Abstract
Understanding the molecular anatomy and neural connectivity of the brain requires imaging technologies that can map the three-dimensional nanoscale distribution of specific proteins in the context of brain ultrastructure. Light and electron microscopy visualize either specific labels or anatomical ultrastructure but combining molecular specificity with anatomical context is challenging. Here we present pan-expansion microscopy of tissue (pan-ExM-t), an all-optical imaging method that combines ~16–24-fold linear expansion with fluorescent pan-stainings of proteins and lipids (providing electron microscopy-like ultrastructural context) and immunolabeling (for molecular imaging). We demonstrate the versatility of this approach by imaging synaptic and cell-specific antibodies in the ultrastructural three-dimensional context of presynaptic and postsynaptic densities, neuropil nanoarchitecture and cellular organelles in dissociated neuron cultures, and mouse brain tissue sections. Furthermore, we demonstrate tracing of neuronal circuitry from pan-ExM-t image volumes, suggesting that any laboratory with access to a confocal microscope can now localize specific molecules within nanoscale cellular and circuit contexts.
Similar content being viewed by others
Main
Three-dimensional (3D) microscopy techniques are instrumental to our understanding of the complex structural and molecular organization of the brain, spanning from the nanoscale to brain-wide neural circuits. Despite substantial advances in imaging techniques, no single microscopy method can reveal the detailed molecular topography of synapses and circuits1. While electron microscopy (EM) is the gold standard for ultrastructural analysis, localizing specific proteins still relies on immunogold labeling or electron-dense peroxidase substrates, neither of which are readily applied for quantitative and routine three-dimensional imaging2.
Fluorescence microscopy, on the other hand, enables highly specific, multicolor labeling of proteins of interest with comparable ease, but is limited in resolution to ~250 nm due to diffraction of light and also does not reveal the underlying ultrastructural context. Super-resolution microscopy, achieving spatial resolutions of ~20 nm or better3,4, overcomes the first limitation. Delineating dense ultrastructural context, however, remains unattainable due to the super-resolution methods’ dependence on fluorescent dyes that are comparatively bulky (~1 nm) and susceptible to quenching when densely packed3,5. To combine specific molecular markers with sample ultrastructure, the community has therefore largely relied on correlative light and EM (CLEM)6. However, the application of CLEM remains limited due to its operational complexity, especially for 3D volumes.
The emergence of expansion microscopy (ExM)7,8, has introduced another path to resolve structures smaller than the diffraction limit of light: ExM physically expands biological samples ~4–20-fold9 in each direction by embedding them in swellable hydrogels. This effectively improves the resolution by the same factor and decrowds the cellular environment. ExM developments and applications initially focused on labeling specific molecules of interest, for example with antibodies, revealing the distribution of these targets at impressive levels of detail10,11,12,13,14. However, like in other fluorescence microscopy techniques, this labeling approach misses the general context of the surrounding subcellular and cellular structures. ExM variants in which proteins are retained through the sample preparation process15,16,17 made possible the introduction of stainings that label the decrowded proteins in bulk, thereby revealing the surrounding tissue context18,19,20,21,22. In particular, when combined with a ~14–20-fold expansion factor achieved by an iterative9 protein-retaining expansion protocol, this bulk-staining approach, which we named pan-ExM, revealed nanoscale ultrastructural details18. The high effective resolution of ~15 nm in pan-ExM makes pan-stainings reminiscent of EM heavy-metal stains and takes light microscopy to the realm of ultrastructural context imaging, as demonstrated by identifying subcellular features such as mitochondria cristae and Golgi cisternae by their anatomical characteristics within adherent monolayer cells18. Moreover, in combination with well-established specific labeling methods in fluorescence microscopy, pan-ExM provides nanoscale context to proteins of interest analogous to CLEM. However, extending this approach to simultaneously investigate the intricate morphologies and complex molecular components of the brain has been hampered by the lack of a suitable protocol to expand not only monolayer cultured mammalian cells but also intact brain tissue. To achieve this, substantial innovations are required to account for differences between tissue and single cells, which profoundly affect critical aspects of pan-ExM protocols, including fixation, denaturation and antibody labeling.
Here we introduce pan-ExM-t, a new protocol adapting the pan-ExM concept to tissue samples with a focus on mouse brain tissue and the ultrastructure of neuronal circuits. Analogous to EM, we discovered that hallmark ultrastructural features such as presynaptic and postsynaptic densities (PSDs) can be identified by their morphological characteristics, and dense neuronal circuits can be traced in 3D, all using light microscopy-based pan-ExM-t without specific labels. Moreover, we show that the addition of specific antibody labels allows localization of specific molecules within the 3D ultrastructural context of the brain. The developments we present in this paper empower neurobiologists to perform routine 3D pan-ExM-t imaging of brain tissue sections using their standard confocal microscope.
Results
Method for ultrastructural imaging of the mouse brain
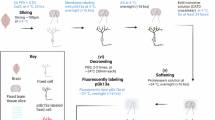
Extended Data Fig. 1 shows an overview of our brain tissue pan-ExM-t protocol. In brief, mice are transcardially perfused with fixative containing both formaldehyde (FA) and acrylamide (AAm) and their brains are extracted surgically and incubated in the same fixative overnight at 4 °C. The brains are then sectioned at 50–100-µm thickness using a vibratome and stored in PBS until future use. Each tissue section to be expanded is embedded in a dense poly(acrylamide/sodium acrylate) copolymer that is crosslinked with N,N′-(1,2-dihydroxyethylene)bis-acrylamide (DHEBA), an AAm crosslinker with a cleavable amidomethylol bond. After polymerization, the now tissue–hydrogel hybrid is denatured with sodium dodecyl sulfate (SDS) in heated buffer (pH 6.8) for 4 hours and expanded roughly fivefold in deionized water. Next, a specific region of interest (~8 × 8 mm2) is cut and re-embedded first in a neutral polyacrylamide hydrogel crosslinked with DHEBA and then in a poly(acrylamide/sodium acrylate) copolymer crosslinked with N,N′-methylenebis(acrylamide) (BIS), a nonhydrolyzable AAm crosslinker. As we previously demonstrated18, no secondary fixation of proteins before the re-embedding is required. The sample is then incubated in 200 mM sodium hydroxide to cleave DHEBA and thereby remove crosslinks of the first and second hydrogel polymer and linearize them. After neutralization with multiple PBS washing steps, the sample is labeled with antibodies, pan-stained with fluorescent dyes to show protein-dense areas, washed with detergents, and expanded to its final size of ~16–24-fold in ultrapure water. The expanded (and, as a side-effect, optically cleared) sample is finally imaged on a standard confocal microscope and can be stored at 4 °C for months.
pan-ExM reveals synapse ultrastructure in dissociated neurons
Before experimenting with expanding mouse brain tissue sections, we tested pan-ExM in dissociated hippocampal rat and mice neurons using our published protocol with modifications in sample fixation (Methods). A visual comparison of N-hydroxysuccinimide (NHS) ester bulk amine pan staining of neurons that are nonexpanded (Fig. 1a) or expanded with pan-ExM (Fig. 1b–k, Supplementary Fig. 1 and Supplementary Videos 1 and 2), illustrates this approach: nonexpanded synapses show essentially uniform staining, revealing little information, whereas ~16-fold expanded synapses can be spatially resolved by their protein density patterns. Analogous to phosphotungstic acid (PTA-)staining of neurons in EM23,24 (Fig. 1l), pan-ExM now resolves nanoscale features such as dense projections (DPs) of the presynaptic bouton (Fig. 1m, lime arrow) and the PSD (Fig. 1m, salmon arrow), allowing for the identification of synapses by their morphological characteristics. We see spines that are stubby (Fig. 1d,f,i), mushroom-shaped (Fig. 1h,j) and thin (Fig. 1g,k). We also observe hexagonal protein-dense patterns formed by presynaptic DPs (Fig. 1e) a discovery made in the 1960s with EM25,26. To determine the achieved linear expansion factors, we imaged SYTOX Green-stained neuron nuclei in nonexpanded samples and samples expanded using our standard protocol and compared the average nuclear cross-sectional area (Supplementary Methods). On average, we obtained an expansion factor of 15.7 ± 0.3 (mean ± s.d.; N = 4 experiments; n = 6–13 nuclei per experiment). Dividing the measured distances between DPs and the PSD (center to center) by the expansion factor determined from nuclei measurements, we obtained a value of 82 ± 26 nm (mean ± s.d.; N = 4 experiments; n = 44 synaptic profiles; Fig. 1n), which is consistent with the range of presynaptic density and PSD distance measurements determined previously by super-resolution microscopy and electron tomography (~70–115 nm)27,28,29,30,31. Similarly, dividing the distance between neighboring DPs by the nuclear expansion factor, we obtained a value of 67 ± 15 nm (mean ± s.d.; N = 6 experiments; n = 78 DP profiles; Fig. 1o), consistent with earlier EM data28,32 (~60–80 nm). In all subsequent experiments, we used the DP–PSD distance as a metric for linear expansion factor calculation (Supplementary Methods).
a, NHS ester pan-stained dendrite in a nonexpanded sample showing dendritic spines. b, Axial view model of a synapse showing DPs and synaptic vesicles (SVs) in the presynaptic bouton, and the PSD, and the active zone (AZ) in the postsynaptic dendritic spine. c, Top view of a synapse showing hexagonal DPs in the presynaptic bouton and SV attachment sites. d,f–k, pan-ExM processed and NHS ester pan-stained spines including mushroom (h,j), stubby (d,f,i) and thin (g,k) shapes. e, NHS ester pan-stained synapse showing hexagonally arranged DPs. l, Transmission EM (TEM) image of a PTA-stained synapse showing prominent DPs (lime arrows) and a PSD (salmon arrow). m, pan-ExM processed and NHS ester pan-stained synapse for comparison, showing similar hallmark ultrastructural features. n, DP–PSD distances (n = 44 measurements from 4 independent experiments). o, DP–DP distances (n = 78 measurements from 6 independent experiments). p, Comparison of Bassoon–PSD and DP–PSD distances (n = 50 measurements from 3 independent samples, Welch’s two-sided t-test, P = 0.035). q, Comparison of DP–Homer1 and DP–PSD distances (n = 85 measurements from 4 independent samples, Welch’s two-sided t-test, P = 0.006). r, Comparison of PSD-95–DP and DP–PSD distances (n = 25 measurements from 2 independent samples, Welch’s two-sided t-test, P = 0.049). *P < 0.05; **P < 0.01. s, Relative spatial distributions of Bassoon, PSD-95 and Homer1 along the trans-synaptic axis. Statistical significance was assessed by Welch’s two-sided t-tests, and medians and interquartile ranges are shown with whiskers drawn down to the minimum and maximum values. t,w,z, Axial (t,w) and top (z) views of synapses pan-stained with NHS ester. u,x,aa, Bassoon immunolabeling of the same areas. v,y,bb, Respective overlays. cc–kk,ll–qq,rr–ww,xx–ccc, Same as t–bb in samples labeled for Homer1, labeled for PSD-95, double-labeled for Homer1 and Bassoon, and labeled for Synaptophysin (SYN), respectively. The inset in ccc shows SYN puncta, representing synaptic vesicles, intercalated between neighboring DPs. ddd, NHS ester image of a mitochondrion in a hippocampal rat neuron. mtDNA, mitochondrial DNA. eee, SYTOX Green (SYX) staining of the same area. fff, Overlay. ggg, NHS ester image of a nuclear pore complex (NPC). hhh, Overlay of ggg with a SYTOX Green image of the same area. iii, NHS ester image of basal bodies in a mouse neuron. The inset shows the familiar centriolar cartwheel structure. jjj, NHS ester image of a cilium in a mouse neuron. Lime and salmon arrows point to the basal body and ciliary tip, respectively. Medians and interquartile ranges are shown with whiskers drawn down to the minimum and maximum values. Panels showing pan staining or TEM (l) are displayed with a white-to-black color table; panels showing immunolabeling or SYTOX Green staining are displayed with a black-to-white color table. Gamma corrections: d,e, γ = 0.8; h,j,k,m,cc,ll,oo,rr,uu,xx,aaa,iii,jjj, γ = 0.7; i, γ = 0.6. jjj is a z-projection (intensity average) of five images. All scale bars are corrected for the expansion factor. Scale bars: 800 nm (a), 200 nm (d–m,t–ccc, jjj), 300 nm (ddd–fff), 50 nm (ggg,hhh), 100 nm (iii). Expansion factor: 16 ± 2.
pan-ExM in dissociated neurons is compatible with immunofluorescence labeling as well as other established chemical stainings, enabling correlative studies that combine specific and contextual pan-staining approaches. Focusing on the synapse, Fig. 1t–ccc and Supplementary Figs. 2–4 show the distributions of synaptic proteins Bassoon, Homer1, PSD-95 and Synaptophysin in the context of synaptic ultrastructure. We observe compartmentalization of active zone protein Bassoon into distinct puncta as DPs (Fig. 1t–bb) with synaptic vesicle protein Synaptophysin intercalating in between neighboring DPs (Fig. 1xx–ccc), supporting the model that DPs represent distinct sites for synaptic vesicle docking and fusion at the active zone. We also observe nanoclustering of PSD proteins Homer1 and PSD-95 along an otherwise macular and dense PSD, with Homer1 slightly offsetting the PSD further into the spine (Fig. 1cc–kk,q) and PSD-95 concentrating directly over the PSD (Fig. 1ll–qq,r), consistent with previous work31,33. The distance between Bassoon and the PSD is 104 ± 24 nm (mean ± s.d.; N = 3 independent samples; n = 50 synaptic profiles; Fig. 1p,s, compared with 90–120 nm reported previously31); the distance between Homer1 and the DP is 83 ± 16 nm (mean ± s.d.; N = 4 independent samples; n = 85 synaptic profiles; Fig. 1q,s) and the distance between PSD-95 and the DP is 68 ± 11 nm (mean ± s.d.; N = 2 independent samples; n = 25 synaptic profiles; Fig. 1r,s). The measured distance between Bassoon and Homer1 of ~90–120 nm is consistent with previous reports using super-resolution imaging (compare with 70–127 nm, ref. 29 or 90–155 nm, ref. 31). Double immunostainings, for example of Bassoon and Homer1 (Fig. 1rr–ww, Supplementary Fig. 5 and Supplementary Video 3), are also compatible with pan-ExM, offering detailed structural information that is inaccessible by conventional confocal microscopy of unexpanded or only roughly fivefold expanded samples (Supplementary Fig. 6).
Similarly, pan-ExM can clearly resolve subcellular structures too small to resolve without expansion, including mitochondrial nucleoids and cristae (Fig. 1ddd–fff), the hollow, circular structure of nuclear pore complexes (Fig. 1ggg–hhh) and the cartwheel structure of basal bodies, their distal appendages and the ciliary tip of cilia (Fig. 1iii–jjj). Furthermore, by combining NHS ester pan staining with metabolic incorporation of palmitic acid azide, it becomes possible to examine the contact sites of membranous organelles, such as between tubules of the endoplasmic reticulum and mitochondria (Supplementary Fig. 8), and other subcellular neuronal features (Supplementary Figs. 9 and 10).
pan-ExM-t reveals tissue ultrastructural features
Having established pan-ExM in dissociated neuron cultures, we adapted our technique to 70-µm-thick mouse brain tissue sections. Because of stark differences in thickness, lipid content and presence of a highly connected extracellular matrix in tissue that is absent in dissociated neurons, brain fixation, sample denaturation and antibody labeling parameters had to be optimized. After comparing many different fixation strategies (Supplementary Methods and Supplementary Figs. 11–14), we found that using 4% formaldehyde + 20% AAm in both the transcardial perfusion and overnight postfixation solutions achieves good neuropil preservation, very little artifactual tissue perforations (Fig. 2a–h), an acceptable extracellular space and lipid membrane (ECS+) fraction of ~38% (Supplementary Methods and Fig. 2j–l) and an expansion factor of 24.1 ± 1.4-fold (mean ± s.d.; N = 10 field of views (FOVs) from 3 independent experiments; n = 254 synaptic profiles, Fig. 2i). We therefore decided to use this fixation strategy for all subsequent experiments.
a, NHS ester pan-stained tissue section of the mouse cortex. b,c, Magnified areas in the black and dotted black boxes in a, respectively. d–h, Magnified areas identified by the correspondingly colored boxes in b,c showing putative excitatory synapses. i, Linear expansion factor (n = 254 measurements from 3 independent experiments, corresponds to data shown in a–h). j, ECS + lipid membrane (ECS+) fraction (n = 14 measurements from 3 independent experiments). k, Image of neuropil in the hippocampus. l, Same area as k where white pixels represent the ECS+ fraction. m–cc, Putative excitatory synapses defined by a prominent PSD. Lime arrows in n and o point to mitochondria in the presynaptic bouton and in the postsynaptic compartment, respectively. The salmon arrow in q points to a spine neck. dd–ii, Putative spine apparatuses (pink arrows) in the postsynaptic compartment defined by a characteristic lamellar arrangement. jj–ll, Classification of synapses based on the patterns and intensity of the PSD. jj, Type I (asymmetric) synapses where the PSD is prominent and macular. kk, Type I (asymmetric) synapses where the PSD is prominent and perforated. ll, Type II (symmetric) synapses, where the PSD, unlike DPs, is thin or barely visible. mm–vv, NHS ester pan-stained multisynaptic boutons and their postsynaptic partners. ww, Lateral view of a centriole showing distal and proximal ends as well as distal appendages (turquoise arrows). xx,yy, Top views of centrioles showing the cartwheel structure (xx) and the ninefold symmetry of microtubule triplets (yy). zz–nnn, Mitochondria with vesicular cristae (zz–ccc,ggg,iii), lamellar cristae (ddd–fff) and teardrop-shaped with tubular extensions (yellow arrows) (jjj–nnn). Salmon arrow in lll points to putative myelinated sheaths. ooo, Brain capillary showing endothelial cells (lavender arrow heads), putative tight junctions (TJs; teal arrows) that link neighboring endothelial cells, putative pericyte branch (salmon arrow) and the basement membrane (yellow arrow). Medians and interquartile ranges are shown with whiskers drawn down to the minimum and maximum values. Panels showing pan staining are displayed with a white-to-black color table. Gamma corrections: a,ww–yy,ooo, γ = 0.7; m–vv,zz–nnn, γ = 0.8. All scale bars are corrected for the expansion factor. Scale bars: 5 µm (a), 1 µm (b,c,ooo), 200 nm (d–h,m–cc,jj–vv,xx,ww,zz–ccc,ggg,jjj,kkk), 3 µm (k,l), 100 nm (dd–ii,xx,yy,ddd–fff,iii,mmm; 400 nm (hhh,lll,nnn). Expansion factor for m–ooo: 21.3 ± 1.5.
Equipped with this pan-ExM-t protocol, we imaged a wide variety of tissue nanostructures across both hippocampal and cortical regions of the mouse brain. Figure 2a shows a tiled ~1 × 1-mm2 image (corresponding to ~42 × 42-µm2 after correction for the expansion factor) of NHS ester pan-stained cortical tissue at synaptic resolution. Hallmark synaptic features such as presynaptic densities and PSDs within a brain tissue section were revealed by standard confocal microscopy (Fig. 2d–h, Supplementary Figs. 15–17 and Supplementary Videos 4 and 5). For example, we could observe that putative excitatory synapses, defined by a prominent PSD (Fig. 2m–cc), often featured mitochondria in the vicinity of axonal boutons (Fig. 2n, arrow) and sometimes in the postsynaptic partner (Fig. 2o, arrow). We can discern densely stained stacked structures in some postsynaptic compartments, suggestive of spine apparatuses34 (Fig. 2dd–ii). Moreover, the obtained resolution allowed us to classify synapses based on variations in PSD patterns and intensities according to the previously established34,35,36,37 Gray classification of synapses: type I (asymmetric, excitatory) synapses with a thick PSD that can be either macular (Fig. 2jj) or perforated (Fig. 2kk); and type II (symmetric, inhibitory) synapses, where the PSD is thin or barely visible, despite prominent DPs in the presynaptic bouton (Fig. 2ll). Furthermore, pan-ExM-t images reveal the spine neck (Fig. 2q, arrow) and multisynaptic boutons (Fig. 2mm–vv).
Centrioles in the perikarya are distinguished by a bright NHS ester pan staining showing clear distal appendages (Fig. 2ww) and, in top views, the cartwheel structure (Fig. 2xx) and ninefold symmetry (Fig. 2zz). The ninefold symmetry is also visible in multiciliated ependymal epithelia lining the lateral ventricles of the mouse brain (Fig. 4l, Supplementary Video 6 and Supplementary Fig. 18). Moreover, we also observe that mitochondria morphologies vary strongly across neuropil (Fig. 2zz–nnn), featuring clearly resolvable lamellar as well as putative tubular cristae, and shapes ranging from vesicular to teardrops with tubular extensions (Fig. 2lll–nnn, arrows). These structures are likely not the result of fixation artifacts, as EM images of tissue preserved with similar protocols resolve mitochondrial ultrastructure well (Extended Data Fig. 4), but reflect native mitochondrial morphology that has been reported to correlate with disease and synaptic performance38,39. A closer look at brain capillaries (Fig. 2ooo and Supplementary Fig. 19) reveals clearly discernible endothelial cells, tight junctions, pericytes and the basement membrane37.
pan-ExM-t is compatible with antibody labeling in thick brain tissue sections
A particular strength of pan-ExM-t is its ability to localize specific proteins to subcompartments revealed in the contextual pan-stained channel. Our protocol must therefore allow for efficient antibody staining, while also maintaining good ultrastructural preservation. Because hydrogels with high monomer concentrations are known to impede antibody diffusion, we investigated whether lowering the monomer concentration in the different interpenetrating hydrogels would result in better signal-to-noise antibody stainings (Supplementary Methods). We found that a combination of 19% (w/v) sodium acrylate (SA) for the first expansion hydrogel followed by 9% (w/v) SA for the second expansion resulted in efficient labeling and low background following synaptic protein immunolabeling (Supplementary Fig. 20).
Using this robust pan-ExM-t protocol, we performed several immunostainings against commonly studied synaptic proteins Bassoon, Homer1, PSD-95 and Synaptophysin (Fig. 3 and Supplementary Video 6). The expansion-corrected distributions of Homer1, PSD-95 and Bassoon within the axial DP–PSD distances are all in agreement with published values29,31,33 as well as with values obtained from cultured neuron measurements in this work. For example, the distance between Bassoon and the PSD is 94 ± 17 nm (mean ± s.d.; N = 3 FOVs; n = 74 synaptic profiles; Fig. 3mmm, compared with 90–120 nm reported previously31); the distance between Homer1 and DP is 93 ± 16 nm (mean ± s.d.; N = 5 fields on views; n = 120 synaptic profiles; Fig. 3nnn) and the distance between PSD-95 and the DP is 76 ± 12 nm (mean ± s.d.; N = 3 FOVs; n = 113 synaptic profiles, Fig. 3ooo). Figure 3ppp shows a plot of the axial positions of Homer1, PSD-95 and Bassoon along the trans-synaptic axis (defined as the axis through the center of the DP pattern and the PSD center). In agreement with previous work, Homer1 is on average localized further inside the spine and away from the cleft than the PSD center, while PSD-95 is concentrated directly onto the PSD33. The measured distance between Bassoon and Homer1 of ~90–120 nm is consistent with previous reports using super-resolution imaging (compare with 70–127 nm, ref. 29 or 90–155 nm, ref. 31). Consistent with previous findings, synapses with thin or absent protein-dense PSDs were colocalized with Bassoon scaffold protein (Fig. 3q–s), but not PSD proteins Homer1 (Figs. 3jj–ll) and PSD-95 (Figs. 3ccc–eee), suggesting that they are inhibitory synapses34.
a, NHS ester (grayscale) pan-stained and Bassoon (red) immunolabeled brain tissue section. b, Magnified area in the red box in a showing a synapse. c, Bassoon immunolabeling of the same area as in b. d, NHS ester pan staining of the same area as in b. e–p, Further examples similar to b–d showing lateral (e–m) and top (n–p) views of Bassoon-immunolabeled synapses. q–s, Lateral view of a type II synapse pan-stained with NHS ester and immunolabeled with Bassoon antibody indicating that type II synapses are positive for Bassoon protein. t, NHS ester (grayscale) pan-stained and Homer1 (magenta) immunolabeled brain tissue section. u, Magnified area in the magenta box in t showing a synapse. v, Homer1 immunolabeling of the same area as in u. w, NHS ester pan staining of the same area as in u. x–ii, Further examples similar to u–w showing lateral (x–ff) and top (gg–ii) views of Homer1-immunolabeled synapses. jj–ll, Lateral view of a type II synapse pan-stained with NHS ester and immunolabeled with Homer1 antibody indicating that type II synapses are negative for Homer1 protein. mm, NHS ester (grayscale) pan-stained and PSD-95 (cyan) immunolabeled brain tissue section. nn, Magnified area in the cyan box in mm showing a synapse. oo, PSD-95 immunolabeling of the same area as in nn. pp, NHS ester pan staining of the same area as in nn. qq–bbb, Further examples similar to nn–pp showing lateral (qq–yy) and top (zz–bbb) views of PSD-95-immunolabeled synapses. ccc–eee, Lateral view of a type II synapse pan-stained with NHS ester and immunolabeled with PSD-95 antibody indicating that type II synapses are negative for PSD-95 protein. fff, NHS ester (grayscale) pan-stained and Synaptophysin (SYN; yellow) immunolabeled brain tissue section. ggg, Magnified area in the yellow box in fff showing a synapse. hhh, Synaptophysin immunolabeling of the same area as in ggg. iii, NHS ester pan staining of the same area as in ggg. jjj–lll, Further example similar to ggg–iii. mmm, Comparison between Bassoon–PSD and DP–PSD distances (n = 74 measurements from 3 FOVs in 1 independent experiment, Welch’s two-sided t-test, P = 1.13 × 10−6). nnn, Comparison between Homer1–PSD and DP–PSD distances (n = 120 measurements from 5 FOVs in 2 independent experiments, Welch’s two-sided t-test, P = 1.64 × 10−14). ooo, Comparison between PSD-95–DP and DP–PSD distances (n = 113 measurements from 3 FOVs in 1 independent experiment, Welch’s two-sided t-test, P = 3.00 × 10−10). ****P < 0.0001. ppp, Relative spatial distributions of Bassoon, PSD-95 and Homer1 along the trans-synaptic axis. Medians and interquartile ranges are shown with whiskers drawn down to the minimum and maximum values. Panels showing pan staining are displayed with a white-to-black color table; panels showing immunolabeling are displayed with a black-to-white color table. All images (with the exception of fff–lll) are z-projections (intensity average) of two images. All NHS ester images were Gamma-corrected with γ = 0.7 with the exception of the Synaptophysin images (γ = 0.6). All scale bars are corrected for the expansion factor. Scale bars: 2 µm (a,t,mm), 200 nm (b–m,q–s,u–ff,jj–ll,nn–yy,ccc–eee,ggg–lll), 100 nm (n–p,gg–ii,zz–bbb), 1 µm (fff). Expansion factor: 21.3 ± 1.5.
pan-ExM-t is not limited to providing context to synaptic proteins. We found that SYTOX Green produces a bright nuclear staining that overlays perfectly with NHS ester pan-stained cell nuclei (Fig. 4a–d). Furthermore, we can resolve individual filaments of glial fibrillary acidic protein (GFAP) in astrocytes (Fig. 4e–k) including near multiciliated ependymal epithelia (Supplementary Fig. 18) and engulfing blood vessels (Fig. 4k and Supplementary Video 7). The distinctive pan-stain pattern of astrocyte nuclei and their thick and fibrous cytoplasmic branches allow for their identification without GFAP, analogous to heavy-metal stains in EM40,41 (Supplementary Fig. 21 and Supplementary Video 8). We also observe that structures enriched in myelin basic protein (MBP) show lighter pan staining, suggesting these represent myelinated axons (Fig. 4m–s). Moreover, when we immunolabeled GFP in Thy1-GFP transgenic mice, we noticed the sparsity of this marker in both neuropil and neuron somas (Fig. 4t–ii, Extended Data Fig. 2 and Supplementary Video 9). Originally designed to resolve individual dendrites and axons in densely packed neuropil42, GFP-Thy1 imaged with pan-ExM-t reveals these transgenic neurons in their ultrastructural context. pan-ExM-t clearly shows the GFP surrounding, but being excluded from, mitochondria (Fig. 4v–x). The ability to distinguish GFP-positive dendritic spine heads (Fig. 4y–aa) and axonal boutons (Fig. 4bb–gg) from their synaptic partners suggests that pan-ExM-t is well-suited for light-based neural tracing and connectomics.
a, Cortical neurons pan-stained with NHS ester (grayscale) and stained with SYTOX Green (teal) showing a neuron nucleus. b–d, Magnified view of the area in the green box in a (b) and the SYTOX Green (c) and NHS ester (d) channels shown separately. e, Astrocyte pan-stained with NHS ester (grayscale) and immunolabeled with GFAP antibody (α-GFAP; magenta). f–h, Magnified view of the area in the magenta box in e (f) showing GFAP filaments surrounding mitochondria, and the α-GFAP (g) and NHS ester (h) channels shown separately. i, 3D rendering of α-GFAP (magenta), SYTOX Green (green) and NHS ester (grayscale). j,k, Two example 3D renderings of α-GFAP (magenta) and blood vessels (BV) pan-stained with NHS ester (grayscale). l, Multiciliated ependymal epithelia pan-stained with NHS ester (grayscale). Inset: ninefold symmetry of cilia. m–r, Lateral (m–o) and top (p–r) views of axons pan-stained with NHS ester (grayscale) and immunolabeled with MBP antibody (α-MBP; red). α-MBP (n,q) and NHS ester (o,r) channels of the areas shown in m and p. s, 3D rendering of anti-MBP showing multiple axons. t,u, Neuropil pan-stained with NHS ester (grayscale) and anti-GFP (green) showing that only a sparse subset of neurites (t) and cell bodies (u) express GFP-Thy1. v–x, Magnified area in the black box in t (v) showing a neurite pan-stained with NHS ester (x) and immunolabeled with anti-GFP (w) where GFP-Thy1 is not expressed inside mitochondria. y–gg, A dendritic spine (y–aa) and axonal boutons (bb–gg) pan-stained with NHS ester and immunolabeled with anti-GFP. z,cc,ff, Anti-GFP channels of the areas shown in y, bb and ee, respectively. aa,dd,gg, NHS ester channels of the areas shown in y, bb and ee, respectively. hh, 3D representation of a surface-rendered dendrite immunolabeled with anti-GFP (green) and NHS ester pan staining (grayscale). ii, 3D rendering of a dendrite immunolabeled with anti-GFP (green) and NHS ester pan staining (grayscale). Panels showing pan staining are displayed with a white-to-black color table; panels showing immunolabeling or SYTOX Green staining are displayed with a black-to-white color table. Gamma corrections: a,e,m,p,t,u,x,aa,dd,gg, γ = 0.7. 3D renderings were processed with Imaris with Gamma corrections applied. 3D image processing details for images i–l, s, hh and ii are found in the Methods section. Scale bars in the 3D renderings i–l, s, hh and ii are not corrected for the expansion factor. All other scale bars are corrected for the expansion factor. Scale bars: 5 µm (a), 2 µm (b–d,e,m–o,u), 500 nm (f–h,v–x), 1 µm (p–r,t), 200 nm (y–gg). Expansion factor: 22 ± 3.
Synaptic circuit tracing with pan-ExM-t
Mapping the fine morphology and connectivity of 3D neuronal networks typically requires specialized EM systems43, but pan-ExM-t makes it possible to achieve this using conventional light microscopy. To assess the feasibility of using pan-ExM-t to trace neuronal wiring diagrams (connectomes), we acquired large-scale datasets of tiled pan-ExM-t image volumes and evaluated circuit tracing based on these data. We found that tissue sections encompassing entire mouse brain sections could be expanded and labeled, and that regions containing circuits of interest could be rapidly imaged using spinning disk confocal microscopy (Fig. 5a–e and Extended Data Fig. 3). For example, we imaged a 2.2 × 2.2 × 0.14-mm (~125 × 128 × 8 µm pre-expansion) large circuit volume at 0.1 × 0.1 × 0.5 µm voxel size (~6 × 6 × 28 nm pre-expansion) in less than 5 hours (total volume size of ~250 gigavoxels, Methods). For a given section, the imageable volume is limited in the Z direction by the working distance of the objective. However, we demonstrate that this limitation can be overcome by sectioning the expanded tissue using a tissue slicer (Methods). Serial sections can be imaged and aligned back together to form a continuous 3D volume, enabling tracing of neuronal wiring across the interfaces (Fig. 5f,g).
a–e, Progressively zoomed-in images of a large pan-ExM-t sample from mouse cerebellum. a, Phase contrast, wide-field image of expanded sample encompassing the entirety of the cerebellum within a brain slice. b, High-resolution (spinning disk confocal) montage of a subvolume (indicated in a, green) spanning granule cell (GC), Purkinje cell (PC) and molecular layers (MLs). Portions of a Purkinje cell visible in this plane are indicated in red outlines. c, Zoomed-in field of view corresponding to subvolume indicated in b (orange). d, Further zoom-in view corresponding to subvolume indicated in c (cyan). Arrow indicates a synapse between a granule cell (parallel fiber) axon and a Purkinje cell spine. e, Zoom-in view of Purkinje cell body also shown in b,h. f,g, Demonstration of serial sectioning approach for large-volume imaging. f, Side view of three serial sections imaged and aligned. Large features (myelinated axons and blood vessels) crossing the cut boundaries are highlighted in color. g, Plan view of aligned surfaces from adjacent sections. Individual neuronal processes can be matched across both images, with several examples highlighted in color. h, Visualization of a Purkinje cell morphology whose processes have been traced throughout the entirety of the pan-ExM-t volume (also shown in b–e in red). i, 3D reconstruction of a section of the Purkinje Cell dendrite, showing dense dendritic spines (red) that terminate in synapses from parallel fiber axons (colors). j, Section of original pan-ExM-t data intersecting the structures visualized in i. k, Segmentation of structures visualized in i superimposed on pan-ExM-t data shown in j. l, Comparison of dendritic spines reconstructed from the pan channel only (magenta, inset top), versus with the aid of a neuron-specific GFP marker (GFP, inset bottom). m, 3D visualization of dense neuronal morphologies from a pan-ExM-t volume in mouse hippocampus. Inset: exploded view of dense cellular fragments shown in m, where space between fragments has been added to aid visualization. n, Virtual section of pan-ExM-t data corresponding to dense segmentations shown in m. o, Dense segmentation of cellular structures visualized in m superimposed on pan-ExM-t data shown in n. Gamma corrections: a, γ = 1.2; b–e, γ = 0.8; all others γ = 1. Expansion factor 17.9 ± 1.0.
By annotating these large 3D image volumes, we show that the large-scale morphology of neurons (Fig. 5h), as well as detailed 3D morphology of neuronal branches (Fig. 5i–k) can be reconstructed based on pan-ExM-t images. The high effective resolution achieved by pan-ExM-t made it possible to distinguish true synaptic contacts from closely passing neurites. We found that pan-ExM-t data was amenable to circuit tracing in a variety of brain areas, including the cortex and hippocampus (Extended Data Fig. 3). Critically, the ~20-fold expansion of pan-ExM-t attains the high resolution needed to trace the smallest neuronal wires, such as spine necks and thin axons, and unambiguously identify individual synaptic connections between neurons. To quantify the resolution, we estimated the point-spread function from small cellular features labeled by GFP (Methods). These measurements placed an upper bound on imaging resolution of 290 ± 60 nm and 840 ± 180 nm in the lateral and axial directions, respectively, corresponding to 14 ± 3 nm lateral and 42 ± 9 nm axial resolution in 20-times expanded samples (Extended Data Fig. 5). To test whether this resolution in combination with the pan-staining contrast is sufficient for connectomic data, we traced spiny dendrites from the pan channel of pan-ExM-t image volumes and compared them to the ‘ground truth’ based on transgenic expression of GFP (Figs. 4ii,hh and 5l). Based on the pan channel only, we were able to successfully trace 77 ± 7% of spines (mean ± s.d. of recall, n = 4 datasets, 199 spines), suggesting that the high-resolution pan channel is sufficient to accurately trace most spine necks (Fig. 5l and Supplementary Fig. 34, Methods). A major advantage of the pan-stain is that it labels all cells, rather than a sparse subset. To illustrate this distinction, we manually reconstructed all cellular structures within a small region of tissue (2 × 1 × 1 μm pre-expansion, Fig. 5m–o and Methods). We found that most (95%) of the reconstructed structures extended beyond the end of the bounding box, and that 40% of the tissue volume was excluded from cellular structures, which is consistent with our previously estimated ECS+ fraction of 38 ± 5% (Fig. 2j and Supplementary Fig. 13). This suggests that cellular morphologies can indeed be densely and accurately reconstructed from pan-ExM-t data.
pacSph enables lipid labeling of tissue with pan-ExM-t
Neural connectomics analysis in EM generally relies on contrast from lipid membranes to delineate cellular boundaries. One way to achieve this in pan-ExM-t would be to preserve the lipid content. We therefore developed a new pan-ExM-t lipid labeling strategy where we use a commercially available photocrosslinkable and clickable sphingosine pacSph44 as a lipid-intercalating reagent (Fig. 6, Supplementary Figs. 30–32 and Methods). This strategy worked in our hands better than alternative approaches (Supplementary Methods). In the perikarya of brain tissue fixed with 4% formaldehyde + 20% AAm and pan-stained with pacSph, we can clearly discern both sides of endoplasmic reticulum tubules (Fig. 6a–f), endoplasmic reticulum and mitochondria contact sites (Fig. 6g–i), and both sides of the nuclear envelope (Fig. 6j–l). In neuropil, lipid membrane boundaries are now discernable (Fig. 6o–oo), revealing the boundaries of neurites and synaptic compartments. In brain capillaries, the different membranes are also differentially highlighted relative to NHS ester pan staining (Fig. 6pp–rr). Moreover, when we experimented with irradiating pacSph pan-stained tissue with ultraviolet (UV) light before or after the first hydrogel embedding, we found that UV irradiation after hydrogel embedding results in significantly higher probe retention, suggesting that diazirine is being photo-crosslinked to the dense hydrogel mesh in addition to surrounding proteins (Fig. 6m and Supplementary Fig. 31). In fact, a recent study showed that diazirine has an affinity for reacting with proteins containing large negative electrostatic surfaces in cells (for example, carboxyl groups)45. We suspect that diazirine in our probe is preferentially photo-crosslinked to carboxyl groups on the poly(acrylamide/acrylate) mesh, offering better retention of the lipid content. While benefiting from future investigation, our data show that photo-crosslinking lipid-intercalating ceramides to the expansion hydrogel represents a promising strategy for labeling lipids in ~20-fold expanded and formaldehyde fixed brain tissue sections.
a–c, Perikaryon pan-stained both with NHS ester (a) and pacSph (b) (overlay shown in c). d–l, Magnified areas of the solid boxes in a–c showing the two sides of endoplasmic reticulum tubules (magenta arrow; d–f), the dashed boxes in a–c showing a mitochondrion (green arrow) in contact with the endoplasmic reticulum (magenta arrow; g–i) and the dotted boxes in a–c showing the nuclear envelope (j–l). m, Mitochondria signal levels in tissue pan-stained with pacSph and not irradiated with UV (white; n = 35 measurements from 5 FOVs in 1 independent experiment); pan-stained with pacSph and irradiated with UV before hydrogel embedding (green; n = 37 measurements from 5 FOVs in 1 independent experiment) and pan-stained with pacSph and irradiated with UV after hydrogel embedding (pink; n = 39 measurements from 5 FOVs in 1 independent experiment). Statistical significance was assessed by Welch’s two-sided t-tests with Bonferroni correction (three comparisons): no UV versus +UV, before, P = 1.37 × 10⁻5; no UV versus +UV, after, P = 5.8 × 10⁻7; +UV, before versus +UV, after, P = 4.5 × 10⁻19). ****P < 0.0001. Medians and interquartile ranges are shown with whiskers drawn down to the minimum and maximum values. n, Chemical structure of pacSph. The probe is photo-crosslinked via its diazirine group (magenta) and labeled via its alkyne group (green). o–t, Synapses pan-stained both with NHS ester (o,r) and pacSph (p,s) (overlays shown in q,t). u–w, Neuropil pan-stained both with NHS ester (u) and pacSph (v) (overlay shown in w). x–cc, Magnified areas of the solid (x,z,bb) and dashed (y,aa,cc) boxes in u–w showing synapses. dd–oo, Neurites and synapses pan-stained both with NHS ester (dd,gg,jj,mm) and pacSph (ee,hh,kk,nn) (overlays shown in ff,ii,ll,oo). pp–rr, Brain capillary pan-stained both with NHS ester (pp) and pacSph (qq) (overlay shown in rr). Green arrows point to mitochondria. Panels showing individual color channels are displayed with a white-to-black color table. Gamma corrections: a, γ = 0.7; p,r,u,dd,gg,jj,mm,pp, γ = 0.5. All scale bars are corrected for the expansion factor. Scale bars:3 µm (a–c), 500 nm (d–i,dd–ii), (2 µm) u–w, 250 nm (j–l,o–t,x–cc,jj–oo),1 µm (pp–rr). Expansion factor 18.1 ± 1.3.
Discussion
The data we present in this paper demonstrate that pan-ExM-t can clearly resolve molecular targets in the context of brain tissue ultrastructure. By combining ~16–24-fold linear sample expansion with pan-stainings, we were able to (1) resolve and identify synapses and their substructures by their morphological characteristics, (2) localize protein markers to their subcellular compartments, (3) delineate cellular membranes and (4) trace neuronal wiring and connectivity within circuits.
Brain tissue structures such as neurons, glia, axons, dendrites, blood capillaries, pericytes, the endothelium and basal bodies, as well as subcellular features such as DPs, PSDs, mitochondria cristae, endoplasmic reticulum tubules, myelin sheaths, multisynaptic boutons, spine necks, spine apparatuses and the cartwheel structure of centrioles can all be identified in the pan-stain channel alone, analogous to classical EM techniques. Chemical synapses can now be distinguished in light microscopic images by their characteristic protein density patterns, analogous to the Gray classification established for PTA and osmium-stained synapses in EM34,35,36. Taking advantage of pan-ExM-t’s strength of directly correlating synapse ultrastructure with immunolabeling against specific proteins, we observed that type I but not type II synapses are positive for Homer1 and PSD-95, supporting the functional Gray synapse concept that synapses with dense PSDs are likely excitatory. This suggests that pan-ExM-t can be used to map the molecular makeup of these synapse types in the context of their distinct ultrastructural properties.
The impact of pan-ExM-t is not limited to addressing questions concerning synapse morphology and composition. For example, cells can be identified by cell-type specific markers such as the demonstrated GFAP labeling (Fig. 4e–k); organelle 3D morphology can be correlated with one or more specific protein distributions, which is of particular interest for poorly understood structures such as the spine apparatus (Fig. 2dd–ii) and pharmacologically important anatomical features such as the blood–brain barrier (Fig. 2ooo) can be investigated in situ both at the intricate structural as well as molecular level46. Moreover, pan-ExM-t works well on other tissues beyond the brain such as liver and kidney (Supplementary Fig. 33) and can offer unprecedented resolution and comprehensive cellular labeling for a wide variety of experimental and clinical applications.
pan-ExM-t achieves the demonstrated level of detail by taking advantage of an opportunity inaccessible to optical super-resolution techniques; the relative size of labels becomes less relevant since labels appear shrunken by the linear expansion factor and the relative radius of a fluorescent dye (~1 nm) approaches ~1 nm/20 = ~50 pm, a size comparable to that of an osmium atom used in heavy-metal EM stainings (~200 pm). The distance between two proteins that originally were in close proximity (~2 nm) grows to ~40 nm, well beyond the Förster radius (~5 nm) of typical fluorescent dyes. Bulk fluorescence staining of decrowded neuropil is therefore no longer limited by size, sampling and quenching restrictions of fluorescent dyes, and ultrastructural details, previously accessible only by EM, can now be examined on standard light microscopes. The localization of labels within protein structures should be interpreted with caution, however, since the denaturation of proteins could alter the location of antigen sites within the volume of the denatured proteins (typically ~10 nm radius).
To achieve the highest ultrastructural preservation of tissue while maximizing sample expansion, we have shown that it is important to optimize fixation and gelation parameters. NHS ester pan staining in this context serves as a readily accessible readout to assess the preservation and retention of proteins. We have demonstrated that reducing interprotein crosslinking enables up to 26-fold expansion without resorting to protease treatments or harsh denaturation conditions. We have also shown that hydrogels of high monomer concentrations (~30% w/v) and sufficient crosslinker density (~0.1% w/v) must be used to ensure that proteins are adequately sampled by the polymer matrix. In this context, it is not surprising that pan-ExM-t gels are stiffer than the gels prepared using the standard ExM protocol7. Moreover, we found that ECS preservation is sensitive to the strength of fixatives used, with higher fixation resulting in lower ECS fractions. In all cases, the ECS is substantially more prominent in pan-ExM-t than typical in EM, and is consistent with a previous live-stimulated emission depletion (STED) microscopy characterization of brain ECS47. While ECS preservation is possible with a variety of expansion approaches48, the ~16–24-times expansion enabled by pan-ExM-t provides critical increases in effective resolution, making it possible to resolve the smallest neuronal processes.
Compared with unexpanded tissues, pan-ExM-t immunostaining tends to appear punctate. This can be largely explained by (1) the improved lateral resolution that can distinguish proteins that were only ~14 nm apart before expansion and (2) optical sections in pan-ExM-t samples being effectively ~20 times thinner, such that each image incorporates signal from only one-twentieth of the volume of a conventional confocal microscope image. To illustrate this point, we computationally blurred the 3D image stack of an expanded Thy1-GFP mouse brain sample as shown in Fig. 4 to simulate how the sample would have looked like before expansion with the same density of fluorescent labels (Extended Data Fig. 2). The resulting images resemble high-quality diffraction-limited images of GFP-labeled dendrites suggesting that the labeling density is not substantially compromised in pan-ExM-t compared to conventional immunolabeling and that the above explanation sufficiently explains the perceived low density of fluorescent labels. However, we point out that not all antibodies work equally well, including some that produce no usable labeling.
We also explored the feasibility of imaging lipid membranes in ~20-times expanded brain tissue sections. We reasoned that attempts to preserve the endogenous lipid content and probe it after expansion are counterproductive. After all, since the free energy of lipid bilayers must be minimized, we hypothesized that even if we were to avoid detergent extraction, lipid membranes would eventually collapse into micelles following sample expansion. Our data show that although lipophilic dyes in protein-denatured samples produce a distinctive pan-stain, these likely also stain the hydrophobic core of proteins. Our strategy was therefore to imprint lipid membranes onto the hydrogel before mechanical homogenization of the sample. We have shown here that pacSph, a photocrosslinkable and clickable sphingosine, enables neuronal membrane imaging in tissue fixed only with formaldehyde. We were able to delineate cellular organelles, synaptic anatomy, as well as membrane boundaries, complementing our established protein pan-stain.
The higher throughput and accessibility of pan-ExM-t compared to 3D CLEM opens the door to the systematic exploration of synapse diversity and a determination of synapse subtypes that are vulnerable to diseases. For example, pan-ExM-t can be used to reveal in the same experiment how presynaptic and postsynaptic ultrastructure degrades during the progression of neurodegenerative diseases while synaptic markers are lost. This is likely to provide insights into how synapses degenerate on structural and molecular levels, step-by-step as the disease proceeds. Moreover, pan-ExM-t can be used to identify which synaptic proteins mark vulnerable and resilient synapses in animal models of neurodegenerative diseases.
We showed that pan-ExM-t allows tracing of the neuronal wires that combine with synapses to form circuits. We demonstrated that rapid, montaged imaging using a spinning disk confocal microscope allows imaging of 3D volumes large enough to incorporate substantial lengths of neuronal axons and dendrites, and accurate reconstruction of their complex branching morphologies. This suggests that pan-ExM-t can be used to map entire neuronal circuits, and even potentially whole brains. While tracing accuracy in pan-ExM-t data is not perfect, it should be noted that EM-based connectomes also contain tracing or segmentation errors, and circuit analyses can be robust to these errors49,50. We also anticipate that the application of deep-learning based segmentation algorithms to pan-ExM-t data will both increase reconstruction accuracy and accelerate circuit analysis. Additional automation in sample preparation and handling will likely be necessary to pursue connectomics at these scales, but our initial results indicate that pan-ExM-t is an extremely promising technology for connectomics, and a potential alternative to volume EM. These conclusions are further supported by related study in ref. 40 that was published during the revision phase of this publication.
We anticipate that many future optimizations will enhance pan-ExM-t. Sample preservation could be further improved, for example by using cryo-fixation51. Maleimide pan-stains18 did not show qualitatively different labeling in tissue from the NHS ester pan staining we generally used in this work, but many alternative pan-staining approaches remain to be explored. In addition, lipid membrane delineation would benefit from systematic optimizations and new dyes, such as pGk13a52 (published during the revision phase of this paper). Future experiments, building on characterizations such as those presented in ref. 22, to characterize the multitude of ultrastructural features revealed by the presented pan-stainings could further maximize the information extracted from pan-ExM-t data. In particular, labeling the plasma membrane, analogous to its successful use in EM-based connectomics41,53, holds great promise to facilitate segmentation and neuronal tracing algorithms for connectomics analysis of pan-ExM-t data. Supported by a theoretical study42, combining this approach with in situ molecular barcoding techniques54,55 could further facilitate the reconstruction of entire connectomes.
New light microscopes that achieve higher volume-imaging speeds without compromising the 3D spatial resolution can substantially increase the throughput and provide access to imaging larger samples. We have already experienced a ~12-fold improvement in imaging speed in this study when switching from a beam-scanning confocal to a spinning disk confocal microscope and anticipate that next-generation light-sheet microscopes in particular have the potential to revolutionize the throughput with which one can image large volumes for connectomics analysis.
Regardless of future optimizations, pan-ExM-t already now enables researchers to investigate proteins, organelles, synapses or cell types of interest highlighted by immunofluorescence in context of dense neuronal processes and neuropil within the brain, providing a molecular dimension to the underlying structures they embody.
Methods
Please see Supplementary Tables 1–6 for an overview of the reagents and materials used in this work.
Neuron culture
Rat and mouse neuron cultures used to generate pan-ExM images shown in Fig. 1 NHS ester, anti-Homer1, anti-Bassoon and anti-PSD-95 images were cultured as described previously56 and expanded and stained using protocols detailed in the Supplementary Methods. Neurons immunolabeled with Homer1, Bassoon or PDS-95 were fixed with 4% formaldehyde in 1× PBS (ThermoFisher; 10010023) for 15 min at room temperature. Neurons immunolabeled with Synaptophysin were fixed with 3% formaldehyde and 0.1% glutaraldehyde in 1× PBS for 15 min at room temperature. Images showing nucleoids in mitochondria were from samples fixed with 3% formaldehyde + 0.1% glutaraldehyde in 1× PBS for 15 min at room temperature. Images showing centrioles and nuclear pore complexes were from samples fixed with 4% formaldehyde in 1× PBS for 15 min at room temperature. After fixation, all samples were rinsed 3 times with 1× PBS and processed according to the pan-ExM protocol immediately after. Formaldehyde (cat. no. 15710) and glutaraldehyde (16019) were purchased from Electron Microscopy Sciences.
Mouse brain tissue
Brain tissue experiments were conducted in two wild-type (C57BL/6) adult male mice purchased from the Charles River Laboratories and two wild-type (C57BL/6) adult male mice and one Thy1-EGFP mouse (strain Tg(Thy1-EGFP)MJrs/J, 4–8 weeks), purchased from The Jackson Laboratory. All experiments were carried out in accordance with National Institutes of Health (NIH) guidelines and approved by the Yale Institutional Animal Care and Use Committee (protocol number 2022-20286) and detailed in the Supplementary Methods. Briefly, mice were anesthetized with a ketamine/xylazine mixture and transcardially perfused first with ice-cold 1× PBS and then with either 4% formaldehyde + 20% AAm (Sigma; A9099) (all in 1× PBS, pH 7.4). Brains were isolated and postfixed overnight in the same perfusion solution at 4 °C, then washed in PBS and sliced using a vibrating microtome. Brain sections were stored in PBS at 4 °C for up to 3 months before further processing.
pan-ExM-t reagents
AAm (A9099) and BIS (146072) were purchased from Sigma-Aldrich. DHEBA was purchased from Sigma-Aldrich (294381) and Santa Cruz Biotechnologies (sc-215503), with the latter being of better purity. SA was purchased from both Sigma-Aldrich (408220) and Santa Cruz Biotechnologies (sc-236893C). To verify that SA was of acceptable purity, 38% (w/v) solutions were made in water and checked for quality as previously reported57. Only solutions that were light yellow were used. Solutions that were yellow and/or had a substantial precipitate were discarded. Solutions with a minimum precipitate were centrifuged at 2,574g for 5 min and the supernatant was transferred to a new bottle and stored at 4 °C until use. Ammonium persulfate (APS) was purchased from both American Bio (AB00112) and Sigma-Aldrich (A3678). N,N,N′,N′-tetramethylethylenediamine (TEMED; AB02020) was purchased from American Bio. Next, 10× phosphate buffered saline (10X PBS; 70011044) was purchased from ThermoFisher. SDS (AB01922) was purchased from American Bio and Guanidine hydrochloride (G-HCl;G3272) was purchased from Sigma-Aldrich.
First round of expansion for brain tissue sections
Here 70-µm-thick brain tissue sections were first incubated in inactivated first expansion gel solution (19% (w/v) SA + 10% AAm (w/v) + 0.1% (w/v) DHEBA in 1× PBS) for 30–45 min on ice and then in activated first expansion gel solution (19% (w/v) SA + 10% AAm (w/v) + 0.1% (w/v) DHEBA + 0.075% (v/v) TEMED + 0.075% (w/v) APS in 1× PBS) for 15–20 min on ice before placing in gelation chamber. The tissue sections were gelled for 15 min at room temperature and 2 h at 37 °C in a humidified chamber. Next, the tissue-gel hybrids were peeled off of the gelation chamber and incubated in ~4 ml of denaturation buffer (200 mM SDS + 200 mM NaCl + 50 mM Tris in MilliQ water, pH 6.8) in 6-well plates for 15 min at 37 °C. Gels were then transferred to denaturation buffer-filled 1.5-ml Eppendorf tubes and incubated at 75 °C for 4 h. The gels were washed twice with 1× PBS for 20 min each and once overnight at room temperature. Gels were optionally stored in 1× PBS at 4 °C. The samples were then placed in six-well plates filled with MilliQ water for the first expansion. Water was exchanged 3 times every 1 h. Gels expanded around five times according to SA purity (‘pan-ExM-t reagents’).
Re-embedding in neutral hydrogel
Expanded hydrogels (of both dissociated neuron and tissue samples) were incubated in fresh re-embedding neutral gel solution (10% (w/v) AAm + 0.05% (w/v) DHEBA + 0.05% (v/v) TEMED + 0.05% (w/v) APS) twice for 20 min each on a rocking platform at room temperature. Immediately after, the residual gel solution was removed by gentle pressing with Kimwipes. Each gel was then sandwiched between 1 no. 1.5 coverslip and 1 glass microscope slide. Gels were incubated for 1.5 h at 37 °C in a nitrogen-filled and humidified chamber. Next, the gels were detached from the coverslips and washed twice with 1× PBS for 20 min each on a rocking platform at room temperature. The samples were stored in 1× PBS at 4 °C. No additional postfixation of samples after the re-embedding step was performed.
Second round of expansion
Re-embedded hydrogels (of both dissociated neuron and tissue samples) were incubated in fresh second hydrogel gel solution (19% (w/v) SA + 10% AAm (w/v) + 0.1% (w/v) BIS + 0.05% (v/v) TEMED + 0.05% (w/v) APS in 1× PBS) twice for 15 min each on a rocking platform in 4 °C. Each gel was then sandwiched between 1 no. 1.5 coverslip and 1 glass microscope slide. Gels were incubated for 1.5 h at 37 °C in a nitrogen-filled and humidified chamber. Next, to dissolve DHEBA, gels were detached from the coverslips and incubated in 200 mM NaOH for 1 h on a rocking platform at room temperature. Gels were washed 3–4 times afterward with 1× PBS for 30 min each on a rocking platform at room temperature or until the solution pH reached 7.4. The gels were optionally stored in 1× PBS at 4 °C. Subsequently, the gels were labeled with antibodies and pan-stained with NHS ester dyes. Finally, the gels were placed in six-well plates filled with MilliQ water for the second expansion. Water was exchanged at least 3 times every 1 h at room temperature. Gels expanded ~4.5× according to SA purity (‘pan-ExM-t reagents’) for a final expansion factor of ~16× (dissociated neurons) and ~24× (brain tissue). Expanded samples show a remarkable stability; when stored in the refrigerator and prevented from drying out, we did not observe any obvious structural degradation or a substantial change in expansion factor over a 7-month time span (CA3 region of hippocampus; expansion factor 20 ± 3 for a freshly prepared sample versus 19 ± 3 after ~7 months).
Antibody labeling of brain tissue samples postexpansion
Brain tissue samples immunolabeled with Homer1, Bassoon, PSD-95 and Synaptophysin were previously fixed with 4% formaldehyde + 20% AAm in 1× PBS overnight at 4 °C and sectioned to 70 µm as described in the ‘Mouse brain tissue’ section. Next, they were washed 3 times with 1× PBS for 30 min each on a rocking platform at room temperature and processed with the pan-ExM-t protocol with this modification: the monomer composition of the second expansion hydrogel was 10% (w/v) AAm + 9% SA (w/v). Samples were subsequently incubated for ~30 h with monoclonal anti-Homer1 antibody (Abcam, ab184955), monoclonal anti-Bassoon antibody (Abcam, ab82958), monoclonal anti-PSD-95 antibody (Antibodies, 75-028) or anti-Synaptophysin antibody (Protein Tech., 17785-1-AP) diluted 1:250 in antibody dilution buffer (0.05% TX-100 + 0.05% NP-40 + 0.2% bovine serum albumin (BSA) in 1× PBS). All primary antibody incubations were performed on a rocking platform at 4 °C. Next, samples were incubated for ~30 h with ATTO594-conjugated anti-mouse antibodies (Sigma-Aldrich, 76085) or ATTO594-conjugated anti-rabbit antibodies (Sigma-Aldrich, 77671) diluted 1:250 in antibody dilution buffer. Gels were then washed in 0.1% (v/v) TX-100 in 1× PBS 4 times for 30 min to 1 h each on a rocking platform at room temperature and once overnight at 4 °C. All secondary antibody incubations were performed on a rocking platform at 4 °C. The gels were subsequently washed in 0.1% (v/v) TX-100 in 1× PBS 4 times for 30 min to 1 h each on a rocking platform at room temperature and once overnight at 4 °C. Gels were stored in PBS at 4 °C until subsequent treatments.
Brain tissue samples immunolabeled with GFAP, MBP and GFP were previously fixed with 4% formaldehyde + 20% AAm in 1× PBS overnight at 4 °C and sectioned to 70 µm as described in the ‘Mouse brain tissue’ section. Next, they were washed 3 times with 1× PBS for 30 min each on a rocking platform at room temperature and processed with the pan-ExM-t protocol. Samples were subsequently incubated for ~30 h with polyclonal anti-GFAP antibody (ThermoFisher, PA1-10019), monoclonal anti-MBP (BioLegend, 808401), or polyclonal anti-GFP (ThermoFisher, A-1122) diluted 1:500 in antibody dilution buffer (0.05% TX-100 + 0.05% NP-40 + 0.2% BSA in 1× PBS). All primary antibody incubations were performed on a rocking platform at 4 °C. Gels were then washed in 0.1% (v/v) TX-100 in 1× PBS 4 times for 30 min to 1 h each on a rocking platform at room temperature and once overnight at 4 °C. Next, samples were incubated for ~30 h with ATTO594-conjugated anti-mouse antibodies (Sigma-Aldrich, 76085) or ATTO594-conjugated anti-rabbit antibodies (Sigma-Aldrich, 77671) diluted 1:500 in antibody dilution buffer. Gels were then washed in 0.1% (v/v) TX-100 in 1× PBS four times for 30 min to 1 h each on a rocking platform at room temperature and once overnight at 4 °C. All secondary antibody incubations were performed on a rocking platform at 4 °C. The gels were subsequently washed in 0.1% (v/v) TX-100 in 1× PBS 4 times for 30 min to 1 h each on a rocking platform at room temperature and once overnight at 4 °C. Gels were stored in PBS at 4 °C until subsequent treatments.
NHS ester pan staining of brain tissue sections
Gels were incubated for 2 h with either 30 µg ml−1 NHS ester-ATTO594 (Sigma-Aldrich, 08741) (ultrastructural samples in Fig. 2) or 30 µg ml−1 NHS ester-ATTO532 (Sigma-Aldrich, cat. no. 88793) (all immunolabeled and lipid-stained samples), dissolved in 100 mM sodium hydrogencarbonate solution on a rocking platform at room temperature. The gels were subsequently washed 4–6 times in either 1× PBS or PBS-T for 20 min each on a rocking platform at room temperature. We also tested NHS ester labeling at neutral pH anticipating that it could stain amino AAm groups on the polymer chains, but did not observe any apparent differences to our standard conditions at pH 8.3.
SYTOX Green staining postexpansion
The pan-ExM and pan-ExM-t processed gels were incubated with SYTOX Green (Invitrogen, S7020) diluted 1:5,000 (dissociated neuron samples) and 1:3,000 (brain tissue samples) in 1× PBS for 30 min on a rocking platform at room temperature. The gels were then washed 3 times with PBS-T for 20 min each on a rocking platform at room temperature.
pan-ExM and pan-ExM-t sample mounting
After expansion, the gels were mounted on glass-bottom dishes (35 mm; no. 1.5; MatTek). A clean 18-mm-diameter coverslip (Marienfeld, 0117580) was put on top of the gels after draining excess water using Kimwipes. The samples were then sealed with two-component silicone glue (Picodent Twinsil, Picodent). After the silicone mold hardened (typically 15–20 min), the samples were stored in the dark at 4 °C until they were imaged.
Image acquisition
Confocal images were acquired using a Leica SP8 STED ×3 confocal microscope or an Andor Dragonfly 600 High Speed Confocal Microscope. The Leica SP8 instrument was equipped with a SuperK Extreme EXW-12 (NKT Photonics) pulsed white light laser as an excitation source. The STED imaging mode of the instrument was not used in this work. Images were acquired using either an HC FLUOTAR L ×25/0.95 numerical aperture (NA) water objective, APO ×63/1.2 NA water objective, or an HC PL APO ×86/1.2 NA water CS2 objective. Application Suite X software (LAS X; Leica Microsystems) was used to control imaging parameters. ATTO532 was imaged with 532-nm excitation. ATTO594 was imaged with 585-nm excitation. CF568 was imaged with 568-nm excitation. ATTO647N was imaged with 647-nm excitation. ATTO488 and SYTOX Green were imaged with 488-nm excitation. The Andor Dragonfly 600 microscope used an inverted Nikon Eclipse Ti2 microscope (Nikon Instruments) as the microscope stand; an integrated laser engine equipped with 488-nm, 561-nm and 637-nm laser lines; and Sona-6 sCMOS cameras. Images were acquired in spinning disk confocal mode using a CFI Plan Apo VC ×60/1.2 NA water-immersion objective or an Apo LWD Lambda S ×40/1.15 NA water-immersion objective. The instrument was controlled by Andor’s Fusion Control Software.
Image processing
Images were visualized, smoothed, gamma-adjusted and contrast-adjusted using FIJI/ImageJ software. STED and confocal images were smoothed for display with a 0.4-pixel to 1.5-pixel sigma Gaussian blur. Minimum and maximum brightness were adjusted linearly for optimal contrast.
All line profiles were extracted from the images using the Plot Profile tool in FIJI/ImageJ. Stitching of tiled images was performed using the Pairwise Stitching tool in FIJI/ImageJ.
3D data visualization
The 3D data stacks were processed with the microscopy image analysis software Imaris (Oxford Instruments; Imaris x64 versions 9.6 or 9.8). Datasets with low signal-to-noise ratios were smoothed with Gaussian filters. Gamma correction and displayed color range were adjusted for contrast to highlight features of interest. Generated videos were further processed with Adobe Premiere Pro (version 22.2.0) to add headers and link videos.
Lipid staining with pacSph
Detailed methods for optimizations of pacSph staining are available in the Supplementary Methods. Briefly, brain sections were incubated in 50 µM pacSph (PhotoClick Sphingosine; Avanti Polar Lipids, 900600) in 1× PBS overnight at 4 °C, washed three times in PBS for 1 h each at 4 °C and either stored at 4 °C (no UV) or photo-crosslinked. Samples that were photo-crosslinked were kept on ice and irradiated with an 8-W, 365-nm UV light source (Analytik Jena, UVL-18) 1 cm away from a lamp surface for 30 min on both section surface sides, either before hydrogel embedding (UV before) or immediately after the first round of expansion for brain tissue while the tissue–hydrogel hybrid was still sandwiched between a glass microscope slide and glass coverslip (UV after). Samples were processed with the remaining steps of the pan-ExM-t protocol with this modification: before NHS ester pan staining, copper-catalyzed azide–alkyne cycloaddition was performed using the Click-iT Protein Reaction Buffer Kit (ThermoFisher, C10276) according to manufacturer instructions. Azide-functionalized ATTO590 dye (ATTO-TEC, cat. no. AD 590-101) was used at a concentration of 5 µM. After copper-catalyzed azide–alkyne cycloaddition, the gels were washed once with 2% (w/v) delipidated BSA (Sigma-Aldrich, A4612) in 1× PBS for 20 min each and three times with 1× PBS for 20 min each on a rocking platform at room temperature.
Synaptic circuit analysis
Large-volume imaging
pan-ExM-t samples were imaged by using a spinning disk confocal microscope (Andor Dragonfly) to montage images across large areas. The individual image tiles were stitched together using Imaris Stitcher. Stitched datasets were converted to a chunked format (Neuroglancer precomputed) using the CloudVolume Python library (https://pypi.org/project/cloud-volume/0.10.4/) to enable viewing and annotation with browser-based software (Neuroglancer and WebKnossos).
Serial sectioning
Fully expanded samples were embedded in Agarose and sectioned into serial 200-μm-thick sections using a Compresstome (Precisionary) tissue slicer. Each section was then imaged serially, ensuring that the axial field of view (FOV) included both top and bottom surfaces of the sections. Serial sections were aligned to one another semi-manually by identifying correspondence points on matching faces of adjacent sections (based on cellular structures that could be unambiguously identified). These correspondence points were then used to calculate affine transformation matrices (MATLAB) that were applied using Neuroglancer.
Neuron tracing
Neuron morphologies were traced using the web-based annotation software WebKnossos (webknossos.org). pan-ExM-t image volumes were converted to the Neuroglancer precomputed format (a chunked 3D image format) using the CloudVolume Python library (https://github.com/seung-lab/cloud-volume) and the image data were made accessible to WebKnossos via https on a server hosted at Yale. Annotators performed two different types of tracing: skeleton-based tracing, in which neurons are abstracted as trees of connected points (Fig. 5h and Supplementary Fig. 34), and volumetric tracing, in which all voxels within a given neuron are painted with the same label (Fig. 5i,k–m,o). To quantify the accuracy of spine tracing (Fig. 5l and Supplementary Fig. 34), two expert annotators traced dendrites from the pan channel, blinded to the GFP channel. Separately, ground truth skeletons were traced from the GFP channel. The pan channel was also used to aid in the tracing of ground truth skeletons to distinguish spines actually connected to the dendrite from stray fluorescence from other cells. The pan-channel skeletons were compared to the ground truth to quantify the number of correctly traced spines (true positive, TP), incorrectly traced spines (false positive, FP) and missed spines (false negative, FN). For this, spines with multiple spine heads were counted as separate spines. Since the rate of false negatives was very low (3 out of 199 spines), we quantified recall (TP/(TP + FN)) as a relevant measure of tracing accuracy.
Statistics and reproducibility
For all quantitative experiments, the number of samples and independent reproductions are listed in the figure legends. GraphPad Prism 9 was used to plot statistical data, and statistical tests (Welch’s two-sided t-test) were performed using Microsoft Excel’s t-test function. Measurements were taken from distinct samples as indicated in figure captions.
The following images are representatives of at least three independent experiments: Figs. 1d–k,m,xx–jjj, 2a–h,k–ooo and 4a–d,l, Supplementary Figs. 1, 9, 10, 11f, 12m–o, 13oo–xx, 14a–c, 15–17, 18a–c, 19, 20a and 21 and Extended Data Fig. 3. The following images are representatives of at least two independent experiments: Figs. 3t–ll and 4e–k and Supplementary Figs. 2, 3, 6, 18d, 20b–e, 22 and 32. The experiments represented in Figs. 1l,p–ww, 3a–s,mm–lll, 4i,m–ii, 5 and 6a–l,o–rr, Supplementary Figs. 4, 5, 7, 8, 11a–e, 12a–l, 13a–nn, 14d–i, 23, 24–29, 31, 33 and 34, Extended Data Figs. 2, 4 and 5 have been performed once.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request. Public links to view these data are also available via GitHub at https://github.com/kuan-lab/pan-exm-t_paper/.
Code availability
The code created during the current study is available via GitHub at https://github.com/kuan-lab/pan-exm-t_paper.
References
Kavalali, E. T. & Jorgensen, E. M. Visualizing presynaptic function. Nat. Neurosci. 17, 10–16 (2014).
Gonda, M. A. in Encyclopedia of Immunology (eds, Delves, P. J. & Roitt, I. M.) 790–795 (Elsevier, 1998).
Baddeley, D. & Bewersdorf, J. Biological insight from super-resolution microscopy: what we can learn from localization-based images. Annu. Rev. Biochem. 87, 965–989 (2018).
Liu, S., Hoess, P. & Ries, J. Super-resolution microscopy for structural cell biology. Annu. Rev. Biophys. 51, 301–326 (2022).
Conroy, E. M. et al. Self-quenching, dimerization, and homo-FRET in hetero-FRET assemblies with quantum dot donors and multiple dye acceptors. J. Phys. Chem. C 120, 17817–17828 (2016).
Hoffman, D. P. et al. Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science 367, eaaz5357 (2020).
Chen, F. et al. Optical imaging. Expansion microscopy. Science 347, 543–548 (2015).
Gallagher, B. R. & Zhao, Y. Expansion microscopy: a powerful nanoscale imaging tool for neuroscientists. Neurobiol. Dis. 154, 105362 (2021).
Chang, J.-B. et al. Iterative expansion microscopy. Nat. Methods 14, 593–599 (2017).
Chozinski, T. J. et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods 13, 485–488 (2016).
Truckenbrodt, S. et al. X10 expansion microscopy enables 25-nm resolution on conventional microscopes. EMBO Rep. 19, e45836 (2018).
Zhao, Y. et al. Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nat. Biotechnol. 35, 757–764 (2017).
Jiang, N. et al. Superresolution imaging of Drosophila tissues using expansion microscopy. Mol. Biol. Cell 29, 1413–1421 (2018).
Freifeld, L. et al. Expansion microscopy of zebrafish for neuroscience and developmental biology studies. Proc. Natl Acad. Sci. USA 114, E10799–E10808 (2017).
Tillberg, P. W. et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 34, 987–992 (2016).
Ku, T. et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol. 34, 973–981 (2016).
Gambarotto, D. et al. Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat. Methods 16, 71–74 (2018).
M'Saad, O. & Bewersdorf, J. Light microscopy of proteins in their ultrastructural context. Nat. Commun. 11, 3850 (2020).
Yu, C.-C. J. et al. Expansion microscopy of C. elegans. eLife 9, e46249 (2020).
Mao, C. et al. Feature-rich covalent stains for super-resolution and cleared tissue fluorescence microscopy. Sci. Adv. 6, eaba4542 (2020).
Nijenhuis, W. Optical nanoscopy reveals SARS-CoV-2-induced remodeling of human airway cells. Preprint at bioRxiv https://doi.org/10.1101/2021.08.05.455126 (2021).
Sim, J. et al. Nanoscale resolution imaging of whole mouse embryos using expansion microscopy. ACS Nano 19, 7910–7927 (2025).
Bloom, F. E. & Aghajanian, G. K. Cytochemistry of synapses: selective staining for electron microscopy. Science 154, 1575–1577 (1966).
Aghajanian, G. K. & Bloom, F. E. The formation of synaptic junctions in developing rat brain: a quantitative electron microscopic study. Brain Res. 6, 716–727 (1967).
Gray, E. G. Electron microscopy of presynaptic organelles of the spinal cord. J. Anat. 97, 101–106 (1963).
Pfenninger, K., Akert, K., Moor, H. & Sandri, C. The fine structure of freeze-fractured presynaptic membranes. J. Neurocytol. 1, 129–149 (1972).
Valtschanoff, J. G. & Weinberg, R. J. Laminar organization of the NMDA receptor complex within the postsynaptic density. J. Neurosci. 21, 1211–1217 (2001).
Claudio, A. et al. How to make an active zone: unexpected universal functional redundancy between RIMs and RIM-BPs. Neuron 91, 792–807 (2016).
Hao, X. et al. Three-dimensional adaptive optical nanoscopy for thick specimen imaging at sub-50-nm resolution. Nat. Methods 18, 688–693 (2021).
Yang, X. & Annaert, W. The nanoscopic organization of synapse structures: a common basis for cell communication. Membranes 11, 248 (2021).
Dani, A., Huang, B., Bergan, J., Dulac, C. & Zhuang, X. Superresolution imaging of chemical synapses in the brain. Neuron 68, 843–856 (2010).
Harlow, M. L., Ress, D., Stoschek, A., Marshall, R. M. & McMahan, U. J. The architecture of active zone material at the frog’s neuromuscular junction. Nature 409, 479–484 (2001).
Xiao, B. et al. Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of homer-related, synaptic proteins. Neuron 21, 707–716 (1998).
Harris, K. M. & Weinberg, R. J. Ultrastructure of synapses in the mammalian brain. Cold Spring Harb. Perspect. Biol. 4, a005587 (2012).
Gray, E. G. Axo-somatic and axo-dendritic synapses of the cerebral cortex: an electron microscope study. J. Anat. 93, 420–433 (1959).
Klemann, C. J. H. M. & Roubos, E. W. The gray area between synapse structure and function-Gray’s synapse types I and II revisited. Synapse 65, 1222–1230 (2011).
Nahirney, P. C. & Tremblay, M.-È Brain ultrastructure: putting the pieces together. Front. Cell Dev. Biol. 9, 629503 (2021).
Zhang, L. et al. Altered brain energetics induces mitochondrial fission arrest in Alzheimer’s Disease. Sci. Rep. 6, 18725 (2016).
Cserép, C. et al. Mitochondrial ultrastructure is coupled to synaptic performance at axonal release sites. eNeuro 5, ENEURO.0390-17.2018 (2018).
Tavakoli, M. R. et al. Light-microscopy-based connectomic reconstruction of mammalian brain tissue. Nature 642, 398–410 (2025).
MICrONS Consortium. Functional connectomics spanning multiple areas of mouse visual cortex. Nature 640, 435–447 (2025).
Yoon, Y.-G. et al. Feasibility of 3D reconstruction of neural morphology using expansion microscopy and barcode-guided agglomeration. Front. Comput. Neurosci. 11, 97 (2017).
Collinson, L. M. et al. Volume EM: a quiet revolution takes shape. Nat. Methods 20, 777–782 (2023).
Haberkant, P. et al. Bifunctional sphingosine for cell-based analysis of protein-sphingolipid interactions. ACS Chem. Biol. 11, 222–230 (2016).
West, A. V. et al. Labeling preferences of diazirines with protein biomolecules. J. Am. Chem. Soc. 143, 19039–19044 (2021).
Sasson, E. et al. Nano-scale architecture of blood-brain barrier tight-junctions. eLife 10, e63253 (2021).
Tønnesen, J. et al. Super-resolution imaging of the extracellular space in living brain tissue. Cell 172, 1108–1121 (2018).
Damstra, H. G. J. et al. Correction: Visualizing cellular and tissue ultrastructure using ten-fold robust expansion microscopy (TREx). eLife 11, e85169 (2022).
Schneider-Mizell, C. M. et al. Quantitative neuroanatomy for connectomics in Drosophila. eLife 5, 1133–1145 (2016).
Kuan, A. T. et al. Synaptic wiring motifs in posterior parietal cortex support decision-making. Nature 627, 367–373 (2024).
Laporte, M. H., Klena, N., Hamel, V. & Guichard, P. Visualizing the native cellular organization by coupling cryofixation with expansion microscopy (Cryo-ExM). Nat. Methods 19, 216–222 (2022).
Shin, T. W. et al. Dense, continuous membrane labeling and expansion microscopy visualization of ultrastructure in tissues. Nat. Commun. 16, 1579 (2025).
Dorkenwald, S. et al. Neuronal wiring diagram of an adult brain. Nature 634, 124–138 (2024).
Askary, A. et al. In situ readout of DNA barcodes and single base edits facilitated by in vitro transcription. Nat. Biotechnol. 38, 66–75 (2020).
Wroblewska, A. et al. Protein barcodes enable high-dimensional single-cell CRISPR screens. Cell 175, 1141–1155 (2018).
Biederer, T. & Scheiffele, P. Mixed-culture assays for analyzing neuronal synapse formation. Nat. Protoc. 2, 670–676 (2007).
Asano, S. et al. Expansion microscopy: protocols for imaging proteins and RNA in cells and tissues. Curr. Protoc. Pharmacol. 80, e56 (2018).
Acknowledgements
We thank M. Lessard for assisting with vibratome sectioning, S. Kayani for assisting with EM sectioning and imaging and M. Nandy for performing neuron tracing. This work was supported by grants from the Wellcome Trust (grant no. 203285/B/16/Z), NIH (grant nos. P30 DK045735; S10 OD020142; R56MH122449 and MH115939 to A.J.K.; NS036251 to P.D.C; R01 DA01892 to T.B.; R43MH128999 and R44MH128999 to O.M.), the Yale Graduate School of Arts and Sciences (A.C.), the Yale Program in Physics, Engineering and Biology (A.C.), the Brain Research Fund (A.T.K.), and the Yale Kavli Institute for Neuroscience (A.J.K., A.T.K., J.B.). H.F. was supported by a HHMI/LSRF postdoctoral fellowship. R.C.S. was supported by an NIH Individual Predoctoral MD/PhD Fellowship (grant no. MH124284).
Author information
Authors and Affiliations
Contributions
O.M. developed and optimized the sample preparation protocols. I.K., H.F., J.G., R.F.N., R.C.S. and S.L. cultured and fixed dissociated neurons. I.K., H.F., R.F.N. and K.W. provided fixed mouse brain samples. X.G. provided fixed mouse kidney and liver samples. O.M., R.K.K. and I.K. expanded, labeled and imaged neuron samples. O.M., A.C., J.E.G. and E.H.B. expanded and imaged brain tissue samples. O.M., J.E.G., E.H.B. and P.K. labeled brain tissue samples. X.L. and A.T.K. provided EM images. O.M., A.C., J.L., A.T.K. and J.B. visualized the data. O.M., A.C., J.L. and A.T.K. analyzed and quantified the data. G.D., P.D.C., J.E.R., A.J.K., T.B., A.T.K. and J.B. supervised the study. All authors interpreted the data. O.M., A.C., A.T.K. and J.B. wrote the paper with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
J.B. has financial interests in Bruker Corp. and Hamamatsu Photonics. O.M., J.E.R. and J.B. filed patent applications with the US patent office covering the presented method. O.M. and J.B. are cofounders and J.E.G. and E.H.B. are employees of panluminate Inc., which is developing related products. The other authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 pan-ExM-t workflow for mouse brain tissue sections.
(a-f) Experimental workflow. (g): Timeline summarizing the protocol. Abbreviations: FA: formaldehyde; AAm: acrylamide; NaOH: sodium hydroxide; DHEBA: N,N′-(1,2-dihydroxyethylene)bis-acrylamide; SDS: sodium dodecyl sulfate; PBS-T: 0.1% (v/v) TX-100 in PBS; ROI: region of interest. Figure created using BioRender.com.
Extended Data Fig. 2 Comparison of pan-ExM-t immunolabeling with simulated conventional (non-expanded) microscopy.
pan-ExM-t images were computationally blurred to simulate imaging of equivalent non-expanded samples as described in the Methods Section under the heading Simulations of non-expanded sample images. Contrast was enhanced using ImageJ’s “Enhance Contrast” function (0.35% saturation), and color channels were rendered inverted (pan) and green (Thy1-eGFP) for visualization. Due to the high effective resolution and optical sectioning resulting from 20x expansion, antibody labeling in pan-ExM samples appears much sparser than in standard, non-expanded samples. The simulations illustrate how these samples would appear without expansion, yielding labeling that appears smoother and denser due to the inferior resolution. a-l, anti-GFP labeling (Thy-1 GFP). a-f, pan-ExM-t images. a, c, d, pan channel overlaid with GFP channel (green). b, e, f, GFP channel only. g-l, simulated non-expanded anti-GFP images. g,i,j, pan channel overlaid with GFP channel (cyan). h, k, l, GFP channel only. m-x, anti-PSD-95 labeling. m-r, pan-ExM-t images. m, o, p, pan channel overlaid with PSD-95 channel (cyan). n, q, r, PSD-95 channel only. s-x, simulated non-expanded anti-PSD-95 images. s, u, v, pan channel overlaid with PSD-95 channel (cyan). t, w, x, PSD-95 channel only. All scale bars: 10 μm, not corrected for expansion factor.
Extended Data Fig. 3 Large-area montages of pan-ExM-t data from cortical and hippocampal regions.
a, Schematic of sample preparation. A coronal hemi-brain section (100 µm thick) was expanded and stained using the pan-ExM-t protocol. b, Wide-field fluorescence image of Thy-1 GFP expression. c, Wide-field phase contrast image after ~10x expansion. d, Wide-field, phase contrast image after ~20x expansion. Sample ROI corresponds to the red box shown in b,c. Letters indicate approximate regions corresponding to high-resolution confocal imaging shown in e-l. e, f, g, pan-ExM montage of cortical neuropil. Progressive zooms shown in f,g, show details. h, pan-ExM image of cortical white matter region. i, pan-ExM image of pyramidal cell layer of the hippocampus. j,k,l, pan-ExM montage of hippocampal neuropil. Progressive zooms shown in k,l, show details. All scale bars: not corrected for the expansion factor.
Extended Data Fig. 4 Electron microscopy of brain tissue prepared with pan-ExM-t fixatives.
a, Experimental approach: mice were transcardially perfused following the pan-ExM-t protocol (Methods), brains were extracted and sliced. Brain slices were divided in half; respective halves were prepared for electron microscopy (EM) and pan-ExM-t, respectively. b,c Comparison of EM (b) and pan-ExM-t (c) images for equivalent brain tissue regions taken from the same brain slice. Orange arrows indicate synaptic clefts. d,e, EM images of brain tissue samples from mouse neocortex. f, g, h, EM images of brain tissue samples from cerebellum. Please note the preservation of subcellular features such as cellular membranes, synaptic vesicles, mitochondrial cristae, and Golgi apparatus. These results suggest that pan-ExM-t based perfusion protocols are sufficient to preserve these features.
Extended Data Fig. 5 Quantification of microscope resolution.
a,b Example images of small, sub-diffraction sized features labeled by anti-GFP antibodies in hippocampal brain tissue from a Thy1 mouse line. a, xy plane. b, yz plane. c, Example gaussian fits to line profiles as indicated in a,b. d,e, Histogram of full-width half-maximum (FWHM) measurements of the fitted gaussian profiles across n = 50 sub-diffraction sized features. d, Distribution of lateral resolutions with mean ± std = 290 ± 60 nm (14 ± 3 nm corrected for 20x expansion, n = 50 features). e, Distribution of axial resolutions with mean ± std = 840 ± 180 nm (42 ± 9 nm corrected for 20x expansion, n = 50 features). Please note that these estimates are upper bounds since cellular features may be larger than ideal point sources.
Supplementary information
Supplementary Information
Supplementary Figs. 1–34, Tables 1–6 and Methods.
Supplementary Video 1
3D pan-ExM-t of a dissociated neuron culture showing active zones, PSDs, mitochondrial cristae and other ultrastructural features.
Supplementary Video 2
3D pan-ExM-t of a dissociated neuron culture showing active zones, PSDs, mitochondrial cristae, nuclear pore complexes, nucleoli, centrioles and other ultrastructural features.
Supplementary Video 3
3D pan-ExM-t of a dissociated neuron culture showing active zones and PSDs.
Supplementary Video 4
3D pan-ExM-t of mouse brain vibratome sections showing neuropil at synaptic resolution.
Supplementary Video 5
3D pan-ExM-t of a mouse brain vibratome section showing ultrastructural features including synapses, mitochondria and putative spine apparatuses.
Supplementary Video 6
3D pan-ExM-t of a mouse brain vibratome section showing ultrastructural features of synapses and localization of PSD-95.
Supplementary Video 7
3D pan-ExM-t of mouse brain vibratome sections showing GFAP labeling in the ultrastructural context of the brain.
Supplementary Video 8
3D pan-ExM-t of a mouse brain vibratome section showing anti-GFAP immunolabeling in a protoplasmic astrocyte and SYTOX Green staining in the ultrastructural context of the brain.
Supplementary Video 9
3D pan-ExM-t of Thy1-GFP transgenic mouse brain vibratome sections showing anti-GFP immunolabeling in dendrites in the ultrastructural context of the brain.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
M’Saad, O., Cairns, A., Gulcicek, J. et al. All-optical visualization of specific molecules in the ultrastructural context of brain tissue. Nat Biotechnol (2025). https://doi.org/10.1038/s41587-025-02905-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41587-025-02905-4