Abstract
Adipose tissue maintains energy homeostasis by storing lipids during nutrient surplus and releasing them through lipolysis in times of energy demand1,2. While lipolysis is essential for short-term metabolic adaptation, prolonged metabolic stress requires adaptive changes that preserve energy reserves2,3. Here we report that β3-adrenergic activation of adipocytes induces a transient and depot-specific infiltration of neutrophils into white adipose tissue (WAT), particularly in lipid-rich visceral WAT. Neutrophil recruitment requires the stimulation of both lipolysis and p38 MAPK in adipocytes, and is mediated by the secretion of leukotriene B4. Recruited neutrophils undergo activation in situ, and locally secrete IL-1β, which suppresses lipolysis and limits excessive energy loss. Neutrophil depletion or blockade of IL-1β production increases lipolysis, leading to reduced WAT mass after repeated β3-adrenergic stimulation. Together, these findings reveal a role of neutrophil-derived IL-1β in preserving lipid stores during metabolic stress, highlighting a physiological function of innate immune cells in limiting lipid loss and maintaining energy homeostasis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the Article and its Supplementary Information. The Supplementary Information includes uncropped western blot scans, qPCR primer sequences and flow cytometry gating strategies. RNA-seq data generated in this paper have been deposited at the NCBI SRA (BioProject: PRJNA1345413). Mouse genome reference sequences used for read alignment were based on the mm10 (GRCm38) build. Publicly available human adipose tissue transcriptomic datasets were obtained from the Adipose Tissue Gene Expression Database (https://adiposetissue.org). Source data are provided with this paper.
Code availability
No custom code was used in this study.
References
Sakers, A., De Siqueira, M. K., Seale, P. & Villanueva, C. J. Adipose-tissue plasticity in health and disease. Cell 185, 419–446 (2022).
Reilly, S. M. & Saltiel, A. R. Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol. 13, 633–643 (2017).
Saltiel, A. R. & Olefsky, J. M. Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Invest. 127, 1–4 (2017).
Rohm, T. V., Meier, D. T., Olefsky, J. M. & Donath, M. Y. Inflammation in obesity, diabetes, and related disorders. Immunity 55, 31–55 (2022).
Man, K., Kallies, A. & Vasanthakumar, A. Resident and migratory adipose immune cells control systemic metabolism and thermogenesis. Cell. Mol. Immunol. 19, 421–431 (2022).
Wernstedt Asterholm, I. et al. Adipocyte inflammation is essential for healthy adipose tissue expansion and remodeling. Cell Metab. 20, 103–118 (2014).
Burn, G. L., Foti, A., Marsman, G., Patel, D. F. & Zychlinsky, A. The neutrophil. Immunity 54, 1377–1391 (2021).
Nemeth, T., Sperandio, M. & Mocsai, A. Neutrophils as emerging therapeutic targets. Nat. Rev. Drug Discov. 19, 253–275 (2020).
Herro, R. & Grimes, H. L. The diverse roles of neutrophils from protection to pathogenesis. Nat. Immunol. 25, 2209–2219 (2024).
Kaplan, M. J. Of larks and owls. Nat. Immunol. 21, 104–105 (2020).
Janssen, H. et al. Monocytes re-enter the bone marrow during fasting and alter the host response to infection. Immunity 56, 783–796 (2023).
Brenner, I. K. et al. Immune changes in humans during cold exposure: effects of prior heating and exercise. J. Appl. Physiol. 87, 699–710 (1999).
Cho, J. S. et al. Neutrophil-derived IL-1β is sufficient for abscess formation in immunity against Staphylococcus aureus in mice. PLoS Pathog. 8, e1003047 (2012).
Swanson, K. V., Deng, M. & Ting, J. P. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 19, 477–489 (2019).
Vandanmagsar, B. et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat. Med. 17, 179–188 (2011).
Dror, E. et al. Postprandial macrophage-derived IL-1β stimulates insulin, and both synergistically promote glucose disposal and inflammation. Nat. Immunol. 18, 283–292 (2017).
Zhou, H. et al. IL-1 induces mitochondrial translocation of IRAK2 to suppress oxidative metabolism in adipocytes. Nat. Immunol. 21, 1219–1231 (2020).
Hofwimmer, K. et al. IL-1β promotes adipogenesis by directly targeting adipocyte precursors. Nat. Commun. 15, 7957 (2024).
Roth Flach, R. J. et al. β3-adrenergic receptor stimulation induces E-selectin-mediated adipose tissue inflammation. J. Biol. Chem. 288, 2882–2892 (2013).
Silva, H. M. et al. Vasculature-associated fat macrophages readily adapt to inflammatory and metabolic challenges. J. Exp. Med. 216, 786–806 (2019).
Watanabe, Y. et al. Bidirectional crosstalk between neutrophils and adipocytes promotes adipose tissue inflammation. FASEB J. 33, 11821–11835 (2019).
Young, J. B., Saville, E., Rothwell, N. J., Stock, M. J. & Landsberg, L. Effect of diet and cold exposure on norepinephrine turnover in brown adipose tissue of the rat. J. Clin. Invest. 69, 1061–1071 (1982).
King, V. L., Dwoskin, L. P. & Cassis, L. A. Cold exposure regulates the norepinephrine uptake transporter in rat brown adipose tissue. Am. J. Physiol. 276, R143–R151 (1999).
Liu, X., Perusse, F. & Bukowiecki, L. J. Mechanisms of the antidiabetic effects of the β3-adrenergic agonist CL-316243 in obese Zucker-ZDF rats. Am. J. Physiol. 274, R1212–R1219 (1998).
Weyer, C., Gautier, J. F. & Danforth, E. Jr. Development of β3-adrenoceptor agonists for the treatment of obesity and diabetes-an update. Diabetes Metab. 25, 11–21 (1999).
Valentine, J. M. et al. β3-Adrenergic receptor downregulation leads to adipocyte catecholamine resistance in obesity. J. Clin. Invest. https://doi.org/10.1172/JCI153357 (2022).
Muzzin, P. et al. An adipose tissue-specific beta-adrenergic receptor. Molecular cloning and down-regulation in obesity. J. Biol. Chem. 266, 24053–24058 (1991).
Nahmias, C. et al. Molecular characterization of the mouse beta 3-adrenergic receptor: relationship with the atypical receptor of adipocytes. EMBO J. 10, 3721–3727 (1991).
Carmen, G. Y. & Victor, S. M. Signalling mechanisms regulating lipolysis. Cell Signal. 18, 401–408 (2006).
Mottillo, E. P., Shen, X. J. & Granneman, J. G. β3-adrenergic receptor induction of adipocyte inflammation requires lipolytic activation of stress kinases p38 and JNK. Biochim. Biophys. Acta 1801, 1048–1055 (2010).
Cao, W. et al. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell. Biol. 24, 3057–3067 (2004).
Reilly, S. M. et al. Catecholamines suppress fatty acid re-esterification and increase oxidation in white adipocytes via STAT3. Nat. Metab. 2, 620–634 (2020).
Metzemaekers, M., Gouwy, M. & Proost, P. Neutrophil chemoattractant receptors in health and disease: double-edged swords. Cell. Mol. Immunol. 17, 433–450 (2020).
Reber, L. L. et al. Neutrophil myeloperoxidase diminishes the toxic effects and mortality induced by lipopolysaccharide. J. Exp. Med. 214, 1249–1258 (2017).
Flores, J., Noel, A., Foveau, B., Beauchet, O. & LeBlanc, A. C. Pre-symptomatic caspase-1 inhibitor delays cognitive decline in a mouse model of Alzheimer disease and aging. Nat. Commun. 11, 4571 (2020).
Yang, X. et al. The G0/G1 switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 11, 194–205 (2010).
Mowers, J. et al. Inflammation produces catecholamine resistance in obesity via activation of PDE3B by the protein kinases IKKepsilon and TBK1. eLife 2, e01119 (2013).
Reilly, S. M. et al. An inhibitor of the protein kinases TBK1 and IKK-varepsilon improves obesity-related metabolic dysfunctions in mice. Nat. Med. 19, 313–321 (2013).
Koprulu, M. et al. Identification of rare loss-of-function genetic variation regulating body fat distribution. J. Clin. Endocrinol. Metab. 107, 1065–1077 (2022).
Ortega-Gomez, A., Perretti, M. & Soehnlein, O. Resolution of inflammation: an integrated view. EMBO Mol. Med. 5, 661–674 (2013).
Soehnlein, O., Steffens, S., Hidalgo, A. & Weber, C. Neutrophils as protagonists and targets in chronic inflammation. Nat. Rev. Immunol. 17, 248–261 (2017).
Herrero-Cervera, A., Soehnlein, O. & Kenne, E. Neutrophils in chronic inflammatory diseases. Cell. Mol. Immunol. 19, 177–191 (2022).
Marzano, A. V. et al. Mechanisms of inflammation in neutrophil-mediated skin diseases. Front. Immunol. 10, 1059 (2019).
Tang, H. N. et al. Plasticity of adipose tissue in response to fasting and refeeding in male mice. Nutr. Metab. 14, 3 (2017).
Ding, H. et al. Fasting induces a subcutaneous-to-visceral fat switch mediated by microRNA-149-3p and suppression of PRDM16. Nat. Commun. 7, 11533 (2016).
Xia, W. et al. Obesity causes mitochondrial fragmentation and dysfunction in white adipocytes due to RalA activation. Nat. Metab. 6, 273–289 (2024).
Shin, H. et al. Lipolysis in brown adipocytes is not essential for cold-induced thermogenesis in mice. Cell Metab. 26, 764–777 (2017).
Yang, X. et al. Switching harmful visceral fat to beneficial energy combustion improves metabolic dysfunctions. JCI Insight 2, e89044 (2017).
Griffin, M. J. On the immunometabolic role of NF-κB in adipocytes. Immunometabolism https://doi.org/10.20900/immunometab20220003 (2022).
Gao, D. et al. Interleukin-1β mediates macrophage-induced impairment of insulin signaling in human primary adipocytes. Am. J. Physiol. Endocrinol. Metab. 307, E289–E304 (2014).
Guo, Q. et al. NF-kappaB in biology and targeted therapy: new insights and translational implications. Signal Transduct. Target. Ther. 9, 53 (2024).
Kahnt, A. S., Hafner, A. K. & Steinhilber, D. The role of human 5-Lipoxygenase (5-LO) in carcinogenesis—a question of canonical and non-canonical functions. Oncogene 43, 1319–1327 (2024).
Oyoshi, M. K. et al. Leukotriene B4-driven neutrophil recruitment to the skin is essential for allergic skin inflammation. Immunity 37, 747–758 (2012).
Afonso, P. V. et al. LTB4 is a signal-relay molecule during neutrophil chemotaxis. Dev. Cell 22, 1079–1091 (2012).
Werz, O., Klemm, J., Samuelsson, B. & Radmark, O. 5-Lipoxygenase is phosphorylated by p38 kinase-dependent MAPKAP kinases. Proc. Natl Acad. Sci. USA 97, 5261–5266 (2000).
Lee, Y. S., Wollam, J. & Olefsky, J. M. An integrated view of immunometabolism. Cell 172, 22–40 (2018).
Stienstra, R. et al. Inflammasome is a central player in the induction of obesity and insulin resistance. Proc. Natl Acad. Sci. USA 108, 15324–15329 (2011).
Hildebrandt, X., Ibrahim, M. & Peltzer, N. Cell death and inflammation during obesity: “know my methods, WAT(son)”. Cell Death Differ. 30, 279–292 (2023).
Sakamoto, T. et al. Macrophage infiltration into obese adipose tissues suppresses the induction of UCP1 level in mice. Am. J. Physiol. Endocrinol. Metab. 310, E676–E687 (2016).
Zhao, P. et al. TBK1 at the crossroads of inflammation and energy homeostasis in adipose tissue. Cell 172, 731–743 (2018).
Wisessaowapak, C. et al. Protocol for differentiating murine 3T3-L1 and SVF-derived preadipocytes and isolating crude mitochondrial fractions. STAR Protoc. 6, 104045 (2025).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Zhong, J. et al. adiposetissue.org: a knowledge portal integrating clinical and experimental data from human adipose tissue. Cell Metab. 37, 566–569 (2025).
Acknowledgements
We thank the members of the Saltiel laboratory for discussions. S.S. was supported by American Diabetes Association Postdoc Fellowship 1-25-PDF-15. H.F. was supported by American Diabetes Association Postdoc Fellowship 1-25-PDF-21. C.X. was supported by a Maryam Ahmadian Fellowship. R.M.E. was supported by the NOMIS Foundation and NIH/NIDDK grants R01DK057978 and P30DK063491. This work was supported by a grant from the Larry L. Hillblom Foundation to R.M.E. and A.R.S., and US National Institute of Health (NIH/NIDDK) grants P30DK063491, R01DK122804, R01DK124496, R01DK125820 and R01DK128796 to A.R.S.
Author information
Authors and Affiliations
Contributions
S.S. performed experimental design, experiments, data acquisition and interpretation, and wrote the manuscript. C.X., J.J. and M. Dinj performed the experiments. H.F. and C.W. performed experimental design and experiments. J.M.V. prepared primary adipocyte samples for RNA-seq. G.A. and W.Y. performed and analysed cell sorting of SVF cells from adipose tissue. Y.D., W.F., R.T.Y., M. Downes and R.M.E. performed and analysed RNA-seq experiments. Y.S. performed experimental design and provided critical data interpretation. A.R.S. conceptualized the study, performed data interpretation and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Lawrence Kazak and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 CL-316,243 injection or cold exposure induces neutrophil infiltration and inflammatory cytokine expression in white adipose tissue.
a. Absolute cell numbers of neutrophils, monocytes and macrophages in SVF of eWAT (top) or iWAT (bottom) from WT mice after CL-316,243 injection (0.5 mg kg−1) for the indicated times (0 h: n = 10,5; 1 h: n = 4,3; 12 h: n = 9,6; 24 h: n = 5,4). P values vs 0 h. b. Inflammatory genes in eWAT after CL for the indicated times (0 h: n = 8; 1 h: n = 6; 3 h: n = 7; 6 h: n = 5; 12 h: n = 8; 24 h: n = 5). P values vs 0 h. c. Quantification of cells in SVF of iWAT from WT mice exposed to cold for 18 h. (n = 3 per time point; each replicate pooled from two mice). P values vs 0 h. d-f. (d) Inflammatory genes in iWAT, (e) serum IL-6, (f) serum glycerol and NEFA levels after cold exposure for the indicated times. (n = 3,4 per time point). P values vs 0 h. Data are mean ± s.e.m. Each dot (n) represents one biological replicate (except c). Data are merged from three (a, b) or two (c, d-f) independent experiments. Statistics: one-way ANOVA with Tukey’s post hoc test (d, e, f (glycerol)); Welch’s one-way ANOVA with Dunnett’s T3 (f (NEFA)); Kruskal-Wallis one-way ANOVA with Dunn’s (a, b); unpaired two-sided Student’s t-test (c). Abbreviations. iWAT, inguinal white adipose tissue.
Extended Data Fig. 2 Repeated CL-316,243 injection does not induce sustained neutrophil infiltration.
a-b. (a) Representative flow cytometry plots (top: neutrophils; middle: monocytes; bottom: macrophages. gated on CD45 + CD11b+Thy1.2-CD19-) and (b) quantification of neutrophils, monocytes and macrophages in SVF of eWAT (right) or iWAT (left) from WT mice 12 h after the final CL-316,243 injection (0.5 mg kg−1 day−1) (CLx1: 1 day; CLx2: 2 days; CLx4: 4 days; n = 4 per group). P values vs 0 h. c. Inflammatory genes expression in eWAT (left) or iWAT (right) from WT mice 12 h after the final CL injection (CLx1: 1 day; CLx2: 2 days; CLx4: 4 days). (left Y axis: IL-1β; right Y axis: IL-6, dotted line). (CLx0,x1,x2: n = 3; CLx4: n = 4 per time point). P values vs 0 h. Data are mean ± s.e.m. Each dot (n) represents one biological replicate. Data are merged from two (a-c) independent experiments. Statistics: one-way ANOVA with Tukey’s post hoc test (b, c); Welch’s one-way ANOVA with Dunnett’s T3 (b (neutrophil)).
Extended Data Fig. 3 Lipolysis-dependent signals recruit neutrophils into white adipose tissue upon β3-adrenergic stimulation.
a. Serum glycerol in WT and ad-ATGL KO mice 0.5 h after CL-316,243 injection (0.5 mg kg−1). (n = 3 per group). P values WT vs ad-ATGL KO. b. Representative flow cytometry plots (top: neutrophils; middle: monocytes; bottom: macrophages. gated on CD45 + CD11b+Thy1.2-CD19-) and quantification of each population in iWAT SVF 12 h after CL injection. (CL 0 h: n = 3,2; CL 12 h: n = 3). P values WT vs ad-ATGL KO. c. Representative Ly6G (top) and F4/80 (bottom) staining of eWAT 12 h after CL injection. Scale bar: 100 µm. d. Inflammatory gene expression in iWAT after CL injection for the indicated times. (n = 4 per time point, except IL-1β, IL-6 at 0 h: n = 2). (normalized to WT 0 h). P values WT vs ad-ATGL KO or vs CL 0 h. e-g. (e, f) Inflammatory gene expression and (g) LTB4 levels in the supernatants from WT and ATGL KO PPDIVs treated with CL-316,243 (1 µM, 3 h) or TNFa (5 ng ml−1, 3 h). (n = 3 per group). (normalized to WT veh). P values WT vs ATGL KO. h-i. LTB4 levels in supernatants from (h) WT PPDIVs and bone marrow cells (BM), or (i) mature adipocytes (mAD) and stromal vascular fraction (SVF) isolated from eWAT of WT mice, treated with CL (1 µM, 3 h). (n = 3 per group). P values compare vehicle vs. CL. Data are mean ± s.e.m. Each dot (n) represents one biological replicate. Data are merged from two (b, d) or representative of two (a, c, e-g, h-i) independent experiments. Statistics: two-way ANOVA with Sidak’s post hoc test (a-i).
Extended Data Fig. 4 LTB4 and p38 pathways mediate neutrophil recruitment into white adipose tissue upon β3-adrenergic stimulation.
a. Chemokines expression in WT PPDIVs pretreated for 30 min with SB203580 (10 µM) or SP600125 (10 µM) inhibitor, followed by CL-316,243 (1 µM, 3 h). (n = 4 per group). P values vs CL alone. b-c. Genes expression in WT PPDIVs pretreated for 30 min with SB203580 (10 µM), followed by CL (1 µM, 3 h). d-g. (d) IL-1β and IL-6 levels, (e) chemokines in eWAT, (f) plasma LTB4 (left), serum glycerol (right) and (g) immunoblots of eWAT from WT mice 6 h after Zileuton (5-LOX inhibitor, 50 mg kg−1) with CL injection. (n = 4 per group). P values veh+CL vs Zileuton+CL. h. Representative flow cytometry plots (top: neutrophils; bottom: monocytes. gated on CD45 + CD11b+Thy1.2-CD19-). i. Chemokines and cytokines in eWAT 3 h after MK886 (10 mg kg−1) with CL injection. (n = 4 per group). P values Veh+CL vs MK886 + CL. j-m. Analysis of WT mice 3 h or 6 h after CL injection with SB203580 (25 mg kg−1). (j) Serum LTB4 (3 h) and serum NEFA (6 h). (k) IL-6 levels in eWAT (3 h). (l) IL-1β gene (6 h) and (m) chemokines in eWAT (3 h and 6 h). (m. left Y axis: cxcl1; right Y axis: Cxcl2, Ccl2). (veh: n = 4; CL 3 h groups: n = 4; veh+CL 6 h: n = 7,8; SB + CL 6 h: n = 8). P values veh+CL vs SB + CL. Data are mean ± s.e.m. Each dot (n) represents one biological replicate. Data are merged from two (i, j (right), l, m) or representative of three (a), two (d-g, j (left), k) independent experiments. Statistics: two-way ANOVA with Sidak’s post hoc test (c, i); one-way ANOVA with Tukey’s (a, b, d, e (Ccl2), f, j, k, m (cxcl1)); Welch’s one-way ANOVA with Dunnett’s T3 (a (Ccl2), e, l, m).
Extended Data Fig. 5 Infiltrated neutrophils are the major source of Interleukin-1β and limit excessive energy loss upon β3-adrenergic activation.
a. Representative flow cytometry plots (CD45 + CD11b+Thy1.2-CD19- cells) and quantification of each cells in spleen from diphtheria toxin (DT, 20 µg kg−1) treated Neu-WT and Neu-DTR mice 12 h after CL injection (0.5 mg kg−1). (Neu-WT: n = 8; Neu-DTR: n = 10). Quantification of cells in bone marrow 24 h after DT injection. (n = 3 per group). P values Neu-WT vs Neu-DTR. b. Serum NEFA and IL-6 in DT-treated mice after CL injection. (glycerol, 0 h: n = 3,4; 2 h: n = 6; 12 h: n = 10,7; 24 h: n = 6,10. IL-6, 0 h:n = 3,2; 2 h: n = 4,3; 12 h: n = 9,7; 24 h: n = 6,9). P values Neu-WT vs Neu-DTR. c. Inflammatory genes in eWAT (Adgre1) or iWAT (IL-1β, IL-6) from DT-treated mice 12 h after CL injection. (CL 0 h: n = 3,4; CL 12 h: n = 7,8. normalized to Neu-WT 0 h). P values Neu-WT vs Neu-DTR. d. IL-1β levels in supernatants from WT PPDIVs treated with CL-316,243 (1 µM, 12 h) or LPS (0.5 µg ml−1, 12 h), followed by ATP (5 mM, 1 h). (n = 2 per group). e-f. (e) Body weight and (f) food intake from DT-treated mice at indicated time points or 6 h after final CL injection. (CLx2: 2 days; CLx7: 7 days). Food intake was measured for 24 h before and after CL injection. (f, n = 6 per group). P values indicate comparisons between Neu-WT and Neu-DTR. g. Tissue weight of eWAT 6 h after second CL injection with or without DT. (n = 3,4 per group). P values Neu-WT vs Neu-DTR. h. IL-1β levels in eWAT from WT mice after CL injection for the indicated times (CL x2: 2 days injection). (0 h, 6 h: n = 4; 12 h, 24 h: n = 5; x2: n = 6). P values vs 0 h. i. IL-1β levels in eWAT from DT-treated mice 1 h after second CL injection. (CL 0 h: n = 2; CL x2: n = 4). P values Neu-WT vs Neu-DTR or relative vs 0 h. Data are mean ± s.e.m. Each dot (n) represents one biological replicate. Data are merged from three (a, b, c, h) or representative of two (d, f, g, i) independent experiments. Statistics: two-way ANOVA with Sidak’s post hoc test (a (bone marrow), b, c, e, f, g, i); one-way ANOVA with Tukey’s (h); unpaired two-sided Welch’s t-test (a).
Extended Data Fig. 6 Neutrophil infiltration upon β3-adrenergic activation does not affect thermogenic gene expression in white adipose tissue.
a-b. Thermogenic genes in (a) iWAT and (b) eWAT from DT-treated Neu-WT and Neu-DTR mice 6 h after the final CL-316,243 injection (0.5 mg kg−1 day−1). DT was administered for 7 days (DT x7), CL was injected for 2 or 7 days (CLx2: 2 days; CLx7: 7 days). (DT x7: n = 4; CLx2: n = 4,6; CLx7: n = 6,7. normalized to Neu-WT DT x7). P values Neu-WT vs Neu-DTR. c-d. Thermogenic genes in (c) iWAT and (d) eWAT from WT mice 6 h after the final CL injection (0.5 mg kg−1 day−1, for 4 days), with vehicle or VX765 (50 mg kg−1, twice per day). (Veh: n = 7; Veh and VX765 + CL group: n = 8). P values vehicle vs VX765. e. Representative UCP1 immunohistochemistry images of iWAT from DT-treated mice 7 days after CL injection. Scale bar: 1 mm. Data are mean ± s.e.m. Each dot (n) represents one biological replicate. Data are merged from two (a-b, c-d) independent experiments. Statistics: two-way ANOVA with Sidak’s post hoc test (a, b); one-way ANOVA with Tukey’s (c, d); Welch’s one-way ANOVA with Dunnett’s T3 (c (UCP1), d (Dio2)). Abbreviations. UCP1, uncoupling protein 1.
Extended Data Fig. 7 Neutrophil-derived IL-1β restrains lipolysis through catecholamine resistance in adipocytes.
a. Immunoblots of eWAT from WT mice 12 h after CL-316,243 injection (0.5 mg kg−1) with vehicle or VX765 (50 mg kg−1, twice per day) or IL-1β levels in eWAT from WT mice 3 h after the second CL injection (0.5 mg kg−1 day−1) with vehicle or VX765. (n = 4 per group). P values vehicle vs VX765. b. Chemokines and LTB4 levels from WT PPDIVs pretreated 30 min with VX765 (40 µM), followed by CL-316,243 (1 µM, 3 h). (veh: n = 3, VX765: n = 2, CL: n = 4; VX + CL: n = 3)). P values vehicle vs VX765. c-e. (c) Serum NEFA and glycerol, (d) IL-6 levels and (e) body weight, iWAT tissue weight after CL injection with vehicle or VX765 for the indicated times (CLx2-4: 2-4 days) (NEFA, 0 h/6 h/12 h: n = 4; 24 h: n = 8,7. Glycerol, 0 h: n = 3; 6 h: n = 7). P values vehicle vs VX765. f. Paired serum glycerol levels from the same mice at 30 min and 1 h after the second CL injection with vehicle or VX765 for the indicated times (n = 8 per group). P values 30 min vs 1 h. g. Analysis of DT-treated mice after the second CL injection. Serum glycerol and immunoblots of eWAT from 15 min after second CL injection. (0 min: n = 5,4; 15 min: n = 6,5; 30 min: n = 7,5; 60 min: n = 6,5). P values Neu-WT vs Neu-DTR. h. Glycerol release from ex vivo cultured eWAT of WT mice. eWAT was collected 24 h after CL injection with vehicle or VX765. Then treated in culture with vehicle or CL-316,243 (1 µM, 30 min). (0 h: n = 6,5; 0.5 h: n = 6,5). P values vehicle vs VX765. i. Fatty acid oxidation genes in both eWAT and iWAT 3 h after the second CL-injection with vehicle or VX765. (veh: n = 7,8; VX: n = 7,8). P values vehicle vs VX765. j-k. WT PPDIVs were pretreated 18 h with IL-1β (5 ng ml−1) or vehicle, followed by CL-316,243 (1 µM, 0.5 h). (j) Immunoblots and (k) glycerol release and fold change compared to vehicle or IL-1β. (n = 4 per group). P values vs groups. l-p. Genes expression in (l) 3T3-L1 adipocytes or (m, p) WT PPDIVs treated with IL-1β (5 ng ml−1, 3 h) or CL-316,243 (1 µM, 3 h) or TNFa (5 ng ml−1, 3 h). (veh: n = 2, IL-1β: n = 3, TNFa: n = 2; CL: n = 3). P values vs vehicle. n. Beta 3 adrenergic receptor gene in eWAT from WT mice 3 h after the second CL injection with vehicle or VX765. (n = 4 per group). P values vehicle vs VX765. o. Beta 3 adrenergic receptor gene expression in eWAT from DT-treated Neu-WT and Neu-DTR mice 6 h after the second CL injection. (n = 3,4 per group). P values Neu-WT vs Neu-DTR. Data are mean ± s.e.m. Each dot (n) represents one biological replicate. Data are merged from three (c-d), two (f-i) or representative of two (a, b, g (blot), j-p) independent experiments. Statistics: two-way ANOVA with Sidak’s post hoc test (c, d, g, h); two-way ANOVA with Fisher’s (e); one-way ANOVA with Tukey’s (b, n, o); Welch’s one-way ANOVA with Dunnett’s T3 (b (cxcl1), k); unpaired two-sided Student’s t-test (f); unpaired two-sided Welch’s t-test (a); two-sided Mann Whitney U test (i).
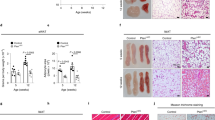
Extended Data Fig. 8 Neutrophil-derived IL-1β limits lipolysis in vivo to prevent excessive energy loss.
a-c. Analysis of IgG or Ly6G-treated WT mice 1 h after the second CL-316,243 injection (0.5 mg kg−1 day−1). (a) eWAT tissue weight. (b) Serum glycerol (left) and NEFA (right) levels from the same mice over time. (c) Immunoblotting of spleen. (IgG: n = 3; Ly6G: n = 4). P values IgG vs Ly6G. d-g. Analysis of WT mice 1 h after the second CL injection with vehicle or MK886 (10 mg kg−1 day−1). (d) eWAT tissue weight. (e) Serum glycerol (left) and NEFA (right) levels from the same mice over time. (f) IL-1β level in eWAT. (g) immunoblotting of eWAT. (n = 4 per group). P values vehicle vs MK886. h-i. Analysis of WT mice under cold exposure (4 °C) for 24 h with vehicle or VX765 (50 mg kg−1, twice per day). (h) Serum glycerol (left) and NEFA (right) levels at indicated time. (4 h: n = 3,4; 24 h: n = 7,8). (i) Core rectal temperature measured at indicated time. (n = 4 per group). P vehicle vs VX765. Data are mean ± s.e.m. Each dot (n) represents one biological replicate. Data are merged from two (h) or representative of two (a-c, d-g, i) independent experiments. Statistics: two-way ANOVA with Sidak’s post hoc test (b, e, h, i); unpaired two-sided Student’s t-test (a, d, g, h (left)). Abbreviations. MK886, 5-Lipoxygenase-activating protein (FLAP) inhibitor.
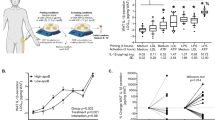
Extended Data Fig. 9 Inflammasome gene expression correlates with obesity-related traits and is increased in obese individuals.
a-d. (top) Spearman correlation of (a) IL1B, (b) IL18, (c) NLRP3 and (d) CASPASE 1 expression with obesity-related parameters (BMI, HOMA-IR, waist, hip, age) and circulating metabolic/inflammatory markers (insulin, glucose, triglycerides, LDL, CRP, HDL, cholesterol, HbA1c). Colours indicate -log10 P values. (n=up to 16 biologically independent cohorts). (bottom) Meta-analysis of genes expression comparing obese and non-obese cohorts. Data are shown as standardized mean difference (SMD) with 95% CI. Diamonds indicate pooled estimates from random-effects models. (n = 9 biologically independent datasets). Data was analysed through AT knowledge portal63. Statistics: two-sided Spearman correlation. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05.
Extended Data Fig. 10 Working model of neutrophil-derived IL-1β regulating lipolysis during β3-adrenergic stimulation.
β3-adrenergic activation transiently recruits neutrophils into white adipose tissue through secreting a lipolysis-p38-dependent Leukotriene B4. Infiltrated neutrophils secrete Interleukin-1β, which suppresses lipolysis and limits excessive energy loss by inducing catecholamine resistance in adipocytes. Created in BioRender. Son, S. (2025) https://BioRender.com/dq8j6v7.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 and Supplementary Table 1, providing uncropped immunoblots, flow cytometry gating and primer sequences.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Son, S., Xu, C., Fu, H. et al. Neutrophils preserve energy storage in sympathetically activated adipocytes. Nature (2025). https://doi.org/10.1038/s41586-025-09839-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41586-025-09839-6