Abstract
As lifelong interphase cells, neurons face an array of unique challenges. A key challenge is regulating nuclear pore complex (NPC) biogenesis and localization, the mechanisms of which are largely unknown. Here we identify neuronal maturation as a period of strongly upregulated NPC biogenesis. We demonstrate that the AAA+ protein torsinA, whose dysfunction causes the neurodevelopmental movement disorder DYT-TOR1A dystonia, co-ordinates NPC spatial organization without impacting total NPC density. We generated an endogenous Nup107-HaloTag mouse line to directly visualize NPC organization in developing neurons and find that torsinA is essential for proper NPC localization. In the absence of torsinA, the inner nuclear membrane buds excessively at sites of mislocalized nascent NPCs, and the formation of complete NPCs is delayed. Our work demonstrates that NPC spatial organization and number are independently determined and identifies NPC biogenesis as a process vulnerable to neurodevelopmental disease insults.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
STEM volumes are available on the Cell Image Library (http://www.cellimagelibrary.org/groups/57193). Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code availability
FIJI, MATLAB and Python scripts for SIM and confocal analyses are available on GitHub at https://github.com/suminkim/NPC_NE_measurements. Scripts for dSTORM analyses are available on GitHub at https://github.com/VeatchLab/smlm-analysis.
References
Knockenhauer, K. E. & Schwartz, T. U. The nuclear pore complex as a flexible and dynamic gate. Cell 164, 1162–1171 (2016).
Beck, M. & Hurt, E. The nuclear pore complex: understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 18, 73–89 (2016).
Coyne, A. N. & Rothstein, J. D. Nuclear pore complexes—a doorway to neural injury in neurodegeneration. Nat. Rev. Neurol. 18, 348–362 (2022).
Cho, U. H. & Hetzer, M. W. Nuclear periphery takes center stage: the role of nuclear pore complexes in cell identity and aging. Neuron 106, 899–911 (2020).
Kandel, E. R. The molecular biology of memory storage: a dialogue between genes and synapses. Science 294, 1030–1038 (2001).
Cohen, S. & Greenberg, M. E. Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annu. Rev. Cell Dev. Biol. 24, 183–209 (2008).
Karpova, A. et al. Encoding and transducing the synaptic or extrasynaptic origin of NMDA receptor signals to the nucleus. Cell 152, 1119–1133 (2013).
Kaushik, R., Grochowska, K. M., Butnaru, I. & Kreutz, M. R. Protein trafficking from synapse to nucleus in control of activity-dependent gene expression. Neuroscience 280, 340–350 (2014).
Sutton, M. A. & Schuman, E. M. Local translational control in dendrites and its role in long-term synaptic plasticity. J. Neurobiol. 64, 116–131 (2005).
Sutton, M. A. & Schuman, E. M. Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127, 49–58 (2006).
Jung, H., Gkogkas, C. G., Sonenberg, N. & Holt, C. E. Remote control of gene function by local translation. Cell 157, 26–40 (2014).
Batista, A. F. R. & Hengst, U. Intra-axonal protein synthesis in development and beyond. Int. J. Dev. Neurosci. 55, 140–149 (2016).
Holt, C. E., Martin, K. C. & Schuman, E. M. Local translation in neurons: visualization and function. Nat. Struct. Mol. Biol. 26, 557–566 (2019).
Grima, J. C. et al. Mutant Huntingtin disrupts the nuclear pore complex. Neuron https://doi.org/10.1016/j.neuron.2017.03.023 (2017).
Pappas, S. S., Liang, C. C., Kim, S., Rivera, C. A. O. & Dauer, W. T. TorsinA dysfunction causes persistent neuronal nuclear pore defects. Hum. Mol. Genet. 27, 407–420 (2018).
Eftekharzadeh, B. et al. Tau protein disrupts nucleocytoplasmic transport in Alzheimer’s disease. Neuron https://doi.org/10.1016/j.neuron.2018.07.039 (2018).
Coyne, A. N. et al. G4C2 repeat RNA initiates a POM121-mediated reduction in specific nucleoporins in C9orf72 ALS/FTD. Neuron 107, 1124–1140.e11 (2020).
Lin, Y.-C. et al. Interactions between ALS-linked FUS and nucleoporins are associated with defects in the nucleocytoplasmic transport pathway. Nat. Neurosci. 24, 1077–1088 (2021).
Basel-Vanagaite, L. et al. Mutated nup62 causes autosomal recessive infantile bilateral striatal necrosis. Ann. Neurol. 60, 214–222 (2006).
Harrer, P. et al. Recessive NUP54 variants underlie early-onset dystonia with striatal lesions. Ann. Neurol. 93, 330–335 (2023).
Savas, J. N., Toyama, B. H., Xu, T., Yates, J. R. & Hetzer, M. W. Extremely long-lived nuclear pore proteins in the rat brain. Science 335, 942 (2012).
Toyama, B. H. et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 154, 971–982 (2013).
Ozelius, L. J. et al. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat. Genet. 17, 40–48 (1997).
Kustedjo, K., Bracey, M. H. & Cravatt, B. F. Torsin A and its torsion dystonia-associated mutant forms are lumenal glycoproteins that exhibit distinct subcellular localizations. J. Biol. Chem. 275, 27933–27939 (2000).
Hewett, J. et al. TorsinA in PC12 cells: localization in the endoplasmic reticulum and response to stress. J. Neurosci. Res. 72, 158–168 (2003).
Goodchild, R. E. & Dauer, W. T. Mislocalization to the nuclear envelope: an effect of the dystonia-causing torsinA mutation. Proc. Natl Acad. Sci. USA 101, 847–852 (2004).
Naismith, T. V., Heuser, J. E., Breakefield, X. O. & Hanson, P. I. TorsinA in the nuclear envelope. Proc. Natl Acad. Sci. USA 101, 7612–7617 (2004).
Gonzalez-Alegre, P. & Paulson, H. L. Aberrant cellular behavior of mutant torsinA implicates nuclear envelope dysfunction in DYT1 dystonia. J. Neurosci. 24, 2593–2601 (2004).
Gerace, L. TorsinA and torsion dystonia: unraveling the architecture of the nuclear envelope. Proc. Natl Acad. Sci. USA 101, 8839–8840 (2004).
Goodchild, R. E. & Dauer, W. T. The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J. Cell Biol. 168, 855–862 (2005).
Callan, A. C., Bunning, S., Jones, O. T., High, S. & Swanton, E. Biosynthesis of the dystonia-associated AAA+ ATPase torsinA at the endoplasmic reticulum. Biochem. J. 401, 607–612 (2006).
Ozelius, L. J. et al. Fine localization of the torsion dystonia gene (DYT1) on human chromosome 9q34: YAC map and linkage disequilibrium. Genome Res. 7, 483–494 (1997).
Ozelius, L. J. et al. The TOR1A (DYT1) gene family and its role in early onset torsion dystonia. Genomics 62, 377–384 (1999).
Nery, F. C. et al. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J. Cell Sci. 121, 3476–3486 (2008).
Vander Heyden, A. B., Naismith, T. V., Snapp, E. L., Hodzic, D. & Hanson, P. I. LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol. Biol. Cell 20, 2661–2672 (2009).
VanGompel, M. J. W., Nguyen, K. C. Q., Hall, D. H., Dauer, W. T. & Rose, L. S. A novel function for the Caenorhabditis elegans torsin OOC-5 in nucleoporin localization and nuclear import. Mol. Biol. Cell 26, 1752–1763 (2015).
Goodchild, R. E., Kim, C. E. & Dauer, W. T. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron 48, 923–932 (2005).
Zhao, C., Brown, R. S. H., Chase, A. R., Eisele, M. R. & Schlieker, C. Regulation of torsin ATPases by LAP1 and LULL1. Proc. Natl Acad. Sci. USA 110, E1545–E1554 (2013).
Rose, A. E., Brown, R. S. H. & Schlieker, C. Torsins: not your typical AAA+ ATPases. Crit. Rev. Biochem. Mol. Biol. 50, 532–549 (2015).
Pappas, S. S. et al. Forebrain deletion of the dystonia protein torsinA causes dystonic-like movements and loss of striatal cholinergic neurons. eLife 4, e08352 (2015).
Tanabe, L. M., Liang, C. C. & Dauer, W. T. Neuronal nuclear membrane budding occurs during a developmental window modulated by torsin paralogs. Cell Rep. 16, 3322–3333 (2016).
Laudermilch, E. et al. Dissecting torsin/cofactor function at the nuclear envelope: a genetic study. Mol. Biol. Cell 27, 3964–3971 (2016).
Kim, C. E., Perez, A., Perkins, G., Ellisman, M. H. & Dauer, W. T. A molecular mechanism underlying the neural-specific defect in torsinA mutant mice. Proc. Natl Acad. Sci. USA 107, 9861–9866 (2010).
Jacquemyn, J. et al. Torsin and NEP1R1-CTDNEP1 phosphatase affect interphase nuclear pore complex insertion by lipid-dependent and lipid-independent mechanisms. EMBO J. 40, e106914 (2021).
Bressman, S. B. et al. The DYT1 phenotype and guidelines for diagnostic testing. Neurology 54, 1746–1752 (2000).
Li, J., Levin, D. S., Kim, A. J., Pappas, S. S. & Dauer, W. T. TorsinA restoration in a mouse model identifies a critical therapeutic window for DYT1 dystonia. J. Clin. Invest. 131, e139606 (2021).
Otsuka, S. et al. Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. eLife 5, e19071 (2016).
Otsuka, S. & Ellenberg, J. Mechanisms of nuclear pore complex assembly—two different ways of building one molecular machine. FEBS Lett. 592, 475–488 (2018).
Otsuka, S. et al. A quantitative map of nuclear pore assembly reveals two distinct mechanisms. Nature 613, 575–581 (2023).
Rampello, A. J. et al. Torsin ATPase deficiency leads to defects in nuclear pore biogenesis and sequestration of MLF2. J. Cell Biol. 219, e201910185 (2020).
Lesuisse, C. & Martin, L. J. Long-term culture of mouse cortical neurons as a model for neuronal development, aging, and death. J. Neurobiol. 51, 9–23 (2002).
Mosalaganti, S. et al. In situ architecture of the algal nuclear pore complex. Nat. Commun. 9, 2361 (2018).
Lin, D. H. & Hoelz, A. The structure of the nuclear pore complex (an update). Annu. Rev. Biochem. 88, 725–783 (2019).
Los, G. V. et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. https://doi.org/10.1021/cb800025k (2008).
Rabut, G., Doye, V. & Ellenberg, J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6, 1114–1121 (2004).
Dultz, E. & Ellenberg, J. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J. Cell Biol. 191, 15–22 (2010).
Lodin, Z., Blumajer, J. & Mares, V. Nuclear pore complexes in cells of the developing mouse cerebral cortex. Acta Histochem. 63, 74–79 (1978).
Jokhi, V. et al. Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep. 3, 988–995 (2013).
Liang, C. C., Tanabe, L. M., Jou, S., Chi, F. & Dauer, W. T. TorsinA hypofunction causes abnormal twisting movements and sensorimotor circuit neurodegeneration. J. Clin. Invest. 124, 3080–3092 (2014).
Maul, G. G., Price, J. W. & Lieberman, M. W. Formation and distribution of nuclear pore complexes in interphase. J. Cell Biol. 51, 405–418 (1971).
Maul, G. & Deaven, L. Quantitative determination of nuclear pore complexes in cycling cells with differing DNA content. J. Cell Biol. 73, 748–760 (1977).
Maul, G. G., Deaven, L. L., Freed, J. J., Campbell, L. M. & Beçak, W. Investigation of the determinants of nuclear pore number. Cytogenet. Genome Res. 26, 175–190 (1980).
Maul, G. G. et al. Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J. Cell Biol. 55, 433–447 (1972).
D’Angelo, M. A., Anderson, D. J., Richard, E. & Hetzer, M. W. Nuclear pores form de novo from both sides of the nuclear envelope. Science 312, 440–443 (2006).
Maeshima, K. et al. Nuclear pore formation but not nuclear growth is governed by cyclin-dependent kinases (Cdks) during interphase. Nat. Struct. Mol. Biol. 17, 1065–1071 (2010).
Varberg, J. M., Unruh, J. R., Bestul, A. J., Khan, A. A. & Jaspersen, S. L. Quantitative analysis of nuclear pore complex organization in Schizosaccharomyces pombe. Life Sci. Alliance 5, e202201423 (2022).
Doucet, C. M., Talamas, J. A. & Hetzer, M. W. Cell cycle-dependent differences in nuclear pore complex assembly in Metazoa. Cell https://doi.org/10.1016/j.cell.2010.04.036 (2010).
Doucet, C. M. & Hetzer, M. W. Nuclear pore biogenesis into an intact nuclear envelope. Chromosoma 119, 469–477 (2010).
Maul, G. G. The nuclear and the cytoplasmic pore complex: structure, dynamics, distribution, and evolution. Int. Rev. Cytol. Suppl. 6, 75–186 (1977).
Bucci, M. & Wente, S. R. In vivo dynamics of nuclear pore complexes in yeast. J. Cell Biol. 136, 1185–1199 (1997).
Belgareh, N. & Doye, V. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J. Cell Biol. 136, 747–759 (1997).
Romanauska, A. & Köhler, A. The inner nuclear membrane is a metabolically active territory that generates nuclear lipid droplets. Cell 174, 700–715.e18 (2018).
Ohsaki, Y. et al. PML isoform II plays a critical role in nuclear lipid droplet formation. J. Cell Biol. 212, 29–38 (2016).
Merta, H. & Bahmanyar, S. The inner nuclear membrane takes on lipid metabolism. Dev. Cell 47, 397–399 (2018).
Sołtysik, K., Ohsaki, Y., Tatematsu, T., Cheng, J. & Fujimoto, T. Nuclear lipid droplets derive from a lipoprotein precursor and regulate phosphatidylcholine synthesis. Nat. Commun. 10, 473 (2019).
Sołtysik, K. et al. Nuclear lipid droplets form in the inner nuclear membrane in a seipin-independent manner. J. Cell Biol. 220, e202005026 (2021).
Lee, S., Carrasquillo Rodrı́guez, J. W., Merta, H. & Bahmanyar, S. A membrane-sensing mechanism links lipid metabolism to protein degradation at the nuclear envelope. J. Cell Biol. 222, e202304026 (2023).
Mathiowetz, A. J. et al. CLCC1 promotes hepatic neutral lipid flux and nuclear pore complex assembly. Preprint at bioRxiv https://doi.org/10.1101/2024.06.07.597858 (2024).
Grillet, M. et al. Torsins are essential regulators of cellular lipid metabolism. Dev. Cell 38, 235–247 (2016).
Cascalho, A. et al. Excess Lipin enzyme activity contributes to TOR1A recessive disease and DYT-TOR1A dystonia. Brain 143, 1746–1765 (2020).
Shin, J.-Y. et al. Nuclear envelope-localized torsinA–LAP1 complex regulates hepatic VLDL secretion and steatosis. J. Clin. Invest. 129, 4885–4900 (2019).
Prophet, S. M. et al. Atypical nuclear envelope condensates linked to neurological disorders reveal nucleoporin-directed chaperone activities. Nat. Cell Biol. 24, 1630–1641 (2022).
Kuiper, E. F. E. et al. The chaperone DNAJB6 surveils FG-nucleoporins and is required for interphase nuclear pore complex biogenesis. Nat. Cell Biol. 24, 1584–1594 (2022).
Keuenhof, K. S. et al. Nuclear envelope budding and its cellular functions. Nucleus 14, 2178184 (2023).
Li, J., Liang, C.-C., Pappas, S. S. & Dauer, W. T. TorsinB overexpression prevents abnormal twisting in DYT1 dystonia mouse models. eLife 9, e54285 (2020).
Quartarone, A. & Hallett, M. Emerging concepts in the physiological basis of dystonia. Mov. Disord. 28, 958–967 (2013).
Quartarone, A. & Ghilardi, M. F. in Handbook of Clinical Neurology Vol. 184 (eds Quartarone, A., Ghilardi, M. F. & Boller, F.) 207–218 (Elsevier, 2022).
Yellajoshyula, D., Opeyemi, S., Dauer, W. T. & Pappas, S. S. Genetic evidence of aberrant striatal synaptic maturation and secretory pathway alteration in a dystonia mouse model. Dystonia 1, 10892 (2022).
Song, C.-H. et al. Subtle microstructural changes of the striatum in a DYT1 knock-in mouse model of dystonia. Neurobiol. Dis. 54, 362–371 (2013).
Song, C.-H., Bernhard, D., Hess, E. J. & Jinnah, H. A. Subtle microstructural changes of the cerebellum in a knock-in mouse model of DYT1 dystonia. Neurobiol. Dis. 62, 372–380 (2014).
Vanni, V. et al. Cerebellar synaptogenesis is compromised in mouse models of DYT1 dystonia. Exp. Neurol. 271, 457–467 (2015).
Maltese, M. et al. Early structural and functional plasticity alterations in a susceptibility period of DYT1 dystonia mouse striatum. eLife 7, e33331 (2018).
Capelson, M. et al. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140, 372–383 (2010).
Ibarra, A. & Hetzer, M. W. Nuclear pore proteins and the control of genome functions. Genes Dev. 29, 337–349 (2015).
Pascual-Garcia, P. et al. Metazoan nuclear pores provide a scaffold for poised genes and mediate induced enhancer-promoter contacts. Mol. Cell 66, 63–76.e6 (2017).
Sun, J., Shi, Y. & Yildirim, E. The nuclear pore complex in cell type-specific chromatin structure and gene regulation. Trends Genet. 35, 579–588 (2019).
Kadota, S. et al. Nucleoporin 153 links nuclear pore complex to chromatin architecture by mediating CTCF and cohesin binding. Nat. Commun. 11, 2606 (2020).
Toda, T. et al. Nup153 Interacts with Sox2 to enable bimodal gene regulation and maintenance of neural progenitor cells. Cell Stem Cell 21, 618–634.e7 (2017).
Orniacki, C. et al. Y-complex nucleoporins independently contribute to nuclear pore assembly and gene regulation in neuronal progenitors. J. Cell Sci. 136, jcs261151 (2023).
Jacinto, F. V., Benner, C. & Hetzer, M. W. The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 29, 1224–1238 (2015).
D’Angelo, M. A., Gomez-Cavazos, J. S., Mei, A., Lackner, D. H. & Hetzer, M. W. A change in nuclear pore complex composition regulates cell differentiation. Dev. Cell 22, 446–458 (2012).
Lupu, F., Alves, A., Anderson, K., Doye, V. & Lacy, E. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev. Cell 14, 831–842 (2008).
Li, J., Kim, S., Pappas, S. S. & Dauer, W. T. CNS critical periods: implications for dystonia and other neurodevelopmental disorders. JCI Insight 6, e142483 (2021).
Grimm, J. B. et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250 (2015).
Grimm, J. B. et al. A general method to improve fluorophores using deuterated auxochromes. JACS Au 1, 690–696 (2021).
Shaw, T. R. et al. Estimating the localization spread function of static single-molecule localization microscopy images. Biophys. J. 121, 2906–2920 (2022).
Chozinski, T. J. et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods 13, 485–488 (2016).
Asano, S. M. et al. Expansion microscopy: protocols for imaging proteins and RNA in cells and tissues. Curr. Protoc. Cell Biol. 80, e56 (2018).
Gambarotto, D. et al. Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat. Methods 16, 71–74 (2019).
Phan, S. et al. 3D reconstruction of biological structures: automated procedures for alignment and reconstruction of multiple tilt series in electron tomography. Adv. Struct. Chem. Imaging 2, 8 (2016).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Veatch, S. L. et al. Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS ONE 7, e31457 (2012).
Ester, M., Kriegel, H.-P., Sander, J. & Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. in Proc. of the Second International Conference on Knowledge Discovery and Data Mining 226–231 (AAAI Press, 1996).
Heydarian, H. et al. Template-free 2D particle fusion in localization microscopy. Nat. Methods 15, 781–784 (2018).
Lord, S. J., Velle, K. B., Mullins, R. D. & Fritz-Laylin, L. K. SuperPlots: communicating reproducibility and variability in cell biology. J. Cell Biol. 219, e202001064 (2020).
Acknowledgements
We thank the University of Michigan Microscopy Core facility; D. Levin and X. Li for animal husbandry; A. Taylor for assistance with MATLAB codes; S. Mosalaganti, A. Erwin, D. Boassa and A. Moore for feedback and experimental discussions; A. Diehl for illustrations; and H. Worman and D. Yellajoshyula for critical reviews. This work was supported by NIH R01 NS110853, R01 DK118480, R01 NS077730 to W.T.D., NIH R01 NS097542, R01 NS113943, R56 NS128110 to S.J.B., NIH GM129347 and NSF MCB1552439 to S.L.V., NIH T32 GM007315 and UM Rackham Predoctoral Fellowship to S.K. and NIH U24 NS120055, 1S10OD021784, R01 GM082949 and NSF2014862-UTA20-000890 to M.H.E.
Author information
Authors and Affiliations
Contributions
S.K., S.S.P., S.J.B. and W.T.D. conceived the study. S.K., H.T.T. and S.S.P. performed experiments. STEM tomograms were acquired by S.P. in M.H.E. laboratory. S.K., S.L.V., T.R.S. and S.P. analysed data. S.K. and S.S.P. wrote the manuscript. S.K. prepared the figures. S.L.V., S.J.B., S.H.S. and W.T.D. contributed to revisions. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Identification and analysis of nucleoporin puncta.
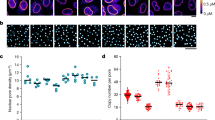
a, SIM images of primary neurons aged DIV4, 6, 8, 10, 14, 18 and 24 labeled with anti-Nup153 and anti-Nup98 antibodies. Peaks column shows peaks identified from Nup153 (green) and Nup98 (magenta) puncta. Scale bar = 2μm. b, Schematic of image analysis pipeline. The flattest part of the nuclear envelope was imaged. Nuclear ROIs were determined by manually outlining each nucleus. ROIs were eroded to account for nuclear envelope curvature at the edge, then overlayed onto Otsu thresholded images. Local maxima were identified with extended-maxima transform and used to establish peaks corresponding to each Nup. c, Superplots of nuclear ROI area as determined by Nup153/Nup98 staining in maturing neurons aged DIV4, 6, 8, 10, 14, 18 and 24. Plots show mean ± SD, with color coding indicative of biological replicates. One-way ANOVA with Dunnett’s multiple comparisons test on 3 biological replicates was performed, with DIV4 as the reference condition. Only DIV14 showed statistical significance; *P = 0.0331. d,e, Frequency distribution of nearest neighbor distances of Nup153 (d) and Nup98 (e) puncta. For each Nup153 and Nup98 peak, the distance to its nearest neighbor was calculated. The results from three biological replicates were combined for each timepoint. Source numerical data are available in source data.
Extended Data Fig. 2 Multiple NPC components exhibit density increase during neuronal maturation.
a, Confocal images of DIV4 and DIV10 primary neurons labeled with anti-Nup153 and anti-Nup98 antibodies. Scale bar = 10μm. b, c, Normalized nuclear rim fluorescence intensity of Nup153 and Nup98 in DIV4 and DIV10 primary neurons. Plots show mean ± SD, with color coding indicative of biological replicates. *P = 0.0196 (b), *P = 0.0139 (c) using two-tailed paired t-test from 4 biological replicates. d, Confocal images of DIV4 and DIV10 primary neurons labeled with anti-FG nucleoporin (mab414) and anti-Nup210 antibodies. Scale bar = 10μm. e, f, Normalized nuclear rim fluorescence intensity of FG-Nups (mab414) and Nup210 in DIV4 and DIV10 primary neurons. Plots show mean ± SD, with color coding indicative of biological replicates. **P = 0.0076 (e), ****P < 0.0001(f), two-tailed paired t-test from 4 biological replicates. Source numerical data are available in source data.
Extended Data Fig. 3 Comparison of WT and torsinA-KO neuronal NPC spatial organization and nuclear morphology.
a, Autocorrelation plot of WT and torsinA-KO neurons calculated from Nup98 WT and torsinA-KO SIM images. b, Normalized autocorrelation plot of WT and torsinA-KO neurons calculated from Nup98 WT and torsinA-KO SIM images. Autocorrelations were normalized to account for amplitude dependency on NPC density. c, Autocorrelation plot of DIV10 WT and torsinA-KO neurons calculated from Nup210 dSTORM images. d, Nearest neighbor distance of segmented DIV10 WT and torsinA-KO NPC centroids. e, f, WT and torsinA-KO Nup153 (e) and Nup98 (f) puncta density in maturing primary neurons aged DIV4, 6, 8, 10, 14. Plots show mean ± SD, with color coding indicative of biological replicates. Timepoints from the same biological replicate were matched. Repeated measures two-way ANOVA with Dunnett’s multiple comparisons test was performed on 3 biological replicates, with DIV4 as the reference condition. ns, not significant; P values are reported on the plots, ****P < 0.0001. g, Nuclear area of WT and torsinA-KO neurons calculated from manually drawn ROIs from SIM images. Plots show mean ± SD. Repeated measures two-way ANOVA with Sidak’s multiple comparisons test was used to compare genotypes at each timepoint, and no comparisons reached statistical significance. Repeated measures two-way ANOVA with Dunnett’s multiple comparisons test was used to compare all timepoints to DIV4 within each genotype, and only torsinA-KO DIV4 versus DIV14 reached significance (*P=0.0102). h, Nuclear ROI circularity of WT and torsinA-KO neurons calculated from manually drawn ROIs from SIM images. Plots show mean ± SD. Repeated measures two-way ANOVA with Sidak’s multiple comparisons test was used to compare genotypes at each timepoint. No statistical significance was detected. i, Expansion microscopy of DIV10 WT and torsinA-KO neurons labeled with Nup210 and Lamin A/C antibodies. Representative images from 3 biological replicates per genotype are shown. Scale bar = 20μm. Source numerical data are available in source data.
Extended Data Fig. 4 Validation of HaloTag-Nup107 mouse line.
a, Schematic of HaloTag-Nup107 fusion, including 5’ untranslated region (UTR, light grey), HaloTag open reading frame (magenta), flexible linker (dark grey), and Nup107 coding sequence (cyan). b, Immunoblot of P0 cortical lysates from Nup107+/+, Nup107KI/+, and Nup107KI/KI mice probed with anti-Nup107 and anti-HaloTag antibodies. GAPDH was blotted as a loading control. Each band represents an independent biological sample (3 animals each). c, Distribution of genotypes in litters derived from intercrossing Nup107KI/+ mice. No deviation from the expected Mendelian ratio was observed (P = 0.3729, Chi-squared test). 74 pups from 9 litters were analyzed. d, Superplots of nuclear ROI area from Nup153 and JFX554 SIM images. Plots show mean ± SD, with color coding indicative of biological replicates. ns, not significant; repeated measures one-way ANOVA with Dunnett’s multiple comparisons test on 3 biological replicates, with DIV4 as a reference condition. e, Representative image of Nup153 and HaloTag-Nup107 colocalization in DIV10 HaloTag-Nup107 neurons from 3 biological replicates. Scale bar = 2 μm. f, Violin plot showing the percent of Nup153 and JFX554 puncta that colocalize with JFX554 and Nup153, respectively, in DIV10 neurons. Nup153 colocalization with JFX554 was measured by calculating the % of Nup153 peaks that overlap with thresholded JFX554 puncta, and vice versa. g, Frequency distribution of nearest neighbor distances of JFX554 puncta. For each JFX554 peak, the distance to its nearest neighbor was calculated. Results from three biological replicates were combined. h, Frequency distribution of JFX554 puncta within two-pore diameter (240 nm) distance. For each JFX554 peak, the number of neighbouring puncta within a radius of 240 nm was calculated. Results from three biological replicates were combined. i, Distribution of genotypes in litters derived from intercrossing Tor1a+/-; Nup107KI/KI mice. All genotypes were born at the expected Mendelian ratio (P-0.4880, Chi-squared test). 46 pups from 6 litters were analyzed. Source numerical data and unprocessed western blot are available in source data.
Extended Data Fig. 5 Comparison of NPC density and distribution from HaloTag-Nup107 pulse-chase.
a, b, c, Nup153 (a), JFX554 (b), and new JF646 (c) puncta density in maturing WT and torsinA-KO primary neurons aged DIV4, 6, 8, 10. Plots show mean ± SD from 3 biological replicates. Repeated measures two-way ANOVA with Sidak’s multiple comparisons test was used to compare genotypes at each timepoint. No comparisons reached statistical significance. Repeated measures two-way ANOVA with Dunnett’s multiple comparisons test was used to compare all timepoints to DIV4, with ****P < 0.0001. d, Autocorrelation of JFX554 images over 0-500 nm separation distance. Similar starting amplitudes reflect constant JFX554 density. e, Autocorrelation of JF646 images over 0-500 nm separation distance. Decreasing starting amplitudes over neuronal maturation reflect increasing JF646 density. Broadening of the curve in torsinA-KO neurons indicates spatial correlation of NPCs over larger distances. Source numerical data are available in source data.
Extended Data Fig. 6 Bleb-associated pores are narrow and exhibit nucleoplasmic central plugs.
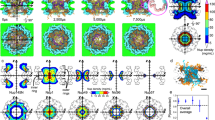
a, Slices from a DIV10 torsinA-KO tomogram oriented as an en-face view of the neuronal nuclear membrane. Pores at clusters of blebs, isolated blebs, or regular NPCs without blebs are marked by color-coded spheres centred around the middle plane of the pore channel, denoted by larger circles. Tomogram shown here is representative of 8 tomograms from two biological replicates. b, Projection of all marked pores from the tomogram shown in (a). c, Superplot of WT, KO regular (non-bleb-associated), and KO bleb-associated pore diameter (nm). Plots show mean ± SD, with color coding indicative of biological replicates. Swarmplot of individual pore diameters (small points) is overlayed with the mean of pore diameters for each biological replicate (larger points). Pores from two WT and two torsinA-KO tomograms were analyzed. KO regular and bleb-associated pores were identified from the same torsinA-KO tomograms. *P = 0.0128, **P = 0.0068; two-tailed unpaired t-test from two biological replicates. d, Blinded quantitation of the percentage of pores that have dense central plugs. Plots show mean ± SD with each point representing a cell. For KO regular and bleb-associated pores, points are color-coded to reflect quantitation from the same tomogram. Blinded quantitation was performed on 593 pores from 8 WT and 8 torsinA-KO tomograms obtained from two biological replicates each, with each tomogram from a different cell. *P = 0.0302; two-tailed unpaired t-test with Welch’s correction, ****P < 0.0001 two-tailed paired t-test, using values from 8 tomograms. e, Slices showing individual pores from DIV10 WT and torsinA-KO tomograms. Regular and bleb-associated torsinA-KO pores were sampled from the same tomogram. From top to bottom, the slices progress from the cytoplasmic side of the pore channel towards the nucleoplasm. Center plane of the pore channel is marked with a horizontal arrow. Scale bar = 100 nm. Source numerical data are available in source data.
Extended Data Fig. 7 NE blebs spatially correlate with NPC clusters.
Line scan analyses of 8 WT and 8 torsinA-KO DIV10 nuclei from two biological replicates labeled with anti-Nup153 and anti-Ubiquitin-K48 antibodies.
Extended Data Fig. 8 Nup358 is recruited to persisting NPC clusters in DIV18 torsinA-KO neurons.
a, SIM images of DIV10 and DIV18 WT and torsinA-KO neurons labeled with Nup153 and Nup358 antibodies. Scale bar = 2μm. b, dSTORM images of Nup210 in DIV18 WT and torsinA-KO neurons. Scale bar = 2 μm. Right panels show zoomed in view. Scale bar for right panels = 200 nm. c, Autocorrelation plot of DIV18 WT and torsinA-KO neurons, determined from Nup210 dSTORM images. 18 WT and 19 torsinA-KO cells from 3 biological replicates each were analyzed. d, Normalized localization density along radial distance in averaged WT and torsinA-KO NPCs from DIV18 dSTORM images. Plots show mean ± SEM from eight bootstrapping rounds with 250 randomly selected pores each. *P = 0.019. e, Confocal images from DIV10–18 pulse-chase of WT and torsinA-KO neurons expressing HaloTag-Nup107. Scale bar = 5μm. f, Plot of Nup358 nuclear rim fluorescence intensity from DIV10–18 pulse-chase. Plots show mean ± SD from two biological replicates with color coding to indicate replicates. All values were normalized to the mean DIV10 WT fluorescence intensity. Reported P values are from a two-way ANOVA test. “n” denotes the number of analyzed nuclei. g, Plot of JFX554 nuclear rim fluorescence intensity from DIV10–18 pulse-chase. Plots show mean ± SD from two biological replicates with color coding to indicate replicates. All values were normalized to the mean DIV10 WT fluorescence intensity. Reported P values are from a two-way ANOVA test. Same number of nuclei were analyzed as in (f). Source numerical data are available in source data.
Extended Data Fig. 9 Summary of the effects of torsinA loss on NPC spatial organization and maturation.
a, Model of interphase NPC assembly in WT and torsinA-KO neurons. Onset of NPC assembly is not affected by torsinA deletion. Nuclear basket, inner ring, and transmembrane nucleoporins are recruited to the nascent NPC as the INM starts to bud. Instead of the normal INM-ONM fusion found in WT neurons, excessive INM extrusion causes NE blebs to emerge and enlarge in torsinA-KO neurons. These blebs stall torsinA-KO NPCs at an intermediate stage while NPC assembly completes in WT neurons. As torsinA-KO neurons continue to mature, NE blebs resolve and INM-ONM fusion occurs. Completion of NPC biogenesis is delayed in torsinA-KO neurons. b, Model of NPC localization in maturing WT (top) and torsinA-KO (bottom) neurons. In WT neurons, newly forming NPCs (blue) localize to empty spaces between existing NPCs (red), thereby maintaining uniform spatial organization. In maturing torsinA-KO neurons, newly forming NPCs (blue) localize abnormally close to each other or to existing NPCs (red), causing aberrant clusters. NPC biogenesis is upregulated in both genotypes and total NPC number is not affected by the absence of torsinA.
Supplementary information
Supplementary Information
Supplementary video legends.
Supplementary Video 1
A 3D volumetric tomogram of torsinA-KO DIV10 nuclear membrane. Movie starts from the cytoplasm and progresses towards the nucleoplasm. Pores occurring where clusters of blebs, single isolated blebs or regular NPCs connect to the nuclear membrane are colour-coded and annotated. Slices from the same tomogram are shown in Extended Data Fig. 6a.
Supplementary Video 2
A 3D volume of the torsinA-KO DIV10 tomogram shown in Fig. 5d–f. INM (magenta), ONM (cyan), membranes inside the blebs (green) and central plugs of pores (yellow) are segmented.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1/Table 1
Statistical source data.
Source Data Extended Data Fig. 2/Table 2
Statistical source data.
Source Data Extended Data Fig. 3/Table 3
Statistical source data.
Source Data Extended Data Fig. 4/Table 4
Statistical source data.
Source Data Extended Data Fig. 4/Table 4
Unprocessed western blot.
Source Data Extended Data Fig. 5/Table 5
Statistical source data.
Source Data Extended Data Fig. 6/Table 6
Statistical source data.
Source Data Extended Data Fig. 8/Table 8
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kim, S., Phan, S., Tran, H.T. et al. TorsinA is essential for neuronal nuclear pore complex localization and maturation. Nat Cell Biol 26, 1482–1495 (2024). https://doi.org/10.1038/s41556-024-01480-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41556-024-01480-1
This article is cited by
-
Structure, function and assembly of nuclear pore complexes
Nature Reviews Molecular Cell Biology (2026)
-
Transcriptomic profiling of DYT-TOR1A patients-derived iPSC reveals dysregulation in extracellular matrix, lipid metabolism, and Chr22q11.23
BMC Medical Genomics (2025)