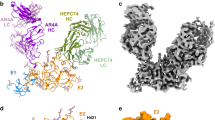
Abstract
Fifty-eight million individuals worldwide are affected by chronic hepatitis C virus (HCV) infection, a primary driver of liver cancer for which no vaccine is available1. The HCV envelope proteins E1 and E2 form a heterodimer (E1/E2), which is the target for neutralizing antibodies2. However, the higher-order organization of these E1/E2 heterodimers, as well as that of any Hepacivirus envelope protein complex, remains unknown. Here we determined the cryo-electron microscopy structure of two E1/E2 heterodimers in a homodimeric arrangement. We reveal how the homodimer is established at the molecular level and provide insights into neutralizing antibody evasion and membrane fusion by HCV, as orchestrated by E2 motifs such as hypervariable region 1 and antigenic site 412, as well as the organization of the transmembrane helices, including two internal to E1. This study addresses long-standing questions on the higher-order oligomeric arrangement of Hepacivirus envelope proteins and provides a critical framework in the design of novel HCV vaccine antigens.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Cryo-EM maps and atomic coordinates have been deposited to the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB), under the EMDB accession codes EMD-19243 (E1/E2 with transmembrane helices) and EMD-19254 (E1/E2 ectodomain) for the cryo-EM maps, and PDB IDs 8RJJ (E1/E2 with transmembrane helices) and 8RK0 (E1/E2 ectodomain) for the atomic coordinates.
References
Hepatitis C Fact sheet. World Health Organization http://www.who.int/mediacentre/factsheets/fs164/en/ (2024).
Torrents de la Pena, A. et al. Structure of the hepatitis C virus E1E2 glycoprotein complex. Science 378, 263–269 (2022).
Law, J. L. et al. A hepatitis C virus (HCV) vaccine comprising envelope glycoproteins gpE1/gpE2 derived from a single isolate elicits broad cross-genotype neutralizing antibodies in humans. PLoS ONE 8, e59776 (2013).
Duncan, J. D., Urbanowicz, R. A., Tarr, A. W. & Ball, J. K. Hepatitis C virus vaccine: challenges and prospects. Vaccines 8, 90 (2020).
Capella-Pujol, J. et al. Signatures of VH1–69-derived hepatitis C virus neutralizing antibody precursors defined by binding to envelope glycoproteins. Nat. Commun. 14, 4036 (2023).
Freedman, H. et al. Computational prediction of the heterodimeric and higher-order structure of gpE1/gpE2 envelope glycoproteins encoded by hepatitis C virus. J. Virol. 91, e02309–e02316 (2017).
Falson, P. et al. Hepatitis C virus envelope glycoprotein E1 forms trimers at the surface of the virion. J. Virol. 89, 10333–10346 (2015).
Gerold, G., Moeller, R. & Pietschmann, T. Hepatitis C virus entry: protein interactions and fusion determinants governing productive hepatocyte invasion. Cold Spring Harb. Perspect. Med. 10, a036830 (2020).
Augestad, E. H. et al. Global and local envelope protein dynamics of hepatitis C virus determine broad antibody sensitivity. Sci. Adv. 6, eabb5938 (2020).
Prentoe, J., Velázquez-Moctezuma, R., Foung, S. K., Law, M. & Bukh, J. Hypervariable region 1 shielding of hepatitis C virus is a main contributor to genotypic differences in neutralization sensitivity. Hepatology 64, 1881–1892 (2016).
Tong, Y., Lavillette, D., Li, Q. & Zhong, J. Role of hepatitis C virus envelope glycoprotein E1 in virus entry and assembly. Front. Immunol. 9, 1411 (2018).
Law, M. et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 14, 25–27 (2008).
Giang, E. et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc. Natl Acad. Sci. USA 109, 6205–6210 (2012).
Yanagi, M., Purcell, R. H., Emerson, S. U. & Bukh, J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl Acad. Sci. USA 94, 8738–8743 (1997).
Meunier, J. C. et al. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc. Natl Acad. Sci. USA 102, 4560–4565 (2005).
Khan, A. G. et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 509, 381–384 (2014).
Flyak, A. I. et al. HCV broadly neutralizing antibodies use a CDRH3 disulfide motif to recognize an E2 glycoprotein site that can be targeted for vaccine design. Cell Host Microbe 24, 703–716.e703 (2018).
Tzarum, N. et al. An alternate conformation of HCV E2 neutralizing face as an additional vaccine target. Sci. Adv. 6, eabb5642 (2020).
Tzarum, N. et al. Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies. Sci. Adv. 5, eaav1882 (2019).
Kumar, A. et al. Structural insights into hepatitis C virus receptor binding and entry. Nature 598, 521–525 (2021).
Kumar, A. et al. Regions of hepatitis C virus E2 required for membrane association. Nat. Commun. 14, 433 (2023).
Kong, L. et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science 342, 1090–1094 (2013).
Pfaff-Kilgore, J. M. et al. Sites of vulnerability in HCV E1E2 identified by comprehensive functional screening. Cell Rep. 39, 110859 (2022).
Olesen, C. H., Augestad, E. H., Troise, F., Bukh, J. & Prentoe, J. In vitro adaptation and characterization of attenuated hypervariable region 1 swap chimeras of hepatitis C virus. PLoS Pathog. 17, e1009720 (2021).
Kong, L. et al. Structure of hepatitis C virus envelope glycoprotein E2 antigenic site 412 to 423 in complex with antibody AP33. J. Virol. 86, 13085–13088 (2012).
Kong, L. et al. Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc. Natl Acad. Sci. USA 109, 9499–9504 (2012).
Kong, L. et al. Structural flexibility at a major conserved antibody target on hepatitis C virus E2 antigen. Proc. Natl. Acad Sci. USA 113, 12768–12773 (2016).
Oliver, M. R. et al. Structures of the Hepaci-, Pegi-, and Pestiviruses envelope proteins suggest a novel membrane fusion mechanism. PLoS Biol. 21, e3002174 (2023).
Perin, P. M. et al. Flunarizine prevents hepatitis C virus membrane fusion in a genotype-dependent manner by targeting the potential fusion peptide within E1. Hepatology 63, 49–62 (2016).
Lavillette, D. et al. Characterization of fusion determinants points to the involvement of three discrete regions of both E1 and E2 glycoproteins in the membrane fusion process of hepatitis C virus. J. Virol. 81, 8752–8765 (2007).
Catanese, M. T. et al. Ultrastructural analysis of hepatitis C virus particles. Proc. Natl Acad. Sci. USA 110, 9505–9510 (2013).
Rey, F. A. & Lok, S. M. Common features of enveloped viruses and implications for immunogen design for next-generation vaccines. Cell 172, 1319–1334 (2018).
Augestad, E. H., Bukh, J. & Prentoe, J. Hepatitis C virus envelope protein dynamics and the link to hypervariable region 1. Curr. Opin. Virol. 50, 69–75 (2021).
Matsuura, Y. et al. Processing of E1 and E2 glycoproteins of hepatitis C virus expressed in mammalian and insect cells. Virology 205, 141–150 (1994).
Hu, T., Wu, Z., Wu, S., Chen, S. & Cheng, A. The key amino acids of E protein involved in early flavivirus infection: viral entry. Virol. J. 18, 136 (2021).
Carlsen, T. H., Scheel, T. K., Ramirez, S., Foung, S. K. & Bukh, J. Characterization of hepatitis C virus recombinants with chimeric E1/E2 envelope proteins and identification of single amino acids in the E2 stem region important for entry. J. Virol. 87, 1385–1399 (2013).
Serre, S. B., Krarup, H. B., Bukh, J. & Gottwein, J. M. Identification of alpha interferon-induced envelope mutations of hepatitis C virus in vitro associated with increased viral fitness and interferon resistance. J .Virol. 87, 12776–12793 (2013).
Prentoe, J. et al. Hypervariable region 1 deletion and required adaptive envelope mutations confer decreased dependency on scavenger receptor class B type I and low-density lipoprotein receptor for hepatitis C virus. J. Virol. 88, 1725–1739 (2014).
Dodev, T. S. et al. A tool kit for rapid cloning and expression of recombinant antibodies. Sci. Rep. 4, 5885 (2014).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D 74, 531–544 (2018).
He, J., Li, T. & Huang, S.-Y. Improvement of cryo-EM maps by simultaneous local and non-local deep learning. Nat. Commun. 14, 3217 (2023).
Williams, C. J. et al. MolProbity: more and better reference data for improved all‐atom structure validation. Protein Sci. 27, 293–315 (2018).
DeLano, W. L. Pymol: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 40, 82–92 (2002).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Yariv, B. et al. Using evolutionary data to make sense of macromolecules with a “face-lifted” ConSurf. Protein Sci. 32, e4582 (2023).
Rothwangl, K. B., Manicassamy, B., Uprichard, S. L. & Rong, L. Dissecting the role of putative CD81 binding regions of E2 in mediating HCV entry: putative CD81 binding region 1 is not involved in CD81 binding. Virol. J. 5, 46 (2008).
Owsianka, A. M. et al. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J. Virol. 80, 8695–8704 (2006).
Acknowledgements
The authors thank T. H. Pape for assistance with sample screening and data collection; J. Conrad, K. Wallden, D. Morado and M. Carroni for sample screening and data collection; L. Mikkelsen, A. L. Sørensen and L. Nielsen for general laboratory support; B. Ø. Lindhardt and C. M. Bonefeld for support of the project; and T. Krey, M. Law, A. Patel and C. Rice for providing reagents. This work was supported by the Novo Nordisk BRIDGE grant NNF20SA0064340 (E.H.A.), Lundbeck Foundation Experiment grant R324-2019-1375 (J.P.), Lundbeck Foundation Experiment R346-2020-2019 (P.G.), Lundbeck Foundation Fellowship R335-2019-2052 (J.P.), Lundbeck Foundation Fellowship R133-A12689 (P.G.), Lundbeck Ascending Investigator R313-2019-774 (P.G.), Lundbeck Foundation Experiment R324-2019-1855 (K.W.), Lundbeck Foundation Postdoc R303-2018-3396 (R.V.-M.), Candys Foundation PhD grant 2016-195 (E.H.A., J.B. and J.P.), Candys Foundation PhD grant 2019-317 (C.H.O., J.B. and J.P.), Sapere Aude Advanced grant 0602-02366B (J.B.), Distinguished Investigator Grant from the Novo Nordisk Foundation NNF19OC0054518 (J.B.), The Danish Council for Independent Research 9039-00273 (P.G.), The Swedish Research Council grants 2016-04474 and 2022-01315 (P.G.), Knut and Alice Wallenberg Foundation Prolongation Fellow grant 2020.0194 (P.G.) and Knut and Alice Wallenberg 2015.0131 (P.G.). The Danish Cryo-EM Facility at CFIM, University of Copenhagen, is supported by Novo-Nordisk Foundation grant no. NNF14CC0001. The Cryo-EM Swedish National Facility at SciLifeLab is funded by the Knut and Alice Wallenberg, Family Erling Persson and Kempe Foundations, SciLifeLab, Stockholm University and Umeå University.
Author information
Authors and Affiliations
Contributions
E.H.A., C.H.O., C.G. and J.P. carried out the protein purification and the cryo-EM sample preparation. E.H.A., C.H.O., C.G. and J.P. optimized the protein purification strategy. E.H.A. and C.H.O. cloned the constructs and performed blue native PAGE analysis. K.W. optimized the cryo-EM sample freezing strategy and video collection. K.W. and J.P. processed the cryo-EM data in Cryosparc and built and refined the atomic models into the cryo-EM maps. E.H.A., C.G., K.W., P.G. and J.P. conceived the protein purification strategy. E.H.A., P.G. and J.P. conceived the construct design. A.S. performed ELISA experiments. E.H.A., C.H.O., A.S., R.V.-M. and M.F. performed protein production. J.B. provided materials and guidance. E.H.A. wrote the paper with contributions from C.H.O. E.H.A. made most of the figures, with contributions from C.H.O. (Fig. 1a and Extended Data Figs. 1, 6d and 7b) and J.P. (Extended Data Figs. 5a and 10a,b). C.H.O. made Fig. 2b,c and Extended Data Fig. 6c, and K.W. made Extended Data Figs. 2–4. All authors contributed to the manuscript text by assisting with writing or providing feedback. Supervision of the research was conducted by E.H.A., C.G., K.W., P.G. and J.P.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Félix Rey and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer review reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Purification of HCV E1/E2 in a higher-order complex.
(a-e) Extraction and detergent solubilization of HCV E1/E2 analyzed by BN PAGE western blots or total-protein Coomassie stain. a, Solubilized HCV E1/E2 of different isolates of genotype 1–6. Red arrows indicate putative higher-order E1/E2 complexes. b, Solubilized E1/E2 from HCV isolate S52 (genotype 3a) with C-terminal E1-TM and E2-TM from H77 (genotype 1a) (termed S52mod). c, Solubilized E1/E2 from H77 and S52mod with internal twinstrep tags (termed H77-TS and S52mod-TS). d, Strep-purification of solubilized E1/E2 from cells expressing HIS- and strep-tagged S52mod E1/E2. e, Coomassie stain of purified H77-TS and S52mod-TS E1/E2 complex. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 2 Cryo-EM data processing scheme of the E1/E2 in a homodimeric complex for S52mod.
a, Representative cryo-EM image of S52mod. b, Selected 2D averages of the particles. c, Cryosparc video processing pipeline. d, Density of the final map used for model building colored by resolution. Extended Data Table 1 contains specific information and parameters regarding data collection.
Extended Data Fig. 3 Structure and density fit.
Selected regions are shown with the density around the final structure. a, Model fit into the original map without sharpening. b, Model fit into the EMReady refined map. Main chains are shown in cartoons and side chains in sticks. The glycosylation is shown as sticks colored in magenta. Glycosylated residues are labeled in magenta.
Extended Data Fig. 4 Structure comparison of H77 and S52mod.
a, Particle count and GSFSC graph for H77. b, Comparison of 2D averages between H77 and S52mod. c, Overlay of the low-resolution map of H77 (shown as red mesh) with S52mod (shown as a grey isosurface). The final map of H77 only reached 8.3 Å resolution, likely because of low sample stability and limited particle numbers, but both the 2D averages and the overlay of the low-resolution map of H77 fit well with the high-resolution map of S52mod with almost identical arrangement.
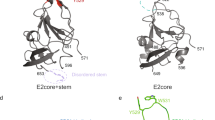
Extended Data Fig. 5 Homodimer surface analysis and alignment with E1/E2 heterodimer structure.
a, Surface analysis of the ectodomain part of the E1/E2 homodimeric complex. Display of glycans (left), sequence conservation (consurf48; middle), and hydrophobicity (right) of surface exposed amino acids on the ectodomain of the E1/E2 homodimeric complex (8rk0). The orange box highlights a conserved, glycan-free patch exposed on the surface of the complex, which may be involved in the formation of higher oligomeric states of E1/E2 homodimers. The top row is rotated 90 degrees compared to the bottom row. Glycan density is represented in dark grey. b-c, Alignment of E1 and E2 from our modeled structure (8rjj; depicted in blue) with the recently solved E1/E2 monomer structure (7t6x; regions that agree across the two structures are depicted in red and regions that do not agree are depicted in transparent red). Unresolved regions are depicted in grey in the schematics and indicated by broken lines in the structures. The locations of disulfide bonds are numbered and highlighted in yellow.
Extended Data Fig. 6 Analysis of the E1/E2 homodimer interface.
a, A single E1/E2 protomer showing the buried surface area of the E2-E2 interface of the homodimeric E1/E2 complex (left) or the Fab 2A12 epitope (right)16. The same view of the ectodomain of the E1/E2 heterodimer (8rk0) is shown with E1 in steel blue and E2 in green. b, Binding of the non-neutralizing antibody 2A12 to purified E1/E2 homodimer complex (2xE1/E2) and sE2 measured by ELISA. Data is based on two technical replicates with error bars representing standard deviation. c, Sequence logo representing amino acid conservation within genotype 1–8 for each E2-E2 contact residue (Los Alamos HCV sequence database). d, Extracted HCV E1/E2 (S52mod) with alanine mutations in the contact residues of the homodimer interface, analyzed by BN PAGE western blot.
Extended Data Fig. 7 Alphafold predictions of unresolved or CD81 binding regions, epitope characterization, and epitope clash analysis on the E1/E2 homodimeric complex.
a, A comparison between selected residues (HVR1 + AS412: 384–423, E2-FL: 422–460, CD81bl: 519–535; in the case of the CD81bl, additional backbone residues are shown in white as these were used to correctly orient the CD81bl structures) of our modeled E2 structure and the AlphaFold predicted structure. b, Key epitopes targeted by cross-reactive antibodies are highlighted on the soluble part of the E1/E2 homodimeric complex (8rk0). The glycan density is depicted in dark grey. The epitope of AR3A is partly flexible in our complex, likely due to not having Fabs bound. The epitopes of IGH505, HCV1, AR3A, and AR4A have been directly visualized in solved structures, whereas those of AR2A and AR5A are defined indirectly through alanine scanning. (c-e) Structure of Fabs bound to E2 (c: 6bkb, d: 7t6x, e: 4dgv) superimposed on the on the soluble part of E1/E2 homodimer structure (8rk0). E1 is depicted in steel blue, E2 in green, HVR1 in purple, AS412 in blue and Fabs in yellow. In panel d, only the variable domain of the light chain and the heavy chain is displayed. Zoom-ins depict the clash of the AR4A heavy chain with HVR1 in red (d) or the HCV1 heavy chain with E2-FL in green (e). Unresolved regions are indicated by broken lines.
Extended Data Fig. 8 Characterization of the CD81 binding site.
Interaction regions and critical contact residues between CD81 and E2. a, The E1/E2 homodimer (8rjj) and the E2-CD81 complex (7mwx) were superimposed and are shown side by side in these aligned front and top views. Residues forming the CD81-E2 interface20 are highlighted on both the homodimeric E1/E2 complex (left, 8rjj) and the E2 structure bound to CD81 (right, 7mwx) and color-coded as detailed in the legend. In 8rjj, one E1/E2 heterodimer is represented in gray and the other in light gray. In 7mwx, E2 is represented in gray. b, A close-up view on the part of AS412/E2-FL (top panel, blue) and the CD81bl (lower panel, yellow) adopting different conformations, with residues critical for CD81 binding and virus entry highlighted49,50. c, A comparison between the aligned residues (E2-FL: 422–460, CD81bl: 519–535; in the case of the CD81bl, additional backbone residues are shown in white as these were used to correctly orient the CD81bl structures) of our modeled E2 structure, and structures elucidated through Fab-based methods. E2 structures with E2-FL in conformation A (4mwf, 6mei, 6uyg, 6meh, 6mej, 7t6x) and conformation B (6wo5)18. E2 structures with CD81bl in retracted (7mww) and bent (8dk6) conformation. Unresolved regions are indicated by broken lines.
Extended Data Fig. 9 Characterization of the E1/E2 membrane helices and flexible hinge regions.
a, Cryo-EM map (transparent) overlayed with the predicted model of the different TM helices of E1 and E2. The helix positioned perpendicular to the C-terminal TM of E1, MX, is depicted in grey. b, Cryo-EM map showing the density of the E1/E2 homodimeric complex embedded in a detergent micelle. The left view is rotated 90 degrees compared to the right view. The micelle corresponds to the empty part of the volume, which is depicted in transparent grey. E1 is colored in steel blue and E2 is colored green. c-e, Comparison of flexible hinge regions, connecting the ectodomains to the TM helices, of our modeled structure (8rjj; depicted in light grey, top panel) with the recently solved E1/E2 heterodimer structure (7t6x; depicted in red, middle panel). The highlighted sections (dark grey) of E1 and E2 are aligned in a close-up view (bottom panel). c, Hinge regions connected to the internal TM helices of E1. d, Hinge regions connected to the C-terminal membrane-associated helices of E1. e, Hinge regions connected to the C-terminal TM helices of E2. Although the residue numbers differ between 8rjj (S52mod) and 7t6x (AMS0232) in the E2 alignment, the sequence in the highlighted section is highly conserved (e, bottom panel). The unresolved regions are indicated by broken lines.
Extended Data Fig. 10 Comparison with in silico predicted HCV E1/E2 TM structures.
a, Comparison of the TM domain bundles of our modeled structure (depicted in light grey) with the recently in silico predicted HCV E1/E2 heterodimer28 (NC_004102; depicted in blue). b, Comparison of individual helices. The kink at H261 (helix above depicted in dark grey) observed in our internal E1-TM is not present in the predicted structure.
Supplementary information
Supplementary Figure 1
Uncropped immunoblots from Extended Data Figs. 1 and 6. a,b, Immunoblots for Extended Data Fig. 1a showing solubilized HCV E1/E2 of different isolates of genotype 1–6 against AR3A (a) and AR4A (b). c, Immunoblots for Extended Data Fig. 1b showing solubilized E1/E2 from HCV isolate S52 (genotype 3a) with TMs from H77 (genotype 1a) (termed S52mod). d,e, Immunoblots for Extended Data Fig.1c showing solubilized E1/E2 from H77 (d) and S52mod (e) with internal TwinStrep tags (termed H77-TS and S52mod-TS). f,g, Immunoblots for Extended Data Fig. 1d showing strep purification of solubilized E1/E2 from cells expressing His- and Strep-tagged S52mod E1/E2, stained with AR3A (f) and Penta-His (g). h–k, Immunoblots for Extended Data Fig. 6d showing extracted HCV E1/E2 (S52mod) with alanine mutations in the contact residues of the homodimer interface. Samples run on the same gel originated from the same transfection. Proteins were detected by western blotting with the specified antibodies. The ladder is stained with WesternSure Pen (Licorebio). The red stapled rectangles indicate the cropping.
Supplementary Table
Cryo-EM data collection, refinement and validation statistics
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Augestad, E.H., Holmboe Olesen, C., Grønberg, C. et al. The hepatitis C virus envelope protein complex is a dimer of heterodimers. Nature 633, 704–709 (2024). https://doi.org/10.1038/s41586-024-07783-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-024-07783-5
This article is cited by
-
Cryo-EM structures of HCV E2 glycoprotein bound to neutralizing and non-neutralizing antibodies determined using bivalent Fabs as fiducial markers
Communications Biology (2025)
-
GLYCO-BUILD: an enzymatic pipeline for the synthesis of peptides carrying eukaryotic N-glycans
Nature Communications (2025)