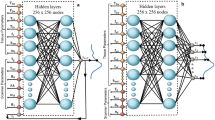
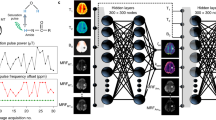
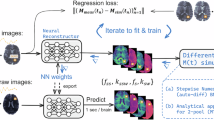
Abstract
Deep learning-based saturation transfer magnetic resonance fingerprinting (MRF) is an emerging approach for noninvasive in vivo imaging of proteins, metabolites and pH. It involves a series of steps, including sample/participant preparation, image acquisition schedule design, biophysical model formulation and artificial intelligence and computational model training, followed by image acquisition, deep reconstruction and analysis. Saturation transfer-based molecular MRI has been slow to reach clinical maturity and adoption for clinical practice due to its technical complexity, semi-quantitative contrast-weighted nature and long scan times needed for the extraction of quantitative molecular biomarkers. Deep MRF provides solutions to these challenges by providing a quantitative and rapid framework for extracting biologically and clinically meaningful molecular information. Here we define a complete protocol for quantitative molecular MRI using deep MRF. We describe in vitro sample preparation and animal and human scan considerations, and provide intuition behind the acquisition protocol design and optimization of chemical exchange saturation transfer (CEST) and semi-solid magnetization transfer (MT) quantitative imaging. We then extensively describe the building blocks for several artificial intelligence models and demonstrate their performance for different applications, including cancer monitoring, brain myelin imaging and pH quantification. Finally, we provide guidelines to further modify and expand the pipeline for imaging a variety of other pathologies (such as neurodegeneration, stroke and cardiac disease), accompanied by the related open-source code and sample data. The procedure takes between 48 min (for two proton pools or in vitro imaging) and 57 h (for complex multi-proton pool in vivo imaging) to complete and is suitable for graduate student-level users.
Key points
-
The procedure includes in vitro sample preparation, animal and human scan considerations, acquisition protocol design, and optimization of chemical exchange saturation transfer and semi-solid magnetization transfer quantitative imaging. We include artificial intelligence models for diagnostic applications.
-
Deep magnetic resonance fingerprinting does not require steady-state imaging conditions, enabling a reduction in scan time when compared with QUESP/QUEST, QUESTRA, Omega Plot, BM fitting, multi-pool Lorentzian fitting or chemical exchange saturation transfer-weighted imaging.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
All the data used in this work are available at https://github.com/momentum-laboratory/deep-molecular-mrf and https://doi.org/10.5281/zenodo.14211516. They include raw MRF data, quantitative parameter maps (Figs. 6–9), a CAD file for 3D printing a six-vial (phantom) holder and pulse sequence files (Table 2). A complete preclinical CEST–MRF pulse sequence for Bruker scanners is available at https://osf.io/52bsg (Paravision 6) and https://github.com/dkorenchan/cest-mrf-image-recon/tree/main/Bruker_PulseSequenceFiles/PV360_3_5 (Paravision 360). The .seq format files used in this work were also deposited at the pulseq CEST open library at https://github.com/kherz/pulseq-cest-library/tree/master/seq-library.
Code availability
All code is available on https://github.com/momentum-laboratory/deep-molecular-mrf and https://doi.org/10.5281/zenodo.14211516 in the format of Python scripts and Jupyter notebooks.
References
Ward, K., Aletras, A. & Balaban, R. S. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST). J. Magn. Reson. 143, 79–87 (2000).
Zaiss, M. & Bachert, P. Chemical exchange saturation transfer (CEST) and MR Z-spectroscopy in vivo: a review of theoretical approaches and methods. Phys. Med. Biol. 58, R221 (2013).
Van Zijl, P. C. & Yadav, N. N. Chemical exchange saturation transfer (CEST): what is in a name and what isn’t? Magn. Reson. Med. 65, 927–948 (2011).
Zhou, J., Payen, J.-F., Wilson, D. A., Traystman, R. J. & Van Zijl, P. C. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat. Med. 9, 1085–1090 (2003).
Wang, E. et al. Mapping tissue pH in an experimental model of acute stroke—determination of graded regional tissue pH changes with non-invasive quantitative amide proton transfer MRI. Neuroimage 191, 610–617 (2019).
Wang, E. et al. pH imaging reveals worsened tissue acidification in diffusion kurtosis lesion than the kurtosis/diffusion lesion mismatch in an animal model of acute stroke. J. Cereb. Blood Flow Metab. 37, 3325–3333 (2017).
Heo, H.-Y. et al. Improving the detection sensitivity of pH-weighted amide proton transfer MRI in acute stroke patients using extrapolated semisolid magnetization transfer reference signals. Magn. Reson. Med. 78, 871–880 (2017).
Heo, H.-Y., Tee, Y. K., Harston, G., Leigh, R. & Chappell, M. A. Amide proton transfer imaging in stroke. NMR Biomed. 36, e4734 (2023).
Ma, B. et al. Applying amide proton transfer-weighted MRI to distinguish pseudoprogression from true progression in malignant gliomas. J. Magn. Reson. Imaging 44, 456–462 (2016).
Mehrabian, H., Desmond, K. L., Soliman, H., Sahgal, A. & Stanisz, G. J. Differentiation between radiation necrosis and tumor progression using chemical exchange saturation transfer. Clin. Cancer Res. 23, 3667–3675 (2017).
Zhou, J., Heo, H.-Y., Knutsson, L., van Zijl, P. C. & Jiang, S. APT-weighted MRI: techniques, current neuro applications, and challenging issues. J. Magn. Reson. Imaging 50, 347–364 (2019).
Cai, K. et al. Magnetic resonance imaging of glutamate. Nat. Med. 18, 302–306 (2012).
Crescenzi, R. et al. In vivo measurement of glutamate loss is associated with synapse loss in a mouse model of tauopathy. Neuroimage 101, 185–192 (2014).
Haris, M. et al. Imaging of glutamate neurotransmitter alterations in Alzheimer’s disease. NMR Biomed. 26, 386–391 (2013).
Wang, K. et al. Lateralization of temporal lobe epileptic foci with automated chemical exchange saturation transfer measurements at 3 Tesla. eBioMedicine 89, 104460 (2023).
Neal, A. et al. Glutamate-weighted imaging contrast in gliomas with 7 Tesla magnetic resonance imaging. Neuroimage Clin. 22, 101694 (2019).
Haris, M. et al. A technique for in vivo mapping of myocardial creatine kinase metabolism. Nat. Med. 20, 209–214 (2014).
AlGhuraibawi, W. et al. CEST MRI reveals a correlation between visceral fat mass and reduced myocardial creatine in obese individuals despite preserved ventricular structure and function. NMR Biomed. 32, e4104 (2019).
Gilad, A. A. et al. Artificial reporter gene providing MRI contrast based on proton exchange. Nat. Biotechnol. 25, 217–219 (2007).
Gilad, A. A., Bar-Shir, A., Bricco, A. R., Mohanta, Z. & McMahon, M. T. Protein and peptide engineering for chemical exchange saturation transfer imaging in the age of synthetic biology. NMR Biomed. 36, e4712 (2023).
Perlman, O. et al. Redesigned reporter gene for improved proton exchange-based molecular MRI contrast. Sci. Rep. 10, 20664 (2020).
Farrar, C. T. et al. Establishing the lysine-rich protein CEST reporter gene as a CEST MR imaging detector for oncolytic virotherapy. Radiology 275, 746–754 (2015).
Meier, S. et al. Non-invasive detection of adeno-associated viral gene transfer using a genetically encoded CEST–MRI reporter gene in the murine heart. Sci. Rep. 8, 4638 (2018).
Wike-Hooley, J., Haveman, J. & Reinhold, H. The relevance of tumour pH to the treatment of malignant disease. Radiother. Oncol. 2, 343–366 (1984).
Webb, B. A., Chimenti, M., Jacobson, M. P. & Barber, D. L. Dysregulated pH: a perfect storm for cancer progression. Nat. Rev. Cancer 11, 671–677 (2011).
Corbet, C. & Feron, O. Tumour acidosis: from the passenger to the driver’s seat. Nat. Rev. Cancer 17, 577–593 (2017).
Lim, H., Albatany, M., Martı́nez-Santiesteban, F., Bartha, R. & Scholl, T. J. Longitudinal measurements of intra-and extracellular pH gradient in a rat model of glioma. Tomography 4, 46–54 (2018).
Walsh, J. J. et al. Imaging hallmarks of the tumor microenvironment in glioblastoma progression. Front. Oncol. 11, 692650 (2021).
Nilsson, C., Johansson, U., Johansson, A.-C., Kaagedal, K. & Öllinger, K. Cytosolic acidification and lysosomal alkalinization during TNF-α induced apoptosis in U937 cells. Apoptosis 11, 1149–1159 (2006).
Perlman, O. et al. Quantitative imaging of apoptosis following oncolytic virotherapy by magnetic resonance fingerprinting aided by deep learning. Nat. Biomed. Eng. 6, 648–657 (2022).
Ma, D. et al. Magnetic resonance fingerprinting. Nature 495, 187–192 (2013).
Cohen, O., Huang, S., McMahon, M. T., Rosen, M. S. & Farrar, C. T. Rapid and quantitative chemical exchange saturation transfer (CEST) imaging with magnetic resonance fingerprinting (MRF). Magn. Reson. Med. 80, 2449–2463 (2018).
Zhou, Z. et al. Chemical exchange saturation transfer fingerprinting for exchange rate quantification. Magn. Reson. Med. 80, 1352–1363 (2018).
Heo, H.-Y. et al. Quantifying amide proton exchange rate and concentration in chemical exchange saturation transfer imaging of the human brain. Neuroimage 189, 202–213 (2019).
Bipin Mehta, B. et al. Magnetic resonance fingerprinting: a technical review. Magn. Reson. Med. 81, 25–46 (2019).
McGivney, D. F. et al. SVD compression for magnetic resonance fingerprinting in the time domain. IEEE Trans. Med. Imaging 33, 2311–2322 (2014).
Cauley, S. F. et al. Fast group matching for MR fingerprinting reconstruction. Magn. Reson. Med. 74, 523–528 (2015).
Yang, M. et al. Low rank approximation methods for MR fingerprinting with large scale dictionaries. Magn. Reson. Med. 79, 2392–2400 (2018).
Cohen, O., Zhu, B. & Rosen, M. S. MR fingerprinting deep reconstruction network (DRONE). Magn. Reson. Med. 80, 885–894 (2018).
Kim, B., Schär, M., Park, H. & Heo, H.-Y. A deep learning approach for magnetization transfer contrast MR fingerprinting and chemical exchange saturation transfer imaging. Neuroimage 221, 117165 (2020).
Kang, B., Kim, B., Schär, M., Park, H. & Heo, H.-Y. Unsupervised learning for magnetization transfer contrast MR fingerprinting: application to CEST and nuclear Overhauser enhancement imaging. Magn. Reson. Med. 85, 2040–2054 (2021).
Cohen, O. et al. CEST MR fingerprinting (CEST–MRF) for brain tumor quantification using EPI readout and deep learning reconstruction. Magn. Reson. Med. 89, 233–249 (2023).
Weigand-Whittier, J. et al. Accelerated and quantitative three-dimensional molecular MRI using a generative adversarial network. Magn. Reson. Med. 89, 1901–1914 (2023).
Singh, M. et al. Bloch simulator–driven deep recurrent neural network for magnetization transfer contrast MR fingerprinting and CEST imaging. Magn. Reson. Med. 90, 1518–1536 (2023).
Kang, B., Singh, M., Park, H. & Heo, H.-Y. Only-train-once MR fingerprinting for B0 and B1 inhomogeneity correction in quantitative magnetization-transfer contrast. Magn. Reson. Med. 90, 90–102 (2023).
Perlman, O. & Azhari, H. in Nanotechnology Characterization Tools for Biosensing and Medical Diagnosis (ed. Kumar, C. S. S. R.) 333–365 (Springer, 2018).
James, M. L. & Gambhir, S. S. A molecular imaging primer: modalities, imaging agents, and applications. Physiol. Rev. 92, 897–965 (2012).
Vladimirov, N. & Perlman, O. Molecular MRI-based monitoring of cancer immunotherapy treatment response. Int. J. Mol. Sci. 24, 3151 (2023).
Rowe, S. P. & Pomper, M. G. Molecular imaging in oncology: current impact and future directions. CA Cancer J. Clin. 72, 333–352 (2021).
Woessner, D. E., Zhang, S., Merritt, M. E. & Sherry, A. D. Numerical solution of the Bloch equations provides insights into the optimum design of PARACEST agents for MRI. Magn. Reson. Med. 53, 790–799 (2005).
Zhou, J. et al. Quantitative description of proton exchange processes between water and endogenous and exogenous agents for WEX, CEST, and APT experiments. Magn. Reson. Med. 51, 945–952 (2004).
Ji, Y. et al. Progress toward quantitative in vivo chemical exchange saturation transfer (CEST) MRI. Isr. J. Chem. 57, 809–824 (2017).
Kim, J., Wu, Y., Guo, Y., Zheng, H. & Sun, P. Z. A review of optimization and quantification techniques for chemical exchange saturation transfer MRI toward sensitive in vivo imaging. Contrast Media Mol. Imaging 10, 163–178 (2015).
Ward, K. M. & Balaban, R. S. Determination of pH using water protons and chemical exchange dependent saturation transfer (CEST). Magn. Reson. Med. 44, 799–802 (2000).
Ji, Y. et al. In vivo pH mapping with omega plot‐based quantitative chemical exchange saturation transfer MRI. Magn. Reson. Med. 89, 299–307 (2023).
Wu, R. et al. Quantitative chemical exchange saturation transfer (qCEST) MRI–omega plot analysis of RF‐spillover‐corrected inverse CEST ratio asymmetry for simultaneous determination of labile proton ratio and exchange rate. NMR Biomed. 28, 376–383 (2015).
Perlman, O., Farrar, C. T. & Heo, H.-Y. MR fingerprinting for semisolid magnetization transfer and chemical exchange saturation transfer quantification. NMR Biomed. 36, e4710 (2023).
Heo, H.-Y. et al. Quantitative assessment of amide proton transfer (APT) and nuclear Overhauser enhancement (NOE) imaging with extrapolated semi-solid magnetization transfer reference (EMR) signals: application to a rat glioma model at 4.7 Tesla. Magn. Reson. Med. 75, 137–149 (2016).
McMahon, M. T. et al. Quantifying exchange rates in chemical exchange saturation transfer agents using the saturation time and saturation power dependencies of the magnetization transfer effect on the magnetic resonance imaging signal (QUEST and QUESP): pH calibration for poly‐l‐lysine and a starburst dendrimer. Magn. Reson. Med. 55, 836–847 (2006).
Zaiss, M. et al. QUESP and QUEST revisited–fast and accurate quantitative CEST experiments. Magn. Reson. Med. 79, 1708–1721 (2018).
Zaiß, M., Schmitt, B. & Bachert, P. Quantitative separation of CEST effect from magnetization transfer and spillover effects by Lorentzian-line-fit analysis of z-spectra. J. Magn. Reson. 211, 149–155 (2011).
Zhou, I. et al. Quantitative chemical exchange saturation transfer (CEST) MRI of glioma using Image Downsampling Expedited Adaptive Least-squares (IDEAL) fitting. Sci. Rep. 7, 84 (2017).
Roeloffs, V., Meyer, C., Bachert, P. & Zaiss, M. Towards quantification of pulsed spinlock and CEST at clinical MR scanners: an analytical interleaved saturation–relaxation (ISAR) approach. NMR Biomed. 28, 40–53 (2014).
Perlman, O., Zhu, B., Zaiss, M., Rosen, M. S. & Farrar, C. T. An end-to-end AI-based framework for automated discovery of rapid CEST/MT MRI acquisition protocols and molecular parameter quantification (AutoCEST). Magn. Reson. Med. 87, 2792–2810 (2022).
Perlman, O. et al. CEST MR-fingerprinting: practical considerations and insights for acquisition schedule design and improved reconstruction. Magn. Reson. Med. 83, 462–478 (2020).
Jeffrey, I. W., Bushell, M., Tilleray, V. J., Morley, S. & Clemens, M. J. Inhibition of protein synthesis in apoptosis: differential requirements by the tumor necrosis factor α family and a DNA-damaging agent for caspases and the double-stranded RNA-dependent protein kinase. Cancer Res. 62, 2272–2280 (2002).
Chen, L. et al. In vivo imaging of phosphocreatine with artificial neural networks. Nat. Commun. 11, 1072 (2020).
Anemone, A., Consolino, L. & Longo, D. L. MRI–CEST assessment of tumour perfusion using X-ray iodinated agents: comparison with a conventional Gd-based agent. Eur. Radiol. 27, 2170–2179 (2017).
Zaiss, M., Kunz, P., Goerke, S., Radbruch, A. & Bachert, P. MR imaging of protein folding in vitro employing nuclear-Overhauser-mediated saturation transfer. NMR Biomed. 26, 1815–1822 (2013).
Goerke, S. et al. Aggregation-induced changes in the chemical exchange saturation transfer (CEST) signals of proteins. NMR Biomed. 30, e3665 (2017).
Zhou, Y. et al. Magnetic resonance imaging of glycogen using its magnetic coupling with water. Proc. Natl Acad. Sci. USA 117, 3144–3149 (2020).
Zaiss, M. et al. Relaxation-compensated CEST–MRI of the human brain at 7 T: unbiased insight into NOE and amide signal changes in human glioblastoma. Neuroimage 112, 180–188 (2015).
Layton, K. J. et al. Pulseq: a rapid and hardware-independent pulse sequence prototyping framework. Magn. Reson. Med. 77, 1544–1552 (2017).
Herz, K. et al. Pulseq-CEST: towards multi-site multi-vendor compatibility and reproducibility of CEST experiments using an open-source sequence standard. Magn. Reson. Med. 86, 1845–1858 (2021).
Roos, T. H. M. et al. Open-source Pulseq sequences on Philips MRI scanners. Preprint at http://arxiv.org/abs/2310.06962 (2023).
Kogan, F. et al. In vivo chemical exchange saturation transfer imaging of creatine (CrCEST) in skeletal muscle at 3T. J. Magn. Reson. Imaging 40, 596–602 (2014).
Wu, B. et al. An overview of CEST MRI for non-MR physicists. EJNMMI Phys. 3, 1–21 (2016).
Khlebnikov, V., van der Kemp, W. J., Hoogduin, H., Klomp, D. W. & Prompers, J. J. Analysis of chemical exchange saturation transfer contributions from brain metabolites to the Z-spectra at various field strengths and pH. Sci. Rep. 9, 1089 (2019).
Vinogradov, E., Sherry, A. D. & Lenkinski, R. E. CEST: from basic principles to applications, challenges and opportunities. J. Magn. Reson. 229, 155–172 (2013).
Liu, G., Gilad, A. A., Bulte, J. W., Van Zijl, P. C. & McMahon, M. T. High-throughput screening of chemical exchange saturation transfer MR contrast agents. Contrast Media Mol. Imaging 5, 162–170 (2010).
Yao, J. et al. A physical phantom for amine chemical exchange saturation transfer (CEST) MRI. Magn. Reson. Mater. Phys. Biol. Med. 34, 569–580 (2021).
Cember, A. T., Nanga, R. P. R. & Reddy, R. Glutamate-weighted CEST (gluCEST) imaging for mapping neurometabolism: An update on the state of the art and emerging findings from in vivo applications. NMR Biomed. 36, e4780 (2023).
Zaiss, M., Jin, T., Kim, S.-G. & Gochberg, D. F. Theory of chemical exchange saturation transfer MRI in the context of different magnetic fields. NMR Biomed. 35, e4789 (2022).
Ladd, M. E. et al. Pros and cons of ultra-high-field MRI/MRS for human application. Prog. Nuclear Magn. Reson. Spectrosc. 109, 1–50 (2018).
Van Zijl, P. C., Lam, W. W., Xu, J., Knutsson, L. & Stanisz, G. J. Magnetization transfer contrast and chemical exchange saturation transfer MRI. Features and analysis of the field-dependent saturation spectrum. Neuroimage 168, 222–241 (2018).
Cohen, O. & Rosen, M. S. Algorithm comparison for schedule optimization in MR fingerprinting. Magn. Reson. Imaging 41, 15–21 (2017).
Zhao, B. et al. Optimal experiment design for magnetic resonance fingerprinting: Cramér-Rao bound meets spin dynamics. IEEE Trans. Med. Imaging 38, 844–861 (2018).
Sommer, K. et al. Towards predicting the encoding capability of MR fingerprinting sequences. Magn. Reson. Imaging 41, 7–14 (2017).
Kara, D. et al. Parameter map error due to normal noise and aliasing artifacts in MR fingerprinting. Magn. Reson. Med. 81, 3108–3123 (2019).
Cohen, O. & Otazo, R. Global deep learning optimization of chemical exchange saturation transfer magnetic resonance fingerprinting acquisition schedule. NMR Biomed. 36, e4954 (2023).
Heo, H.-Y., Singh, M., Yedavalli, V., Jiang, S. & Zhou, J. CEST and nuclear Overhauser enhancement imaging with deep learning–extrapolated semisolid magnetization transfer reference: Scan-rescan reproducibility and reliability studies. Magn. Reson. Med. 91, 1002–1015 (2024).
Heo, H.-Y. et al. Prospective acceleration of parallel RF transmission-based 3D chemical exchange saturation transfer imaging with compressed sensing. Magn. Reson. Med. 82, 1812–1821 (2019).
Mueller, S. et al. Whole brain snapshot CEST at 3T using 3D-EPI: aiming for speed, volume, and homogeneity. Magn. Reson. Med. 84, 2469–2483 (2020).
Zaiss, M., Ehses, P. & Scheffler, K. Snapshot-CEST: optimizing spiral-centric-reordered gradient echo acquisition for fast and robust 3D CEST MRI at 9.4 T. NMR Biomed. 31, e3879 (2018).
Liu, G., Song, X., Chan, K. W. & McMahon, M. T. Nuts and bolts of CEST MR imaging. NMR Biomed. 26, 810 (2013).
Liu, J. et al. Encoding capability prediction of acquisition schedules in CEST MR fingerprinting for pH quantification. Magn. Reson. Med. 87, 2044–2052 (2022).
Kang, B., Kim, B., Park, H. & Heo, H.-Y. Learning-based optimization of acquisition schedule for magnetization transfer contrast MR fingerprinting. NMR Biomed. 35, e4662 (2022).
McConnell, H. M. Reaction rates by nuclear magnetic resonance. J. Chem. Phys. 28, 430–431 (1958).
Zaiss, M. et al. A combined analytical solution for chemical exchange saturation transfer and semi-solid magnetization transfer. NMR Biomed. 28, 217–230 (2015).
Golub, G. H. & Van Loan, C. F. Matrix Computations (Johns Hopkins Univ. Press, 2013).
Van Zijl, P. C. et al. Mechanism of magnetization transfer during on-resonance water saturation. A new approach to detect mobile proteins, peptides, and lipids. Magn. Reson. Med. 49, 440–449 (2003).
Stanisz, G. J. et al. T1, T2 relaxation and magnetization transfer in tissue at 3T. Magn. Reson. Med. 54, 507–512 (2005).
Geades, N. et al. Quantitative analysis of the z-spectrum using a numerically simulated look-up table: application to the healthy human brain at 7T. Magn. Reson. Med. 78, 645–655 (2017).
Liu, D. et al. Quantitative characterization of nuclear overhauser enhancement and amide proton transfer effects in the human brain at 7 tesla. Magn. Reson. Med. 70, 1070–1081 (2013).
Yarnykh, V. L. et al. Fast whole-brain three-dimensional macromolecular proton fraction mapping in multiple sclerosis. Radiology 274, 210–220 (2015).
Samsonov, A. et al. Quantitative MR imaging of two-pool magnetization transfer model parameters in myelin mutant shaking pup. Neuroimage 62, 1390–1398 (2012).
Xu, J., Chung, J. J. & Jin, T. Chemical exchange saturation transfer imaging of creatine, phosphocreatine, and protein arginine residue in tissues. NMR Biomed. 36, e4671 (2023).
Kazemi, K. & Noorizadeh, N. Quantitative comparison of SPM, FSL, and brainsuite for brain MR image segmentation. J. Biomed. Phys. Eng. 4, 13 (2014).
Glang, F. et al. DeepCEST 3T: robust MRI parameter determination and uncertainty quantifcation with neural networks—application to CEST imaging of the human brain at 3T. Magn. Reson. Med. 84, 450–466 (2020).
Assländer, J. et al. Rapid quantitative magnetization transfer imaging: utilizing the hybrid state and the generalized Bloch model. Magn. Reson. Med. 91, 1478–1497 (2024).
Nagar, D., Vladimirov, N., Farrar, C. T. & Perlman, O. Dynamic and rapid deep synthesis of chemical exchange saturation transfer and semisolid magnetization transfer MRI signals. Sci. Rep. 13, 18291 (2023).
Heo, H. Y. et al. Unraveling contributions to the Z‐spectrum signal at 3.5 ppm of human brain tumors. Magn. Reson. Med. 92, 2641–2651 (2024).
Zaiss, M. et al. Downfield-NOE-suppressed amide-CEST–MRI at 7 Tesla provides a unique contrast in human glioblastoma. Magn. Reson. Med. 77, 196–208 (2017).
Cohen-Adad, J. et al. Generic acquisition protocol for quantitative MRI of the spinal cord. Nat. Protocols 16, 4611–4632 (2021).
Bar-Shir, A., Liu, G., Greenberg, M. M., Bulte, J. W. & Gilad, A. A. Synthesis of a probe for monitoring HSV1-tk reporter gene expression using chemical exchange saturation transfer MRI. Nat. Protocols 8, 2380–2391 (2013).
Krupnick, A. S. et al. Quantitative monitoring of mouse lung tumors by magnetic resonance imaging. Nat. Protocols 7, 128–142 (2012).
Kim, M., Gillen, J., Landman, B. A., Zhou, J. & Van Zijl, P. C. Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn. Reson. Med. 61, 1441–1450 (2009).
Schuenke, P. et al. Simultaneous mapping of water shift and B1 (WASABI)—application to field-inhomogeneity correction of CEST MRI data. Magn. Reson. Med. 77, 571–580 (2017).
Zhou, J. et al. Review and consensus recommendations on clinical APT-weighted imaging approaches at 3T: application to brain tumors. Magn. Reson. Med. 88, 546–574 (2022).
Sun, P. Z. Simplified quantification of labile proton concentration-weighted chemical exchange rate (kws) with RF saturation time dependent ratiometric analysis (QUESTRA): normalization of relaxation and RF irradiation spillover effects for improved quantitative chemical exchange saturation transfer (CEST) MRI. Magn. Reson. Med. 67, 936–942 (2012).
Acknowledgements
The authors would like to thank K. Herz for his valuable work on pulseq–CEST standard development, D. Korenchan for his work on the Paravision 360 protocol, and B. Kang, H. Shmueli and A. Finkelstein for their technical assistance and feedback. This work was supported by the Ministry of Innovation, Science and Technology, Israel, and the Tel Aviv University Center for AI and Data Science (TAD). The authors acknowledge financial support from the NIH/NIBIB grants R01EB031008, R37-CA262662 and R01EB029974. This project was funded by the European Union (ERC, BabyMagnet, project no. 101115639). Views and opinions expressed are, however, those of the authors only and do not necessarily reflect those of the European Union or the European Research Council. Neither the European Union nor the granting authority can be held responsible for them.
Author information
Authors and Affiliations
Contributions
Conceptualization: N.V., C.T.F. and O.P. Methodology: N.V., O.C., H.-Y.H., M.Z., C.T.F. and O.P. Data curation: O.C., H.-Y.H., M.Z., C.T.F. and O.P. Writing: N.V., H.-Y.H., C.T.F. and O.P. Reviewing and editing: N.V., O.C., H.-Y.H., M.Z., C.T.F. and O.P. Supervision: O.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing interests: C.T.F. and O.C. hold a patent for the CEST–MRF method (patent no. US10,605,877).
Peer review
Peer review information
Nature Protocols thanks Jakob Asslander and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key references
Perlman, O. et al. Nat. Biomed. Eng. 6, 648–657 (2022): https://doi.org/10.1038/s41551-021-00809-7
Cohen, O. et al. Magn. Reson. Med. 89, 233–249 (2023): https://doi.org/10.1002/mrm.29448
Kang, B. et al. Magn. Reson. Med. 85, 2040–2054 (2021): https://doi.org/10.1002/mrm.28573
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2, Discussion 1 and Table 1–5.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Vladimirov, N., Cohen, O., Heo, HY. et al. Quantitative molecular imaging using deep magnetic resonance fingerprinting. Nat Protoc 20, 3024–3054 (2025). https://doi.org/10.1038/s41596-025-01152-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41596-025-01152-w
This article is cited by
-
Multi-contrast generation and quantitative MRI using a transformer-based framework with RF excitation embeddings
Communications Biology (2025)
-
Multi-parameter molecular MRI quantification using physics-informed self-supervised learning
Communications Physics (2025)
-
Quantitative multi-metabolite imaging of Parkinson’s disease using AI boosted molecular MRI
npj Imaging (2025)