 μl of mRNA vaccine at the base of the dorsal fin. The fish were fasted for 3 days prior to the experiment. The mRNA vaccine was administered at doses of 1, 10, 20, 30, and 40 μg per fish. A blank control group was established, receiving 50 μl of PBS per fish. Considering that LNP may also stimulate an immune response in fish, a negative control group was included, receiving mFLUC at 40 μg per fish.
After 28 days, fish were anesthetized by MS-222 and then intraperitoneally challenged with 100 μl of 103 TCID50 GS21 per fish (n = 30 per group) at a temperature of (12 ± 1) °C. Immune and challenge experiments were performed in accordance with the Animal Experiment Committee of the East China University of Science and Technology (Protocol No. 2006272). The survival rates of rainbow trout were monitored for 14 consecutive days. All the surviving rainbow trout were euthanized by MS-222 at the predetermined clinical endpoints.
Determination of indirect ELISA for serum IgM antibody
Serum samples were collected from fish (n = 6) at 14 and 28 dpv. After incubating the samples at room temperature for at least 2 h, they were centrifuged at 3000 × g for 10 min.
IgM levels were quantified using a conventional indirect ELISA method. A 96-well ELISA plate was coated overnight at 4 °C with 10⁶ TCID₅₀/well of the GS21 strain. After washing three times with PBST, the wells were blocked with 1% (w/v) BSA solution (300 μl/well) at 22 °C for 2 h. After additional washes with PBST, 100 μl of diluted serum samples were added in triplicate to each well and incubated at 22 °C for 3 h. PBS was used as a negative control. After washing with PBST, 100 μl diluted anti-rainbow trout IgM monoclonal antibody (Aquatic Diagnostics Ltd, F11) was added and incubated at 22 °C for 1 h. The wells were washed again with PBST, and 100 μl of diluted anti-mouse IgG-HRP (HUABIO, HA1008) was added, followed by another incubation at 22 °C for 1 h. After a final wash with PBST, 100 µl/well of TMB substrate was added and incubated at 22 °C for 10 min. The reaction was stopped by adding 50 μl/well of 2 M H₂SO₄, and absorbance at 450 nm was immediately measured.
Detection of neutralizing antibody (NAb) titer
Serum samples were collected as described above. The samples were heat-inactivated at 56 °C for 30 min (n = 4). Serum samples were then diluted two-fold from 1:10 to 1:320 and incubated with an equal volume of 100 TCID50 of IHNV at 15 °C for 1 h. Following this incubation, the mixed samples were added to a monolayer of CHSE-214 cells and incubated at 15 °C for 7–10 days, until no further CPE was observed. The neutralization titer was defined as the highest dilution at which 50% of the wells showed no cytopathic effect. The neutralizing titer of the serum was calculated using the Reed-Muench method.
Detection of mRNA vaccine retention time
Muscle and liver samples were collected at 14 and 28 dpv (n = 4) to assess the retention of the vaccine. As described above, RT-qPCR was carried out, and the mRNA vaccine content in the tissues was analyzed using absolute quantification methods. The primers used are listed in Table 1.
Detection of immune gene expression
Head kidney and spleen samples were collected from each group (n = 4) at 14 and 28 dpv to analyze the relative expression profiles of Mx-1, IFN-γ, CD4, CD8, and IgM. β-actin was used as the reference gene. Relative gene expressions were calculated using the 2−ΔΔCt method in RT-qPCR. The primers used are listed in Table 1.
Statistical analysis
Statistical data were analyzed using GraphPad Prism 9.5 software (La Jolla). The differences in detection of the expression and vaccine retention in cells among multiple groups were calculated using one-way ANOVA with Kruskal–Wallis test. Two-way ANOVA was employed to assess the differences in serum IgM levels, neutralizing antibody (NAb) titers, vaccine retention in fish, and immune gene expression across multiple groups. Significance was indicated as follows: (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant).
Data Availability
Data is provided within the manuscript. Plasmids and strains generated in this study are available from the corresponding author upon reasonable request. The sequences for mEs and mGs have been deposited in NCBI under accession numbers of PV867280-PV867285.
References
Hogan, M. J. & Pardi, N. mRNA vaccines in the COVID-19 pandemic and beyond. Annu. Rev. Med. 73, 17–39 (2022).
Pulido, M. R. et al. RNA immunization can protect mice against foot-and-mouth disease virus. Antivir. Res 85, 556–558 (2010).
Tseng, H. F., Bradley, K. & Ackerson, S. mRNA-1273 bivalent (original and Omicron) COVID-19 vaccine effectiveness against COVID-19 outcomes in the United States. Nat. Commun. 14, 5851 (2023).
Fernando, P., Polack, M. D. & Stephen, J. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020).
Zhang, G. et al. mRNA vaccines in disease prevention and treatment. Signal Transduct. Target. Ther. 8, 1–30 (2023).
Pardi, N. et al. Development of a pentavalent broadly protective nucleoside-modified mRNA vaccine against influenza B viruses. Nat. Commun. 13, 4677 (2022).
Xu, S. et al. mRNA vaccine era - mechanisms, drug platform and clinical prospection. Int. J. Mol. Sci. 21, 6582 (2020).
Saxena, S. et al. Induction of immune responses and protection in mice against rabies using a self-replicating RNA vaccine encoding rabies virus glycoprotein. Vet. Microbiol. 136, 36–44 (2009).
Hajam, I. A., Senevirathne, A. & Hewawaduge, C. Intranasally administered protein coated chitosan nanoparticles encapsulating influenza H9N2 HA2 and M2e mRNA molecules elicit protective immunity against avian influenza viruses in chickens. Vet. Res. 51, 37 (2020).
Li, J. et al. An mRNA-based rabies vaccine induces strong protective immune responses in mice and dogs. Virol. J. 19, 184 (2022).
Gong, L. et al. Advancing vaccine development: evaluation of a mannose-modified lipid nanoparticle-based candidate for African swine fever p30 mRNA vaccine eliciting robust immune response in mice. Int. J. Biol. Macromol. 270, 132432 (2024).
Snoeck, J. et al. Evaluation of a self-amplifying mRNA reporter vaccine in explant models of broiler chickens. Poult. Sci. 102, 103078 (2023).
Eygeris, Y. et al. Chemistry of lipid nanoparticles for RNA delivery. Acc. Chem. Res. 55, 2–12 (2022).
Mokhtar, D. M. et al. Main components of fish immunity: An overview of the fish immune system. Fishes 8, 93 (2023).
Martinez-Alonso, S. et al. The introduction of multi-copy CpG motifs into an antiviral DNA vaccine strongly up-regulates its immunogenicity in fish. Vaccine 29, 1289–1296 (2011).
Kang, D. D., Li, H. Y. & Dong, Y. Z. Advancements of in vitro transcribed mRNA (IVT mRNA) to enable translation into the clinics. Adv. Drug Deliv. Rev. 199, 114961 (2023).
Leppek, K. et al. Combinatorial optimization of mRNA structure, stability, and translation for RNA-based therapeutics. Nat. Commun. 13, 1536 (2022).
Vogel, A. B. et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 592, 283–289 (2021).
Sahin, U. et al. COVID-19 vaccine BNT162b1 elicits human antibody and Th1 T cell responses. Nature 586, 594–599 (2020).
Fang, E. et al. Advances in COVID-19 mRNA vaccine development. Signal Transduct. Target. Ther. 7, 94 (2022).
Zhang, H. et al. Algorithm for optimized mRNA design improves stability and immunogenicity. Nature 621, 396–403 (2023).
Wu, D. et al. Protection of a CpG-adjuvanted DNA vaccine against infectious hematopoietic necrosis virus (IHNV) nature infection in rainbow trout (Oncorhynchus mykiss). Aquaculture 572, 739555 (2023).
Ammayappan, A., Lapatra, S. E. & Vakharia, V. N. Molecular characterization of the virulent infectious hematopoietic necrosis virus (IHNV) strain 220-90. Virol. J. 7, 10–20 (2010).
Gebre, M. S. et al. Optimization of non-coding regions for a non-modified mRNA COVID-19 vaccine. Nature 601, 410–414 (2022).
Lima, S. A. et al. Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol. 24, 1057–1063 (2018).
Engelking, H. M. & Leong, J. A. C. The glycoprotein of infectious hematopoietic necrosis virus elicits neutralizing antibody and protective responses. Virus Res. 13, 213–230 (1989).
Lindsey, R. et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 384, 403–416 (2021).
Pardi, N. et al. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 9, 3361 (2018).
Lutz, J. et al. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. npj Vaccines 2, 1–9 (2017).
Bitounis, D. et al. Strategies to reduce the risks of mRNA drug and vaccine toxicity. Nat. Rev. Drug Discov. 23, 281–300 (2024).
Zhang, B. et al. Neutralizing antibody immune correlates in COVAIL trial recipients of an mRNA second COVID-19 vaccine boost. Nat. Commun. 16, 759 (2025).
LaPatra, S. E., Corbeil, S. & Jones, G. R. Protection of rainbow trout against infectious hematopoietic necrosis virus four days after specific or semi-specific DNA vaccination. Vaccine 19, 4011–4019 (2000).
Food and Drug Administration. Public assessment report authorisation for temporary supply COVID-19 mRNA vaccine BNT162. (FDA, 2024).
European Medicines Agency. Comirnaty: EPAR - Public assessment report. (European Medicines Agency, 2023).
Wolff, J. A. et al. Direct gene transfer into mouse muscle in vivo. Science 247, 1465–1468 (1990).
Lorenzen, E. et al. The protective mechanisms induced by a fish rhabdovirus DNA vaccine depend on temperature. Vaccine 27, 3870–3880 (2009).
Pardi, N. & Krammer, F. mRNA vaccines for infectious diseases - advances, challenges and opportunities. Nat. Rev. Drug Discov. 23, 838–861 (2024).
Kim, S. et al. Innate immune responses against mRNA vaccine promote cellular immunity through IFN-β at the injection site. Nat. Commun. 15, 7226 (2024).
David, M. M. et al. mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. USA 116, 24075–24083 (2019).
Chaudhary, N., Weissman, D. & Whitehead, K. A. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat. Rev. Drug Discov. 20, 817–838 (2021).
Buschmann, M. D. et al. Nanomaterial delivery systems for mRNA vaccines. Vaccines 9, 65 (2021).
Zhang, Z. et al. Production and evaluation of a bivalent adjuvant inactivated vaccine against infectious hematopoietic necrosis virus and infectious pancreatic necrosis virus. Aquaculture 597, 741914 (2025).
Acknowledgements
This work was supported by grants from the National Key Research and Development Program (2023YFD2400702), Qingdao Marine Science and Technology Center (11–04), and Shanghai Agricultural Science and Technology Innovation Program (K2023014).
Author information
Authors and Affiliations
Contributions
Conceptualization: S.S. and L.Q. Data curation: W.J., W.K.X., L.Y., and S.S. Funding acquisition: S.S. and L.Q. Investigation: W.J., W.K.X., L.Y., W.S.Y., and Z.L. Project administration: R.Y.H., Z.D.H., and L.R.H. Supervision: S.S. and L.Q. Validation: S.S. Visualization: W.J. and W.K.X. Writing—original draft: W.J., W.K.X., and S.S. Writing – review and editing: S.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, J., Wu, K., Liu, Y. et al. Development of an mRNA vaccine encoding IHNV glycoprotein protects rainbow trout (Oncorhynchus mykiss) from infection. npj Vaccines 10, 162 (2025). https://doi.org/10.1038/s41541-025-01222-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41541-025-01222-w