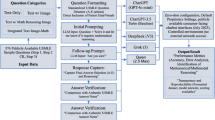
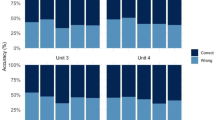
Abstract
DeepSeek is a newly introduced large language model (LLM) designed for enhanced reasoning, but its medical-domain capabilities have not yet been evaluated. Here we assessed the capabilities of three LLMs— DeepSeek-R1, ChatGPT-o1 and Llama 3.1-405B—in performing four different medical tasks: answering questions from the United States Medical Licensing Examination (USMLE), interpreting and reasoning on the basis of text-based diagnostic and management cases, providing tumor classification according to RECIST 1.1 criteria and providing summaries of diagnostic imaging reports across multiple modalities. In the USMLE test, the performance of DeepSeek-R1 (accuracy 0.92) was slightly inferior to that of ChatGPT-o1 (accuracy 0.95; P = 0.04) but better than that of Llama 3.1-405B (accuracy 0.83; P < 10−3). For text-based case challenges, DeepSeek-R1 performed similarly to ChatGPT-o1 (accuracy of 0.57 versus 0.55; P = 0.76 and 0.74 versus 0.76; P = 0.06, using New England Journal of Medicine and Médicilline databases, respectively). For RECIST classifications, DeepSeek-R1 also performed similarly to ChatGPT-o1 (0.74 versus 0.81; P = 0.10). Diagnostic reasoning steps provided by DeepSeek were deemed more accurate than those provided by ChatGPT and Llama 3.1-405B (average Likert score of 3.61, 3.22 and 3.13, respectively, P = 0.005 and P < 10−3). However, summarized imaging reports provided by DeepSeek-R1 exhibited lower global quality than those provided by ChatGPT-o1 (5-point Likert score: 4.5 versus 4.8; P < 10−3). This study highlights the potential of DeepSeek-R1 LLM for medical applications but also underlines areas needing improvements.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The USMLE dataset is available at https://www.usmle.org/exam-resources. The NEJM case challenges are available at https://www.nejm.org/case-challenges. The BMJ endgames are available at https://www.bmj.com/specialties/endgames. The MIMIC-III dataset can be requested at https://physionet.org/content/mimiciii/1.4/. The Médicilline dataset (http://www.medicilline.com/) of multiple-choice questions for clinical diagnosis and management translated in English is available upon request for private or research use only, after agreement of Médicilline. Médicilline data requests should be sent to Mickael.Tordjman@mssm.edu. In response to the inquiry, the timeframe for responding to requests is approximately within 2 weeks of the request. The radiological reports used for summarization and RECIST classification are not available due to privacy issues.
Code availability
All studies were conducted using Azure OpenAI service (ChatGPT), Azure AI Foundry (Llama) and Azure AI service (DeepSeek). The algorithms used in this study are based on the official guidelines provided by the developers of the LLMs evaluated—DeepSeek, ChatGPT and Llama. The implementation followed these guidelines strictly to ensure consistency and reproducibility of results. As the study did not involve the development of new code but rather the application of existing, officially provided algorithms, specific source codes referenced are proprietary and maintained by their respective developers.
References
The Lancet Digital Health. Large language models: a new chapter in digital health. Lancet Digit. Health 6, e1 (2024).
Gibney, E. Scientists flock to DeepSeek: how they’re using the blockbuster AI model. Nature https://doi.org/10.1038/d41586-025-00275-0 (2025).
Conroy, G. & Mallapaty, S. How China created AI model DeepSeek and shocked the world. Nature https://doi.org/10.1038/d41586-025-00259-0 (2025).
OpenAI. GPT-4 technical report. Preprint at http://arxiv.org/abs/2303.08774 (2023).
Grattafiori, A. et al. The Llama 3 Herd of Models. Preprint at https://arxiv.org/abs/2407.21783 (2024).
Johnson, A. E. W. et al. MIMIC-III, a freely accessible critical care database. Sci. Data 3, 160035 (2016).
Zhang, T., Kishore, V., Wu, F., Weinberger, K. Q. & Artzi, Y. BERTScore: evaluating text generation with BERT. Preprint at https://arxiv.org/abs/1904.09675 (2020).
Fleiss, J. L. Measuring nominal scale agreement among many raters. Psychol. Bull. 76, 378–382 (1971).
Exam resources. United States Medical Licensing Examination https://www.usmle.org/exam-resources (2025).
Case challenges. N. Engl. J. Med. https://www.nejm.org/case-challenges (2025).
Endgames. BMJ https://www.bmj.com/education/endgames (2025).
Médicilline dataset. Médicilline http://www.medicilline.com/13-externe-ecni (2025).
Laurent, G., Craynest, F., Thobois, M. & Hajjaji, N. Automatic classification of tumor response from radiology reports with rule-based natural language processing integrated into the clinical oncology workflow. JCO Clin. Cancer Inform. 7, e2200139 (2023).
Penny, P., Bane, R. & Riddle, V. Advancements in AI medical education: assessing ChatGPT’s performance on USMLE-style questions across topics and difficulty levels. Cureus 16, e76309 (2024).
Zhao, W. X. et al. A survey of large language models. Preprint at https://arxiv.org/abs/2303.18223 (2024).
Learning to reason with LLMs. OpenAI https://openai.com/index/learning-to-reason-with-llms/ (2024).
Temsah, M.-H., Jamal, A., Alhasan, K., Temsah, A. A. & Malki, K. H. OpenAI o1-Preview vs. ChatGPT in Healthcare: A New Frontier in Medical AI Reasoning. Cureus 16, e70640 (2024).
DeepSeek-AI et al. DeepSeek-R1: incentivizing reasoning capability in LLMs via reinforcement learning. Preprint at https://arxiv.org/abs/2501.12948 (2025).
Suh, P. S. et al. Comparing large language model and human reader accuracy with New England Journal of Medicine image challenge case image inputs. Radiology 313, e241668 (2024).
Tenner, Z. M., Cottone, M. C. & Chavez, M. R. Harnessing the open access version of ChatGPT for enhanced clinical opinions. PLoS Digit. Health 3, e0000355 (2024).
Cabral, S. et al. Clinical reasoning of a generative artificial intelligence model compared with physicians. JAMA Intern. Med. 184, 581–583 (2024).
Zhang, L. et al. Constructing a large language model to generate impressions from findings in radiology reports. Radiology 312, e240885 (2024).
Park, J., Oh, K., Han, K. & Lee, Y. H. Patient-centered radiology reports with generative artificial intelligence: adding value to radiology reporting. Sci. Rep. 14, 13218 (2024).
Eisenhauer, E. A. et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2009).
Acknowledgements
We thank G. Zagury and Médicilline Editions for sharing their dataset. We thank N. Venturelli, L. Chiche, J. Beaziz and R. Lejoyeux for their help updating these questions based on the latest medical knowledge. This project is supported by the Eric and Wendy Schmidt AI in Human Health Fellowship, a program of Schmidt Sciences. M.T. is supported by the French Society of Radiology and the French Musculoskeletal Imaging Society. Figure 1 was created with BioRender.com.
Author information
Authors and Affiliations
Contributions
Concept and design: M.T., Z.L., H.-C.L., Z.A.F. and X.M. Acquisition, analysis or interpretation of data: M.T., Z.L., M.Y., V.F., Y.M., J.H., I.B., H.A., C.H., A.S.P., A.G., A.M., N.Y., N.N., P.R., A.Z., S.L., M.H., T.D., B.T., H.-C.L., Z.A.F. and X.M. Drafting of the paper: M.T., Z.L., M.Y., V.F., A.M., I.B., S.L., B.T., H.-C.L., Z.A.F. and X.M. Critical revision of the paper and final draft: all authors.
Corresponding authors
Ethics declarations
Competing interests
T.D. is the managing partner of RadImageNet LLC and a paid consultant to GEHC and AirsMedical. X.M. is a paid consultant to RadImageNet LLC. The other authors declare no competing interests.
Peer review
Peer review information
Nature Medicine thanks Ahmed Alaa, Kirk Roberts and Jie Yang for their contribution to the peer review of this work. Primary Handling Editors: Michael Basson, Lorenzo Righetto and Saheli Sadanand, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tordjman, M., Liu, Z., Yuce, M. et al. Comparative benchmarking of the DeepSeek large language model on medical tasks and clinical reasoning. Nat Med 31, 2550–2555 (2025). https://doi.org/10.1038/s41591-025-03726-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41591-025-03726-3
This article is cited by
-
Benchmark evaluation of deepseek AI models in antibacterial clinical decision-making for infectious diseases
BMC Medical Informatics and Decision Making (2026)
-
Evaluation of large language models in nutrition risk screening: a comparative analysis across 8 LLMs based on real-world EHR datasets
BMC Medical Informatics and Decision Making (2026)
-
The impact of DeepSeek’s perceived interactivity on medical students’ self-directed learning ability
Scientific Reports (2026)
-
DermaGPT a federated multimodal framework with a meta learned trust function for interpretable dermatology diagnostics
Scientific Reports (2026)
-
Transforming clinical medicine with multimodal artificial intelligence, agentic systems, and the model-context protocol: a perspective on future directions
Discover Health Systems (2026)