Abstract
Research on the morphology, physiology and genomics of Asgard archaea has provided valuable insights into the evolutionary history of eukaryotes1,2,3. A previous study suggested that eukaryotes are nested within Heimdallarchaeia4, but their exact phylogenetic placement within Asgard archaea remains controversial4,5. This debate complicates understanding of the metabolic features and timescales of early eukaryotic ancestors. Here we generated 223 metagenome-assembled nearly complete genomes of Asgard archaea that have not previously been documented. We identify 16 new lineages at the genus level or higher, which substantially expands the known phylogenetic diversity of Asgard archaea. Through sophisticated phylogenomic analysis of this expanded genomic dataset involving several marker sets we infer that eukaryotes evolved before the diversification of all sampled Heimdallarchaeia, rather than branching with Hodarchaeales within the Heimdallarchaeia. This difference in the placement of eukaryotes is probably caused by the previously underappreciated chimeric nature of Njordarchaeales genomes, which we find are composed of sequences of both Asgard and TACK archaea (Asgard’s sister phylum). Using ancestral reconstruction and molecular dating, we infer that the last Asgard archaea and eukaryote common ancestor emerged before the Great Oxidation Event and was probably an anaerobic H2-dependent acetogen. Our findings support the hydrogen hypothesis of eukaryogenesis, which posits that eukaryotes arose from the fusion of a H2-consuming archaeal host and a H2-producing protomitochondrion.
Similar content being viewed by others
Main
The origin of eukaryotic cells has long remained a mystery6. Asgard archaea contain more genes encoding eukaryotic signature proteins (ESPs) than other archaea and are deemed to be the closest relatives of eukaryotes4,5,7,8,9. Isolation and cultivation of the first Asgard archaeon (‘Candidatus Prometheoarchaeum (Ca. P.) syntrophicum’ MK-D1), belonging to Lokiarchaeia, revealed that the archaeon has long and branching protrusions and is able to degrade amino acids syntrophically1. These morphological and metabolic features provide new insights into the episodes of eukaryogenesis. For example, it is assumed that cell protrusion may mediate cell–cell interactions, leading to engulfment of its partner and gradual formation of mitochondrion. Subsequently, the second Asgard archaeon (‘Candidatus Lokiarchaeum (Ca. L.) ossiferum’) was recently enriched, and cryo-electron tomography and immunostaining revealed the presence of the actin-like cytoskeletal filament and Lokiactin expression2. It is inferred that the Lokiactin may have served as a scaffold for the Asgard archaeal cell and played a role in maintenance of cell shape, cell division and molecular trafficking, similar to actin filaments in eukaryotic cells10,11. The results and hypotheses implicate that the eukaryotic nuclear lineages evolved from within the archaea.
Although some eukaryotic-like features have been identified in pure culture or enrichment of Asgard archaea, with such a limited number of strains, it is challenging to infer the exact phylogenetic position, timescales and metabolic characteristics of early eukaryotic ancestors. In contrast, phylogenomic analyses based on a wealth of archaeal genomic data can compensate for these shortcomings12. By using supertree and coalescent methods using more than 3,000 gene families in archaea and eukaryotes, the Heimdallarchaeota have been proposed as the closest relatives of eukaryotes13. However, the study used only six Asgard archaeal genomes and failed to represent the currently expanded diversity of Asgard archaea. Recently, two studies obtained inconsistent results revolving around the relationship between eukaryotes and Asgard archaea based on an expanded genomic sampling of Asgard archaea. One suggested that eukaryotes are more likely to have branched from within Asgard archaea as a sister group to the Heimdallarchaeota–Wukongarchaeota branch or are a deeper branch within archaea5, whereas another proposed that eukaryotes branch as a sister lineage to Hodarchaeales within Heimdallarchaeia4. The former used only 29 concatenated markers to infer phylogenies, whereas the latter used non-ribosomal proteins as markers in combination with extensive exclusion of the sites from the alignments. The exact relationship between eukaryotes and archaea remains to be further clarified.
Here we present 223 new Asgard archaeal genomes, including the identification of 16 additional order-, family- or genus-level lineages, recovered from metagenomic samples generated from 14 sites across coastal wetlands of China, which significantly expand the diversity of Asgard archaea. By analysing the expanded genomic sampling of Asgard archaea leveraging sophisticated phylogenomic approaches, including recoding of alignments, use of complex site-heterogeneous evolution models in maximum likelihood (ML) and Bayesian inferences and reduction of rate heterogeneity, we robustly place eukaryotes within the Asgard archaea as a sister clade to Heimdallarchaeia. By applying ancestral reconstruction together with molecular dating, the timescale and metabolic traits of the last Asgard archaea and eukaryote common ancestor (LAECA) were delineated and found to differ significantly from previous studies1,4.
Expanded diversity of Asgard archaea
The increase in genomic diversity of Asgard contributes to resolving the evolutionary relationship between eukaryotes and Asgard archaea4,5,7. To this end, we collected 40 sediment samples from salt marsh and mangrove wetlands across China and performed metagenomic sequencing (Supplementary Table 1 and Supplementary Fig. 1a). After de novo assembly and binning of scaffolds, we reconstructed 11,878 metagenome-assembled genomes (MAGs) (Supplementary Fig. 1b). Of these MAGs, 223 belonged to the Asgard archaea, with greater than 70% completeness and less than 10% contamination (Supplementary Table 2 and Supplementary Fig. 1c). These MAGs were combined with 395 publicly available genomes with high quality. By using dRep14 to select representatives at species level (ANI at least 95%), we finally obtained a set of 411 Asgard genomes, of which 136 are derived from this study (Supplementary Table 2). Among these, two are from cultured Asgard archaea (Ca. P. syntrophicum and Ca. L. ossiferum) while the remaining genomes are MAGs recovered from environmental samples (Fig. 1). These genomes have a mean completeness of 85.3% and a mean contamination of 3.6%.
ML tree inferred using IQ-TREE under the LG + C60 + F + G + PMSF model, based on a set of 579 archaeal taxa (411 Asgard archaea, 51 DPANN archaea, 47 Euryarchaea and 70 representatives of TACK archaea). Only Asgard archaeal lineages are shown in the tree. Two cultured Asgard archaeal strains are highlighted with a red star. Newly identified genomes in this study are displayed by coloured bars in the outermost ring. Bootstrap support values of at least 95% are represented by black dots. The scale bar denotes the average expected number of substitutions per site.
To resolve the phylogenetic position of these clades, we inferred an ML tree of a concatenated set of 53 archaeal-specific marker proteins in the Genome Taxonomy Database (GTDB)15 (GTDB.ar53) for a phylogenetically diverse set of archaeal genomes (411 Asgard archaea, 51 DPANN (Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota and Nanohaloarchaeota) archaea, 47 Euryarchaea and 70 TACK (Thaumarchaeota, Aigarchaeota, Crenarchaeota, Korarchaeota) archaea representatives, Supplementary Table 3). The tree confirmed the existence of ten class-level Asgard archaeal lineages (Fig. 1). Based on the relative evolutionary distance16, we identified two putative order-level lineages in Odinarchaeia and Heimdallarchaeia (relative evolutionary distance less than 0.53), which were provisionally named Yangjianarchaeales and Wenzhongarchaeales. In addition, four new family-level clades (represented by six MAGs) and ten new genus-level clades (represented by 18 MAGs) were identified within Sifarchaeia, Hodarchaeales, Hermodarchaeia and Lokiarchaeia. In six out of the ten Asgard archaea classes, we presented 108 species-level new genomes (Fig. 1). Currently, the placement of the recently proposed Njordarchaeales in archaeal phylogeny is intensely debated4,17,18. Recently, it was placed within Heimdallarchaeia in the tree inferred using new alternative markers (NM57), forming a clade with Gerdarchaeales, Kariarchaeaceae and Heimdallarchaeaceae4. We selected 12 sets of frequently used marker proteins from previous studies (Supplementary Table 4)4,5,7,8,13,19,20,21,22,23 that are conserved in most of archaeal clades (Methods). ML phylogenomic analyses of these marker sets, based on a set of 579 archaeal representatives including the enlarged genomic sampling of Asgard archaea, were performed under a sophisticated evolutionary model (LG + C60 + F + G + PMSF). These results showed that, except for in the NM57 tree, Njordarchaeales was placed within the TACK superphylum and branched as a sister lineage to Korarchaeota with high support in the other 11 trees (Supplementary Fig. 2). Given that Njordarchaeales often branched with eukaryotes in previous phylogenomic analyses4,18, it is essential to identify the factors underlying the conflicting topologies among different trees for position of Njordarchaeales within archaea.
Revisit of Njordarchaeales placement
Eme et al.4 argued that the monophyly of Njordarchaeales and Korarchaeota in the RP56 tree may be caused by high compositional similarity in RP56 ribosomal marker protein sequences, which is connected with thermophilic lifestyle of the two lineages. Based on this hypothesis, they proposed a new marker dataset consisting of 57 proteins of archaeal origin that excluded ribosomal proteins to infer the phylogeny of archaea. The NM57 markers are selected from a set of 200 archaeal conserved marker proteins (NM200) identified by Petitjean et al.21 and most of them are functional proteins involved in metabolic and cellular processes. We observed that, despite the absence of ribosomal proteins, the ML tree of the NM200 dataset positioned Njordarchaeales as a sister clade to Korarchaeota within the TACK superphylum (Supplementary Fig. 2k). This suggests that the clustering of Njordarchaeales and Korarchaeota is not necessarily related to ribosomal proteins. The inconsistent topologies may be attributed to the differences in the number and nature of amino acid positions in the alignments of the two datasets. In contrast to the NM57 topology, the NM200 tree, with more amino acid sites, is more likely to represent the true evolutionary relationship between Njordarchaeales and other archaea.
In phylogenomic analysis, the even distribution of marker proteins in the different archaeal phyla or clades is crucial for clarifying the phylogenetic relationships between archaeal clades. To this end, we integrated the 12 sets of frequently used marker proteins from previous studies (Supplementary Table 4) and selected 67 markers (S67) that are conserved across all sampled archaeal genomes (identified in at least 60% of representatives of each of the archaeal clades; Methods). The S67 marker set was comprised of 39 ribosomal proteins and 28 functional proteins involved in diverse cellular activities (Supplementary Table 4). ML phylogenomic analyses of the S67 dataset under sophisticated evolutionary models confirmed Njordarchaeales as a sister to Korarchaeota and resolved all nodes of the Asgard archaea with high support (Bootstrap values at least 95) (Extended Data Fig. 1).
Given that concatenation of large amount of data may strengthen non-phylogenetic signal and lead to artifactual trees with high support, we next evaluated the effect of data exclusion on phylogenetic inference by removing mutationally saturated or rate-biased sites24,25,26,27. We tracked the evolution of bootstrap support for the monophyly of either (1) Njordarchaeales and Korarchaeota or (2) Njordarchaeales and Heimdallarchaeia in trees inferred from the S67 alignments using complex site-heterogeneous evolution model, as fast-evolving sites are progressively removed. The grouping of Njordarchaeales and Korarchaeota consistently received strong support until 80% or more of the fastest-evolving sites were removed, whereas the monophyly of Njordarchaeales and Heimdallarchaeia was never supported (Extended Data Fig. 2a). The significant loss of phylogenetic signal probably accounted for the decline in support observed after the removal of 80% or more of the fastest-evolving sites.
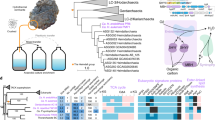
Nevertheless, it is observed that the incremental exclusion of the fastest-evolving sites from the NM57 alignments also strongly supported the grouping of Njordarchaeales and Heimdallarchaeia (Extended Data Fig. 2b). To identify the cause of this effect, we determined the taxonomic profiles of contigs of the ten Njordarchaeales representatives using CAT28,29 and MMseqs2 (refs. 30,31) taxonomy tools. Although the two tools use different algorithms, they generated comparable results. In each genome, approximately 24–60% of the contigs (Fig. 2a,b) were assigned to TACK archaea, which collectively accounted for 22–51% of the genome size (Supplementary Fig. 3). In contrast, 3–35% of the contigs were assigned to Asgard archaea, whose combined lengths comprised 1–34% of the genome size (Supplementary Fig. 3). The results indicate that these Njordarchaeales MAGs may contain high levels of contamination stemming from misbinning, or alternatively, that extensive horizontal gene transfers (HGT) have occurred between TACK and Asgard archaea, specifically into the Njordarchaeales. Subsequently, we examined the distribution patterns of contigs of these MAGs across several metagenomes from the same sampling location4,32,33 using read recruitment. We found that contigs from four out of the ten MAGs were mapped by reads from at least two samples. Based on their sequence composition and differential coverage patterns across different metagenomes, the contigs of each of the four MAGs were partitioned into two to four distinct clusters (Fig. 2c,d). For the B7_G17_GCA_029856635, contigs in its two larger clusters were used separately to infer ML phylogeny based on the NM57 markers. The contigs in one cluster were placed as a sister to Korarchaeota, while those in another cluster branched with Asgardarchaeia within Asgard archaea (Extended Data Fig. 3). The results indicate that these Njordarchaeales MAGs may represent chimeric assemblies derived from two or three distinct populations. Compared with other datasets, The NM57 dataset may harbour more robust phylogenetic signals that support the Asgard part of these MAGs, possibly because the NM57 markers were selected based on their distribution across Asgard archaea4. The taxonomic profiles of Njordarchaeales and its phylogenetic trees inferred using ribosomal proteins-containing marker sets (Fig. 2, Supplementary Figs. 2 and 4 and Supplementary Discussion) revealed that the bulk of these MAGs appeared to be affiliated with the TACK archaea. The inclusion of such chimeras in tree reconstruction can be expected to impact the evolutionary position of eukaryotes within archaea.
a,b, Percentage of contigs/scaffolds assigned to Thermoproteota (formerly known as the TACK superphylum) or Asgardarchaeota based on count for each of the ten Njordarchaeales representatives; classification of contigs/scaffolds was determined using CAT (a) or MMseqs2 (b) tools. c,d, Hierarchical clustering of contigs/scaffolds in Njordarchaeales B7_G17 and B20_G9 (c) and B62_G16 and S143_49 (d) genomes based on their sequence composition and differential mean coverage across different metagenomes.
Eukarya emerged outside Heimdallarchaeia
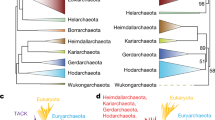
Our expanded Asgard archaeal genomes help to resolve the placement of eukaryotes among archaea. To this end, we added 14 commonly used eukaryotic taxa4,5,7 (Supplementary Table 3) into the S67 dataset to generate a supermatrix: ES67. The ML tree of the ES67 dataset placed eukaryotes within the TACK superphylum as a sister clade to Njordarchaeales and Korarchaeota (Supplementary Fig. 5a), which had never been observed previously in previous studies4,17. This may be caused by the high compositional similarity between the Asgard archaeal sequences in the Njordarchaeales genomes and eukaryotic sequences. To mitigate the effect of distant outgroups on the placement of eukaryotes, we removed DPANN, Euryarchaea, Korarchaeota and Njordarchaeales sequences from the ES67 dataset while retaining 50 representatives of TACK archaea as the outgroup; alternatively, all outgroup sequences were omitted. The resulting supermatrices were named tES67 and AsES67, respectively. ML analysis of the tES67 dataset showed that eukaryotes branched with Heimdallarchaeia with high support (Heimdallarchaeia-sister) (Fig. 3a and Supplementary Fig. 5b). The unrooted ML phylogeny of Asgard archaea and eukaryotes on the basis of the AsES67 dataset also strongly supported the Heimdallarchaeia-sister topology (Supplementary Fig. 5c).
a, ML phylogenetic analysis of 67 concatenated marker proteins based on 461 archaeal taxa and 14 eukaryotes (inferred using IQ- IQ-TREE under LG + C60 + F + G + PMSF model), using 50 TACK archaea as the outgroup (tES67 alignment; 13,348 sites). The number below Lokiarchaeales represents 150 MAGs and the genomes of two cultured Asgard strains. Meanwhile, the alignment was SR4-recoded and its Bayesian inference was performed using the CAT + GTR model (Supplementary Fig. 6a; two chains; 50,000 generations). PP support for the node of eukaryotes and the closest Asgard relatives is shown (0.69). b, ML phylogenetic analysis of 97 concatenated marker proteins based on 411 Asgard archaeal taxa and 14 eukaryotes (inferred using IQ- IQ-TREE under the LG + C60 + F + G + PMSF model) (S97 alignment; 20,067 sites). This tree is unrooted. In addition, Bayesian inferences of the SR4-recoded S97, S150 and NM57 supermatrices were performed using the CAT + GTR model (Supplementary Figs. 7 and 8b). PP support for the node of eukaryotes and the closest Asgard relatives is shown (PP = 1 for S97, PP = 0.8 for S150 and PP = 1 for NM57). c, Evolution of bootstrap support for the grouping of eukaryotes with Heimdallarchaeia (Heimdallarchaeia-sister, EHeim) or Hodarchaeales (Hodarchaeales-sister, EHod) in phylogenetic trees inferred from S97 and NM57 datasets, as the fastest-evolving sites were progressively removed.
Based on the tES67 and AsES67 datasets, we examined the effect of rate heterogeneity on the position of eukaryotes by progressively removing fast-evolving sites of the two datasets. Along the decreasing heterogeneity gradient, ML phylogenies were computed to assess the support for Heimdallarchaeia-sister and the monophyly of eukaryotes and Hodarchaeales (Hodarchaeales-sister). Support for Hodarchaeales-sister was never observed, whereas support for Heimdallarchaeia-sister fluctuated as rate heterogeneity decreased (Extended Data Fig. 4). We inferred that the fluctuation in support for Heimdallarchaeia-sister may be ascribed to the dynamic relationship between the loss of phylogenetic signals and the level of rate heterogeneity. Although Bayesian inferences of the SR4-recoded34 tES67 and AsES67 datasets displayed the Heimdallarchaeia-sister topology, support was weak (posterior probability (PP) support values of 0.69 and 0.5, respectively) (Fig. 3a and Supplementary Fig. 6). The results suggest that the S67 marker set does not contain enough phylogenetic signal to resolve the node between eukaryotes and Asgard clades.
To address the issue, we relaxed our marker selection criterion. Based on conservation of marker proteins in the expanded Asgard genomes, we selected two additional marker sets from previously reported marker proteins that are conserved in archaea4,5,7,13,19,21. The first marker set consisted of 97 markers (S97) that were found in at least 60% of representatives of each of the Asgard clades, whereas the second marker set included 150 markers (S150) that were identified in at least 80% of Asgard archaeal genomes (Supplementary Table 4). ML trees inferred from the untreated S97 dataset and its fastest-evolving site-excluded subsets based on Asgard archaea and eukaryotic taxa provided significant support for the Heimdallarchaeia-sister topology (bootstrap support greater than 95) (Fig. 3b,c). In contrast, an ML tree inferred from the untreated S150 dataset strongly supported the clustering of eukaryotes and Hodarchaeales (Extended Data Fig. 5a). Given that the larger amount of missing data contained in the S150 supermatrix could bias the estimation of the site evolutionary rates, a site-by-site desaturation strategy was also applied to the S150 supermatrix. We observed that support for the Hodarchaeales-sister was replaced by support for the Heimdallarchaeia-sister after the 40% fastest-evolving sites were removed (Extended Data Fig. 5b). The grouping of eukaryotes with Heimdallarchaeia was also confirmed by the Bayesian inferences of the SR4-recoded S97 and S150 datasets (PP values of 1.0 and 0.8, respectively; Fig. 3b, Supplementary Fig. 7).
In the study by Eme et al.4, based on the NM57 marker set, eukaryotes were positioned within Heimdallarchaeia, as a sister lineage to Hodarchaeales. We redid phylogenomic analyses of the NM57 dataset using the expanded Asgard archaea and eukaryotes. Although the ML tree of the untreated NM57 supported the Hodarchaeales-sister topology (Supplementary Fig. 8a), the support for Hodarchaeales-sister decreased dramatically, whereas support for Heimdallarchaeia-sister rose markedly as fastest-evolving sites were progressively reduced (Fig. 3c). The Bayesian inferences of the SR4-recoded34 NM57 dataset also confidently placed eukaryotes outside Heimdallarchaeia (PP, 1.0) (Supplementary Fig. 8b). The result indicated that the relationship between eukaryotes and Asgard archaea reported4 was probably artifactual. Additional tests using vertically evolving markers that support the Heimdallarchaeia-sister topology are delineated in Supplementary Information (Supplementary Table 4 and Supplementary Figs. 9–12). In addition, we examined the distribution of ESPs in the expanded sampling of Asgard archaea and found that the previously delineated ESPs specific to Hodarchaeales were also present in other Asgard clades (Supplementary Results and Discussion and Supplementary Figs. 13–16).
Taken together, our phylogenetic analyses indicate that eukaryotes may have evolved before the diversification of all sampled Heimdallarchaeia. The results were consistent with one of scenarios inferred using the 29-marker tree reported previously by Liu et al.5, but challenged the recently proposed Hodarchaeales-sister hypothesis4. Compared with the study of Liu et al.5, our results were more robust as they were generated using an expanded genomic sampling of Asgard archaea, in combination with exclusion of outgroups, removal of fast-evolving sites and application of complex site-heterogeneous evolution models and several marker sets. Our finding helps to infer the nature of the eukaryotic ancestors.
Timescale and traits of the LAECA
We used MCMCTree35 to estimate the divergence times of key nodes within the Asgard phylum using a relaxed molecular clock model. An archaeal root age range of 4.29–3.8 billion years ago (Ga) and uniform age priors for the other fossil calibrations were applied in the molecular dating analysis (Supplementary Table 5). The last Asgard archaea common ancestor (LAsCA) emerged at around 3.38 Ga (confidence interval (CI), 3.72–3.06 Ga) in the Archean (Supplementary Fig. 17). The estimate for the age of the Heimdallarchaeia and Lokiarchaeia common ancestor was about 3.16 Ga (CI, 3.54–2.82 Ga) in the Mesoarchean. As the sister of eukaryotes, crown Heimdallarchaeia diverged 3.12–2.26 Ga, predating the Great Oxidation Event36. The result indicates that eukaryotic stem lineages evolved before the Great Oxidation Event, which is basically consistent with the timescale of stem eukaryotes reported by Betts et al.3. Recently, based on the evolutionary history of ATP synthases, it was suggested that eukaryotes diverged from Hodarchaeales within Heimdallarchaeia 2.67–2.19 Ga37. This partially overlaps in time with our estimates for divergence of Heimdallarchaeia and eukaryotes.
We used a probabilistic gene–tree species-tree reconciliation method38 (amalgamated likelihood estimation (ALE)) to infer ancestral events and gene content on key nodes of the Asgard archaeal species tree. To refine the metabolic reconstruction, a subset of the aforementioned archaeal genomes with more than 80% completeness and less than 5% contamination was selected for ALE analyses. This subset, which included 235 Asgard archaea, 47 Euryarchaea and 60 TACK archaea, had a mean completeness of 88.5% and a mean contamination of 2.7% (Supplementary Tables 2 and 3). In the species tree, a gradual increase in ancestral gene content was observed during the evolution from LAsCA to Heimdallarchaeia ancestor (Supplementary Fig. 18). Along the evolutionary route, rates of gene loss also increased gradually, implying that a large amount of gene gain occurred before Heimdallarchaeia radiation.
Next, we reconstructed metabolic traits of key ancestors of Asgard archaea based on presence probability of genes in nodes reported by ALE (Supplementary Table 6) and gene frequencies across Asgard archaeal genomes (Supplementary Table 7). Recent studies on two cultured Asgard archaea, Ca. P. syntrophicum and Ca. L. ossiferum, have shown their ability to degrade amino acids anaerobically by syntrophy1,2. Based on these physiological features, it was inferred that the LAsCA was probably an amino-acid-degrading anaerobe that produced H2 and fatty acids as byproducts39. However, our analysis revealed that the two cultivated Asgard strains and their three closely related MAGs lacked most of the key genes for the H4MPT methyl branch of the archaeal Wood–Ljungdahl pathway40 (WLP), but encoded three key enzymes in the H4F methyl branch of bacterial WLP: formyltetrahydrofolate synthetase (fhs), methylenetetrahydrofolate dehydrogenase (folD) and methylenetetrahydrofolate reductase (metF) (Supplementary Fig. 19). This partial H4F branch can convert glycine, serine and histidine to formate that mediates interspecies electron transfer1 (Supplementary Fig. 20). In contrast, the LAsCA was inferred to harbour a complete archaeal WLP but lacked Fhs in the bacterial WLP (Fig. 4a, Supplementary Tables 6 and 7 and Supplementary Figs. 19 and 20). It also encoded various NiFe hydrogenases (Supplementary Table 6 and Supplementary Fig. 21). These results suggest that the LAsCA was a H2-dependent chemolithoautotroph. Abundant CO2 in the Archean ocean41 and marine H2 from serpentinization42 would have provided ample substrates for these organisms.
a, Transition from the Asgard common ancestor to Lokiarchaeia and Heimdallarchaeia ancestors. Based on ALE results, it is inferred that the LAsCA was a H2-dependent chemolithoautotroph. The archaeal WLP was inferred to be present in all four ancestors; thus, it could also have been present in the LAECA. Four genes related to acetogenesis (pta, ack, acs and acd) were predicted to be present in the Heimdallarchaeia ancestor, suggesting that the LAECA may have been an anaerobic H2-dependent acetogen. b, Distribution of key enzymes of the WLP in Heimdallarchaeia. As a basal branch within Heimdallarchaeia, Hodarchaeales possessed a complete WLP, supporting presence of the pathway in the Heimdallarchaeia ancestor. During the transition from Hodarchaeales to Heimdallarchaeaceae and Kariarchaeaceae, these key enzymes of the WLP appeared to be progressively lost. Fully filled circles indicate that the gene was detected in at least half of the genomes of the clade. Half-filled circles indicate that the gene was detected in fewer than half of the genomes of the clade. EMP, Embden–Meyerhof–Parnas; NOPPP, nonoxidative pentose phosphate pathway; RuMP, ribulose monophosphate pathway; sp/np-ED, semi/non-phosphorylative Entner–Doudoroff pathway; TCA cycle, citrate cycle; AA, amino acid metabolism; β, β-oxidation; H4F, tetrahydrofolate methyl branch; H4MPT, H4MPT methyl branch tetrahydromethanopterin; Fdh, formate dehydrogenase; Mvh/Hdr, F420-non-reducing hydrogenase and heterodisulfide reductase complex; Pyr, pyruvate; Ech, energy-converting hydrogenase; F420, coenzyme F420-reducing hydrogenase; Mcr, methyl-CoM reductase; Nucl, nucleotide; H, Heimdallarchaeaceae; K, Kariarchaeaceae; G, Gerdarchaeales; Hod, Hodarchaeales; W, Wukongarchaeia; fwd, formylmethanofuran dehydrogenase; ftr, formylmethanofuran-tetrahydromethanopterin N-formyltransferase; mch, methenyltetrahydromethanopterin cyclohydrolase; mtd, methylenetetrahydromethanopterin dehydrogenase; mer, 5,10-methylenetetrahydromethanopterin reductase; cdh, acetyl-CoA decarbonylase/synthase, CODH/ACS complex.
Lokiarchaeia included both Helarchaeales and Lokiarchaeales. All members of Helarchaeales (13 MAGs) are found to encode a methyl-CoM reductase-like enzyme (Supplementary Table 7) that is similar to that found in butane-oxidizing archaea43,44. ALE analysis inferred the presence of the methyl-CoM reductase-like enzyme in Lokiarchaeia ancestor (Supplementary Table 6), suggesting that this ancestor had the potential to degrade hydrocarbon anaerobically by coupling the β-oxidation of fatty acids with reverse WLP (Fig. 4a and Supplementary Fig. 20).
Contrary to the result by Eme et al.4, our inference revealed that the Heimdallarchaeia and Hodarchaeales ancestors also possessed a complete archaeal WLP pathway (Fig. 4a, Supplementary Fig. 20 and Supplementary Table 6). We found that, in the MAGs of Heimdallarchaeia (including three Hodarchaeales genomes) used by Eme et al.4, genes encoding several key enzymes in the archaeal WLP are missing. However, we presented 22 new genomes of Hodarchaeales, representing two new families and seven new genera, in which several genomes or lineages contained a complete archaeal WLP (Fig. 4b). As a basal-branching group in Heimdallarchaeia, the presence of archaeal WLP in Hodarchaeales supported an autotrophic lifestyle of crown Heimdallarchaeia. Except for the WLP, the crown Heimdallarchaeia was inferred to encode a phosphotransacetylase (pta) and an acetate kinase (ack) that are required for the conversion from acetyl-CoA to acetate, as well as an energy-converting hydrogenase (ech) that can potentially serve as an ion-translocating chemiosmotic coupling site, and an ATP synthase (Supplementary Tables 6 and 7 and Supplementary Fig. 20). Gene frequency analysis revealed that ack genes were found exclusively in members of Heimdallarchaeia (Supplementary Fig. 21a). These data inspired the inference that Heimdallarchaeia ancestor was an anaerobic H2-dependent acetogen, as shown in a previous study45.
As a parent of crown Heimdallarchaeia, the LAECA may also be a H2-dependent acetogenic archaeon. Our metabolic evidence supported the hydrogen hypothesis, which posits that eukaryogenesis was mediated initially by a syntrophic partnership between a H2-consuming archaeal host and a H2-producing protomitochondrion living under anoxic conditions, with the protomitochondrion being a facultative aerobe46,47. We identified an incomplete cytochrome bd-I ubiquinol oxidase and an incomplete cytochrome o ubiquinol oxidase in 38 members of Hodarchaeales and 16 members of Kariarchaeaceae (Supplementary Fig. 21b and Supplementary Table 7). However, only two subunits of the cytochrome o ubiquinol oxidase were inferred to be present in the Heimdallarchaeia ancestor (Supplementary Fig. 20 and Supplementary Table 6), which may have been acquired through HGT from a bacterial partner. These incomplete complexes probably lacked the capacity to transport electrons or reduce molecular oxygen to water. The inferred presence of extremely O2 sensitive enzymes, such as acetyl-CoA decarbonylase/synthase (cdh), 2-oxoacid: ferredoxin oxidoreductases (kor, vor and por) and NiFe hydrogenases in the Heimdallarchaeia ancestor (Supplementary Fig. 20 and Supplementary Table 6 and 7), together with the anoxic deep ocean environment at that time46, suggests that this ancestor must have grown anaerobically.
In summary, we used sophisticated phylogenomic methods in combination with a significantly expanded sampling of Asgard archaea to re-evaluate relationships among Asgard archaea, Njordarchaeales and eukaryotes. We revealed that the Njordarchaeales genomes probably represent chimeric assemblies comprising mainly TACK and Asgard archaeal sequences. These weird assembled genomes would significantly affect phylogenetic position of eukaryotes in archaea. After excluding outgroups, using several marker sets, eukaryotes were placed confidently within Asgard archaea as a sister to Heimdallarchaeia instead of being nested within Heimdallarchaeia branching with Hodarchaeales. Ancestral reconstructions inferred that the host lineage at eukaryotic origin was an anaerobic, H2-dependent chemolithoautotroph. Our findings rectified the existing knowledge and filled some gaps in episodes of the early evolution of eukaryotes. The further expansion of Heimdallarchaeia diversity and their isolation will help to refine evolutionary reconstructions48 of eukaryogenesis.
Methods
Sample acquisition and DNA sequencing
We collected 40 sediment samples from six different locations in coastal wetlands across China: Techeng Island (mangrove swamp); Qingmei Port (mangrove swamp); Tongming Port (mangrove swamp); Dongzai Port (mangrove swamp); Changjiang Estuary (salt marsh) and Luchao Port (salt marsh) (Supplementary Table 1 and Supplementary Fig. 1). A peat sampler was employed to collect sediment cores. After collection, sediment samples were sealed in sterile plastic bags, placed immediately in a pre-chilled icebox and transported to the laboratory as quickly as possible, where they were stored at −80 °C until DNA extraction. Detailed information of the sampling is presented in the following sections.
Samples from Techeng Island
Two 1-m-deep sediment cores were collected from a mangrove swamp on Techeng Island (Zhanjiang) on November 25, 2018. Three sections with depths of 15–25, 40–45 and 95–100 cm each core were taken in an anoxic glove box. Genomic DNA was extracted from 5–10 g sediment using a PowerSoil DNA Isolation Kit (MoBio Laboratories) according to the manufacturer’s instructions. Sequencing libraries were prepared using the NEBNext Ultra DNA Library Prep Kit (New England Biolabs) and sequenced on Illumina HiSeq 2500 instruments at Guangdong MagiGene Technology Corporation, generating approximately 60 Gbp of raw sequencing data (2 × 150 bp) for each sample.
Samples from Qingmei Port
Two 0.5-m-deep sediment cores were collected from the Qingmeigang Mangrove Reserve (Sanya) in September 2020. Four fractions taken from depths of 0–5, 10–15, 25–30 and 45–50 cm, respectively, for each core were used for DNA extraction. Metagenomic sequencing was carried out on Illumina HiSeq 2500 platform at Suzhou Genewiz Biotechnology Company. Each sample yielded 80–120 Gbp of sequencing data in the form of 2 × 150-bp paired-end reads.
Samples from Tongming Port
Three 1-m-deep sediment cores were acquired from a mangrove swamp near Tongming Harbour (Zhanjiang) in June 2021. Samples for DNA extraction were taken from the layers of 15–20 and 45–50 cm of each core. Metagenomic sequencing was performed on Illumina HiSeq 2500 platform at Novogene Biotechnology Company (Tianjin, China), yielding 80–120 Gbp of sequencing data for each sample.
Samples from Dongzai Port
Three sediment cores were obtained from the Dongzhai Port Mangrove Nature Reserve (Haikou) in September 2021. Six samples of each of these cores were taken from depths of 15–20 and 45–50 cm. One additional sample was collected from the 95–100 cm layer of one of the cores. Metagenomic sequencing were conducted as described for samples of Tongming Harbour.
Samples from Changjiang Estuary
Two sediment cores were obtained in November 2018 from the DongTan salt marsh wetland (Shanghai), covered with reeds, located at the estuary of the Yangtze River. Six samples of the two cores were taken from depths of 15–20, 40–55 and 95–100 cm for DNA extraction. Metagenomic sequencing was generated using Illumina HiSeq 2500 by Novogene, yielding 80–120 Gbp of sequencing data for each sample.
Samples from Luchao Port
In November 2021, two sediment cores were obtained from a salt marsh wetland covered with Spartina alterniflora near the Luchao Port of Shanghai. Samples for DNA extraction were collected from the layers of 20–25, 40–45 and 95–100 cm of each core. Nucleic acids were sequenced using Illumina HiSeq 2500 by Novogene. Each sample produced 100 Gbp of sequencing data.
Metagenomic assembly and genome reconstruction
A total of 3.54 Tbp of raw reads was obtained from metagenomes generated from the 40 sediment samples. These reads were trimmed using Trimmomatic49 (v.0.39) to remove the adaptors and low-quality regions. The resulting clean reads were single-sample de novo assembled using SPAdes50 (v.3.15.3) with the parameters: ‘-k 21,33,55,77,99 -meta’. The assembled scaffolds were used to recruit reads from their own metagenomes and other metagenomes from the same geographical location using Bowtie2 (ref. 51) (v.2.4.4). The SAM files generated were converted to BAM files using Samtools52 (v.1.9). The coverage for each of BAM files was then calculated with the jgi_summarize_bam_contig_depth script in MetaBAT2 (ref. 53) (v.2.15). These coverage files were applied as input files for Metabat2 (ref. 53) binning analysis. For each sample, binning was performed using both single-coverage and multi-coverage methods54, combined with eight different combinations of specificity and sensitivity parameters: --maxP 95 or 60, --minS 95 or 60, and --maxEdges 200 or 500. As for a single assembly, an optimized, non-redundant set of bins was chosen by using DAS-Tool55 (v.1.1.6). Scaffolds in each bin with divergent GC content or tetranucleotide signatures or coverage profiles were removed with mmgenome56 and RefineM57. In the end, 11,878 genome bins were obtained from these samples (Supplementary Fig. 1b) and their quality including the completeness, contamination and strain heterogeneity were assessed using CheckM58 (v.1.1.3).
Asgard archaeal genome set
A total of 223 high-quality Asgard MAGs (completeness at least 70%, contamination less than or equal to 10%) were reconstructed in this study. An additional 395 high-quality Asgard MAGs plus 15 Njordarchaeales MAGs were downloaded from publicly available databases (NCBI and GTDB15) as of September 23, 2023. Redundant genomes at species level (ANI at least 95%) were then removed using dRep14 (v.3.4.3) with parameters ‘-comp 70 -con 10 --S_ani 0.95’, resulting in 411 Asgard genome representatives. Of these, 136 were derived from this study, constituting approximately 32% of the total (Supplementary Table 2). Protein coding genes, ribosomal RNA genes and transfer RNA genes in these genomes were identified using Prodigal59 (v.2.6.3), Barrnap (v.0.9) (https://github.com/tseemann/barrnap) and tRNAscan-SE60 (v.2.0.9), respectively.
Phylogenomic analysis
Selection of other archaeal genomes outside Asgardarchaeota
Taxa in the GTDB61 archaeal reference tree (gtdb_r207_ar53_decorated_fullids_unrooted.tree) were clustered using TreeCluster62 with a distance threshold of length 1 (parameter: --threshold 1). After excluding these singleton clusters, the highest quality genome in a cluster was retained. To balance the taxon sampling, 10–15 genomes were selected randomly from each order. For orders with fewer than ten genomes, all available genomes were used. Finally, we obtained 51 DPANN archaea, 47 Euryarchaea and 70 TACK archaea representatives as the outgroup of the following Asgard phylogenies (Supplementary Table 3).
Phylogenomic analyses of previously used archaeal marker sets
A phylogeny of 53 archaeal marker proteins from the GTDB Toolkit20 (GTDB-Tk) was inferred to examine phylogenetic diversity of Asgard archaea. In addition, to resolve the phylogenetic position of Njordarchaeales in archaea, an additional 12 sets of pre-existing markers that are conserved across archaea were used for inference of trees. Information on these markers is as follows: (1) the RP55 set comprising 55 ribosomal protein sequences7,8, (2) the Dacunha_35uni set consisting of 21 ribosomal proteins and 14 functional proteins19, (3) the Zare_48uni set consisting of 31 ribosomal proteins and 17 functional proteins7, (4) the Spang_udin56ar set comprising 25 ribosomal proteins and 31 functional proteins23, (5) the Spang_udin28ar set consisting of ten ribosomal proteins and 18 functional proteins23, (6) the TomNEE_21 set consisting of nine ribosomal proteins and 12 functional proteins13, (7) the Liu_209 set consisting of 45 ribosomal proteins and 164 functional proteins5, (8) the RP15 set consisting of 15 ribosomal proteins25, (9) the PV4 set consisting of 30 ribosomal proteins and seven functional proteins22, (10) the Liu_29 set consisting of 23 ribosomal proteins and six functional proteins5, (11) the NM200 set consisting of 200 non-ribosomal proteins21 and (12) the NM57 set consisting of 57 non-ribosomal proteins4 (Supplementary Table 4). These markers were identified in genomes of 411 Asgard archaea and the outgroup by searching against a self-built database composed of all arCOG63 sequences as well as partial sequences from COG64, AsCOG5, Pfam65 and TIGRFAMs65, using either BLASTP66 or HMMER67. For the RP15 set, these ribosomal proteins were adopted if their genes were located on a scaffold harbouring at least five out of 15 ribosomal protein genes. The marker sequences were aligned with MAFFT68 (v.7.487, -linsi) and pruned using BMGE69 (v.1.12) (-m BLOSUM30). IQ-TREE70 (v.2.2.2.6) was used to infer phylogenetic trees under the LG + C60 + F + G + PMSF model. Branch support was assessed with 1,000 ultrafast bootstrap approximations71 (-B 1,000). Phylogenetic trees were visualized using iTol72 (https://itol.embl.de). Phylogenomic analyses of all marker sets placed Njordarchaeales within TACK superphylum as a sister clade to Korarchaeota, except for the NM57 dataset.
Construction and phylogenomic analyses of new marker sets
The expanded diversity of Asgard genomes necessitates new sets of markers that are distributed evenly across Asgard lineages or other archaeal phyla. To further resolve phylogenetic position of Njordarchaeales and eukaryotes within archaea, we integrated these previously reported marker sets used for confidently inferring the archaeal phylogeny4,5,13,21 and selected 67 markers (S67) that are conserved across all sampled archaeal genomes. The 67 markers were selected because they were identified in at least 60% of representatives of each of the archaeal clades as well as in 60% of eukaryotic taxa. The S67 marker set was comprised of 39 ribosomal proteins and 28 functional proteins involved in diverse cellular activities (Supplementary Table 4). We used the 67 markers to build four supermatrices: S67, ES67, tES67 and AsES67. The S67 supermatrix was constructed by including all sampled archaeal genomes (411 Asgard archaea, 51 DPANN archaea, 47 Euryarchaea, and 70 TACK archaea representatives), whereas the ES67 supermatrix was built by adding 14 representative eukaryotic genomes into the S67 dataset. These eukaryotic taxa were chosen according to previous studies4,7. The tES67 supermatrix was created by removing DPANN, Euryarchaea, Korarchaeota and Njordarchaeales sequences from the ES67 supermatrix, whereas the AsES67 supermatrix was built by including only 411 Asgard archaea and 14 eukaryotic taxa.
To increase the phylogenetic signal of supermatrices when outgroups were excluded, we relaxed our marker selection criterion and generated two additional marker sets: S150 and S97 (Supplementary Table 4). Both of these were extracted from the aforementioned 13 set of marker proteins4,5,7,8,13,15,19,21,22. The S150 set comprises 150 proteins, including 52 ribosomal proteins and 98 functional proteins, which were selected based on presence of these markers in at least 80% of Asgard archaeal genomes. The S97 set consists of 97 proteins, including 41 ribosomal proteins and 56 functional proteins, which were selected based on their presence of at least 60% of representatives of each of the Asgard clades. The two marker sets were used to build two supermatrices: S150 and S97, by including 411 Asgard archaea and 14 eukaryotic taxa.
Before inferring phylogenies of these marker sets, preliminary trees of each individual marker, inferred using FastTree2 (ref. 73) under the LG + CAT model, were checked manually to identify the correct orthologues for each taxon and to detect any instances of paralogues. Any identified paralogous sequences were removed manually from the remaining trees. Single-gene trees with paralogous sequences removed have been uploaded to Figshare (https://figshare.com/s/6e523322b0b647b91dda)74. In addition, by excluding markers that failed to recover archaea and bacteria as reciprocally monophyletic domains, we obtained 47, 60 and 99 markers (S47, S60 and S99) from the S67, S97 and S150 marker sets, respectively (Supplementary Table 4). Detailed analyses of these marker sets are provided in the Supplementary Information. ML trees of these amino acid supermatrices were inferred using IQ-TREE75 with the LG + C60 + F + G + PMSF model. The model was selected because it can capture variations in the substitution or replacement process across sites. The robustness of the resulting trees was assessed with 1,000 ultrafast bootstrap approximations71 (-B 1,000). Bayesian inferences were performed using PhyloBayes-MPI76 (v.1.9) with CAT + GTR after the alignments were recoded into four categories using the SR4 scheme34. Two independent Markov chains were run until a sufficient effective sample size (greater than 300) was achieved. For consensus tree reconstruction, the first 5,000 cycles (for the run with 20,000 iteration times) or the first 10,000 cycles (for the run with 50,000 iteration times) were discarded as burn-in. In all Bayesian inferences, the two chains did not converge, probably due to the large size of the supermatrices and computational limitations.
To examine effect of mutationally saturated or rate-biased sites on tree reconstruction24,26,77, we tracked the bootstrap support values of branches of interest. The evolutionary rate of sites (evolving-rate score) was estimated using IQ-TREE70 with the empirical Bayesian algorithm78 (--rate). The sites were classified into ten categories from the fastest to the slowest evolving. A series of alignments were generated by removing 10% to 90% of the data in a stepwise fashion. The resulting alignments were used to compute 1,000 ultrafast bootstrap71 support values in IQ-TREE75, using the LG + C60 + F + G + PMSF model. In the end, the bootstrap support for each bipartition of interest was calculated and tracked as the fastest-evolving sites were progressively removed.
Assessment of Njordarchaeales representative MAGs
The taxonomic profiles of contigs of Njordarchaeales MAGs were determined using two state-of-the-art tools: CAT28,29 and MMseqs2 (refs. 30,31), by comparison with TACK and Asgard archaeal sequences from the GTDB database79 (r220) excluding Njordarchaeales-related sequences. CAT is a tool for taxonomic annotation of contigs based on protein homologies to a reference database, while MMseqs2 determines the taxonomic identity of contigs through a weighted voting mechanism. The two tools both rely on a well-defined reference database. Given that the position of Njordarchaeales moved between TACK superphylum and Asgardarchaeota in species trees, we retrieved representative genomes of TACK and Asgard archaea only from the GTDB15 database (r220) to construct the reference database. This strategy aims to minimize the impact of other archaeal sequences on taxonomic classification. As shown in Fig. 2a,b, 20–73% of the contigs in each MAG were not assigned to a specific classification, probably indicating that these contigs contain sequences that are too divergent from those in the database.
Metagenomic samples from which Njordarchaeales MAGs were reconstructed were downloaded from publicly available databases4,32,33. These metagenomes were generated from deep-sea hydrothermal vent sediments. Metagenomic reads were mapped to these Njordarchaeales MAGs using Bowtie2 (ref. 80) (v.2.3.5). The resulting SAM files were converted to BAM files with Samtools52 (v.1.9), which were used to create an input index for Anvi’o80. Coverage and GC content of contigs were recorded. Contigs of these MAGs were clustered according to their sequence composition and coverage of contigs. Contigs originating from the same organism typically exhibit similar sequence coverage. Therefore, those with divergent coverage profiles are often considered as potential contaminants81,82. These contaminating contigs are particularly prone to displaying differential coverage patterns across different metagenomic samples81. In this study, four Njordarchaeales MAGs were completely matched by reads from several metagenomes generated from similar habitats. The average coverages of contigs in the clusters differed across metagenomes.
Phylogenetic divergence time estimation
Molecular dating analysis was carried out with the MCMCtree program in PAML83 using the WAG model. The analysis was run iteratively until convergence was achieved. Seven nodes in the phylogenetic tree were calibrated using four types of constraint84: (1) the Archaeal root constraint was set at 4.29–3.80 Ga, (2) the chitin age had a minimum constraint of 1.58 Ga, (3) the oxygen age had a maximum constraint of 2.32 Ga and (4) the HGT from Viridiplantae to Thaumarchaeota was constrained to 1.49–0.75 Ga. Further details are provided in Supplementary Table 5.
Ancestral metabolic reconstruction
For analyses of amalgamated likelihood estimation38,85, a set of archaeal genomes with more than 80% completeness and less than 5% contamination (mean completeness of 88.5% and mean contamination of 2.7%) was selected, comprising 235 Asgard archaea, 47 Euryarchaea and 60 TACK archaea representatives (Supplementary Table 2 and 3). These genomes had a mean quality score of 75%, with the lowest quality value57 being 51%. All predicted protein sequences from these genomes were annotated by searching against arCOGs63 and nr database using BLASTP86 (E-value less than 1 × 10−5) or Pfam65 and TIGRFAMs87 databases using HMMER88 with a GA bitscore cutoff. The SwissProt database89 was further used to validate protein functions. Protein sequences with identical functional descriptions were clustered to infer individual gene trees. Sequences were aligned and pruned using MAFFT68 (-linsi) and BMGE69 (-m BLOSUM30). Gene tree profiles were then inferred using FastTree2 (ref. 73) with the LG + CAT model. Subsequently, the species tree of the S47 dataset, along with the gene tree profiles, were applied to perform gene–tree species tree reconciliations using ALEobserve (v.1.0) and ALEml_undated. Events of loss, transfer, origination or duplication as well as presence/absence of gene (copies) were considered only if they had a raw reconciliation frequency of at least 0.1. The copy number (proteome size) and the events are summarized in Supplementary Table 6 and 8, and visualized on the branches of the species tree (Supplementary Fig. 18). The resulting data were then used for metabolic reconstruction. To support the inferences of ALE, gene frequencies for key metabolic pathways across Asgard archaea are provided in Supplementary Table 7.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All sequence data are archived in the NCBI database under BioProject ID PRJNA1162170. All raw data underlying phylogenomic analyses (raw and processed alignments and corresponding phylogenetic trees) are available at Figshare (https://figshare.com/s/6e523322b0b647b91dda)74.
References
Imachi, H. et al. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 577, 519–525 (2020).
Rodrigues-Oliveira, T. et al. Actin cytoskeleton and complex cell architecture in an Asgard archaeon. Nature 613, 332–339 (2023).
Betts, H. C. et al. Integrated genomic and fossil evidence illuminates life’s early evolution and eukaryote origin. Nat. Ecol. Evol. 2, 1556–1562 (2018).
Eme, L. et al. Inference and reconstruction of the heimdallarchaeial ancestry of eukaryotes. Nature 618, 992–999 (2023).
Liu, Y. et al. Expanded diversity of Asgard archaea and their relationships with eukaryotes. Nature 593, 553–557 (2021).
Lopez-Garcia, P. & Moreira, D. Open questions on the origin of eukaryotes. Trends Ecol. Evol. 30, 697–708 (2015).
Zaremba-Niedzwiedzka, K. et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017).
Spang, A. et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 (2015).
Lu, Z., Zhang, S., Liu, Y., Xia, R. & Li, M. Origin of eukaryotic-like Vps23 shapes an ancient functional interplay between ESCRT and ubiquitin system in Asgard archaea. Cell Rep. 43, 113781 (2024).
Akil, C. & Robinson, R. C. Genomes of Asgard archaea encode profilins that regulate actin. Nature 562, 439–443 (2018).
Izore, T., Kureisaite-Ciziene, D., McLaughlin, S. H. & Lowe, J. Crenactin forms actin-like double helical filaments regulated by arcadin-2. eLife 5, e21600 (2016).
Spang, A., Caceres, E. F. & Ettema, T. J. Genomic exploration of the diversity, ecology, and evolution of the archaeal domain of life. Science 357, eaaf3883 (2017).
Williams, T. A., Cox, C. J., Foster, P. G., Szollosi, G. J. & Embley, T. M. Phylogenomics provides robust support for a two-domains tree of life. Nat. Ecol. Evol. 4, 138–147 (2020).
Olm, M. R., Brown, C. T., Brooks, B. & Banfield, J. F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868 (2017).
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics 38, 5315–5316 (2022).
Parks, D. H. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004 (2018).
Xie, R. et al. Expanding Asgard members in the domain of Archaea sheds new light on the origin of eukaryotes. Sci. China Life Sci. 65, 818–829 (2022).
Liu, Y. & Li, M. The unstable evolutionary position of Korarchaeota and its relationship with other TACK and Asgard archaea. mLife 1, 218–222 (2022).
Da Cunha, V., Gaia, M., Gadelle, D., Nasir, A. & Forterre, P. Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genet. 13, e1006810 (2017).
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36, 1925–1927 (2019).
Petitjean, C., Deschamps, P., Lopez-Garcia, P., Moreira, D. & Brochier-Armanet, C. Extending the conserved phylogenetic core of archaea disentangles the evolution of the third domain of life. Mol. Biol. Evol. 32, 1242–1254 (2015).
Darling, A. E. et al. PhyloSift: phylogenetic analysis of genomes and metagenomes. Peerj 2, e243 (2014).
Dombrowski, N. et al. Undinarchaeota illuminate DPANN phylogeny and the impact of gene transfer on archaeal evolution. Nat. Commun. 11, 3939 (2020).
Fan, L. et al. Phylogenetic analyses with systematic taxon sampling show that mitochondria branch within Alphaproteobacteria. Nat. Ecol. Evol. 4, 1213–1219 (2020).
Martijn, J., Vosseberg, J., Guy, L., Offre, P. & Ettema, T. J. G. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 557, 101–105 (2018).
Viklund, J., Ettema, T. J. & Andersson, S. G. Independent genome reduction and phylogenetic reclassification of the oceanic SAR11 clade. Mol. Biol. Evol. 29, 599–615 (2012).
Ramulu, H. G. et al. Ribosomal proteins: toward a next generation standard for prokaryotic systematics? Mol. Phylogenet. Evol. 75, 103–117 (2014).
von Meijenfeldt, F. A. B., Arkhipova, K., Cambuy, D. D., Coutinho, F. H. & Dutilh, B. E. Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol. 20, 217 (2019).
Hauptfeld, E. et al. Integrating taxonomic signals from MAGs and contigs improves read annotation and taxonomic profiling of metagenomes. Nat. Commun. 15, 3373 (2024).
Steinegger, M. & Soding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 35, 1026–1028 (2017).
Mirdita, M., Steinegger, M., Breitwieser, F., Soding, J. & Levy Karin, E. Fast and sensitive taxonomic assignment to metagenomic contigs. Bioinformatics 37, 3029–3031 (2021).
Dombrowski, N., Seitz, K. W., Teske, A. P. & Baker, B. J. Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments. Microbiome 5, 106 (2017).
Zhou, Z., St John, E., Anantharaman, K. & Reysenbach, A. L. Global patterns of diversity and metabolism of microbial communities in deep-sea hydrothermal vent deposits. Microbiome 10, 241 (2022).
Susko, E. & Roger, A. J. On reduced amino acid alphabets for phylogenetic inference. Mol. Biol. Evol. 24, 2139–2150 (2007).
Yang, Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 (1997).
Kump, L. R. The rise of atmospheric oxygen. Nature 451, 277–278 (2008).
Mahendrarajah, T. A. et al. ATP synthase evolution on a cross-braced dated tree of life. Nat. Commun. 14, 7456 (2023).
Szollosi, G. J., Rosikiewicz, W., Boussau, B., Tannier, E. & Daubin, V. Efficient exploration of the space of reconciled gene trees. Syst. Biol. 62, 901–912 (2013).
Sousa, F. L., Neukirchen, S., Allen, J. F., Lane, N. & Martin, W. F. Lokiarchaeon is hydrogen dependent. Nat. Microbiol. 1, 1–3 (2016).
Adam, P. S., Borrel, G. & Gribaldo, S. An archaeal origin of the Wood-Ljungdahl H(4)MPT branch and the emergence of bacterial methylotrophy. Nat. Microbiol. 4, 2155–2163 (2019).
Catling, D. C. & Zahnle, K. J. The Archean atmosphere. Sci. Adv. 6, eaax1420 (2020).
Tamblyn, R. & Hermann, J. Geological evidence for high H2 production from komatiites in the Archaean. Nat. Geosci. 16, 1194–1199 (2023).
Seitz, K. W. et al. Asgard archaea capable of anaerobic hydrocarbon cycling. Nat. Commun. 10, 1822 (2019).
Laso-Pérez, R. et al. Thermophilic archaea activate butane via alkyl-coenzyme M formation. Nature 539, 396–401 (2016).
Martin, W. F., Neukirchen, S., Zimorski, V., Gould, S. B. & Sousa, F. L. Energy for two: New archaeal lineages and the origin of mitochondria. Bioessays 38, 850–856 (2016).
Mills, D. B. et al. Eukaryogenesis and oxygen in Earth history. Nat. Ecol. Evol. 6, 520–532 (2022).
Martin, W. & Müller, M. J. N. The hydrogen hypothesis for the first eukaryote. Nature 392, 37–41 (1998).
Giger, G. H. et al. Inducing novel endosymbioses by implanting bacteria in fungi. Nature 635, 415–422 (2024).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357 (2012).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Kang, D. W. D., Froula, J., Egan, R. & Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. Peerj 3, e1165 (2015).
Mattock, J. & Watson, M. A comparison of single-coverage and multi-coverage metagenomic binning reveals extensive hidden contamination. Nat. Methods 20, 1170–1173 (2023).
Sieber, C. M. K. et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843 (2018).
Albertsen, M. et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 31, 533–538 (2013).
Parks, D. H. et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2, 1533–1542 (2017).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 11, 119 (2010).
Chan, P. P., Lin, B. Y., Mak, A. J. & Lowe, T. M. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 49, 9077–9096 (2021).
Parks, D. H. et al. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 38, 1079–1086 (2020).
Balaban, M., Moshiri, N., Mai, U., Jia, X. & Mirarab, S. TreeCluster: clustering biological sequences using phylogenetic trees. PLoS ONE 14, e0221068 (2019).
Makarova, K. S., Wolf, Y. I. & Koonin, E. V. Archaeal clusters of orthologous genes (arCOGs): an update and application for analysis of shared features between Thermococcales, Methanococcales, and Methanobacteriales. Life (Basel) 5, 818–840 (2015).
Galperin, M. Y. et al. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 49, D274–D281 (2021).
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 (2014).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A. & Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled–coil regions. Nucleic Acids Res. 41, e121 (2013).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Criscuolo, A. & Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10, 210 (2010).
Minh, B. Q., Trifinopoulos, J., Schrempf, D., Schmidt, H. & Lanfear, R. IQ-TREE version 2.0: tutorials and manual phylogenomic software by maximum likelihood. Nucleic Acids Res. 44, W232–W235 (2022).
Hoang, D. T., Chernomor, O., Von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52, W78–W82 (2024).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Zhang, J. et al. Phylogenomic analyses with expanded taxon sampling reveal deep origin of eukaryotes outside Heimdallarchaeia. Figshare https://figshare.com/s/6e523322b0b647b91dda (2025).
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Lartillot, N., Rodrigue, N., Stubbs, D. & Richer, J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 62, 611–615 (2013).
Munoz-Gomez, S. A. et al. Site-and-branch-heterogeneous analyses of an expanded dataset favour mitochondria as sister to known Alphaproteobacteria. Nat. Ecol. Evol. 6, 253–262 (2022).
Mayrose, I., Graur, D., Ben-Tal, N. & Pupko, T. Comparison of site-specific rate-inference methods for protein sequences: empirical Bayesian methods are superior. Mol. Biol. Evol. 21, 1781–1791 (2004).
Parks, D. H. et al. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 50, D785–D794 (2022).
Eren, A. M. et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. Peerj 3, e1319 (2015).
Chen, L. X., Anantharaman, K., Shaiber, A., Eren, A. M. & Banfield, J. F. Accurate and complete genomes from metagenomes. Genome Res. 30, 315–333 (2020).
Alneberg, J. et al. Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146 (2014).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Yang, Y. et al. The evolution pathway of ammonia-oxidizing archaea shaped by major geological events. Mol. Biol. Evol. 38, 3637–3648 (2021).
Szollosi, G. J., Davin, A. A., Tannier, E., Daubin, V. & Boussau, B. Genome-scale phylogenetic analysis finds extensive gene transfer among fungi. Philos. Trans. R. Soc. B Biol. Sci. 370, 20140335 (2015).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Haft, D. H., Selengut, J. D. & White, O. The TIGRFAMs database of protein families. Nucleic Acids Res. 31, 371–373 (2003).
Eddy, S. R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 7, e1002195 (2011).
Bairoch, A. & Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28, 45–48 (2000).
Acknowledgements
H.-P.D. and L.-J.H. were supported by the Chinese National Key Programmes for Fundamental Research and Development (2024YFF0808800), the National Natural Science Foundation of China (42476110, 42030411, 41971125) and the Ocean Negative Carbon Emissions (ONCE) Program. M. Li was supported by the National Natural Science Foundation of China (32393971, 32225003, 32393970, 92251306), Guangdong Major Project of Basic and Applied Basic Research (2023B0303000017) and Shenzhen University 2035 Program for Excellent Research (2022B002). X.F. was supported by Guangdong Basic and Applied Basic Research (2025A1515012817).
Author information
Authors and Affiliations
Contributions
This study was conceived by H.-P.D. with input from J.Z. and L.-J.H. Metagenomic assembly, binning and phylogenetic analyses were conducted primarily by J.Z. X.F. provided bioinformatics support, with input from M. Li and Y.L. H.-P.D., L.-J.H. and M. Liu contributed data. H.-P.D. wrote the manuscript and Supplementary Information, with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks William Martin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 The maximum likelihood phylogenomic analysis of the S67 dataset.
The tree was inferred using IQ-TREE under the LG + C60 + F + G + PMSF model, based on 579 archaeal taxa (11,860 sites, 67 concatenated proteins, 579 taxa). This tree was rooted to DPANN and Euryarchaeota. Bootstrap support values ≥ 95% are represented by black dots. The scale bar denotes the average expected number of substitutions per site.
Extended Data Fig. 2 Evolution of ultrafast bootstrap support for either the monophyly of Njordarchaeales and Heimdallarchaeia (NHeim, red line) or the monophyly of Njordarchaeales and Korarchaeia (NKor, blue line).
The bootstrap values were obtained from phylogenomic trees inferred from the S67 (a) and NM57 (b) datasets, based on 579 archaeal taxa, with the fastest-evolving sites progressively removed. The trees were inferred using IQ-TREE under the LG + C60 + F + G + PMSF model.
Extended Data Fig. 3 The maximum likelihood phylogenomic analysis of NM57 marker set, showing the position of contigs in two larger clusters of Njordarchaeales B7_G17_GCA_029856635.
Contigs of Njordarchaeales B7_G17_GCA_029856635 were grouped into three clusters (Fig. 2c). a, Phylogenetic position of contigs in cluster 1. b, Phylogenetic position of contigs in cluster 2. The trees were inferred using IQ-TREE under the LG + C60 + F + G + PMSF model and rooted to DPANN and Euryarchaeota. Bootstrap support values ≥ 95% are represented by black dots. The scale bar denotes the average expected number of substitutions per site.
Extended Data Fig. 4 Evolution of ultrafast bootstrap support for the monophyly of either eukaryotes and Heimdallarchaeia (EHeim, red line) or the monophyly of eukaryotes and Hodarchaeales (EHod, blue line).
The bootstrap values were obtained from the phylogenies inferred from the tES67 dataset (a) and AsES67 dataset (b), as the fastest-evolving sites were progressively removed. The trees were inferred using IQ-TREE under the LG + C60 + F + G + PMSF model.
Extended Data Fig. 5 Phylogenetic analyses and site-exclusion treatment of the S150 dataset for 411 Asgard archaeal taxa and 14 eukaryotic taxa.
a, The maximum likelihood phylogenomic analysis based on the S150 dataset (32,277 sites, 150 concatenated proteins, 425 taxa). The trees were inferred using IQ-TREE under the LG + C60 + F + G + PMSF model. Bootstrap support values ≥ 95% are represented by black dots. The scale bar denotes the average expected number of substitutions per site. b, Evolution of ultrafast bootstrap support for the monophyly of either eukaryotes and Heimdallarchaeia (EHeim, red line) or the monophyly of eukaryotes and Hodarchaeales (EHod, blue line), in the phylogenies inferred from the S150 dataset. The trees were inferred using IQ-TREE under the LG + C60 + F + G + PMSF model.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Results and Discussion 1–4, References, Figs. 1–21 and legends for Tables 1–9.
Supplementary Table 1
Sampling information.
Supplementary Table 2
Information of Asgard archaeal MAGs, proposed taxonomy.
Supplementary Table 3
Information on reference genomes used for phylogenetic trees in this study.
Supplementary Table 4
Marker sets used in this study.
Supplementary Table 5
Genomes and calibration nodes used for phylogenetic divergence time estimation.
Supplementary Table 6
Reconciliation frequency of genes in key Asgard ancestors inferred by ALE.
Supplementary Table 7
Main metabolic pathways in Asgard archaea and their corresponding gene frequencies.
Supplementary Table 8
Results of ALE analyses.
Supplementary Table 9
Distribution of ESPs in Asgard lineages.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, J., Feng, X., Li, M. et al. Deep origin of eukaryotes outside Heimdallarchaeia within Asgardarchaeota. Nature 642, 990–998 (2025). https://doi.org/10.1038/s41586-025-08955-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-08955-7
This article is cited by
-
Serial innovations by Asgard archaea shaped the DNA replication machinery of the early eukaryotic ancestor
Nature Ecology & Evolution (2025)
-
Distinct filament morphology and membrane tethering features of the dual FtsZ paralogs in Odinarchaeota
The EMBO Journal (2025)
-
Insights into Phylum Asgardarchaeota: Analysis of Two Novel MAGs Within Class Njordarchaeia
Indian Journal of Microbiology (2025)