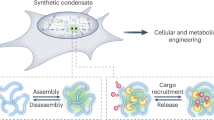
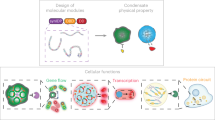
Abstract
Engineering synthetic intrinsically disordered proteins (synIDPs) enables regulation of biomolecular condensation and protein solubility. However, limited understanding of how sequence-dependent interaction cooperativity relates to the fitness impacts of synIDPs on endogenous cellular processes constrains our design capability. Here, to circumvent this design challenge, we present a systematic directed evolution method for the evolution of synIDPs capable of mediating diverse phase behaviors in living cells. The selection methods allow us to evolve a toolbox of synIDPs with distinct phase behaviors and thermoresponsive features in living cells, leading to the evolution of synthetic condensates. The reverse-selection method further allows us to select synIDPs as solubility tags. We demonstrate the applications of the evolved synIDPs in protein circuits to (1) regulate intracellular protein activity and (2) reverse antibiotic resistance. Our systematic evolution and selection strategies provide a versatile platform for developing synIDPs for broad applications in synthetic biology and biotechnology.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data supporting the findings of this work are provided in the manuscript and its related Supplementary Information. Source data are provided with this paper.
References
Holehouse, A. S. & Kragelund, B. B. The molecular basis for cellular function of intrinsically disordered protein regions. Nat. Rev. Mol. Cell Biol. 25, 187–211 (2024).
Pappu, R. V., Cohen, S. R., Dar, F., Farag, M. & Kar, M. Phase transitions of associative biomacromolecules. Chem. Rev. 123, 8945–8987 (2023).
van der Lee, R. et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631 (2014).
Wright, P. E. & Dyson, H. J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 (2015).
Garabedian, M. V. et al. Designer membraneless organelles sequester native factors for control of cell behavior. Nat. Chem. Biol. 17, 998–1007 (2021).
Dai, Y., You, L. & Chilkoti, A. Engineering synthetic biomolecular condensates. Nat. Rev. Bioeng. 1, 466–480 (2023).
Dai, Y. et al. Programmable synthetic biomolecular condensates for cellular control. Nat. Chem. Biol. 19, 518–528 (2023).
Guo, H. et al. Spatial engineering of E. coli with addressable phase-separated RNAs. Cell 185, 3823–3837 (2022).
Shin, Y. et al. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168, 159–171 (2017).
Reinkemeier, C. D., Girona, G. E. & Lemke, E. A. Designer membraneless organelles enable codon reassignment of selected mRNAs in eukaryotes. Science 363, eaaw2644 (2019).
Rana, U. et al. Asymmetric oligomerization state and sequence patterning can tune multiphase condensate miscibility. Nat. Chem. 16, 1073–1082 (2024).
Welles, R. M. et al. Determinants that enable disordered protein assembly into discrete condensed phases. Nat. Chem. 16, 1062–1072 (2024).
Shapiro, D. M. et al. Synthetic biomolecular condensates enhance translation from a target mRNA in living cells. Nat. Chem. 17, 448–456 (2025).
Mitrea, D. M., Mittasch, M., Gomes, B. F., Klein, I. A. & Murcko, M. A. Modulating biomolecular condensates: a novel approach to drug discovery. Nat. Rev. Drug Discov. 21, 841–862 (2022).
Roden, C. A. et al. Double-stranded RNA drives SARS-CoV-2 nucleocapsid protein to undergo phase separation at specific temperatures. Nucleic Acids Res. 50, 8168–8192 (2022).
Zhang, Y. et al. Probing condensate microenvironments with a micropeptide killswitch. Nature 643, 1107–1116 (2025).
Bremer, A. et al. Deciphering how naturally occurring sequence features impact the phase behaviours of disordered prion-like domains. Nat. Chem. 14, 196–207 (2022).
Wake, N. et al. Expanding the molecular grammar of polar residues and arginine in FUS phase separation. Nat. Chem. Biol. 21, 1076–1088 (2025).
Hilditch, A. T. et al. Assembling membraneless organelles from de novo designed proteins. Nat. Chem. 16, 89–97 (2024).
Miki, T. et al. De novo designed YK peptides forming reversible amyloid for synthetic protein condensates in mammalian cells. Nat. Commun. 15, 8503 (2024).
Yu, W. et al. De novo engineering of programmable and multi-functional biomolecular condensates for controlled biosynthesis. Nat. Commun. 15, 7989 (2024).
Dzuricky, M., Rogers, B. A., Shahid, A., Cremer, P. S. & Chilkoti, A. De novo engineering of intracellular condensates using artificial disordered proteins. Nat. Chem. 12, 814–825 (2020).
Lasker, K. et al. The material properties of a bacterial-derived biomolecular condensate tune biological function in natural and synthetic systems. Nat. Commun. 13, 5643 (2022).
Tang, N. C. et al. Synthetic intrinsically disordered protein fusion tags that enhance protein solubility. Nat. Commun. 15, 3727 (2024).
Molina, R. S. et al. In vivo hypermutation and continuous evolution. Nat. Rev. Methods Primers 2, 36 (2022).
Packer, M. S. & Liu, D. R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 16, 379–394 (2015).
Wörsdörfer, B., Woycechowsky, K. J. & Hilvert, D. Directed evolution of a protein container. Science 331, 589–592 (2011).
Wang, Y. et al. Directed evolution: methodologies and applications. Chem. Rev. 121, 12384–12444 (2021).
Piraner, D. I., Abedi, M. H., Moser, B. A., Lee-Gosselin, A. & Shapiro, M. G. Tunable thermal bioswitches for in vivo control of microbial therapeutics. Nat. Chem. Biol. 13, 75–80 (2017).
Touzé, T. et al. Colicin M, a peptidoglycan lipid-II-degrading enzyme: potential use for antibacterial means? Biochem. Soc. Trans. 40, 1522–1527 (2012).
You, L., Cox, R. S., Weiss, R. & Arnold, F. H. Programmed population control by cell–cell communication and regulated killing. Nature 428, 868–871 (2004).
Ashley, C. E. et al. Cell-specific delivery of diverse cargos by bacteriophage MS2 virus-like particles. ACS Nano 5, 5729–5745 (2011).
Yan, G. et al. The N-terminal and central domain of colicin A enables phage lysin to lyse Escherichia coli extracellularly. Antonie Van Leeuwenhoek 110, 1627–1635 (2017).
Rottinghaus, A. G., Ferreiro, A., Fishbein, S. R. S., Dantas, G. & Moon, T. S. Genetically stable CRISPR-based kill switches for engineered microbes. Nat. Commun. 13, 672 (2022).
Bernhardt, T. G., Roof, W. D. & Young, R. Genetic evidence that the bacteriophage φX174 lysis protein inhibits cell wall synthesis. Proc. Natl Acad. Sci. USA 97, 4297–4302 (2000).
Chang, J., Lenhoff, A. M. & Sandler, S. I. Solvation free energy of amino acids and side-chain analogues. J. Phys. Chem. B 111, 2098–2106 (2007).
Martin, E. W. et al. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699 (2020).
Rana, U., Brangwynne, C. P. & Panagiotopoulos, A. Z. Phase separation vs aggregation behavior for model disordered proteins. J. Chem. Phys. 155, 125101 (2021).
Del Conte, A. et al. CAID prediction portal: a comprehensive service for predicting intrinsic disorder and binding regions in proteins. Nucleic Acids Res. 51, W62–W69 (2023).
Sun, J. et al. Precise prediction of phase-separation key residues by machine learning. Nat. Commun. 15, 2662 (2024).
Vernon, R. M. et al. Pi–pi contacts are an overlooked protein feature relevant to phase separation. elife 7, e31486 (2018).
Rekhi, S. et al. Expanding the molecular language of protein liquid–liquid phase separation. Nat. Chem. 16, 1113–1124 (2024).
Liao, S.-M., Du, Q.-S., Meng, J.-Z., Pang, Z.-W. & Huang, R.-B. The multiple roles of histidine in protein interactions. Chem. Cent. J. 7, 44 (2013).
Tsang, B., Pritišanac, I., Scherer, S. W., Moses, A. M. & Forman-Kay, J. D. Phase separation as a missing mechanism for interpretation of disease mutations. Cell 183, 1742–1756 (2020).
Liu, X. et al. Phase separation drives decision making in cell division. J. Biol. Chem. 295, 13419–13431 (2020).
Zhuang, Y. et al. Circadian clocks are modulated by compartmentalized oscillating translation. Cell 186, 3245–3260 (2023).
Brophy, J. A. N. & Voigt, C. A. Principles of genetic circuit design. Nat. Methods 11, 508–520 (2014).
Meyer, A. J., Segall-Shapiro, T. H., Glassey, E., Zhang, J. & Voigt, C. A. Escherichia coli ‘Marionette’ strains with 12 highly optimized small-molecule sensors. Nat. Chem. Biol. 15, 196–204 (2019).
Ruff, K. M., Roberts, S., Chilkoti, A. & Pappu, R. V. Advances in understanding stimulus-responsive phase behavior of intrinsically disordered protein polymers. J. Mol. Biol. 430, 4619–4635 (2018).
Quiroz, F. G. & Chilkoti, A. Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat. Mater. 14, 1164–1171 (2015).
Meyer, D. E. & Chilkoti, A. Purification of recombinant proteins by fusion with thermally-responsive polypeptides. Nat. Biotechnol. 17, 1112–1115 (1999).
Zeng, X. & Lin, J. β-Lactamase induction and cell wall metabolism in Gram-negative bacteria. Front. Microbiol. 4, 128 (2013).
Ma, H. R., Xu, H. Z., Kim, K., Anderson, D. J. & You, L. Private benefit of β-lactamase dictates selection dynamics of combination antibiotic treatment. Nat. Commun. 15, 8337 (2024).
Gao, X. J., Chong, L. S., Kim, M. S. & Elowitz, M. B. Programmable protein circuits in living cells. Science 361, 1252–1258 (2018).
Chen, Z. et al. A synthetic protein-level neural network in mammalian cells. Science 386, 1243–1250 (2024).
Schein, C. H. Solubility as a function of protein structure and solvent components. Bio/technology 8, 308–317 (1990).
Han, X., Ning, W., Ma, X., Wang, X. & Zhou, K. Improving protein solubility and activity by introducing small peptide tags designed with machine learning models. Metab. Eng. Commun. 11, e00138 (2020).
Fernandez-Rodriguez, J. & Voigt, C. A. Post-translational control of genetic circuits using Potyvirus proteases. Nucleic Acids Res. 44, 6493–6502 (2016).
Nathan, C. & Cars, O. Antibiotic resistance—problems, progress, and prospects. N. Engl. J. Med. 371, 1761–1763 (2014).
Xu, X. et al. In silico screening of protein-binding peptides with an application to developing peptide inhibitors against antibiotic resistance. PNAS Nexus 3, pgae541 (2024).
Reinkemeier, C. D. & Lemke, E. A. Synthetic biomolecular condensates to engineer eukaryotic cells. Curr. Opin. Chem. Biol. 64, 174–181 (2021).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Risso-Ballester, J. et al. A condensate-hardening drug blocks RSV replication in vivo. Nature 595, 596–599 (2021).
Jain, A. & Vale, R. D. RNA phase transitions in repeat expansion disorders. Nature 546, 243–247 (2017).
Chen, M. W. et al. Condenzymes: biomolecular condensates with inherent catalytic activities. bioRxiv, Preprint at https://doi.org/10.1101/2024.07.06.602359 (2025).
Dai, Y. et al. Biomolecular condensates regulate cellular electrochemical equilibria. Cell 187, 5951–5966 (2024).
Dai, Y., Wang, Z.-G. & Zare, R. N. Unlocking the electrochemical functions of biomolecular condensates. Nat. Chem. Biol. 20, 1420–1433 (2024).
Yu, W. et al. Aging-dependent evolving electrochemical potentials of biomolecular condensates regulate their physicochemical activities. Nat. Chem. 17, 756–766 (2025).
Chen, M. W. et al. Transition-state-dependent spontaneous generation of reactive oxygen species by Aβ assemblies encodes a self-regulated positive feedback loop for aggregate formation. J. Am. Chem. Soc. 147, 8267–8279 (2025).
Posey, A. E. et al. Biomolecular condensates are characterized by interphase electric potentials. J. Am. Chem. Soc. 146, 28268–28281 (2024).
Boulos, L., Prevost, M., Barbeau, B., Coallier, J. & Desjardins, R. LIVE/DEAD BacLight: application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J. Microbiol. Methods 37, 77–86 (1999).
Acknowledgements
Y.D. acknowledges the experimental and funding support from the Center for Biomolecular Condensates and the McKelvey School of Engineering at Washington University. We acknowledge the experimental and material support and discussion with J. Su of the A. Chilkoti Lab at Duke University.
Author information
Authors and Affiliations
Contributions
Y.D. conceptualized the study and acquired funding. Y.D. and Y.M. designed the experiments. Y.M., L.Y., Y.C., M.W.C. and W.Y. performed the evolution experiments. L.Y. performed the computational algorithm analysis. Y.M., Y.C. and W.Y. purified the proteins. Y.M. and L.Y. performed the synthetic biology experiments. Y.M., L.Y. and M.W.C. analyzed the data. Y.D. supervised the work. Y.D. and Y.M. wrote the manuscript. All authors read and/or edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
Y.D. and Y.M. are coinventors on a US provisional patent application (application number: 63/884,618) that incorporates the methods described in this paper. Their interests are reviewed and managed by Washington University in St. Louis in accordance with their conflict-of-interest policies. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Verification of genetic elements for the development of selection methods for directed evolution of synIDPs.
a, Evaluation of the killing efficiency of toxic proteins using plate-based assay. Data is represented as mean ± SD. n = 3 biological replicates. b, Evaluation of the killing efficiency of protein E against different strains using plate-based assay. Data is represented as mean ± SD. n = 3 biological replicates. c, Representative images of the growth conditions (based on turbidity) for cells expressing protein E-RLPWT and protein E alone. Time points correspond to durations after induction. d, Confocal images of cells expressing Protein E-RLPWT at different time points after dyeing with mixture of SYTO 9 and propidium iodide (PI). When used alone, the SYTO 9 stain generally labels all bacteria in a population. In contrast, PI penetrates only bacteria with damaged membranes, causing a reduction in the SYTO 9 stain fluorescence when both dyes are present. The arrows mark the positions of the biomolecular condensates in the cell. Scale bar represents 5 µm. e, Evaluation of the fraction of live DH5α cells expressing Protein E–RLPWT at different time points post-induction, measured using a plate reader. Data is represented as mean ± SD. n = 3 biological replicates. f, SDS-PAGE analysis of whole-cell lysates. Whole-cell lysates were analyzed by SDS-PAGE to evaluate the expression level of protein E and protein E-RLP (MW: 27.6 kDa). The results showed that protein E-RLP was expressed at substantially higher levels than protein E, which confirmed that the restored cell survival was not due to differences in protein expression levels. The arrows mark the positions of the target proteins on the bands.
Extended Data Fig. 2 Verification of selection methods for the directed evolution of synIDPs.
a, Design principle to transform a soluble promoting residue into a hydrophobic or a “sticker” residue through a single nucleotide mutation. This approach leverages the minimal genetic alteration needed to induce significant changes in the physicochemical properties of the protein, enabling a decrease in chain solvation with minimal sequence modification. b, Sequence alignment of Protein E–synIDP variants generated using SnapGene. Aligned regions are highlighted in red, while unaligned or mutated residues are shown in white. c, Representative fluorescence microscopy images of cells expressing fusions of sfGFP-synIDPs in BL21(DE3) before and after phase transition. Scale bar represents 3 μm. d, Summary of phase separation scores (PScore) for evolved IDPs selected from plate-based selection based on different rounds of evolution. The dashed line indicates the mean PScore for each round and the solid black line represents the fitted trend curve using an asymmetric sigmoidal model. Statistical significance was determined using an ordinary one-way ANOVA test. e, Summary of the key mutations of amino acids across four rounds of evolution.
Extended Data Fig. 3 A selection method of the evolved IDPs exhibiting different thermoresponsive phase behaviors.
a. Schematics of the selection strategies for the evolution of synIDPs with hysteric behaviors. To identify synIDP variants exhibiting reversible thermoresponsive phase transitions, a three-round replica plating strategy was employed. Following the LCST- and UCST- selection described above, colonies from the first round that displayed temperature-dependent are considered potential candidates for reversible phase behavior. Each of the two induced plates incubated at 30 °C and 45 °C are then independently replicated onto fresh plates containing the same arabinose concentration but incubated under the opposite temperature condition for at least 12 h. This second round of selection aimes to assess the reversibility of the observed temperature-dependent growth phenotype, distinguishing reversible or irreversible phase transitions. A third round of replica plating is transferring colonies from the second set of plates back to the original temperature condition (30 °C or 45 °C). b. Evaluation of CFUs of DH5α under different temperature. Statistical significance was determined using two sample T-test. Data is represented as mean ± SD. n = 3 biological replicates. c. Representative confocal images of an rIDP, and an irIDP illustrating differences in thermal hysteresis. RLPWT and ELPWT serve as controls exhibiting reversible UCST- and LCST-type phase behavior, respectively. Scale bars represent 3 μm.
Extended Data Fig. 4 Evaluation of the performance of soluble IDPs (sIDPs).
a. The evolution landscape of evolved soluble IDPs. The panel shows fitness curves of the selected evolved soluble IDPs (sIDPs) from two rounds of evolution. selected: variants with growth curves within the blue-shaded area were chosen for further characterization. non-selected: variants with growth curves within the red-shaded area were excluded from further confocal microscopy characterizations. Selected variants were subcloned and fused with sfGFP for confocal characterization. The non-phase separable variants identified by confocal microscopy were termed sIDP1-6, respectively. n = 1. Scale bars represent 3 μm. b. Kyte-Doolittle hydropathy scores of the evolved soluble synIDPs. The hydropathy parameter is a rescaled Kyte-Doolittle hydropathy value ranging from 0 (least hydrophobic) to 9 (most hydrophobic). c. Evaluation of the expression of the TEV-sIDPs by SDS-PAGE from the soluble and insoluble fraction of the cell lysates. The arrows mark the positions of the target proteins on the bands. Prestained Kaleidoscope™ ladder was used as a molecular weight standard for all gels.
Extended Data Fig. 5 Evaluation of the effect of condensate formation by lsIDPs for ampicillin resistance.
a. Representative fluorescence microscopy images of cells expressing fusions of TF7-sfGFP-lsIDPs in DH5α. Scale bars represent 3 μm. b. Plate-based cellular survival assay for the evaluation of the functional capacity of different lsIDPs fusions. Statistical significance was determined using an ordinary one-way ANOVA test. Data is represented as mean ± SD. n = 3 biological replicates. p = 3.04 × 10⁻⁶ for 500 mg/L ampicillin. c. Evaluation of growth conditions of DH5α expressing TF7-sfGFP-lsIDPs fusion proteins under different concentrations of ampicillin. nc: DH5α cells harboring a bla plasmid and a plasmid encoding sfGFP-lsIDP6. Data is represented as mean ± SD. n = 3 biological replicates.
Supplementary information
Supplementary Information
Supplementary Figs. 1–4 and Tables 1–11.
Supplementary Table
Predicted disordered scores of evolved synIDPs in this study.
Source data
Source Data Figs. 1–5 and Extended Data Figs. 1–5
Source data for all the main and extended figures.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, Y., Yang, L., Chen, Y. et al. Directed evolution of functional intrinsically disordered proteins. Nat Chem Biol (2026). https://doi.org/10.1038/s41589-025-02128-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41589-025-02128-3