Abstract
Long-lived memory CD8+ T cells are essential for the control of persistent viral infections. The mechanisms that preserve memory cells are poorly understood. Fate mapping of the transcriptional repressor GFI1 identified that GFI1 was differentially regulated in virus-specific CD8+ T cells and was selectively expressed in stem cell memory and central memory cells. Deletion of GFI1 led to reduced proliferation and progressive loss of memory T cells, which in turn resulted in failure to maintain antigen-specific CD8+ T cell populations following infection with chronic lymphocytic choriomeningitis virus or murine cytomegalovirus. Ablation of GFI1 resulted in downregulation of the transcription factors EOMES and BCL-2 in memory CD8+ T cells. Ectopic expression of EOMES rescued the expression of BCL-2, but the persistence of memory CD8+ T cells was only partially rescued. These findings highlight the critical role of GFI1 in the long-term maintenance of memory CD8+ T cells in persistent infections by sustaining their proliferative potential.
Similar content being viewed by others
Main
Antigen recognition by naive CD8+ T (TN) cells initiates clonal expansion and a hierarchical differentiation program that generates heterogeneous populations, including short-lived effector CD8+ T (TSLEC) cells with potent cytotoxicity and memory CD8+ T (TM) cells that confer durable immunity. CD8+ TM cells can persist for decades, even without re-exposure to cognate antigens1,2,3. These long-term responses are sustained by central memory CD8+ T (TCM) cells and stem cell memory CD8+ T (TSCM) cells that retain both self-renewal and immune reconstitution potential4,5. CD8+ TCM cells and CD8+ TSCM cells emerge early in viral infection6,7, while dysfunctional T cell emergence in chronic infections and cancers has been associated with loss of CD8+ TSCM cells8,9.
CD8+ TSCM cells are a heterogeneous population with distinct functional and developmental properties. However, persistent antigenic stimulation in chronic viral infection can disrupt memory programming. CD8+ TSCM cells comprise two subsets: one that expresses the transcription factor TCF1 with remarkable longevity and a second progenitor type that also expresses elevated levels of the transcription factor TOX and the inhibitory receptor PD-1 (refs. 5,10,11). Both subsets have been identified in mice and humans, with the latter subset now recognized as precursors of exhausted CD8+ T cells (TPEX)12,13. The CD8+ TPEX cell population propagates exhausted CD8+ T cells (TEX) that express high levels of multiple inhibitory immune checkpoint receptors such as PD-1, LAG3, 2B4 and TIM3. Furthermore, CD8+ TEX cells gradually lose effector functions, including interferon-γ (IFNγ) secretion4. The transcription factors TCF1, ID3, BACH2 and BATF have all been shown to regulate the generation of CD8+ TSCM cells4,14. However, the determinants that maintain the essential self-renewal potential of CD8+ TSCM cells to form long-lived populations are not fully resolved.
Although readily cleared acute viral infections give rise to effector (TEFF) and TM cells, including TCM and TSCM cells, persistent infections such as hepatitis virus, human immunodeficiency virus in humans and certain strains of lymphocytic choriomeningitis virus (LCMV) in mice also give rise to CD8+ TSCM cells, even though CD8+ TPEX cell populations contribute significantly to the propagation of CD8+ TEX cells in these chronic infections10,15. However, in latent herpesvirus infections, pathogen-specific T cell populations remain functional. For instance, cytomegalovirus infection can generate significantly expanded long-term stable CD8+ T cell populations in a phenomenon known as memory inflation16. The factors sustaining inflationary CD8+ T cells remain unclear.
The transcriptional repressor GFI1 is highly expressed in thymocytes following T cell receptor (TCR) activation, and loss of GFI1 impairs early T cell development in the thymus17. In mature T cells, GFI1 deficiency differentially affects CD4+ and CD8+ T cell subsets, with GFI1-deficient CD4+ T cells exhibiting aberrant proliferation, while Gfi1–/– CD8+ T cells are not impaired18. Antiviral CD8+ T cells in Gfi1–/– mice show elevated IL-7R expression19, suggesting that a lack of GFI1 may enhance IL-7-dependent proliferation. However, the exact role of GFI1 in maintaining TM cells following acute and chronic viral infection is not understood.
Here, we examined the role of GFI1 in CD8+ TM cell development after acute and chronic viral infection. GFI1 was differentially expressed across TM and TEFF cells resolving GFI1hi and GFI1lo populations. GFI1hi cells were transcriptionally distinct from GFI1lo cells, expressing a memory phenotype and a superior ability to generate long-lived TM cells. Ablation of GFI1 early in chronic infection showed that antiviral CD8+ T cells could still develop, whereas CD8+ TSCM cells were lost. Altogether, our data identify that GFI1 is a key regulator of the fitness of CD8+ TSCM cells, which maintain memory compartments in chronic infection and enable replenishment of effector populations.
Results
GFI1 is selectively expressed by TM cells
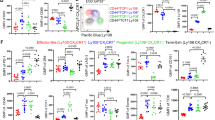
To understand how GFI1 is regulated in CD8+ T cells, Gfi1tdTomato/+ reporter mice, which express tdTomato under the Gfi1 promotor in all cells20,21,22, were infected with LCMV that induces either acute infection (Armstrong strain, LCMVArm) or chronic infection (clone 13 strain, LCMVc13). In naive Gfi1tdTomato/+ mice, GFI1-tdTomato was uniformly highly expressed in CD8+ T cells (Extended Data Fig. 1a). Following infection, CD8+ T cells showed a significant downregulation of GFI1-tdTomato expression (Extended Data Fig. 1b,c). Further analyses revealed a graded expression of GFI1-tdTomato whereby CD8+ TCM and CD8+ TSCM cells exhibited high expression of GFI1-tdTomato (Fig. 1a,b and Extended Data Fig. 1d). TCF1−CX3CR1+ TEFF cells showed the lowest GFI1-tdTomato expression (Fig. 1a,b and Extended Data Fig. 1e), whereas high expression was maintained in CD8+ TSCM cells (Extended Data Fig. 1e). TCF1+TOX+ CD8+ TPEX cells that maintain stem-like proliferation following LCMVc13 infection4 showed significantly lower expression of GFI1-tdTomato than CD8+ TCM and CD8+ TSCM cells (Fig. 1b). Further resolving the CD8+ TPEX cell population revealed that CD62L+CD8+ TPEX cells, which were reported to exhibit superior proliferative capacity23, had higher GFI1-tdTomato expression than CD62L−CD8+ TPEX cells (Extended Data Fig. 1f). Following the initial downregulation of GFI1-tdTomato expression after LCMV infection, GFI1-tdTomato subsequently increased between day 7 and day 21 for LCMVArm infection but remained low following LCMVc13 infection (Fig. 1c). This pattern persisted in LCMV-specific gp33+CD8+ T cells during late LCMVc13 infection (Fig. 1d and Extended Data Fig. 1g,h). Tracking the temporal dynamics of GFI1 expression early in infection using Gfi1tdTomato/+ P14 CD8+ T cells showed that GFI1-tdTomato expression was significantly downregulated on day 2 and day 3 in P14 T cells isolated from spleen and mesenteric lymph nodes (mLN), respectively (Fig. 1e and Extended Data Fig. 1i,j). FK506, a potent inhibitor of calcineurin that blocks TCR-dependent signaling, showed that although TOX expression was impaired, GFI1-tdTomato was not affected (Fig. 1f). Thus, calcineurin signaling was not essential to GFI1 regulation, and infection-induced inflammation was sufficient for GFI1 downregulation. Together, these data show that GFI1 is rapidly downregulated in activated CD8+ T cells after infection and is selectively maintained in TM cell subsets.
a, Representative histogram (left) and quantification (right) of GFI1-tdTomato expression in splenic CD11a–CD44–CD8+ TN cells from C57BL/6 mice (control) and CD8+ TN, CD11a+CD44+CD62L+CD27+CD8+ TCM cells, CD11a+CD44+TCF1+CX3CR1–CD8+ TSCM cells, TCF1-CX3CR1+CD8+ TEFF cells and gp33+CD8+ T cells from the spleen of Gfi1tdTomato/+ mice at day 7 (D7) post infection with LCMVArm. Data are pooled from two experiments (n = 7 mice). gMFI, geometric mean fluorescence intensity. b, Representative histogram (left) and quantification (right) of GFI1-tdTomato expression in splenic CD8+ TN cells from C57BL/6 mice (control) and CD8+ TN, CD8+ TCM, CD8+ TSCM, CD8+ TEFF, CD8+gp33+ and TCF1+TOX+CD8+ TPEX cells from the spleen of LCMVc13-infected Gfi1tdTomato/+ mice at D7. Pooled from four experiments (n = 13 mice). c,d, GFI1-tdTomato in total CD8+ T cells (c) and gp33+CD8+ T cells (d) from the spleen of LCMVArm and LCMVc13-infected Gfi1tdTomato/+ mice at D7 and D21. Pooled from two experiments (n = 8 or 17 mice per time point). e,f, Expression of GFI1-tdTomato in splenic CD8+ P14 T cells at D0–D7 post LCMVc13 infection (n = 8 mice per time point) (e) and GFI1-tdTomato and TOX in splenic CD8+ P14 T cells at D7 post LCMVc13 infection (f) in C57BL/6 mice transferred with congenically labeled Gfi1tdTomato/+ CD8+ P14 TN cells, infected with LCMVc13 1 day later (e,f) and treated with FK506 or vehicle daily from D4–D6 post infection (f). Pooled from two experiments (n = 6 mice). g–i, TCF1 (g), EOMES (h) and T-BET (i) expression in activated CD11a+CD44+GFI1hi and CD11a+CD44+GFI1lo CD8+ T cells from LCMVArm-infected and LCMVc13-infected Gfi1tdTomato/+ mice at D7 and D21 post infection. Data pooled from two experiments (n = 7 or 8 mice). Individual values and means are shown; error bars, s.e.m. Error bars not shown in e. Statistical significance using a one-way ANOVA and Dunnett’s post hoc test for a, b and e; two-tailed Student’s t-test for c, d and f; Wilcoxon signed-rank two-tailed paired t-test for g–i.
Given the emergence of distinct high and low GFI1-tdTomato-expressing populations among activated CD11a+CD44+ CD8+ T cells, the GFI1hi and GFI1lo CD8+ T cells (Extended Data Fig. 1b) were further analyzed to determine the expression of other transcription factors. GFI1hiCD8+ T cells showed increased expression of TCF1 and EOMES compared to GFI1loCD8+ T cells at day 7 post LCMVArm or LCMVc13 infection (Fig. 1g,h). TCF1 remained high in GFI1hiCD8+ T cells at day 21, while EOMES expression was similar in GFI1hi and GFI1lo CD8+ T cells at day 21 (Fig. 1h). T-BET expression was low in GFI1hiCD8+ T cells at day 7 and day 21 after LCMVArm and LCMVc13 infection (Fig. 1i). Therefore, GFI1 was differentially regulated among TM and TEFF cells and was associated with memory CD8+ T cell formation.
GFI1 identifies transcriptionally distinct CD8+ T cells
RNA sequencing (RNA-seq) of GFI1hi and GFI1lo CD8+ T cells isolated from LCMVArm-infected or LCMVc13-infected mice (Extended Data Fig. 2a) showed that GFI1hiCD8+ T cells responding to both infections exhibited a similar transcriptional profile, which was distinct from that of GFI1loCD8+ T cells (Extended Data Fig. 2b). We identified 929 and 220 unique differentially expressed genes (DEGs) in GFI1hiCD8+ T cells isolated from LCMVArm-infected and LCMVc13-infected mice, respectively (Fig. 2a and Supplementary Tables 1 and 2). GFI1hiCD8+ T cells showed upregulation of transcription factors associated with T cell memory4,24, including Eomes, Id3 and Tcf7, whereas transcription factors linked with TEFF cell programs such as Id2, Tbx21 and Zeb2 were downregulated24,25 (Fig. 2b,c). GFI1hiCD8+ T cells showed higher Gfi1 expression than GFI1lo CD8+ T cells (Fig. 2c and Extended Data Fig. 2c), suggesting that the reporter levels reflect GFI1 gene expression. GFI1hiCD8+ T cells showed upregulation of genes associated with cell proliferation and cell cycle, including Cdk1, Top2a and Myc (Fig. 2c and Extended Data Fig. 2d) and significant enrichment of memory signature genes, such as Tcf7, Eomes and Il7r (Fig. 2d and Extended Data Fig. 2e), suggesting that GFI1hiCD8+ T cells had enhanced proliferative potential. This premise was further supported by upregulation of E2f target genes and the G2M checkpoint pathway in GFI1hiCD8+ T cells (Extended Data Fig. 2f). To understand whether the memory signature26 observed in the transcriptome of GFI1hiCD8+ T cells was solely caused by enrichment of CD8+ TCM and CD8+ TSCM cell subsets or whether GFI1-tdTomato expression regulated memory gene expression within CD8+ TCM and CD8+ TSCM cell populations, the transcriptional profile of GFI1hi and GFI1lo subsets from CD44+CD62L+ and CD44+Ly108+ (Ly108 was used as a surrogate marker for TCF1+ memory cells27) CD8+ T cell populations were analyzed. We found that GFI1hiCD8+ T cell fractions within CD8+ TCM and CD8+ TSCM cells expressed higher levels of Id3, Ikzf2 and Tcf7, whereas Gzmb, Havcr2 and Zeb2 showed lower expression (Fig. 2e, Extended Data Fig. 3a,b and Supplementary Tables 3 and 4). These data strongly suggest that high expression of GFI1 identifies key features of memory programmed CD8+ T cells, indicating a superior capacity to respond to infection.
a, Shared and unique DEGs (P < 0.05) in activated CD11a+CD44+GFI1hi CD8+ T cells isolated from the spleen of Gfi1tdTomato/+ mice at D7 post infection with LCMVc13 or LCMVArm and analyzed by RNA-seq. b, Volcano plot of DEGs in GFI1hi CD8+ T cells from LCMVc13-infected mice as in a. Blue and red indicate genes upregulated in activated GFI1lo and GFI1hi CD8+ T cells, respectively. Gray dots indicate P > 0.05. c, DEseq2-normalized expression of selected genes in GFI1hi and GFI1lo CD8+ T cells isolated from LCMVc13-infected mice as in a. d, Gene set enrichment analysis of GFI1hi and GFI1lo CD8+ T cells isolated from LCMVc13-infected mice as in a, using a memory CD8+ T cells gene signature26. NES, normalized enrichment score. e, Shared and unique DEGs (P < 0.05) in CD44+CD62L+GFI1hi and CD44+Ly108+GFI1hi CD8+ TM cells isolated from the spleen of Gfi1tdTomato/+ mice at D7 post infection with LCMVc13 and analyzed by RNA-seq. f,g, Representative flow cytometry plots (f) and quantification (g) of spleen donor CD45.2+CD8+ T cells at D7 post LCMVArm infection in congenic C57BL/6 mice transferred intravenously (i.v.) with splenic CD8+ T cells isolated at D21 post LCMVArm infection from CD45.2+ Gfi1tdTomato/+ mice and infected with LCMVArm 24 h later. Data pooled from two experiments (n = 7 mice per group). h, Total donor CD45.2+CD8+ T cells at D14 post LCMVc13 infection in the spleen of congenic C57BL/6 recipients infected with LCMVc13 7 days before transfer of infection-matched (D7) CD8+ T cells isolated from LCMVc13-infected CD45.2+ Gfi1tdTomato/+ mice. Data pooled from two experiments (n = 5 or 7 mice per group). P values were calculated using a Wald’s test (b, d and e) or a two-tailed Student’s t-test (g and h). Data in g and h show means; error bars, s.e.m.
To determine whether GFI1hi and GFI1lo CD8+ T cells differed in their capacity to respond to a secondary infection, GFI1hi and GFI1lo CD8+ T cells were isolated from the spleen of LCMVArm-infected wild-type (WT) mice on day 21 after infection and adoptively transferred into secondary recipients that were challenged with LCMVArm the next day (Extended Data Fig. 3c). In this acute challenge model, GFI1hi CD8+ T cells expanded approximately fourfold more than GFI1lo CD8+ T cells by day 7 post LCMVArm infection (Fig. 2f,g and Extended Data Fig. 3d), demonstrating the enhanced proliferative potential of GFI1hi CD8+ T cells in response to a secondary infection. Next, we adoptively transferred CD44+CD62L+GFI1hi, CD44+Ly108+GFI1hi or CD44+GFI1lo CD8+ T cells isolated from spleen and mLN of LCMVArm-infected mice at day 21 after infection into congenically marked secondary recipient mice that were subsequently infected with LCMVArm the next day. This procedure showed that both CD44+CD62L+GFI1hi and CD44+Ly108+GFI1hi CD8+ T cell populations exhibited superior expansion compared with that of CD44+GFI1loCD8+ T cells (Extended Data Fig. 3e). In line with these observations, adoptive transfer of GFI1hi and GFI1lo CD8+ T cells isolated from spleen of Gfi1tdTomato/+ mice infected with LCMVc13 at day 7 after infection into infection-matched recipients (Extended Data Fig. 3f) showed that GFI1hiCD8+ T cells had more enhanced capacity to proliferate than GFI1lo CD8+ T cells (Fig. 2h). Collectively, these data showed that GFI1hi CD8+ T cells identified a TM cell population with superior expansion and recall response capacity.
GFI1 epigenetically regulates CD8+ T cell function and persistence
To study the role of GFI1, Gfi1fl/fl mice were crossed with CD8acre/+ (E8I-Cre)28 to create GFI1ΔCD8 mice, in which GFI1 was deleted in mature peripheral CD8+ T cells. Unlike T cells in the Gfi1–/– strain18, the numbers of CD4+ and CD8+ T cells in the thymus (Extended Data Fig. 4a,b) and spleen (Extended Data Fig. 4c,d) of naive GFI1ΔCD8 mice were similar to WT mice. Moreover, selective ablation of GFI1 did not alter TCF1, EOMES, T-BET or TOX expression in naive CD8+ T cells (Extended Data Fig. 4e). When equal numbers of WT (CD45.1+) and GFI1ΔCD8 (CD45.1+CD45.2+) P14 CD8+ T cells were co-transferred into congenic recipients, both WT and GFI1ΔCD8 P14 CD8+ T cells expanded equivalently by day 5 after LCMVc13 infection (Extended Data Fig. 5a). However, the frequency and number of GFI1ΔCD8 CD8+ T cells declined from day 7 and were largely lost by day 21 after LCMVc13 infection in spleen, blood, mLN and lungs (Fig. 3a,b and Extended Data Fig. 5a). This decline mirrored a reduction in P14 CD8+ TSCM cells (Fig. 3c,d and Extended Data Fig. 5b) and P14 CD8+ TCM cells (Fig. 3e). CD8+ TEFF cells were also decreased at day 7 (Fig. 3c and Extended Data Fig. 5c). GFI1ΔCD8 P14 CD8+ T cells showed lower expression of Ki-67, TCF1, EOMES, TOX and CX3CR1 (Fig. 3f,g and Extended Data Fig. 5d) and increased expression of FOXO1, CD127 and TIM3 (Extended Data Fig. 5d) compared to WT P14 CD8+ T cells. These results demonstrate that GFI1 is needed for antigen-specific memory CD8+ T cell persistence in chronic infection.
a,b, Representative flow cytometry plots (a) and total number (b) of WT and GFI1ΔCD8 live CD8+ P14 T cells at D5–D60 post infection in the spleen of C57BL/6 mice that received a 1:1 mix of congenically labeled WT and GFI1ΔCD8 TN cells followed by LCMVc13 infection 24 h later. Data are pooled from two (n = 10 mice for D7 and D21) or one experiment (n = 5 for D5, D14, D28 and D60). c, Representative plots showing frequency of CD8+ TSCM cells at D7 in the spleen of LCMVc13-infected mice as in a. d,e, Total number of CD8+ P14 TSCM (d) and CD8+ P14 TCM (e) cells in the spleen of LCMVc13-infected mice as in a. Data are pooled from two experiments (n = 10 mice per time point). f, Representative histograms at D7 (right) and quantification (left) of Ki-67 expression in WT and GFI1ΔCD8 CD8+P14 T cells as in a. D7 data are representative of two experiments (n = 5 mice); D14 data are representative of one experiment. g, TCF1, EOMES and TOX expression in CD8+ P14 T cells at D7 in the spleen of LCMVc13-infected mice. Data are representative of two experiments (n = 6 mice). h, Expression of selected genes in WT and GFI1ΔCD8 CD8+ P14 T cells from the spleen of C57BL/6 mice transferred i.v. with congenically marked WT or GFI1ΔCD8 CD8+ P14 TN cells, infected with LCMVc13 24 h post transfer and analyzed by RNA-seq at D7 and D21 post LCMVc13 infection. i, PCA plot of WT or GFI1ΔCD8 CD8+P14 T cells at D7 and D21 post LCMVc13 infection based on RNA-seq as in h. j, Shared and unique DEGs at D7 and D21in GFI1ΔCD8 CD8+ T cells as in h. k, ATAC-seq PCA plot of splenic WT and GFI1ΔCD8 CD8+ T cells at D0, D3, D5 and D7 post LCMVc13 infection in C57BL/6 mice transferred i.v. with congenically labeled WT or GFI1ΔCD8 CD8+ P14 T cells 24 h before infection. l, Total number of DARs detected in GFI1ΔCD8 CD8+ P14 T cells versus WT CD8+ P14 T cells at D0, D3, D5 and D7 post LCMVc13 infection as in k. m, Number of DARs annotated in promotor or exon regions in GFI1ΔCD8 CD8+ P14 T cells compared with WT CD8+ P14 T cells as in l. n, Predicted GFI1 binding motif by HOMER analysis using DEGs from GFI1ΔCD8 CD8+ T cells isolated at D7 post LCMVc13 infection as in h. Statistical significance was calculated using a two-tailed paired t-test (b and d–g) or Wald’s test (j, l and m).
To identify the transcriptional program regulated by GFI1 following CD8+ T cell activation, WT and GFI1ΔCD8 P14 CD8+ T cells were analyzed by RNA-seq on day 7 and day 21 after LCMVc13 infection. Loss of GFI1 impaired expression of proliferation and cell cycle genes such as Cdk1, Cdk2, E2f2, Mki67 and Top2a (Fig. 3h) and altered transcription of memory genes including Eomes, Id2 and Tox (Fig. 3h,i and Supplementary Table 5). RNA-seq identified 2,832 unique DEGs in GFI1ΔCD8 P14 CD8+ T cells at day 7 and 302 DEGs at day 21 (Fig. 3j). Pathway analyses showed downregulation of DNA replication and cell division pathways (Extended Data Fig. 5e), indicating that GFI1 had a key role in proliferation. GFI1 loss also disrupted metabolic gene expression associated with glucose and lipid uptake such as Acss1, Pgk1 and Uqcrh (Extended Data Fig. 6a,b), increased mitochondrial dysfunction (Extended Data Fig. 6c,d) and reduced granzyme B production (Extended Data Fig. 6e).
GFI1 modifies chromatin to repress transcription29. Assay for transposase-accessible chromatin using sequencing (ATAC-seq) showed that WT and GFI1ΔCD8 P14 CD8+ TN cells had similar epigenetic profiles (Fig. 3k,l and Extended Data Fig. 6f). By contrast, CD8+ T cell activation following LCMVc13 infection resulted in a significant change in the epigenetic profile of LCMV-activated CD8+ T cells compared with CD8+ TN cells (Fig. 3k,l). Both WT and GFI1ΔCD8 CD8+ T cells showed a similar epigenetic profile on day 3 and day 5 after infection, whereas at day 7, the profile of GFI1ΔCD8 CD8+ T cells was substantially different from WT CD8+ T cells (Fig. 3k,l and Supplementary Table 6). Differentially accessible chromatin regions (DARs) were upregulated in GFI1ΔCD8 CD8+ T cells following activation but not in CD8+ TN cells (Fig. 3l,m and Extended Data Fig. 6g). The HOMER motif discovery analysis predicted that the GFI1 binding motif in the promotor region of various targets is important for CD8+ T cell proliferation and differentiation, such as E2f7, Eomes, Foxo1, Havcr2 and Il7r (Fig. 3n and Supplementary Table 7). Altogether, these data show that GFI1-mediated epigenetic silencing regulates the CD8+ T cell transcriptional program to promote their persistence during chronic viral infection.
Loss of GFI1 abrogates inflationary CD8+ T cell responses
Murine cytomegalovirus (MCMV) infection is characterized by the development of ‘memory inflation’ driven by TSCM cell precursors6,30,31. To understand the role of GFI1 in the development of these unusual T cell populations, we generated mixed bone marrow chimeras with WT or GFI1ΔCD8 bone marrow cells. Then, 8 weeks after bone marrow reconstitution, mice were infected with MCMV, and antigen-specific CD8+ T cells were monitored longitudinally in blood (Extended Data Fig. 7a). The number of WT and GFI1ΔCD8 CD8+ T cells in peripheral blood of chimeric mice was similar before virus infection (Extended Data Fig. 7b), suggesting that CD8+ TN cell development from GFI1ΔCD8 bone marrow was not impaired. Following MCMV infection, WT CD8+ T cells mounted a strong response to the non-inflationary M45 epitope at day 7, followed by a sharp contraction of the response (Fig. 4a,b). No change in CD4+ T cell frequency was observed (Fig. 4c). WT M38-specific CD8+ T cells accumulated gradually to produce ‘memory inflation’ during virus latency (Fig. 4a,b)31,32. By contrast, GFI1ΔCD8 CD8+ T cells had impaired responses for both epitopes and contracted prematurely (Fig. 4a,b). The M38 and m139 inflationary epitope-specific GFI1ΔCD8 CD8+ T cells were significantly lower in spleen during latency (Fig. 4d). The inflationary T cell responses have been previously found to be maintained by continuous production of KLRG1+CD27− CD8+ TSLEC cells, which arise from KLRG1−CD27+ memory precursor CD8+ T (TMPEC) cells30,31. The GFI1ΔCD8 M38-specific and total activated CD11a+CD44+CD8+ T cell population exhibited a lower frequency of CD8+ TSLEC cells (Fig. 4e and Extended Data Fig. 7c). Thus, GFI1 deficiency resulted in loss of long-term CD8+ T cell responses, in particular inflationary responses, following latent virus infection. GFI1ΔCD8 P14 CD8+ T cells mounted an impaired inflationary response following infection with a recombinant MCMV that expressed LCMV gp33 epitope (MCMV-ie2-gp33) (Fig. 4f,g). This included reduced CD8+ TSCM cells, CD8+ TCM cells (Fig. 4h) and CD8+ TSLEC cells (Extended Data Fig. 7d) and lower Ki-67, TCF1, EOMES and TOX expression (Extended Data Fig. 7e). Altogether, these data demonstrate the essential role of GFI1 in mounting inflationary CD8+ T cell responses in MCMV infection.
a,b, Representative flow cytometry plots at D7 and D90 (a) and total number at D0–D90 (b) of CD45.1+M45+ and CD45.1+M38+ CD8+ T cells post infection with MCMV in the peripheral blood of chimeric C57BL/6 mice that were reconstituted with a 1:1 mix of congenically labeled Cd8acre/+ (WT) CD45.1+ and WT CD45.2+ (WT) or Cd8acre/+Gfi1fl/fl (GFI1ΔCD8) CD45.1+ and WT CD45.2+ (GFI1ΔCD8) bone marrow (BM) after lethal irradiation and infected with MCMV 8 weeks post BM reconstitution. c, Frequency of blood CD45.1+CD8+ T cells and CD45.1+CD4+ T cells among CD8+ and CD4+ T cells in WT and GFI1ΔCD8 mice as in a. d, Total splenic M38+CD45.1+CD8+ and m139+CD45.1+CD8+ T cells at D90 post infection with MCMV in WT and GFI1ΔCD8 mice as in a. e, Representative plot (left) and quantification (right) of CD45.1+KLRG1+CD27− CD8+ TSLEC cells and CD45.1+KLRG1−CD27+ CD8+ TMPEC cells in spleen at D90 post infection with MCMV in WT and GFI1ΔCD8 mice as in a. f, Kinetics of WT or GFI1ΔCD8 CD8+ P14 T cells at D0, D14, D30, D60 and D90 post MCMV-ie2-gp33 infection in the peripheral blood of C57BL/6 mice that received 1:1 mix of congenically labeled WT and GFI1ΔCD8 CD8+ P14 TN cells followed by infection with MCMV-ie2-gp33 24 h post transfer. g, Number of WT and GFI1ΔCD8 CD8+ T cells in the spleen, mLN and lung at D90 post infection with MCMV-ie2-gp33 as in f. h, Number of CD8+ TSCM and CD8+ TCM cells in the spleen at D90 post infection with MCMV-ie2-gp33 as in f. Data are shown as means; error bars, s.e.m. i, Normalized gene expression in splenic WT and GFI1ΔCD8 CD8+ P14 T cells at D7 post infection with MCMV-ie2-gp33 as in f. j, Frequency of upregulated and downregulated DARs in splenic GFI1ΔCD8 CD8+ P14 T cells at D7 post infection with MCMV-ie2-gp33 as in f. Outer donut, distribution of DARs within the exonic regions (3′-UTR, 5′-UTR, CDS and other exons). P values: two-tailed Student’s t-test (b–e); two-tailed paired t-test (f–h). Data in b–e are pooled from three experiments; mean values are shown; error bars, s.e.m. (n = 10 mice per group); data in f–h are pooled from two experiments (n = 10 mice). CDS, coding DNA sequence; UTR, untranslated region.
Transcriptional analysis showed that GFI1ΔCD8 CD8+ T cells downregulated TEFF cell surface receptor genes such as Cx3cr1 and Klrg1 (Fig. 4i and Supplementary Table 8). Furthermore, the transcriptional profile of GFI1ΔCD8 P14 CD8+ T cells was similar following infection with either MCMV-ie2-gp33 or chronic LCMVc13 (Fig. 3i), with GFI1ΔCD8 cells showing both Eomes and Tox downregulation (Fig. 4i). We observed that Tnf expression was upregulated in GFI1ΔCD8 P14 CD8+ T cells (Fig. 4i). Peptide stimulation confirmed a higher frequency of IFNγ+ and TNFα+ cells among GFI1ΔCD8 CD8+ T cells compared to WT CD8+ T cells (Extended Data Fig. 7f). Following MCMV infection, ATAC-seq revealed that GFI1 deficiency resulted in increased chromatin accessibility in GFI1ΔCD8 CD8+ T cells (Fig. 4j and Extended Data Fig. 7g), with 54,137 upregulated DARs and 646 downregulated DARs (Supplementary Table 9) compared to WT CD8+ T cells. Thus, GFI1 acted to epigenetically repress the transcriptional landscape of CD8+ T cells and promote inflationary T cell responses to cytomegalovirus infection.
GFI1 epigenetically regulates TSCM CD8+ T cell transcription
To map the epigenetic and transcriptional landscape at single-cell level, we performed single-cell multiome sequencing (scMultiome-seq), which combines ATAC and gene expression analyses on WT and GFI1ΔCD8 P14 CD8+ T cells isolated at day 7 after LCMVc13 infection (Extended Data Fig. 8a). Uniform manifold approximation and projection (UMAP) and unsupervised clustering of integrated WT and GFI1ΔCD8 CD8+ T cells divided them into seven clusters based on their epigenome and transcriptome (Fig. 5a,b and Extended Data Fig. 8b). Cluster 1 was identified as CD8+ TSCM cell precursors and clusters 2 and 3 were proliferating cells, respectively (Extended Data Fig. 8c). Cluster 1 cells expressed high levels of Tcf7, Slamf6 and Id3 and were thus annotated as CD8+ TSCM cells (Extended Data Fig. 8d), while cluster 2 and cluster 3 cells expressed high levels of Cdk1, Birc5 and Mki67 (Extended Data Fig. 8d). The GFI1ΔCD8 CD8+ T cell population had fewer cells in clusters 1 and 2 (Fig. 5c and Extended Data Fig. 8b). By contrast, a higher fraction of the GFI1ΔCD8 CD8+ T population was located in cluster 5 (Fig. 5c). This population exhibited reduced expression of genes associated with proliferation and high expression of Btg1 (Extended Data Fig. 8d). GFI1ΔCD8 CD8+ T cells had high Btg1, while E2f2, Eomes and Tcf7 was downregulated (Fig. 5d,e). scATAC-seq data confirmed increased chromatin accessibility at the Btg1 locus in clusters 1 and 5 (Extended Data Fig. 8e). To identify DEGs and DARs within the CD8+ TSCM cell cluster, we performed pseudobulk RNA-seq (Supplementary Table 10) and ATAC-seq (Supplementary Table 11) analyses on cluster 1 cells. These analyses showed that loss of GFI1 led to increased Btg1 and Btg2 expression, whereas the expression of Tcf7, Eomes, Bcl2 and Mki67 was reduced in CD8+ TSCM cells (Fig. 5f). These transcriptional changes were linked to upregulated DARs in the Btg1 and Btg2 gene loci and downregulated DARs in E2f1 and Mki67 genes (Fig. 5f). Thus, GFI1ΔCD8 CD8+ TSCM cells had reduced proliferation gene expression and increased quiescence gene expression.
a, UMAP showing unsupervised clustering of 7,629 WT and 5,805 GFI1ΔCD8 CD8+ T cells isolated at D7 post infection with LCMVc13 from the spleen of C57BL/6 mice transferred i.v. with congenically labeled WT or GFI1ΔCD8 CD8+ TN cells 24 h before infection, analyzed using scMultiome-seq (scRNA-seq + scATAC-seq Seurat-integrated data). b, UMAP showing WT and GFI1ΔCD8 CD8+ P14 T cell distribution in integrated data clusters as in a. c, Percentage of WT and GFI1ΔCD8 CD8+ P14 T cells in each cluster as in a. d, UMAP showing normalized expression of Btg1, E2f2, Eomes and Tcf7 in WT and GFI1ΔCD8 CD8+ P14 T cells as in a. e, Dot plot showing expression of selected genes in WT and GFI1ΔCD8 CD8+ P14 T cells in clusters 1–7 as in a. Dot size indicates fraction of cells expressing gene; color represents mean expression. f, DEGs (top) and DARs (bottom) from cluster 1 as in a, using pseudobulk analysis of scRNA-seq and scATAC-seq, respectively. g, Selected top-ranking transcription-factor-linked eRegulons predicted by SCENIC+ analysis using scRNA-seq and scATAC-seq as in a. Color scale shows gene expression-based enrichment score; dot size illustrates chromatin accessibility-based enrichment score for each eRegulon and cell cluster. h, WT and GFI1ΔCD8 CD8+ T cell chromatin accessibility and gene expression at Tcf7 gene locus in cluster 1 as in a. Dashed boxes highlight differentially accessible chromatin regions.
To further understand the gene regulatory networks of each cluster, SCENIC+ analysis33 identified enhancer-driven gene regulatory networks and candidate upstream transcription factors for each cell cluster (Fig. 5g). The top-ranked eRegulon for cluster 1 was regulated by Tcf7 (Fig. 5 and Extended Data Fig. 8f), in line with the high expression of memory-related transcription factors such as Eomes, Id3 and Myb within that cluster. The chromatin region enrichment score of Tcf7-driven eRegulon was higher for cluster 1 GFI1ΔCD8 CD8+ T cells owing to upregulated DARs in the Tcf7 gene body, while the chromatin accessibility at the 5′-untranslated region of Tcf7 was reduced (Fig. 5h), in agreement with reduced Tcf7 expression in GFI1ΔCD8 CD8+ T cells (Fig. 5e). Additionally, gene regulatory network analyses identified that E2f1-driven and E2f7-driven direct gene networks important for regulating cell proliferation were disrupted in GFI1ΔCD8 CD8+ T cells (Fig. 5g and Extended Data Fig. 8g). These findings suggest that GFI1 is a key epigenetic modulator of transcriptional networks critical for memory CD8+ T cell proliferation.
GFI1 enhances CD8+ T cell recall responses and survival
Given the changes in gene expression affecting cell proliferation, TCR and cytokine-mediated proliferation of GFI1ΔCD8 CD8+ TM cells, isolated from spleen of LCMVc13-infected mice at day 7 after infection, was quantitated by culturing CD8+ TM cells in vitro with IL-2 + IL-7 or IL-2 + CD3/CD28 stimulation beads (Extended Data Fig. 9a). This showed that fewer GFI1ΔCD8 CD8+ T cells entered division when exposed to cytokines IL-2 + IL-7 and exhibited extremely limited expansion compared with WT CD8+ T cells (Extended Data Fig. 9b,c). Although TCR activation was able to drive strong proliferation by day 4 post-stimulation in all conditions, GFI1ΔCD8 CD8+ T cells showed significantly less proliferation than WT CD8+ T cells (Extended Data Fig. 9b,c). To investigate whether the impaired proliferation of GFI1ΔCD8 CD8+ T cells observed following primary infection also impacted CD8+ T cell recall responses, we isolated WT and GFI1ΔCD8 CD8+ TM cells from LCMVc13-infected mice at day 5 after infection and co-transferred them to a secondary host followed by heterologous infection with LCMVArm or MCMV-ie2-gp33 (Extended Data Fig. 9d). This procedure showed that recall responses by GFI1ΔCD8 CD8+ TM cells were severely impaired (Fig. 6a,b). To test the antiviral capacity of GFI1ΔCD8 CD8+ TM cells, WT or GFI1ΔCD8-activated P14 CD8+ T cells were transferred to Rag2–/–Il2rγ–/– mice, which lack natural killer cells and CD4+ T cell responses to MCMV34. Although WT CD8+ T cells were able to mediate effective protection against MCMV infection, mice receiving GFI1ΔCD8 CD8+ T cells exhibited severe weight loss (Fig. 6c). This finding correlated with impaired virus control in the lungs and liver in mice reconstituted with GFI1ΔCD8 CD8+ T cells (Fig. 6d). These results demonstrate that GFI1 is crucial for recall responses and antiviral functions of memory CD8+ T cells.
a,b, Representative plot (left) and quantification (right) of donor WT and GFI1ΔCD8 CD8+ T cells at D7 post infection with LCMVArm (a) or MCMV-ie2-gp33 (b) in the spleen of secondary C57BL/6 recipients transferred with 1:1 mix of congenically labeled activated WT and GFI1ΔCD8 CD8+ T cells isolated at D7 post infection with LCMVc13 from the spleen of primary C57BL/6 recipients that received congenically labeled WT or GFI1ΔCD8 CD8+ TN cells 24 h before infection with LCMVc13. Data are pooled from two experiments (n = 9 or 10 mice); mean values are shown; error bars, s.e.m. c, Weight-loss kinetics of Rag2–/–Il2rγ–/– mice that received activated WT or GFI1ΔCD8 CD8+ P14 T cells from C57BL/6 mice that received WT or GFI1ΔCD8 CD8+ P14 TN cells 1 day before infection with LCMVc13. d, Virus titer in the lungs and liver of Rag2–/–Il2rγ–/– at D14 post transfer of activated WT or GFI1ΔCD8 CD8+ P14 T cells as in c. Dashed line, assay detection limit. e, scMultiome-seq data showing expression of Bcl2 in splenic WT and GFI1ΔCD8 CD8+ T cells isolated at D7 post infection from C57BL/6 mice that received congenically labeled WT or GFI1ΔCD8 CD8+ TN cells 24 h before LCMVc13 infection. f, Representative histogram (top) and quantification (bottom) of BCL-2 expression at D7 in splenic CD8+ P14 T cells isolated from C57BL/6 mice transferred with a 1:1 mix of congenically labeled WT and GFI1ΔCD8 TN cells 24 h before LCMVc13 infection. Data are representative of two experiments (n = 6 mice). g, Cluster 1 WT (blue) and GFI1ΔCD8 (red) CD8+ T cell chromatin accessibility and gene expression at the Bcl2 locus as in e. Dashed boxes, DARs. h, Representative plots (left) and quantification (right) of splenic WT and GFI1ΔCD8 caspase-3+CD8+ T cells at D7 post LCMVc13 infection as in f. Control, FMO staining of WT CD8+ T cells. Data are representative of two experiments (n = 5 mice). i, BCL-2 expression in CD8+ TN cells from the spleen of naive WT and GFI1ΔCD8 mice; mean ± s.e.m. Data are pooled from two experiments (n = 6 or 7 mice per genotype). P values were calculated using a two-tailed paired t-test (a, b, f and h) or two-tailed Student’s t-test (c, d and i). Data in c and d are representative of two experiments; mean values are shown; error bars, s.e.m. (n = 4, 5 or 6 mice per condition).
scRNA-seq of spleen CD8+ TM cells isolated day 7 after LCMVc13 infection showed that anti-apoptotic genes such as Bcl2, Mcl1 and Xiap were diminished, while pro-apoptotic genes like Bax and Bid35,36 were elevated in GFI1ΔCD8 CD8+ T cells compared to WT CD8+ T cells (Extended Data Fig. 9e). Bcl2 transcript levels were notably lower in clusters 1 and 2 in GFI1ΔCD8 CD8+ T cells compared to WT CD8+ T cells (Fig. 6e and Extended Data Fig. 9e). This paralleled lower BCL-2 expression (Fig. 6f) and reduced accessibility at the Bcl2 locus (Fig. 6g) in GFI1ΔCD8 CD8+ TSCM cells. Elevated caspase-3 activation indicated increased apoptosis in GFI1ΔCD8 CD8+ T cells (Fig. 6h). Similar patterns were observed following MCMV infection (Extended Data Fig. 9f,g). However, GFI1ΔCD8 CD8+ TN cells had normal BCL-2 expression (Fig. 6i), indicating the specific role of GFI1 in CD8+ T cell survival post activation and its action to inhibit activation-induced T cell death. Altogether, these data show that GFI1 promotes CD8+ T cell proliferation, survival and virus control capacity.
EOMES rescues GFI1ΔCD8 CD8+ T cell persistence
TCF1 promotes memory CD8+ T cell persistence through EOMES37. Given the downregulation of EOMES in CD8+ TSCM cells (Fig. 7a,b) and reduced chromatin accessibility at the Eomes locus in GFI1ΔCD8 CD8+ TM cells (Fig. 7c), we examined the requirement for GFI1-mediated EOMES in maintaining the persistence of CD8+ T cells following chronic viral infection. WT and GFI1ΔCD8 P14 CD8+ T cells transduced with Eomes were adoptively transferred into congenic recipient mice that were infected with LCMVc13. EOMES overexpression partially rescued the number of GFI1ΔCD8 CD8+ T cells (Fig. 7d,e) but did not impact proliferation, despite increased BCL-2 expression (Fig. 7f,g), suggesting that GFI1 drove CD8+ T cell survival partly by regulating EOMES but requires additional regulators to fully support proliferation.
a, Representative histogram (left) and quantification (right) of EOMES expression in splenic WT and GFI1ΔCD8 CD8+ P14 T cells at D7 post infection in C57BL/6 mice that received a 1:1 mix of congenically labeled WT and GFI1ΔCD8 CD8+ P14 T cells 24 h before infection with LCMVc13. Data are pooled from two experiments (n = 10 mice per time point). b, Representative histogram (left) and quantification (right) of EOMES expression in splenic WT and GFI1ΔCD8 CD8+ TSCM cells at D7 post LCMVc13 infection as in a. One of two experiments is shown (n = 5 mice). c, Chromatin accessibility at Eomes locus in WT (blue) and GFI1ΔCD8 (red) CD8+ T cells isolated at D7 post infection from the spleen of C57BL/6 mice that received congenically labeled WT or GFI1ΔCD8 CD8+ TN cells 24 h before infection with LCMVc13; analyzed with scMultiome-seq. Dashed boxes, DARs. d,e Representative plots (d) and quantification (e) of WT and GFI1ΔCD8 CD45.1+CD8+ P14 T cells at D14 in the spleen of CD45.2+ C57BL/6 mice transferred with WT or GFI1ΔCD8 CD45.1+CD8+ P14 T cells transduced with control or EOMES-expressing (EOMES-OE) lentivirus and infected with LCMVc13 24 h later. f, Ki-67 expression in splenic WT and GFI1ΔCD8 CD8+ P14 T cells as in d. g, Representative histograms (left) and quantification (right) of BCL-2 expression in splenic WT and GFI1ΔCD8 CD8+ P14 T cells as in d. h, Representative flow cytometry of splenic EOMEShiCD8+ P14 T cells at D7 post infection in C57BL/6 mice that received congenically labeled EomesmCherry/+ CD8+ P14 TN cells 24 h before infection with LCMVc13. Data are representative of two independent experiments. i, Representative flow cytometry plot (left) and quantification (right) of splenic CD45.1+CD8+ P14 T cells at D21 post LCMVc13 infection in secondary infection recipients that at D7 post infection received matched (D7 post infection) activated CD45.1+EOMEShi or CD45.1+EOMESlo CD8+ T cells isolated from primary-infected C57BL/6 mice, which were adoptively transferred with EomesmCherry/+ CD8+ P14 TN cells 1 day before primary infection with LCMVc13. Data are from one experiment; mean values are shown; error bars, s.e.m. (n = 5 mice per condition). P values were calculated using a two-tailed paired t-test (a, b and j) or two-tailed Student’s t-test (e–g). Data in d–g are pooled from two experiments (n = 4 or 6 mice per condition); mean values are shown; error bars, s.e.m.
Examination of P14 CD8+ T cells from EomesmCherry reporter mice revealed approximately 5–10% of CD8+ T cells had high expression of EOMES on day 7 after LCMVc13 primary infection (Fig. 7h). Adoptive transfer of these EOMEShi T cells into infection-matched secondary recipients showed that these EOMEShiCD8+ T cells had higher re-population capacity than EOMESlo CD8+ T cells (Fig. 7i). These results indicate that early emerging EOMES-expressing CD8+ T cells are crucial for long-term persistence during chronic infection, highlighting the importance of GFI1-driven EOMES expression for antiviral T cell memory.
Continuous GFI1 expression maintains CD8+ T cell persistence
To assess whether GFI1 is essential for long-term CD8+ T cell maintenance after chronic infection, congenically labeled naive R26creERT2/+Gfi1fl/+ and R26creERT2/+Gfi1fl/fl P14 CD8+ T cells were co-transferred into C57BL/6 recipients and infected with MCMV-ie2-gp33 virus (Extended Data Fig. 10a). Tamoxifen-induced GFI1 ablation resulted in a reduction of R26creERT2/+Gfi1fl/fl CD8+ T cell numbers in blood and tissues compared to R26creERT2/+Gfi1fl/+ CD8+ T cells (Fig. 8a,b and Extended Data Fig. 10b), significantly decreasing the GFI1-deficient TSCM cell population by day 28 (Extended Data Fig. 10c). TCF1, EOMES and T-BET expression were impaired at day 14 (Extended Data Fig. 10d) and remained low at day 28 (Fig. 8c), whereas BCL-2 levels were unchanged (Fig. 8c) in R26creERT2/+Gfi1fl/fl CD8+ T cells. This was accompanied by reduced proliferation of R26creERT2/+Gfi1fl/fl P14 CD8+ T cells at day 28 but not at day 14 (Fig. 8d). Similarly, tamoxifen-mediated GFI1 ablation in R26creERT2/+Gfi1fl/fl at day 15 post LCMVc13 infection led to reduced R26creERT2/+Gfi1fl/fl CD8+ T cell persistence (Extended Data Fig. 10e). Thus, continuous expression of GFI1 is crucial for sustaining persistent CD8+ T cell responses in chronic and latent infections.
a, Representative flow cytometry plot (left) and quantification (right) of R26Cre-ERT2/+Gfi1fl/+ and R26Cre-ERT2/+Gfi1fl/fl CD8+ P14 T cells in peripheral blood of MCMV-ie2-gp33-infected C57BL/6 mice that received 1:1 mix of congenically labeled R26Cre-ERT2/+Gfi1fl/+ and R26Cre-ERT2/+Gfi1fl/fl CD8+ P14 T cells 30 days before tamoxifen treatment and were infected with MCMV-ie2-gp33 1 day post CD8+ T cell adoptive transfer; mean values are shown; error bars, s.e.m. b, Number of R26Cre-ERT2/+Gfi1fl/+ and R26Cre-ERT2/+Gfi1fl/fl CD8+ P14 T cells in spleen at D14 and D28 after tamoxifen treatment as in a; mean values are shown; error bars, s.e.m. c, Expression of TCF1, EOMES and BCL-2 in R26Cre-ERT2/+Gfi1fl/+ and R26Cre-ERT2/+Gfi1fl/fl CD8+ P14 T cells at D28 after tamoxifen treatment as in a. d, Representative histogram at D28 (right) and quantification at D14 and D28 (left) of Ki-67 expression in splenic R26Cre-ERT2/+Gfi1fl/+ and R26Cre-ERT2/+Gfi1fl/fl CD8+ P14 T cells as in a. In b–d, data are pooled from two independent experiments (n = 9 or 10 mice per time point). P values were calculated using a two-tailed paired t-test.
Discussion
Here, we defined a role of GFI1 in regulating CD8+ TSCM cell persistence. CD8+ TN cells had high expression of GFI1, and although GFI1 was rapidly downregulated in effector cells following activation, the CD8+ TSCM cell population selectively maintained high GFI1 expression. GFI1 ablation markedly impaired TM cells during chronic infection owing to reduced proliferation and increased GFI1-deficient CD8+ T cell death. Furthermore, we showed that continuous GFI1 expression was required to sustain CD8+ T cell proliferation during chronic infection. Collectively, these observations indicated that GFI1 maintained CD8+ TSCM cell persistence by promoting proliferation and by inhibiting EOMES-dependent activation-induced cell death.
CD4+CD8+ (double-positive) thymic T cells express high levels of GFI1 (ref. 22), which evicts autoreactive T cells from the thymus to induce tolerance38. CD8+ T cells have been reported to transiently upregulate GFI1 after in vitro stimulation with concanavalin A22. We found that GFI1 was downregulated in CD8+ TEFF cells following chronic infection but was maintained in memory subsets, including CD8+ TSCM and CD8+ TCM cells. Mapping GFI1 expression amongst CD8+ TPEX populations showed that CD62L+CD8+ TPEX cells maintained higher proliferation capacity23 and exhibited higher GFI1 than CD62L−CD8+ TPEX cells, suggesting that GFI1 allowed CD62L+CD8+ TPEX cells to retain proliferation characteristics.
GFI1 was found to be crucial for maintaining CD8+ T cell persistence during chronic viral infection. This lack of CD8+ T cell persistence could be attributed to reduced proliferation of GFI1-deficient CD8+ TSCM cells, especially cytokine-driven proliferation. We observed impaired cell cycle regulation in GFI1-deficient CD8+ TSCM cells, consistent with a previous observation that GFI1 is required for thymic T cell proliferation during the development17 and proliferation of other cell types, including hematopoietic stem cells39. GFI1 maintained the proliferation of CD8+ TSCM cells by promoting gene regulatory networks driven by the E2F family, notably E2f1 and E2f7. HOMER analysis showed a GFI1 binding motif in the promotor of E2f7, suggesting direct epigenetic regulation. Moreover, GFI1 promoted CD8+ TSCM cell persistence by epigenetically silencing the expression of Btg1 and Btg2, which has been shown to inhibit T cell quiescence40. We found that continuous expression of GFI1 was necessary to maintain CD8+ TSCM cell proliferation, indicating that stable GFI1 expression in TSCM cells was required to sustain the cell cycle regulatory circuits necessary for memory CD8+ T cell proliferation.
Transcriptional mapping showed that GFI1 loss impaired the expression of the transcriptional regulators Id2 and Zeb2, which are important for TEFF cell differentiation and CD8+ TSLEC cell formation25,41. Reduced proliferation of CD8+ TCM cells during latent MCMV infection was shown to impair CD8+ TSLEC cell development, resulting in loss of MCMV-specific memory inflation30. Here, the impaired proliferation and subsequent loss of CD8+ TSCM cells in the absence of GFI1 provides an explanation for the impaired MCMV-specific memory inflation in the GFI1-deficient CD8+ T cells. This highlights the requirement for GFI1 expression in CD8+ T cells throughout the course of chronic viral infection to generate immune protection.
We showed that GFI1 promoted BCL-2 expression in CD8+ TSCM cells in an EOMES-dependent manner. Although GFI1 loss in CD8+ TN cells did not alter EOMES or BCL-2 expression, GFI1-deficient CD8+ TSCM cells exhibited reduced BCL-2. This reduction paralleled reduced chromatin accessibility at the Bcl2 locus in CD8+ TSCM cells and elevated death in these cells. This GFI1-mediated BCL-2 expression in CD8+ TSCM cells is consistent with previous reports that BCL-2 overexpression protects GFI1-deficient thymic T cells from apoptosis17,42. The loss of GFI1 in CD8+ TN cells did not impact the chromatin accessibility, but it altered the epigenetic profile of activated CD8+ T cells, indicating that GFI1 regulated CD8+ T cell chromatin accessibility following T cell activation. Thus, GFI1-mediated epigenetic gene regulation of CD8+ TSCM cells promotes BCL-2 expression following CD8+ T cell activation but not in CD8+ TN cells. The transcription factor EOMES was dysregulated in activated T cells following GFI1 ablation. EOMES inhibits activation-induced T cell death by promoting BCL-2 expression37,43. Overexpression of EOMES induced BCL-2 expression and partially rescued GFI1-deficient CD8+ T cell persistence. Impaired BCL-2 expression has also been reported in TCF1-deficient CD8+ T cells, and similar to GFI1-deficient CD8+ T cells, this phenotype was rescued by EOMES overexpression37. A binding motif for GFI1 in the Eomes promotor suggests that GFI1 directly regulates EOMES expression to support TM cell but not TN cell survival. Collectively, these findings demonstrate that GFI1 drives EOMES-dependent BCL-2 expression in CD8+ T cells to generate ‘fit’ TM cells.
In conclusion, this study reveals an important function for GFI1 in maintaining CD8+ T cell persistence following chronic viral infection. We demonstrate that GFI1 is a key regulator of memory CD8+ T cells that sculpts their gene regulatory network by modulating epigenetic repression. Finally, we demonstrate that selective GFI1 expression in CD8+ TSCM cells ensures their long-term persistence by promoting enduring proliferative potential required for self-renewal and maintenance of memory populations.
Methods
Mice
C57BL/6 (CD45.2+/+), B6.SJL-PtprcaPep3b/BoyJ (CD45.1+/+), Gfi1tdTomato/+ (refs. 20,21), EomesmCherry/+ (ref. 44), Rag2–/–Il2rγ–/–, B6.Tg(Cd8a-cre)1Itan (Cd8acre/+)28, B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J (ref. 45) (R26cre/ERT2/+) and Gfi1fl/fl (ref. 46) mice have been described previously. B6.Cg-Tcratm1MomTg(TcrLCMV)327Sdz/TacMmjax (P14) mice carry CD8+ P14 T cells transgenic for the T cell antigen receptor specific for the LCMV-derived gp33-41 epitope47. Gfi1tdTomato/+ mice were backcrossed to the C57BL/6 background for at least ten generations. Gfi1fl/fl and EomesmCherry/+ were crossed with CD45.1+/+ and P14 strains to generate congenically labeled strains. The Cd8acre/+ strain was crossed to Gfi1fl/flCD45.1+/+ mice to generate congenically labeled mice selectively lacking GFI1 in CD8+ T cells. Male and female mice were used at 6–16 weeks old unless otherwise indicated. All mice were bred and maintained under specific-pathogen-free conditions at the animal facility of the University of Queensland. Mice were housed under a 12 h light/12 h dark cycle at 22 ± 2°C and 55 ± 15% humidity. All animals were handled according to the guidelines of the Australian Code for the Care and Use of Animals of the National Health and Medical Research Council of Australia. Experimental procedures were approved by the Animal Ethics Committees of the University of Queensland.
Cell lines and virus infection
BHK-21 (CCL-10), M2-10B4 (CRL-1972) and Vero E6 (CRL-1586) cells were obtained from ATCC. All cell lines were maintained in DMEM supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 U ml−1 penicillin and 100 μg ml−1 streptomycin.
LCMVArm and LCMVc13 were propagated and titrated on BHK-21 and Vero E6 cells, respectively. MCMV was derived from pSM3fr-MCK-2fl clone 3.3 BAC. MCMV-ie2-gp33 was kindly provided by L. Cicin-Sain48. MCMV strains were reconstituted by BAC transfection of M2-10B4 cells. After reconstitution, the virus was propagated on M2-10B4 cells. Virus stocks were prepared according to a previously described protocol49. Mice were infected intraperitoneally with 2 × 105 plaque-forming units (PFU) of LCMVArm for acute viral infection. For chronic LCMV infection, mice were infected intravenously with 2 × 106 PFU of LCMVc13. For MCMV infection, animals were infected intraperitoneally with 2 × 105 PFU of cell culture grown virus. To determine MCMV replication, tissues were homogenized and titrated on M2-10B4 cells as described previously30.
Tamoxifen and FK506 treatment
Tamoxifen was dissolved in corn oil at a concentration of 20 mg ml−1. Infected C57BL/6 mice were treated by injecting 100 mg tamoxifen per kg body weight by intraperitoneal injection. Tamoxifen was administered once every 24 h for four consecutive days. MCMV-infected mice were treated with tamoxifen at 30 days after infection, followed by a rest period of 14–28 days before analyses. LCMVc13-infected mice were treated 15 days after infection, rested and then analyzed at 30 days after infection. FK506 was dissolved in 30% PEG400 and 2% Tween 80 in PBS. For FK506 treatment, 10 mg FK506 per kg body weight or vehicle was injected intraperitoneally daily from day 4–6 after virus infection.
Cell isolation and flow cytometric analyses
Single-cell suspensions were generated by forcing tissues through 70 μm cell strainers, and red blood cells were removed using hypotonic lysis using ACK buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA pH 7.4). Peripheral blood samples were collected by retro-orbital bleeding, and red blood cells were lysed using ACK buffer. Lungs were perfused with approximately 5 ml PBS through the right ventricle to remove circulating blood. The lungs were placed in collagenase type IV (1 mg ml−1; Worthington), deoxyribonuclease I (200 μg ml−1; Roche) and dispase (0.4 U ml−1; Gibco) in complete RPMI medium (RPMI 1640 medium containing 10% heat-inactivated FCS, 1 mM l-glutamine, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and 50 μM β-mercaptoethanol) and then dissociated and homogenized using the gentleMACS Dissociator (Miltenyi Biotec) mouse lung digestion protocol setting. After dissociation, mononuclear cells were purified by gradient centrifugation using a 40–80% Percoll gradient. Cell suspensions were blocked with PBS containing 5 μg ml−1 anti-CD16/CD32 (2.4G2) and stained (30 min on ice) with fluorophore-conjugated antibodies or reagents in FACS buffer (PBS containing 2.5% heat-inactivated FCS and 50 mM EDTA), unless stated otherwise. Cell suspensions were incubated with fluorophore-conjugated MHCI tetramers for 30 min at 25 °C. M45-specific (H-2Db restricted peptide HGIRNASFI) and M38-specific (H-2Kb restricted peptide SSPPMFRV) MHCI–biotin monomers were provided by the National Institutes of Health Tetramer Core Facility, and tetramers were generated using streptavidin–fluorophore conjugates. Antigen-specific CD8+ T cell cytokine analysis was performed by incubating single-cell suspensions with 1 µg ml−1 KAVYNFATM (H-2Db-restricted) peptide in complete RPMI medium for 1 h at 37 °C, followed by 10 μg ml−1 brefeldin A (Golgiplug; BD Pharmingen) addition and further 5 h incubation. For intracellular staining, surface-labeled cells were fixed using eBioscience Foxp3/Transcription Factor Staining Buffer (Thermo Fisher) and then stained for intracellular cytokines or transcription factors. Live cells were identified by exclusion staining with a fixable viability dye (BD Biosciences or BioLegend) or 7-AAD (BD Biosciences). All antibodies and staining reagents used in the study are outlined in Supplementary Table 12. Flow cytometry analysis was performed on a Cytek Aurora (Cytek Biosciences) or LSRFortessa X-20 (BD Biosciences), and analysis was performed using FlowJo software (v.10.10) (BD Biosciences).
Adoptive CD8+ T cell transfer
For primary population transfer experiments, naive Cd8acre/+Gfi1+/+ P14 (WT P14), Cd8acre/+Gfi1fl/fl P14 (GFI1ΔCD8 P14) or Gfi1tdTomato/+ CD8+ T cells were isolated from naive mice spleens using the TN CD8a+ T Cell Isolation Kit (Miltenyi Biotec) according to the manufacturer’s instructions. Congenically labeled naive CD8+ P14 WT (CD45.1+ or CD45.1+CD45.2+) and GFI1-deficient (GFI1ΔCD8, CD45.1+ or CD45.1+CD45.2+) P14 T cells mixed at a 1:1 ratio (5 × 103 cells of each type) were adoptively transferred into C57BL/6 (CD45.2+) recipient mice. Then, 1 day later, recipient mice were infected with the specified virus. For secondary transfer of transgenic cells, P14 T cells were first enriched from the spleen and lymph nodes of primary recipients using the CD8a+ T Cell Isolation Kit (Miltenyi Biotec). Cells were then stained with anti-mouse CD3ε, anti-mouse CD8a, anti-mouse CD45.1, anti-mouse CD45.2 and 7-AAD to allow discrimination of live and dead cells. Cells were then flow-cytometrically sorted on a BD FACSAria II (BD Biosciences) or Aurora CS Cell Sorter (Cytek Biosciences), and 1 × 104 cells were transferred into each recipient. The secondary response and expansion capacity of GFI1hi and GFI1lo CD8+ T cells was evaluated following adoptive transfer of 1 × 105 cells into the secondary host. For evaluating virus control capacity of CD8+ T cells, 1 × 105 activated WT or GFI1ΔCD8 P14 T cells were transferred into Rag2–/–Il2rγ–/– mice followed by MCMV-ie2-gp33 infection the next day. Viral titer was determined at day 14 after infection or at the time of death in liver and lungs. Unless specified otherwise, equal numbers of cells of each CD8+ T cell subset were injected for secondary transfer into naive or infection-matched secondary hosts.
In vitro T cell culture
Naive WT and GFI1ΔCD8 P14 T cells were isolated using the Naive CD8a+ T Cell Isolation Kit (Miltenyi Biotec). Enriched naive congenically labeled P14 T cells were adoptively transferred into C57BL/6 (CD45.2+) recipients, which were infected with LCMVc13 24 h later. P14 T cells were isolated from the spleen of infected animals 5 days after infection using the BD FACSAria II (BD Biosciences) or Aurora CS Cell Sorter (Cytek Biosciences). The CD8+ T cells were labeled with 5 μM CellTrace Violet dye (Thermo Fisher) according to the manufacturer’s recommended protocol, and 5 × 103 T cells were cultured in complete RPMI 1640 medium. Complete RPMI medium was supplemented with either IL-2 (30 U ml−1; Thermo Fisher), IL-2 + IL-7 (10 ng ml−1 IL-7; Thermo Fisher) or IL-2 + CD3/CD28 beads (Dynabeads Mouse T-Activator CD3/CD28, Thermo Fisher). Dynabeads Mouse T-Activator CD3/CD28 beads were added at a 1:1 ratio to cells per well. CellTrace Violet expression and T cell expansion were quantified using Cytek Aurora (Cytek Biosciences).
Generation of bone marrow chimeric mice
C57BL/6 recipient mice (6–10 weeks old) were lethally irradiated with two doses of 5.5 Gy (3 h apart). Bone marrow cells were isolated from Cd8acre/+Gfi1+/+ or Cd8acre/+Gfi1fl/fl (CD45.1+) and C57BL/6 (CD45.2+) donor mice by flushing the femoral and tibial bones with 3 × 1 ml sterile FACS buffer to create a single-cell suspension. Red blood cells were lysed using ACK buffer and then washed twice with FACS buffer. Live cells were enumerated using Trypan blue exclusion. Cd8acre/+Gfi1fl/fl or Cd8acre/+Gfi1+/+ (CD45.1+ or CD45.1+CD45.2+) bone marrow cells were mixed in a 1:1 ratio with C57BL/6 (CD45.2+) bone marrow cells, and 2–4 × 106 mixed bone marrow cells were then adoptively transferred into the irradiated recipients. Chimeric mice were allowed 6–10 weeks to fully reconstitute their hematopoietic system with donor bone marrow cells before viral infection.
Lentivirus transduction
An EOMES overexpression construct was generated by conjugating the EOMES open reading frame with an EF1a-driven enhanced green fluorescent protein (EGFP) using a T2A linker (pLV-EF1a-EGFP-T2A-EOMES). Lentiviruses were produced by the University of Queensland Viral Vector Core. CD3/CD28 bead-activated P14 T cells were spinoculated with lentiviruses carrying a control (pLV-EF1a-EGFP-T2A-Puro) or the EOMES overexpression construct. In brief, 200 µl of the lentivirus suspension was centrifuged at 3,000g at 32 °C for 2 h in a 48-well plate coated with RetroNectin (Takara Bio) according to the manufacturer’s instructions. Next, 5 × 105 P14 T cells resuspended in complete RPMI medium supplemented with 100 ng ml−1 of mIL-2 was added to 400 µl per well. The cells were then centrifuged at 800g at 32 °C for 1.5 h. After 2 d in vitro culture, transduced CD8+ T cells were sorted by flow cytometry to enrich for GFP+-transduced P14 cells. 1–5 × 103 GFP+P14 (CD45.1+ or CD45.1+CD45.2+) CD8+ T cells were adoptively transferred into C57BL/6 recipient mice, which were then infected with LCMVc13.
RNA isolation and bulk RNA-seq
P14 CD8+ T cells were sorted from splenocytes isolated from mice on day 7 or day 21 after viral infection. Total RNA was extracted using RNeasy Plus Micro kit (Qiagen) according to the manufacturer’s instructions. The quality and integrity of total RNA was measured using Bioanalyzer or TapeStation systems (Agilent Technologies). Libraries were prepared using the TruSeq Stranded Total RNA Kit (Illumina) or NEBNext Single Cell/Low Input Library Prep Kit (New England Biolabs) and sequenced using a NovaSeq S1 PE100 flow cell (Illumina) or NovaSeq SP 100 flow cell (Illumina).
RNA-seq read quality was assessed, and low-quality reads were trimmed with fastp (v.0.22.0)50. Reads were mapped to the mouse genome (mm10) using STAR (v.2.7.10)51 and quantified with featureCounts (v.2.0.1)52. Read counts were normalized, and differential gene expression was quantified with DESeq2 (v.1.4.0). A log(fold change) larger than one and a false discovery rate cutoff of 5% was used to select significantly over-represented and under-represented genes. Gene set enrichment analysis was performed using clusterProfiler (v.4.8.3). Volcano plots and heatmaps were plotted using EnhancedVolcano (v.1.18.0) and pheatmap (v.1.0.12) packages, respectively.
Promotor motif discovery analysis
A list of DEGs, expressed in GFI1ΔCD8 at day 7 after LCMVc13 infection (Supplementary Table 5), was used to discover the presence of the GFI1 binding motif in the promotor and enhancer regions of sequences using the findMotifs.pl command (HOMER v.5.1) with default parameters. The GFI1 binding motif weight matrix was downloaded from the SwissRegulon Portal or JASPAR databases.
Bulk ATAC-seq
ATAC-seq was performed using the Omni-ATAC protocol53 with minor modifications. P14 CD8+ T cells were flow-cytometrically sorted from mice on day 3, 5 or 7 after virus infection. A total of 50,000 sorted P14 CD8+ T cells were lysed to extract nuclei using cell lysis buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.1% Tween-20, 0.1% IGEPAL CA-630, 0.01% digitonin and 1% BSA). Nuclei were washed and resuspended in 50 μl 1× TDE1 buffer (Illumina) containing 2.5 μL TDE1 transposase (Illumina). The transposase reaction was conducted at 37 °C for 30 min with mild shaking. Library amplification and barcoding were performed with NEBNext Ultra II Q5 Master Mix (New England Biolabs) using IDT dual index primer set (Integrated DNA Technologies). PCR was conducted for 10–11 cycles. Library purification was performed with the MinElute PCR Purification Kit (Qiagen), and library size distribution was assessed using the TapeStation High Sensitivity DNA Kit (Agilent). ATAC-seq libraries were quantified before pooling and sequencing using the real-time NEBNext Library Quant Kit for Illumina (New England Biolabs). Paired-end sequencing was performed on a NovaSeq SP 100 flow cell (Illumina) with 50 cycles for each read.
Sequencing read quality was assessed, and low-quality reads were trimmed with fastp (v.0.22.0)50. These trimmed reads were mapped to the mouse genome (mm10) using bowtie2 (v.2.4.2) with standard parameters54. Picard (v.2.26.4) was used to remove PCR duplicates. The deduplicated reads were then filtered to remove mitochondrial chromosome, Y chromosome, improperly paired and non-mapping reads using samtools flags. Peak summits were called using macs2 (v.2.2.9.1) using parameters --nomodel, --keep-dup all and --call-summits. ATAC-seq library normalization was performed using the trimmed mean of M values method. Identification of differentially accessible regions (fold change of at least one and a false discovery rate of <0.05) was performed using DiffBind (v.3.12.0). Peaks were annotated using ChIPpeakAnno (v.3.36.1)55.
scMultiome-seq
scmultiome-seq was performed using the 10× Single Cell Multiome ATAC + GEX analyses kit (10× Genomics). P14 CD8+ T cells were FAC-sorted from mice at day 7 after LCMVc13 infection. CD8+ T cells were pooled from 3–5 mice, and ~1 × 105 cells were used for nuclei isolation. Cells were pelleted for 5 min at 300g followed by nuclei isolation by incubating cells with chilled cell lysis buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.1% Tween-20, 0.1% IGEPAL CA-630, 0.01% digitonin, 1 mM dithiothreitol, 1 U μl−1 RNase inhibitor and 1% BSA) for 3 min. Nuclei were washed twice in 1 ml of wash buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% Tween-20, 1 mM dithiothreitol, 1 U µl−1 RNase inhibitor and 1% BSA) by centrifuging at 500g for 5 min. After centrifugation, cells were resuspended in chilled Nuclei Buffer (1× Nuclei Buffer, 1 mM dithiothreitol and 1 U µl−1 RNase inhibitor). Nuclei were incubated in a transposition mix according to the Chromium Next GEM Single Cell Multiome ATAC + GEX user guide (protocol CG000338 Rev F). Following transposition, GEMs were generated using 10× Chip J. Sample cleanup and amplification PCR were performed as per the user guide. For the ATAC-seq library, eight PCR cycles were run, and seven PCR cycles were used for cDNA amplification. ATAC libraries were sequenced using the NovaSeq SP 100 flow cell (Illumina) with the following read protocol: 50 cycles (read 1), 8 cycles (i7 index read), 24 cycles (i5 index read) and 49 cycles (read 2). RNA libraries were sequenced on a NovaSeq SP 100 flow cell (Illumina) with the following settings: 28 cycles (read 1), 10 cycles (i7 index read), 10 cycles (i5 index read) and 90 cycles (read 2).
Single-cell multiome data processing
RNA and ATAC raw reads from WT and GFI1ΔCD8 T cell samples were processed with CellRanger-arc (v.2.0.2) to map RNA transcripts and ATAC peaks to the mm10 reference genome. Seurat objects (WT and GFI1ΔCD8 T cells) were created using RNA matrix files using Seurat (v.5.0.3)56. The ATAC data were added to the Seurat object using CreateChromatinAssay (Seurat). Transcription start site enrichment and nucleosome signal scores were calculated using Signac (v.1.12.0)57. Quality control was performed by filtering cells with the following criteria: transcription start site enrichment score of >1, a nucleosome signal score of <2, between 100 and 15,000 total RNA counts, between 2,000 and 30,000 total ATAC counts and percent mitochondrial counts of <20. Cell cycle scores were assigned based on G2/M and S phase variability scores using the Seurat CellCycleScoring function. The Seurat object was split into RNA and ATAC objects for individual processing.
RNA gene expression unique molecular identifier count data were normalized using SCTransform, and principal component analysis was performed on the SCTransformed Pearson residual matrix using the RunPCA function in Seurat. We found the 50 nearest neighbors for each cell using the Louvain algorithm with the FindNeighbors function. After preprocessing, WT and GFI1ΔCD8 T cell RNA data were merged and integrated using the IntegrateLayers (RPCAIntegration) function of Seurat.
For WT and GFI1ΔCD8 T cell ATAC data, consensus peaks were called using the Signac CallPeaks function. ATAC data were processed by computing term-frequency inverse-document-frequency and running singular value decomposition using the Signac RunTFIDF and RunSVD function. Dimension reduction was performed on the ATAC dataset using latent semantic indexing (LSI) and UMAP. Then, graph-based clustering was performed on LSI components 2 to 30 by first computing a shared nearest neighbor graph using LSI low-dimensional space and then applying the Louvain algorithm using the FindNeighbors function followed by FindClusters with algorithm = 2 in Seurat. WT and GFI1ΔCD8 T cell ATAC data were integrated using low-dimensional cell embeddings across datasets using the Signac IntegrateEmbeddings function. Finally, UMAP dimensional reduction was performed using integrated LSI to visualize the integrated data.
The Seurat FindMultiModalNeighbors function was used to compute a joint neighbor graph that represented both the gene expression and DNA accessibility measurements using the weighted nearest neighbor methods. UMAP was performed using a joint weighted nearest neighbor map followed by unsupervised clustering using the Seurat FindClusters function. Integrated UMAP plots were produced using the DimPlot function with a Viridis color scale. Marker genes were identified by the Seurat FindAllMarkers function. DEGs and DARs between groups of cells were calculated using the FindMarkers function. DEGs and DARS were classified as an adjusted P value (Bonferroni-corrected) of <0.01 and absolute log2(fold change) of >1. Chromatin accessibility track plots were generated using the Signac CoveragePlot function, and gene expression was taken from normalized non-SCtransformed RNA data.
SCENIC+ analysis
Integrated single-cell transcriptomic and single-cell chromatin accessibility data from the Seurat analysis was used to identify gene regulatory networks using the SCENIC+ (v.1.0a1) algorithm as described previously33. Topic modeling, dimensionality reduction, dropout imputation and differential accessibility region inference were performed using pycisTopic (v.2.0) with default parameters. A serial latent Dirichlet allocation model with collapsed Gibbs sampler (500 iterations) was used for topic modeling. Topics ranged from 2 to 500, with the final model comprising 200 topics. PycisTarget (v.1.0) was used with default settings to incorporate cisTarget and differential enrichment of motifs using bulk consensus peaks motif database. SCENIC+ was run with default parameters, and http://ensembl.org/biomart was used as the biomaRt host. eRegulon results were filtered based on both the correlation between gene-based regulon area under the curve and region-based regulon area under the curve with a cutoff of >0.7. Gene regulatory networks identified by SCENIC+ analysis were plotted using Cytoscape (v.3.10.0).
Quantification and statistical analysis
Statistical analysis was performed using Prism (v.10.0) software (GraphPad). Data are shown as the mean ± s.e.m. Data distribution was assumed to be normal, but this was not formally tested.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
RNA-seq and ATAC-seq data have been deposited in the Gene Expression Omnibus repository with accession number GSE271885. All other data generated or analyzed in this study are included in the paper and Supplementary Information files.
Code availability
No custom code or algorithms were used in this study.
References
Gattinoni, L., Speiser, D. E., Lichterfeld, M. & Bonini, C. T memory stem cells in health and disease. Nat. Med. 23, 18–27 (2017).
Hammarlund, E. et al. Duration of antiviral immunity after smallpox vaccination. Nat. Med. 9, 1131–1137 (2003).
Soerens, A. G. et al. Functional T cells are capable of supernumerary cell division and longevity. Nature 614, 762–766 (2023).
Utzschneider, D. T. et al. T cell factor 1-expressing memory-like CD8+ T cells sustain the immune response to chronic viral infections. Immunity 45, 415–427 (2016).
Zehn, D., Thimme, R., Lugli, E., de Almeida, G. P. & Oxenius, A. ‘Stem-like’ precursors are the fount to sustain persistent CD8+ T cell responses. Nat. Immunol. 23, 836–847 (2022).
Grassmann, S. et al. Early emergence of T central memory precursors programs clonal dominance during chronic viral infection. Nat. Immunol. 21, 1563–1573 (2020).
Utzschneider, D. T. et al. Early precursor T cells establish and propagate T cell exhaustion in chronic infection. Nat. Immunol. 21, 1256–1266 (2020).
Angelosanto, J. M., Blackburn, S. D., Crawford, A. & Wherry, E. J. Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J. Virol. 86, 8161–8170 (2012).
Schietinger, A. et al. Tumor-specific T cell dysfunction is a dynamic antigen-driven differentiation program initiated early during tumorigenesis. Immunity 45, 389–401 (2016).
McLane, L. M., Abdel-Hakeem, M. S. & Wherry, E. J. CD8 T cell exhaustion during chronic viral infection and cancer. Annu Rev. Immunol. 37, 457–495 (2019).
Lugli, E., Galletti, G., Boi, S. K. & Youngblood, B. A. Stem, effector, and hybrid states of memory CD8+ T cells. Trends Immunol. 41, 17–28 (2020).
Galletti, G. et al. Two subsets of stem-like CD8+ memory T cell progenitors with distinct fate commitments in humans. Nat. Immunol. 21, 1552–1562 (2020).
Costa del Amo, P. et al. Human TSCM cell dynamics in vivo are compatible with long-lived immunological memory and stemness. PLoS Biol. 16, e2005523 (2018).
Silva, J. G. et al. Emergence and fate of stem cell-like Tcf7+ CD8+ T cells during a primary immune response to viral infection. Sci. Immunol. 8, eadh3113 (2023).
Blank, C. U. et al. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 19, 665–674 (2019).
Klenerman, P. & Oxenius, A. T cell responses to cytomegalovirus. Nat. Rev. Immunol. 16, 367–377 (2016).
Yucel, R., Karsunky, H., Klein-Hitpass, L. & Moroy, T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J. Exp. Med. 197, 831–844 (2003).
Pargmann, D. et al. Differential impact of the transcriptional repressor Gfi1 on mature CD4+ and CD8+ T lymphocyte function. Eur. J. Immunol. 37, 3551–3563 (2007).
Chandele, A. et al. Formation of IL-7Rαhigh and IL-7Rαlow CD8 T cells during infection is regulated by the opposing functions of GABPα and Gfi-1. J. Immunol. 180, 5309–5319 (2008).
Thambyrajah, R. et al. GFI1 proteins orchestrate the emergence of haematopoietic stem cells through recruitment of LSD1. Nat. Cell Biol. 18, 21–32 (2016).
Wilson, N. K. et al. Gfi1 expression is controlled by five distinct regulatory regions spread over 100 kilobases, with Scl/Tal1, Gata2, PU.1, Erg, Meis1, and Runx1 acting as upstream regulators in early hematopoietic cells. Mol. Cell. Biol. 30, 3853–3863 (2010).
Yucel, R., Kosan, C., Heyd, F. & Moroy, T. Gfi1:green fluorescent protein knock-in mutant reveals differential expression and autoregulation of the growth factor independence 1 (Gfi1) gene during lymphocyte development. J. Biol. Chem. 279, 40906–40917 (2004).
Tsui, C. et al. MYB orchestrates T cell exhaustion and response to checkpoint inhibition. Nature 609, 354–360 (2022).
Belz, G. T. & Kallies, A. Effector and memory CD8+ T cell differentiation: towards a molecular understanding of fate determination. Curr. Opin. Immunol. 22, 279–285 (2010).
Masson, F. et al. Id2-mediated inhibition of E2A represses memory CD8+ T cell differentiation. J. Immunol. 190, 4585–4594 (2013).
Kaech, S. M., Hemby, S., Kersh, E. & Ahmed, R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111, 837–851 (2002).
Chen, Z. et al. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity 51, 840–855.e5 (2019).
Maekawa, Y. et al. Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat. Immunol. 9, 1140–1147 (2008).
Grimes, H. L., Chan, T. O., Zweidler-McKay, P. A., Tong, B. & Tsichlis, P. N. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol. 16, 6263–6272 (1996).
Chaudhry, M. Z., Borkner, L., Kulkarni, U., Berberich-Siebelt, F. & Cicin-Sain, L. NFAT signaling is indispensable for persistent memory responses of MCMV-specific CD8+ T cells. PLoS Pathog. 20, e1012025 (2024).
Snyder, C. M. et al. Memory inflation during chronic viral infection is maintained by continuous production of short-lived, functional T cells. Immunity 29, 650–659 (2008).
Munks, M. W. et al. Four distinct patterns of memory CD8 T cell responses to chronic murine cytomegalovirus infection. J. Immunol. 177, 450–458 (2006).
Bravo Gonzalez-Blas, C. et al. SCENIC+: single-cell multiomic inference of enhancers and gene regulatory networks. Nat. Methods 20, 1355–1367 (2023).
Krmpotic, A., Bubic, I., Polic, B., Lucin, P. & Jonjic, S. Pathogenesis of murine cytomegalovirus infection. Microbes Infect. 5, 1263–1277 (2003).
Masson, F., Kupresanin, F., Mount, A., Strasser, A. & Belz, G. T. Bid and Bim collaborate during induction of T cell death in persistent infection. J. Immunol. 186, 4059–4066 (2011).
Fischer, S. F., Belz, G. T. & Strasser, A. BH3-only protein Puma contributes to death of antigen-specific T cells during shutdown of an immune response to acute viral infection. Proc. Natl Acad. Sci. USA 105, 3035–3040 (2008).
Zhou, X. et al. Differentiation and persistence of memory CD8+ T cells depend on T cell factor 1. Immunity 33, 229–240 (2010).
Badr, M. E., Zhang, Z., Tai, X. & Singer, A. CD8 T cell tolerance results from eviction of immature autoreactive cells from the thymus. Science 382, 534–541 (2023).
Zeng, H., Yucel, R., Kosan, C., Klein-Hitpass, L. & Moroy, T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 23, 4116–4125 (2004).
Hwang, S. S. et al. mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science 367, 1255–1260 (2020).
Omilusik, K. D. et al. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med. 212, 2027–2039 (2015).
Vadnais, C. et al. GFI1 facilitates efficient DNA repair by regulating PRMT1 dependent methylation of MRE11 and 53BP1. Nat. Commun. 9, 1418 (2018).
Kavazovic, I. et al. Eomes broadens the scope of CD8 T-cell memory by inhibiting apoptosis in cells of low affinity. PLoS Biol. 18, e3000648 (2020).
Kara, E. E. et al. CCR2 defines in vivo development and homing of IL-23-driven GM-CSF-producing Th17 cells. Nat. Commun. 6, 8644 (2015).
Ventura, A. et al. Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665 (2007).
Zhu, J., Jankovic, D., Grinberg, A., Guo, L. & Paul, W. E. Gfi-1 plays an important role in IL-2-mediated Th2 cell expansion. Proc. Natl Acad. Sci. USA 103, 18214–18219 (2006).
Pircher, H., Burki, K., Lang, R., Hengartner, H. & Zinkernagel, R. M. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature 342, 559–561 (1989).
Welten, S. P. M. et al. Tcf1+ cells are required to maintain the inflationary T cell pool upon MCMV infection. Nat. Commun. 11, 2295 (2020).
Chaudhry, M. Z. et al. Cytomegalovirus inhibition of extrinsic apoptosis determines fitness and resistance to cytotoxic CD8 T cells. Proc. Natl Acad. Sci. USA 117, 12961–12968 (2020).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Grandi, F. C., Modi, H., Kampman, L. & Corces, M. R. Chromatin accessibility profiling by ATAC-seq. Nat. Protoc. 17, 1518–1552 (2022).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Zhu, L. J. et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP–seq and ChIP–chip data. BMC Bioinformatics 11, 237 (2010).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Stuart, T., Srivastava, A., Madad, S., Lareau, C. A. & Satija, R. Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021).
Acknowledgements
We thank L. Cicin-Sain for providing the MCMV-ie2-gp33 virus, members of flow cytometry facility at Translational Research Institute and the Biological Resources Facility of The University of Queensland for technical assistance and discussions; we also thank V. Coyne at Central Analytical Research Facility, Queensland University of Technology for technical assistance. This work was supported by grants and fellowships from the Deutsche Forschungsgemeinschaft, Walter Benjamin Fellowship (CH2805-1 to M.Z.C.), the National Health and Medical Research Council (NHMRC) of Australia (1165443, 1122277, 1054925, 1135898 and 2008542 to G.T.B.), the Australian Research Council (DP200101058, DP230101156 and FL240100130 to G.T.B.) and a grant to The University of Queensland Chair of Immunology (Frazer Institute, to G.T.B.). The Translational Research Institute is supported by a grant from the Australian Government.
Author information
Authors and Affiliations
Contributions
G.T.B. and M.Z.C. conceived the original idea and designed experiments. G.T.B. and M.Z.C. supervised the study and wrote the manuscript with input from all co-authors. M.Z.C., E.C., A.J., R.D., H.Y., Q.H. and J.S. performed experiments and analyzed data. M.Z.C., H.O.M., A.I. and Z.K.T. performed bioinformatics analyses. S.N. and Z.K.T. supervised the bioinformatic analyses. All authors reviewed, edited, provided input and approved the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Ioana Staicu, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 GFI1 expression in antiviral CD8+ T cell following virus infection.
a, Expression of GFI1-tdTomato in naïve CD8+ T cells (live CD45+CD3+TCRb+CD8a+) isolated from different tissues of naïve Gfi1tdTomato mice. Control CD8+ T cells were isolated from the spleen of a naïve C57BL/6 mouse. Histograms show geometric mean florescence intensity (gMFI) of GFI1-tdTomato expression. Data representative of three independent experiments except for the bone marrow (BM) data which is from a single experiment. b, Flow cytometric dot plots showing frequency of GFI1hi and GFI1lo cell among total CD8+ T cells (live CD3+TCRb+CD8a+) isolated from spleen of naïve, LCMVArm- or LCMVc13-infected Gfi1tdtomato mice at D7 after infection. c, GFI1-tdTomato expression in total CD8+ T cells isolated from spleen of naïve, LCMVArm- or LCMVc13-infected Gfi1tdtomato mice at D7 after infection. Data are pooled from two experiments (n = 7, 10 or 15 mice/condition). p values were calculated using a two-tailed unpaired Student’s t test. d, Gating strategy for naïve (TN, CD11a–CD44–) and activated (CD11a+CD44+) CD8+ T cells among total CD8+ T cells (live CD45+CD3+CD8a+). Activated CD8+ T cells were further separated into TCM (CD27+CD62L+), TSCM (TCF1+CX3CR1-), TEFF (TCF1-CX3CR1+) and TPEX (TCF1+TOX+) populations. e, Flow cytometric dot plots showing TSCM (red dots) and TEFF (blue dots) cells isolated from spleen at D7 after LCMVc13 infection (left panel). GFI1-tdTomato expression in TSCM and TEFF CD8+ T cells isolated from spleen of LCMVc13 infected mice (right panel). Data pooled from two experiments (D0, D7 and D21, n = 6, 7 or 13 mice/timepoint), or a single experiment (D14, n = 4 mice) and show individual responses. p values were determined using the Wilcoxon signed-rank paired t test. f, Frequency of CD62L+ TPEX (blue dots) and CD62L– TPEX (red dots) among total CD8+ TPEX T cells isolated from spleen at D7 after LCMVc13 infection (left panel). GFI1-tdTomato expression in CD62L+ and CD62L– TPEX CD8+ T cells isolated from spleen at D7 (right panel). p values were calculated using a Wilcoxon signed-rank paired t test. Data are pooled from two independent experiments (n = 13 mice). g, h, GFI1-tdTomato expression in gp33+CD8+ T cells in spleen, mLN and lungs following LCMVArm (g) or LCMVc13 (h) infection. Data pooled from two experiments (D7 and D21, n = 6, 7 or 8 mice/timepoint), or a single experiment (D14 for panel h, n = 4 mice) and show individual responses. p values were calculated using a two-tailed unpaired Student’s t test. i,j, Naïve Gfi1tdTomato P14 T cells (CD45.1+CD45.2+, 1×105 cells) were transferred to congenic recipients (CD45.2+) and the next day recipients were infected with LCMVc13. i, Flow cytometric plots showing P14 T cells frequency among total CD8+ T cells following LCMVc13 infection. j, Expression of GFI1-tdTomato by P14 T cells isolated from mLN following infection. p values were calculated using one-way ANOVA and Dunnett’s post-test. Data pooled from two independent experiments (n = 8 mice/ timepoint).
Extended Data Fig. 2 Transcriptional profile of antiviral GFI1high and GFI1low CD8+ T cells following chronic or acute LCMV infection.
Gfi1tdTomato/+ mice were infected with LCMVc13 or LCMVArm virus via tail vein or intraperitoneal injection, respectively. Activated (CD11a+CD44+) GFI1hi and GFI1lo CD8+ T cells were flow cytometrically sorted at D7 after infection and RNA-seq was performed. a, Gating strategy used to sort GFI1hi and GFI1lo CD8+ T cells from spleen of infected Gfi1tdTomato/+ mice at D7. b, Principal component analyses (PCA) plot showing clustering of CD8+ T cells. Replicates of the same group are indicated by the same colour and shape. c, DEseq2 normalized gene expression for Gfi1 in GFI1hi and GFI1lo CD8+ T cells following LCMVc13 infection. p value was calculated using two-tailed Student’s t test. d, DEseq2 normalized expression of selected genes in CD8+ T cells isolated from LCMVc13 or LCMVArm infected Gfi1tdTomato mice. e, GSEA analysis of CD8+ T cells isolated from LCMVArm-infected mice tested on memory CD8+ T cells gene signature26. p value was calculated using Wald’s test. f, GSEA Hallmark pathway analysis. Dot plots show the changes in GSEA hallmark pathways, where size of dot represents absolute pathway enrichment in GFI1hi vs GFI1lo comparison. p values were calculated using Wald’s test.
Extended Data Fig. 3 Transcriptional profile of GFI1high central memory and stem cell memory CD8+ T cells.
a,b, Gfi1tdTomato/+ mice were infected with LCMVc13 via tail vein injection. GFI1hi and GFI1lo CD8+ T cells from CD44+CD62L+ (TCM) and CD44+Ly108+ (TSCM precursor) CD8+ T cells were flow cytometrically sorted from spleen and mLN at D7 after infection and RNA-seq was performed. p values were calculated using Wald’s test. a, Volcano plot showing DEGs (p < 0.05) in CD44+CD62L+ and CD44+Ly108+ GFI1hi CD8+ T cells isolated from LCMVc13-infected mice. Blue and red dots show genes upregulated in GFI1lo and GFI1hi CD8+ T cells, respectively. b, DEseq2 normalized expression of selected genes in CD44+CD62L+ and CD44+Ly108+ CD8+ T cells isolated from LCMVc13-infected Gfi1tdTomato mice. c, Schematic of Gfi1tdTomato/+ CD8+ T cell isolation and transfer (1×105 cells/recipient) to congenic mice followed by challenge with LCMVArm infection. d, Total donor cells in mLN of recipients at D7 after LCMVArm infection. Data pooled from two experiments (n = 7 mice/group). p value calculated using two-tailed Student’s t test. e, CD44+Ly108+GFI1hi, CD44+CD62L+GFI1hi or CD44+GFI1lo CD8+ T cells were flow cytometrically isolated from spleen of Gfi1tdTomato mice (CD45.2+) at D21 after LCMVArm infection. Each population was adoptively transferred (5×104 cells) into congenic recipient mice (CD45.1+CD45.2+ or CD45.1+), then infected with LCMVArm. Flow cytometric dot plots show frequency of donor cells (CD45.2+) among total CD8+ T cells isolated from spleen at D7 after LCMVArm infection (left plots). Total number of donor cells in spleen of recipients at D7 after LCMVArm infection. Data are pooled from two independent experiments (n = 6 or 7 mice/group). p values were calculated using Kruskal-Wallis test followed by Benjamini–Hochberg FDR test for individual comparison. f, Schematic of Gfi1tdTomato/+ CD8+ T cell isolation and transfer (1×105 cells/recipient) to infection matched mice.
Extended Data Fig. 4 Phenotypic characterisation of naïve GFI1ΔCD8 CD8+ T cells.
a, Frequency of CD4+ single positive (SP) and CD8+ SP cells among total live cells isolated from thymus of naïve age matched Cd8acre/+Gfi1+/+ (WT) and Cd8acre/+Gfi1fl/fl (GFI1ΔCD8) mice. b, Total number of CD8+ (live TCRb+CD4-CD8+) and CD4+ (live TCRb+CD4+CD8-) T cells in thymus of naïve Cd8acre/+Gfi1+/+ (WT) and Cd8acre/+Gfi1fl/fl (GFI1ΔCD8) mice. c, Frequency of CD4+ and CD8+ T cells among total T cells (live TCRb+CD3+) isolated from spleen of naïve Cd8acre/+Gfi1+/+ (WT) and Cd8acre/+Gfi1fl/fl (GFI1ΔCD8) mice. d, Total number of CD8+ (live CD3+CD8+) and CD4+ (live CD3+CD4+) T cells in spleen of naïve Cd8acre/+Gfi1+/+ (WT) and Cd8acre/+Gfi1fl/fl (GFI1ΔCD8) mice. e, TCF1, EOMES, T-BET and TOX expression in naïve CD8+ T cells isolated from spleen of naïve Cd8acre/+Gfi1+/+ (WT) and Cd8acre/+Gfi1fl/fl (GFI1ΔCD8) mice. b, d, and e, data pooled from two independent experiments (n = 6 or 7 mice/genotype). p values were determined using a two-tailed Student’s t test.
Extended Data Fig. 5 CD8+ T cell responses mounted by GFI1-deficient following chronic LCMV infection.
a-e, Congenically-labelled naïve WT (CD45.1+) and GFI1ΔCD8 (CD45.1+CD45.2+) P14 T cells were co-transferred (5×103 cells each) to C57BL/6 mice that were subsequently infected with LCMVc13 24 h later. a, Total number of WT and GFI1ΔCD8 P14 T cells in peripheral blood, mLN and Lungs. b, Total number of TSCM P14 T cells in spleen. a, b, Data pooled from two (n = 10 mice for D7 and D21) or one experiment (n = 5 for D5, D14, D28 and D60). c, Total number of TCF1-CX3CR1+ and TCF1-CX3CR1- P14 T cells in spleen at D7 after infection. Data are pooled from two independent experiments (n = 11 mice). d, CD127, CX3CR1, FOXO1, KLRG1, PD-1, TIM3 and T-BET expression in WT and GFI1ΔCD8 P14 T cells at D7 after LCMVc13 infection. Data show expression from one of two independent experiments (n = 6 mice). e, Genes differentially expressed between GFI1ΔCD8 and WT P14 T cells were subjected to GO pathway enrichment analysis. Bar plots show top 10 significantly upregulated and downregulated pathways. p values calculated using Wald’s test followed by Benjamini–Hochberg FDR testing. a, b, data show mean ± s.e.m. b-e p values were determined with a two-tailed paired t test.
Extended Data Fig. 6 Characterization of transcriptional and epigenetic profile of GFI1-deficient CD8+ T cells following chronic LCMV infection.
a, Naïve congenically labelled WT or GFI1ΔCD8 P14 CD8+ T cells (CD45.1+) were adoptively transferred into C57BL/6 mice that were subsequently infected with LCMVc13. P14 T cells were isolated from spleen at D7 after infection and analysed using RNA-seq. DEseq2 normalized expression of selected genes is shown. b-e, Congenically-labelled naïve WT (CD45.1+) and GFI1ΔCD8 (CD45.1+CD45.2+) P14 T cells were mixed 1:1 and co-transferred to C57BL/6 mice that were subsequently infected with LCMVc13 24 h later. Splenic P14 T cells were analysed at D7 after infection. b, Uptake of fluorescent glucose analog 2-NBDG and fluorescent fatty acid (BODIPY C16) in P14 T cells isolated from spleen at D7. c, Mitochondrial mass and membrane potential determined by Mitotracker Green and Mitotracker Red (CMX ROS) labeling, respectively, in P14 T cells isolated at D7. d, Plots showing frequency of P14 cells with dysfunctional mitochondria (left panels). Data showing frequency of P14 cells with dysfunctional mitochondria (right panel). e, Histograms showing granzyme B expression in WT (blue line) and GFI1ΔCD8 (red line) P14 CD8+ T cells following gp33 peptide stimulation. Control (black dotted line) shows unstimulated WT P14 CD8+ T cells (left panel). Data show granzyme B expression in P14 T cells at D7. b-e Data are representative of two experiments (n = 6 mice). p values calculated using a two-tailed paired t test. f,g, Naïve congenically-labelled WT or GFI1ΔCD8 P14 CD8+ T cells (CD45.1+) were adoptively transferred into C57BL/6 mice that were infected with LCMVc13 24 h later. Nuclei from activated splenic P14 T cells (D7 after infection) and naïve P14 T cells were isolated to perform bulk ATAC-seq. f, Chromatin accessibility read density enrichment around peak centres for P14 T cells. Upper panels show the average profile around detected peak centres. Lower panels show read intensity heatmaps ranked by total read intensity. Reads are centred on the middle of the accessible peak ±1.5 Kbp. Data pooled from three samples to generate read density map. g, Number of differentially accessible regions detected in naïve GFI1ΔCD8 P14 CD8+ T cells and annotated in and around promotor or exon regions.
Extended Data Fig. 7 CD8+ T cell responses mounted by GFI1-deficient following latent MCMV infection.
a, Schematic showing irradiation, BM cells injection, MCMV infection, peripheral blood and tissue analysis. b,c, Mixed bone marrow chimeras were generated by adoptive transfer of equal mix of WT CD45.1+ and WT CD45.2+ (WT + WT) or GFI1ΔCD8 CD45.1+ and WT CD45.2+ (WT + GFI1ΔCD8) bone marrow cells into irradiated C57BL/6 mice that were infected with MCMV intraperitoneally 60 days after reconstitution. b, Total number of CD45.1+CD8+ T cells in peripheral blood 8 weeks following bone marrow reconstitution. c, Flow cytometric dot plots showing frequency of TSLEC (KLRG1+CD27-) and TMPEC (KLRG1-CD27+) among activated (CD11a+CD44+) CD8+ T cells isolated from the spleen of bone marrow chimeric mice at D90 after infection (left panels). Total number of TSLEC and TMPEC CD45.1+CD8+ T cells in spleen at D90 (right panels). b, c Data pooled from 2 experiments (n = 9 or mice/condition) is shown as mean ± s.e.m. p values were calculated using a two-tailed unpaired Student’s t test. d,e, Naïve congenically-labelled WT (CD45.1+) and GFI1ΔCD8 (CD45.1+CD45.2+) P14 T cells were mixed 1:1 and co-transferred to C57BL/6 mice that were subsequently infected with MCMV-ie2-gp33 the next day. Mice were euthanized at D7 after infection to assess CD8+ T cell responses in tissues. d, Frequency of TSELC and TMPEC among WT and GFI1ΔCD8 P14 CD8+ T cells isolated from spleen at D7 after infection. Data pooled from two experiments (n = 10 mice). e, Data show Ki-67, TCF1, EOMES, T-BET and TOX expression from one of two independent experiments (n = 6 mice). f, Frequency of IFN-γ+ and IFN-γ+TNFα+ among total WT and GFI1ΔCD8 P14 CD8+ T cells isolated from spleen following gp33 peptide stimulation (left panels). Data showing frequency of IFN-γ+ and TNFα+ P14 CD8+ T cells from one of two independent experiments (n = 5 mice). d-f, p values calculated using a two-tailed paired t test. g, Chromatin accessibility read density enrichment around peak centres for adoptively transferred congenically-labelled WT or GFI1ΔCD8 P14 CD8+ T cells (CD45.1+) in response to MCMV-ie2-gp33 at D7. Upper panels show the average profile around detected peak centres. Lower panels show read intensity heatmaps ranked by total read intensity. Reads are centred on the middle of the accessible peak ±1.5 Kbp. Data pooled from three samples to generate read density map.
Extended Data Fig. 8 scMultiome-seq analysis of virus-specific CD8+ T cells following chronic virus infection.
Naïve congenically-labelled WT or GFI1ΔCD8 P14 CD8+ T cells (CD45.1+) were transferred to C57BL/6 mice that were subsequently infected with LCMVc13 the next day. P14 T cells were isolated on D7 after infection for scMutiomic-seq. a, Schematics showing scMutiomic-seq workflow. b, UMAP projection showing unsupervised clustering of WT and GFI1ΔCD8 CD8+ T cells (WT + GFI1ΔCD8 Seurat integrated data). c, UMAP projection showing cell-cycle phases of WT and GFI1ΔCD8 CD8+ T cells based on expression of G2/M and S phase genes (scRNA-seq data). d, Aggregate gene expression of top 20 upregulated genes for each cluster. e, Cluster 1 and 5 WT (blue histogram and violin plots) and GFI1ΔCD8 (red histogram and violin plots) CD8+ T cell chromatin accessibility and gene expression at Btg1 gene locus. Dashed boxes highlight differentially assessable chromatin regions. f, Transcription factor linked gene regulatory networks identified by SCENIC+ analysis. Core regulatory genes shown in red and target genes in blue. g, Cluster 1 and 2 WT (blue histogram and violin plots) and GFI1ΔCD8 (red histogram and violin plots) CD8+ T cell chromatin accessibility at E2f1 (left panel) and E2f7 (right panel) gene locus. Dashed boxes highlight differentially assessable chromatin regions.
Extended Data Fig. 9 GFI1-deficient CD8+ T cells exhibit reduced proliferation and are prone to activation induced T cell death.
a, Schematic representation showing naïve P14 CD8+ T cells transfer to C57BL/6 mice that were infected with LCMVc13 after 24 h. P14 T cells were isolated at D5 after infection, labelled with CTV then cultured in vitro for 4 days with IL-2, IL-2 + IL-7 or IL-2 + CD3/CD28 beads. b, Flow cytometric histograms showing WT (blue) and GFI1ΔCD8 (red) CD8+ T cell proliferation. Dotted lines mark cell divisions. c, Total number of WT and GFI1ΔCD8 CD8+ T cells per well after 4 days in vitro culture shown as mean ± s.e.m. Data pooled from two independent experiments (n = 6 mice). p values were calculated using a two-tailed Student’s t test. d, Schematics showing P14 T cell activation with primary infection, sorting, and transfer to secondary recipients and virus infection. e, Naïve congenically-labelled WT or GFI1ΔCD8 P14 CD8+ T cells (CD45.1+) were transferred to C57BL/6 mice that were subsequently infected with LCMVc13 the next day. P14 T cells were isolated on D7 after infection for scMutiomic-seq. Dot plot showing expression of selected cell death related genes in WT and GFI1ΔCD8 P14 T cells, with dot size indicating fraction of cells expressing gene and colour representing mean expression. f, Mixed bone marrow chimeras were generated by adoptive transfer of equal mix of WT CD45.1+ and WT CD45.2+ (WT + WT) or GFI1ΔCD8 CD45.1+ and WT CD45.2+ (WT + GFI1ΔCD8) bone marrow into irradiated C57BL/6 mice that were infected with MCMV intraperitoneally at D60 after reconstitution. Histograms (left panel) and bar plots (right panels) showing expression of BCL-2 in CD8+ T cells isolated from spleen at D7 after infection shown as mean ± s.e.m. g, Flow cytometric plot showing frequency of caspase-3+ (casp-3) CD45.1+CD8+ T cells isolated from spleen 7 days after infection (left panels). Frequency of casp-3+ CD45.1+ CD8+ T cells in spleen 7 days after infection shown as mean ± s.e.m. (right panel). f, g, Data pooled from two experiments (n = 5 or 6mice). p values were determined using a two-tailed unpaired Student’s t test.
Extended Data Fig. 10 Continuous expression of GFI1 is required to maintain persistence of antiviral CD8+ T cells.
a-d, Congenically-labelled naïve R26Cre-ERT2/+Gfi1fl/+ and R26Cre-ERT2/+Gfi1fl/fl P14 T cells were co-transferred to C57BL/6 mice that were subsequently infected with MCMV-ie2-gp33 the next day. Mice were treated with tamoxifen daily for 4 days from day 30 after infection and CD8+ T cell responses analysed at D14 or D28 after tamoxifen. a, Schematic showing P14 T cell transfer, virus infection, tamoxifen treatment and analysis. b, Total number of P14 T cells in lungs and liver at D14 and D28 after tamoxifen treatment. c, Total number of TSCM P14 T cells in spleen at D14 and D28 after tamoxifen treatment. d, Expression of TCF1, EOMES, T-BET, FOXO1 and BCL-2 in P14 T cells isolated from spleen of tamoxifen-treated animals at D14 after treatment. b-d Data pooled from two experiments/timepoint (n = 9 or 10 mice/timepoint). e, Congenically-labelled naïve R26Cre-ERT2/+Gfi1fl/+ and R26Cre-ERT2/+Gfi1fl/fl P14 T cells were co-transferred to C57BL/6 mice that were subsequently infected with LCMVc13 the next day. Mice were treated with tamoxifen at D15 after infection for 4 days and CD8+ T cell responses were analysed on D30 after infection. Plot showing frequency of TSCM P14 T cells in spleen at D30 (left panel). Total number of P14 and TSCM T cells in spleen and lungs (left panel). Data pooled from two experiments (n = 11 mice). b-e Data show mean ± s.e.m. p values were determined using a two-tailed paired t test.
Supplementary information
Supplementary Table
Supplementary Tables 1–12.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chaudhry, M.Z., Chen, E., Man, H.O. et al. GFI1-driven transcriptional and epigenetic programs maintain CD8+ T cell stemness and persistence. Nat Immunol 26, 894–907 (2025). https://doi.org/10.1038/s41590-025-02151-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41590-025-02151-5