Abstract
Dendritic cells are essential for establishing thymic central tolerance; however, mechanisms supporting their homeostasis and activation remain unresolved. Through single-cell transcriptomics and functional assays, we identify seven thymic conventional dendritic cell (cDC) subsets and discriminate their abilities to present self-antigens and induce regulatory T cells. Mice blocked at different stages of T cell development revealed that CD4+ single-positive (CD4SP) and CD8SP thymocytes differentially support homeostasis and activation of type 1 cDCs (cDC1s) versus cDC2s/plasmacytoid DCs (pDCs), respectively. CD8SP thymocytes indirectly support pDC survival and cDC2 thymic migration, and they induce interferon signaling in cDCs, partly by promoting type 3 interferon expression by medullary thymic epithelial cells. By contrast, CD4SP thymocytes undergo cognate interactions with cDCs, inducing CD40 signaling required for activation of cDC1s. Activated cDC1s make nonredundant contributions to central tolerance. Together, this study comprehensively identifies distinct thymic DC subsets and elucidates requirements for cross-talk with thymocyte subsets that support their homeostasis, activation and function.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
scRNA-seq data have been deposited at the Gene Expression Omnibus under accession number GSE239780. Any additional information required to reanalyze the data reported in this paper is available upon request from the corresponding author, L.I.R.E. (lehrlich@austin.utexas.edu).
Code availability
This study generated original code for histocytometry analyses, which can be found at GitHub (https://github.com/EhrlichLab/histocytometry_Srinivasan_2023). Scripts used for p-Creode analyses are also available at GitHub (github.com/Ken-Lau-Lab/pCreode).
References
Ashby, K. M. & Hogquist, K. A. A guide to thymic selection of T cells. Nat. Rev. Immunol. 24, 103–117 (2024).
Klein, L. & Petrozziello, E. Antigen presentation for central tolerance induction. Nat. Rev. Immunol. 25, 57–72 (2025).
Ohnmacht, C. et al. Constitutive ablation of dendritic cells breaks self-tolerance of CD4 T cells and results in spontaneous fatal autoimmunity. J. Exp. Med. 206, 549–559 (2009).
Březina, J., Vobořil, M. & Filipp, D. Mechanisms of direct and indirect presentation of self-antigens in the thymus. Front. Immunol. 13, 926625 (2022).
Li, J., Park, J., Foss, D. & Goldschneider, I. Thymus-homing peripheral dendritic cells constitute two of the three major subsets of dendritic cells in the steady-state thymus. J. Exp. Med. 206, 607–622 (2009).
Wu, L. & Shortman, K. Heterogeneity of thymic dendritic cells. Semin. Immunol. 17, 304–312 (2005).
Hu, Z. et al. CCR7 modulates the generation of thymic regulatory T cells by altering the composition of the thymic dendritic cell compartment. Cell Rep. 21, 168–180 (2017).
Ardouin, L. et al. Broad and largely concordant molecular changes characterize tolerogenic and immunogenic dendritic cell maturation in thymus and periphery. Immunity 45, 305–318 (2016).
Oh, J. et al. CD40 mediates maturation of thymic dendritic cells driven by self-reactive CD4+ thymocytes and supports development of natural regulatory T cells. J. Immunol. 200, 1399–1412 (2018).
Vobořil, M. et al. A model of preferential pairing between epithelial and dendritic cells in thymic antigen transfer. eLife 11, e71578 (2022).
Breed, E. R. et al. Type 2 cytokines in the thymus activate SIRPα+ dendritic cells to promote clonal deletion. Nat. Immunol. 23, 1042–1051 (2022).
Spidale, N. A., Wang, B. & Tisch, R. Cutting edge: Antigen-specific thymocyte feedback regulates homeostatic thymic conventional dendritic cell maturation. J. Immunol. 193, 21–25 (2014).
Ashby, K. M. et al. Sterile production of interferons in the thymus affects T cell repertoire selection. Sci. Immunol. 9, eadp1139 (2024).
Guilliams, M. et al. Unsupervised high-dimensional analysis aligns dendritic cells across tissues and species. Immunity 45, 669–684 (2016).
Zilionis, R. et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 50, 1317–1334 (2019).
Eferl, R. & Wagner, E. F. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3, 859–868 (2003).
Leylek, R. et al. Integrated cross-species analysis identifies a conserved transitional dendritic cell population. Cell Rep. 29, 3736–3750 (2019).
Vobořil, M. et al. Thymic myeloid cells are heterogenous and include a novel population of transitional dendritic cells. J. Exp. Med. 223, e20250733 (2026).
Vollmann, E. H. et al. Specialized transendothelial dendritic cells mediate thymic T-cell selection against blood-borne macromolecules. Nat. Commun. 12, 6230 (2021).
Zegarra-Ruiz, D. F. et al. Thymic development of gut-microbiota-specific T cells. Nature 594, 413–417 (2021).
Vobořil, M. et al. Toll-like receptor signaling in thymic epithelium controls monocyte-derived dendritic cell recruitment and Treg generation. Nat. Commun. 11, 2361 (2020).
Lancaster, J. N., Li, Y. & Ehrlich, L. I. R. Chemokine-mediated choreography of thymocyte development and selection. Trends Immunol. 39, 86–98 (2018).
Zhou, T.-A. et al. Thymic macrophages consist of two populations with distinct localization and origin. eLife 11, 2021.11.04.467238 (2022).
Li, Y. et al. CCR4 and CCR7 differentially regulate thymocyte localization with distinct outcomes for central tolerance. eLife 12, e80443 (2023).
Daley, S. R., Hu, D. Y. & Goodnow, C. C. Helios marks strongly autoreactive CD4+ T cells in two major waves of thymic deletion distinguished by induction of PD-1 or NF-κB. J. Exp. Med. 210, 269–285 (2013).
Stritesky, G. L. et al. Murine thymic selection quantified using a unique method to capture deleted T cells. Proc. Natl Acad. Sci. USA 110, 4679–4684 (2013).
Kurts, C. et al. Constitutive class I-restricted exogenous presentation of self antigens in vivo. J. Exp. Med. 184, 923–930 (1996).
Kurts, C., Miller, J. F. A. P., Subramaniam, R. M., Carbone, F. R. & Heath, W. R. Major histocompatibility complex class I-restricted cross-presentation is biased towards high dose antigens and those released during cellular destruction. J. Exp. Med. 188, 409–414 (1998).
Humblet, C., Rudensky, A. Y. & Kyewski, B. Presentation and intercellular transfer of self antigen within the thymic microenvironment: expression of the Eα peptide–l-Ab complex by isolated thymic stromal cells. Int. Immunol. 6, 1949–1958 (1994).
Owen, D. L. et al. Thymic regulatory T cells arise via two distinct developmental programs. Nat. Immunol. 20, 195–205 (2019).
Proietto, A. I. et al. Dendritic cells in the thymus contribute to T-regulatory cell induction. Proc. Natl Acad. Sci. USA 105, 19869–19874 (2008).
Roche, P. A. & Furuta, K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 15, 203–216 (2015).
Watanabe, M., Lu, Y., Breen, M. & Hodes, R. J. B7–CD28 co-stimulation modulates central tolerance via thymic clonal deletion and Treg generation through distinct mechanisms. Nat. Commun. 11, 6264 (2020).
Laurent, J., Bosco, N., Marche, P. N. & Ceredig, R. New insights into the proliferation and differentiation of early mouse thymocytes. Int. Immunol. 16, 1069–1080 (2004).
Mombaerts, P. et al. Mutations in T-cell antigen receptor genes α and β block thymocyte development at different stages. Nature 360, 225–231 (1992).
Bendelac, A., Killeen, N., Littman, D. R. & Schwartz, R. H. A subset of CD4+ thymocytes selected by MHC class I molecules. Science 263, 1774–1778 (1994).
Hadeiba, H. et al. Plasmacytoid dendritic cells transport peripheral antigens to the thymus to promote central tolerance. Immunity 36, 438–450 (2012).
Bonasio, R. et al. Clonal deletion of thymocytes by circulating dendritic cells homing to the thymus. Nat. Immunol. 7, 1092–1100 (2006).
Lopes, N., Charaix, J., Cédile, O., Sergé, A. & Irla, M. Lymphotoxin α fine-tunes T cell clonal deletion by regulating thymic entry of antigen-presenting cells. Nat. Commun. 9, 1262 (2018).
Hikosaka, Y. et al. The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity 29, 438–450 (2008).
Irla, M. et al. Autoantigen-specific interactions with CD4+ thymocytes control mature medullary thymic epithelial cell cellularity. Immunity 29, 451–463 (2008).
Benhammadi, M. et al. IFN-λ enhances constitutive expression of MHC class I molecules on thymic epithelial cells. J. Immunol. 205, 1268–1280 (2020).
Barnden, M. J., Allison, J., Heath, W. R. & Carbone, F. R. Defective TCR expression in transgenic mice constructed using cDNA- based α- and β-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76, 34–40 (1998).
Edilova, M. I., Abdul-Sater, A. A. & Watts, T. H. TRAF1 signaling in human health and disease. Front. Immunol. 9, 2969 (2018).
Herring, C. A. et al. Unsupervised trajectory analysis of single-cell RNA-seq and imaging data reveals alternative tuft cell origins in the gut. Cell Syst. 6, 37–51 (2018).
Gulati, G. S. et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 367, 405–411 (2020).
Wu, R. & Murphy, K. M. DCs at the center of help: origins and evolution of the three-cell-type hypothesis. J. Exp. Med. 219, e20211519 (2022).
Van den Berge, K. et al. Trajectory-based differential expression analysis for single-cell sequencing data. Nat. Commun. 11, 1201 (2020).
Lazear, H. M., Schoggins, J. W. & Diamond, M. S. Shared and distinct functions of type I and type III interferons. Immunity 50, 907–923 (2019).
Xing, Y., Wang, X., Jameson, S. C. & Hogquist, K. A. Late stages of T cell maturation in the thymus involve NF-κB and tonic type I interferon signaling. Nat. Immunol. 17, 565–573 (2016).
Ferris, S. T. et al. cDC1 prime and are licensed by CD4+ T cells to induce anti-tumour immunity. Nature 584, 624–629 (2020).
Breed, E. R., Watanabe, M. & Hogquist, K. A. Measuring thymic clonal deletion at the population level. J. Immunol. 202, 3226–3233 (2019).
Calindi, A. & Ehrlich, L. I. R. Intrathymic regulation of dendritic cell subsets and their contributions to central tolerance. Immunol. Rev. 332, e70039 (2025).
Park, J.-E. et al. A cell atlas of human thymic development defines T cell repertoire formation. Science 367, eaay3224 (2020).
Perry, J. S. A. et al. CD36 mediates cell-surface antigens to promote thymic development of the regulatory T cell receptor repertoire and allo-tolerance. Immunity 48, 923–936 (2018).
Herbin, O. et al. Medullary thymic epithelial cells and CD8α+ dendritic cells coordinately regulate central tolerance but CD8α+ cells are dispensable for thymic regulatory T cell production. J. Autoimmun. 75, 141–149 (2016).
Perry, J. S. A. et al. Distinct contributions of AIRE and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity 41, 414–426 (2014).
Gallucci, S., Lolkema, M. & Matzinger, P. Natural adjuvants: endogenous activators of dendritic cells. Nat. Med. 5, 1249–1255 (1999).
Montoya, M. et al. Type I interferons produced by dendritic cells promote their phenotypic and functional activation. Blood 99, 3263–3271 (2002).
Lienenklaus, S. et al. Novel reporter mouse reveals constitutive and inflammatory expression of IFN-β in vivo. J. Immunol. 183, 3229–3236 (2009).
Sakref, C. et al. Type III interferon primes pDCs for TLR7 activation and antagonizes immune suppression mediated by TGF-β and PGE2. Nat. Commun. 16, 3045 (2025).
Baba, T., Nakamoto, Y. & Mukaida, N. Crucial contribution of thymic SIRPα+ conventional dendritic cells to central tolerance against blood-borne antigens in a CCR2-dependent manner. J. Immunol. 183, 3053–3063 (2009).
Joyee, A. G., Uzonna, J. & Yang, X. Invariant NKT cells preferentially modulate the function of CD8α+ dendritic cell subset in inducing type 1 immunity against infection. J. Immunol. 184, 2095–2106 (2010).
Leventhal, D. S. et al. Dendritic cells coordinate the development and homeostasis of organ-specific regulatory T cells. Immunity 44, 847–859 (2016).
Wu, R. et al. Mechanisms of CD40-dependent cDC1 licensing beyond costimulation. Nat. Immunol. 23, 1536–1550 (2022).
Seach, N., Wong, K., Hammett, M., Boyd, R. L. & Chidgey, A. P. Purified enzymes improve isolation and characterization of the adult thymic epithelium. J. Immunol. Methods 385, 23–34 (2012).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2025).
Miller, J. C. et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13, 888–899 (2012).
Seita, J. et al. Gene expression commons: an open platform for absolute gene expression profiling. PLoS ONE 7, e40321 (2012).
Wang, J., Vasaikar, S., Shi, Z., Greer, M. & Zhang, B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45, W130–W137 (2017).
Chen, B. et al. Differential pre-malignant programs and microenvironment chart distinct paths to malignancy in human colorectal polyps. Cell 184, 6262–6280 (2021).
Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Lutz, K. et al. Ly6D+Siglec-H+ precursors contribute to conventional dendritic cells via a ZBTB46+Ly6D+ intermediary stage. Nat. Commun. 13, 3456 (2022).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Pachitariu, M. & Stringer, C. Cellpose 2.0: how to train your own model. Nat. Methods 19, 1634–1641 (2022).
Schapiro, D. et al. MCMICRO: a scalable, modular image-processing pipeline for multiplexed tissue imaging. Nat. Methods 19, 311–315 (2022).
Acknowledgements
We thank the Animal Resource Center staff at the University of Texas at Austin for mouse maintenance support and R. Salinas for maintenance of FACS instruments. scRNA-seq library preparation and sequencing was performed by H. Stevenson at the GSAF at The University of Texas at Austin, Center for Biomedical Research Support. Xcr1cre/+Cd40fl/− and Cd40−/− mice were generously provided by K. Murphy (Washington University School of Medicine). We thank N. Manley, E. Richie and L. Hale for constructive feedback and S. Yi for initial bioinformatics analyses. This research was supported by a grant from the National Institutes of Health (P01AI139449) to L.I.R.E., K.S.L. and Q.L. Servers contributed by Advanced Micro Devices to L.I.R.E. were used for imaging data analysis.
Author information
Authors and Affiliations
Contributions
L.I.R.E., J.S., C.R.M. and A.C. conceptualized the study and wrote the paper. J.S., C.R.M. and A.C. designed and performed the experiments and analyzed data with supervision from L.I.R.E. K.S.L., Q.L., B.R.H., Y.Y. and C.N.H. analyzed scRNA-seq datasets. J.F.M. performed the histocytometry analyses. H.J.S. assisted with FACS isolation of cells for functional assays. L.I.R.E. acquired funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: S. Houston, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Identification of transcriptionally distinct HAPC subsets.
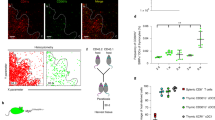
Related to Fig. 1. (a) Flow cytometric gating strategy used to sort HAPCs for scRNA-seq analysis. (b) UMAP visualization showing major HAPC populations identified from 8404 FACS-sorted HAPCs (and residual thymocytes) from adult C57BL6/J thymi- B cells (Cd19), cDCs (Zbtb46, Itgax), pDCs (Itgax, Siglech), macrophages/monocytes (Csf1r, C1qa), granulocytes (Camp), and thymocytes (Lck) from n = 2 mice. (c) Feature plots showing expression levels of select genes associated with the major HAPC subsets overlaid on the UMAP visualization from (b). (d) Heatmap displaying normalized expression of the top 10 differentially expressed genes in each transcriptionally distinct cDC cluster from Fig. 1a. Arrows highlight select genes specific to the transcriptionally distinct cDC1, cDC2, and aDC clusters. (e) Violin plot displaying normalized expression levels of Zbtb46 gene in Ly6d+ cDCs and pDCs. (f) To compare our transcriptionally distinct DC clusters to those reported in the literature, we carried out Spearman correlation analysis of thymic cDC clusters from our dataset with cDC and other clusters in the dataset from11.
Extended Data Fig. 2 Identification of transcriptionally distinct cDC clusters and their correlation with FACS-sorted cDC subsets.
Related to Fig. 1. (a) Flow cytometric gating strategy to identify distinct DC subsets shown in Fig. 1d. (b-e) To determine if DC subsets identified by flow cytometry correlate with the expected transcriptional signatures, DC subsets were FACS-purified, hashtagged to informatically distinguish the sorted subsets, pooled, and evaluated by scRNA-seq. (b) UMAP visualization of scRNA-seq transcriptional data from pooled FACS-sorted cDC subsets, colored by their hashtag labels. (c) Same UMAP plot as (b), colored by transcriptionally distinct cDC clusters defined by the Seurat pipeline. (d) Dot plot showing normalized expression levels (z-score) of select genes associated with the hashtagged FACS-sorted cDC subsets. Expression is normalized per row (representing a gene) across different FACS-sorted cDC subsets. (e) Frequency of each transcriptionally distinct cDC cluster contained within the FACS-sorted hashtagged cDC subsets. (f-i) To determine intrathymic localization of DC subsets, immunofluorescence staining and histocytometry were carried out on N = 3 independent biological thymic replicates. (f) Representative immunofluorescence image of MertK+ macrophages and CD63+SIRPa+MerTK− aDC2s in thymic sections. Centroids are placed over CD63+ SIRPα+ cells that also express MertK: the frequency of MertK-expressing cells among CD63+SIRPα+ myeloid cells was used to correct aDC2 densities from data as in (h). Scale bar, 250 µm. Lines demarcate the cortex (C), medulla (M), and CMJ in the thymic sections. Centroids with 40% opacity were overlaid on the quantified cells. (g) Representative immunofluorescence images of DC subsets identified by the indicated markers. White boxes indicate the regions of interest displayed in Fig. 1e. Scale bars, 1,000 µm. (h) Quantification of cell densities of each of the indicated DC subsets in distinct anatomical regions of the thymus, based on histocytometry analyses of data as in (f-g). Dots represent biological thymic replicates. Statistical analysis was performed using Tukey’s Honest Significant Difference test. (i) Quantification of cortical cell densities of each of the indicated DC subsets, based on histocytometry analyses of data as in (f-g).
Extended Data Fig. 3 aDC1s, followed by aDC2s, are highly efficient at mTEC-derived antigen presentation on MHC-I and MHC-II.
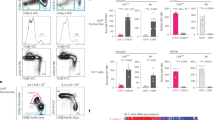
Related to Fig. 2. (a-c) Thymic DC subsets isolated from RIP-OVAhi mice were tested for their capacity to cross-present the mTEC-derived OVA TRA on MHC-I and induce OT-I proliferation. (a) Representative histograms showing the percentage of OT-I CD8+ T cells that underwent at least 1 cell division after co-culture for 3.5 days with the specified DC subsets isolated from RIP-OVAhi mice. OT-I CD8+ T cells incubated only with IL-2 served as negative controls, while those co-cultured with 50 nM OVA257-264-pulsed splenocytes served as positive controls. For (a-c), data are representative of N = 3 independent experiments. (b) Quantification of OT-I CD8+ T cells that underwent proliferation from the experiment described in (a). Bars represent mean ± SEM. Dots represent replicate wells from one representative experiment. Statistical analysis was performed using one-way ANOVA with Dunnett’s multiple comparisons test. Significance is relative to “IL-2 only” control wells. (c) Modeling, based on data from the experiment in (a), of the frequency of OT-I CD8+ T cells that underwent the indicated number of cell divisions. Bars represent mean ± SEM. (d-g) To determine if the different functional capacities of DCs subsets could reflect differences in their ability to process and present antigens or co-stimulate thymocytes/T cells, differential expression of genes and proteins were evaluated. (d, f) Heatmaps displaying average normalized gene expression values of select (d) genes associated with antigen processing and presentation and (f) costimulatory genes in each cDC cluster from Fig. 1a. Expression is z-score normalized per gene across different cDC clusters. (e, g) Relative cell-surface expression levels of (e) MHC-I and MHC-II, and (g) CD80 and CD86 proteins by cDC subsets, as determined by flow cytometry. Data are normalized to average MFI levels of aDC1s for each protein within an individual experiment. Data are compiled from N = 3 independent experiments; n = 8 total WT mice. Bars represent mean ± SEM and symbols represent individual mice. Statistical analysis was performed using one-way ANOVA with Dunnett’s multiple comparisons test. Significance is relative to the aDC1 subset.
Extended Data Fig. 4 Thymic cDC1 and cDC2 homeostasis and activation depend on the presence of distinct thymocyte subsets.
Related to Fig. 3. To determine if thymic DC homeostasis/activation requires the presence of different thymocyte subsets, the frequencies and numbers of thymic DCs were evaluated in genetically modified mouse strains in which T-cell development was blocked at different stages of differentiation. (a-c) Quantification of (left to right) frequencies of pDCs, total cDC subsets, cDC1 subsets, cDC2 subsets, and aDC subsets in the thymi of the following littermate genotypes (a) Rag2+/+(n = 10) and Rag2−/− (n = 10); (b) Tcra+/+ (n = 9) and Tcra−/− (n = 9); and (c) B2m+/+ (n = 11) and B2m−/− (n = 11). The population from which the frequencies were calculated is indicated on the respective Y-axes. (d) Quantification of (left to right) total thymus cellularity and cell numbers of pDCs, cDC1 subsets, cDC2 subsets, and aDC subsets from mouse thymi of Cd1d−/− relative to littermate control mice; n = 5 Cd1d+/+ and n = 5 Cd1d−/− mice. (e) Quantification of (left to right) frequencies of pDCs, total cDC subsets, cDC1 subsets, cDC2 subsets, and aDC subsets in the thymi of MHC-II−/− relative to littermate control mice; n = 9 MHC-II+/+ and n = 8 MHC-II−/− mice. Data are compiled from (a,b,d,e) N = 3 and (c) N = 4 independent experiments. Bars represent mean ± SEM and symbols represent individual mice. (f-h) To test whether the absence of thymocyte crosstalk altered the APC activity of DC subsets, in vitro Treg generation assays were carried out using sorted DC subsets from the indicated genotypes. Quantification of frequencies of Foxp3+ CD25+ Tregs induced after co-culturing CD73− CD25− Foxp3− CD4SP thymocytes with the indicated thymic DC subsets from WT littermate versus (f) Rag2−/−, (g) B2m−/−, and (h) MHC-II−/− mice for 4 days. Data are compiled from N = 3 independent experiments. Bars represent mean ± SEM and symbols represent data from individual wells. Statistical analysis was performed using two-tailed unpaired Student t-test.
Extended Data Fig. 5 Quantification of DC subsets from lymph nodes and spleens of mice with impaired T cell development.
Related to Fig. 3. To determine if altered numbers of thymic cDC2s and pDCs in genetic mouse models could reflect peripheral changes in these DC subsets, which migrate into the thymus, DC composition was analyzed in secondary lymphoid organs. (a-f) Quantification of frequencies of (left to right) pDCs, cDC subsets, cDC1 subsets, cDC2 subsets and aDC subsets in (a,c,e) pooled axillary, brachial, and inguinal lymph nodes and (b,d,f) spleens of the following littermate genotypes (a-b) Rag2+/+ (n = 10) and Rag2−/− (n = 12); (c-d) B2m+/+ (n = 12) and B2m−/− (n = 10); and (e-f) MHC-II−+/+ (n = 9) and MHC-II−/− (n = 12). The populations from which the frequencies were calculated are indicated on the Y-axes. (a-f) Data are compiled from N = 3 independent experiments. Bars represent mean ± SEM and symbols represent individual mice. Statistical tests were performed using two-tailed unpaired Student t-test.
Extended Data Fig. 6 Transcriptional profiling of thymic HAPCs in mice devoid of distinct thymocyte subsets reveals the impact on DC subset transcriptomes and frequencies.
Related to Fig. 5. To determine how absence of crosstalk with different thymocyte subsets alters the transcriptomes of DCs, FACS-purified HAPCs from WT, Rag2−/−, B2m−/−, and MHC-II−/− thymi were subjected to scRNA-seq analysis. (a) Merged UMAP visualization of 34,750 cells depicting major cell continents in WT, MHC-II−/−, B2m−/− and Rag2−/− HAPC scRNA-seq datasets. Data are compiled from N = 2 independent experiments (n = 2 mice) per genotype. (b) Feature plots displaying expression levels of select genes associated with distinct HAPC subtypes – B cells (Cd19), cDC (Itgax, zbtb46), pDC (Siglech, Itgax), macrophages and monocytes (MertK, Csf1r) and granulocytes (Camp). Residual thymocytes included in the sorted cells were identified by Lck expression. (c) Heatmap displaying normalized expression of the top 10 enriched genes in each transcriptionally distinct cDC cluster from Fig. 5a. (d) Feature plots showing scRNA-seq expression levels of (top) B2m transcripts in WT versus B2m−/− cDCs and (bottom) H2-Aa transcripts in WT versus MHC-II−/− cDCs overlaid on the UMAP visualization from Fig. 5a, confirming expected genotypes of B2m−/− and MHC-II−/− datasets. (e) Feature plots showing expression levels of Cd4 and Cd8a transcripts in thymocytes from WT versus Rag2−/− mice overlaid on the UMAP visualization in (a), confirming expected absence of thymocytes expressing these transcripts in Rag2−/− mice. (f) Quantification of the frequencies of cDC1, cDC2, aDC1, aDC2, and Il15rahi aDC subsets identified in Fig. 5a from scRNA-seq transcriptional profiling of thymic DCs in the four indicated genotypes. Data are compiled from 2 independent experiments (n = 2 mice) per genotype. Bars represent mean and symbols represent individual mice.
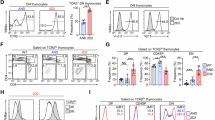
Extended Data Fig. 7 SP thymocytes are necessary for full cDC activation.
Related to Fig. 6. (a) RNA velocity analysis with arrows showing relative transition probabilities derived from ratio calculations of unspliced over spliced transcripts (cDCs -> aDCs). These analyses are consistent with differentiation trajectories of cDC1 to aDC1 and cDC2 to aDC2. (b) p-Creode analysis depicting cell-state transition trajectories for (left) cDC1 and (right) cDC2 transcriptional subsets, as in Fig. 6a,b. The topology was constructed with hierarchical placement, which connects the two transcriptionally closest end-states with path nodes. Overlay represents CytoTrace score to indicate cells with a high degree of stemness and transcriptional diversity (score closer to 1) versus those with a low degree of stemness, indicative of a more differentiated transcriptional state (score closer to 0). CytoTrace scores are consistent with more differentiated states of aDC1 and aDC2 relative to non-activated cDC1 and cDC2, respectively. (c) p-Creode (left) cDC1 and (right) cDC2 differentiation trajectories from WT and Rag2−/− scRNA-seq datasets overlaid with normalized expression values of select genes associated with cDC activation and licensing. (d) Violin plots showing normalized expression levels of the indicated genes, associated with DC activation or co-stimulation of T cells, from scRNA-seq data of WT versus Rag2−/− aDCs (combined aDC1, aDC2, and Il15rahi aDC clusters).
Extended Data Fig. 8 CD8SP thymocytes promote expression of genes associated with interferon signaling in DCs, while DCs activated in the absence of SP thymocytes express elevated levels of genes associated with canonical NF-kB signaling.
Related to Fig. 6. (a-c) Heatmaps show normalized expression (z-score) values of genes associated with the indicated pathways in cDC1s, cDC2s, aDC1s and aDC2s. Genes shown represent the union of differentially regulated genes identified via tradeSeq analysis along the p-Creode cDC1 and cDC2 differentiation trajectories from pairwise WT versus KO comparisons (Fig. 6c-d). Pathways were identified via gene enrichment analysis of the differentially regulated genes along the cDC1/cDC2 trajectories, carried out using WebgestaltR with pathways from KEGG and MSigDB databases.
Extended Data Fig. 9 Cognate interactions with CD4SP thymocytes and CD40-CD40L interactions are indispensable for cDC1 activation and central tolerance.
Related to Fig. 8. (a) Cd40 gene expression levels overlaid on the UMAP representation of scRNA-seq datasets from WT and KO thymic cDCs, as in Fig. 5a, demonstrating highest CD40 expression in aDCs. (b) CD40 RFI levels in flow cytometry-defined cDC subsets demonstrate that aDC1s express the highest levels of CD40, followed by aDC2s. Data are normalized to average MFI levels of aDC1s per experimental repeat. Data are compiled from N = 3 independent experiments; n = 8 WT mice. (c-d) (c) Representative flow cytometry plots and (d) quantification of the frequencies of CD40L expression by the indicated major thymocyte subsets, demonstrating highest CD40L expression by CD4SP thymocytes. DN, DP, CD4SP, and CD8SP thymocyte populations are plotted from the same experiments as for Fig. 8b with N = 2 independent repeats; n = 6 Cd40+/+ and n = 5 Cd40−/− mice. (e) Quantification of cell numbers (top) and frequencies (bottom) of the indicated (left to right) DP, CD4SP, and CD8SP thymocyte subsets in Xcr1+/+CD40f/− versus Xcr1cre/+CD40f/−littermate controls demonstrate an increased number of CD4SP SM cells in thymi devoid of CD40-activated cDC1s. Data are compiled from N = 5 independent experiments; n = 14 Xcr1+/+Cd40f/− and n = 13 Xcr1cre/+Cd40f/− mice. (f) Expression of Treg effector markers by CD25−Foxp3− CD4SP conventional thymocytes, CD25+Foxp3− TregP, CD25−Foxp3lo TregP, and CD25+Foxp3+ Tregs in Xcr1+/+CD40f/− versus Xcr1cre/+CD40f/− thymi. Data are compiled from N = 4 independent experiments; n = 11 Xcr1+/+Cd40f/− and n = 12 Xcr1cre/+Cd40f/− mice. (b,d,e,f) Bars represent mean ± SEM and symbols represent individual mice. Statistical tests were performed using (b) one-way ANOVA with Dunnett’s multiple comparisons test relative to aDC1 and (d-f) two-tailed unpaired Student t-test.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Srinivasan, J., Moore, C.R., Calindi, A. et al. The differentiation and function of heterogeneous thymic dendritic cell subsets require signals provided by distinct thymocyte cell types. Nat Immunol (2026). https://doi.org/10.1038/s41590-025-02371-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41590-025-02371-9