Abstract
Intestinal intraepithelial lymphocytes (IELs) exhibit prompt innate-like responses to microenvironmental cues and require strict control of effector functions. Here we showed that Aiolos, an Ikaros zinc-finger family member encoded by Ikzf3, acted as a regulator of IEL activation. Ikzf3−/− CD8αα+ IELs had elevated expression of NK receptors, cytotoxic enzymes, cytokines and chemokines. Single-cell RNA sequencing of Ikzf3−/− and Ikzf3+/+ IELs showed an amplified effector machinery in Ikzf3−/− CD8αα+ IELs compared to Ikzf3+/+ counterparts. Ikzf3−/− CD8αα+ IELs had increased responsiveness to interleukin-15, which explained a substantial part, but not all, of the observed phenotypes. Aiolos binding sites were close to those for the transcription factors STAT5 and RUNX, which promote interleukin-15 signaling and cytolytic programs, and Ikzf3 deficiency partially increased chromatin accessibility and histone acetylation in these regions. Ikzf3 deficiency in mice enhanced susceptibility to colitis, underscoring the relevance of Aiolos in regulating the effector function in IELs.
Similar content being viewed by others
Main
The intestinal epithelium is home to a large population of T cells, known as intestinal intraepithelial lymphocytes (IELs), as well as fewer intraepithelial innate lymphoid cells (ILCs)1,2,3. IELs encompass both conventional and unconventional T cell populations (hereafter cIELs and uIELs, respectively). cIELs include T cell antigen receptor (TCR) αβ+ T cells that express CD4 (CD4+ cIELs) or CD8αβ (CD8αβ+ cIELs) heterodimers, which home to the gut epithelium in response to antigenic activation in the periphery4,5. A fraction of CD4+ cIELs acquire cell surface expression of CD8αα and a cytolytic program, and are referred to as double-positive T cells (DP IELs). uIELs encompass γδ T cells (γδ uIELs) and TCRαβ+ cells that express CD8αα homodimers, but lack CD4 and CD8αβ and are referred to as double-negative IELs (DN uIELs)2,4,5. Intraepithelial ILCs mainly include type 1 ILCs (ILC1s), which produce interferon (IFN)-γ and release secretory lysosomes containing perforin and granzymes, particularly granzyme C6. ILC1s include a major subset of NKp46+NK1.1+ cells and a smaller subset of NKp46−NK1.1lo/− cells, which express Ly49E and are highly dependent on interleukin (IL)-15 (ref. 7). IELs and ILC1s maintain tolerance to nutritional and commensal antigens, protect the host from infections through release of cytolytic molecules, cytokines and chemokines, and facilitate repair of damaged intestinal epithelium1,4.
uIELs recognize ligands through TCRs5, but also exhibit innate-like features, and display rapid and stereotypical responses to tissue alarmins and stress-induced ligands of NK receptors independent of antigen recognition8. This binary status of IELs, often defined as ‘resting yet poised to activation’4,9, is essential to maintain homeostasis and host defense without causing unwarranted inflammation and intestinal diseases. In fact, the dysregulated activation of IELs contributes to the pathogenesis of celiac disease and inflammatory bowel diseases10,11,12. While mechanisms that induce activation of IELs have been extensively studied, those modulating IELs remain incompletely undefined.
The Ikaros zinc-finger (IKZF) family encompasses five transcription factors with roles in development, maturation and functions of immune cells13. These transcription factors form homodimerization or heterodimerization within the IKZF family members13. IKZF transcription factors interact with the nucleosome remodeling and deacetylase complex (NuRD), which deacetylates histones and restrains gene expression through histone deacetylase complex14,15. The IKZF family member Ikaros, encoded by Ikzf1, controls early lymphoid cell development. Ikaros-deficient mice have marked reduction of T cell, B cell and natural killer (NK) cell numbers and succumb to lymphoproliferative disorders16,17. Aiolos, encoded by Ikzf3, forms homodimers or heterodimers with Ikaros. Aiolos deficiency impacts NK cell maturation, B cell activation, type 17 helper T (TH17) cell development, cytokine production by type 2 ILCs (ILC2s) and eosinophil migration18,19,20,21,22,23. In humans, Aiolos represses the production of IL-22, the signature cytokine of type 3 ILCs (ILC3s), and facilitates ILC3 differentiation into intestinal intraepithelial ILC1s24,25.
In this Article, we found that Aiolos controlled the expression of genes encoding NK1.1 and other NK receptors, cytotoxic mediators and IFN-γ in uIELs, in part by attenuating IL-15 signaling. Epigenetic analyses showed that Aiolos bound to regulatory regions of these genes close to STAT5 and RUNX binding sites, modifying chromatin accessibility and histone acetylation. Thus, Aiolos controlled intestinal inflammation by modulating the effector programs of uIELs through epigenetic modifications likely coordinated with STAT5 and RUNX family members.
Results
Aiolos deficiency enhances uIEL activation program
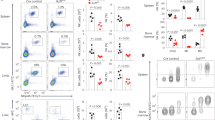
We initially compared the abundance and phenotype of intestinal intraepithelial ILC1s (live CD45+CD3−NK1.1+NKp46+ cells) in Ikzf3−/− and Ikzf3+/+ mice. The overall percentage of ILC1s was similar in the two groups (Fig. 1a). A more detailed analysis indicated a selective expansion of the NK1.1+NKp46− ILC1 subset in Ikzf3−/− compared to Ikzf3+/+ mice, whereas the NK1.1+NKp46+ ILC1 subset was unchanged (Fig. 1a). Because the NK1.1+NKp46− ILC1 subset contained very few cells, it was no longer considered for subsequent analyses. The total cell counts of cIELs and uIELs were similar in Ikzf3+/+ and Ikzf3−/− mice (Extended Data Fig. 1), but Ikzf3−/− uIELs acquired higher expression of the NK receptor NK1.1 compared to Ikzf3+/+ IELs (Fig. 1b). Phenotyping of IEL subsets in Ikzf3−/− mice indicated that NK1.1 was mainly induced in CD45+CD3+TCRγδ+ cells (hereafter γδ uIELs unless otherwise specified) and CD45+CD3+TCRγδ−TCRαβ+CD4−CD8β−CD8α+ cells (hereafter DN uIELs), as well as DP IELs (Fig. 1c). Intracellular staining in Ikzf3+/+ mice indicated that uIELs and DP IELs had the highest expression of Aiolos (Fig. 1d).
a, Representative flow cytometry plots showing expression of NK1.1 and NKp46 along with quantification of live CD45+CD3−NK1.1+NKp46+ and live CD45+CD3−NK1.1+NKp46− cells isolated from small intestine epithelium of Ikzf3+/+ and Ikzf3−/− mice (n = 6). b, Representative flow cytometry plots showing expression of NK1.1 and CD3 on CD45+ cells isolated from small intestine epithelium of Ikzf3+/+ and Ikzf3−/− mice. c, Representative flow cytometry plots and quantification of NK1.1 expression on live CD45+CD3+TCRγδ+ cells (γδ uIELs), live CD45+CD3+TCRγδ−TCRαβ+CD4−CD8β−CD8α+ cells (DN uIELs), live CD45+CD3+TCRγδ−TCRαβ+CD4+CD8β−CD8α+ cells (DP uIELs), live CD45+CD3+TCRγδ-TCRαβ+CD4+CD8β−CD8α− cells (CD4+ cIELs) and live CD45+CD3+TCRγδ−TCRαβ+CD4−CD8β−CD8α+ cells (CD8αβ+ cIELs) from small intestine epithelium of Ikzf3+/+ and Ikzf3−/− mice (n = 4). d, Expression (upper) and quantification (MFI, lower) of Aiolos in the indicated cell types as in c (n = 4). a–d, Data are representative of two independent experiments. All samples are biological replicates. Data represent mean ± s.d. FMO, fluorescence minus one; MFI, mean fluorescence intensity. P values were determined by two-tailed, unpaired Student’s t-tests in a and c, and by one-way analysis of variance (ANOVA) with Tukey’s multiple-comparisons test in d.
We used bulk RNA-sequencing (RNA-seq) analysis to investigate broader changes in the transcriptional program of Ikzf3−/− γδ uIELs and DN uIELs. DP IELs were not included because of their very low numbers. Ikzf3−/− γδ uIELs and Ikzf3−/− DN uIELs exhibited broad reprogramming and showed upregulation of NK receptors (encoded by Klra1, Klra3, Klra9, Klra13-ps, Klrb1b, Klrb1c, Klrc1, Klrk1), granzymes (encoded by Gzmc, Gzmk, Gzmf), various cytokines and chemokines (encoded by Ifng, Ccl4, Spp1) and the antiapoptotic molecule Bcl2 compared to Ikzf3+/+ counterparts (Fig. 2a). Ikzf3−/− γδ uIELs and DN uIELs also exhibited increased expression of checkpoint inhibitors and intracellular modulators that are induced by activation, such as Lair1, Lag3 and Socs3, compared to Ikzf3+/+ counterparts (Fig. 2a). We conclude that Aiolos represses the activation program of uIELs.
a, Volcano plots comparing gene expression in γδ uIELs and DN uIELs isolated from small intestine epithelium of Ikzf3+/+ and Ikzf3−/− mice. b. Representative flow cytometry plots and quantification of NK1.1 (n = 10), NKG2D (n = 4), NKG2A (n = 4), Ly49A (n = 4), Ly49H (n = 3), granzyme C (n = 10) and IFN-γ (n = 10) in γδ uIELs and DN uIELs isolated from small intestine epithelium of sub-lethally irradiated Rag1−/− intravenously injected with an equal mix of bone marrow from Ikzf3+/+ (CD45.1+) and Ikzf3−/− (CD45.2+) mice and analyzed 6 weeks after reconstitution. Data are representative of two independent experiments. All samples are biological replicates. P values were determined by two-tailed, unpaired Student’s t-test. In RNA-seq, FDR was determined by two-sided Wald test (DESeq2). Data represent the mean ± s.d. FDR, false discovery rate.
Aiolos modulation of uIELs is cell intrinsic
To investigate whether the effect of Aiolos deficiency in uIELs was cell intrinsic, we co-injected bone marrow from congenically marked Ikzf3+/+ mice (CD45.1+) and Ikzf3−/− mice (CD45.2+) into sub-lethally irradiated Rag1−/− mice. At week 6 after transplant, expression of the NK receptors NK1.1, NKG2A and NKG2D, as well as the intracellular content of granzyme C and IFN-γ were higher in Ikzf3−/− uIELs than Ikzf3+/+ uIELs (Fig. 2b). Ly49A was more expressed in Ikzf3−/− DN uIELs compared with Ikzf3+/+ uIELs, but not expressed in Ikzf3−/− and Ikzf3+/+ γδ uIELs (Fig. 2b). Other Ly49 receptors, such as Ly49D and Ly49H, were absent in both Ikzf3+/+ and Ikzf3−/− uIELs (Fig. 2b). Thus, Aiolos restrained the expression of NK receptors, cytotoxic enzymes and IFN-γ in γδ uIELs and DN uIELs in a cell-intrinsic manner. Ikzf3−/− uIELs also had enhanced expression of NKG2D (encoded by Klrk1) compared to Ikzf3+/+ uIELs (Fig. 2b and Extended Data Fig. 2a). To test whether enhanced NKG2D expression mediated uIEL cytotoxicity, we sorted γδ and DN uIELs from Ikzf3+/+ and Ikzf3−/− mice and incubated them with RMA-S cells or RMA-S cells overexpressing Rae1γ, a ligand for NKG2D. Ikzf3+/+ or Ikzf3−/− uIELs did not lyse the RMA-S cells in an NKG2D-dependent manner (Extended Data Fig. 2b). We conclude that enhanced NKG2D expression in Ikzf3−/− uIELs was insufficient to induce NKG2D-mediated cytotoxicity, consistent with reports that NKG2D only mediates costimulation in skin γδ T cells26.
Aiolos reprograms the Tcf7 − Il2rb hi subset of CD8αα+ uIELs
We sought to obtain a high-resolution view of the impact of Aiolos deficiency on individual IEL subsets by single-cell RNA-seq (scRNA-seq). We sorted live CD45+ cells from the small intestine epithelium in Ikzf3+/+ and Ikzf3−/− mice and performed scRNA-seq. We profiled a total of 49,205 quality-controlled cells that were visualized using the uniform manifold approximation and projection (UMAP) method. Based on differentially expressed genes (DEGs), we annotated clusters corresponding to CD8αα+ uIELs, CD4+ cIELs, CD8αβ+ cIELs, NK-ILC1s, plasma cells, B cells and plasmacytoid dendritic cells (Extended Data Fig. 3a,b). Trac and Trbc1, encoding TCRα constant gene segment and TCR beta constant 1, respectively, and Tcrg-V1 and Tcrg-V7, encoding the most prevalent TCRγ chains among γδ uIELs were expressed in the same CD8αα+ uIEL clusters (Extended Data Fig. 3a,c), indicating a substantial overlap of γδ uIEL and DN uIEL programs, despite the expression of different TCRs.
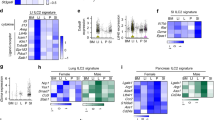
After CD8αα+ uIELs computational reclustering, we could distinguish three major clusters: Tcf7+ uIELs, Tcf7−Il2rbhi uIELs and Tcf7−Il2rblo uIELs (Fig. 3a). The profile of Tcf7+ uIELs was biased toward the expression of Id3, Ltb, Kit27, Cd160 (ref. 28 and Xcl1 (ref. 29) (Fig. 3b and Extended Data Fig. 4a), indicating a functional program typical of stem-like or memory CD8+ T cells30. Conversely, Tcf7− uIELs expressed genes indicative of effector function, such as Prdm1, Gzma, Gzmb and Ifng, as well as Klrk1 and Hcst (encoding NKG2D and its adaptor DAP10, respectively31; Fig. 3b). Tcf7− uIELs were distinguished into Il2rbhi and Il2rblo uIELs based on expression of Il2rb, which encodes CD122, the β-subunit of IL-2 receptor shared by the receptor for IL-15 and IL-2 (Fig. 3b and Extended Data Fig. 4a). Il2ra was not expressed (Extended Data Fig. 4b). Tcf7−Il2rblo uIELs preferentially expressed Il7r, Ccl3 and Ccl4, and the signaling adaptors encoded by Lat, Lat2 and Gzmk (Fig. 3b and Extended Data Fig. 4a,b). Genes preferentially expressed in Tcf7−Il2rbhi uIELs included Tyrobp32, which encodes the signaling adaptor DAP12, utilized by activating NK and myeloid receptors, Gzmc, which encodes granzyme C, a marker of CD49a+CD103+ ILC1s in gut and tumors33,34,35 and various genes, including Lag3, Cd200r4, Cd200r1 and Cd200r2 (Fig. 3b and Extended Data Fig. 4a, b). Conversely, Thy1 expression was relatively low in Tcf7−Il2rbhi uIELs compared to Tcf7−Il2rblo or Tcf7+ uIELs (Fig. 3b and Extended Data Fig. 4b). Because IL-15 is known to induce DAP12 (ref. 36) and granzyme C33, and to downregulate Thy1 (ref. 37), the profile of Tcf7−Il2rbhi uIELs was consistent with a strong IL-15 imprinting. We also identified four small CD8αα+ clusters: lipolytic CD8αα+ uIELs expressed fatty acid metabolic genes (Fabp1, Fabp2, Rbp2); proliferating CD8αα+ uIELs were enriched in genes associated with cell cycling (Mki67, Tuba1b, Tubb5); IFN-stimulated CD8αα+ uIELs expressed IFN-stimulated genes (Ifit1, Ifit3, Isg15, Irf7, Stat1); and stressed CD8αα+ uIELs were enriched in stress-induced genes (mt-Co2, Hsph1, Hst1h1c, Hspa1b; Fig. 3b and Extended Data Fig. 4a).
a, UMAP plots of combined CD8αα+ uIELs isolated from Ikzf3+/+ and Ikzf3−/− cells (Ikzf3+/+.1, 11,843 cells; Ikzf3+/+.2, 14,123 cells; Ikzf3−/−.1, 9,730 cells; Ikzf3−/−.2, 13,509 cells). b, Representative UMAP plots depicting expression of Trac, Trbc1, Tcrg-V7, Tcf7, Id3, Kit, Cd160, Xcl1, Il2rb, Prdm1, Gzmb, Ifng, Klrk1, Hcst, Ccl3, Ccl4, Gzmk, Tyrobp, Gzmc, Thy1 and Klra5. c, Separate UMAP plots of Ikzf3+/+ CD8αα+ uIELs and Ikzf3−/− CD8αα+ uIELs. Clusters of color codes as in a. d, Cluster representation in Ikzf3+/+ CD8αα+ uIELs and Ikzf3−/− CD8αα uIELs from four biological samples (mice) analyzed. e, Volcano plots of genes differentially expressed in Ikzf3+/+ and Ikzf3−/− Tcf7−Il2rbhi uIELs, Tcf7−IL2rblo uIELs andTcf7+ uIELs. FDR was determined by two-sided Wald test (DESeq2).
Comparison of scRNA-seq data from Ikzf3+/+ and Ikzf3−/− CD8αα+ uIELs revealed that Tcf7−Il2rbhi formed a distinct cluster in each genotype (Fig. 3c,d). Additionally, Tcf7−Il2rblo were less abundant in Ikzf3−/− than in Ikzf3+/+ mice (Fig. 3c,d), whereas other clusters were less affected by Aiolos deficiency (Fig. 3d). Comparison of Tcf7−Il2rbhi uIELs from Ikzf3−/− mice versus Ikzf3+/+ mice indicated 431 unique DEGs (Fig. 3e). Upregulated genes in Ikzf3−/− uIELs included Ccl4, Ifng, granzyme genes (Gzmc, Gzmd, Gzmf, Gzmk) and NK receptor genes (Klra1, Klra3, Klrb1b, Klrb1c; Fig. 3e). Comparison of Tcf7−−Il2rblo uIELs from Ikzf3−/− and Ikzf3+/+ mice showed 75 DEGs (Fig. 3e). In total, 102 genes were differentially expressed in Ikzf3−/− versus Ikzf3+/+ Tcf7+ uIELs (Fig. 3e). In all three clusters, effector genes such as granzymes and NK receptors were upregulated in Ikzf3−/− uIELs compared to Ikzf3+/+ uIELs although the impact of Ikzf3 deletion was most pronounced in the Tcf7−Il2rbhi cluster (Fig. 3e). In summary, Aiolos deficiency preferentially affected Tcf7−Il2rbhi CD8αα+ uIELs, enhancing gene expression programs related to activating receptors and effector functions.
Aiolos-deficient IELs are hyperresponsive to IL-15
We analyzed intestinal CD45+ cells by flow cytometry to validate the protein expression of TCF1 (encoded by Tcf7) and CD122 (encoded by Il2rb) in CD8αα+ uIELs in Ikzf3+/+ and Ikzf3−/− mice. Gated CD8αα+ uIELs of both genotypes encompassed three cell subsets: TCF1+ uIELs, TCF1−CD122hi uIELs, and TCF1−CD122lo uIELs (Fig. 4a). The highest differential expression of NK1.1 between Ikzf3−/− and Ikzf3+/+ uIELs was detected on TCF1−CD122hi uIELs (Fig. 4b). This observation, together with the fact that exposure to IL-15 induces expression of NK cell receptors on uIELs38, indicates that loss of Aiolos impacts uIEL responsiveness to IL-15.
a, Representative flow cytometry plots showing CD122 and TCF1 expression in γδ uIELs and DN uIELs isolated from the small intestine epithelium of Ikzf3+/+ mice (n = 5). b, Flow cytometry plots and quantification of NK1.1+ γδ uIELs and NK1.1+ DN uIELs in the small intestine epithelium of Ikzf3+/+ and Ikzf3−/− mice (n = 5). c, Flow cytometry plots and quantification of NK1.1+ γδ uIELs and NK1.1+ DN uIELs in the duodenum, jejunum and ileum of Ikzf3+/+ and Ikzf3−/− mice (n = 5). d, Flow cytometry plots and quantification of NK1.1+ γδ uIELs and NK1.1+ DN uIELs isolated from the small intestine epithelium of Ikzf3+/+ and Ikzf3−/− mice 72 h after in vitro culture with 10 ng ml−1 or 100 ng ml−1 of IL-15. a–d, Data are representative of two independent experiments and all samples are biological replicates. Data represent the mean ± s.d. P values were determined by one-way ANOVA with Tukey’s multiple-comparisons test in b and c, and by two-way ANOVA with Tukey’s multiple-comparisons test in d.
Next, we purified CD45+ cells from different segments of the small intestines of Ikzf3+/+ and Ikzf3−/− mice and analyzed differential expression of NK1.1 on gated uIELs. The expression of NK1.1 in Ikzf3−/− mice was highest in uIELs of the duodenum, and progressively declined in uIELs of distal segments (Fig. 4c). This pattern directly correlated with the expression of mRNA for IL-15 (encoded by Il15) and IL-15Rα (encoded by Il15ra), which are required for IL-15 signaling39, in the segments of the small intestine (Extended Data Fig. 5a), and suggested increased trans-presentation of IL-15 and IL-15 signaling in the duodenum compared to the jejunum and ileum.
To test whether Aiolos deficiency enhanced the responsiveness to IL-15, we purified CD45+ cells from the whole small intestine of Ikzf3+/+ and Ikzf3−/− mice, and cultured these cells with IL-15. Flow cytometric analysis of γδ uIELs and DN uIELs showed that IL-15 induced the expression of NK1.1 in a dose-dependent fashion, and Ikzf3−/− uIELs expressed higher NK1.1 than Ikzf3+/+ uIELs at all IL-15 concentrations (Fig. 4d). IL-15 induced the expression of NKG2A in Ikzf3−/− uIELs, but not in Ikzf3+/+ uIELs (Extended Data Fig. 5b). IL-15 also induced Ly49A in Ikzf3−/− DN uIELs but not in Ikzf3−/− γδ uIELs, Ikzf3+/+ DN uIELs or Ikzf3+/+ γδ uIELs (Extended Data Fig. 5c). IL-15 did not induce expression of NKG2D on any subset of Ikzf3+/+ or Ikzf3−/− uIELs (Extended Data Fig. 5d). In summary, Ikzf3−/− uIELs acquired a heightened response to IL-15, which might account for some of the observed phenotype in vivo, although some phenotypes were probably independent of IL-15 responsiveness.
Aiolos-deficient uIELs are detrimental in colitis
We asked whether inflammatory conditions that activate IELs coordinately reduced Aiolos expression. We used intraperitoneal injection of polyinosinic:polycytidylic acid (poly-I:C) as a model of inflammation. Poly-I:C causes systemic inflammation, colitis and IEL activation38,40 by inducing massive release of proinflammatory cytokines, including type I interferon (IFN) and IL-12 (ref. 41). At day 1 after injection of poly-I:C in Ikzf3+/+ mice, Aiolos expression was significantly downregulated in γδ uIELs and DN uIELs compared to uIELs from vehicle-treated mice (Fig. 5a). To further define the impact of inflammatory cytokines on Aiolos expression, sorted γδ uIELs and DN uIELs were cultured with IL-2 and IL-15 to support survival in vitro and stimulated with IL-12 or IFN-β for 3 d. Both stimuli downregulated Aiolos expression in uIELs compared to the uIELs cultured with IL-2 and IL-15 alone (Fig. 5b), indicating that inflammatory conditions downregulated Aiolos expression.
a, Flow cytometry histograms and quantification of Aiolos protein in γδ uIELs and DN uIELs isolated from the small intestine epithelium of Ikzf3+/+ mice (n = 7) 24 h after injection with either PBS or poly-I:C. b, Flow cytometry histograms and quantification of Aiolos protein in γδ uIELs and DN uIELs isolated from the small intestine of Ikzf3+/+ mice (n = 6) and cultured with either IL-2 + IL-15, IL-2 + IL-15 + IL-12, or IL-2 + IL-15 + IFN-β for 72 h before analysis. c,d, Representative flow cytometry plots (c) and quantification (d) of NK1.1 and granzyme C protein expression in γδ uIELs and DN uIELs isolated from the small intestine epithelium of Ikzf3fl/fl and E8ICreIkzf3fl/fl mice (n = 6). e, Frequency of hematochezia-free Ikzf3fl/fl and E8ICreIkzf3fl/fl mice during day 0–5 after administration of DSS in the drinking water. f,g, Quantification of colon length (f) and representative images of H&E-stained sections of the colon (g) from Ikzf3fl/fl and E8ICreIkzf3fl/fl mice at day 4 after DSS administration in the drinking water (n = 9). a–g, Data are representative of two independent experiments and all samples are biological replicates. Data represent the mean ± s.d. Scale bar, 100 μm (g). P values were determined by two-tailed, unpaired Student’s t-test in a, b, d and f, and log-rank test in e.
To test whether Aiolos-deficient uIELs altered host responses to local inflammation in the gut, we used a classical model of acute colitis induced by administration of dextran sulfate sodium (DSS) in drinking water. DSS damages the epithelium and induces colitis42. To selectively delete Ikzf3 in CD8α+ T cells, we crossed Ikzf3fl/fl mice (Extended Data Fig. 6a) with E8ICre mice, which express Cre recombinase under the core E8I enhancer of Cd8a gene. We validated that E8I-driven Cre activity preferentially occurred in cells that express CD8α in the periphery43 by crossing E8ICre mice with Rosa26RtdTomato mice. E8ICre released tdTomato expression in γδ uIELs, DN uIELs, CD8αβ+ cIELs and DP IELs, but not in CD4+ cIELs, B cells or NK/ILC1s isolated from small intestine of E8ICreRosa26RtdTomato mice (Extended Data Fig. 6b), validating that E8ICre preferentially deleted the genes from uIELs. We then analyzed the phenotype of uIELs in E8ICre Ikzf3fl/fl mice. E8ICre Ikzf3fl/fl uIELs had higher expression of NK1.1 and granzyme C compared to Ikzf3fl/fl uIELs (Fig. 5c,d), recapitulating the phenotype of Ikzf3−/− uIELs. After Ikzf3fl/fl and E8ICreIkzf3fl/fl mice were administered DSS in the drinking water, E8ICreIkzf3fl/fl mice exhibited more blood in stools (Fig. 5e), shorter colon length (Fig. 5f and Extended Data Fig. 6c), more prominent ulcers and higher crypt damage (Fig. 5g) than Ikzf3fl/fl mice at day 4 of treatment, indicating that Aiolos deficiency in CD8α+ cells aggravates chemically induced colitis.
Aiolos binds regulatory regions of IEL effector genes
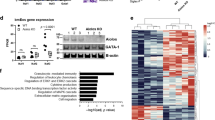
To elucidate the mechanisms underpinning the transcriptional reprogramming of Ikzf3−/− uIELs, we profiled the Aiolos binding sites in γδ uIELs and DN uIELs isolated from Ikzf3+/+ and Ikzf3−/− mice using CUT&RUN-seq (cleavage under targets and release using nuclease sequencing). Around 64% and 66% of Aiolos binding sites in γδ uIELs and DN uIELs, respectively, were found in promoter regions (Extended Data Fig. 7a). We identified a total of 7,284 and 3,899 Aiolos binding peaks in γδ uIELs and DN uIELs, respectively, with 3,512 of those peaks shared between the two cell types (Extended Data Fig. 7b). Pathway analysis of genes associated with Aiolos binding sites showed enrichment in TCR signaling and NK cell-mediated cytotoxicity pathways (Fig. 6a). Characterization of motifs enriched in Aiolos binding sites using Homer de novo motif analysis exposed the IKZF common binding GGGAA motif (Fig. 6b)44,45, which was similar to the previously reported ETS motif46. Comparison of genes associated with Aiolos binding sites in the CUT&RUN-seq analysis with the DEGs in the RNA-seq analysis showed that Aiolos bound 155 genes of the 343 DEGs in γδ uIELs, and 70 genes of the 396 DEGs in DN uIELs (Fig. 6c). These genes included NK cell receptors (Klrb1b, Klrb1c), cytotoxic enzymes (Gzmc), chemokines (Ccl3, Ccl4) and previously known Aiolos target genes such as Irf8 (ref. 47,48; Fig. 6c).
a, Pathway analysis of Aiolos-bound genes derived from CUT&RUN-seq for Aiolos using γδ uIELs and DN IELs isolated from small intestine epithelium of Ikzf3+/+ mice. b, Homer de novo motif prediction in genome-wide Aiolos peaks. Left, de novo motifs; middle, closest match in the HOMER database; right, P value of motif enrichment. c, Venn diagrams showing the overlap between Aiolos-bound genes detected by CUT&RUN-seq and DEGs detected by RNA-seq in γδ uIELs and DN uIELs isolated from the small intestine epithelium of Ikzf3+/+ mice. d, Venn diagrams showing the number of overlapping peaks between Aiolos and STAT5. Aiolos peaks were derived from CUT&RUN-seq for Aiolos using γδ uIELs and DN IELs isolated from small intestine epithelium of Ikzf3+/+ mice. STAT5 peaks were sourced from GSE100674. e, IGV snapshots depicting Aiolos peaks derived from CUT&RUN-seq for Aiolos in γδ uIELs and DN IELs isolated from the small intestine epithelium of Ikzf3+/+ mice and STAT5 peaks sourced from GSE100674. f, Venn diagram showing the numbers of overlapping peaks between Aiolos peaks derived from CUT&RUN-seq for Aiolos in γδ uIELs and DN IELs isolated from small intestine epithelium of Ikzf3+/+ mice and either RUNX1 or RUNX3 peaks sourced from GSE93755 or GSE50131, respectively. IGV, Integrative Genomics Viewer. P values were calculated by hypergeometric enrichment calculations in a and b.
Because Aiolos restrains IL-15-driven IEL activation and IL-15 signaling triggers the activation of STAT5, we inquired whether STAT5 and Aiolos bound DNA in close proximity of each other. Comparison of genome-wide distribution of STAT5 and Aiolos to identify regions of overlap indicated that 41% and 39% of the Aiolos binding regions were also bound by STAT5 in γδ uIELs and DN uIELs, respectively, with 2,958 overlapping peaks among 7,284 Aiolos binding peaks in γδ uIELs and 1,527 peaks among 3,899 Aiolos binding peaks in DN uIELs (Fig. 6d). These overlaps were statistically significant (Extended Data Fig. 7d). Aiolos-STAT5 overlapping regions were located in the proximity of NK receptors (Klrb1b, Klrb1c), granzyme B (Gzmb) and chemokines (Ccl3, Ccl5; Fig. 6e). The intracellular content of phosphorylated STAT5 (p-STAT5) was not significantly different between Ikzf3−/− and Ikzf3+/+ γδ uIELs and DN uIELs (Extended Data Fig. 7c), suggesting Aiolos affected STAT5-driven transcription, but not STAT5 phosphorylation. Moreover, Aiolos binding regions overlapped with the binding sites of other transcription factors and were specifically enriched in RUNX motifs (Fig. 6b). Comparison of the genome-wide distribution of Aiolos and RUNX binding sites showed that among total Aiolos binding peaks, 4,250 and 2,289 overlapped with RUNX1 in γδ uIELs and DN uIELs respectively, while 726 and 351 overlapped with RUNX3 in γδ uIELs and DN uIELs, respectively (Fig. 6f). The binding regions shared by Aiolos with either RUNX1 or RUNX3 were statistically significant (Extended Data Fig. 7d). In summary, these data suggest that Aiolos might cooperate with STAT5 and RUNX family members in regulating gene expression at certain genomic loci.
Aiolos promotes histones deacetylation of IEL effector genes
Aiolos homodimers and heterodimers with Ikaros19 interact with the NuRD complex to recruit histone deacetylases14,15. Thus, we used CUT&RUN-seq to profile acetylated histone H3 Lys27 (H3K27ac), a histone marker associated with active transcription. In parallel, we also determined global changes of chromatin accessibility in the same uIEL subsets using the assay for transposase-accessible chromatin sequencing (ATAC-seq). Principal component analysis of these epigenetic features showed a segregation of γδ uIELs and DN uIELs based on the Ikzf3 genotype (Fig. 7a), indicating that Aiolos deficiency impacted the epigenetic landscape. Analysis of H3K27ac peaks showed significantly higher acetylation (5,704 higher peaks versus 72 lower peaks) in Ikzf3−/− compared to Ikzf3+/+ γδ uIELs (Fig. 7b). Among the differentially acetylated peaks, 1,888 were located in promoter regions (8 peaks in Ikzf3+/+ γδ uIELs compared to 1,880 peaks in Ikzf3−/− γδ uIELs) and 3,688 in the enhancer regions (64 peaks in Ikzf3+/+ γδ uIELs versus 3,624 peaks in Ikzf3−/− γδ uIELs; Fig. 7b). A similar trend was observed in H3K27ac peaks in Ikzf3+/+ and Ikzf3−/− DN uIELs, with 57 differentially acetylated peaks in the promoter regions (1 peak in Ikzf3+/+ DN uIELs compared to 56 peaks in Ikzf3−/− DN uIELs) and 442 peaks in the enhancer regions (36 peaks in Ikzf3+/+ DN uIELs compared to 406 peaks in Ikzf3−/− DN uIELs; Fig. 7b). Differential H3K27ac peaks were more frequent in Ikzf3−/− γδ uIELs than in Ikzf3−/− DN uIELs (Fig. 7b), probably due to a compensatory upregulation of Helios, encoded by Ikzf2, in Ikzf3−/− DN uIELs (Extended Data Fig. 8a) that could attenuate the effects of Aiolos deficiency.
a, Principal component analysis of the ATAC-seq peaks and H3K27ac peaks in γδ uIELs and DN uIELs isolated from the small intestine epithelium of Ikzf3+/+ and Ikzf3−/− mice. b, Total numbers of differential H3K27ac regions in γδ uIELs and DN uIELs as in a. c, Total numbers of differentially accessible (DA) regions in γδ uIELs and DN uIELs in the small intestine epithelium in Ikzf3+/+ and Ikzf3−/− mice. d, IGV snapshots of representative loci depicting CUT&RUN-seq for H3K27ac and ATAC-seq in Ikzf3+/+ and Ikzf3−/− γδ uIELs and DN uIELs. e, Heatmap showing the transcription factor motif enrichment in differentially acetylated or differentially accessible regions in Ikzf3+/+ and Ikzf3−/− γδ uIELs and DN uIELs. P values were calculated by hypergeometric enrichment calculations and the log-transformed P values were z-scored. f,g, Plots showing fold changes in gene expression in RNA-seq versus fold changes in H3K27ac peaks (f) or ATAC peaks (g) in the promoter and enhancer regions of γδ uIELs and DN uIELs isolated from the small intestine epithelium of Ikzf3+/+ and Ikzf3−/− mice. Highlighted points indicate FDR < 0.05 and fold change > 1.5 or < −1.5 in both RNA-seq and H3K27ac peaks/ATAC peaks. The line represents a linear regression fit to the highlighted points, with the Pearson correlation coefficient (r) indicating the strength and direction of the linear relationship between the two variables. FDR was calculated by Wald test (DEseq2). P, promoter; E, enhancer; FC, fold change.
Next, we examined the differentially accessible chromatin regions in Ikzf3+/+ and Ikzf3−/− γδ uIELs and DN uIELs. A total of 2,311 peaks were differentially accessible between Ikzf3+/+ and Ikzf3−/− γδ uIELs (Fig. 7c), of which 437 were located in the promoter regions (28 peaks in Ikzf3+/+ γδ uIELs versus 409 peaks in Ikzf3−/− γδ uIELs), and 1,874 in the enhancer regions (311 peaks in Ikzf3+/+ γδ uIELs versus 1,563 peaks in Ikzf3−/− γδ uIELs; Fig. 7c). In DN uIELs, we observed 2,423 differential accessible peaks, of which 261 were in promoter regions (103 peaks in Ikzf3+/+ DN uIELs versus 158 peaks in Ikzf3−/− DN uIELs) and 2,162 in enhancer regions (921 peaks in Ikzf3+/+ DN uIELs versus 1,241 peaks in Ikzf3−/− DN uIELs; Fig. 7c).
To explore the relationship between the differential H3K27ac peaks, differential accessibility regions (DARs) and Aiolos binding regions, we performed an integrated analysis. Significant overlaps were observed between differential H3K27ac peaks and DARs in γδ uIELs and DN uIELs (Fig. 7d and Extended Data Fig. 8b). To address whether Aiolos affected the accessibility of other transcription factors, thereby influencing gene expression indirectly, we determined which transcription factor motifs were enriched in the DARs and differential H3K27ac peaks. Transcription factor motifs including RUNX, BATF and Tbx21 were more highly represented in Ikzf3−/− γδ uIELs and DN uIELs compared to their Ikzf3+/+ counterparts (Fig. 7e).
Lastly, to determine how changes in the epigenetic landscape due to Aiolos deficiency contributed to differential gene expression, we annotated the genes associated with differentially accessible and acetylated peaks, and analyzed their correlations with DEGs in RNA-seq. Positive correlations were observed between DEGs and differential H3K27ac peaks both in the promoter and enhancer regions (Fig. 7f). H3K27ac peaks in Ikzf3−/− γδ uIELs and DN uIELs correlated with genes upregulated in Ikzf3−/− γδ uIELs and DN uIELs, including those for NK receptors (Klrb1b, Klrb1c, Klrk1), chemokines (Ccl1, Ccl3) and granzyme C (Gzmc; Fig. 7f). Chromatin regions more accessible in Ikzf3−/− uIELs correlated with NK receptors (Klrb1b, Klrb1c), chemokines (Ccl3) and known Aiolos target genes, such as Irf8 (Fig. 7g), indicating that differential ATAC peaks in the promoter and enhancer regions also correlated with DEGs. Integration of Aiolos, STAT5, RUNX1 and RUNX3 binding data, DARs, and differential H3K27ac indicated that these features overlapped in multiple genes expressed in IELs, including NK receptors, chemokines and granzymes (Supplementary Table 2). Taken together, Aiolos modulated chromatin accessibility, promoted histone deacetylation and likely interfered with other transcription factors to regulate expression of IEL effector genes including NK receptors, chemokines and granzymes.
Discussion
Intestinal IELs are often described as innate-like cells that are resting, yet poised for activation, and therefore able to provide an immediate response to tissue injury. Here we showed that Aiolos had a critical role in restraining uIEL expression of NK receptors, cytotoxic mediators, chemokine and IFN-γ, in part by interfering with their responsiveness to IL-15 signaling. This phenotype was particularly evident in uIELs marked by high expression of CD122, the receptor for IL-15, which also expressed high amounts of Aiolos. We also showed that inflammatory cytokines that activate IELs, such as IL-12 and type I IFN, attenuated Aiolos expression. Thus, we predict that downregulation of Aiolos in response to these cytokines might contribute to activating poised regulatory elements and unleash IEL effector programs. These observations may help explain the reported association between inflammatory bowel disease and Aiolos expression49.
Epigenetic analyses showed that Aiolos bound the regulatory regions of genes that encode NK receptors, effector molecules and inhibitory checkpoints, reducing accessibility and H3K27 acetylation of these chromatin regions. Aiolos DNA binding sites significantly overlapped with those of STAT5, the major signaling effector of IL-15. Thus, Aiolos may attenuate STAT5-mediated transcriptional activation. This possibility is consistent with reports showing that Ikaros, a heteromeric partner of Aiolos, antagonizes the STAT5 network50,51 and a report showing that Aiolos mitigates CD4+ T cell effector functions by antagonizing IL-2–STAT5 signaling52. However, Aiolos DNA binding sites also overlapped with RUNX binding elements, suggesting that Aiolos may additionally interfere with the activity of RUNX transcription factors, such as RUNX3, which also promotes cytotoxic T cell effector programs.
Of note, despite its increased expression in Ikzf3−/− uIELs, the NKG2D receptor did not trigger uIEL cytotoxicity alone. Further studies are warranted to explore the impact of Aiolos on NK receptors in uIELs beyond their expression, including their signaling pathways. The ability of Aiolos to interfere with IL-15 signaling is singularly appropriate to maintain intestinal immune homeostasis, as IL-15 is produced in high amounts in the intestinal mucosa and unrestricted IL-15 signaling can cause pathogenic intestinal inflammation as typified by celiac disease53.
This study highlights Aiolos-dependent regulatory mechanisms in uIEL function through epigenetic modification likely coordinated with STAT5 and RUNX family members, and its pivotal roles in maintaining intestinal homeostasis and preventing inflammation.
Methods
Mice
Aiolos germline deletion mice (Ikzf3−/−) were obtained from RIKEN. Aiolos conditional knockout mice (Ikzf3fl/fl) were generated at the Mouse Genetic Core at Washington University School of Medicine. Briefly, LoxP sites were inserting upstream of exon 4 and downstream of exon 6, to delete exons 4–6. Rag knockout mice (strain no. 002216) and CD45.1 mice (strain no. 002014), E8ICre mice (strain no. 008766), Cg-Gt(Rosa)26Sorm14(CAG-tdTomato)Hze/J (CAG promoter driven tdTomato expression is prevented by loxP-flanked stop cassette and this construct is inserted into the Rosa26 locus; strain number 007914) were purchased from The Jackson Laboratory. For all experiments, mice were maintained in a specific pathogen-free facility at Washington University in St. Louis (ambient temperature of 68–74 °F, 30–70% humidity, 12-h light/dark cycle). All the experiments on mice were approved by and performed in accordance with the Institutional Animal Care and Use Commnittee at Washington University in St. Louis.
DSS colitis
Mice were fed with 3% DSS in the drinking water and occult blood in stool was checked by Hemoccult II Sensa kit (Fisher Scientific SK-64130). DSS was purchased from TdB Labs. Dissected full-length colons were cleaned of luminal content, open lengthwise and pinned out. Tissues for paraffin embedding were fixed in 10% neutral buffered formalin solution (MilliporeSigma) overnight at 4 °C. Fixed tissues were washed three times in 70% ethanol and embedded in 2% agar (MilliporeSigma), followed by paraffin embedding, sectioning and H&E staining.
Flow cytometry and antibodies
Single-cell suspensions were incubated with Fc block for 15 min then stained with antibodies. For intracellular cytokine staining, BD Bioscience Fixation/Permeabilization Solution kit was used according to the manufacturer’s instructions. For transcription factor staining, True-Nuclear Transcription factor staining kit (BioLegend) was used according to the manufacturer’s instructions. For pSTAT5 staining, stimulated IELs were first fixed with 4% paraformaldehyde then permeabilized with 90% methanol. Data were acquired on a BD FACSymphony and analyzed using FlowJo. The following antibodies were used: anti-CD3 (145-2C11, BioLegend), anti-CD4 (GK1.5, BioLegend), anti-CD8α (53-6.7, BioLegend), anti-CD8b (YTS156.7.7, BioLegend), anti-CD122 (TM-B1, BioLegend), anti-NK1.1 (PK136, BioLegend), anti-NKG2A (16A11, BioLegend), anti-NKG2D (CX5, BioLegend), anti-granzyme C (SFC1D8, BioLegend), anti-IFN-γ (XMG1.2, BioLegend), anti-TCRb (H57-597, BioLegend), anti-TCRd (GL3, BioLegend), anti-TCF1 (C63D9, Cell Signaling), anti-Rabbit IgG (Poly4064, BioLegend), pSTAT5 (clone 47/Stat5 (p694) BD Biosciences), Ly49D (4E5, BioLegend), Ly49A/D (12A8, eBioscience), Ly49E/F (CM4, eBioscience) and Ly49H (3D10, BioLegend). For flow cytometry, all antibodies were diluted 1 to 100.
Tissue preparation
IELs were prepared as previously described54. Briefly, small intestines were flushed and Peyer’s patches were removed. Then, small intestines were sliced and agitated in HBSS, HEPES, dithioerythritol (Sigma) and bovine calf serum. The supernatant was filtered through 100-μm mesh and subjected to density gradient using 40% and 70% Percoll.
Poly-I:C treatment
Poly-I:C was purchased from Invivogen (tlrl-picw) and dissolved in pathogen-free PBS. Around 20 μg per gram body weight of Poly-I:C was given intraperitoneally. Twenty-four hours after the treatment, IELs were harvested.
IELs in vitro culture
When cultured in vitro to analyze cytokine production and response to IL-12, IL-15 or IFN-β, IELs were purified with EasySep Mouse CD45 Positive Selection kit (STEMCELL Technologies, 18945). For cytokine stimulation, a 96-well flat plate was coated with anti-CD3 antibody (5 μg ml−1) overnight, then washed three times with sterile PBS. Isolated IELs were stimulated with plate-bound CD3 for 20 h, and the last 4 h with Brefeldin A (BD GolgiPlug). When analyzing the IEL response to IL-12 (10 ng ml−1), IL-15 (10 or 100 ng ml−1) and IFN-β (1,000 units/ml), cytokines were added to the RPMI medium and purified IELs were co-cultured for 72 h. When analyzing pSTAT5, purified IELs were cultured with IL-15 (100 ng ml−1) in RPMI medium for 15 min.
Mixed bone marrow chimera
Recipient Rag-deficient mice underwent 6-Gy irradiation. Bone marrow was harvested from Aiolos-deficient mice and CD45.1 mice, then mixed at a 1:1 ratio. In total, 1 × 106 mixed bone marrow cells were injected into sub-lethally irradiated Rag-deficient mice through the retro-orbital route. At weeks 6–8 after the injection, IELs were harvested from the recipient mice.
CUT&RUN-seq
CUT&RUN-seq was performed using CUTANA ChIC/CUT&RUN kit (EpiCypher, 14-1048) following the manufacturer’s instructions. For each procedure, 1 μg of antibodies and sorted 0.5 × 106 cells were used. Anti-H3K27 (EpiCypher, 13-0045), anti-Aiolos (Sigma Aldrich, 9D10) and rabbit-anti-mouse immunoglobulin G (included in the kit, EpiCypher, 13-0045) were used. Libraries are generated using NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB, E7645S) and NEBNext Multiplex oligonucleotides for Illumina (NEB, E7600S) following the manufacturer’s protocol. Total DNA integrity was determined using an Agilent Bioanalyzer. Then, paired-end sequencing of 75 base pairs in length was performed using a NovaSeq 6000.
ATAC-seq
A total of 50,000 cells were sorted and processed as per the manufacturer’s instructions (Nextera DNA Library Preparation Kit, Illumina) and previously described55. Total DNA integrity was determined using an Agilent Bioanalyzer. Paired-end sequencing of 150 base pairs in length was performed using a NovaSeq 6000.
Epigenome analysis
For STAT5, RUNX1 and RUNX3 binding analysis, published data from chromatin immunoprecipitation coupled with sequencing (ChIP–seq) were used (GSE100674, GSE93755, GSE50131). Sequencing data from ATAC-seq, CUT&RUN-seq and ChIP–seq were mapped onto mouse genome build mm9 using Bowtie2. Duplicate reads were removed using Picard’s MarkDuplicates. MACS2 was used for peak calling using a P-value threshold of 1 × 10−5. For CUT&RUN-seq for Aiolos, Aiolos-deficient cells were used as a control input source, and irreproducible discovery rate peaks were identified using a threshold of 0.05. BigWig tracks were visualized by IGV. A permutation test was performed using the ChIPpeakAnno library, and for each transcription factor, DAR and differential H3K27ac, we executed 1,000 permutations. As the background for the permutation, we utilized the total ATAC peak counts of wild-type cells. Differential accessible peaks and differential H3K27ac peaks were calculated, and principal component analysis was conducted using Diffbind R package. Promoter regions were defined as peaks that overlapped with a region that was +1 kb to −1kb from the transcription start sites. Peak annotation was conducted using the ‘annotatePeaks’ module in the HOMER package. De novo motif analysis and KEGG pathway analysis using CUT&RUN-seq for Aiolos were performed by the ‘findMotifsGenome’ module and the ‘annotatePeaks’ module respectively. Overlapping peaks were identified using BedTools intersect with settings --v and --wa. Graphs were generated using custom R code in Rstudio. Quality-control metrics are presented in Supplementary Table 2.
Single-cell RNA-seq
CD45+ cells were sorted and then cDNA was prepared after the GEM generation and barcoding, followed by the GEM-RT reaction and bead cleanup steps. Purified cDNA was amplified for 10–14 cycles before being cleaned up using SPRIselect beads. Samples were then run on a Bioanalyzer to determine the cDNA concentration. GEX libraries were prepared as recommended by the 10x Genomics Chromium Single Cell V(D)J Reagent Kits (v1 Chemistry) user guide with appropriate modifications to the PCR cycles based on the calculated cDNA concentration. For sample preparation on the 10x Genomics platform, the Chromium Single Cell 5′ Library and Gel Bead Kit v1 (PN-1000006), Chromium Single Cell A Chip Kit (PN-1000152) and Chromium Single Index Kit T (PN-1000213) were used. The concentration of each library was accurately determined through qPCR utilizing the KAPA library Quantification Kit according to the manufacturer’s protocol (KAPA Biosystems/Roche) to produce cluster counts appropriate for the Illumina NovaSeq 6000 instrument. Normalized libraries were sequenced on a NovaSeq 6000 S4 Flow Cell using the XP workflow and a 151 × 10 × 10 × 151 sequencing recipe according to the manufacturer’s protocol. A median sequencing depth of 50,000 reads per cell was targeted for each sample. Cell Ranger was used to align samples to the reference mm10 genome and generate gene–cell expression matrices. Downstream analysis including normalization, scaling and clustering was performed using Seurat package (v.4.0.3). Cells with fewer than 200 or more than 5,000 detected genes, or cells with the percentage of mitochondrial reads greater than 5% were excluded from the analysis. Filtered data were scaled by library size, and then log-transformed. Highly variable genes were selected using the FindVariableFeatures, and clustering was performed using FindClusters function. UMAP was used to project cells into two dimensions using the first 20 principal components. Differential gene expression of Il2rbhi CD8αα or IL2rblo CD8αα clusters from Ikzf3+/+ and Ikzf3−/− was analyzed using DESeq2 package.
RNA-seq
In total, 50,000 cells of Ikzf3+/+ or Ikzf3−/− γδ IELs and DN IELs were sorted, and RNA was extracted with an RNeasy Micro Kit (QIAGEN) following manufacturer’s instructions. Total RNA integrity was determined using an Agilent Bioanalyzer or 4200 Tapestation. Double-stranded cDNA was prepared using the SMARTer Ultra Low RNA kit for Illumina Sequencing (Takara-Clontech) according to the manufacturer’s protocol. cDNA was fragmented using a Covaris E220 sonicator using the following settings: duty cycle, 10; intensity, 5; cycles/burst, 200; time, 180 s. cDNA was blunt ended, had an A base added to the 3′ ends, and then had Illumina sequencing adaptors ligated to the ends. Ligated fragments were then amplified for 12 cycles using primers incorporating unique index tags. Fragments were sequenced on an Illumina HiSeq 3000 using single reads extending 50 bases. Sequences were aligned and gene count tables were produced using STAR aligner. DESeq2 R package was used to normalize and identify DEGs.
Microarray
A small portion of duodenum and ileum were harvested and washed with PBS. RNA was extracted with an RNeasy Mini Kit (QIAGEN) following the manufacturer’s instructions. Total RNA integrity was determined using an Agilent Bioanalyzer. Biotinylated cDNA was prepared according to the Affymetrix WT Plus reagent kit from 100–500 ng total RNA. Following fragmentation and labeling, 3.5 µg of cDNA was hybridized onto the Mouse Gene 2.0 arrays and placed in a GeneChip Hybridization Oven 640 for 18 h at 45 °C. Microarrays were washed and stained in the Affymetrix Fluidics Station 450. GeneChips were scanned using the Affymetrix GeneChip 7 G 3000 Scanner. Raw data were normalized with RMA using Affy R package, and differential expression was identified using limma R package.
Statistics and reproducibility
Statistical analysis was performed with R or Prism 9 software (GraphPad). For statistical comparisons between two groups, we used Student’s two-tailed t-test. When analyzing more than three groups, we performed either one-way or two-way ANOVA followed by Tukey’s multiple-comparison test for pairwise group comparisons. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications22,52. Data distribution was assumed to be normal, but this was not formally tested. Age-matched and sex-matched Ikzf3+/+ littermates were used as controls, and mice were randomly assigned in each group. No data were excluded. Data collection and analysis were not performed blind to the conditions of the experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Code availability
Codes will be provided upon reasonable request. Please contact M. Colonna for further information.
References
Mayassi, T. & Jabri, B. Human intraepithelial lymphocytes. Mucosal Immunol. 11, 1281–1289 (2018).
Olivares-Villagomez, D. & Van Kaer, L. Intestinal intraepithelial lymphocytes: sentinels of the mucosal barrier. Trends Immunol. 39, 264–275 (2018).
Colonna, M. Innate lymphoid cells: diversity, plasticity, and unique functions in immunity. Immunity 48, 1104–1117 (2018).
Cheroutre, H., Lambolez, F. & Mucida, D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol. 11, 445–456 (2011).
McDonald, B. D., Jabri, B. & Bendelac, A. Diverse developmental pathways of intestinal intraepithelial lymphocytes. Nat. Rev. Immunol. 18, 514–525 (2018).
McFarland, A. P. et al. Multi-tissue single-cell analysis deconstructs the complex programs of mouse natural killer and type 1 innate lymphoid cells in tissues and circulation. Immunity 54, 1320–1337 (2021).
Van Acker, A. et al. A murine intestinal intraepithelial NKp46-negative innate lymphoid cell population characterized by group 1 properties. Cell Rep. 19, 1431–1443 (2017).
Mayassi, T., Barreiro, L. B., Rossjohn, J. & Jabri, B. A multilayered immune system through the lens of unconventional T cells. Nature 595, 501–510 (2021).
Hayday, A., Theodoridis, E., Ramsburg, E. & Shires, J. Intraepithelial lymphocytes: exploring the Third Way in immunology. Nat. Immunol. 2, 997–1003 (2001).
Abadie, V., Discepolo, V. & Jabri, B. Intraepithelial lymphocytes in celiac disease immunopathology. Semin. Immunopathol. 34, 551–566 (2012).
Catalan-Serra, I., Sandvik, A. K., Bruland, T. & Andreu-Ballester, J. C. Gammadelta T cells in Crohn’s disease: a new player in the disease pathogenesis? J. Crohns Colitis 11, 1135–1145 (2017).
Atlasy, N. et al. Single cell transcriptomic analysis of the immune cell compartment in the human small intestine and in Celiac disease. Nat. Commun. 13, 4920 (2022).
Heizmann, B., Kastner, P. & Chan, S. The Ikaros family in lymphocyte development. Curr. Opin. Immunol. 51, 14–23 (2018).
Koipally, J., Renold, A., Kim, J. & Georgopoulos, K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 18, 3090–3100 (1999).
Zhang, J. et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat. Immunol. 13, 86–94 (2011).
Molnar, A. & Georgopoulos, K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol. Cell Biol. 14, 8292–8303 (1994).
Georgopoulos, K. et al. The Ikaros gene is required for the development of all lymphoid lineages. Cell 79, 143–156 (1994).
Quintana, F. J. et al. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat. Immunol. 13, 770–777 (2012).
Morgan, B. et al. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 16, 2004–2013 (1997).
Wang, J. H. et al. Aiolos regulates B cell activation and maturation to effector state. Immunity 9, 543–553 (1998).
Qiu, J. et al. Tissue signals imprint Aiolos expression in ILC2s to modulate type 2 immunity. Mucosal Immunol. 14, 1306–1322 (2021).
Felton, J. M. et al. Aiolos regulates eosinophil migration into tissues. Mucosal Immunol. 14, 1271–1281 (2021).
Holmes, M. L. et al. Peripheral natural killer cell maturation depends on the transcription factor Aiolos. EMBO J. 33, 2721–2734 (2014).
Cella, M. et al. Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat. Immunol. 20, 980–991 (2019).
Mazzurana, L. et al. Suppression of Aiolos and Ikaros expression by lenalidomide reduces human ILC3-ILC1/NK cell transdifferentiation. Eur. J. Immunol. 49, 1344–1355 (2019).
Whang, M. I., Guerra, N. & Raulet, D. H. Costimulation of dendritic epidermal gammadelta T cells by a new NKG2D ligand expressed specifically in the skin. J. Immunol. 182, 4557–4564 (2009).
Puddington, L., Olson, S. & Lefrancois, L. Interactions between stem cell factor and c-Kit are required for intestinal immune system homeostasis. Immunity 1, 733–739 (1994).
Shui, J. W. et al. HVEM signalling at mucosal barriers provides host defence against pathogenic bacteria. Nature 488, 222–225 (2012).
Rezende, R. M. et al. gammadelta T cell-secreted XCL1 mediates anti-CD3-induced oral tolerance. J. Immunol. 203, 2621–2629 (2019).
Zehn, D. et al. ‘Stem-like’ precursors are the fount to sustain persistent CD8+ T cell responses. Nat. Immunol. 23, 836–847 (2022).
Wu, J. et al. An activating immunoreceptor complex formed by NKG2D and DAP10. Science 285, 730–732 (1999).
Lanier, L. L. DAP10- and DAP12-associated receptors in innate immunity. Immunol. Rev. 227, 150–160 (2009).
Nixon, B. G. et al. Cytotoxic granzyme C-expressing ILC1s contribute to antitumor immunity and neonatal autoimmunity. Sci. Immunol. 7, eabi8642 (2022).
Yomogida, K. et al. Hobit confers tissue-dependent programs to type 1 innate lymphoid cells. Proc. Natl Acad. Sci. USA 118, e2117965118 (2021).
Friedrich, C. et al. Effector differentiation downstream of lineage commitment in ILC1s is driven by Hobit across tissues. Nat. Immunol. 22, 1256–1267 (2021).
Gilfillan, S. et al. NKG2D recruits two distinct adapters to trigger NK cell activation and costimulation. Nat. Immunol. 3, 1150–1155 (2002).
Ma, L. J., Acero, L. F., Zal, T. & Schluns, K. S. Trans-presentation of IL-15 by intestinal epithelial cells drives development of CD8alphaalpha IELs. J. Immunol. 183, 1044–1054 (2009).
Zhou, R., Wei, H., Sun, R. & Tian, Z. Recognition of double-stranded RNA by TLR3 induces severe small intestinal injury in mice. J. Immunol. 178, 4548–4556 (2007).
Lodolce, J. P. et al. T cell-independent interleukin 15rα signals are required for bystander proliferation. J. Exp. Med. 194, 1187–1194 (2001).
Zhou, R. et al. NKG2D recognition mediates Toll-like receptor 3 signaling-induced breakdown of epithelial homeostasis in the small intestines of mice. Proc. Natl Acad. Sci. USA 104, 7512–7515 (2007).
Longhi, M. P. et al. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J. Exp. Med 206, 1589–1602 (2009).
Chassaing, B., Aitken, J. D., Malleshappa, M. & Vijay-Kumar, M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Immunol. 104, 152511–152514 (2014).
Maekawa, Y. et al. Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat. Immunol. 9, 1140–1147 (2008).
Hahm, K. et al. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev. 12, 782–796 (1998).
Schjerven, H. et al. Selective regulation of lymphopoiesis and leukemogenesis by individual zinc fingers of Ikaros. Nat. Immunol. 14, 1073–1083 (2013).
Hollenhorst, P. C., Shah, A. A., Hopkins, C. & Graves, B. J. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 21, 1882–1894 (2007).
Ma, S. et al. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol. Cell Biol. 30, 4149–4158 (2010).
Bjorklund, C. C. et al. Rate of CRL4(CRBN) substrate Ikaros and Aiolos degradation underlies differential activity of lenalidomide and pomalidomide in multiple myeloma cells by regulation of c-Myc and IRF4. Blood Cancer J. 5, e354 (2015).
Peloquin, J. M. et al. Characterization of candidate genes in inflammatory bowel disease-associated risk loci. JCI Insight 1, e87899 (2016).
Katerndahl, C. D. S. et al. Antagonism of B cell enhancer networks by STAT5 drives leukemia and poor patient survival. Nat. Immunol. 18, 694–704 (2017).
Heizmann, B. et al. Ikaros antagonizes DNA binding by STAT5 in pre-B cells. PLoS ONE 15, e0242211 (2020).
Read, K. A. et al. Aiolos represses CD4+ T cell cytotoxic programming via reciprocal regulation of TFH transcription factors and IL-2 sensitivity. Nat. Commun. 14, 1652 (2023).
Abadie, V. et al. IL-15, gluten and HLA-DQ8 drive tissue destruction in coeliac disease. Nature 578, 600–604 (2020).
Lefrancois, L. & Lycke, N. Isolation of mouse small intestinal intraepithelial lymphocytes, Peyer’s patch, and lamina propria cells. Curr. Protoc. Immunol. 3, 19 (2001).
Buenrostro, J. D. et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Acknowledgements
We thank the Genome Technology Access Center at the McDonnell Genome Institute at Washington University for RNA-seq, ATAC-seq, CUT&RUN-seq and scRNA-seq. The center is supported in part by the Alvin J. Siteman Cancer Center and Barnes-Jewish Hospital, and the Institute of Clinical and Translational Sciences (ICTS). The Siteman Cancer Center is supported in part by an NCI Cancer Center Support Grant (P30 CA091842) and the ICTS is funded by the National Institutes of Health’s NCATS Clinical and Translational Science Award (CTSA) program grant (UL1 TR002345). We thank E. Lantelme and D. Brinja and the Pathology and Immunology Flow Cytometry Core for cell sorting. This work was supported by grants UO1 AI095542, RO1 DK126969, RO1 DK132327, RO1 AI134035, RO1 DK124699 and U19 AI142733 (to M. Colonna). K.Y. was supported by the Rheumatology Research Foundation Tobé and Stephen E. Malawista, MD Endowment in Academic Rheumatology, CHRC K12 and K08DK128544.
Author information
Authors and Affiliations
Contributions
K.Y., T.T., R.S., P.F.R., A.U.A., H.I., B.D. and P.L.C. performed experiments and analyzed the data. K.Y., H.I., P.L.C., M. Cella, E.M.O., M.T.B. and M. Colonna designed experiments. H.I. and M.T.B. provided crucial reagents. S.G. maintained the mouse colony. K.Y. and M. Colonna wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Bana Jabri and Pandurangan Vijayanand for their contribution to the peer review of this work. Primary Handling Editor: Ioana Visan, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Aiolos does not alter abundance of IELs.
Cell counts of γδ IEL, DN IEL, DP IEL, CD4+ IEL and CD8αβ+ IEL from small intestine in Ikzf3+/+ and Ikzf3−/− mice. Data are representative of two independent experiments (n = 4) and all samples are biological replicates. DN = double negative, DP = double positive (CD4 cytotoxic), Data represent mean ± SD. P values were determined by two-tailed, unpaired Student’s t-test.
Extended Data Fig. 2 NKG2D does not directly induce cytotoxicity in unconventional IELs.
a, Gene expression of NK receptors associated with DAP12 (Klrk1, Klrc2 and Klra8) in γδ IEL and DN IEL from small intestine in Ikzf3+/+ and Ikzf3−/− mice based on RNA-seq. b, γδ IEL and DN IEL from small intestine in Ikzf3+/+ and Ikzf3−/− mice were sorted, and co-cultured with RMA-S or RMA-S overexpressing Rae1γ. Frequency of live target cells are shown. Data showed one experiment in b and all samples are biological replicates. P values in a were calculated by Wald test (DEseq2). WT=Ikzf3 +/+ ; KO=Ikzf3 –/–.
Extended Data Fig. 3 scRNA-seq demonstrate heterogeneity of IELs.
a, UMAP plots of 25,966 cells of Ikzf3+/+ small intestine CD45+ IELs. b, Representative UMAP plots depicting expression of Trac, Cd8a, Tcrg-V7 and Tcrg-V1. c, Heatmap displaying top differentially expressed genes in clusters designated as CD8aa-1, CD8αα-2, CD8αα-3, CD8αα-4, CD8αα-5, CD8αα-6, CD8αβ+, CD4+, pDC, B cells and plasma cells.
Extended Data Fig. 4 CD8αα+ IELs comprise diverse groups of cells.
a, Heatmap displaying top differentially expressed genes in clusters designated as Ikzf3−/−Tcf7−Il2rbhi, Ikzf3+/+Tcf7−Il2rbhi, Tcf7−Il2rblo, Tcf7+, stressed CD8αα, lipidolytic CD8αα, proliferating CD8αα and IFN-stimulated CD8αα. b, Representative UMAP plots depicting expression of Klra4, Klra8, Klrc2, Il2ra, Cd200r1 Cd200r2, Lat, Lat2, Lag3, Cd200r4, Pdcd1, Havcr2, Lair2 and Il7r.
Extended Data Fig. 5 Ikzf3-deficiency enhances responses to IL-15.
a, Heatmap showing differentially expressed cytokine genes in the duodenum and ileum from Ikzf3+/+ mice. b-d, Flow cytometry plots and quantification of NKG2A (b), Ly49A and D (c) and NKG2D (d) on γδ IEL and DN IEL from small intestine in Ikzf3+/+ and Ikzf3−/− mice 72 hours after culturing with either 10 ng/ml or 100 ng/ml of IL-15. Data are representative of two independent experiments and all samples are biological replicates. n=8 in b, c and n=5 (Ikzf3+/+) and 6 (Ikzf3−/−) in d. Data represent mean ± SD. P values were calculated by two-way ANOVA with Tukey’s multiple comparisons test in b, c and d.
Extended Data Fig. 6 Generation and phenotype of Aiolos conditional deficient mice.
a, Diagram of the construct for conditional Ikzf3fl/fl mice showing LoxP sites flanking exon 4 and exon 6. b, Representative histograms show tdTomato expression in γδ IEL and DN IEL, DP IEL, CD4+ IEL, CD8αβ+ IEL, NK cells and B cells from small intestine in E8ICre R26RtdTomato mice. c, Representative image of colon from Ikzf3fl/fl and E8ICreIkzf3fl/fl mice at day 4 post DSS administration in the drinking water. Data are representative of two independent experiments.
Extended Data Fig. 7 Interactions of Aiolos with STAT5 and RUNX members.
a, A pie chart showing the chromosomal location of Aiolos binding regions in the indicated cell types. b, A Venn diagram depicting Aiolos binding peaks in γδ IELs and DN IELs. c, Represenative histogram plots and quantification of pSTAT5 expression in γδ IEL, DN IEL from the small intestine of Ikzf3+/+ and Ikzf3−/− mice. Cells were either not stimulated or stimulated for 15 minutes with IL-15 (100 ng/ml). d, The results of permutation test designed to analyze the significance of overlap of Aiolos with either STAT5, RUNX1 or RUNX3. Evperm: the number of permutation events, Evobs: the number of observed events (overlapped peak counts). Data are representative of two independent experiments and all samples are biological replicates (n = 4 in c). P values were calculated by unpaired, two-tailed Student’s t-test in c and Permutation test in d (ChIPpeakAnno). Data represent mean ± SD. pSTAT5 = phosphorylated STAT5, UTR = untranslated region.
Extended Data Fig. 8 Aiolos shapes a distinct epigenetic landscape.
a, Gene expression of IKZF family members in γδ IEL and DN IEL from the small intestine in Ikzf3+/+ and Ikzf3−/− mice based on RNAseq (see Fig. 2). The expression of Ikzf3 in Ikzf3–/– IEL reflects a portion of the nonfuntional transcript present in the knockout mouse. b, Venn diagrams depicting overlap of Aiolos binding regions, differential H3K27ac peaks and DAR, and the result of permutation test designed to analyze the significance of overlap of differential H3K27ac peaks with DAR in γδ IELs and DN IELs. FDR were calculated by Wald test (DESeq2) in a and P values were calculated Permutation test in b (ChIPpeakAnno). DAR = differential accessible regions.
Supplementary information
Supplementary Tables 1 and 2
Supplementary Table 1 Comprehensive list of DEGs in the indicated cell types, along with their respective associations with DAR, differential H3K27ac, binding of Aiolos, STAT5, RUN1 or RUNX3. ‘+’ indicates positive association, whereas ‘–’ denotes no association. Supplementary Table 2 Quality-control metrics of ATAC-seq and CUT&RUN-seq for Aiolos and H3K27ac.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yomogida, K., Trsan, T., Sudan, R. et al. The transcription factor Aiolos restrains the activation of intestinal intraepithelial lymphocytes. Nat Immunol 25, 77–87 (2024). https://doi.org/10.1038/s41590-023-01693-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41590-023-01693-w
This article is cited by
-
Aiolos restricts the generation of antigen-inexperienced, virtual memory CD8+ T cells in mice
Nature Communications (2025)