Abstract
Realizing the full potential of organoids and assembloids to model neural development and disease will require improved methods for long-term, minimally invasive recording of electrical activity. Current technologies, such as patch clamp, penetrating microelectrodes, planar electrode arrays and substrate-attached flexible electrodes, do not allow chronic recording of organoids in suspension, which is necessary to preserve architecture. Inspired by kirigami art, we developed flexible electronics that transition from a two-dimensional to a three-dimensional basket-like configuration with either spiral or honeycomb patterns to accommodate the long-term culture of organoids in suspension. Here we show that this platform, named kirigami electronics (KiriE), integrates with and enables chronic recording of cortical organoids for up to 120 days while preserving their morphology, cytoarchitecture and cell composition. We demonstrate integration of KiriE with optogenetic and pharmacological manipulation and modeling phenotypes related to a genetic disease. Moreover, KiriE can capture corticostriatal connectivity in assembloids following optogenetic stimulation. Thus, KiriE will enable investigation of disease and activity patterns underlying nervous system assembly.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Single-cell gene expression data are deposited to the Gene Expression Omnibus under the accession number GSE221032. The data that support the other findings of this study are available from the corresponding authors.
Code availability
The custom code used in this study is available on GitHub at https://github.com/cforro/KiriE_2023_NatureBiotech.
References
Pașca, S. P. The rise of three-dimensional human brain cultures. Nature 553, 437–445 (2018).
Kelley, K. W. & Pașca, S. P. Human brain organogenesis: toward a cellular understanding of development and disease. Cell 185, 42–61 (2022).
Zhang, Y. et al. Fast and sensitive GCaMP calcium indicators for imaging neural populations. Nature 615, 884–891 (2023).
Evans, S. W. et al. A positively tuned voltage indicator for extended electrical recordings in the brain. Nat. Methods 20, 1104–1113 (2023).
Birey, F. et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017).
Samarasinghe, R. A. et al. Identification of neural oscillations and epileptiform changes in human brain organoids. Nat. Neurosci. 24, 1488–1500 (2021).
Quadrato, G. et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53 (2017).
Giandomenico, S. L. et al. Cerebral organoids at the air–liquid interface generate diverse nerve tracts with functional output. Nat. Neurosci. 22, 669–679 (2019).
Trujillo, C. A. et al. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell 25, 558–569 (2019).
Zafeiriou, M.-P. et al. Developmental GABA polarity switch and neuronal plasticity in bioengineered neuronal organoids. Nat. Commun. 11, 3791 (2020).
Sharf, T. et al. Functional neuronal circuitry and oscillatory dynamics in human brain organoids. Nat. Commun. 13, 4403 (2022).
Park, Y. et al. Three-dimensional, multifunctional neural interfaces for cortical spheroids and engineered assembloids. Sci. Adv. 7, eabf9153 (2021).
Huang, Q. et al. Shell microelectrode arrays (MEAs) for brain organoids. Sci. Adv. 8, eabq5031 (2022).
Floch, P. L. et al. Stretchable mesh nanoelectronics for three-dimensional single-cell chronic electrophysiology from developing brain organoids. Adv. Mater. 34, 2106829 (2022).
McDonald, M. et al. A mesh microelectrode array for non-invasive electrophysiology within neural organoids. Biosens. Bioelectron. 228, 115223 (2023).
Xu, T. et al. Characterization of the mechanical behavior of SU-8 at microscale by viscoelastic analysis. J. Micromech. Microeng. 26, 105001 (2016).
Widlund, T., Yang, S., Hsu, Y.-Y. & Lu, N. Stretchability and compliance of freestanding serpentine-shaped ribbons. Int. J. Solids Struct. 51, 4026–4037 (2014).
Yang, X. et al. Bioinspired neuron-like electronics. Nat. Mater. 18, 510–517 (2019).
Paşca, A. M. et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678 (2015).
Yoon, S.-J. et al. Reliability of human cortical organoid generation. Nat. Methods 16, 75–78 (2019).
Lee, J. M. et al. Nanoenabled direct contact interfacing of syringe-injectable mesh electronics. Nano Lett. 19, 5818–5826 (2019).
Sloan, S. A., Andersen, J., Pașca, A. M., Birey, F. & Pașca, S. P. Generation and assembly of human brain region–specific three-dimensional cultures. Nat. Protoc. 13, 2062–2085 (2018).
Klapoetke, N. C. et al. Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014).
Khan, T. A. et al. Neuronal defects in a human cellular model of 22q11.2 deletion syndrome. Nat. Med. 26, 1888–1898 (2020).
Miura, Y. et al. Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat. Biotechnol. 38, 1421–1430 (2020).
Shepherd, G. M. G. Corticostriatal connectivity and its role in disease. Nat. Rev. Neurosci. 14, 278–291 (2013).
Li, T. L. et al. Stretchable mesh microelectronics for the biointegration and stimulation of human neural organoids. Biomaterials 290, 121825 (2022).
Canales, A. et al. Multifunctional fibers for simultaneous optical, electrical and chemical interrogation of neural circuits in vivo. Nat. Biotechnol. 33, 277–284 (2015).
Sahasrabudhe, A. et al. Multifunctional microelectronic fibers enable wireless modulation of gut and brain neural circuits. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01833-5 (2023).
McCaughan, A. N. et al. PHIDL: Python-based layout and geometry creation for nanolithography. J. Vac. Sci. Technol. B 39, 062601 (2021).
Geuzaine, C. & Remacle, J.-F. Gmsh: a 3-D finite element mesh generator with built-in pre- and post-processing facilities. Int. J. Numer. Methods Eng. 79, 1309–1331 (2009).
Hale, J. S., Brunetti, M., Bordas, S. P. A. & Maurini, C. Simple and extensible plate and shell finite element models through automatic code generation tools. Comput. Struct. 209, 163–181 (2018).
Alnæs, M. S. et al. The FEniCS Project Version 1.5. Arch. Numer. Software 3, 9–23 (2015).
Yang, X. et al. Laminin-coated electronic scaffolds with vascular topography for tracking and promoting the migration of brain cells after injury. Nat. Biomed. Eng. 7, 1282–1292 (2023).
Andersen, J. et al. Generation of functional human 3D cortico-motor assembloids. Cell 183, 1913–1929 (2020).
Jahed, Z. et al. Nanocrown electrodes for parallel and robust intracellular recording of cardiomyocytes. Nat. Commun. 13, 2253 (2022).
Santoro, F. et al. Revealing the cell–material interface with nanometer resolution by focused ion beam/scanning electron microscopy. ACS Nano 11, 8320–8328 (2017).
Iandolo, D. et al. Electron microscopy for 3D scaffolds—cell biointerface characterization. Adv. Biosyst. 3, 1800103 (2019).
Birey, F. et al. Dissecting the molecular basis of human interneuron migration in forebrain assembloids from Timothy syndrome. Cell Stem Cell 29, 248–264 (2022).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Kim, J. et al. Human assembloids reveal the consequences of CACNA1G gene variants in the thalamocortical pathway. Preprint at bioRxiv https://doi.org/10.1101/2023.03.15.530726 (2023).
Acknowledgements
We thank members of the Pașca and Cui labs for helpful discussions and technical support. In particular, we thank S. J. Yoon for sharing the hCOs for initial testing, Y. Yang for help with initial testing, and J. I. Kim, M. Y. Li and O. Revah for discussions on electrophysiological recordings. This work was financially supported by the National Institutes of Health (R01NS121934, R01HL165491 and R35GM141598; to B.C.), the Stanford Big Idea Project on Brain Organogenesis (Wu Tsai Neurosciences Institute) (to S.P.P. and B.C.), the David and Lucile Packard Foundation (to B.C.), the Kwan Funds (to S.P.P.), the Senkut Funds (to S.P.P.), the Coates Foundation (to S.P.P.), the Ludwig Family Foundation (to S.P.P.), the Alfred E. Mann Foundation (to S.P.P.), the Marie Skłodowska-Curie Fellowship (846567; to C.F. and F.S.) and an EMBO Postdoctoral Fellowship (ALTF 321-2021; to S.K.). S.P.P. is a New York Stem Cell Foundation (NYSCF) Robertson Stem Cell Investigator, a CZ BioHub Investigator and a CZI Ben Barres Investigator. Part of this work was performed in the nano@stanford, supported by the National Science Foundation as part of the National Nanotechnology Coordinated Infrastructure under the award ECCS-2026822.
Author information
Authors and Affiliations
Contributions
B.C., S.P.P., X.Y. and C.F. conceived and designed the project. C.F. developed the Python code for scripted pattern design, performed the FEM simulations and optimized the KiriE parameters. X.Y. and T.L.L. maintained and differentiated hiPS cells into hCOs. Y.M. maintained and differentiated hiPS cells into hStrOs. C.F., C.-T.T., X.Y. and T.J.Z. fabricated the devices. X.Y., B.C., C.F. and T.L.L. designed the culture chamber and X.Y. characterized the media exchange system. X.Y., C.F. and T.L.L. developed the device assembly protocol and X.Y. developed direct contact interfacing protocol. T.J.Z., X.Y., T.L.L. and C.F. assembled the devices. X.Y. and T.L.L. integrated neural organoids with devices and maintained the cultures. X.Y., S.K. and X.C. performed scRNA-seq experiments. X.Y., S.K. and Y.M. analyzed scRNA-seq data. X.Y. and J.P.M. performed quantitative polymerase chain reaction experiments. J.P.M. characterized the morphology of hCOs. X.C. generated the CAG::eGFP hiPS cell line. X.Y. performed immunocytochemistry, confocal imaging and data analysis. X.Y. and T.L.L. performed electrophysiological recordings. V.M., F.S. and C.-T.T. performed electron microscopy imaging. C.F. wrote the code and analyzed the electrophysiological data. X.Y., C.F., B.C. and S.P.P. wrote the manuscript with input from all authors. B.C. and S.P.P. supervised the work.
Corresponding authors
Ethics declarations
Competing interests
Stanford University holds the patent for the generation of cortical organoids/spheroids (US11279914B2, listing S.P.P. as an inventor). Stanford University has filed a patent application that covers the devices, systems and methods of KiriE (S22-514 63/435,939, listing B.C., S.P.P., X.Y. and C.F. as inventors). The other authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks B. Tian for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Script-assisted design to generate KiriE patterns and automated electrode routing.
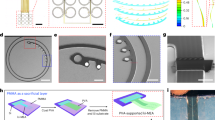
a, Pattern generation for lithography. First, (x,y) coordinates are generated and connected in a clock-wise order. Utilizing a Python geometry library, PHIDL, the coordinates are converted to filled polygons compatible with GDS-II file format. b, Latch design. The starting points (p), the normal vectors at those points (\(\vec{n}\)) and their rotation (θ) are input parameters to build a latch connecting two polygons. The magnitude (m) is adjustable. c, The starting points and the normal vectors (rotated normal in green) define four points p1, p2, c1, c2 which control a NURBS curve. d, The curve is given a width and its ends are smoothly snapped onto the polygons. e, The attachment is adjusted to smoothen sharp joins (see Fig. 1e). f, The resulting latch is integrated into the design. g, Different latches can be produced by changing the script parameters. h, The spiral KiriE is programmatically generated by inputting the number of latches, rings, size, latch parameters, etc. i, The simulation workflow. The design file is triangulated with pygmsh for FEM simulations. j, The spiral KiriE pattern shows the device area and the mesh outside the device area for attaching to the supporting structure. k, To automatically route the electrode lines, nodes and paths were generated for each polygon to provide possible routes for metal lines. l, Electrodes (yellow) are routed through individual non-intersecting metal lines toward the contact pads. m, The nodes (red) define a graph and are connected if they are close enough. n, Inset of the outer wavy ring of the device showing the implicit nodes and paths inside the polygon. o, A node-disjoint flow through the graph was computed using the Networkx package, which corresponds to non-intersecting routes from electrode to pad. The implicit paths connected to red nodes were used to outline the exact shape of the metal line route (yellow paths). The right inset shows metal paths from electrodes to pads as they travel through the outer ring toward the pads without intersecting.
Extended Data Fig. 2 Influence of latch shape, latch length, and organoid size on vertical elongation in the spiral KiriE design.
a, Two latch shapes of equal length connect two rings with identical connecting points. b, The bending strain from unfolding occurs at the outer rim of the design. The outer rim is fixed, while the rest deforms under load. Therefore, the latch closest to the rim supports the most load. c, A side view of the extended KiriE shows that both latches bend downward close to the rim, where the bending strain is important (see b). The red latch extends half as deep as the black latch under equal load. d, Exaggerated representation of a red latch and a black latch for explanatory purposes. The red latch starts with a sharp turn and achieves less extension than the black latch. The initial segment of the black latch bends and points downwards, imposing a downward direction for the rest of the latch. In contrast, after its initial turn, the rest of the red latch does not point downward, preventing vertical extension. e, The extension depends on geometrical arguments, since the strains for black and red latches are nearly identical. f, FEM simulation shows that for fixed attachment points and lengths, the vertical deformation depends on latch shape. The asterisks correspond to the latch shapes. g, The three highlighted latches from f are compared: design 2 vertically extends ∼15% more than the optimized latch, but has a higher curve near the rim, which increases local strain. The optimized shape is designed according to Extended Data Fig. 1j–o (Methods) to minimize total curvature for a given length. This produces a smooth latch that extends enough with low maximal strain. h, The vertical deformation depends on the latch length. i, Schematic of KiriE vertically deformed by a neural organoid with a radius of 0.1, 0.2, 0.3, 0.4, 0.5, and 0.6 mm, respectively. j, The strain along the metal interconnect of a microelectrode. k, Elongation along Z of the metal interconnect of a microelectrode.
Extended Data Fig. 3 Honeycomb KiriE design and nanofabrication of KiriEs.
a, A serpentine line connected to a 3-way connector is the core of honeycomb pattern. The algorithm for automated electrode routing is the same as spiral design in Extended Data Fig. 1j–o. The only differences are locations of implicit nodes and paths for the basic polygons used to make the device. b, The tiling of the serpentine is done via the 3-way connector and tiled until it reaches a preset perimeter. c, The final honeycomb pattern with the motif tiled hexagonally within the confines of the outer ring (1 cm diameter). d, The yellow lines and circles are the automatically routed lines and electrodes, respectively. e, Strain distribution in a simplified honeycomb KiriE under the weight of KiriE itself and a 1.2 mm-diameter organoid in medium. The strain is appreciable at the center because of the way the design gains vertical extension. The serpentine loop stretches to gain vertical elongation, which strains at the central part of the serpentines. f, The plot of the strain along the blue interconnect line in e. The strain increases as the line gets closer to the center. g, Left: the cross-sectional schematic of the Pt/Au microelectrode (150 nm thick) supported on a layer of SU-8 (450 nm thick). Right: the cross-sectional schematic of an interconnecting electrode line that shows Pt/Au is sandwiched between two insulating layers of SU-8 (900 nm thick). h, A photograph showing 8 fabricated spiral KiriEs on a 4 inch silicon wafer before etching the Ni sacrificial layer. i and j, Optical images showing the entire pattern of (i) spiral KiriE design and (j) honeycomb KiriE design. The mesh lines outside the 1-cm wavy ring are designed to adhere the device to a PDMS top support. k,l, Zoomed-in optical images of the 32 electrodes located in the center of spiral KiriE. m, 32 channel-indexing barcodes incorporated into the KiriE design. n, A confocal image of an electrode with its associated channel-indexing barcode, a 3-digit geometrical shape (arrows). Barcodes were imaged across 32 channels.
Extended Data Fig. 4 Generation and characterization of hCOs, and single-cell transcriptomic characterization of KiriE-integrated hCOs.
a, Representative images of hCOs generated in the AggreWell microwell plate at day 0, and hCOs cultured in ultra-low attachment plates at days 2, 6 and 25 of differentiation. Scale bars, 500 µm. Imaging was repeated in hCOs from three differentiation experiments with similar results. b, Gene expression of FOXG1, NKX2-1, EN1, FOXA2, RAX, HOXB4, SOX17, and BRACH relative to housekeeping gene RPL13A in hCOs at day 25 of differentiation (n = 6 samples from three hiPS cell lines). Data are presented as mean ± s.e.m. c, UMAP visualization of expression of selected genes in the hCO scRNA-seq data at day 100 of differentiation (n = 25,546 cells from n = 4 samples of control hCOs and KiriE-integrated hCOs from two hiPS cell lines). d, UMAP visualization of expression of forebrain (FOXG1), dorsal forebrain (EMX1), midbrain (EN1), hypothalamus (RAX), endoderm (SOX17) markers in hCOs. e, UMAP visualization of annotated clusters in scRNA-seq data from control hCOs and KiriE-integrated hCOs. f, Heatmap of the top 10 genes in each cluster. g, Pearson correlations of the normalized average gene expression of 1208-2 hCO/KiriE vs. 1208-2 hCO (r2 = 0.990), 0524-1 hCO/KiriE vs. 0524-1 hCO (r2 = 0.993), and 1208-2 hCO vs. 0524-1 hCO.
Extended Data Fig. 5 Platform for long-term KiriE-organoid integration and demonstration of KiriE reusability.
a, Schematics of the KiriE-organoid culture and media exchange system. Inlet and outlet of the culture chamber are connected to two syringes that are attached to a syringe pump through custom-designed 3D printed adapters. b, An illustration of how the medium exchange efficiency is measured. c, Photograph of fluid collected from the culture chamber after replacing 0.5, 1, 2 and 4 times of the chamber volume of clear water with trypan blue solution, as well as negative (clear water) and positive (trypan blue solution) controls. d, UV-Vis absorption spectra of trypan blue in the fluid normalized against positive control. e, Maximum absorption as a function of media exchange volume. f–j, Illustrations of the KiriE device assembly process. f, Glue an FFC on the surface of the culture chamber. g, Transfer the KiriE device with a PDMS support onto the center of the culture chamber and align the contact pads of KiriE to FFC. Inset shows the top view and side view of the direct contact interface. h, Glue a 3D printed bar to the opening of the chamber with Metabond cement to seal the chamber. i, Aligned contact pads were fully dried by surgical spears (Braintree Scientific) and fixed in place using epoxy adhesive. j, The interface was covered with Metabond cement. k, An image of the contact pads on the KiriE tail aligned with the electrode pads of the FFC through direct contact interfacing. l–p, KiriE is reusable after a two-step cleaning. The transmission images and corresponding red fluorescence images of KiriE (l) before integration with an hCO, (m) after integration with the 1st hCO, (n) after SDS treatment and organoid removal, (o) after Ultrazyme® enzymatic cleaner treatment, and finally (p) after integrating with the 2nd hCO illustrate the complete cycle of KiriE reuse. Scale bars for l–p, 200 µm. KiriE cleaning was repeated in two hCOs with similar results.
Extended Data Fig. 6 Integration of hCOs with KiriE.
a, The top view and the side view of CAG::eGFP hCOs (green) on the honeycomb KiriE (red) reconstructed from live-cell confocal imaging. The level of the medium was lowered to better visualize the deformation of KiriE. Scale bars, 500 µm. b, Normalized fluorescence intensity (mean ± s.d.) of eGFP in the hCOs as a function of distance from spiral KiriEs (n = 3 hCOs) and honeycomb KiriEs (n = 3 hCOs), respectively. c, 3D interface of CAG::eGFP hCOs (green) and spiral KiriE (red) reconstructed from confocal imaging. The hCO–KiriE assembly was fixed and excised from the culture chamber before imaging. Scale bar, 500 µm. d, Normalized number of voxels of KiriE embedded within hCOs as a function of distance from the surface of hCOs after 26 days of integration. The KiriE is embedded as deep as 300–500 µm inside hCOs. e, Images of an hCO integrated with a honeycomb KiriE at different stages of differentiation. Scale bars, 1 mm. f, A FIB-SEM image showing the deformation of a KiriE line as it penetrates into the hCO. Scale bar, 2 µm. g, Immunostaining of SOX2+ neural progenitors in or near neural ventricular-like zones in KiriE-hCO cryosection. Scale bar, 100 µm. h, Immunostaining of deep layer (TBR1) and superficial layer (SATB2) cortical markers in KiriE-hCO cryosection. Scale bar, 100 µm. Experiments were repeated in three (c), three (f), five (g), and four (h) hCOs with similar results.
Extended Data Fig. 7 Morphological and cell survival characterizations of hCOs cultured in suspension (control), on KiriE, and on the adhesive substrate (Matrigel-coated glass-bottom plates).
a, Time-lapse live-cell confocal images showing the flattening of a LMNB1-mEGFP hCO after being plated on an adhesive substrate. Scale bars, 100 µm. Time-lapse confocal imaging was repeated in six hCOs with similar results. b, Live-cell confocal images using SYTOX™ deep red dead cell stain in day 95–109 LMNB1-mEGFP hCOs maintained in suspension (control), on KiriE, or on an adhesive substrate for 40–45 days. Scale bars, 200 µm. c, Quantification of cell viability across these conditions (n = 8 for control, n = 8 for KiriE, and n = 5 for adhesive substrate). One-way ANOVA test, F2,18 = 101.29, P = 1.60 × 10−10. P = 0.881 (control versus KiriE), ****P = 1.95 × 10−10 (control versus adhesive substrate), ****P = 1.65 × 10−10 (KiriE versus adhesive substrate). d, Immunostaining of c-Cas3+ cells in cryosections of day 91–102 hCOs maintained in suspension (control), on KiriE, and on the adhesive substrate for 41–48 days. Scale bars, 50 µm. e, Quantification of the ratio of c-Cas3 / Hoechst cells across these conditions (n = 20 for control, n = 19 for KiriE, and n = 21 for adhesive substrate). One-way ANOVA test, F2,57 = 13.69, P = 1.39 × 10−5. P = 0.829 (control versus KiriE), ****P = 2.10 × 10−5 (control versus adhesive substrate), ****P = 5.56 × 10−5 (KiriE versus adhesive substrate). f,g, Normalized fluorescence intensity (mean ± s.d.) as a function of distance from KiriE for Hoechst and SOX2 in day 40 hCOs (n = 3 for f) and Hoechst, MAP2 and GFAP in day 190 hCOs (n = 4 for g) in KiriE-hCO cryosections shown in Fig. 2i,j. Data are presented as mean ± s.e.m. (c,e).
Extended Data Fig. 8 Electrophysiological recordings of hCOs and data analysis pipeline.
a, Electrophysiological recordings of spontaneous activity of the hCO shown in Fig. 3a on days 137, 164, and 179. Scale bars, 100 ms (lateral) and 200 µV (vertical). b, For each channel, a sliding standard deviation with a 1 s window is computed. Any signal with a magnitude larger than 5 s.d. (dashed blue lines) is extracted (1.5 ms before and 1.5 ms after the peak value; marked in red). c, An overlay of all the detected signals shows a diversity of signal waveforms. d, UMAP dimensionality reduction is performed on the signal into two components. Density-based clustering (DBSCAN) is performed on the component space. e, Signals from the numbered clusters in d are shown. The red line is point-wise average of all signals within the detected clusters, and the blue shading is one point-wise s.d. Clusters 1–3 are artifacts while clusters 4–6 are electrophysiological signals. The signal waveforms are used as templates to find spikes of similar shape. f, Sample traces to illustrate how template matching enables the detection of small signals within the initial 5 s.d. threshold shown in b. The red line indicates the position where a template was detected and the number corresponds to clusters in e. g, Distributions of the spike amplitudes at different days of hCO differentiation. The average spike amplitudes across these days are 52.69, 56.82, 46.54, 45.49, 46.73, and 59.05 µV, respectively. h, Time-dependent electrode impedance at 1 kHz across n = 26 electrodes in a KiriE-hCO. Data are presented as mean ± s.d. i, Quantification of firing rates of day 116–125 hCOs measured by acutely inserted silicon shank electrodes (n = 32 channels from an hCO) and by KiriEs (n = 72 channels from three hCOs). P = 0.99; two-tailed t-test. j, A photograph of the optogenetic stimulation setup. k, The heatmap and quantification of firing rates showing the recovery of neural activity upon TTX washout (n = 25 channels). ****P = 7.52 × 10−7; two-tailed t-test. Data are presented as mean ± s.e.m (i,k).
Extended Data Fig. 9 Electrophysiological recordings of hCOs, hStrOs, and cortico-striatal assembloids with integrated KiriE.
a, Electrophysiological recordings of spontaneous activity of the DGCR8+/− hCO and isogenic control hCO. Scale bars, 100 ms (lateral) and 200 µV (vertical). b, Immunostaining of MAP2+ neurons and GFAP+ glial lineage cells in KiriE-hStrO cryosections at day 271 of differentiation. Blue, green, magenta, and red colors represent Hoechst, MAP2, GFAP, and KiriE, respectively. Scale bar, 20 µm. c, Representative recordings of an hStrO on days 128, 129, and 131. Scale bars, 100 ms (lateral) and 200 µV (vertical). d, Single units from 4 representative channels in the hStrO on days 128, 129, and 131. Scale bars, 1 ms (lateral) and 50 µV (vertical). e, Live-cell confocal fluorescence image of a day 80 + 31 cortico-striatal assembloid on a KiriE. Scale bar, 100 µm. f, Immunostaining of neurofilament (NF) marker in cryosections of hCO–hStrO assembloids. Scale bar, 50 µm. g, Live-cell confocal fluorescence image of the mCherry channel of the cortico-striatal assembloid shown in Fig. 4e, showing that there are minimal projections from hStrO to hCO. Scale bar, 400 µm. h, Electrophysiological recordings and the corresponding raster plot of spiking activity during optogenetic stimulation of the cortico-striatal assembloid shown in Fig. 4f. The orange horizontal lines indicate light stimulation. Scale bars, 1 s (lateral) and 200 µV (vertical). i, Quantification of hStrO firing rates in the hCO–hStrO assembloid during the light-off (baseline) and the light-on (stimulation) phases at days 23, 35, 46, and 55 post-fusion (n = 27 channels over 18–30 light pulses). **P = 1.04 × 10−3; two-tailed t-test. j, Quantification of hStrO firing rates during the light-on phases in a cortico-striatal assembloid before and after the application of NBQX and APV (n = 22 channels analyzed over 18 light pulses). *P = 1.62 × 10−2; two-tailed t-test. Data are presented as mean ± s.e.m. (i,j). Experiments were repeated in two hStrOs (b), two independent hCO–hStrO assembloids (e), three independent hCO–hStrO assembloids (f), and two independent hCO–hStrO assembloids (g) with similar results.
Supplementary information
Supplementary Table 1
List of genes and primer sequences used for RT–qPCR.
Supplementary Video 1
An animation that shows the transformation of spiral KiriE from a flat 2D pattern to a 3D basket-like geometry and the integration with a neural organoid for long-term electrophysiology.
Supplementary Video 2
A Z-stack confocal image video of a 3D KiriE–hCO after 26 days of integration shows that KiriE is embedded within the hCO.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, X., Forró, C., Li, T.L. et al. Kirigami electronics for long-term electrophysiological recording of human neural organoids and assembloids. Nat Biotechnol 42, 1836–1843 (2024). https://doi.org/10.1038/s41587-023-02081-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41587-023-02081-3
This article is cited by
-
Flexible kirigami microelectrode arrays for neuronal activity recordings in non-human primate brains
Nature Electronics (2026)
-
From organoid culture to manufacturing: technologies for reproducible and scalable organoid production
npj Biomedical Innovations (2026)
-
Organoid analytical toolkits
Nature Reviews Bioengineering (2026)
-
Mapping the Evolution of Brain Organoid Research: A Decade of Progress and Future Perspectives (2015–2025)
Molecular Neurobiology (2026)
-
Modelling human brain development and disease with organoids
Nature Reviews Molecular Cell Biology (2025)