Abstract
Heat shock factor 1 (HSF1) is the critical orchestrator of cell responses to heat shock, and its dysfunction is linked to various diseases. HSF1 undergoes phase separation upon heat shock, and its activity is regulated by post-translational modifications (PTMs). The molecular details underlying HSF1 phase separation, temperature sensing and PTM regulation remain poorly understood. Here, we discovered that HSF1 exhibits temperature-dependent phase separation with a lower critical solution temperature behavior, providing a new conceptual mechanism accounting for HSF1 activation. We revealed the residue-level molecular details of the interactions driving the phase separation of wild-type HSF1 and its distinct PTM patterns at various temperatures. The mapped interfaces were validated experimentally and accounted for the reported HSF1 functions. Importantly, the molecular grammar of temperature-dependent HSF1 phase separation is species specific and physiologically relevant. These findings delineate a chemical code that integrates accurate phase separation with physiological body temperature control in animals.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The authors declare that all the data associated with the figures are provided as Supplementary Information. Source data are provided with this paper. All the experimental and simulation raw data are available at https://doi.org/10.5281/zenodo.13895417 (ref. 79). Should any raw data files be needed in another format, they are available from the corresponding author upon reasonable request.
Code availability
The code can be downloaded from https://github.com/carolinge/HSF_MD.
References
Anckar, J. & Sistonen, L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu. Rev. Biochem. 80, 1089–1115 (2011).
Zheng, X. et al. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. eLife 5, e18638 (2016).
Kijima, T. et al. HSP90 inhibitors disrupt a transient HSP90−HSF1 interaction and identify a noncanonical model of HSP90-mediated HSF1 regulation. Sci. Rep. 8, 6976 (2018).
Li, J., Labbadia, J. & Morimoto, R. I. Rethinking HSF1 in stress, development, and organismal health. Trends Cell Biol. 27, 895–905 (2017).
Mendillo, M. L. et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 150, 549–562 (2012).
Meng, L., Gabai, V. L. & Sherman, M. Y. Heat-shock transcription factor HSF1 has a critical role in human epidermal growth factor receptor-2-induced cellular transformation and tumorigenesis. Oncogene 29, 5204–5213 (2010).
Dai, C., Whitesell, L., Rogers, A. B. & Lindquist, S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130, 1005–1018 (2007).
Goetzl, E. J. et al. Low neural exosomal levels of cellular survival factors in Alzheimer’s disease. Ann. Clin. Transl. Neurol. 2, 769–773 (2015).
Gomez-Pastor, R. et al. Abnormal degradation of the neuronal stress-protective transcription factor HSF1 in Huntington’s disease. Nat. Commun. 8, 14405 (2017).
Gomez-Pastor, R., Burchfiel, E. T. & Thiele, D. J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 4–19 (2018).
Hentze, N., Le Breton, L., Wiesner, J., Kempf, G. & Mayer, M. P. Molecular mechanism of thermosensory function of human heat shock transcription factor Hsf1. eLife 5, e11576 (2016).
Xu, Y. M., Huang, D. Y., Chiu, J. F. & Lau, A. T. Post-translational modification of human heat shock factors and their functions: a recent update by proteomic approach. J. Proteome Res. 11, 2625–2634 (2012).
Zhang, H. et al. Reversible phase separation of HSF1 is required for an acute transcriptional response during heat shock. Nat. Cell Biol. 24, 340–352 (2022).
Lafontaine, D. L. J., Riback, J. A., Bascetin, R. & Brangwynne, C. P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 22, 165–182 (2021).
Gibson, B. A. et al. Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484 (2019).
Chong, S. et al. Tuning levels of low-complexity domain interactions to modulate endogenous oncogenic transcription. Mol. Cell 82, 2084–2097 (2022).
Cotto, J., Fox, S. & Morimoto, R. HSF1 granules: a novel stress-induced nuclear compartment of human cells. J. Cell Sci. 110, 2925–2934 (1997).
Jolly, C., Morimoto, R., Robert-Nicoud, M. & Vourc’h, C. HSF1 transcription factor concentrates in nuclear foci during heat shock: relationship with transcription sites. J. Cell Sci. 110, 2935–2941 (1997).
Gaglia, G. et al. HSF1 phase transition mediates stress adaptation and cell fate decisions. Nat. Cell Biol. 22, 151–158 (2020).
Martin, E. W. et al. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699 (2020).
Kim, T. H. et al. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 365, 825–829 (2019).
Ibáñez de Opakua, A. et al. Molecular interactions of FG nucleoporin repeats at high resolution. Nat. Chem. 14, 1278–1285 (2022).
Paloni, M., Bailly, R., Ciandrini, L. & Barducci, A. Unraveling molecular interactions in liquid–liquid phase separation of disordered proteins by atomistic simulations. J. Phys. Chem. B 124, 9009–9016 (2020).
Collepardo-Guevara, R. et al. Chromatin unfolding by epigenetic modifications explained by dramatic impairment of internucleosome interactions: a multiscale computational study. J. Am. Chem. Soc. 137, 10205–10215 (2015).
Guilln-Boixet, J. et al. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361 (2020).
McCarty, J., Delaney, K. T., Danielsen, S. P. O., Fredrickson, G. H. & Shea, J. E. Complete phase diagram for liquid−liquid phase separation of intrinsically disordered proteins. J. Phys. Chem. Lett. 10, 1644–1652 (2019).
Lin, Y. H. & Chan, H. S. Phase separation and single-chain compactness of charged disordered proteins are strongly correlated. Biophys. J. 112, 2043–2046 (2017).
Das, S., Lin, Y.-H., Vernon, R. M., Forman-Kay, J. D. & Chan, H. S. Comparative roles of charge, π, and hydrophobic interactions in sequence-dependent phase separation of intrinsically disordered proteins. Proc. Natl Acad. Sci. USA 117, 28795–28805 (2020).
Li, L. G. & Hou, Z. Theoretical modelling of liquid−liquid phase separation: from particle-based to field-based simulation. Biophys. Rep. 8, 55–67 (2022).
Regy, R. M., Thompson, J., Kim, Y. C. & Mittal, J. Improved coarse-grained model for studying sequence dependent phase separation of disordered proteins. Protein Sci. 30, 1371–1379 (2021).
Tesei, G., Schulze, T. K., Crehuet, R. & Lindorff-Larsen, K. Accurate model of liquid–liquid phase behavior of intrinsically disordered proteins from optimization of single-chain properties. Proc. Natl Acad. Sci. USA 118, e2111696118 (2021).
Tejedor, A. R. et al. Protein structural transitions critically transform the network connectivity and viscoelasticity of RNA-binding protein condensates but RNA can prevent it. Nat. Commun. 13, 5717 (2022).
Alexander, E. C. et al. TDP-43 α-helical structure tunes liquid–liquid phase separation and function. Proc. Natl Acad. Sci. USA 117, 5883–5894 (2020).
Bremer, A. et al. Deciphering how naturally occurring sequence features impact the phase behaviours of disordered prion-like domains. Nat. Chem. 14, 196–207 (2022).
Najbauer, E. E., Ng, S. C., Griesinger, C., Gorlich, D. & Andreas, L. B. Atomic resolution dynamics of cohesive interactions in phase-separated Nup98 FG domains. Nat. Commun. 13, 1494 (2022).
Babu, M., Favretto, F., Rankovic, M. & Zweckstetter, M. Peptidyl prolyl isomerase A modulates the liquid−liquid phase separation of proline-rich IDPs. J. Am. Chem. Soc. 144, 16157–16163 (2022).
Murthy, A. C. et al. Molecular interactions underlying liquid−liquid phase separation of the FUS low-complexity domain. Nat. Struct. Mol. Biol. 26, 637–648 (2019).
Gomes, G.-N. W. et al. Conformational ensembles of an intrinsically disordered protein consistent with NMR, SAXS, and single-molecule FRET. J. Am. Chem. Soc. 142, 15697–15710 (2020).
Refinetti, R. The circadian rhythm of body temperature. Front. Biosci. 15, 564–594 (2010).
Piestun, Y., Druyan, S., Brake, J. & Yahav, S. Thermal manipulations during broiler incubation alter performance of broilers to 70 days of age. Poult. Sci. 92, 1155–1163 (2013).
López-Olmeda, J. F. & Sánchez-Vázquez, F. J. Thermal biology of zebrafish (Danio rerio). J. Therm. Biol. 36, 91–104 (2011).
Li, L. & Hou, Z. Crosslink-induced conformation change of intrinsically disordered proteins have a nontrivial effect on phase separation dynamics and thermodynamics. J. Phys. Chem. B 127, 5018–5026 (2023).
Quiroz, F. G. & Chilkoti, A. Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat. Mat.14, 1164–1171 (2015).
Quiroz, F. G. et al. Intrinsically disordered proteins access a range of hysteretic phase separation behaviors. Sci. Adv. 5, eaax5177 (2019).
Da Vela, S. et al. Kinetics of liquid−liquid phase separation in protein solutions exhibiting LCST phase behavior studied by time-resolved USAXS and VSANS. Soft Matter 12, 9334–9341 (2016).
Matsuyama, A. & Tanaka, F. Theory of solvation-induced reentrant phase separation in polymer solutions. Phys. Rev. Lett. 65, 341 (1990).
Türkaydin, B. et al. Atomistic mechanism of coupling between cytosolic sensor domain and selectivity filter in TREK K2P channels. Nat. Commun. 15, 4628 (2024).
Winter, H., Huber, J. L. & Huber, S. C. Membrane association of sucrose synthase: changes during the graviresponse and possible control by protein phosphorylation. FEBS Lett. 420, 151–155 (1997).
Martin, E. W. et al. Sequence determinants of the conformational properties of an intrinsically disordered protein prior to and upon multisite phosphorylation. J. Am. Chem. Soc. 138, 15323–15335 (2016).
Das, R. K. & Pappu, R. V. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl Acad. Sci. USA 110, 13392–13397 (2013).
Ma, Y. et al. Nucleobase clustering contributes to the formation and hollowing of repeat-expansion RNA condensate. J. Am. Chem. Soc. 144, 4716–4720 (2022).
Mammen Regy, R., Zheng, W. & Mittal, J. Chapter one—Using a sequence-specific coarse-grained model for studying protein liquid–liquid phase separation. In Methods in Enzymology (ed. Keating, C. D.), Vol. 646, 1−17 (Academic Press, 2021).
Shrestha, U. R. et al. Generation of the configurational ensemble of an intrinsically disordered protein from unbiased molecular dynamics simulation. Proc. Natl Acad. Sci. USA 116, 20446–20452 (2019).
Kulkarni, P. et al. Intrinsically disordered proteins: ensembles at the limits of Anfinsen’s dogma. Biophys. Rev. 3, 011306 (2022).
Dannenhoffer-Lafage, T. & Best, R. B. A data-driven hydrophobicity scale for predicting liquid–liquid phase separation of proteins. J. Phys. Chem. B 125, 4046–4056 (2021).
Go, N. Theoretical studies of protein folding. Annu. Rev. Biophys. Bioeng. 12, 183–210 (1983).
Riback, J. A. et al. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168, 1028–1040 (2017).
Chen, S. & Wang, Z. G. Using implicit-solvent potentials to extract water contributions to enthalpy−entropy compensation in biomolecular associations. J. Phys. Chem. B 127, 6825–6832 (2023).
Chowdhary, S., Kainth, A. S., Paracha, S., Gross, D. S. & Pincus, D. Inducible transcriptional condensates drive 3D genome reorganization in the heat shock response. Mol. Cell 82, 4386–4399 (2022).
Kmiecik, S. W., Le Breton, L. & Mayer, M. P. Feedback regulation of heat shock factor 1 (Hsf1) activity by Hsp70-mediated trimer unzipping and dissociation from DNA. EMBO J. 39, e104096 (2020).
Guettouche, T., Boellmann, F., Lane, W. S. & Voellmy, R. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 6, 4 (2005).
Murshid, A. et al. Protein kinase A binds and activates heat shock factor 1. PLoS ONE 5, e13830 (2010).
Sourbier, C. et al. Englerin A stimulates PKCθ to inhibit insulin signaling and to simultaneously activate HSF1: pharmacologically induced synthetic lethality. Cancer Cell 23, 228–237 (2013).
Matsarskaia, O. et al. Tuning phase transitions of aqueous protein solutions by multivalent cations. Phys. Chem. Chem. Phys. 20, 27214–27225 (2018).
Du, Z., et al. Phosphorylation modulates estrogen receptor disorder by altering long-range hydrophobic interactions. Preprint at bioRxiv https://doi.org/10.1101/2023.07.14.548966 (2023).
Kolakofsky, D., Kowalinski, E. & Cusack, S. A structure-based model of RIG-I activation. RNA 18, 2118–2127 (2012).
Lane, T. J., Shukla, D., Beauchamp, K. A. & Pande, V. S. To milliseconds and beyond: challenges in the simulation of protein folding. Curr. Opin. Struct. Biol. 23, 58–65 (2013).
Fawzi, N. L., Ying, J., Torchia, D. A. & Clore, G. M. Kinetics of amyloid beta monomer-to-oligomer exchange by NMR relaxation. J. Am. Chem. Soc. 132, 9948–9951 (2010).
Fawzi, N. L. et al. Dark-state exchange saturation transfer. Nat. Methods 8, 997 (2011).
Clore, G. M., Tang, C. & Iwahara, J. Elucidating transient macromolecular interactions using paramagnetic relaxation enhancement. Curr. Opin. Struct. Biol. 17, 603–616 (2007).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Hopkins, J. B., Gillilan, R. E. & Skou, S. BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Crystallogr. 50, 1545–1553 (2017).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Lee, W., Tonelli, M. & Markley, J. L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015).
Kjaergaard, M. & Poulsen, F. M. Sequence correction of random coil chemical shifts: correlation between neighbor correction factors and changes in the Ramachandran distribution. J. Biomol. NMR 50, 157–165 (2011).
Camilloni, C., De Simone, A., Vranken, W. F. & Vendruscolo, M. Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry 51, 2224–2231 (2012).
Ashbaugh, H. S. & Hatch, H. W. Natively unfolded protein stability as a coil-to-globule transition in charge/hydropathy space. J. Am. Chem. Soc. 130, 9536–9542 (2008).
Li, L. & Ren, Q. Experimental and Simulation data for the molecular mechanism of temperature-dependent phase separation of HSF1. Zenodo https://doi.org/10.5281/zenodo.13895417 (2024).
Acknowledgements
We thank the staff and G. Liu from the BL19U2 beamline(https://cstr.cn/31129.02.NFPS.BL19U2) of the National Facility for Protein Science in Shanghai (https://cstr.cn/31129.02.NFPS) at the Shanghai Synchrotron Radiation Facility for providing technical support and assistance in SAXS data collection and analysis. This work was supported by grants from the National Natural Science Foundation of China (32090040 to S.X., Z.H. and Y.X.; 31971128 to S.X.; 92254302 to Y.X.) and the National Key R&D Program (2019YFA0508403 to S.X.; 2022YFA1303100 to Z.H. and Y.X.) and the Strategic Priority Research Program of the Chinese Academy of Sciences Grant no. XDB 37040202 to S.X. and CAS Project for Young Scientists in Basic Research, Grant no. YSBR-068 to S.X.
Author information
Authors and Affiliations
Contributions
Q.R., L. Li, X.Y., Z.H. and S.X. conceived and designed the experiments. Q.R. performed most of the biochemical and NMR experiments. L. Li performed the coarse-grained simulation. L. Liu assisted with purification of the IDR of HSF1. J.L. and C.S. helped with the NMR experiments. Y.S. provided the original plasmids of HSF1. Q.R., L. Li, X.Y., Z.H. and S.X. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
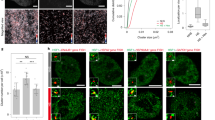
Extended Data Fig. 1 Temperature effects on phase transition and aggregation of HSF1.
a. The correlation between species body temperature and the middle point temperature of the phase transition curve. b. The box-plots of droplet sizes in Fig. 1e. n = 79 for WT-25 °C, n = 100 for WT-37 °C, n = 71 for M1-5 °C, n = 94 for M1-25 °C and n = 85 for M1-37 °C across 3 independent experiments. ND, not determined. c. The peak with a retention time of 10 ml was the LZ1-3 oligomers by SEC-MALS. The UV absorbance values were in red, and the light scattering values were in gray. The molar mass values (g/mol) were in blue. d. DSS cross-linked lz1-3 at different temperatures were detected by SDS‒PAGE, which is representative of 3 independent experiments. e. The box-plots of droplet sizes in Fig. 1f. n = 86 for WT-25 °C, n = 190 for WT-37 °C, n = 153 for M1-5 °C, n = 153 for M1-25 °C and n = 151 for M1-37 °C across 3 independent experiments. ND, not determined. In the box plots, data are presented as mean values, box plots: 25th to 75th percentiles, median line, 1.5× interquartile as whiskers and points.
Extended Data Fig. 2 The backbone assignments of RD(WT), RD(M1) and RD(M2).
The backbone assignments were plotted on the 1H-15N HSQC, which were recorded at 5 °C.
Extended Data Fig. 3 Comparative NMR analysis of wild-type and mutant RD domains.
a. The secondary chemical shifts of WT RD, M1 and M2. b. Secondary structure populations of WT RD, M1 and M2 obtained from the delta2D method using experimentally assigned backbone chemical shifts. c. The hetNOE values of the backbone amide groups of WT RD, M1 and M2. The light orange columns represent the WT, while the red and blue lines depict M1 and M2, respectively. d. T1 and e. T2 of WT RD, M1 and M2. WT, gray line. M1, red line. M2, blue line. NMR data were collected at 5 °C.
Extended Data Fig. 4 Temperature-dependent NMR chemical shifts and SAXS-derived Rg values of RD protein variants: WT, M1, and M2.
a-c.The temperature coefficient of the 1HN chemical shift values of the backbone amide groups of WT(a), M1(b), and M2(c) at different temperature ranges. d. Rg values of WT RD, M1, and M2 mutants were measured by SAXS at 5 °C and 37 °C. At each temperature for each sample, a set of 20 sample curves and their corresponding 20 buffer curves were collected. The software processed these data by averaging the 20 sample curves and then subtracting the average background curve to eliminate any baseline effects. Subsequently, the mean radii of gyration (Rg), along with their associated errors, were calculated through multiple Guinier fits. Consequently, the figure presents the result of the collective average curve rather than plotting individual data points.
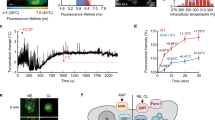
Extended Data Fig. 5 Analysis of Mean-Field Model parameters and solution viscosity of different conditions.
a. Fitting parameters for the mean-field model. b, c. The diffusion coefficient of 1,4-Dioxane in 0.1 mM and 1 mM RD at 5 °C and 30 °C using DOSY.
Extended Data Fig. 6 NMR intensities ratios reveal the intermolecular interactions between RD domains.
The NMR signal intensity ratio of every residue of the WT RD(a), M1(b) and M2(c) when it was alone or in the presence of 10-fold excess unlabeled counterpart at 15 °C and 25 °C. All error bars were calculated according to the signal-to-noise ratio of the spectra, as explained in the methods section. d-e. The NMR spectra slices along 1HN-dimension of M212 and A329 of WT RD from 5 °C to 30 °C.
Extended Data Fig. 7 The linewidth distribution of peaks in 15N-RD and 15N + 14N-RD samples at 5 °C (a) and 30 °C (b).
In both correlation plots, the dots on the upper right are linewidth values in the 1H dimension, and the dots on the lower left are linewidth values in the 15N dimension. The linear regression analysis results were shown on each plot.
Extended Data Fig. 8 Sequence analysis with localCIDER and phase separation simulation of RD protein variants with different force fields.
a. Sequence analysis results of some commonly applied sequence parameters, calculated with localCIDER b. The simulation using existing force fields failed to reproduce the phase separation. The experimental data of WT and M1 are shown in gray and red dots. The current NMR-driven CG simulation predicted values at the 4 temperatures (5 °C, 15 °C, 25 °C and 30 °C) are shown in gray and red crosses.
Extended Data Fig. 9 Force field optimization and Coarse-Grained simulation of protein phase separation.
a. Normalized difference between simulated and experimental HSQC values \(\langle \tfrac{|NM{R}_{\exp }^{i}-NM{R}_{{\rm{sim}}}^{i}|}{NM{R}_{\exp }^{i}}\rangle\) as a function of the number of iterations for force field optimization. b. Comparison between experimental HSQC and simulated HSQC after force field optimization. c. Snapshots of the CG simulation, which reproduced the experimental phase separation observation of LZ1-3-RD at 15 °C and 25 °C. d. Distributions of the reduced density ρ of WT, M1, and M2 in the CG simulation at 15 °C and 25 °C. e. Changes in the protein amounts in the largest condensate ρ of WT, M1, and M2 during CG simulation at 15 °C and 25 °C.
Extended Data Fig. 10 Schematic representation of peptide mutants in biochemical assays and interspecies sequence variation in HSF1 LZ1-3-RD-LZ4 domains.
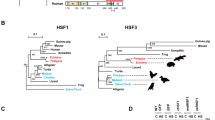
Sequence schemes of RD(WT) aa224-248-S(a) and RD(M1) aa320-343-S(b) mutants. The positively charged residues were labeled in red and the negatively charged residues were labeled in blue, while the hydrophilic mutations are in black. c. The sequence alignment of the LZ1-3-RD-LZ4 regions from HSF1 of humans, mice, chicken, and zebrafish showed the main species-specific differences between residue 320 and 360, matching with the regions with a temperature-dependent interaction pattern in Fig. 5a.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data for the turbidity curves of LZ1−3-RD-LZ4 from different species.
Source Data Fig. 2
Source data for phase diagrams and water and entropy statistics for a,c,d. Statistical source data for droplet sizes of RD(M1)-L.
Source Data Fig. 3
Source data for the NMR signal intensity ratio of every residue at 5 °C and 30 °C.
Source Data Fig. 4
Statistical source data for density, cluster growth and Rg distributions for panels c−e.
Source Data Fig. 5
Statistical source data for droplet sizes.
Source Data Fig. 6
Statistical source data for the turbidity−temperature curves.
Source Data Extended Data Fig. 1
Statistical source data for droplet sizes and SEC−MALS of LZ1−3.
Source Data Extended Data Fig. 2
Source data for the backbone assignments of WT, M1 and M2 RD.
Source Data Extended Data Fig. 3
Source data for the secondary chemical shifts, the secondary structure populations, the hetNOE values, T1 and T2 of WT, M1 and M2 RD.
Source Data Extended Data Fig. 4
Source data for temperature-dependent NMR chemical shifts and SAXS-derived Rg values of WT, M1 and M2 RD.
Source Data Extended Data Fig. 5
Source data for the diffusion coefficient of 1,4-dioxane in 0.1 mM and 1 mM RD at 5 °C and 30 °C.
Source Data Extended Data Fig. 6
Source data for the NMR signal intensity ratio of every residue at 15 °C and 25 °C.
Source Data Extended Data Fig. 7
Statistical source data for the linewidth distribution of peaks in 15N-labeled RD and 15N + 14N-labeled RD samples at 5 °C and 30 °C.
Source Data Extended Data Fig. 8
Source data for phase diagrams of other force fields.
Source Data Extended Data Fig. 9
Statistical source data for simulated HSQC data and density, cluster growth and distributions for b,d,e.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ren, Q., Li, L., Liu, L. et al. The molecular mechanism of temperature-dependent phase separation of heat shock factor 1. Nat Chem Biol 21, 831–842 (2025). https://doi.org/10.1038/s41589-024-01806-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41589-024-01806-y
This article is cited by
-
OsLEML2 Regulates Rice Thermotolerance Through Auxin and ROS Homeostasis
Rice (2025)
-
Heat stress response factor ZmHSF10 positively regulates heat tolerance in maize
BMC Plant Biology (2025)