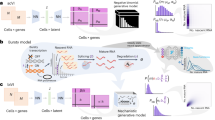
Abstract
Single-molecule imaging techniques have provided unprecedented insights into functional changes in composition and conformation across diverse biological systems. As with other biophysical methods, single-molecule fluorescence and Förster resonance energy transfer investigations are typically limited to examination of one sample at a time. Consequently, experimental throughput is restricted, and experimental variances are introduced that can obscure functional distinctions in closely related systems. Here, to address these limitations, we introduce parallel rapid exchange single-molecule fluorescence and single-molecule Förster resonance energy transfer to enable simultaneous steady-state and pre-steady-state interrogations of diverse systems. Using this approach, we elucidate the timing of distinct conformational events underpinning β-arrestin1 activation, unmask antibiotic-induced impacts on messenger RNA decoding fidelity and demonstrate that endogenously encoded ribosomal RNA sequence variation modulates antibiotic sensitivity. This generalizable and scalable method promises to broaden the scope and reproducibility of quantitative single-molecule interrogations of biomolecular function.
Similar content being viewed by others
Main
Single-molecule fluorescence (smF) and Förster resonance energy transfer (FRET) imaging enable real-time measurements of fluctuations in the composition and conformation that underpin the function of individual biomolecules1. This approach enables direct detection of rare, transient events otherwise obscured at the ensemble scale while requiring orders-of-magnitude less material than traditional biophysics investigations. As a consequence, smF(RET) investigations have provided critical insights into regulation in diverse biological systems2,3, including those derived from mammals4,5,6,7.
The range of biological questions that can be interrogated at the single-molecule scale, along with the accuracy, precision and experimental throughput of smF(RET) measurements, has expanded in parallel with advances in organic fluorophore performance8, site-specific labeling9, camera technologies and imaging strategies10. Recent advancements in smF(RET) imaging throughput include zero-mode-waveguide technologies11,12, automation of confocal smFRET data acquisition for screening initiatives13, multiplexed examination of mixed sample populations14,15,16 and objective-based total internal reflection fluorescence (TIRF) imaging combined with next-generation sequencing17,18.
Despite the power and sensitivity of smF(RET), comparative investigations on wild-type versus mutant systems7,19 or studies of an individual system from multiple structural perspectives20 are generally limited to independent measurements. Consequently, variabilities in component concentrations as well as experimental and instrumental noise can obscure structural or kinetic differences of functional significance, including subtle yet consequential perturbations, which may have disproportionate impacts on processive systems or signaling cascades. One path to circumvent such uncertainties is to collect data on distinct samples in parallel followed by a posteriori parsing methods. Steady-state investigations of sample mixtures have been parsed based on spectral properties21,22, FRET signatures23,24 and position17,18,25,26. A posteriori methods also require processing procedures that may result in the obfuscation of sample identity and corresponding losses in information recovery. An alternative path for parallelized measurements is to know the identity of each sample before data acquisition. However, a generalizable approach for a priori parallel investigations of diverse biological specimens has yet to be achieved.
Here, we introduce parallel rapid exchange smF (PRE-smF) and FRET (PRE-smFRET) to enable simultaneous imaging of diverse biological systems under identical steady-state and pre-steady-state conditions. PRE-smF(RET) uses wide-field, prism-based TIRF (pTIRF) imaging implemented with flat-field illumination. pTIRF uniquely supports pre-steady-state measurements in which stimulus can be rapidly introduced to surface-tethered biomolecules by solution exchange (analogous to stopped-flow) or by photo-induced uncaging27. Additionally, pTIRF generally offers higher experimental throughput than objective-based approaches and allows high time resolution measurements requiring increased excitation power density28,29. To enable parallelized a priori investigations, we integrate pTIRF with automated liquid-handling systems using microcontact-printed microfluidic devices that enable DNA-barcoded immobilization with epitaxial removal aided by strand exchange (ERASE)30. We highlight the sensitivity of this method by differentiating the photophysical characteristics of organic fluorophores linked to DNA, the order and timing of conformational events underpinning β-arrestin1 activation processes, kinetic dissimilarities in mRNA-decoding rates in protein synthesis and the distinct drug sensitivities of endogenously encoded ribosome subtypes linked to the nutrient limitation-induced stress response in bacteria31. Our findings demonstrate PRE-smF(RET) to be a generalizable and scalable solution for comparative steady-state and pre-steady-state single-molecule measurements of diverse biological systems at high spatial and temporal resolution with reliability and sensitivity. By introducing this method, we seek to facilitate an era of discovery in which subtle functional distinctions can be revealed in diverse biological systems with unprecedented resolution.
Results
Microfluidic chips with microarrays for single-molecule imaging
For PRE-smF(RET), we used a pTIRF platform equipped with one scientific complementary metal oxide semiconductor camera per spectral channel28 to image one 250 µm × 250 µm field of view (FOV) at a high signal-to-noise ratio (Extended Data Fig. 1). We surface tethered biomolecules in custom-built quartz microfluidic devices (Methods and Extended Data Fig. 2), passivated with a 1:100 mixture of biotin-polyethylene glycol (PEG):methoxy-PEG32, using ERASE30. Low-density biotin-PEGylation ensures that molecules can be tethered via a biotin–streptavidin bridge with spacing that prevents their interaction (Supplementary Note 1). ERASE enables an isothermal hybridization approach to specifically pull down a ‘tether’ oligonucleotide-linked biomolecules of interest with a surface-tethered complementary ‘anchor’ oligonucleotide (Fig. 1a and the Methods). A key feature of ERASE is that each tethered biomolecule can be rapidly released from the imaging chamber via toehold-mediated strand displacement (TMSD)33 using ‘switch’ oligonucleotides complementary to the tether sequence. In this manner, ERASE enables fine-tuning of the tethered biomolecule surface density as well as complete resetting of the microfluidic device for reuse.
a, Schematic of surface tethering of a fluorescently labeled biomolecule using ERASE. Biomolecules are specifically surface tethered through the hybridization of a tether oligonucleotide (conjugated to the biomolecule) and an anchor oligonucleotide (immobilized on the surface). The addition of a switch strand removes tethered biomolecules through toehold-mediated strand displacement. b, Schematic of microarray printing on a biotin-PEG-coated surface with a pin-based microarrayer. The anchor DNA is tethered to the surface via a biotin–streptavidin bridge. c, Layout of a 25 × 76 mm arrayed quartz microfluidic device with ten 3 µl fluidic chambers, each measuring 19 × 3 × 0.05 mm. d, Bright-field microscopy image of a printed microarray with multiple microdroplet groups, each containing four distinct anchor DNA sequences. Inset shows a schematic of a microscope FOV with a 60× objective lens. Green rounded square shows the area of the channel illuminated by flat-top illumination during image collection. e, Cross-sectional schematic of one imaging chamber. Each chamber is independently connected to a fluidics handling system that enables buffer exchange during imaging. Panels a and b created with BioRender.com.
To engender a priori parallelized PRE-smF(RET) investigations, we microcontact printed four orthogonal anchor oligonucleotides in 2 × 2 arrays to enable location-specific pulldown of four tether-barcoded biomolecules (Fig. 1b and the Methods). We spaced 60 arrays in ten individual microfluidic chambers (3 µl volume) such that each 2 × 2 array could be interrogated independently and in rapid sequence via automated stage movements (Fig. 1c,d and the Methods). We precision drilled inlet and outlet ports at each end of each microfluidic chamber and coupled them via PEEK tubing (Fig. 1e and the Methods) to a computer-controlled liquid-handling system to enable rapid and reproducible pre-steady-state measurements via solution exchange without the need for a mixing chamber, the pTIRF equivalent of stopped-flow.
Binding to microarrays is specific and tunable
To demonstrate the binding specificity to these microarrayed devices, we monitored the tethering of four FRET-labeled (LD555 and LD655) DNA duplexes, each with a unique FRET signature and an associated single-stranded tether barcode (Fig. 2a, Methods and Supplementary Video 1). We introduced orthogonal switch oligonucleotides to independently tune the tethering density of each sample to ensure optimal separation of each single-molecule emitter for imaging28 (Fig. 2a). We also demonstrated the ability to completely strip each specific molecule from the surface (Fig. 2a).
a, Demonstration of control over biomolecule binding density afforded by ERASE. Top, snapshots taken at various times throughout an smF movie; bottom, the number of molecules in each printed spot over time; t0 = 20 s, t1 = 160 s, t2 = 310 s, t3 = 420 s, t4 = 500 s. At t0, we injected a mixture of four distinct LD555- and/or LD655-labeled DNA duplexes (samples A–D), each with a unique single-stranded tether domain, and sample A was purposefully added at a higher concentration to achieve a tethering density that was too high. At t1, 40 nM switch A was added to reduce sample A density to a usable level. Simultaneously, 1 µM switch D was added to completely remove sample D. At t2, fresh sample D was injected. Finally, at t3, we added 1 µM switches A, B and D to selectively remove all samples except sample C. After a brief switch incubation, no molecules could be observed in spots A, B or D (t4). b, Measurement of binding specificity using four FRET-labeled duplexes, each with a unique interdye spacing and an associated single-stranded tether barcode. Population histograms of FRET efficiency in each quadrant show a sharp single peak. n, number of traces; nt, nucleotides; sp., specificity of binding, defined as the fraction of molecules in a printed circle with the expected FRET efficiency value. c, PRE-smFRET enables head-to-head comparison of fluorophore photophysical properties under identical conditions. Plots show mean and s.d. of signal-to-noise ratio (SNR; Methods) and photon yield (cts, photon counts) from 16 (ambient) or 15 (deoxygenated) independent repeats (over 12,000 individual molecules per fluorophore).
After achieving a tethering density of ~2,000 molecules per printed spot, we extracted ~300–400 well-defined single molecules from each area to compile population FRET histograms for individual FOVs. Consistent with highly specific tethering, each set of extracted molecules exhibited a narrow FRET efficiency distribution, the maxima of which was consistent with the expected interdye distances of each tethered species (Fig. 2b). By quantifying the total area of the dominant FRET distribution compared to the total FRET distribution area, we estimated the binding specificity in each microcontact-printed spot to be ~97.5% ± 1.8% (Methods). Similar experiments on diverse biological samples demonstrated specificities between 95% and 98% (Extended Data Fig. 3).
Parallelized quantification of fluorophore performance
Bright and photostable fluorophores are the foundation of both single-molecule imaging and ensemble fluorescence investigations across a broad range of life sciences8,10. Single-molecule analyses of fluorophore performance provide quantitative information for optimizing new fluorophore designs8,34. Such studies are typically performed both in the presence and absence of oxygen to capture the range of conditions typical for smF(RET) investigations. Comparative analyses of this kind are challenging because brightness, photon emission dynamics and total photon budget are highly sensitive to pH, illumination intensity, the efficiency of enzymatic oxygen scavenging35, redox chemistry34 and, if employed, the effective concentrations of stabilizing agents in solution29.
Here, we employed PRE-smF to perform head-to-head comparison of fluorophore performance under identical conditions by imaging eight tether oligonucleotide-barcoded DNA duplexes, each linked to a single fluorophore at the 3′ terminus (Methods). We compared the performance of four donor and four acceptor fluorophores simultaneously in a single FOV by preparing each set with four unique DNA constructs such that each spot in the 2 × 2 array contained a roughly equal mixture of one donor- and one acceptor-labeled DNA duplex (Extended Data Fig. 4 and Supplementary Table 1). In each FOV, donor and acceptor fluorophores were excited independently using alternating-laser excitation36 illumination at 532 nm and 640 nm (Methods). We collected data in rapid sequence from oxygenated (16 FOVs) and deoxygenated (15 FOVs) conditions to robustly characterize fluorophore performance in terms of signal-to-noise ratio and photon budget. These studies showed that self-healing LD555 and LD655 fluorophores globally outperformed their donor and acceptor counterparts under both conditions (Fig. 2c and Supplementary Video 2). ATTO fluorophores, while 15–30% brighter in oxygenated conditions, exhibited substantial reductions in brightness under deoxygenated environments: ATTO 532 brightness decreased fourfold, while ATTO 647N brightness decreased twofold, accompanied by a nearly fivefold reduction in total photon yield (Supplementary Table 2). We attribute this behavior to the propensity of ATTO fluorophores to exhibit increased occupancy of nonfluorescing triplet and radical dark states29,34. Parallel performance evaluations of this kind demonstrate an eightfold increase in experimental throughput while guaranteeing identical conditions and thus greater reproducibility. To demonstrate this latter point, we show that the deviations in photon yield in each quadrant of a single FOV are ≥4-fold lower than those from four independent experiments (Extended Data Fig. 4b and the Methods).
Computer-controlled, stopped-flow PRE-smF(RET) investigations
We enabled pre-steady-state smF(RET) imaging by implementing a two-injector port, computer-controlled fluidics system (Fig. 3a and the Methods) that ensures reproducible solution exchange rates down to 60 ms (Fig. 3b,c). With this design, solution exchange could be rapidly and repeatedly achieved through alternating injections (Fig. 3c). To validate this system, we studied the clamshell-like closure process accompanying ligand binding using a site specifically labeled isoform of the leucine–isoleucine–valine binding protein (LIV-BPSS)37 (Fig. 3d). The speed and reproducibility of this system were demonstrated by synchronous LIV-BPSS transitions between apo (open state, FRET efficiency = 0.35) and leucine-bound (closed state, FRET efficiency = 0.65) conformations upon injection and washout of 25 µM leucine across 18 distinct FOVs examined in rapid sequence (Fig. 3e and the Methods).
a, Schematic of the microfluidic system for rapid reagent delivery during single-molecule imaging. b, Quantification of solution exchange time using the 10–90% criterion (top) with 250 nM LD555 dye being injected into the observation chamber at different flow rates (bottom). Box plot represents data collected from eight independent repeats. Box boundaries indicate interquartile range, horizontal line represents median, and whiskers show data points within 1.5× interquartile range. c, Demonstration of 18× 40 µl repeated injections of 250 nM Cy5 (directly excited using a 640 nm laser) from a 1 ml loop, resulting in a highly reproducible concentration profile. Right, mean and s.d. from 18 repeated injections. d, Schematic showing the LIV-BPSS protein switching reversibly between open (FRET efficiency = 0.35) and closed (FRET efficiency = 0.65) conformations upon leucine binding and release. e, Demonstration of non-equilibrium smFRET imaging of LIV-BPSS with rapid injection of 25 µM leucine (t = 0.4 s), followed by rapid channel rinsing (t = 0.9 s). This population histogram contains data compiled from 18 computer-controlled injections and was composed without post-synchronizing each smFRET trace. Panel d created with BioRender.com.
Parallelized multiperspective imaging of β-arrestin1 activation
To demonstrate the ability of PRE-smFRET to achieve multiperspective imaging, we investigated the process of β-arrestin activation from four distinct structural perspectives. β-arrestins engage agonist-activated and G protein-coupled receptor (GPCR) kinase-phosphorylated GPCRs to undergo an activation process that facilitates the termination of GPCR-activated heterotrimeric G protein signaling, the initiation of alternative intracellular signaling pathways and GPCR internalization38,39. Structural and molecular dynamics studies show that GPCR-mediated β-arrestin activation entails release of its autoinhibitory C-terminal tail, conformational changes in its finger loop and other central crest elements and a ~20° rotation of the C-terminal domain relative to the N-terminal domain40 (Fig. 4a). Currently, it is not known whether these distinct activating conformational changes occur simultaneously or with a defined order and timing.
a, Activation of β-arrestin1 upon binding to V2Rpp, a phospho-peptide mimetic of the phosphorylated C tail of V2R. One labeling perspective (C-tail sensor 1) is shown. b, Overview of the generic protein immobilization strategy for smFRET microarrays using adaptor molecules. This strategy enabled PRE-smFRET imaging of β-arrestin1 activation upon V2Rpp addition from four structural perspectives: C-tail release sensors 1 and 2, finger loop conformational changes and C-domain rotation. c, Representative smFRET traces showing steady-state conformational changes in β-arrestin1 C-tail sensor 1 at increasing concentrations of V2Rpp. Oligo, oligonucleotide. d, Titration of V2Rpp against arrestin measured from four structural perspectives in parallel. Plotted data represent mean and s.d. from three independent repeats. e, A representative smFRET trace showing the activation and deactivation of β-arrestin1 C-tail sensor 1 upon V2Rpp injection and washout. Binding rates were estimated from the waiting time distribution between V2Rpp injection to first transit to the low FRET active state, ton. Dissociation rates were estimated from the distribution of waiting times between V2Rpp washout and the adoption of the stable high FRET basal state, toff. f, PRE-smFRET imaging of β-arrestin1 activation and deactivation upon V2Rpp injection and rinsing, correspondingly; 10 µM V2Rpp was injected at ti = 5 s and rinsed at tr = 25 s (vertical dashed lines). Contour plots represent data compiled from 15 injections, without post-synchronization.
We therefore sought to use PRE-smFRET to monitor the conformational events in β-arrestin1 upon binding to a phospho-peptide mimicking the phosphorylated C tail of the human V2 vasopressin receptor (V2Rpp)41,42. We visualized β-arrestin activation from multiple structural perspectives by employing two previously reported donor- and acceptor-labeled sensors of C-tail release19 and two new sensors designed to report on finger loop conformational changes and C-domain rotation relative to the N domain (Methods and Supplementary Table 3). We established a generalized workflow for investigations of this kind by using tether oligonucleotide-conjugated antibodies (adaptor molecules) specific to commonly used epitopes for affinity purification (Fig. 4b and the Methods). In so doing, we could spatially tether and image four β-arrestin1 sensors simultaneously in a single FOV using adaptors specific to the N-terminal Strep-II Tag epitope of β-arrestin (Fig. 4b).
As anticipated from previous smFRET investigations and an autoinhibited basal-state structure43,44, all four β-arrestin1 smFRET sensors exhibited highly stable, sensor-specific FRET efficiencies in the absence of V2Rpp (Supplementary Table 3). All four sensors robustly exited the basal state with increasing probability upon titration of the V2Rpp peptide into the imaging chamber to achieve activated conformations with distinct FRET values (Fig. 4c). We measured the activation process by quantifying the loss of basal-state occupancy for the three sensors exhibiting relatively large changes in FRET (for C-tail and finger loop sensors) using hidden Markov modeling (HMM) and likelihood maximization methods (Methods). These analyses yielded apparent dissociation constants (\({K}_{{\rm{D}}}^{\mathrm{app}}\)) of 2.2 ± 0.4 µM for C-tail sensor 1 and 3.1 ± 0.9 µM for C-tail sensor 2 (Fig. 4d, Methods and Supplementary Table 3), consistent with isothermal titration calorimetry measurements of V2Rpp affinity for unlabeled β-arrestin1 (2.4 ± 0.2 µM)19. By contrast, the finger loop sensor exhibited a significantly higher affinity than the C-tail release sensors, with an apparent \({K}_{{\rm{D}}}^{\mathrm{app}}\) of 1.4 ± 0.2 µM. While the dynamic range of the C-domain rotation sensor precluded HMM analysis, we quantified the change in FRET efficiency as a function of V2Rpp concentration to show that it exhibited a \({K}_{{\rm{D}}}^{\mathrm{app}}\) of 4.6 ± 0.45 µM (Fig. 4d, Extended Data Fig. 5a, Supplementary Table 3 and the Methods).
For the C-tail release and finger loop sensors, we could determine the apparent rate of V2Rpp binding (\({k}_{{\rm{on}}}^{{\rm{app}}}\)) from the steady-state transitions as <0.1 µM−1 s−1 (Supplementary Table 3), more than three orders of magnitude below the diffusion limit (~200 µM−1 s−1) and consistent with autoinhibition mechanisms observed in distinct biological systems45. This finding suggests that V2Rpp binding is a multistep process, where activating conformational changes in β-arrestin1 are rate limited by one or more unresolved binding step(s). The bimolecular association (\({k}_{{\rm{on}}}^{{\rm{app}}}\)) and dissociation (\({k}_{{\rm{off}}}^{{\rm{app}}}\)) rates of V2Rpp binding to the C-tail and finger loop sensors revealed by this analysis were in line with the observed affinities toward V2Rpp (Supplementary Table 3).
To directly obtain the \({k}_{{\rm{on}}}^{{\rm{app}}}\) and \({k}_{{\rm{off}}}^{{\rm{app}}}\) of V2Rpp binding to all four β-arrestin1 sensors, we performed >12 independent pre-steady-state measurements at five distinct V2Rpp concentrations (Fig. 4e,f, Methods, Extended Data Fig. 6a,b and Supplementary Table 3). Consistent with our steady-state experiments, \({k}_{{\rm{on}}}^{{\rm{app}}}\) was <0.1 µM−1 s−1 for all four sensors. Furthermore, we observed statistically significant differences in \({k}_{{\rm{on}}}^{{\rm{app}}}\) between sensors, where the finger loop and C-domain rotation sensors exhibited higher and lower rates of binding, respectively, compared to the C-tail sensors (Extended Data Fig. 6b and Supplementary Table 3).
To evaluate whether the observed differences in V2Rpp affinities reflect an ordered sequence of events in the β-arrestin1 activation process or label-specific perturbations, we introduced mutations at the unique labeling sites used for C-domain rotation (L191A) and finger loop (D69A) measurements into the C-tail 1 sensor. We also introduced C-tail 1 (K397A) and finger loop (D69A) mutations into the C-domain rotation sensor (Extended Data Fig. 5b and Supplementary Table 3). Consistent with contributions to a basal-state-stabilizing ionic lock46, the D69A mutation increased the affinity of both β-arrestin1 C-tail 1 and C-domain sensors by approximately twofold. Hence, disruption of the ionic lock caused by the D69A mutation predisposes β-arrestin1 to activation. By contrast, the C-tail 1 (K397A) and C-domain (L191A) sensor mutations had no measurable impact on affinity (Extended Data Fig. 5b and Supplementary Table 3). We conclude from these findings that C-domain rotation is likely to represent the rate-limiting step of the β-arrestin1 activation process. Future PRE-smFRET studies will enable sensitive investigations into whether the sequential β-arrestin1 activation mechanism is conserved in other arrestin isoforms, the extent to which it contributes to specific GPCR signaling outcomes and turnover and how it is affected by the nature of GPCR phosphorylation marks and the phospho-code47 as well as GPCR sequence48 and physiological context49.
Parallelized measurements of mRNA decoding
To demonstrate the application of PRE-smFRET to more complex biological systems and to ascertain whether the ERASE approach was amenable to isothermal RNA hybridization, we investigated the multistep, irreversible process of transfer RNA (tRNA) selection by the bacterial ribosome. Faithful translation of messenger RNA (mRNA) sequence into protein occurs during the elongation phase of protein synthesis catalyzed by the ribosome. Each mRNA codon is decoded by a specific aminoacyl-tRNA species and delivered to the ribosome as a ternary complex with the GTP-bound translation factor EF-Tu (EF-Tu–GTP–aminoacyl-tRNA)50. During tRNA selection, the ternary complex first transiently engages the leading edge of the ribosome at the aminoacyl (A) site in a codon-independent manner51. Improper base pairing between the mRNA codon in the A site and near-cognate and noncognate aminoacyl-tRNAs results in inefficient domain closure within the small subunit (SSU) of the ribosome, which leads to rapid dissociation of the ternary complex. By contrast, proper base pairing facilitates ternary complex capture via SSU domain closure, which forms the codon recognition (CR) state. SSU domain closure facilitates EF-Tu docking to the large subunit of the ribosome, which ultimately forms the GTPase-activated (GA) state resulting in GTP hydrolysis. Subsequently, aminoacyl-tRNA is released from EF-Tu to enter the large subunit peptidyl transferase center, where it achieves a fully accommodated (AC) state capable of peptide bond formation51,52,53,54. In the current kinetic model of mRNA decoding54,55, CR-to-GA transitions represent the initial selection process, while GA-to-AC transitions represent the proofreading process. Structural transitions associated with initial selection and proofreading, quantified by smFRET51 as well as with molecular dynamics simulations56, indicate that the rate and fidelity of mRNA decoding requires geometric alignment of the incoming ternary complex and aminoacyl-tRNA with the GTPase-activating and peptidyl transferase centers, respectively.
Using PRE-smFRET, we simultaneously assessed the rate and fidelity of tRNA selection on the Escherichia coli ribosome for two cognate and two near-cognate codons to the phenylalanine tRNA under pre-steady-state conditions. To perform these studies, we assembled four bacterial ribosome initiation complexes (ICs) programmed with Cy3-labeled tRNAfMet in the P site each with a unique codon in the A site: cognate (phenylalanine, UUU and UUC) or near cognate (leucine, CUC; serine, UCC) (Methods). We achieved spatial localization of four ICs within a single FOV by including a unique tether sequence upstream of the start (AUG) codon of each mRNA to enable mRNA–DNA hybrid formation with its corresponding surface anchor in 2 × 2 array format (Fig. 5a). After IC immobilization, tRNA selection was initiated by stopped-flow delivery of 50 nM ternary complex formed with LD655-labeled L-α-Phe-tRNAPhe (GAA anticodon) (Methods). After data collection, the microfluidic device was reset via TMSD to enable eight experimental replicates within the same microfluidic device.
a, Ribosome complexes are region specifically immobilized through hybridization between a programmed tether domain of each mRNA and a surface-immobilized DNA anchor. Four complexes carrying unique A site codons are measured simultaneously: two cognate (UUU and UUC) and two near cognate (CUC and UCC). Rates of transitions between the key tRNA selection steps (IC, CR, GA and AC states) are measured by smFRET. b, FRET contour plots reveal changes in tRNA selection fidelity with and without streptomycin, a miscoding-inducing antibiotic. c, Streptomycin results in a lower rate of ternary complex binding to the ribosome for both cognate and near-cognate codons. Data points represent mean, error bars, s.e.m.; P values, two-tailed unpaired t-test. d, Log odds of forward transition through initial selection and proofreading in the absence and presence of streptomycin. Data points represent estimated log odds of state transition, error bars denote standard error of estimate from logistic regression, where P values are derived with the Wald test. Data used for c,d represent nine (no streptomycin condition) or 14 (streptomycin condition) independent repeats.
As expected, ribosomes bearing cognate codons rapidly and efficiently incorporated Phe-tRNAPhe (catalytic efficiency for UUC-bearing ICs; 38 ± 1 µM−1 s−1) via efficient passage through CR (FRET efficiency = 0.2), GA (FRET efficiency = 0.5) and AC (FRET efficiency = 0.75) states51,57 (Fig. 5b and Extended Data Fig. 7b). Consistent with a high-fidelity, energy-efficient process51,54, ribosomes programmed with near-cognate codons rapidly and efficiently rejected the same ternary complex, primarily during initial selection. The shorter lifetime of the GA state in near-cognate complexes results in a lower apparent FRET efficiency due to time averaging51. The observed differences in population FRET histograms comparing both cognate codons and both near-cognate codons (Fig. 5b) are consistent with kinetic and fidelity distinctions documented using ensemble stopped-flow investigations58. These findings highlight the potential of PRE-smFRET to differentiate mRNA-decoding rates when only single hydrogen bonds are altered in the mRNA codon–tRNA anticodon pair.
We next performed equivalent experiments in the presence of the miscoding-inducing aminoglycoside antibiotic streptomycin, which binds adjacent to the decoding center through interactions with 16S ribosomal RNA (rRNA)59. As anticipated from ensemble measurements60, streptomycin reduced the overall rate of mRNA decoding (that is, from TC injection to AC; Extended Data Fig. 7a) on cognate codons while efficiently promoting Phe-tRNAPhe misincorporation on ribosomes programmed with both near-cognate codons (Fig. 5b and Extended Data Fig. 7c). Strikingly, and in contrast to previous findings60, streptomycin reduced the apparent rate of ternary complex binding to the ribosome by approximately twofold (Fig. 5c). Also unexpectedly, streptomycin significantly increased the mRNA-decoding rates (that is, from CR to AC; Extended Data Fig. 7b) and efficiencies of initial selection and proofreading (Extended Data Fig. 7d and Fig. 5d) for both cognate and near-cognate mRNA decoding. Based on extant literature61,62, we speculate that streptomycin likely exerts both impacts through the parsimonious mechanism of promoting SSU domain closure. This process would prevent aminoacyl-tRNA access to the mRNA codon, thus slowing the CR process, while promoting productive tRNA incorporation by stabilizing codon–anticodon interactions throughout the decoding process59. Future investigations will be required to ascertain whether this conclusion is specific to streptomycin or general to other aminoglycoside class antibiotics.
PRE-smF(RET) reveals differential antibiotic sensitivity of ribosomes bearing rRNA sequence variation
To demonstrate the application of PRE-smFRET to the emerging frontier of investigating the functional consequences of ribosome heterogeneities31,63, we examined the functions of two evolutionarily conserved ribosome subtypes found in E. coli. Laboratory strains of E. coli, such as MG1655, encode seven ribosomal DNA operons in their genomes that are expressed at different levels under distinct growth conditions31 (Fig. 6a). While all seven ribosomal DNA operons support life64, nutrient limitation-induced stress alters the cellular ribosome pool such that ribosomes formed from operons distal to the origin of replication (rrnD, rrnG and rrnH) become relatively more abundant, while three of the four operons near the origin become relatively less abundant (rrnA, rrnB and rrnE) and others (rrnC) remain unchanged31. The rrnH operon, the most highly enriched upon nutrient limitation and the most evolutionarily conserved across the Enterobacter genus, was further shown to programmatically alter gene expression and phenotype in a manner that potentiated the nutrient limitation-induced stress response31.
a, Map of E. coli K-12 chromosome showing the location of each of the seven rRNA operons. b, Organization of rrnB and rrnHBB. Each transcript is expressed from tandem promoters (P1 and P2) and encodes a 16S rRNA (rrsB for rrnB, rrsH for rrnHBB), a 23S rRNA and a 5S rRNA and is terminated at tandem terminators (t1 and t2). Variant bases in 16S rRNA are highlighted. c, Log odds of tRNA selection in the absence of drug, the presence of 1 µM tetracycline and the presence of 100 nM oxytetracycline for rrnB and rrnHBB ribosomes isolated from ∆7prrn-BBB and ∆7prrn-HBB, respectively. Data comprise five independent repeats for each condition. Data points represent estimated log odds of accommodation, error bars denote standard error of estimate from logistic regression and P values are derived with the Wald test.
Strikingly, these investigations also showed that cells exclusively expressing ribosomes bearing either the SSU head domain sequence of the rrnB operon or the ten SNPs present in the rrsH gene of the rrnH operon (Fig. 6b) exhibited altered sensitivities to tetracycline class antibiotics31, clinically relevant small molecules that bind directly to the SSU head domain to inhibit mRNA decoding65. Specifically, cells expressing ribosomes bearing rrsH variants were relatively resistant to tetracycline and relatively sensitive to oxytetracycline31. Leveraging the power of PRE-smFRET to perform experiments under identical experimental conditions, we conclusively demonstrate that evolutionarily conserved ribosome subtypes bearing rrsH variants show differential sensitivities to drugs in the same antibiotic class (Fig. 6c and Extended Data Fig. 8).
Discussion
smF imaging has revolutionized the study of conformational dynamics in biomolecular systems by providing direct access to the order and timing of events as well as detection of rare and transient species undetectable by ensemble methods2. Another critical advantage is that single-molecule methods require only picomole quantities of material. This relatively modest material demand has been vital to the frontier of in-depth biophysical interrogations of mammalian systems4,7,63,66, which are otherwise difficult to access given the substantially (~100–1000-fold) greater material requirements for ensemble interrogations. However, as for all biophysical methods in which experiments must be carried out in sequence, single-molecule measurements have been hampered by low experimental throughput. Comparative studies are thus time consuming and potentially prone to inherent experimental variabilities. Consequently, confidence in subtle functional distinctions is reduced and potentially important biological insights may be overlooked.
Spurred by this challenge, strategies have been developed to increase the experimental throughput of single-molecule imaging, including multiplexed investigations. Confocal-based single-molecule imaging methods have been introduced with plate reader automation strategies13. However, such methods are inherently sequential in practice, time consuming and limited to ~1 ms bursts. Camera-based methods have been introduced that enable imaging of thousands of distinct molecules over extended periods (multiple seconds) in a single experiment17,18. However, these approaches appear restricted to DNA samples that are stable over long observation periods (reported to be multiple days) and they are currently restricted to steady-state conditions. Such constraints are generally unsuitable for inherently unstable biological samples (that is, mammalian systems) and preclude pre-steady-state measurements of irreversible processes. This method also relies on complex a posteriori methods to identify the molecules that have been imaged, requiring next-generation sequencing and resulting in undesirable assignment lag times and uncertainties.
PRE-smF and PRE-smFRET uniquely enable high spatial and temporal resolution parallel single-molecule investigations of diverse biological specimens under both steady- and pre-steady-state conditions. PRE-smF(RET) leverages a pTIRF imaging platform and highly customizable microfluidic devices, in which microcontact printing, combined with ERASE technology-enabled DNA barcoding, is used to array biomolecules in a manner that provides a priori knowledge of the identity of each sample. This approach can be repeated within a single microfluidic device and requires multiple orders of magnitude less material than ensemble stopped-flow investigations, the current gold standard of kinetic measurements. Our findings demonstrate that PRE-smF(RET) increases experimental throughput, reduces experimental variance and offers an approach that can reveal even subtle kinetic and structural insights across a broad range of diverse biological systems.
In the current embodiment of PRE-smF(RET), we demonstrate simultaneous interrogation of four (eight with spectral multiplexing) distinct samples in parallel. Efforts to further increase parallelization and advance the scalability of this approach will be enabled by computational design of orthogonal TMSD networks, printing smaller features and imaging with larger camera sensors. For instance, by implementing contactless printing methods and cameras with larger chip areas, we can already achieve parallel imaging of nine samples within one FOV without spectral multiplexing (Extended Data Fig. 9 and Supplementary Note 2), thus increasing experimental throughput by an additional twofold.
Careful scaling of printed arrays using direct printing, a larger number of orthogonal ERASE systems and DNA-barcoded protein libraries67, together with automated microscopy and fluidics systems, could ultimately enable high-throughput screening for diverse applications. Alternative means for orthogonal tethering of distinct biomolecules are also foreseeable, such as the direct printing of antibody or nanobody libraries for direct pulldown of molecules of interest from complex mixtures68. Continued progress on this front will advance an era of single-molecule biophysics with enhanced experimental throughput and sensitivity that is capable of affording insights and knowledge vital to understanding biological function and regulation that were previously inaccessible.
Methods
Refer to Supplementary Tables 1 and 5 for the sequences of all DNA oligonucleotides used in this study. Refer to Supplementary Table 4 for a list of materials and equipment used in this study.
DNA labeling
DNA was labeled with fluorophores as described previously29. In brief, amino-modified DNA was labeled with a tenfold molar excess of NHS ester-activated fluorophore in a final volume of 10 µl in labeling buffer (50 mM potassium borate (pH 8.1), 200 mM KCl and 10% DMSO) for 1 h at room temperature. The labeling reaction was quenched with 0.2 µl 1 M Tris-acetate (pH 7.5), and then complementary strands for each dye distance were mixed in an equimolar ratio. Strands were annealed by heating to 90 °C for 2 min and then allowed to passively cool to room temperature. To this, 10 µl of 3 M sodium acetate (pH 6) was added for neutralization, followed by dilution to 100 µl with water. DNA was precipitated with ethanol (300 µl) at –20 °C overnight. Precipitated DNA was collected by centrifugation at 18,400g for 20 min. The pellet was resuspended in 400 µl of buffer A (1.7 M ammonium sulfate, 10 mM ammonium acetate, pH 5.8) and further purified on a phenyl-5PW column (FPLC, Äkta Purifier, GE Healthcare) by eluting over a gradient from buffer A to buffer B (10% methanol, 10 mM ammonium acetate, pH 5.8). The desired peak of interest was collected and used directly for smF(RET) investigations.
Conjugation of tether DNAs to antibody
Tether DNA sequences at equimolar concentrations were individually conjugated to a mouse high-affinity anti-Strep tag IgG1 monoclonal antibody (StrepMAB-Immo, IBA) using the Oligonucleotide Conjugation Kit (Abcam) according to the manufacturer’s instructions.
Preparation of β-arrestin1 reagents and samples
LD555p-MAL and LD655-MAL (Lumidyne Technologies) were reconstituted in DMSO. V2Rpp (synthesized by the peptide synthesis core facility at Tufts University) was resuspended in 20 mM HEPES (pH 7.0) with 100 mM NaCl at a final concentration of 2 mM. Glucose oxidase (Sigma) and catalase (Sigma) were each resuspended in T50 buffer (10 mM Tris-acetate, pH 7.5, 50 mM KCl) with 10 mM 2-mercaptoethanol. Catalase was further purified by size exclusion chromatography.
Sequences for site-specific cysteines were introduced in the previously reported parental pTrcHisB plasmid encoding the Strep-tagged, cysteine-less variants of bovine β-arrestin1 (ref. 69) (Supplementary Table 3) using QuickChange site-directed mutagenesis (Agilent). The β-arrestin variants, each carrying two cysteines, were overexpressed in BL21-Gold competent E. coli cells by induction with 35 μM isopropyl β-D-thiogalactopyranoside for 6 h at 30 °C. The cells were lysed by incubating with lysozyme (Sigma) followed by high-pressure (3.5–7 MPa) homogenization using the EmulsiFlex-C3 instrument (Avestin). β-arrestin variants were purified from the cell lysates by ammonium sulfate precipitation followed by affinity chromatography using Strep-Tactin Superflow high-capacity resin (IBA Lifesciences). The purified proteins were first labeled with an 0.8 M excess of LD555p-MAL (Lumidyne Technologies) followed by a 2 M excess of LD655-MAL (Lumidyne Technologies) for an additional 3 h. Free dye was separated from the labeled proteins using a Zeba 2 ml 7k MWCO Spin Desalting Column (Thermo Fisher)19.
Single-molecule prism-based TIRF imaging platform for rapid solution exchange during imaging
Single-molecule imaging experiments were performed using a custom-built pTIRF microscope, as described previously28. The imaging platform is a Nikon Ti2 microscope equipped with a superresolution water-immersion objective lens (Nikon CFI SR Plan Apo IR 60× 1.27 NA WI), continuous-wave excitation lasers (0.5 W 473 nm Laser Quantum Ciel, 6 W 532 nm Laser Quantum Opus, 1 W 640 nm Coherent Genesis and a 0.5 W 721 nm Laserglow) coupled to the observation chamber via a custom-fabricated quartz prism (Eksma) and synchronized scientific complementary metal oxide semiconductor cameras (Hamamatsu Fusion C14440-20UP).
To ensure uniform illumination intensity, all laser lines were passed through a flat-field beam-shaping element. A flat-top, rectangular illumination profile was achieved using a beam-expanding telescope, a shearing interferometer (Thorlabs, SI050, SIVS) and a Gaussian-to-top-hat beam-shaping lens (Eksma, GTH-3.6-1.75FA-VIS)70 (Extended Data Fig. 1). The high-aspect-ratio rectangular illumination profile was expanded into a square profile with a cylindrical lens telescope consisting of an achromatic doublet (Thorlabs, ACY254-075-A) and a plano-concave singlet lens (Thorlabs, LK1900L1-A). An achromatic doublet laser-focusing lens (Thorlabs, AC254-100-A) was used to minimize chromatic aberration, with the focal length chosen to fill the imaging area.
All smF imaging data were acquired with custom microscope control software written in LabVIEW (National Instruments) and C++. The microfluidic system, controlled by CETONI Elements software, was synchronized with the imaging system via a TTL trigger pulse. Unless otherwise stated, all smFRET data were processed with SPARTAN28 version 3.9.2 to extract and correct FRET traces. Traces were assigned to corresponding printed spots that were precisely delineated using the Canny edge algorithm71 and RANSAC72 fitting of circles to detected arc-shaped edges.
Computer-controlled microfluidic system
The core of the computer-controlled microfluidic system (Fig. 3a) is a motorized ten-port, two-position valve (CETONI) with two PEEK tubing injection loops. Each loop was loaded using a Hamilton syringe via an HPLC port (Rheodyne), and its content was injected into the observation chamber at a user-defined time point during data acquisition. To achieve rapid response times, we minimized both hydraulic resistance and hydraulic capacitance (the ability of the system to store and release fluid due to elastic expansion and compression) by using rigid PEEK tubing, a stainless steel syringe and the shortest feasible connections with relatively large inner diameters. This design balances the competing need to reduce dead volume, critical for conserving material, while maintaining low flow resistance. System performance was optimized through extensive empirical testing, and a pressure-monitoring module was included to detect clogs or leaks based on expected backpressure values. Fluid flow through either injection loop was controlled using a motorized syringe pump (CETONI) driving an 8 ml stainless steel syringe (Harvard Apparatus) filled with the working buffer. The working buffer was selected with a motorized six-port valve (Valco Instruments). The valves and the pump were controlled via CETONI Elements software.
To measure the effective solution exchange (mixing) time within the evanescent field, 250 nM LD555 dye was injected over a range of flow rates while imaging at a time resolution of 5 ms with 532 nm (125 W cm−2) laser illumination. The concentration rise time was quantified using the 10–90% criterion (Fig. 3b). Experiments with Liv-BPSS (Fig. 3d,e) were performed in 30 mM Tris (pH 7.0) with 150 mM NaCl and simultaneous illumination from 532 nm (150 W cm−2) and 721 nm (130 W cm−2) lasers at a time resolution of 10 ms. A mixture of 25 µM leucine and 250 nM Cy7 (to independently measure the effective solution exchange time) was injected at t = 400 ms and rinsed at t = 900 ms from the start of imaging. Data were acquired from 33 independent FOVs under automated computer control, with solution exchange times of 60 ms.
Preparation of microarrayed imaging chambers
PEGylation of quartz slides and assembly into microfluidic imaging chambers are summarized in Extended Data Fig. 2. Quartz slides (25 mm × 76 mm, 1 mm thick) were prepared by drilling ten holes along each long edge using a ball-tipped diamond-coated drilling bit mounted in a high-speed drill press (Extended Data Fig. 2a). A custom indexing slide-holding fixture with spring-loaded slide-securing pins was mounted on the drill press table to aid in precise drilling. The fixture maintains slide position against fixed stops and includes a water reservoir to enable submerged drilling, which reduces chipping and prevents dust adhesion. This fixture was fabricated by 3D printing (Formlabs Form 4 printer with Tough 2000 resin). The parametric CAD design, available on the Blanchard laboratory GitHub (https://github.com/stjude-smc/CAD-indexed-slide-drilling-fixture), enables adjustment of hole pitch and channel number and can be readily adapted for custom layouts. After drilling, slides were laser engraved with a unique identifier and coverslips were engraved to track the passivated sides throughout the process (Extended Data Fig. 2b).
Slides and coverslips were cleaned by sequential sonication: 10 min in water, 20 min in acetone, 20 min in 1 M KOH prepared from semiconductor-grade powder and 10 min in water (Extended Data Fig. 2c). After the final rinse, the slides were briefly dried. To remove residual organics, slides were flame treated for 30–60 s on each side using a propane torch and then cooled down (Extended Data Fig. 2d). Silanization was performed by swirling the slides and coverslips in a mixture of 12 ml VECTABOND in 700 ml dry acetone for 2 min (Extended Data Fig. 2e), followed by quenching in a 1:1 acetone–water mixture for 1 min (Extended Data Fig. 2f). Slides were rinsed twice with water and air-dried.
Surface passivation was performed using a 1:100 mixture of NHS-biotin-PEG and NHS-PEG (both 5 kDa), dissolved at a total concentration of 100 μg µl−1 in 100 mM potassium borate buffer, pH 8.1 (Extended Data Fig. 2f). Sixty-five µl of the PEG solution was applied on each slide, followed by placement of a coverslip to ensure uniform spreading (Extended Data Fig. 2h). Slides were incubated for 1 h at room temperature in a humidified chamber (Extended Data Fig. 2i) and then rinsed thoroughly under water and air-dried (Extended Data Fig. 2j). To cap unreacted amines, 25 mM sulfo-NHS-acetate in 100 mM potassium borate buffer, pH 8.1, was applied to the slides in the same manner as for PEGylation. After 1 h of incubation, slides were rinsed and air-dried.
Microarrays were printed on passivated quartz slides using a pin-based arrayer (Quanterix, 2470) using a 110 µm deposition pin, 60% humidity and an extended pin-rinse duration of 5 s to avoid cross-contamination. For each unique ERASE sequence, anchor–streptavidin complexes were prepared by mixing 1,3 trans-bivalent streptavidin73 and biotinylated ERASE anchor at an equimolar ratio in T50 buffer with 50% (vol/vol) glycerol. Each microarray consisted of 60 identical subarrays, with each subarray containing four unique ERASE anchor–streptavidin complexes, printed at 250 nM, arranged in a 2 × 2 grid with a distance of 160 µm between the closest spots (Fig. 1d). The subarrays were arranged in a 3 × 20 matrix along the channel, with a distance of 400 µm between subarrays.
To create microfluidic chambers on the passivated and microarrayed quartz slides, adhesive gaskets were laser cut from double-sided acrylic adhesive polyester film and applied to form ten independent flow channels, each with precisely positioned inlet and outlet ports. Slides constructed in this manner were stored with desiccant in the dark until use.
Single-molecule imaging of DNA samples
Experiments with fluorescently labeled DNA were performed in 10 mM Tris-HCl (pH 7.5), 200 mM KCl buffer. Samples were introduced at 1 nM and incubated until the desired surface density was reached. Unless otherwise mentioned, we used an enzymatic oxygen-scavenging system74,75 consisting of 1 mM 3,4-dihydroxybenzoic acid and 50 nM protocatechuate 3,4-deoxygenase.
Data in Fig. 2a were collected from LD555- and/or LD655-labeled DNA FRET duplexes (with LD555 at the 5′ end of the tether-barcoded strand and LD655 at 11, 14, 17 or 20 nucleotides away on the complementary strand). We used stroboscopic imaging with a 10% duty cycle to mitigate photobleaching. The FOV was intermittently illuminated for 100 ms by a 532 nm laser (65 W cm−2), an image was saved, and the process was repeated every 1,000 ms. Particles in each quadrant of the Cy5 spectral channel were counted using a custom Python script, which included flat-field correction, thresholding and local maximum detection in each frame of the movie.
FRET data shown in Fig. 2b were acquired using the same DNA FRET duplexes at an exposure time of 500 ms and continuous 532 nm illumination (140 W cm−2). Criteria for selecting traces were: correlation coefficient between donor and acceptor from –1.1 to 0.5, signal-to-noise ratio > 8, background noise < 70 cts and FRET lifetime > 15 frames.
Fluorophore performance (Fig. 2c and Supplementary Table 2) was characterized at a time resolution of 100 ms using 532 nm and 635 nm (both at 130 W cm−2) illumination in alternating-laser excitation mode to avoid indirect excitation of red fluorophores. smF traces were analyzed using SPARTAN28 version 3.9.2 to determine fluorescence intensity, signal-to-noise ratio (mean total fluorescence intensity divided by the s.d. of total intensity before photobleaching) and photon yield.
To quantify the variability of photophysical behavior within a single experiment compared to independent experiments, we performed repeated independent immobilization of LD655-labeled DNA. For each experimental repeat, we extracted traces from each of the four quadrants and calculated the mean and s.d. of photon yield between each quadrant in the same repeat. For comparison, we calculated the mean and s.d. of photon yield between each repeat. These data show that parallelization reduces the standard deviation of the total photon count measured by fourfold (Extended Data Fig. 4b).
Single-molecule FRET experiments with β-arrestin1
Fluorophore-labeled β-arrestin variants and tether-conjugated StrepMAB-Immo were mixed at an equimolar ratio to form antibody–arrestin complexes with distinct tether DNA sequences. After 5 min of incubation, complexes were diluted to 5 nM with imaging buffer (50 mM HEPES-NaOH, pH 7.5, 150 mM NaCl, 0.8% (wt/vol) glucose) containing a block mixture (2 μM 25-nucleotide DNA duplex (IDT) and 2 μM BSA) to reduce any nonspecific binding to the imaging surface. Complexes were sequentially injected into the microfluidic channel and incubated until sufficient confluence had been achieved. Data were collected with increasing concentrations of V2Rpp containing glucose oxidase (2.5 U ml−1) and catalase (1.75 U μl−1). For pre-steady-state experiments, V2Rpp at increasing concentrations was introduced at 5 s and rinsed with imaging buffer at 25 s. Data were acquired from 12–15 independent FOVs under automated computer control, with estimated buffer exchange times of ~100 ms. Between conditions, the molecules were removed from the surface with a mixture of switch oligonucleotides (1 µM each), and fresh arrestin sensors were immobilized.
A 532 nm laser was used for illumination, with a power density of 52 W cm−2 for steady-state experiments and 44 W cm−2 for pre-steady-state experiments using the computer-controlled fluidic system. All data were collected with an integration time of 100 ms. The selection criteria for FRET traces were: correlation coefficient between donor and acceptor from –1.1 to 0.5, signal-to-noise ratio > 20, signal-to-noise (FRET) ratio > 8 for all sensors in non-equilibrium experiments (>6 for C-domain rotation sensor in equilibrium experiments), background noise < 100 and FRET lifetime > 50 frames for equilibrium experiments (>300 for the non-equilibrium experiments).
For steady-state determination of the apparent dissociation constant, single-molecule traces were analyzed for V2Rpp concentration-dependent loss of basal-state occupancy with HMM and likelihood maximization for C-tail 1, C-tail 2 and finger loop sensors. For the C-domain rotation sensor, we quantified the affinity by measuring the change in mean FRET efficiency as a function of V2Rpp concentration, as the magnitude of FRET change (∼0.03) is below the sensitivity threshold of HMM. The apparent association (\({k}_{{\rm{on}}}^{{\rm{app}}}\)) rates of V2Rpp binding to the C-tail and finger loop sensors (the slope of the linear fit) were measured using the observed rates of transition from basal to active states up to 1 μM of V2Rpp before it starts plateauing. Apparent rates of V2Rpp dissociation (\({k}_{{\rm{off}}}^{{\rm{app}}}\)) were independent of V2Rpp concentration, and were determined by linear fitting of the observed rates of transition between 3 and 30 μM V2Rpp.
For pre-equilibrium experiments, the observed binding rate (kobs) at each concentration was estimated by calculating the waiting time for each molecule between injection of V2Rpp and the first transition to the active FRET state, ton (Fig. 4e and Supplementary Table 3). Cumulative distributions of waiting times were fit with a double-exponential function. Observed binding rates (Extended Data Fig. 5) represent the dominant component of the fit. The bimolecular association rate constant \({k}_{{\rm{on}}}^{{\rm{app}}}\) was determined by fitting of the concentration dependence of kobs. To estimate dissociation rates (\({k}_{{\rm{off}}}^{{\rm{app}}}\)), the waiting times between rinse-out of V2Rpp and adoption of stable basal FRET (Fig. 4e and Supplementary Table 3) were calculated. Dissociation rates were then estimated by fitting the cumulative distributions of waiting times with a single-exponential function.
Single-molecule FRET of bacterial ribosome complexes
Bacterial ICs programmed with Cy3-fMet-tRNAfMet in the P site were made as described previously51. Briefly, tight-coupled 70S ribosomes were purified from E. coli MRE600 by cryomilling (MM400, Retsch), resuspension in lysis buffer (20 mM Tris-HCl (pH 7.5), 100 mM NH4Cl, 10 mM MgCl2, 0.1 mM EDTA, 6 mM 2-mercaptoethanol), two rounds of ultracentrifugation at 126,000g for 22 h through a 37% sucrose cushion (20 mM Tris-HCl (pH 7.5), 500 mM NH4Cl, 10 mM MgCl2, 0.1 mM EDTA, 6 mM 2-mercaptoethanol) and isolation by ultracentrifugation at 87,000g for 19 h using an SW28 rotor (Beckman) through a 10–40% sucrose gradient made in lysis buffer. Individual 30S and 50S subunits were purified from 70S ribosomes by dialysis against bacterial polymix buffer (see below) containing 1 mM MgCl2, followed by 10–40% sucrose gradient ultracentrifugation in the same buffer. All steps were performed at 4 °C. Bacterial ICs were initiated on synthesized gene32-derived mRNA by first mixing 30 pmol of each of the 30S and 50S subunits with 100 pmol of each of the recombinantly expressed and purified initiation factors IF-1, IF-2 and IF-3 and 50 pmol of S1 protein, 1 mM GTP and 1 mM myokinase in bacterial polymix buffer (50 mM Tris-OAc (pH 7.5), 100 mM KCl, 5 mM NH4OAc, 5 mM Mg(OAc)2, 0.5 mM Ca(OAc)2, 5 mM putrescine, 1 mM spermidine, 0.1 mM EDTA, 6 mM 2-mercaptoethanol). Next, 100 pmol of in situ aminoacylated and formylated Cy3-fMet-tRNAfMet and 100 pmol mRNA were added to the reaction, the final volume was adjusted to 50 μl, and samples were incubated for 15 min at 37 °C. To isolate bacterial ICs, the reaction was loaded on top of a 10–40% sucrose gradient and then centrifuged at 112,000g for 14 h at 4 °C. All sucrose gradients were fractionated using a Piston Gradient Fractionator (Biocomp). IC-containing fractions were pooled, aliquoted and stored in liquid nitrogen until use. A unique mRNA was used to prepare each complex such that a specified codon was displayed in the A site, with an associated unique tether sequence in the 5′ UTR. All tRNA selection experiments were performed in bacterial polymix buffer supplemented with a cocktail of triplet-state quenchers (1 mM cyclooctatetraene, 1 mM nitrobenzyl alcohol and 1 mM Trolox) and an enzymatic oxygen-scavenging system (2 mM protocatechuic acid and 50 nM protocatechuate 3,4-dioxygenase)74. ICs were site specifically immobilized via hybridization of the mRNA tether with surface-immobilized anchor DNA. To suppress the appearance of hybrid states51, surface-tethered ICs were treated with puromycin-containing polymix (pH 8.5) to deacylated P-site tRNA. ICs were then equilibrated with polymix buffer at pH 7.5, containing either no streptomycin or 20 µM streptomycin. A preformed ternary complex consisting of 50 nM Phe-tRNAPhe-LD655, 500 nM EF-Tu–EF-Ts and 500 µM GTP was injected, and data were collected at either an integration time of 10 ms and an irradiance of 550 W cm−2 with at least ten repeats per condition (Fig. 5b–d and Extended Data Fig. 6b,d) or an integration time of 100 ms and an irradiance of 40 W cm−2 with three repeats per condition (Extended Data Fig. 7a,c).
Extraction of fluorescence intensities and calculation of FRET intensity traces were performed using SPARTAN 3.9.2 (ref. 28). The selection criteria for taking traces forward for further analysis were: correlation coefficient between donor and acceptor from –1.1 to 0.5, signal-to-noise ratio > 8, background noise < 70 cts, FRET at first frame < 0.1 and highest FRET value > 0.2.
Selected traces were idealized using segmental k-means as implemented in SPARTAN with a previously validated kinetic model51. Population histograms (Fig. 5b) were produced by identifying all FRET events by idealization to a simple two-state model and then separating and synchronizing each event to the appearance of FRET. The overall rate of aminoacyl-tRNA accommodation (Extended Data Fig. 7c) was determined from experiments performed at an integration time of 100 ms. Accommodation times, taccommodation (Extended Data Fig. 7a), were calculated from the wait time between TC injection and arrival to AC. Wait times were used to construct a cumulative distribution, which was then fit with a two-exponential function with a delay to account for buffer exchange times. Rates of initial binding (Fig. 5c), the rate of mRNA decoding (Extended Data Fig. 7d) and comparison of tRNA accommodation probabilities (Fig. 5d) were calculated from experiments performed at an integration time of 10 ms.
The rate of initial binding was estimated by identifying the first arrival to FRET for each molecule after injection of ternary complex, tbinding (Extended Data Fig. 7b). A cumulative distribution of observed waiting times was generated, and these distributions were fitted with a double-exponential function with a delay to account for buffer exchange time. The mRNA-decoding rate (that is, from CR to AC) was determined by first identifying all FRET events resulting in accommodation and then calculating the passage time from the first nonzero FRET state to AC for each event, tdecoding. A cumulative distribution was constructed from the passage times and fit with a double-exponential function. Rates of overall aminoacyl-tRNA accommodation (Extended Data Fig. 7c), initial binding (Fig. 5c) and mRNA decoding (Extended Data Fig. 7d) represent the dominant component from double-exponential fitting.
Comparison of tRNA accommodation probability, as shown in Fig. 5d, was performed by first calculating the number of transitions between states from HMM idealizations. State transition probabilities were compared between conditions (with and without streptomycin) using logistic regression, implemented in R version 4.1.2 using the glm function from the stats package.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data generated during this study are available on request from the corresponding author, S.C.B.
Code availability
TIRF data were acquired using in-house-developed software for microscope control. All smFRET data were processed in the SPARTAN software environment, which is available on GitHub (https://github.com/stjude-smc). The microfluidic system was controlled using CETONI Elements (CETONI). Custom scripts for specific analyses and visualizations, which rely on Python (dask, Jupyter, Matplotlib, napari, NumPy, pandas, plotnine, SciPy and uncertainties), R (dplyr, ggplot2, stats, tidyr and zoo), MATLAB and Origin, are available upon request to corresponding author, S.C.B.
References
Henzler-Wildman, K. & Kern, D. Dynamic personalities of proteins. Nature 450, 964–972 (2007).
Weiss, S. Fluorescence spectroscopy of single biomolecules. Science 283, 1676–1683 (1999).
Ha, T. et al. Fluorescence resonance energy transfer at the single-molecule level. Nat. Rev. Methods Primers 4, 21 (2024).
Holm, M. et al. mRNA decoding in human is kinetically and structurally distinct from bacteria. Nature 617, 200–207 (2023).
Jain, A. et al. Probing cellular protein complexes using single-molecule pull-down. Nature 473, 484–488 (2011).
Sungkaworn, T. et al. Single-molecule imaging reveals receptor–G protein interactions at cell surface hot spots. Nature 550, 543–547 (2017).
Levring, J. et al. CFTR function, pathology and pharmacology at single-molecule resolution. Nature 616, 606–614 (2023).
Zheng, Q. et al. Ultra-stable organic fluorophores for single-molecule research. Chem. Soc. Rev. 43, 1044–1056 (2014).
Minoshima, M., Reja, S. I., Hashimoto, R., Iijima, K. & Kikuchi, K. Hybrid small-molecule/protein fluorescent probes. Chem. Rev. 124, 6198–6270 (2024).
Juette, M. F. et al. The bright future of single-molecule fluorescence imaging. Curr. Opin. Chem. Biol. 20, 103–111 (2014).
Chen, K. Y. et al. Fabrication of zero mode waveguides for high concentration single molecule microscopy. J. Vis. Exp. https://doi.org/10.3791/61154 (2020).
Chen, J. et al. High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proc. Natl Acad. Sci. USA 111, 664–669 (2014).
Hartmann, A. et al. An automated single-molecule FRET platform for high-content, multiwell plate screening of biomolecular conformations and dynamics. Nat. Commun. 14, 6511 (2023).
Shema, E. et al. Single-molecule decoding of combinatorially modified nucleosomes. Science 352, 717–721 (2016).
Fedyuk, V. et al. Multiplexed, single-molecule, epigenetic analysis of plasma-isolated nucleosomes for cancer diagnostics. Nat. Biotechnol. 41, 212–221 (2023).
Makasheva, K. et al. Multiplexed single-molecule experiments reveal nucleosome invasion dynamics of the Cas9 genome editor. J. Am. Chem. Soc. 143, 16313–16319 (2021).
Aguirre Rivera, J. et al. Massively parallel analysis of single-molecule dynamics on next-generation sequencing chips. Science 385, 892–898 (2024).
Severins, I. et al. Single-molecule structural and kinetic studies across sequence space. Science 385, 898–904 (2024).
Asher, W. B. et al. GPCR-mediated β-arrestin activation deconvoluted with single-molecule precision. Cell 185, 1661–1675 (2022).
Wasserman, M. R., Alejo, J. L., Altman, R. B. & Blanchard, S. C. Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat. Struct. Mol. Biol. 23, 333–341 (2016).
Yim, S. W. et al. Four-color alternating-laser excitation single-molecule fluorescence spectroscopy for next-generation biodetection assays. Clin. Chem. 58, 707–716 (2012).
Lee, N. K. et al. Three-color alternating-laser excitation of single molecules: monitoring multiple interactions and distances. Biophys. J. 92, 303–312 (2007).
Kaur, A., Sapkota, K. & Dhakal, S. Multiplexed nucleic acid sensing with single-molecule FRET. ACS Sens. 4, 623–633 (2019).
Khanna, K. et al. Rapid kinetic fingerprinting of single nucleic acid molecules by a FRET-based dynamic nanosensor. Biosens. Bioelectron. 190, 113433 (2021).
Zaza, C. et al. Super-resolved FRET imaging by confocal fluorescence-lifetime single-molecule localization microscopy. Small Methods 7, e2201565 (2023).
Butler, C. et al. Multi-dimensional spectral single molecule localization microscopy. Front. Bioinform. 2, 813494 (2022).
Sabantsev, A. et al. Spatiotemporally controlled generation of NTPs for single-molecule studies. Nat. Chem. Biol. 18, 1144–1151 (2022).
Juette, M. F. et al. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat. Methods 13, 341–344 (2016).
Pati, A. K. et al. Tuning the Baird aromatic triplet-state energy of cyclooctatetraene to maximize the self-healing mechanism in organic fluorophores. Proc. Natl Acad. Sci. USA 117, 24305–24315 (2020).
Broadwater, D. W. B., Altman, R. B., Blanchard, S. C. & Kim, H. D. ERASE: a novel surface reconditioning strategy for single-molecule experiments. Nucleic Acids Res. 47, e14 (2019).
Kurylo, C. M. et al. Endogenous rRNA sequence variation can regulate stress response gene expression and phenotype. Cell Rep. 25, 236–248 (2018).
Joo, C. & Ha, T. Preparing sample chambers for single-molecule FRET. Cold Spring Harb. Protoc. 2012, 1104–1108 (2012).
Yurke, B., Turberfield, A. J., Mills, A. P., Simmel, F. C. & Neumann, J. L. A DNA-fuelled molecular machine made of DNA. Nature 406, 605–608 (2000).
Ha, T. & Tinnefeld, P. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu. Rev. Phys. Chem. 63, 595–617 (2012).
Aitken, C. E., Marshall, R. A. & Puglisi, J. D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 94, 1826–1835 (2008).
Kapanidis, A. N. et al. Alternating-laser excitation of single molecules. Acc. Chem. Res. 38, 523–533 (2005).
Fitzgerald, G. A. et al. Quantifying secondary transport at single-molecule resolution. Nature 575, 528–534 (2019).
Lohse, M. J., Benovic, J. L., Codina, J., Caron, M. G. & Lefkowitz, R. J. β-arrestin: a protein that regulates β-adrenergic receptor function. Science 248, 1547–1550 (1990).
DeWire, S. M., Ahn, S., Lefkowitz, R. J. & Shenoy, S. K. β-arrestins and cell signaling. Annu. Rev. Physiol. 69, 483–510 (2007).
Latorraca, N. R. et al. Molecular mechanism of GPCR-mediated arrestin activation. Nature 557, 452–456 (2018).
Shukla, A. K. et al. Structure of active β-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature 497, 137–141 (2013).
Zhai, R. et al. Distinct activation mechanisms of β-arrestin-1 revealed by 19F NMR spectroscopy. Nat. Commun. 14, 7865 (2023).
Milano, S. K., Pace, H. C., Kim, Y.-M., Brenner, C. & Benovic, J. L. Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry 41, 3321–3328 (2002).
Han, M., Gurevich, V. V., Vishnivetskiy, S. A., Sigler, P. B. & Schubert, C. Crystal structure of β-arrestin at 1.9 Å: possible mechanism of receptor binding and membrane translocation. Structure 9, 869–880 (2001).
Nussinov, R. et al. Autoinhibition in Ras effectors Raf, PI3Kα, and RASSF5: a comprehensive review underscoring the challenges in pharmacological intervention. Biophys. Rev. 10, 1263–1282 (2018).
Sente, A. et al. Molecular mechanism of modulating arrestin conformation by GPCR phosphorylation. Nat. Struct. Mol. Biol. 25, 538–545 (2018).
Nobles, K. N. et al. Distinct phosphorylation sites on the β2-adrenergic receptor establish a barcode that encodes differential functions of β-arrestin. Sci. Signal. 4, ra51 (2011).
Marti-Solano, M. et al. Combinatorial expression of GPCR isoforms affects signalling and drug responses. Nature 587, 650–656 (2020).
Marti-Solano, M. A multi-dimensional view of context-dependent G protein-coupled receptor function. Biochem. Soc. Trans. 51, 13–20 (2023).
Ramakrishnan, V. Ribosome structure and the mechanism of translation. Cell 108, 557–572 (2002).
Geggier, P. et al. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J. Mol. Biol. 399, 576–595 (2010).
Morse, J. C. et al. Elongation factor-Tu can repetitively engage aminoacyl-tRNA within the ribosome during the proofreading stage of tRNA selection. Proc. Natl Acad. Sci. USA 117, 3610–3620 (2020).
Valle, M. et al. Locking and unlocking of ribosomal motions. Cell 114, 123–134 (2003).
Gromadski, K. B. & Rodnina, M. V. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell 13, 191–200 (2004).
Hopfield, J. J. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl Acad. Sci. USA 71, 4135–4139 (1974).
Girodat, D., Wieden, H.-J., Blanchard, S. C. & Sanbonmatsu, K. Y. Geometric alignment of aminoacyl-tRNA relative to catalytic centers of the ribosome underpins accurate mRNA decoding. Nat. Commun. 14, 5582 (2023).
Cruz-Navarrete, F. A. et al. β-amino acids reduce ternary complex stability and alter the translation elongation mechanism. ACS Cent. Sci. 10, 1262–1275 (2024).
Zhang, J., Ieong, K.-W., Johansson, M. & Ehrenberg, M. Accuracy of initial codon selection by aminoacyl-tRNAs on the mRNA-programmed bacterial ribosome. Proc. Natl Acad. Sci. USA 112, 9602–9607 (2015).
Demirci, H. et al. A structural basis for streptomycin-induced misreading of the genetic code. Nat. Commun. 4, 1355 (2013).
Gromadski, K. B. & Rodnina, M. V. Streptomycin interferes with conformational coupling between codon recognition and GTPase activation on the ribosome. Nat. Struct. Mol. Biol. 11, 316–322 (2004).
Ogle, J. M. & Ramakrishnan, V. Structural insights into translational fidelity. Annu. Rev. Biochem. 74, 129–177 (2005).
Aitken, C. E., Petrov, A. & Puglisi, J. D. Single ribosome dynamics and the mechanism of translation. Annu. Rev. Biophys. 39, 491–513 (2010).
McMahon, M. et al. A single H/ACA small nucleolar RNA mediates tumor suppression downstream of oncogenic RAS. eLife 8, e48847 (2019).
Condon, C., Philips, J., Fu, Z. Y., Squires, C. & Squires, C. L. Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. EMBO J. 11, 4175–4185 (1992).
Wilson, D. N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 12, 35–48 (2014).
Gregorio, G. G. et al. Single-molecule analysis of ligand efficacy in β2AR–G-protein activation. Nature 547, 68–73 (2017).
Credle, J. J. et al. Unbiased discovery of autoantibodies associated with severe COVID-19 via genome-scale self-assembled DNA-barcoded protein libraries. Nat. Biomed. Eng. 6, 992–1003 (2022).
Liu, B. & Yang, D. Easily established and multifunctional synthetic nanobody libraries as research tools. Int. J. Mol. Sci. 23, 1482 (2022).
Hanson, S. M. et al. Arrestin mobilizes signaling proteins to the cytoskeleton and redirects their activity. J. Mol. Biol. 368, 375–387 (2007).
Rowlands, C. J., Ströhl, F., Ramirez, P. P. V., Scherer, K. M. & Kaminski, C. F. Flat-field super-resolution localization microscopy with a low-cost refractive beam-shaping element. Sci. Rep. 8, 5630 (2018).
Canny, J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 8, 679–698 (1986).
Fischler, M. A. & Bolles, R. C. Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography. Commun. ACM 24, 381–395 (1981).
Fairhead, M., Krndija, D., Lowe, E. D. & Howarth, M. Plug-and-play pairing via defined divalent streptavidins. J. Mol. Biol. 426, 199–214 (2014).
Senavirathne, G., Lopez, M. A., Messer, R., Fishel, R. & Yoder, K. E. Expression and purification of nuclease-free protocatechuate 3,4-dioxygenase for prolonged single-molecule fluorescence imaging. Anal. Biochem. 556, 78–84 (2018).
Blanchard, S. C., Kim, H. D., Gonzalez, R. L., Puglisi, J. D. & Chu, S. tRNA dynamics on the ribosome during translation. Proc. Natl Acad. Sci. USA 101, 12893–12898 (2004).
Acknowledgements
This work was supported by the US National Institutes of Health (grants 5R01GM079238-18 and 5RM1HG011563-04 to S.C.B. and R01MH054137 to J.A.J.). This work was also supported by St. Jude Children’s Research Hospital Collaborative Research Consortium on GPCRs. We thank A. Ansari and C. Adura (Chemical Biology and Therapeutics, St. Jude Children’s Research Hospital) for their support of the Quanterix microarrayer employed in our investigations. We are grateful to M.M. Babu for insightful comments on the manuscript. We thank A. Pati for providing fluorescently labeled DNA samples, M. Holm for early contributions related to mRNA-decoding assays, K. Chattrakun for her site-specific labeling of LIV-BPSS and M. Martin for his input regarding quantification of fluorophore performance. We also thank the Single-molecule Imaging Center and Biomedical Engineering at St. Jude.
Author information
Authors and Affiliations
Contributions
S.C.B. conceived the project, designed the experiments and, together with R.B.A. and D.S.T., supervised the project. R.K., R.A.B., A.M., F.A.C.-N., D.S.T., R.B.A. and S.C.B. wrote the manuscript. All authors contributed to editing the manuscript. R.K. improved the existing microfluidic chip preparation protocol and designed chips with ten chambers and microarrays and conceived, designed, implemented and characterized the rapid, computer-controlled microfluidic system. R.A.B. optimized surface passivation to reduce nonspecific binding. D.S.T. designed and implemented the flat-field illumination optical system for prism-type TIRF microscopy. R.K. and R.A.B. prepared FRET- and single-labeled DNA oligonucleotides, performed experiments and analyzed the data. A.M. conceived and prepared adaptor molecules. W.B.A., A.M. and J.A.J. designed and prepared β-arrestin samples. A.M. performed β-arrestin experiments and analyzed data with assistance from R.K. and R.A.B. R.A.B. and F.A.C.-N. prepared ribosome samples, performed ribosome experiments and analyzed data. J.L.A. prepared ribosome complexes bearing rRNA sequence variation.
Corresponding author
Ethics declarations
Competing interests
S.C.B. and R.B.A. have equity interests in Lumidyne Technologies. The other authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Jong-Bong Lee, Shixin Liu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Allison Doerr and Rita Strack, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Imaging platform for p-TIRF smFRET.
(a) Schematic layout of the single-molecule imaging setup with prism-based flat-field total internal reflection (TIRF) excitation. BD, beam dump; BP, emission bandpass filter; CLT, cylindrical lens beam expanding telescope; CU, laser clean up filter; D, dichroic mirror; FL, achromatic doublet TIRF focusing lens; GTH, Gaussian to top hat refractive beam shaping lens; L, singlet lens; λ/2, half wave plate; λ/4, quarter wave plate; PBS, polarizing beam splitter; M, mirror. (b) TIRF illumination profiles (640 nm), obtained using a 20x air objective imaging a slide filled with LD655 fluorophores free in solution: Gaussian illumination (left), flat top illumination without (middle), and with (right) cylindrical lens telescope. Dashed square overlay shows the 60x magnification microscope field of view used for single molecule imaging. (c) Distribution of fluorescent molecules by their average photon count per 100 ms exposure time demonstrating more homogeneous illumination compared to Gaussian illumination profile.
Extended Data Fig. 2 Preparation of passivated imaging chambers.
Quartz imaging chambers were prepared as described in detail in Methods. First, quartz slides were drilled (a) then slides and coverslips were laser engraved (b), followed by cleaning and degreasing (c,d). Slides and coverslips were then aminosilanized (e,f) and finally PEGylated with a 1:100 mixture of biotinylated PEG and methoxy PEG (h-j).
Extended Data Fig. 3 Microarrayed microfluidic devices for PRE-smFRET display high binding specificity for a range of biological samples.
(a) Sequential addition of bacterial initiation complexes containing unique tether domains demonstrates high localization accuracy. Quantification of binding specificity for: (b) FRET labelled double-stranded DNA, specificity of 98 ± 2% (c) FRET labelled β-arrestin1, specificity of 96 ± 2% (d) and bacterial initiation complexes containing Cy3-labelled fMet- tRNAfMet, specificity of 95 ± 2%. Samples prepared as described in Methods. Created with BioRender.com.
Extended Data Fig. 4 Structures of all fluorophores tested using PRE-smFRET and comparison of variability between experimental repeats of photophysical characterization.
(a) Structures of characterized dyes, as shown in Fig. 2c, and Supplementary Table 2. Each dye is linked to a specific tether oligonucleotide, see Supplementary Table 1. (b) Comparison of variance in photon yield of LD655-labelled DNA (see a, Methods) between parallel and independent experiments. Four independent repeats were collected across two days. To estimate variance within a PRE-smF experiment, photon yield was determined from molecules extracted within each quadrant of one experimental repeat ( > 2000 molecules per quadrant). Data points represent mean and SD of photon yield across quadrants. For comparison, we calculated the mean and S.D. of photon yield across the same 4 independent experimental repeats (Combined; > 8000 molecules per repeat). Standard deviation of measured photon yield is 4-fold lower when measured in parallel (average SD across four PRE- smF repeats) compared to independent experiments.
Extended Data Fig. 5 Affinity of β-arrestin1 sensors towards V2Rpp measured using PRE-smFRET.
Apparent dissociation constant (\({\rm{K}}_{\rm{D}}^{\rm{app}}\)) estimated from steady-state V2Rpp titrations against β-arrestin1 sensors. (a) Comparison of \({\rm{K}}_{\rm{D}}^{\rm{app}}\) for sensors used for steady-state and pre-steady-state studies. (b) Effect of labelling-site mimicking mutations in C-tail 1 and C-domain rotation sensors. For both (a) and (b), plots show mean and SD of \({\rm{K}}_{\rm{D}}^{\rm{app}}\) values determined from 3 (C-tail 2, Finger loop, C-tail 1 D69A, C-tail1 L191A, C-domain rotation D69A, C-domain rotation K397A) or 9 (C-tail 1, C-domain rotation) independent repeats. P values are from independent two-tail t-tests.
Extended Data Fig. 6 Determination of bimolecular association rate of V2Rpp peptide and four distinct β-arrestin sensors from pre- steady state PRE- smFRET measurements.
(a) Observed binding rates, kobs, were determined at concentrations from 2 to 10 µM of V2Rpp from the observed rate of entry into the low FRET (active) state following peptide addition by rapid computer-controlled injection. Dissociation rates, \({k}_{\rm{off}}^{\rm{app}}\), were estimated from rate of return to stable high FRET (basal) state following wash out of V2Rpp. Bimolecular association rate constant, \({k}_{\rm{on}}^{\rm{app}}\), was determined by fitting of the concentration dependance of kobs. Error bars in observed binding rates denote SEM determined from 1000 bootstrap resamples. (b) Measured bimolecular association rates from pre-steady-state PRE-smFRET measurements of V2Rpp induced β-arrestin1 conformational change, determined by linear fitting of (a). Error bars denote associated standard error of slope from linear fitting. P values are from F-test.
Extended Data Fig. 7 Kinetic analysis of tRNA selection.
(a) Representative smFRET trace of tRNA selection measured at 100 ms time-resolution. Apparent rates of aminoacyl-tRNA accommodation (see (c)) were calculated from waiting time from ternary-complex injection to accommodation, taccommodation. (b) Representative smFRET trace of tRNA selection measured at 10 ms time-resolution. Apparent rates of initial binding (see Fig. 5c) were calculated from waiting time from ternary-complex injection to the first detected non-zero FRET event, tbinding. Rates of mRNA decoding (combined rate of decoding steps following ternary complex binding, see (d)) were calculated from the passage time from the first non-zero FRET event to formation of stable accommodated state, tdecoding, for each event which resulted in aminoacyl-tRNA accommodation. (c) Apparent accommodation rate in the presence and absence of streptomycin, determined from smFRET data collected at 100 ms. Waiting times from TC injection to accommodation (taccommodation, see (a)) were used to construct a cumulative distribution, which was fit with a two-exponential function with a delay to account for buffer exchange time. In the presence of streptomycin, the apparent rate of accommodation increases for near-cognate codons (10-fold UCC, 25-fold CUC) and decreases for cognate codons (2.5-fold both UUU, UUC). Error bars denote SEM from 3 experimental replicates. P values are from unpaired two-tailed t-test. (d) Rate of mRNA decoding, determined from smFRET data collected at 10 ms time-resolution. Passage time from CR to AC (tdecoding, see (b)) were used to construct a cumulative distribution, which fit to a double-exponential function to estimate mRNA decoding rate. In the presence of streptomycin, mRNA decoding rate is increased by <2-fold for cognate codons (1.9-fold UUU, 1.6-fold UUC), while increasing dramatically for near-cognate codons (570-fold CUC, 950-fold UUC). Error bars represent SEM from nine (-streptomycin) or fourteen ( + streptomycin) experimental repeats. P values are from unpaired two-tailed t-test.
Extended Data Fig. 8 PRE-smFRET reveals differential impact of tetracycline class antibiotics on ribosomes bearing endogenously encoded rRNA sequence variation.
FRET contour plots of tRNA selection of rrnB and rrnHBB bacterial ribosome complexes isolated from ∆7prrn-BBB and ∆7prrn-HBB respectively in (a) the absence of drug, (b) presence of 1 µM tetracycline, and (c) the presence of 100 nM oxytetracycline.
Extended Data Fig. 9 Increasing scaling of PRE-smFRET to 9 independent samples within one FOV.
(a) Bright field image of arrays printed with contactless array printer. Each microdroplet group contains 9 printed spots, each containing an anchor oligonucleotide with an orthogonal binding domain. (b) Schematic of one microdroplet group as imaged in one FOV using a microscope with a 60x objective lens, projected onto Teledyne Kinetix cameras. (c) Single- molecule fluorescence image of 9 FRET-labelled DNA duplexes, each with a distinct single-stranded tether domain and associated FRET signal, obtained with our pTIRF imaging platform. Image false colored by FRET efficiency. (d) FRET efficiency histogram of pTIRF images as in (c), colored by location as shown in (b). Solid lines show individual Gaussian fits to histograms extracted from each printed spot. (e) FRET efficiency, from the center of Gaussian distributions in (d), against dye distance, estimated from molecular dynamics simulations. Red solid line shows fit to Extended Data Equation 1 resulting in measured Förster distance, R0, of 59.8 ± 1 Å, in agreement with the reported R0 of 60.7 Å for LD555-LD655 on DNA.
Supplementary information
Supplementary Information
Supplementary Notes 1 and 2 and Tables 1–5
Supplementary Video 1
Video demonstrating control over biomolecule binding density to a PRE-smFRET device, as quantified in Fig. 2a.
Supplementary Video 2
Using PRE-smFRET to measure the photophysical performance of 4 green and 4 red fluorophores in parallel under identically deoxygenated conditions.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kiselev, R., Brady, R.A., Modak, A. et al. Parallel stopped-flow interrogation of diverse biological systems at the single-molecule scale. Nat Methods 23, 78–87 (2026). https://doi.org/10.1038/s41592-025-02944-4
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-025-02944-4